ABSTRACT
Sulfolobus spindle-shaped virus 1 represents a model for studying virus-host interaction in harsh environments, and it is so far the only member of the family Fuselloviridae that shows a UV-inducible life cycle. Although the virus has been extensively studied, mechanisms underpinning the maintenance of lysogeny as well as those regulating the UV induction have received little attention. Recently, a novel SSV1 transcription factor, F55, was identified. This factor was able to bind in vitro to several sequences derived from the early and UV-inducible promoters of the SSV1 genome. The location of these binding sites together with the differential affinity of F55 for these sequences led to the hypothesis that this protein might be involved in the maintenance of the SSV1 lysogeny. Here, we report an in vivo survey of the molecular events occurring at the UV-inducible region of the SSV1 genome, with a focus on the binding profile of F55 before and after the UV irradiation. The binding of F55 to the target promoters correlates with transcription repression, whereas its dissociation is paralleled by transcription activation. Therefore, we propose that F55 acts as a molecular switch for the transcriptional regulation of the early viral genes.
IMPORTANCE Functional genomic studies of SSV1 proteins have been hindered by the lack of similarity with other characterized proteins. As a result, few insights into their in vivo roles have been gained throughout the last 3 decades. Here, we report the first in vivo investigation of an SSV1 transcription regulator, F55, that plays a key role in the transition from the lysogenic to the induced state of SSV1. We show that F55 regulates the expression of the UV-inducible as well as the early genes. Moreover, the differential affinity of this transcription factor for these targets allows a fine-tuned and temporal coordinated regulation of transcription of viral genes.
INTRODUCTION
The majority of viruses isolated from Crenarchaea shows virion morphotypes that have not been observed for viruses infecting Bacteria and Eukarya. Consequently, eight novel viral families have been introduced in order to classify these novel viruses (Fuselloviridae, Lipothrixviridae, Rudiviridae, Guttaviridae, Globuloviridae, Bicaudaviridae, Ampullaviridae, and Clavaviridae), but so far, there are still several unique archaeal viruses that remain to be classified (1–3). Many analyses of environmental samples have shown that spindle-shaped viruses are abundant and occupy several niches, including deep sea hydrothermal vents (4, 5), hypersaline environments (6, 7), anoxic freshwaters (8), cold Antarctic lakes (9), terrestrial hot springs (10–12), and acidic mines (13, 14), where they frequently outnumber head-tailed viruses. Notably, this unique virion morphotype seems to be a hallmark of viruses infecting Archaea, since it has never been observed for bacteriophages or eukaryal viruses (15). To date, the family Fuselloviridae comprises nine members (SSV1, SSV2, SSV4, SSV5, SSV6, SSV7, SSV8, SSV9, and ASV1) isolated from several geographic locations (2). The lack of functional characterization of proteins encoded by the fuselloviral genomes has limited the dissection of fundamental processes, such as (i) virion uptake, assembly and release, (ii) transcriptional regulation, (iii) genome replication, and (iv) lysogeny/induction switch. Indeed, with the exception of a few proteins (16–23), structural and functional annotations in main databases are not available for fuselloviral gene products (2, 22).
SSV1 is the most extensively characterized member of this viral family and is the only one showing a UV-inducible life cycle. Upon infecting a host cell, SSV1 integrates one copy of its genome into the host chromosome at an arginyl-tRNA gene, forming a provirus (24). However, unlike the well-characterized lambda phage, for which lysogenic cells harbor only the provirus, SSV1 lysogens carry also ∼5 copies of the episomal DNA per cell. This led to the hypothesis that SSV1 expresses a minimal set of genes to ensure a basal level of replication that is required for maintaining the carrier state (23). Moreover, structural proteins (VP1, VP2, and VP3) are constitutively expressed under conditions for which viral replication is not induced (23), allowing the production of viral particles by the lysogenic cells. Upon exposure to UV light, SSV1 exhibits a temporally coordinated pattern of gene expression. At first, it activates the expression of a UV-inducible transcript (Tind), followed by the transcription of the early (T5, T6, and T9), late (T1/2, T3, Tx, and T4/7), and late-extended (T4/7/8) RNAs. This cascade of events leads, in turn, to the induction of the SSV1 genome replication and eventually to a steep increase of the viral titer (25).
Despite the extensive characterization that SSV1 has received over the last 30 years (25–28), the mechanisms underpinning the transition from lysogenic growth to viral induction are still murky. Very recently, a new mRNA (Tlys) was discovered, which is transcribed in the direction opposite to that of the UV-inducible Tind (23). Since Tlys is one of the most abundant SSV1 transcripts during lysogenic growth, it was suggested that the encoded protein could play a fundamental role in the maintenance of the carrier state. Indeed, Tlys encodes a 55-amino-acid protein (F55) that interacts, in a concentration-dependent manner, with tandem-repeat sequences clustered within the UV-inducible region of the viral genome. Moreover, F55 could act as a transcription repressor, since these operators encompass both the transcription start sites (TSSs) and the B recognition elements (BREs) of the T5, T6, Tind, and Tlys promoters (23). So far, F55 is the only transcription repressor for which a defined role in the regulation of a fuselloviral life cycle has been proposed. Here, we report an in vivo survey of the molecular events occurring upon irradiation at the UV-inducible region of the SSV1 genome, with a focus on the pleiotropic effect of F55 on several SSV1 promoters. We show that F55 acts as the molecular switch controlling the SSV1 life cycle.
MATERIALS AND METHODS
Strains, media, growth conditions, and UV irradiation.
An SSV1 lysogenic strain of Sulfolobus solfataricus (SSV1-InF1) was generated by infecting the uracil auxotrophic mutant InF1 (29), as described elsewhere (28, 30). An aliquot from a frozen sample of SSV1-InF1 culture was thawed, and a few microliters was spotted onto an SCVYU-Gelrite plate and incubated at 75°C. After 3 to 5 days of incubation, local growth areas (spots) were inoculated into 50 ml of SCVYU medium, i.e., a glycine-buffered Brock's basal salt solution, supplemented with 0.2% (wt/vol) sucrose, 0.2% (wt/vol) Casamino Acids, 1× vitamins (31), 0.005% (wt/vol) yeast extract, and 0.02 mg ml−1 uracil, with pH adjusted to 3.5 with concentrated H2SO4. Cultivation of the Sulfolobus strains was conducted in a 250-ml Erlenmeyer flask with a long neck at 75°C with a shaking rate of 180 rpm using a MaxQ 4000 benchtop orbital shaker (Thermo Scientific). Cell growth was monitored spectrophotometrically at 600 nm (optical density at 600 nm [OD600]) by means of a Variant Cary 50 Bio UV/visible spectrophotometer (McKinley Scientific). Once the culture reached the logarithmic phase of growth, it was diluted to an OD600 of 0.05 in 50 ml of fresh SCVYU medium and allowed to grow to an OD600 of 0.5 to 0.8.
The SSV1-InF1 culture was UV-irradiated by using a fluence of 45 J m−2 and an irradiance of 0.5 J m−2 s−1, as described elsewhere (32). Growth was spectrophotometrically monitored before and after the treatment to construct comparative growth curves. Samples were taken at 2, 4, 6, 8, and 10 h posttreatment, and pellets for each time point were obtained through centrifugation at 3,000 × g for 10 min using a 5810R centrifuge (Eppendorf). Pellets were treated for total DNA, RNA, and protein preparations (see below).
