ABSTRACT
Coronaviruses (CoVs) are unique in encoding a 3′→5′ exoribonuclease within nonstructural protein 14 (nsp14-ExoN) that is required for high-fidelity replication, likely via proofreading. nsp14 associates with the CoV RNA-dependent RNA polymerase (nsp12-RdRp), and nsp14-ExoN activity is enhanced by binding nsp10, a small nonenzymatic protein. However, it is not known whether nsp10 functions in the regulation of CoV replication fidelity. To test this, we engineered single and double alanine substitution mutations into the genome of murine hepatitis virus (MHV-A59) containing ExoN activity [ExoN(+)] at positions within nsp10 known to disrupt the nsp10-nsp14 interaction in vitro. We show that an nsp10 mutant, R80A/E82A-ExoN(+), was five to ten times more sensitive to treatment with the RNA mutagen 5-fluorouracil (5-FU) than wild-type (WT)-ExoN(+), suggestive of decreased replication fidelity. This decreased-fidelity phenotype was confirmed using two additional nucleoside analogs, 5-azacytidine and ribavirin. R80A/E82A-ExoN(+) reached a peak titer similar to and demonstrated RNA synthesis kinetics comparable to those seen with WT-ExoN(+). No change in 5-FU sensitivity was observed for R80A/E82A-ExoN(−) relative to MHV-ExoN(−), indicating that the decreased-fidelity phenotype of R80A/E82A-ExoN(−) is linked to the presence of ExoN activity. Our results demonstrate that nsp10 is important for CoV replication fidelity and support the hypothesis that nsp10 functions to regulate nsp14-ExoN activity during virus replication.
IMPORTANCE The adaptive capacity of CoVs, as well as all other RNA viruses, is partially attributed to the presence of extensive population genetic diversity. However, decreased fidelity is detrimental to CoV replication and virulence; mutant CoVs with decreased replication fidelity are attenuated and more sensitive to inhibition by RNA mutagens. Thus, identifying the viral protein determinants of CoV fidelity is important for understanding CoV replication, pathogenesis, and virulence. In this report, we show that nsp10, a small, nonenzymatic viral protein, contributes to CoV replication fidelity. Our data support the hypothesis that CoVs have evolved multiple proteins, in addition to nsp14-ExoN, that are responsible for maintaining the integrity of the largest known RNA genomes.
INTRODUCTION
Viral adaptation is driven by selection and genetic bottlenecks present within the host and during transmission. It is the error rate (fidelity) of the viral replicase that generates the genetic diversity that is subjected to selection. At the core of all RNA virus replicases, with the exception of retroviruses, is the RNA-dependent RNA polymerase (RdRp). Research performed with numerous RNA viruses, including foot-and-mouth disease virus (1–3), poliovirus (4–10), chikungunya virus (11, 12), influenza virus (13), coxsackievirus B3 (14, 15), and human enterovirus 71 (16–18), has demonstrated that mutations within the viral RdRp can either increase or decrease replicase fidelity. In the vast majority of cases, alterations in replication fidelity result in decreased viral fitness and attenuation of virulence (reviewed in reference 19). Also, viruses with altered replication fidelity have potential therapeutic value as live attenuated vaccines (5, 9, 20, 21). Together, these observations underscore the need to elucidate the contributions of individual protein-protein interactions and virus-encoded enzymes to replicase fidelity.
The coronavirus (CoV) replicase is more complex than that of many other RNA viruses due to their large 27-to-32-kb genomes (19, 22) and the presence of multiple RNA-modifying activities (23). In addition to encoding RdRp activity within nonstructural protein 12 (nsp12-RdRp), CoVs encode 3′→5′ exoribonuclease (ExoN) activity within nsp14 (23, 24). Biochemical studies have shown that ExoN is capable of removing 3′ mismatches (25), and inactivation of ExoN activity decreases the fidelity of the CoV replicase by up to 20-fold in tissue culture and in vivo (21, 26, 27). Viruses lacking ExoN activity [ExoN(−)] have increased susceptibility to lethal mutagenesis in the presence of RNA mutagens (28) compared to wild-type (WT) viruses containing ExoN activity [ExoN(+)]. Thus, all available bioinformatic, biochemical, and virological data support the conclusion that CoVs have evolved proofreading capacity. Attempts to reconstitute the CoV replicase using recombinant proteins have been impeded by the limited polymerase activity observed for nsp12-RdRp in vitro (29–31). Recent work by Subissi et al. demonstrated that nsp7 and nsp8 are required for processive RNA synthesis during CoV replication, in part by increasing the binding of nsp12-RdRp to the RNA template (32). nsp12-RdRp is also capable of associating with nsp14-ExoN without disrupting the capacity of ExoN to cleave an RNA template.
Aside from nsp14, no other viral proteins have been demonstrated to affect the fidelity of the CoV replicase. nsp10, a small CoV protein with no known enzymatic function, binds nsp14 and enhances ExoN activity by up to 35-fold in vitro (25). Mutations in nsp10 that alter the nsp10-nsp14 interaction in vitro reduce or abolish the enhancement of ExoN activity (25, 33). In addition to enhancing ExoN activity, nsp10 is an allosteric regulator of nsp16, a 2′-O-methyltransferase (2′-OMT) involved in capping of CoV RNA and evasion of host interferon (IFN)-stimulated genes (34–38). Targeted mutagenesis studies also have implicated nsp10 as a critical regulator of CoV RNA synthesis (39) and proteolytic processing by the nsp5 viral protease (40). These studies indicated that nsp10 is an essential component of the CoV replicase that directly interacts with nsp14 and affects ExoN activity in vitro. However, it is not known whether nsp10 affects the fidelity of the CoV replicase or whether nsp10 affects the activity of nsp14-ExoN during virus replication.
Here we show that mutations in murine hepatitis virus (MHV-A59) nsp10, at positions previously demonstrated to disrupt nsp10-nsp14 interactions in vitro, decrease CoV replication fidelity. The nsp10 R80A/E82A-ExoN(+) double mutant has increased sensitivity to the base and nucleoside analogs 5-fluorouracil (5-FU), 5-azacytidine (AZA), and ribavirin (RBV) compared to WT-ExoN(+), indicative of decreased fidelity. When the nsp10 R80A/E82A substitutions were introduced into MHV-ExoN(−), there was no increase in 5-FU sensitivity beyond that of MHV-ExoN(−). Our results demonstrate that mutations in nsp10 decrease CoV fidelity and support the hypothesis that nsp10-mediated fidelity is dependent on intact ExoN activity. Our results are consistent with recent biochemical studies (25, 32) and support a model in which CoVs use multiple nonstructural proteins, including nsp10, nsp12, and nsp14, to faithfully replicate their large RNA genomes.
MATERIALS AND METHODS
Cell culture.
DBT-9 (delayed brain tumor, murine astrocytoma clone 9) cells (41) were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS; Invitrogen), 100 U/ml penicillin and streptomycin (Gibco), 10 mM HEPES, and 0.25 μg/ml amphotericin B (Corning). Baby hamster kidney (BHK) cells stably expressing the MHV receptor (BHK-R [42]) were maintained in the same medium but supplemented with 0.8 mg/ml of G418 (Mediatech).
