ABSTRACT
Human enterovirus 71 (EV71) and coxsackievirus A16 (CVA16) are the two major causative agents for hand-foot-and-mouth disease (HFMD). Previously, we demonstrated that a virus-like particle (VLP) for EV71 produced from Saccharomyces cerevisiae is a potential vaccine candidate against EV71 infection, and an EV71/CVA16 chimeric VLP can elicit protective immune responses against both virus infections. Here, we presented the crystal structures of both VLPs, showing that both the linear and conformational neutralization epitopes identified in EV71 are mostly preserved on both VLPs. The replacement of only 4 residues in the VP1 GH loop converted strongly negatively charged surface patches formed by portions of the SP70 epitope in EV71 VLP into a relatively neutral surface in the chimeric VLP, which likely accounted for the additional neutralization capability of the chimeric VLP against CVA16 infection. Such local variations in the amino acid sequences and the surface charge potential are also present in different types of polioviruses. In comparison to EV71 VLP, the chimeric VLP exhibits structural changes at the local site of amino acid replacement and the surface loops of all capsid proteins. This is consistent with the observation that the VP1 GH loop located near the pseudo-3-fold junction is involved in extensive interactions with other capsid regions. Furthermore, portions of VP0 and VP1 in EV71 VLP are at least transiently exposed, revealing the structural flexibility of the VLP. Together, our structural analysis provided insights into the structural basis of enterovirus neutralization and novel vaccine design against HFMD and other enterovirus-associated diseases.
IMPORTANCE Our previous studies demonstrated that the enterovirus 71 (EV71) virus-like particle (VLP) produced from yeast is a vaccine candidate against EV71 infection and that a chimeric EV71/coxsackievirus A16 (CVA16) VLP with the replacement of 4 amino acids in the VP1 GH loop can confer protection against both EV71 and CVA16 infections. This study reported the crystal structures of both the EV71 VLP and the chimeric EV71/CVA16 VLP and revealed that the major neutralization epitopes of EV71 are mostly preserved in both VLPs. In addition, the mutated VP1 GH loop in the chimeric VLP is well exposed on the particle surface and exhibits a surface charge potential different from that contributed by the original VP1 GH loop in EV71 VLP. Together, this study provided insights into the structural basis of enterovirus neutralization and evidence that the yeast-produced VLPs can be developed into novel vaccines against hand-foot-and-mouth disease (HFMD) and other enterovirus-associated diseases.
INTRODUCTION
Hand-foot-and-mouth disease (HFMD) is a worldwide infectious disease in infants and young children that may lead to death. In recent years, numerous outbreaks of HFMD have occurred in the Asia-Pacific regions, causing significant morbidity and mortality (1). The number of reported fatal cases of HFMD in mainland China has reached over 3,000 since 2008 (www.chinacdc.cn). However, no vaccines are available at present. Human enterovirus 71 (EV71) and coxsackievirus A16 (CVA16), both of species Enterovirus A, genus Enterovirus, family Picornaviridae, have been identified as the major causative agents. Therefore, for better effective control and prevention of HFMD, a multivalent vaccine is highly desirable.
The mapping of neutralizing epitopes is important for vaccine development. The neutralizing epitopes of EV71 have been screened extensively, including the identification of synthetic peptides that induce neutralizing antibodies and the mapping of antibody neutralization escape mutations. Some major neutralizing epitopes of EV71 have been mapped to residues 97 to 105, 163 to 177, 208 to 222 (SP70 epitope), and 253 to 267 of VP1 (2–4); residues 141 to 150 of VP2 (5); and residues 1 to 15 and 28 to 42 of VP3 (4). Lee et al. identified a strain-specific neutralization epitope of EV71 in a cryo-electron microscopy (cryo-EM) study of EV71 in complex with Fab from a neutralizing antibody (6). The footprint of Fab on EV71 includes residues 98, 145, 242, and 244 of VP1, near the 5-fold axis of the virus. Kiener et al. identified a novel universal neutralizing monoclonal antibody against EV71 that targets the highly conserved “knob” region of VP3, which is a conformational neutralization epitope mapped to residues 59, 62, and 67 of VP3 (7). During our study, Shi et al. identified several linear neutralization epitopes within the VP1 protein of CVA16, including PEP71 (residues 211 to 225), which is similar to SP70, as both map to the GH loop (8).
Virus-like particles (VLPs) have been used extensively in virus studies and vaccine development, as they resemble authentic viral capsids in structure but are not infectious due to the lack of viral genetic material. Importantly, the repetitive ordered arrangement of epitopes on the capsid surface makes VLPs promising candidates as potent immunogens and vaccine products. We previously demonstrated that coexpression of EV71-P1 and 3CD in Saccharomyces cerevisiae led to the cleavage of P1 and the formation of EV71 VLPs (9). Similar in both composition and morphology to the naturally occurring empty particles from EV71-infected cells, these VLPs could elicit humoral and cellular immune responses and protect neonate mice against lethal challenge. Based on our study of EV71-VLP (9) and the crystal structure analysis of EV71 (10–12) and CVA16 (unpublished data), we have produced a novel EV71/CVA16 chimeric VLP by replacement of a neutralizing epitope of EV71 (SP70) with its corresponding region of CVA16 (13). The replacement occurred in the GH loop of VP1, with the replacement of only 4 amino acids (K215L, E217A, K218N, and E221D). This chimeric VLP can elicit protective immune responses against both EV71 and CVA16 infections, thus providing an alternative platform for multivalent HFMD vaccine development.