Semiquantitative PCR and EcoRI total DNA digestions.
SSV1-InF1 cell pellets, obtained as described previously, were treated for preparation of highly pure total DNA using the DNeasy tissue kit (Qiagen), following the manufacturer's instructions. The concentration of the DNA samples was spectrophotometrically determined using a Nanodrop 2000 spectrophotometer (Thermo-Scientific), by performing the measurements in triplicate. Their purity was assessed by the 260-nm/280-nm adsorption ratio, and only DNA samples with a value of 1.8 or higher were used for subsequent experiments.
In order to detect variations of the viral DNA content among the different samples, total DNA samples from irradiated SSV1-InF1 cultures were analyzed by semiquantitative PCR (sqPCR). With this aim, two primer couples were designed using Primer3 software (http://bioinfo.ut.ee/primer3-0.4.0/), in order to amplify (i) a 155-bp region of the SSV1 single-copy gene vp2 and (ii) a 108-bp region of the host single-copy gene orc1 (Table 1). An sqPCR master mix was set up as follows: 1× Taq buffer, 2.5 mM MgCl2, a 0.2 mM concentration of a deoxynucleoside triphosphate (dNTP) mix, 0.6 μM orc1-fw, 0.6 μM orc1-rv, 0.6 μM vp2-fw, 0.6 μM vp2-rv, and 50 U ml−1 of Taq DNA polymerase (Thermo Scientific) in a total volume of 500 μl. The master mix was then separated into seven aliquots of 60 μl each, placed in different tubes, and labeled the control sample (mock treated, collected at an OD600 of 0.5) and the 2-h, 4-h, 6-h, 8-h, and 10-h samples (UV-treated samples). To each tube, 100 ng of total DNA from mock-treated or UV-irradiated samples was added as the template. The aliquots were then split into three subaliquots (20 μl each), labeled (20th, 25th, and 30th), and placed into a personal Mastercycler (Eppendorf). A negative (no-template) control and two positive controls for the amplification conditions were also included in the analysis. The thermal cycling protocol was as follows: an initial denaturation step of 5 min at 95°C, followed by 30 cycles of 40 s at 95°C, 40 s at 62°C, and 1 min at 72°C. A final step at 72°C was carried out for 10 min at the end of the 30th cycle. For each time point, tubes were taken from the thermocycler at the 20th, 25th, and 30th cycles of amplification. PCR products were run on a 2% (wt/vol) agarose gel in 1× TAE buffer (40 mM Tris, 20 mM acetic acid, and 1 mM EDTA).
TABLE 1.
Primers used for semiquantitative PCR assays on ChIP samples
| Name | Sequence (5′–3′) | Length (nt) |
|---|---|---|
| T5-fw | CCCAAACACTGTGTATATAGAG | 22 |
| T5-rv | AGTTTGTGCCATATTCCCAT | 20 |
| T6-fw | ATGATAATATTAAATGATTCACGAT | 25 |
| T6-rv | TTTCGGGTTTGGGGTGAAAC | 20 |
| Tind-fw | CTGCTGTCTGACAAGAGTTT | 20 |
| Tind-rv | GATTTTGCACATCCCATATT | 20 |
| Tlys-fw | ATCGTGAATCATTTAATATTATCAT | 25 |
| Tlys-rv | ATTGGAATCGAAACGGTCAC | 20 |
| orc1-fw | TATAAATTGTTATAGACATAGAACGCTGTA | 30 |
| orc1-rv | TTAAATACTTCTTGTGCCGATAGTCC | 26 |
| vp2-fw | GGAGGGTACATCGCTACCTTATGA | 24 |
| vp2-rv | CAGTAGGGCTGACAGTAAACTACG | 24 |
The same DNA samples were enzymatically digested using the sequence-specific endonuclease EcoRI, whose target sequence recurs four times in the SSV1 genome. In brief, 2 μg of each total DNA sample were digested overnight with 20 units of EcoRI (Roche) at 37°C and run on a 1% (wt/vol) agarose gel.
Northern blot analysis.
Highly pure total RNA samples were prepared from mock-treated and UV-irradiated SSV1-InF1 cultures by means of the TRIzol reagent (Sigma-Aldrich), according to the manufacturer's instructions. RNA pellets were dissolved in nuclease-free water, and concentration was determined by means of a Nanodrop 1000 spectrophotometer (Thermo Scientific). Sample purity was checked through the determination of the 260-nm/280-nm adsorption ratio, and only samples showing a ratio of 1.9 or higher were used for subsequent experiments.
For each sample, 20 μg of total RNA was run on a denaturing, formaldehyde-containing 2.0% (wt/vol) agarose gel and then transferred onto a nylon membrane (Hybond-XL; Amersham-Pharmacia). T4 polynucleotide kinase (Fermentas Life Sciences) was used to label 5′ ends of single-stranded oligonucleotides Tlys-rv (5′-AAGTTCTTCAATGCGTCTTCTGATT-3′), Tind-fw (5′-TCTGAGCTACTAATACTGCTTGAAT-3′), and 16S-rv (5′-CTCTCCTACTCGGGTGGAGCAAC) with radioactive [γ-32P]ATP, following the manufacturer's instructions. Purification of radiolabeled oligonucleotides was achieved by gel filtration chromatography using Illustra Nick columns (Amersham Biosciences), and hybridizations with single-stranded DNA probes were carried out as described elsewhere (33). Probes were eventually removed by boiling in 0.1% (wt/vol) sodium dodecyl sulfate (SDS) for 10 min in order to reuse the membrane for subsequent hybridizations. Radioactive signals were quantified by means of a Molecular Dynamics Bio-Rad PhosphorImager (Quantity One software) to determine the relative abundance of the transcripts (16S, Tlys, and Tind). Tlys/16S and Tind/16S ratios were determined to normalize RNA signals and compensate for operator errors. The size of the Northern blot signals was determined using RNA molecular size markers (Roche) as standards.
Western blot analysis.
Cells from 30 ml of mock-treated and UV-irradiated SSV1-InF1 cultures were harvested by centrifugation, and pellets were suspended in 2 ml of 10 mM Tris-HCl (pH 8.0) (lysis buffer), containing Complete 600 protease inhibitor cocktail tablets (Roche). Cells were lysed by sonication at 20% of the maximal amplitude for 3 min, alternating 20 s of pulse-on and 20 s of pulse-off, by means of an ultrasonic liquid processor (Heat System Ultrasonic, Inc.). Lysates were centrifuged at 30,000 × g (SW41 rotor; Beckman) for 30 min in order to clarify crude protein extracts. Protein concentration was spectrophotometrically determined by standardized Coomassie (Bradford) protein assays and normalized for each sample, as described elsewhere (34). In brief, Bradford assays were performed on bovine serum albumin (BSA; Thermo Scientific) as a standard to construct a calibration curve. In particular, known protein concentrations (1, 2, 4, 6, and 8 μg/ml) were plotted against their absorbance at 595 nm. Concentration of protein samples was determined by means of the standard curve, and sample quality was assessed by SDS-PAGE followed by Coomassie staining.