Site-directed mutagenesis.
To reduce the number of downstream sequencing reactions, a region of the MHV E fragment (positions 2106 to 2982) containing nsp10 was subcloned into pCR-2.1-TOPO TA vector (Invitrogen) according to the manufacturer's instructions. Twenty-five nanograms of template was used to introduce the point mutations within nsp10. QuikChange (Stratagene) site-directed mutagenesis was performed according to the manufacturer's protocol. The nsp16 D130A mutant was generated similarly, except mutagenesis was performed using the F fragment. Each plasmid was sequenced (Genhunter Corporation, Nashville, TN) to ensure that only the intended mutations were present. Once confirmed, the fragment containing nsp10 was excised from TOPO using BamHI and KpnI and ligated into a WT E fragment linearized with BamHI and KpnI.
Assembly and recovery of viruses.
The infectious cDNA clone for MHV-A59 (GenBank accession no. AY910861) has been described previously (42). Briefly, a total of 1.5 μg of linearized WT MHV fragments A to D and fragments F and G was ligated together with the appropriate E fragment containing the nsp10 mutation(s) overnight at 16°C using T4 ligase (New England BioLabs). For the ExoN(−) viruses, the F fragment containing the two ExoN mutations (D89A and E91A) was used (26). A 1/10 volume of 3 M sodium acetate (pH = 5.5) and 1 volume of chloroform were added to the ligation reaction. The aqueous layer was removed to a new tube, and 1 volume of isopropanol was added to precipitate the DNA. This purified product was resuspended in nuclease-free water and was then transcribed using Ambion mMessage mMachine T7 with the following modifications in a total reaction volume of 50 μl: 7.5 μl of 3 mM GTP, 25 μl 2× NTP/cap, 5 μl of 10× reaction buffer, 7.5 μl of template, and 5 μl of the enzyme mix. The reaction was modified in order to efficiently generate longer transcripts at 40.5°C for 30 min, 37°C for 1 h, 40.5°C for 30 min, 37°C for 30 min, and 40.5°C for 30 min. Transcript for the N gene was generated using water instead of the 3 mM GTP. Fifty microliters of the transcription reaction and 25 μl of N transcript were electroporated (Bio-Rad GenePulser Xcell) into 6 × 107 BHK-R cells and added to T-75 flasks seeded with DBT-9 cells. Cells were grown at 37°C, and flasks containing virus were frozen when cytopathic effect (CPE) was present throughout the flask. Flasks were thawed, and debris was pelleted at 4,000 × g (Sorvall RC 3B Plus; HA-6000A rotor) for 10 min at 4°C. The supernatant was then divided into aliquots and considered passage 0 (P0) stock. The virus titer was determined by a plaque assay using DBT-9 cells as described previously (26).
Propagation and confirmation of recombinant viruses.
Subconfluent DBT-9 cells were infected at a multiplicity of infection (MOI) of 0.01 PFU/cell to generate P1 stocks. Virus was harvested as described above. To ensure that the recovered viruses contained only the intended mutations, RNA was harvested from DBT-9 cells infected with P0 stock using TRIzol (Ambion). The RNA was purified according to the manufacturer's protocol and reverse transcribed (RT) using SuperScript III (Invitrogen) as described previously (28). For all viruses, the entire nsp10 coding region was sequenced to ensure the absence of additional mutations. The P0 stocks of both N40A/R80A-ExoN(+) and R80A/E82A-ExoN(+) were sequenced across the entire genome to ensure that no other mutations had arisen during recovery. For full-genome sequencing, 12 overlapping ∼3-kb cDNA amplicons were generated using 2 μl of RT product in a total reaction volume of 50 μl containing 100 ng each of forward and reverse primers, 5 μl of 10× reaction buffer, 1 μl of 10 mM (each) deoxynucleoside triphosphates (dNTPs), and 0.5 μl (5 U/μl) of high-fidelity Easy A polymerase (Agilent). PCR was performed using a Bio-Rad C1000 Touch Thermal Cycler as follows: for step 1, 95°C for 2 min; for step 2, 95°C for 30 s; for step 3, 58°C for 30 s; for step 4, 72°C for 3.5 min; and for step 5, 72°C for 7 min. Steps 2 through 4 were repeated 30 times. All primers generated single bands and were column purified using a Wizard SV Gel and PCR cleanup system (Promega). The nucleotide sequences of the amplicon and sequencing primers are available upon request.
Base and nucleoside analog sensitivity studies.
5-Fluorouracil (5-FU), 5-azacytidine (AZA), and ribavirin (RBV) were purchased from Sigma; stock solutions were made in dimethyl sulfoxide (DMSO) at 200 mM, water at 50 mM, and water at 200 mM, respectively. Sensitivity studies were performed at the indicated concentration(s) of base or nucleoside analog for both low (0.01 PFU/cell)- and high (1 PFU/cell)-MOI infections, as described previously (28). For 5-FU sensitivity experiments at 37°C and 40°C, cells were shifted to the appropriate temperature once the treatment was added back postinfection. We have demonstrated that 5-FU and RBV exhibit minimal cellular toxicity in DBT-9 cells at concentrations up to 400 μM (28). The cytotoxicity of AZA and RBV, at concentrations up to 50 μM, was determined using CellTiter-Glo (Promega) according to the manufacturer's instructions. Briefly, DBT-9 cells were seeded into an opaque tissue-culture-grade 96-well plate approximately 18 h prior to the addition of AZA or RBV. Cells were then incubated with AZA or RBV at the indicated concentrations for 24 h, and cell viability was determined.
Replication curves and RNA synthesis kinetics.
Viral replication kinetics were determined at both a high MOI (1 PFU/cell) and a low MOI (0.01 PFU/cell) as described previously (26). Supernatant (300 μl) was collected at the indicated time points, and the virus titer was determined by a plaque assay. Kinetics of genomic and subgenomic RNA synthesis at an MOI of 1 PFU/cell, with or without 5-FU treatment, were determined by two-step real-time quantitative RT-PCR (qRT-PCR) (28) using RNA harvested at the times indicated. RNA extraction with TRIzol and RT reactions were performed as described above. Primers used to detect genomic (nsp10) and subgenomic (N) RNA, as well as GAPDH (glyceraldehyde-3-phosphate dehydrogenase), have been previously reported (28, 39). The region of nsp10 used to quantify genomic RNA levels does not overlap the mutations introduced within nsp10. Reactions and data analysis were performed as described previously (28) with modifications: the RT product was diluted 1:200, the values determined for duplicate wells of each sample were averaged into one value to minimize well-to-well variation, and data were normalized relative to GAPDH using the threshold cycle (2−ΔCT) method.
IFN-β sensitivity assay.