To elucidate the structural basis of the VLPs produced from yeast as vaccine candidates, we determined the crystal structures of EV71 VLP and the chimeric VLP. Both structures shared similarity with that of the naturally occurring empty particle (14) and showed that both the linear and conformational neutralization epitopes identified in EV71 were structurally preserved on both VLPs. In addition, strongly negatively charged surface patches present in EV71 VLP, contributed by portions of the SP70 epitope, changed into a relatively neutral surface in the chimeric VLP, which was likely responsible for the additional neutralization capability of the chimeric VLP against CVA16 infection. In comparison to EV71 VLP, the chimeric VLP exhibited conformational changes at the local site of amino acid replacement and the loop regions in all capsid proteins. Such structural variations, resulting from the substitutions in the VP1 GH loop near the pseudo-3-fold junction site, were consistent with the observation that the VP1 GH loop is involved in extensive interactions with other capsid regions. Moreover, similarly to what was observed in the empty particle (14), portions of VP1 and VP0 in EV71 VLP were exposed at least transiently on the capsid surface. Together, our structural studies provided insights into the development of yeast-produced VLPs as multivalent vaccines against HFMD.
MATERIALS AND METHODS
Production of VLPs.
A Chinese endemic genotype C4 strain EV71, AH08/06 (GenBank accession no. HQ611148), was used for vector construction for VLP assembly. Production of EV71 VLPs was described in detail elsewhere (9). The CVA16 sequences were derived from a Chinese endemic coxsackievirus A16 strain, CA16/GD09/119 (GenBank accession no. KC117318.1), which was isolated from a severe HFMD patient. A PCR experiment was performed to replace the SP70 epitope (residues 208 to 222, YPTFGEHKQEKDLEY) in VP1 of EV71-P1 with the corresponding region from CVA16-P1 (YPTFGEHLQANDLDY). The PCR experiment, yeast transformation, and production of the chimeric EV71/CVA16 VLPs were described in detail previously (13).
Purification of VLPs.
Cell pellet from 2 liters of yeast culture after induction was dissolved in 300 ml phosphate-buffered saline (PBS) buffer (pH 7.2) (containing 1 mM phenylmethylsulfonyl fluoride [PMSF] and 1 mM β-mercaptoethanol [β-ME]) followed by high-pressure homogenization at about 180 MPa for 3 times to release the intracellular VLPs. Then, NP-40 was added to a concentration of 1% and subjected to centrifugation at 10,000 rpm (12,857 × g) for 30 min at 4°C to remove cell debris. The clarified supernatant was precipitated with 8% polyethylene glycol (PEG) 8000, 200 mM NaCl, overnight. Then, it was subjected to centrifugation at 10,000 × g for 1 h at 4°C. The precipitant was dissolved in 120 ml PBS buffer (pH 7.2) followed by homogenization. Then, the solution was subjected to low-speed centrifugation (10,000 × g, 30 min, 4°C) to remove large cell debris and contaminating proteins. Then, the supernatant was subjected to ultracentrifugation at 28,000 rpm (141,100 × g) for 3 h in a 30% sucrose cushion. The precipitate was dissolved in 5 ml PBS (pH 7.2) overnight, followed by homogenization and sonication. The clarified supernatant was layered onto a discontinuous sucrose gradient (10 to 50%) followed by ultracentrifugation (22,000 rpm, 4 h, SW28 rotor). The result was analyzed by 12% SDS-PAGE. VLPs were collected and further concentrated by ultracentrifugation (28,000 rpm, 4 h, 4°C).
Crystallization and diffraction data collection.
The purified VLPs were subjected to crystallization trials. Crystals of EV71 VLP were obtained by the hanging drop method by mixing 2 μl of VLPs (3.6 mg/ml in PBS) with an equal volume of a reservoir solution containing 0.1 M imidazole (pH 6.8) and 1 M sodium acetate. Crystals of EV71/CVA16 chimeric VLP were obtained by mixing 2 μl of VLPs (3 mg/ml in PBS) with an equal volume of a reservoir solution containing 0.1 M HEPES (pH 7.5) and 0.5 M magnesium formate dehydrate.