A serum sample (15 mg of total protein) from an F55-immunized rabbit (Innovagen AB) was loaded onto a 1-ml HiTrap protein A column (GE Healthcare) connected to a fast-performance liquid chromatography system (ÄKTA; GE Healthcare), and total IgGs were purified following the manufacturer's instructions. Antibody-containing fractions were pooled and dialyzed against 20 mM sodium phosphate buffer (pH 8.0), and protein concentration (0.4 mg ml−1) was determined by the Coomassie (Bradford) protein assay, as described above. Antibody integrity was checked by running 10 μg of total IgG sample on 15% SDS-PAGE. For Western blot hybridizations, total protein samples (10 μg) were run on 15% SDS-PAGE and electrotransferred onto Immobilon polyvinylidene difluoride (PVDF) membranes (Millipore). Subsequently, membranes were (i) incubated for 1 h at room temperature in blocking solution, i.e., 1× TBS-T (50 mM Tris [pH 7.5], 150 mM NaCl, and 0.1% Tween 20) containing 5% (wt/vol) BSA (Sigma); (ii) incubated for 16 h at 4°C with total IgG sample (see above) diluted (1:1,000) in blocking solution; (iii) washed three times for 15 min with 1× TBS-T at room temperature; (iv) incubated for 1 h at room temperature with horseradish peroxidase-conjugated goat anti-rabbit IgG (Roche) diluted (1:10,000) in 1× TBS-T; (v) washed twice for 15 min with TBS-T; and (vi) washed once with TBS. Detection by enzyme-linked chemiluminescence was performed with an Immobilon Western chemiluminescent horseradish peroxidase (HRP) substrate kit (Millipore) and a ChemiDoc XRS+ system (Bio-Rad), according to the manufacturer's instructions. The F55 concentration in the analyzed samples was determined using a calibration curve, which was constructed by plotting known amounts of F55 (from 10 to 35 ng) against densitometric values measured using the Quantity One software (Bio-Rad). The Western blot data were validated by repeating the experiment in triplicate.
ChIP-sqPCR analysis.
SSV1-InF1 cells were cultivated in 900 ml of SCVYU medium in a 2-liter Erlenmeyer flask and grown to an OD600 of 0.6. A 300-ml control sample was harvested before UV treatment of the remaining volume (600 ml). Samples were harvested after 2 and 4 h of incubation and rapidly cooled down at 37°C, before the addition of 1% (wt/vol) formaldehyde (Sigma) for DNA-protein cross-linking. Incubation at 37°C was performed for 5 min with a shaking rate of 120 rpm. The cross-linking reaction was then quenched by the addition of glycine (125 mM final concentration) to the cultures, followed by an additional incubation of 5 min at 37°C. Afterwards, cells were collected by centrifugation at 3,000 × g for 15 min (JA-14 rotor; Beckman). Pellets were washed twice with 1× PBS (137 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4 · 2 H2O, 1.76 mM KH2PO4 [pH 7.4]) and finally resuspended in 6 ml of 1× DNase buffer (400 mM Tris-HCl, 100 mM NaCl, 60 mM MgCl2, and 10 mM 8 CaCl2, pH 7.9). Cells were lysed by sonication at 20% of the maximal amplitude for 18 min, alternating 3 s of pulse-on and 9 s of pulse-off, by means of an ultrasonic liquid processor (Heat System Ultrasonic, Inc.). Lysates containing cross-linked DNA-protein complexes were clarified by centrifuging for 30 min at 30,000 × g (SW41 rotor; Beckman). In order to further narrow down the length of the DNA fragments, RNase-free DNase I (Roche) was added at a final concentration of 30 U/ml, followed by incubation of 30 min at 37°C. To recover DNA-protein complexes, samples were subjected to phenol extraction by the addition of 1 volume of phenol-chloroform-isoamyl alcohol (25:24:1; Sigma) and centrifugation for 10 min at 3,000 × g. The DNA-protein complexes were recovered by ethanol precipitation and centrifugation for 10 min at 15,000 × g. Pellets were dissolved in 20 mM sodium phosphate buffer (pH 8.0) and treated with 100 μg ml−1 of RNase A (Invitrogen) for 1 h at 37°C. Cross-linked F55-DNA complexes were enriched by affinity chromatography using a 1-ml HiTrap protein A column (GE Healthcare) connected to a fast-performance liquid chromatography system (ÄKTA). In brief, the column was equilibrated in binding buffer (20 mM sodium phosphate buffer [pH 8.0]), loaded with purified IgG from an F55-immunized rabbit (Innovagen AB), and washed with 5 ml of binding buffer. DNA-protein samples were loaded onto the column, and 5 ml of binding buffer was used to elute unspecific complexes. F55-DNA specific complexes were then eluted from the column by washing with 0.1 M citrate buffer (pH 6.2) (elution buffer). In order to de-cross-link F55-DNA complexes, the eluted fractions were pooled and incubated at 65°C for 16 h with a shaking rate of 180 rpm. Free-protein DNA was obtained by purification using the MinElute PCR purification kit (Qiagen), according to the manufacturer's instructions. The concentration of the DNA samples was measured using a Nanodrop 2000 spectrophotometer, as described above. To check the enrichment of DNA fragments containing F55 target sequences, chromatin immunoprecipitation assays and semiquantitative PCR (ChIP-sqPCR) were set up using the primer pairs listed in Table 1. Master mixes were prepared as follows: 1× Taq buffer, 2.5 mM MgCl2, 0.2 mM dNTP mix, 0.3 μM forward primer, 0.3 μM reverse primer, and 50 U ml−1 of Taq DNA polymerase (Thermo Scientific) in a total volume of 250 μl. Two aliquots of 50 μl each were used to perform negative and positive controls of the amplification, while to the remaining 150 μl was added 1 ng of enriched DNA obtained by ChIP (see above). The aliquot of 150 μl was then divided into three tubes (50 μl each) which were labeled (20th, 25th, and 30th) and placed in a personal Mastercycler (Eppendorf). The thermal cycling protocol was as follows: an initial denaturation step of 3 min at 95°C, followed by 30 cycles of 40 s at 95°C, 40 s at 50°C, and 1 min at 72°C. For amplification of the host sequence orc1, the annealing temperature was 62°C. Tubes were taken from the thermocycler at the 20th, 25th, and 30th cycles of amplification. PCR products were run on a 2% (wt/vol) agarose gel in 1× TAE buffer. The same procedure was carried out by using as templates the enriched DNA samples from mock-treated and UV-irradiated SSV1-InF1 cells collected after 2 and 4 h postirradiation.
RESULTS
Host response to UV irradiation.
In order to investigate the role of F55 in the SSV1 life cycle, an S. solfataricus InF1 strain carrying the virus (SSV1-InF1) was treated by UV irradiation as described elsewhere (32). The effect of the treatment on its physiology was assessed by measuring the generation time and cell viability of the mock- and UV-treated cultures. We found that, in the immediate aftermath of the treatment, cell viability of the UV-treated culture was approximately 35% of that of the untreated control. Furthermore, the growth rate of the UV-irradiated sample was lower than that of the mock-treated control, probably as a consequence of the stress caused by both the irradiation itself and the viral induction (Fig. 1), as was also observed in previous experiments (32).
FIG 1.