Subconfluent monolayers of DBT-9 cells in 24-well plates were pretreated with either 100 U/ml or 500 U/ml of mouse beta interferon (IFN-β) (PBL Assay Science, Piscataway, NJ) for 18 h prior to infection. The pretreatment reaction mixture was removed, and monolayers were inoculated with the indicated virus at an MOI of 1 PFU/cell for 45 min at 37°C. The inoculum was removed, and fresh DMEM was added to each well. The supernatant was harvested at 12 h postinfection, and the virus titer was determined by a plaque assay.
Statistical analysis.
Statistical tests were applied where noted using GraphPad Prism 6 (La Jolla, CA) software. Significance is denoted (*, P < 0.05; **, P < 0.01; ***, P < 0.0001) and was determined using the test described in the figure legend. In some cases, data were normalized to untreated controls; this was also performed using GraphPad Prism 6. The number of replicate samples is denoted within each figure legend.
RESULTS
Effect of mutations in nsp10 on virus sensitivity to 5-fluorouracil (5-FU).
To test the role of nsp10 in fidelity, we engineered mutations coding for alanine substitutions into the infectious clone of MHV-A59 (42) at conserved amino acid positions reported to disrupt the severe acute respiratory syndrome (SARS)-CoV nsp10-nsp14 interaction in vitro: G69A, N40A, R80A (H80 in SARS-CoV), and E82A (D82 in SARS-CoV) (Fig. 1). With the exception of the G69A mutation, all nsp10 mutations permitted virus recovery; earlier attempts by another group to recover G69A were also unsuccessful (39). We previously demonstrated that the base analog 5-FU is incorporated into the CoV genome by nsp12-RdRp during viral replication, resulting in increased genomic mutations (28). Incorporation of 5-FU is increased in low-fidelity viruses lacking ExoN activity [ExoN(−)], indicating that virus sensitivity to 5-FU treatment can be used to test for decreases in replication fidelity. We compared the replication of the single alanine substitution mutants N40A, R80A, and E82A to that of WT-ExoN(+) in the presence and absence of 5-FU. All three mutant viruses exhibited 5-FU sensitivity phenotypes similar to that of the WT-ExoN(+) virus at both an MOI of 1 PFU/cell and an MOI of 0.01 PFU/cell (Fig. 2A and B). 5-FU exhibited no toxicity in DBT-9 cells at concentrations up to 400 μM (28).
FIG 1.
Alignment of nsp10 and modeled structure. (A) The MHV-A59 genome is shown. Selected examples of nsp's are highlighted: nsp10 (orange), nsp14 (blue) (ExoN domain, solid; N7-MT domain, hatched), and nsp16-2′-OMT (purple). Abbreviations: RdRp, RNA-dependent RNA polymerase; ExoN, 3′→5′ exoribonuclease; N7-MT, N7-methyltransferase; 2′-OMT, 2′-O-methyltransferase. (B) MHV-A59 nsp10 was modeled using Phyre2 (58). A high (>90%)-confidence model was obtained using the structure of SARS-CoV nsp10 (57). A surface representation is shown, with mutated nsp10 residues shown in orange. The image was generated using PyMOL (59). (C) Alignment of nsp10 from MHV-A59 (GenBank accession no. AY910861), SARS-CoV (GenBank accession no. AY278741), and Middle East respiratory syndrome (MERS)-CoV (GenBank accession no. JX869059). Identities are denoted by shaded boxes, while similarities are denoted by white boxes. Mutations made during this study that resulted in viable virus are denoted by an asterisk.
FIG 2.
Virus sensitivity to 5-FU treatment. DBT-9 cells were pretreated with the indicated concentration of 5-FU and infected with the indicated virus at an MOI of 1 PFU/cell (A and C) or 0.01 PFU/cell (B, D, and E). Virus supernatants were harvested at 12 h postinfection (MOI = 1) or 24 h postinfection (MOI = 0.01), and the virus titer was determined by a plaque assay. For each virus, titers were normalized to values obtained from DMSO-treated samples using Prism 6. Data are presented as means ± standard errors of the means (SEM). Statistical significance compared to WT-ExoN(+) or MHV-ExoN(−) (panel E) is denoted (*, P < 0.05; **, P < 0.01; ***, P < 0.0001) and was determined using one-way analysis of variance (ANOVA) with Dunnett's post hoc test for multiple comparisons (n = 4 to 7).
Because formation of a stable nsp10-nsp14 complex likely involves multiple surface-exposed residues, substitution at one position might not be sufficient to alter nsp10-nsp14 interactions during virus infection. Thus, we introduced the alanine double substitutions N40A/R80A and R80A/E82A within nsp10. We then tested whether the alanine double substitutions N40A/R80A and R80A/E82A affected virus sensitivity to 5-FU treatment. No increased sensitivity was detected during infection at an MOI of 1 PFU/cell (Fig. 2C). Infection at a higher MOI could mask a subtle increase in 5-FU sensitivity due to the capacity of the population to tolerate or complement mutations. Thus, we examined the sensitivity of N40A/R80A-ExoN(+) and R80A/E82A-ExoN(+) to 5-FU treatment at a low MOI (0.01 PFU/cell). Both N40A/R80A-ExoN(+) and R80A/E82A-ExoN(+) demonstrated increased sensitivity to 5-FU treatment compared to WT-ExoN(+) (Fig. 2D). The 5-FU sensitivity of R80A/E82A-ExoN(+) was concentration dependent across the range tested, while N40A/R80A-ExoN(+) demonstrated a subtle increase in sensitivity only at 200 μM 5-FU. Because of this, we focused primarily on the R80A/E82A-ExoN(+) virus for subsequent experiments. These data demonstrate that mutations in nsp10 increase 5-FU sensitivity, consistent with decreased replication fidelity. Our data also indicate that the impact of nsp10 mutations on fidelity is less than that seen after inactivation of ExoN activity (28), as the titer of MHV-ExoN(−) was decreased by almost 4 log10 following treatment with 80 μM 5-FU (Fig. 2E).
R80A/E82A-ExoN(+) shows increased sensitivity to nucleoside analogs AZA and RBV.