Growth curves of mock- and UV-treated SSV1-InF1 cultures. The OD600 values were measured every 2 h and plotted versus the incubation time. Before being mock or UV treated, cells were grown exponentially to an OD600 of 0.5 (21st hour of incubation). Afterwards, the culture was split in two and incubated to 75°C to estimate the effect of the treatment. Error bars show standard deviations (n = 3). *, P < 0.05.
Time course of the SSV1 replication induction upon UV irradiation.
Induction of SSV1 replication upon UV irradiation was investigated in a time course experiment with a 10-h window, following a UV irradiation protocol reported recently (32). Samples were withdrawn from both mock-treated and UV-irradiated SSV1-InF1 cultures every 2 h during incubation, and the relative amount of SSV1 was estimated using semiquantitative PCR analysis of total DNAs extracted from cell samples. Two single-copy genes were chosen for this analysis, i.e., the viral gene vp2, representing the SSV1 genome, and orc1 for the host genome. PCR was conducted for 20, 25, and 30 cycles of amplification, and the PCR products were analyzed by agarose gel electrophoresis. We found that, when the genes were amplified for the same numbers of PCR cycles, the intensity of the PCR product of the internal control orc1 (108 bp) was very similar in all the samples. However, the intensity of the vp2 amplicon (155 bp) was significantly increased in the samples after UV irradiation, starting from hour 2 and peaking at hour 8 postirradiation (Fig. 2). We reasoned that the increase of viral DNA, before completion of viral gene expression (Fig. 2, 2 h posttreatment), resulted from the irradiation-mediated cell division arrest (Fig. 1), which did not affect the basal viral replication. Thereafter, transcriptional activation was completed for all viral genes (25), and the induction of the SSV1 replication accounted for the further increase of relative amounts of viral DNA in UV-irradiated cultures (Fig. 2, 6 to 10 h).
FIG 2.
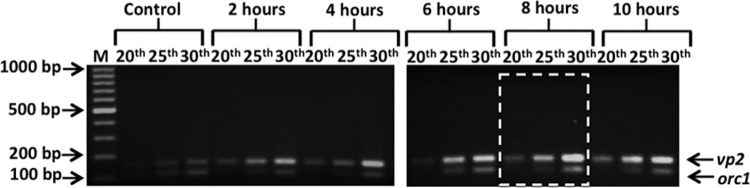
Time course of the viral replication induction after UV irradiation by semiquantitative PCR. Molecular size markers are indicated on the left; host (orc1, 108 bp) and viral (vp2, 155 bp) PCR products are indicated on the right. Total DNA samples were prepared from mock-treated cells (control) and UV-treated cultures collected at 2, 4, 6, 8, and 10 h postirradiation. The maximum amount of SSV1 DNA is detectable for the sample collected 8 h posttreatment (dashed white box).
To confirm the temporal onset of UV-induced viral replication, the total DNA extracted was digested with EcoRI and analyzed by agarose gel electrophoresis. As shown in Fig. 3, the EcoRI fragments of the SSV1 genome (7.9, 2.9, 2.4, and 2.1 kb) became dominant in the UV-irradiated cells, with the largest amount being detectable 8 to 10 h postirradiation. Altogether, the above data indicated that the previously established procedure for UV irradiation was suitable for studying the induction of SSV1 replication (32).
FIG 3.
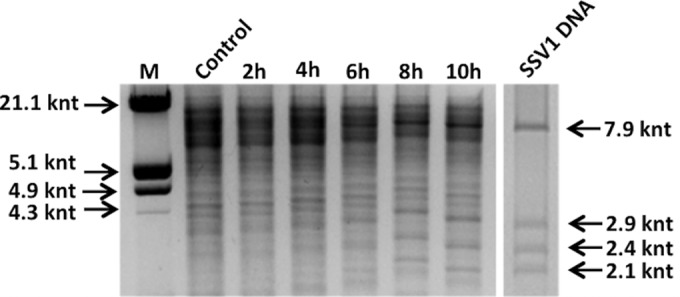
Detection of the viral DNA after UV irradiation by EcoRI restriction analysis. The restriction profiles of total DNA samples from control cultures and SSV1-InF1-irradiated cells and of SSV1 episomal DNA digested with EcoRI are shown. Molecular size markers are indicated on the left; SSV1-derived fragments (7.9, 2.9, 2.4, and 2.1 kbp) are indicated on the right. The intensity of the SSV1 fragments is higher for the sample collected 8 to 10 h postirradiation.
Tind and Tlys transcription patterns in mock- and UV-treated cells.
In order to reveal the molecular circuit regulating the maintenance of SSV1 lysogeny and to identify the molecular switch to the induced state of SSV1, a transcriptional analysis was carried out on the head-to-head-oriented transcripts Tind and Tlys, the main players in SSV1 UV-inducible expression (Fig. 4A). Total RNA samples were prepared from mock- and UV-treated SSV1-InF1 cells and analyzed by Northern hybridization to detect Tind and Tlys transcripts (Fig. 4B). Normalization for all samples was performed by measuring the signal of the housekeeping gene 16S and by calculating the Tind/16S and Tlys/16S signal ratios.
FIG 4.
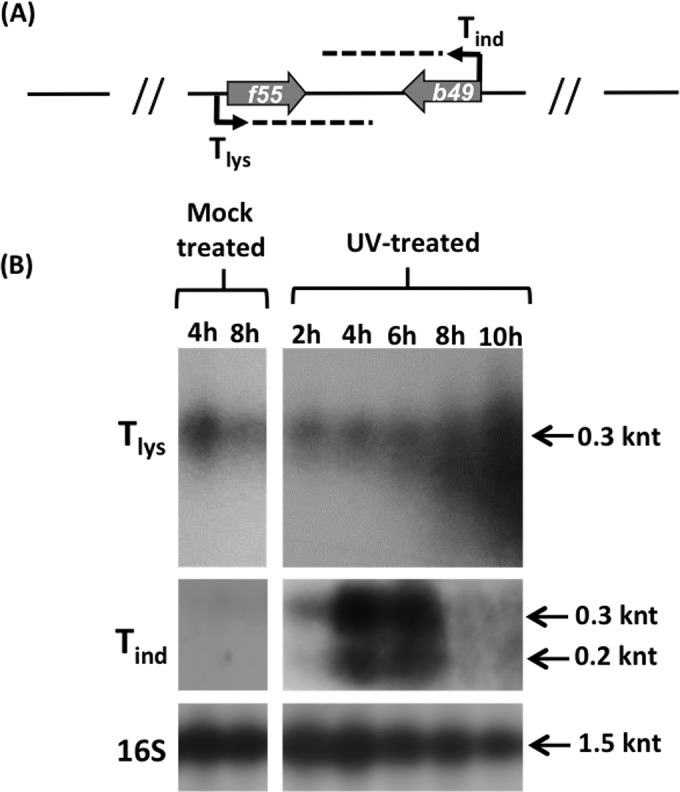
Transcription analysis of Tlys and Tind. (A) Schematic representation of the head-to-head-oriented transcripts Tind and Tlys. (B) SSV1-InF1 total RNAs isolated from mock- and UV-treated cells at different time points were analyzed by Northern hybridization to detect Tlys and Tind transcripts. The hybridization signals were normalized using the 16S housekeeping gene. The Tind transcript was expressed over a short time and only in UV-irradiated cells, whereas Tlys was detectable in both mock-treated and UV-irradiated cells. Degradation occurred around the 8th to 10th hour only for the Tlys transcript.
Tind was not detectable in mock-treated cells, which is consistent with the transcript being repressed in the carrier state (lysogeny). Conversely, soon after UV irradiation (2 h posttreatment) two typical Tind hybridization signals (25) were detected (200 nucleotides [nt] and 300 nt). Then, the Tind/16S ratio increased 3-fold 4 to 6 h after UV irradiation. Thereafter, the intensity of the Tind signal dropped sharply, and the signal became undetectable 6 h posttreatment (Fig. 4B).
On the other hand, the antagonistic transcript Tlys showed a downregulation of its expression in the mock-treated culture between 4 and 8 h, as judged by the 50% decrease of the signal ratio Tlys/16S. Interestingly, this value did not change between the 2nd and the 4th hour postirradiation (Fig. 4B), despite the increase of the SSV1 copy number in the same time window, as evidenced by sqPCR (Fig. 2).This suggests that there could exist a fine-tuning mechanism regulating the expression of these two convergent genes. Furthermore, the 300-nt Tlys signal did not occur as a discrete band but formed a continuous smearing, with smaller RNA products mainly visible between 8 and 10 h posttreatment (Fig. 4B). This indicated that Tlys could be degraded by a posttranscriptional regulation mechanism. If so, the apparent increase of Tlys signal should not reflect the actual change of content of the encoded transcriptional regulator (F55) in the UV-irradiated cells (see below).
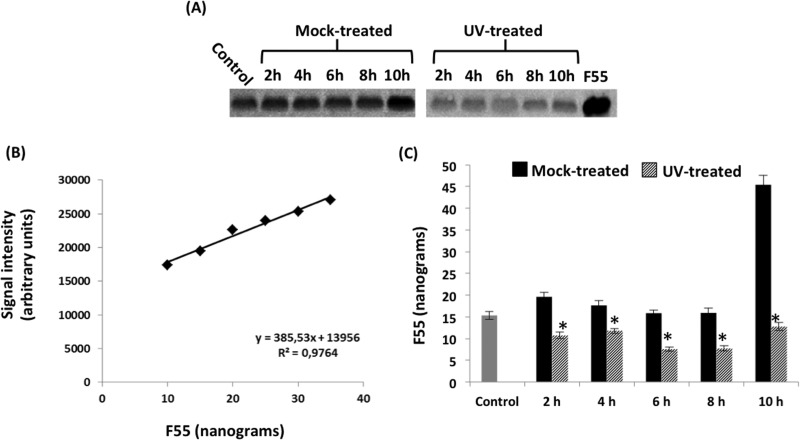
In vivo expression and DNA-binding activity of F55.
To quantify the putative lysogeny regulator F55 in UV-induced and mock-treated cells, its content was determined by densitometric analysis on immunoblots (Fig. 5A) and compared to known amounts of recombinant F55 as a reference (Fig. 5B). The intensity of the chemiluminescent signal obtained from the standards was linear in the range considered. Interestingly, throughout the time window analyzed, the concentration of F55 in UV-treated cells was lower than that in the corresponding mock-treated samples (Fig. 5C). Furthermore, a 50% drop of the F55 cellular concentration (Fig. 5C, 2 h, gray bar) coincided with the onset of the Tind transcription (Fig. 4B, 2 h, UV treated). Together these results support the hypothesis that F55 plays a role in maintaining the lysogeny. Furthermore, the simultaneous reduction of F55 protein and the augmentation of Tlys signal at the 8th to 10th hour (Fig. 4B) suggest that the regulation of Tlys expression must occur at the posttranscriptional level.
FIG 5.
Western blot analysis of F55 in mock- and UV-treated InF1-SSV1 cells. (A) Western blot of cell extracts from control, mock-treated, and UV-treated samples. (B) F55 quantification was performed using a standard curve. (C) Quantitative data are reported as histograms. Error bars show the standard deviations (n = 3), and a control sample (gray bar) is used as a reference. The amount of F55 in the UV-treated culture is significantly lower than that in the mock one at every time point analyzed (*, P < 0.05).
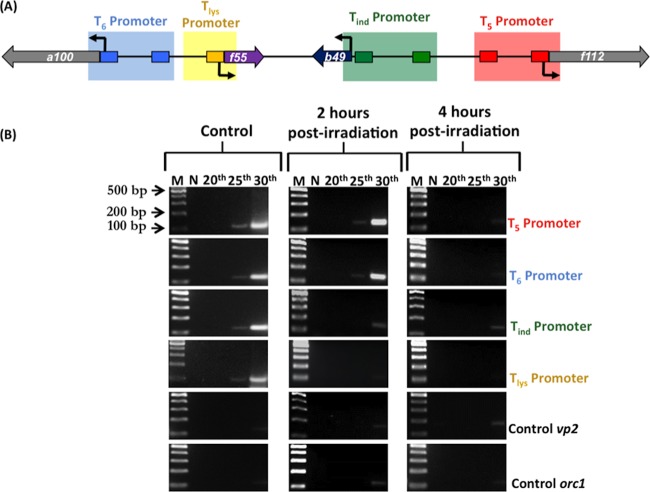
It was previously demonstrated that F55 binds in vitro, with differential affinity, to DNA sequences derived from the promoters of T5, T6, Tind, and Tlys transcripts. Based on the location of the binding sites, i.e., partially overlapping the BRE or encompassing the TSS, it was hypothesized that this regulator could function as a transcriptional repressor (23). To investigate binding of F55 to the aforementioned promoters in vivo (Fig. 5A), DNAs were recovered by ChIP assays before and after the UV treatment and subjected to sqPCR analysis (ChIP-sqPCR) as described above. In lysogenic cells, promoter regions of T5, T6, Tind, and Tlys were detected in ChIP-sqPCR already after 25 cycles (Fig. 6B, control), indicating that F55 binds to these promoters in vivo. Two hours after UV irradiation, binding of F55 to its own promoter was depleted, whereas the interaction with that of Tind was greatly weakened (Fig. 6B, 2 h postirradiation). Subsequently, the promoter regions of the early transcripts T5 and T6 were released by F55, and indeed, the relative ChIP-sqPCR signals fell to an undetectable level (Fig. 6B, 4 h postirradiation). Altogether, these results indicate that the cascade expression of the SSV1 early genes is regulated through a differential binding of F55 to its target operators.
FIG 6.
Chromatin immunoprecipitation (ChIPs) on the UV-inducible region of SSV1 using F55 specific antibodies. (A) Scheme of the UV-inducible region of SSV1 genome. The amplified regions of the target promoters that are recognized by F55 are in blue, yellow, green, and red. All the promoters except that of Tlys contain two F55 binding sites. (B) Total DNA samples prepared from control and UV-treated cultures and collected at 2 and 4 h postirradiation were used as the templates for semiquantitative PCRs. Two negative controls, vp2 (viral) and orc1 (host), were included to assess method specificity. N is the negative control for PCRs.