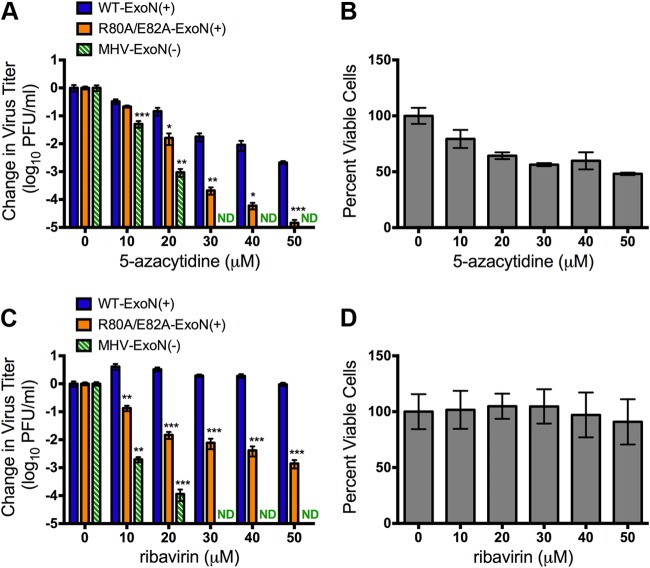
To confirm that the observed 5-FU sensitivity was a result of decreased fidelity, we tested whether R80A/E82A-ExoN(+) was sensitive to the nucleoside analogs 5-azacytidine (AZA) and ribavirin (RBV). AZA and RBV have been shown to induce mutagenesis of RNA virus genomes, most recently for influenza virus (43). We examined the sensitivity of R80A/E82A-ExoN(+) to AZA and RBV during infection at a low MOI (0.01 PFU/cell). Both MHV-ExoN(−) and R80A/E82A-ExoN(+) showed increased sensitivity to AZA compared with WT-ExoN(+) at concentrations at and above 20 μM (Fig. 3A). R80A/E82A-ExoN(+) was between 10-fold and 100-fold more sensitive to AZA treatment than WT-ExoN(+). MHV-ExoN(−) was undetectable by plaque assay at concentrations above 20 μM, consistent with the strong mutator phenotype. Since AZA also inhibited WT-ExoN(+) replication and exhibited cellular toxicity (Fig. 3B), we examined the sensitivity of all three viruses to the guanosine analog RBV. We previously showed that MHV-ExoN(−), but not WT-ExoN(+), is sensitive to treatment with RBV (28). Consistent with our previous data, RBV showed minimal cytotoxicity at concentrations up to 50 μM and had no effect on the WT-ExoN(+) titer (Fig. 3C and D). MHV-ExoN(−) was undetectable by plaque assay at RBV concentrations above 20 μM, and R80A/E82A-ExoN(+) was more than 100-fold more sensitive to RBV treatment than WT-ExoN(+) at higher concentrations. These results indicate that the R80A/E82A substitutions in nsp10 decrease CoV replication fidelity to levels between those seen with WT-ExoN(+) and MHV-ExoN(−).
FIG 3.
Virus sensitivity to the nucleoside analogs AZA and RBV. (A) DBT-9 cells were pretreated with the indicated concentration of AZA and infected with the indicated virus at an MOI of 0.01 PFU/cell. Virus supernatants were harvested at 24 h postinfection, and the virus titer was determined by a plaque assay. Virus titers were normalized to values obtained from vehicle-treated samples using Prism 6. Data are presented as means ± SEM. Statistical significance compared to WT-ExoN(+) is denoted (*, P < 0.05; **, P < 0.01; ***, P < 0.0001) and was determined using one-way ANOVA with Dunnett's post hoc test for multiple comparisons (n = 6). (B) The cytotoxicity of AZA during a 24-h treatment is shown and was determined using CellTiter-Glo (means ± SEM; n = 3). (C) The experiment was performed as described for panel A, except with RBV. ND, not detected. (D) The cytotoxicity of RBV during a 24-h treatment is shown and was determined using CellTiter-Glo (means ± SEM; n = 3).
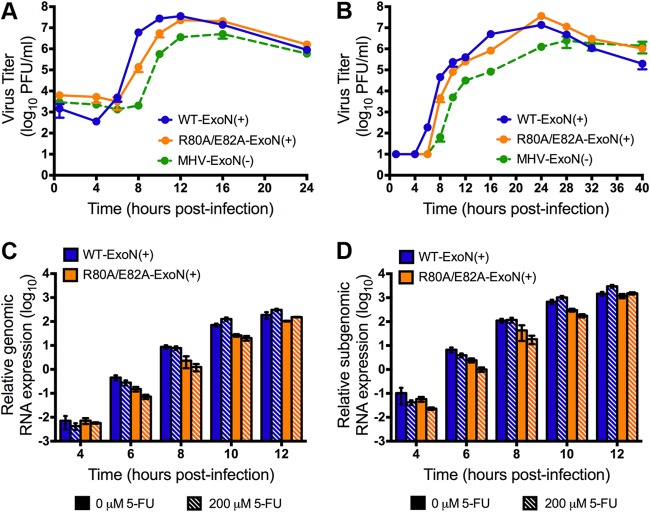
Replication and RNA synthesis kinetics of the R80A/E82A-ExoN(+) mutant.
To determine the impact of these mutations during infection, we examined the replication kinetics of R80A/E82A-ExoN(+) compared to those of WT-ExoN(+) and MHV-ExoN(−). R80A/E82A-ExoN(+) replication kinetics were similar to those seen with WT-ExoN(+) during infection at an MOI of 1 and 0.01 PFU/cell (Fig. 4A and B), though the virus titers were lower than those of WT-ExoN(+) at early time points. No enzymatic activity has been reported or predicted for nsp10. However, mutations in MHV-A59 nsp10 have been reported to impact viral RNA synthesis (39). We therefore measured genomic and subgenomic RNA levels of the R80A/E82A-ExoN(+) mutant during infection at an MOI of 1 PFU/cell using two-step qRT-PCR. Genomic and subgenomic RNA levels were normalized to the endogenous GAPDH control and were measured in both the presence and the absence of 5-FU treatment. WT-ExoN(+) and R80A/E82A-ExoN(+) demonstrated similar RNA synthesis kinetics, and RNA accumulation was not affected by 5-FU treatment (Fig. 4C and D). On the basis of these data, we conclude that the increased 5-FU sensitivity of R80A/E82A-ExoN(+) is not due to impairment of viral RNA synthesis.
FIG 4.
Virus replication and RNA synthesis kinetics. (A and B) DBT-9 cells were infected with the indicated virus at an MOI of 1 PFU/cell (A) or 0.01 PFU/cell (B), and 300 μl of supernatant was collected at the indicated time points. The virus titer was determined by a plaque assay. (C and D) Kinetics of genomic (C) and subgenomic (D) RNA synthesis, with or without 5-FU treatment at an MOI of 1 PFU/cell, were determined by two-step real-time qRT-PCR using RNA harvested at the times indicated. Data were normalized to GAPDH using the 2−ΔCT method. Data are presented as means ± SD for triplicate samples.
R80A/E82A-ExoN(+) does not have increased sensitivity to IFN-β.
In addition to stimulation of nsp14-ExoN activity, nsp10 binding to nsp16 is required for SARS-CoV nsp16–2′-OMT activity (38, 44). Both MHV-A59 and SARS-CoV lacking 2′-OMT activity display increased sensitivity to exogenous interferon beta (IFN-β) treatment due to increased recognition by host interferon-stimulated genes (34–36). Because of the dual functions of nsp10, we tested whether R80A/E82A-ExoN(+) displayed increased sensitivity to IFN-β. As a positive control, we engineered and recovered MHV-A59 with an alanine substitution mutation in nsp16 (nsp16 D130A). This mutation abolishes SARS-CoV 2′-OMT activity and renders both MHV-A59 and SARS-CoV sensitive to exogenous IFN-β treatment (34, 36). Following IFN-β treatment, the nsp16 D130A virus titer was reduced by approximately 5 log10 (Fig. 5). In contrast, titers of both WT-ExoN(+) and R80A/E82A-ExoN(+) were reduced by approximately 1 log10. These results demonstrate that the R80A/E82A substitutions in nsp10 do not significantly impact 2′-OMT activity as measured by sensitivity to IFN-β treatment.
FIG 5.
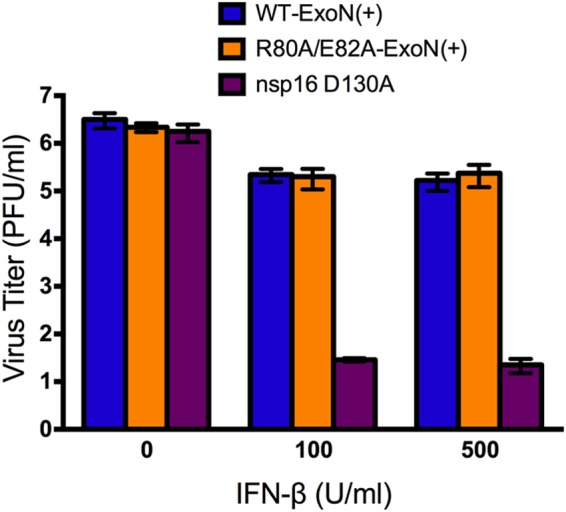
Virus sensitivity to exogenous interferon beta (IFN-β) treatment. DBT-9 cells were pretreated with either 100 or 500 U/ml of IFN-β for 18 h and infected with the indicated virus at an MOI of 1 PFU/cell. Virus supernatant was harvested at 12 h postinfection, and the virus titer was determined by a plaque assay. Data are presented as means ± SEM (n = 4).
R80A and E82A substitutions do not enhance the sensitivity of MHV-ExoN(−) to 5-FU treatment.
If the phenotype of R80A/E82A-ExoN(+) resulted from alterations in the nsp10-nsp14 interaction, as demonstrated by previous biochemical studies (25, 32), then introduction of these mutations into the MHV-ExoN(−) background should not increase 5-FU sensitivity beyond that of MHV-ExoN(−). To test this, we examined the sensitivity of R80A/E82A-ExoN(−) and MHV-ExoN(−) to 5-FU treatment at a low MOI (0.01 PFU/cell), as described above. The R80A/E82A substitutions within nsp10 did not result in increased sensitivity to 5-FU treatment (Fig. 6) compared to that of MHV-ExoN(−), consistent with the observation that nsp10 functions as a stimulatory factor for nsp14-ExoN (25, 32). Our data indicate that the decreased-fidelity phenotype of R80A/E82A-ExoN(+) is dependent upon the presence of intact ExoN activity.
FIG 6.
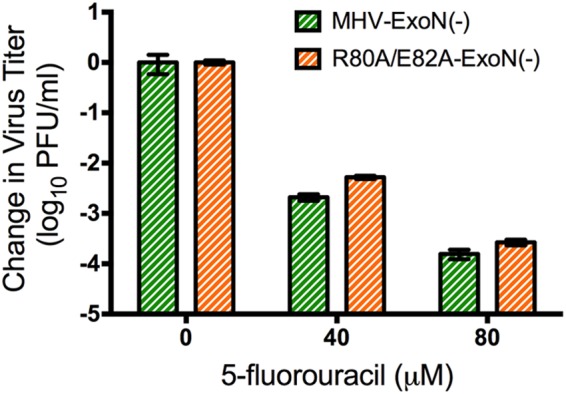
5-FU sensitivity of R80A/E82A-ExoN(−) compared to MHV-ExoN(−). DBT-9 cells were pretreated with the indicated concentration of 5-FU and infected at an MOI of 0.01 PFU/cell. Supernatants were harvested at 24 h postinfection, and the virus titer was determined by a plaque assay. Data are presented as means ± SEM. There was no statistically significant difference between R80A/E82A-ExoN(−) and MHV-ExoN(−) at each concentration as determined by a two-tailed unpaired t test (n = 4).
The 5-FU sensitivity of R80A/E82A-ExoN(+) is increased at 40°C.
The mechanism by which nsp10 stimulates nsp14-ExoN activity is unknown. On the basis of our results and previous biochemical studies (25, 32), we hypothesized that nsp10 functions to stabilize nsp14 or regulate association of nsp14 with other components of the replication complex. To test this hypothesis, we sought to stress protein-protein interactions by increasing the temperature at which we performed the 5-FU sensitivity experiments. In the absence of 5-FU, the titers of both WT-ExoN(+) and R80A/E82A-ExoN(+) decreased by approximately 1 log10 during infection at 40°C compared to 37°C (Fig. 7A), indicating that R80A/E82A-ExoN(+) was not temperature sensitive. Following treatment with either 100 μM or 200 μM 5-FU at 40°C, the titer of WT-ExoN(+) decreased by only 2-fold to 3-fold relative to treatment at 37°C (Fig. 7B). In contrast to WT-ExoN(+) results, the R80A/E82A-ExoN(+) virus titer was decreased by almost 100-fold in the presence of 200 μM 5-FU (Fig. 7B). Combined, our data suggest that the R80A/E82A mutations in nsp10 disrupt protein-protein interactions, potentially, those between nsp10 and nsp14, that are necessary for CoV replication fidelity.
FIG 7.
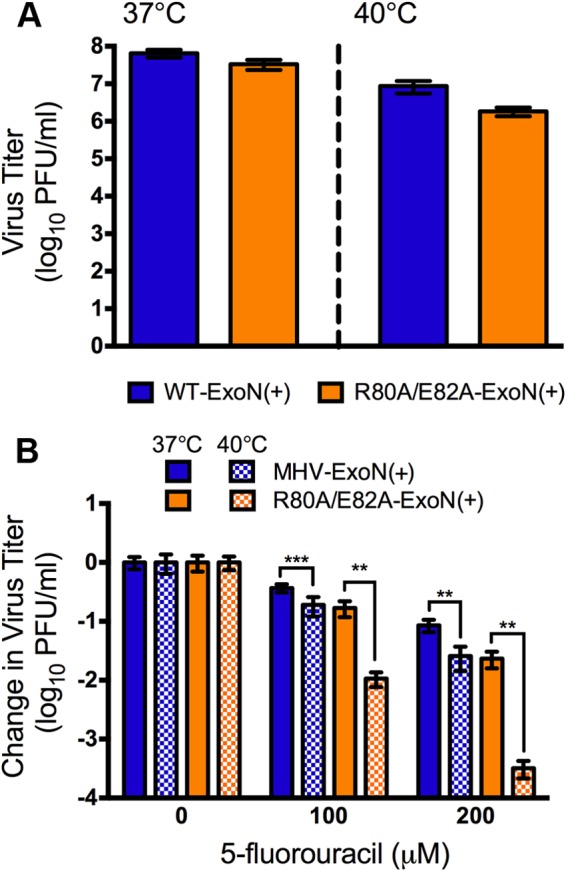
Virus sensitivity to 5-FU treatment at elevated temperature. (A) Virus titers following replication at an MOI of 0.01 PFU/cell at either 37°C or 40°C in the absence of 5-FU are shown. R80A/E82A-ExoN(+) virus titers were not statistically different from WT-ExoN(+) virus titers at either temperature as determined by a two-tailed unpaired t test (n = 6). (B) Virus sensitivity to 5-FU treatment at either 37°C or 40°C is shown. DBT-9 cells were pretreated with the indicated concentration of 5-FU and then infected at an MOI of 0.01 PFU/cell. DMEM containing DMSO or 5-FU was added back, and cells were incubated at either 37°C or 40°C. At 24 h postinfection, the supernatant was harvested and the virus titer was determined by a plaque assay. Data are presented as means ± SEM. The statistical significance of the results of comparisons of the virus titer at 37°C to that at 40°C was determined (*, P < 0.05; **, P < 0.01; ***, P < 0.0001) using a two-tailed unpaired t test (n = 6).