DISCUSSION
Numerous transcription analyses of the UV-inducible fusellovirus SSV1 have been carried out in S. solfataricus to dissect viral gene expression patterns before and after UV irradiation. Nevertheless, it was still unclear how this fusellovirus regulates the transition from the lysogeny to the induced status during its life cycle. Our recent work has assigned a key role in this process to the DNA-binding protein F55, which is encoded by the most abundant RNA (Tlys) expressed by SSV1 in the carrier state (23). In order to shed further light on the role of F55 in the regulation of the SSV1 life cycle, an in vivo analysis of the events occurring soon after the UV irradiation was carried out. Several lines of evidence indicate that F55 is indeed the key regulator of the lysogeny/induction transition of SSV1, including the following: (i) F55 is the only transcriptional regulator expressed by SSV1 in the lysogenic status; (ii) it binds in vivo to the promoters of Tind, Tlys, T5, and T6 in the absence of UV stimulus, as shown by the ChIP-sqPCR experiments; (iii) Tind expression is shut down when F55 is bound to its promoter, as shown by Northern blotting; and (iv) transcription activation of Tind occurs only upon release of F55 in the immediate aftermath of the UV irradiation, i.e., within 2 h. Notably, the transcriptional regulation of Tind is crucial for the lysogeny/induction switch, since it is the first RNA whose expression is unlocked in UV-irradiated cells (25).
ChIP-sqPCR data demonstrated that F55 dissociates upon UV irradiation first from the Tind (2 h posttreatment) and subsequently from the T5 and T6 promoters (4 h posttreatment) (Fig. 6B). This is in perfect agreement with its differential affinity for these regulatory sequences in vitro (T5 = T6 > Tind) (23). Intriguingly, the transcription of T5 and T6 is repressed during the SSV1 carrier state and is activated in UV-irradiated cells at 4 h posttreatment (25). These data strongly indicate that F55 regulates the transcriptional activity of these mRNAs in a fashion similar to that of Tind, i.e., by repressing their expression during the lysogenic growth and allowing their transcription activation through clearance of their target sites upon UV irradiation. Although the release of F55 from its targets coincides with the transcriptional activation of the relative genes, it is not possible to exclude the contribution of other factors to enhance their expression levels.
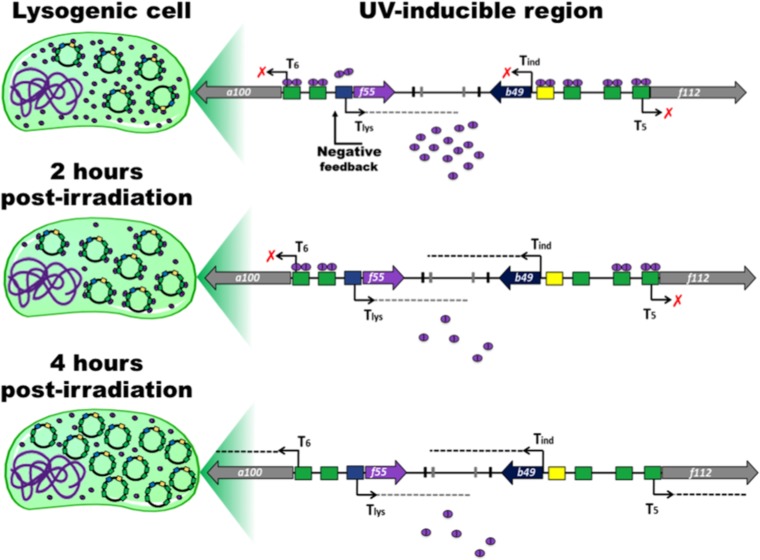
Quantification of the Western blot signals demonstrates that the concentration of F55 drops about 50% in UV-treated cells compared to the control sample and does not dramatically change up to 10 h posttreatment (Fig. 5). How can these target promoters be progressively released by F55 without fluctuation of the protein levels in the aftermath of UV irradiation? A reasonable hypothesis is that the intracellular concentration of F55 becomes progressively suboptimal to saturate all the regulative binding sites in the UV-induced cells. Indeed, in the carrier state, SSV1 constantly replicates in actively dividing cells to keep its copy number around 5 copies per cell (23). Under these conditions, F55 saturates 14 sites per SSV1 genome, i.e., 4 sites for each of the Tind, T5, and T6 promoters and 2 for that of Tlys (Fig. 7), for a total of about 70 binding sites per cell. Upon UV irradiation, whereas cell division slows down (Fig. 1), basal viral replication proceeds, consistently leading to an increase of the SSV1 copy number starting from the 2nd hour posttreatment (Fig. 2). Thereafter, the episomal SSV1 content reaches ∼160 copies/cell (32), and the total number of sites that has to be bound by F55 rises about 32-fold. Since in UV-irradiated cells the intracellular level of F55 is constant, the ratio of the number of binding sites (BS) to F55 concentration (BS/F55) increases. This variation in the BS/F55 ratio allows the progressive dissociation of F55 from SSV1 genome that eventually leads to transcription derepression of the target genes (Fig. 6). The proposed mechanism strictly depends on the maintenance of a low and constant F55 concentration in UV-irradiated cells. In this regard, it has been observed in Northern blot experiments that an additional level of control acts posttranscriptionally by degrading Tlys (Fig. 4B), probably to compensate for the “sloppiness” of the transcriptional control.
FIG 7.
A suboptimal concentration of F55 allows the derepression of the target genes. Schematic representations of the infected cell and of the UV-inducible region of SSV1 genome are presented. The operators recognized by F55 are in green, yellow, and blue. Bent arrows indicate the transcription start sites, and dashed lines represent transcripts. Dimers of F55 are represented by purple ovals. In the lysogenic cell, the amount of F55 is suitable to saturate most of its binding sites and to keep SSV1 in a steady carrier state. At 2 h postirradiation, a decrease of about 50% of the F55 concentration and a concurrent increase of the viral copy number led to the dissociation from the lower-affinity operators in the promoter of Tind and Tlys. Later, at 4 h postirradiation, the dilution effect is enhanced by a further accumulation of the viral DNA, which results in the release of the early promoters (i.e., those of T5 and T6), thus allowing transcription derepression.
Regarding the transcription regulation of Tlys, we did not observe an immediate derepression of its expression, although its promoter, like that of Tind, is released 2 h postirradiation. Indeed, upregulation of Tlys occurs only when Tind transcription is abrogated (8 to 10 h postirradiation) (Fig. 4B). It must be taken into account that Tlys and Tind RNAs are transcribed in a convergent orientation (Fig. 4A), and therefore the relative genes may be susceptible to transcriptional interference, which is typical of head-to-head-oriented gene couples. This architecture strongly resembles that of the nonlambdoid UV-inducible coliphage 186 (35, 36), although a different mechanism may be evoked. Indeed, it has been proved in both eukaryotes (37–39) and prokaryotes (40–42) that transcription of genes separated by a few kilobases is not independent but is coupled by torsional stress, which, by influencing the opening properties of the promoters, facilitates or hinders transcription initiation. This kind of influence reduces the expression of convergent gene couples (43), such as in the case of Tlys and Tind. This hypothesis seems reasonable in the light of the Northern blot and ChIP-sqPCR data reported above. Indeed, the upregulation of the Tlys transcription takes place only after the abrogation of Tind expression.