DISCUSSION
Recent data demonstrate that multiple CoV nsp's are required for processive RNA replication (32) and that nsp10 interacts with this complex. Combined with data from previous studies showing the capacity of nsp10 to enhance nsp14-ExoN activity (25, 32) and the involvement of nsp10 in CoV RNA synthesis (39), these data suggest that nsp10 is a core component of the CoV replicase. Here we show that R80A and E82A mutations within nsp10, positions previously shown to disrupt the SARS-CoV nsp10-nsp14 interaction in vitro, led to decreased replication fidelity (Fig. 2 and 3). During the course of our study, Bouvet et al. reported an extensive panel of SARS-CoV nsp10 mutations that disrupt the nsp10-nsp14 interaction (33). Only two mutations, K43A and Y96F, could be recovered, but the authors observed no increase in virus sensitivity to 5-FU treatment. These data are consistent with the phenotype of our single nsp10 mutants (Fig. 2) and suggest that single point mutations within nsp10 might not be sufficient to disrupt the nsp10-nsp14 interaction during virus replication. This is in contrast with results of studies performed using transiently transfected and recombinant nsp10 and nsp14 (25, 33), in which single point mutations were capable of disrupting nsp10-nsp14 interactions. However, the differences between these previous studies and ours are probably due to the absence of other CoV replicase proteins. Replicase stability is likely determined by a network of protein-protein interactions involving the surfaces of multiple CoV nsp's (45–47). Thus, a slight decrease in the strength of the nsp10-nsp14 interaction might be compensated for by other protein-protein interactions.
Our data support the hypothesis that the phenotype of R80A/E82A-ExoN(+) is due to modulation of ExoN activity. R80A/E82A-ExoN(+) displayed RNA synthesis kinetics similar to those of WT-ExoN(+) (Fig. 4) and did not exhibit increased IFN-β sensitivity compared to WT-ExoN(+) (Fig. 5), indicating that the R80A and E82A substitutions do not impact nsp16–2′-OMT activity. Most importantly, introduction of the R80A and E82A substitutions into the genome of MHV-A59 lacking ExoN activity [R80A/E82A-ExoN(−)] did not further increase virus sensitivity to 5-FU treatment (Fig. 6) compared to that seen with MHV-ExoN(−). By analogy to Escherichia coli DNA polymerase III (Pol III), nsp10 could function by stabilizing nsp14-ExoN, by enhancing removal of 3′ terminal mismatches during virus replication, or by driving the association of nsp14 with other replicase components. The DNA Pol III holoenzyme contains at least 17 subunits, and the catalytic site is composed of a heterotrimeric polymerase core with polymerase (α), exonuclease (ε), and θ subunits (recently reviewed in reference 48). The θ subunit is a small 8-to-9-kDa protein that has no known enzymatic function but has been shown to enhance the proofreading activity of ε by 2-fold to 4-fold (49, 50) and to stabilize ε under thermal inactivation conditions (51, 52). However, ε is not dependent upon the presence of the θ subunit (49, 51, 53, 54). Both the N7-MT and ExoN activities of nsp14 were first reported independently of nsp10 (24, 55), demonstrating that nsp10 is not a requirement for nsp14 enzymatic activity. This is in contrast to the SARS-CoV nsp10-nsp16 interaction, in which binding of nsp10 to nsp16 is required for nsp16–2′-OMT activity (37, 38, 56). The affinity of nsp14 for RNA has not been reported; however, the presence of two zinc finger (ZnF) domains within nsp10, coupled with the capacity of nsp10 to bind RNA, suggests that nsp10 could recruit nsp14 to RNA (57). nsp10 has not been reported to enhance nsp14–N7-MT activity (44); however, the interaction of nsp10 with nsp14 could conceivably dictate whether or not nsp14 functions primarily in viral RNA capping, fidelity, or other uncharacterized functions.
The ∼100-fold increase in 5-FU sensitivity observed for R80A/E82A-ExoN(+) at 40°C indicates that fidelity is likely further decreased at elevated temperature (Fig. 7). Because nsp10 is required for CoV replication, it is unlikely that the phenotype observed at 40°C is due to temperature-dependent misfolding of nsp10. This interpretation is supported by the lack of temperature sensitivity of R80A/E82A-ExoN(+) compared to WT-ExoN(+) at 40°C (Fig. 7A). Elevated temperature could be further disrupting nsp14 association with nsp10, thus either exerting a direct effect on nsp14-ExoN activity or affecting the rate at which nsp14 is associating with the replicase complex. Additionally, this phenotype could be due to destabilization of nsp14 in the absence of nsp10. The development of reagents to examine and detect nsp14 expression in tissue culture will be critical in resolving these important issues.
The results of our studies identify nsp10 as a determinant of CoV replication fidelity and support a model in which CoVs use multiple nonstructural proteins to faithfully replicate their large RNA genomes. Our data also indicate that the mechanism by which CoVs regulate their fidelity might be similar to that of DNA-based organisms. nsp7, nsp8, nsp12, and nsp13 could represent the minimal unit for RNA synthesis, while nsp10, nsp12, and nsp14 would be the primary determinants of replication fidelity. The ability to test these functions in replicating virus and to recapitulate this putative complex biochemically will be critical for a mechanistic understanding of CoV replication.
ACKNOWLEDGMENTS
We thank Nicole Sexton for critical review of the manuscript and Xiaotao Lu for technical assistance. We are grateful to members of the Denison laboratory and to Craig Cameron at Penn State University for helpful discussions.
This work was supported by United States Public Health Service awards T32 AI095202 (E.C.S.), R01 AI108197 (M.R.D.), R01 AI026603 (M.R.D.), and F32 AI108102 (E.C.S.) and NIH NHLBI grant T32 HL007751 (J.B.C.), all from the National Institutes of Health, and by Laboratoire d'Excellence “Integrative Biology of Emerging Infectious Diseases” grant no. ANR-10-LABX-62-IBEID to M.V.
REFERENCES
- 1.Arias A, Arnold JJ, Sierra M, Smidansky ED, Domingo E, Cameron CE. 2008. Determinants of RNA-dependent RNA polymerase (in)fidelity revealed by kinetic analysis of the polymerase encoded by a foot-and-mouth disease virus mutant with reduced sensitivity to ribavirin. J Virol 82:12346–12355. doi: 10.1128/JVI.01297-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeng J, Wang H, Xie X, Yang D, Zhou G, Yu L. 2013. An increased replication fidelity mutant of foot-and-mouth disease virus retains fitness in vitro and virulence in vivo. Antiviral Res 100:1–7. doi: 10.1016/j.antiviral.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 3.Zeng J, Wang H, Xie X, Li C, Zhou G, Yang D, Yu L. 2014. Ribavirin-resistant variants of foot-and-mouth disease virus: the effect of restricted quasispecies diversity on viral virulence. J Virol 88:4008–4020. doi: 10.1128/JVI.03594-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. 2006. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 439:344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vignuzzi M, Wendt E, Andino R. 2008. Engineering attenuated virus vaccines by controlling replication fidelity. Nat Med 14:154–161. doi: 10.1038/nm1726. [DOI] [PubMed] [Google Scholar]
- 6.Pfeiffer JK, Kirkegaard K. 2003. A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. Proc Natl Acad Sci U S A 100:7289–7294. doi: 10.1073/pnas.1232294100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arnold JJ, Vignuzzi M, Stone JK, Andino R, Cameron CE. 2005. Remote site control of an active site fidelity checkpoint in a viral RNA-dependent RNA polymerase. J Biol Chem 280:25706–25716. doi: 10.1074/jbc.M503444200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu X, Yang X, Lee CA, Moustafa IM, Smidansky ED, Lum D, Arnold JJ, Cameron CE, Boehr DD. 2013. Vaccine-derived mutation in motif D of poliovirus RNA-dependent RNA polymerase lowers nucleotide incorporation fidelity. J Biol Chem 288:32753–32765. doi: 10.1074/jbc.M113.484428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weeks SA, Lee CA, Zhao Y, Smidansky ED, August A, Arnold JJ, Cameron CE. 2012. A polymerase mechanism-based strategy for viral attenuation and vaccine development. J Biol Chem 287:31618–31622. doi: 10.1074/jbc.C112.401471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korboukh VK, Lee CA, Acevedo A, Vignuzzi M, Xiao Y, Arnold JJ, Hemperly S, Graci JD, August A, Andino R, Cameron CE. 2014. RNA virus population diversity, an optimum for maximal fitness and virulence. J Biol Chem 289:29531–29544. doi: 10.1074/jbc.M114.592303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coffey LL, Beeharry Y, Borderia AV, Blanc H, Vignuzzi M. 2011. Arbovirus high fidelity variant loses fitness in mosquitoes and mice. Proc Natl Acad Sci U S A 108:16038–16043. doi: 10.1073/pnas.1111650108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rozen-Gagnon K, Stapleford KA, Mongelli V, Blanc H, Failloux A-B, Saleh M-C, Vignuzzi M. 2014. Alphavirus mutator variants present host-specific defects and attenuation in Mammalian and insect models. PLoS Pathog 10:e1003877. doi: 10.1371/journal.ppat.1003877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung PPH, Watson SJ, Choy K-T, Fun Sia S, Wong DDY, Poon LLM, Kellam P, Guan Y, Malik Peiris JS, Yen H-L. 2014. Generation and characterization of influenza A viruses with altered polymerase fidelity. Nat Commun 5:4794. doi: 10.1038/ncomms5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gnädig NF, Beaucourt S, Campagnola G, Bordería AV, Sanz-Ramos M, Gong P, Blanc H, Peersen OB, Vignuzzi M. 2012. Coxsackievirus B3 mutator strains are attenuated in vivo. Proc Natl Acad Sci U S A 109:E2294–E2303. doi: 10.1073/pnas.1204022109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campagnola G, McDonald S, Beaucourt S, Vignuzzi M, Peersen OB. 2015. Structure-function relationships underlying the replication fidelity of viral RNA-dependent RNA polymerases. J Virol 89:275–286. doi: 10.1128/JVI.01574-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sadeghipour S, McMinn PC. 2013. A study of the virulence in mice of high copying fidelity variants of human enterovirus 71. Virus Res 176:265–272. doi: 10.1016/j.virusres.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sadeghipour S, Bek EJ, McMinn PC. 2013. Ribavirin-resistant mutants of human enterovirus 71 express a high replication fidelity phenotype during growth in cell culture. J Virol 87:1759–1769. doi: 10.1128/JVI.02139-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng T, Kwang J. 2014. Attenuation of human enterovirus 71 high-replication-fidelity variants in AG129 mice. J Virol 88:5803–5815. doi: 10.1128/JVI.00289-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith EC, Sexton NR, Denison MR. 2014 Thinking outside the triangle: replication fidelity of the largest RNA viruses. Annu Rev Virol 1:111–132. doi: 10.1146/annurev-virology-031413-085507. [DOI] [PubMed] [Google Scholar]
- 20.Lauring AS, Jones JO, Andino R. 2010. Rationalizing the development of live attenuated virus vaccines. Nat Biotechnol 28:573–579. doi: 10.1038/nbt.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graham RL, Becker MM, Eckerle LD, Bolles M, Denison MR, Baric RS. 2012. A live, impaired-fidelity coronavirus vaccine protects in an aged, immunocompromised mouse model of lethal disease. Nat Med 18:1820–1826. doi: 10.1038/nm.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res 117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, Poon LLM, Guan Y, Rozanov M, Spaan WJM, Gorbalenya AE. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol 331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minskaia E, Hertzig T, Gorbalenya AE, Campanacci V, Cambillau C, Canard B, Ziebuhr J. 2006. Discovery of an RNA virus 3″→5″ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci U S A 103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bouvet M, Imbert I, Subissi L, Gluais L, Canard B, Decroly E. 2012. RNA 3′-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc Natl Acad Sci U S A 109:9372–9377. doi: 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eckerle LD, Lu X, Sperry SM, Choi L, Denison MR. 2007. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J Virol 81:12135–12144. doi: 10.1128/JVI.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eckerle LD, Becker MM, Halpin RA, Li K, Venter E, Lu X, Scherbakova S, Graham RL, Baric RS, Stockwell TB, Spiro DJ, Denison MR. 2010. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog 6:e1000896. doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith EC, Blanc H, Vignuzzi M, Denison MR. 2013. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog 9:e1003565. doi: 10.1371/journal.ppat.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.te Velthuis AJW, Arnold JJ, Cameron CE, van den Worm SHE, Snijder EJ. 2010. The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Res 38:203–214. doi: 10.1093/nar/gkp904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahn D-G, Choi J-K, Taylor DR, Oh J-W. 2012. Biochemical characterization of a recombinant SARS coronavirus nsp12 RNA-dependent RNA polymerase capable of copying viral RNA templates. Arch Virol 157:2095–2104. doi: 10.1007/s00705-012-1404-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng A, Zhang W, Xie Y, Jiang W, Arnold E, Sarafianos SG, Ding J. 2005. Expression, purification, and characterization of SARS coronavirus RNA polymerase. Virology 335:165–176. doi: 10.1016/j.virol.2005.