Altogether, these data demonstrate that F55, like the CI repressor of lambda, acts as the key switch regulator involved in the lysogeny/induction transition of SSV1. However, some intriguing differences between lambda and SSV1 need to be discussed. Lambda lysogens harbor the viral DNA only as provirus, and the only viral gene expressed is cI. The lysogenic/lytic cycle is strictly regulated, since viral progeny is produced only upon UV irradiation and causes cell lysis. In contrast, in SSV1 lysogens, the provirus coexists with some episomal copies, and a constitutive extrusion of the viral particles occurs. Therefore, the lysogeny of SSV1 could be better defined as a carrier state (44). In this case, the UV irradiation only enhances the rate of SSV1 replication and progeny extrusion without causing cell lysis. This difference is mirrored by the expression regulation of F55, which is not as stringent as that for the functional homolog CI of lambda phage. From an evolutionary point of view, this kind of host-virus relationship, which is typical of all Fuselloviridae, could have coevolved with the nonlytic nature of these viruses.
ACKNOWLEDGMENTS
This study was supported by a grant from the Regione Campania, legge 5 (CUP number E69D15000210002), as well as grants from the Danish Independent Research Council (DFF-0602-02196B and DFF-1323-00330) and a grant from the Carlsberg Foundation.
REFERENCES
- 1.Prangishvili D. 2013. The wonderful world of archaeal viruses. Annu Rev Microbiol 8:565–585. doi: 10.1146/annurev-micro-092412-155633. [DOI] [PubMed] [Google Scholar]
- 2.Contursi P, Fusco S, Cannio R, She Q. 2014. Molecular biology of fuselloviruses and their satellites. Extremophiles 3:473–489. doi: 10.1007/s00792-014-0634-0. [DOI] [PubMed] [Google Scholar]
- 3.Wang H, Peng N, Shah SA, Huang L, She Q. 2015. Archaeal extrachromosomal genetic elements. Microbiol Mol Biol Rev 79:117–152. doi: 10.1128/MMBR.00042-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geslin C, Le Romancer M, Gaillard M, Erauso G, Prieur D. 2003. Observation of virus-like particles in high temperature enrichment cultures from deep-sea hydrothermal vents. Res Microbiol 154:303–307. doi: 10.1016/S0923-2508(03)00075-5. [DOI] [PubMed] [Google Scholar]
- 5.Gorlas A, Koonin EV, Bienvenu N, Prieur D, Geslin C. 2012. TPV1, the first virus isolated from the hyperthermophilic genus Thermococcus. Environ Microbiol 14:503–516. doi: 10.1111/j.1462-2920.2011.02662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sime-Ngando T, Lucas S, Robin A, Tucker KP, Colombet J, Bettarel Y, Desmond E, Gribaldo S, Forterre P, Breitbart M, Prangishvili D. 2011. Diversity of virus-host systems in hypersaline Lake Retba, Senegal. Environ Microbiol 13:1956–1972. doi: 10.1111/j.1462-2920.2010.02323.x. [DOI] [PubMed] [Google Scholar]
- 7.Porter K, Russ BE, Dyall-Smith ML. 2007. Virus-host interactions in salt lakes. Curr Opin Microbiol 10:418–424. doi: 10.1016/j.mib.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 8.Borrel G, Colombet J, Robin A, Lehours AC, Prangishvili D, Sime-Ngando T. 2012. Unexpected and novel putative viruses in the sediments of a deep-dark permanently anoxic freshwater habitat. ISME J 6:2119–2127. doi: 10.1038/ismej.2012.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.López-Bueno A, Tamames J, Velazquez D, Moya A, Quesada A, Alcami A. 2009. High diversity of the viral community from an Antarctic lake. Science 326:858–861. doi: 10.1126/science.1179287. [DOI] [PubMed] [Google Scholar]
- 10.Rice G, Stedman K, Snyder J, Wiedenheft B, Willits D, Brumfield S, McDermott T, Young MJ. 2001. Viruses from extreme thermal environments. Proc Natl Acad Sci U S A 98:13341–13345. doi: 10.1073/pnas.231170198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bize A, Peng X, Prokofeva M, MacLellan K, Lucas S, Forterre P, Garrett RA, Bonch-Osmolovskaya EA, Prangishvili D. 2008. Viruses in acidic geothermal environments of the Kamchatka Peninsula. Res Microbiol 159:358–366. doi: 10.1016/j.resmic.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 12.Rachel R, Bettstetter M, Hedlund BP, Haring M, Kessler A, Stetter KO, Prangishvili D. 2002. Remarkable morphological diversity of viruses and virus-like particles in hot terrestrial environments. Arch Virol 147:2419–2429. doi: 10.1007/s00705-002-0895-2. [DOI] [PubMed] [Google Scholar]
- 13.Baker BJ, Comolli LR, Dick GJ, Hauser LJ, Hyatt D, Dill BD, Land ML, Ver Berkmoes NC, Hettich RL, Banfield JF. 2010. Enigmatic, ultrasmall, uncultivated Archaea. Proc Natl Acad Sci U S A 107:8806–8811. doi: 10.1073/pnas.0914470107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Comolli LR, Baker BJ, Downing KH, Siegerist CE, Banfield JF. 2009. Three-dimensional analysis of the structure and ecology of a novel, ultra-small archaeon. ISME J 3:159–167. doi: 10.1038/ismej.2008.99. [DOI] [PubMed] [Google Scholar]
- 15.Krupovic M, Quemin ER, Bamford DH, Forterre P, Prangishvili D. 2014. Unification of the globally distributed spindle-shaped viruses of the Archaea. J Virol 88:2354–2358. doi: 10.1128/JVI.02941-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kraft P, Kümmel D, Oeckinghaus A, Gauss GH, Wiedenheft B, Young M, Lawrence CM. 2004. Structure of D-63 from Sulfolobus spindle-shaped virus 1: surface properties of the dimeric four-helix bundle suggest an adaptor protein function. J Virol 78:7438–7442. doi: 10.1128/JVI.78.14.7438-7442.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kraft P, Oeckinghaus A, Kümmel D, Gauss GH, Gilmore J, Wiedenheft B, Young M, Lawrence CM. 2004. Crystal structure of F-93 from Sulfolobus spindle-shaped virus 1, a winged-helix DNA binding protein. J Virol 78:11544–11550. doi: 10.1128/JVI.78.21.11544-11550.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menon SK, Maaty WS, Corn GJ, Kwok SC, Eilers BJ, Kraft P, Gillitzer E, Young MJ, Bothner B, Lawrencem CM. 2008. Cysteine usage in Sulfolobus spindle-shaped virus 1 and extension to hyperthermophilic viruses in general. Virology 376:270–278. doi: 10.1016/j.virol.2008.03.026. [DOI] [PubMed] [Google Scholar]
- 19.Guillière F, Peixeiro N, Kessler A, Raynal B, Desnoues N, Keller J, Delepierre M, Prangishvili D, Sezonov G, Guijarro JI. 2009. Structure, function, and targets of the transcriptional regulator SvtR from the hyperthermophilic archaeal virus SIRV1. J Biol Chem 284:22222–22237. doi: 10.1074/jbc.M109.029850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Contursi P, D'Ambrosio K, Pirone L, Pedone E, Aucelli T, She Q, De Simone G, Bartolucci S. 2011. C68 from the Sulfolobus islandicus plasmid-virus pSSVx is a novel member of the AbrB-like transcription factor family. Biochem J 435:157–166. doi: 10.1042/BJ20101334. [DOI] [PubMed] [Google Scholar]
- 21.Schlenker C, Goel A, Tripet BP, Menon S, Willi T, Dlakić M, Young MJ, Lawrence CM, Copié V. 2012. Structural studies of E73 from a hyperthermophilic archaeal virus identify the “RH3” domain, an elaborated ribbon-helix-helix motif involved in DNA recognition. Biochemistry 51:2899–2910. doi: 10.1021/bi201791s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Contursi P, Fusco S, Limauro D, Fiorentino G. 2013. Host and viral transcriptional regulators in Sulfolobus: an overview. Extremophiles 17:881–895. doi: 10.1007/s00792-013-0586-9. [DOI] [PubMed] [Google Scholar]
- 23.Fusco S, She Q, Bartolucci S, Contursi P. 2013. Tlys, a newly identified Sulfolobus spindle-shaped virus 1 transcript expressed in the lysogenic state, encodes a DNA-binding protein interacting at the promoters of the early genes. J Virol 87:5926–5936. doi: 10.1128/JVI.00458-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nadal M, Mirambeau G, Forterre P, Reiter WD, Duguet M. 1986. Positively supercoiled DNA in a virus-like particle of an archaebacterium. Nature 321:256–258. doi: 10.1038/321256a0. [DOI] [Google Scholar]
- 25.Fröls S, Gordon PM, Panlilioc MA, Schleper C, Sensen CW. 2007. Elucidating the transcription cycle of the UV-inducible hyperthermophilic archaeal virus SSV1 by DNA microarrays. Virology 365:48–59. doi: 10.1016/j.virol.2007.03.033. [DOI] [PubMed] [Google Scholar]
- 26.Reiter WD, Palm P, Yeats S, Zillig W. 1987. Gene expression in archaebacteria: physical mapping of constitutive and UV-inducible transcripts from the Sulfolobus virus-like particle SSV1. Mol Gen Genet 209:270–275. doi: 10.1007/BF00329653. [DOI] [PubMed] [Google Scholar]
- 27.Palm P, Schleper C, Grampp B, Yeats S, McWilliam P, Reiter WD, Zillig W. 1991. Complete nucleotide sequence of the virus SSV1 of the archaebacterium Sulfolobus shibatae. Virology 185:242–250. doi: 10.1016/0042-6822(91)90771-3. [DOI] [PubMed] [Google Scholar]
- 28.Schleper C, Kubo K, Zillig W. 1992. The particle SSV1 from the extremely thermophilic archaeon Sulfolobus is a virus: demonstration of infectivity and of transfection with viral DNA. Proc Natl Acad Sc U S A 89:7645–7649. doi: 10.1073/pnas.89.16.7645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gudbergsdottir S, Deng L, Chen Z, Jensen JVK, Jensen LR, She Q, Garrett RA. 2011. Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol Microbiol 79:35–49. doi: 10.1111/j.1365-2958.2010.07452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Contursi P, Jensen J, Aucelli T, Rossi M, Bartolucci S, She Q. 2006. Characterization of the Sulfolobus host-SSV2 virus interaction. Extremophiles 10:615–627. doi: 10.1007/s00792-006-0017-2. [DOI] [PubMed] [Google Scholar]
- 31.Wolin EA, Wolin MG, Wolfe RS. 1963. Formation of methane by bacterial extracts. J Biol Chem 238:2882–2886. [PubMed] [Google Scholar]
- 32.Fusco S, Aulitto M, Bartolucci S, Contursi P. 2015. A standardized protocol for the UV induction of Sulfolobus spindle-shaped virus 1. Extremophiles 19:539–546. doi: 10.1007/s00792-014-0717-y. [DOI] [PubMed] [Google Scholar]
- 33.Contursi P, Cannio R, Prato S, She Q, Rossi M, Bartolucci S. 2007. Transcriptional analysis of the genetic element pSSVx: differential and temporal regulation of gene expression reveals correlation between transcription and replication. J Bacteriol 189:6339–6350. doi: 10.1128/JB.00638-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Götz D, Paytubi S, Munro S, Lundgren M, Bernander R, White MF. 2007. Responses of hyperthermophilic crenarchaea to UV irradiation. Genome Biol 8:R220. doi: 10.1186/gb-2007-8-10-r220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Callen BP, Shearwin KE, Egan JB. 2004. Transcriptional interference between convergent promoters caused by elongation over the promoter. Mol Cell 14:647–656. doi: 10.1016/j.molcel.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 36.Dodd IB, Egan JB. 2002. Action at a distance in CI repressor regulation of the bacteriophage 186 genetic switch. Mol Microbiol 45:697–710. doi: 10.1046/j.1365-2958.2002.03038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi CH, Kalosakas G, Rasmussen KØ, Hiromura M, Bishop AR, Usheva A. 2004. DNA dynamically directs its own transcription initiation. Nucleic Acids Res 32:1584–1590. doi: 10.1093/nar/gkh335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leblanc BP, Benham CJ, Clark DJ. 2000. An initiation element in the yeast cup1 promoter is recognized by RNA polymerase ii in the absence of TATA box-binding protein if the DNA is negatively supercoiled. Proc Natl Acad Sci U S A 97:10745–10750. doi: 10.1073/pnas.200365097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alexandrov BS, Gelev V, Yoo SW, Alexandrov LB, Fukuyo Y, Bishop AR, Rasmussen KØ, Usheva A. 2010. DNA dynamics play a role as a basal transcription factor in the positioning and regulation of gene transcription initiation. Nucleic Acids Res 38:1790–1795. doi: 10.1093/nar/gkp1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ouafa ZA, Reverchon S, Lautier T, Muskhelishvili G, Nasser W. 2012. The nucleoid-associated proteins h-ns and fis modulate the DNA supercoiling response of the pel genes, the major virulence factors in the plant pathogen bacterium Dickeya dadantii. Nucleic Acids Res 40:4306–4319. doi: 10.1093/nar/gks014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hatfield GW, Benham CJ. 2002. DNA topology-mediated control of global gene expression in Escherichia coli. Annu Rev Gen 36:175–203. doi: 10.1146/annurev.genet.36.032902.111815. [DOI] [PubMed] [Google Scholar]
- 42.Du X, Wojtowicz D, Bowers AA, Levens D, Benham CJ, Przytycka TM. 2013. The genome-wide distribution of non-b DNA motifs is shaped by operon structure and suggests the transcriptional importance of non-b dna structures in Escherichia coli. Nucleic Acids Res 41:5965–5977. doi: 10.1093/nar/gkt308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyer S, Beslon G. 2014. Torsion-mediated interaction between adjacent genes. PLoS Comput Biol 10:e1003785. doi: 10.1371/journal.pcbi.1003785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fusco S, Liguori R, Limauro D, Bartolucci S, She Q, Contursi P. 17 April 2015. Transcriptome analysis of Sulfolobus solfataricus infected with two related fuselloviruses reveals novel insights into the regulation of CRISPR-Cas system. Biochimie doi: 10.1016/j.biochi.2015.04.006. [DOI] [PubMed] [Google Scholar]