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subissi L, Posthuma CC, Collet A, Zevenhoven-Dobbe JC, Gorbalenya AE, Decroly E, Snijder EJ, Canard B, Imbert I. 2014. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc Natl Acad Sci U S A 111:E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bouvet M, Lugari A, Posthuma CC, Zevenhoven JC, Bernard S, Betzi S, Imbert I, Canard B, Guillemot J-C, Lécine P, Pfefferle S, Drosten C, Snijder EJ, Decroly E, Morelli X. 2014. Coronavirus Nsp10, a critical co-factor for activation of multiple replicative enzymes. J Biol Chem 289:25783–25796. doi: 10.1074/jbc.M114.577353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin T-Y, Schneller S, Züst R, Dong H, Thiel V, Sen GC, Fensterl V, Klimstra WB, Pierson TC, Buller RM, Gale M, Shi P-Y, Diamond MS. 2010. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Züst R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, Siddell SG, Ludewig B, Thiel V. 2011. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol 12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Menachery VD, Yount BL, Josset L, Gralinski LE, Scobey T, Agnihothram S, Katze MG, Baric RS. 2014. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2′-o-methyltransferase activity. J Virol 88:4251–4264. doi: 10.1128/JVI.03571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Decroly E, Debarnot C, Ferron F, Bouvet M, Coutard B, Imbert I, Gluais L, Papageorgiou N, Sharff A, Bricogne G, Ortiz-Lombardia M, Lescar J, Canard B. 2011. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog 7:e1002059. doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Y, Su C, Ke M, Jin X, Xu L, Zhang Z, Wu A, Sun Y, Yang Z, Tien P, Ahola T, Liang Y, Liu X, Guo D. 2011. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2′-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog 7:e1002294. doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Donaldson EF, Sims AC, Graham RL, Denison MR, Baric RS. 2007. Murine hepatitis virus replicase protein nsp10 is a critical regulator of viral RNA synthesis. J Virol 81:6356–6368. doi: 10.1128/JVI.02805-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Donaldson EF, Graham RL, Sims AC, Denison MR, Baric RS. 2007. Analysis of murine hepatitis virus strain A59 temperature-sensitive mutant TS-LA6 suggests that nsp10 plays a critical role in polyprotein processing. J Virol 81:7086–7098. doi: 10.1128/JVI.00049-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen W, Baric RS. 1996. Molecular anatomy of mouse hepatitis virus persistence: coevolution of increased host cell resistance and virus virulence. J Virol 70:3947–3960. 8648732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yount B, Denison MR, Weiss SR, Baric RS. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J Virol 76:11065–11078. doi: 10.1128/JVI.76.21.11065-11078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pauly MD, Lauring AS. 2015. Effective lethal mutagenesis of influenza virus by three nucleoside analogs. J Virol 89:3584–3597. doi: 10.1128/JVI.03483-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bouvet M, Debarnot C, Imbert I, Selisko B, Snijder EJ, Canard B, Decroly E. 2010. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog 6:e1000863. doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.von Brunn A, Teepe C, Simpson JC, Pepperkok R, Friedel CC, Zimmer R, Roberts R, Baric R, Haas J. 2007. Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PLoS One 2:e459. doi: 10.1371/journal.pone.0000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Imbert I, Snijder EJ, Dimitrova M, Guillemot J-C, Lécine P, Canard B. 2008. The SARS-coronavirus PLnc domain of nsp3 as a replication/transcription scaffolding protein. Virus Res 133:136–148. doi: 10.1016/j.virusres.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pan J, Peng X, Gao Y, Li Z, Lu X, Chen Y, Ishaq M, Liu D, Dediego ML, Enjuanes L, Guo D. 2008. Genome-wide analysis of protein-protein interactions and involvement of viral proteins in SARS-CoV replication. PLoS One 3:e3299. doi: 10.1371/journal.pone.0003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johansson E, Dixon N. 2013. Replicative DNA polymerases. Cold Spring Harb Perspect Biol 5:pii=a012799. doi: 10.1101/cshperspect.a012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Studwell-Vaughan PS, O'Donnell M. 1993. DNA polymerase III accessory proteins. V. Theta encoded by holE. J Biol Chem 268:11785–11791. [PubMed] [Google Scholar]
- 50.Perrino FW, Harvey S, McNeill SM. 1999. Two functional domains of the ε subunit of DNA polymerase III. Biochemistry 38:16001–16009. doi: 10.1021/bi991429+. [DOI] [PubMed] [Google Scholar]
- 51.Taft-Benz SA, Schaaper RM. 2004. The theta subunit of Escherichia coli DNA polymerase III: a role in stabilizing the epsilon proofreading subunit. J Bacteriol 186:2774–2780. doi: 10.1128/JB.186.9.2774-2780.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamdan S, Bulloch EM, Thompson PR, Beck JL, Yang JY, Crowther JA, Lilley PE, Carr PD, Ollis DL, Brown SE, Dixon NE. 2002. Hydrolysis of the 5′-p-nitrophenyl ester of TMP by the proofreading exonuclease (ε) subunit of Escherichia coli DNA polymerase III. Biochemistry 41:5266–5275. doi: 10.1021/bi0159480. [DOI] [PubMed] [Google Scholar]
- 53.Scheuermann RH, Echols H. 1984. A separate editing exonuclease for DNA replication: the epsilon subunit of Escherichia coli DNA polymerase III holoenzyme. Proc Natl Acad Sci U S A 81:7747–7751. doi: 10.1073/pnas.81.24.7747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scheuermann R, Tam S, Burgers PM, Lu C, Echols H. 1983. Identification of the epsilon-subunit of Escherichia coli DNA polymerase III holoenzyme as the dnaQ gene product: a fidelity subunit for DNA replication. Proc Natl Acad Sci U S A 80:7085–7089. doi: 10.1073/pnas.80.23.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen Y, Cai H, Pan J, Xiang N, Tien P, Ahola T, Guo D. 2009. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc Natl Acad Sci U S A 106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lugari A, Betzi S, Decroly E, Bonnaud E, Hermant A, Guillemot JC, Debarnot C, Borg JP, Bouvet M, Canard B, Morelli X, Lecine P. 2010. Molecular mapping of the RNA cap 2′-O-methyltransferase activation interface between severe acute respiratory syndrome coronavirus nsp10 and nsp16. J Biol Chem 285:33230–33241. doi: 10.1074/jbc.M110.120014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Joseph JS, Saikatendu KS, Subramanian V, Neuman BW, Brooun A, Griffith M, Moy K, Yadav MK, Velasquez J, Buchmeier MJ, Stevens RC, Kuhn P. 2006. Crystal structure of nonstructural protein 10 from the severe acute respiratory syndrome coronavirus reveals a novel fold with two zinc-binding motifs. J Virol 80:7894–7901. doi: 10.1128/JVI.00467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kelley LA, Sternberg MJE. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 59.Schrodinger LLC. 2010. The PyMOL Molecular Graphics System, Version 1.3r1. Schrodinger LLC, Cambridge, MA. [Google Scholar]