Prior to data collection, cryoprotection of the crystals was achieved by resuspending the crystals in the mother liquor with increasing concentrations of glycerol through six steps: 5%, 10%, 15%, 20%, 25%, and 30% (vol/vol). The equilibration time at each concentration was at least 30 s. These crystals were then flash-frozen in liquid nitrogen and used for diffraction data collection on an ADSC Quantum-315 charge-coupled device (CCD) detector at beamline BL17U1 at the Shanghai Synchrotron Radiation Facility. For EV71 VLP, a data set at 3.65-Å resolution was collected with monochromatic X rays (λ = 0.97853 Å) and a detector-to-crystal distance of 330 mm using an oscillation angle of 0.3° and an exposure time of 1 s. For the EV71/CVA16 chimeric VLP, a data set at 3.8-Å resolution was collected with monochromatic X rays (λ = 1.281579 Å) and a detector-to-crystal distance of 350 mm using an oscillation angle of 0.3° and an exposure time of 1 s.
Structure determination and refinement.
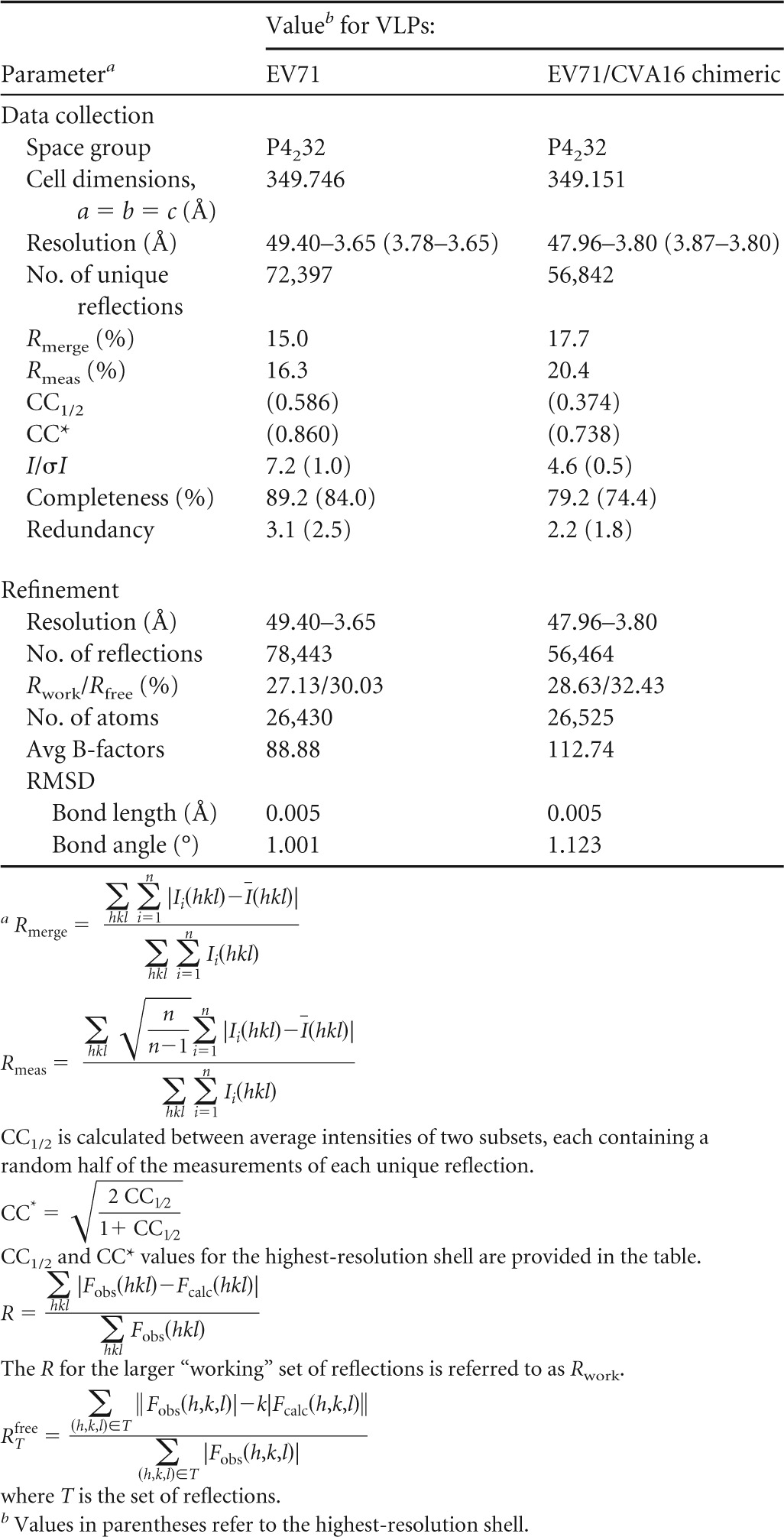
Crystals of both VLPs belong to space group P4232 with 5 copies of the protomer as the asymmetric unit. Thus, 5-fold noncrystallographic symmetry was employed during structure determination and refinement. The program GLRF (15) was used to calculate the self-rotation function and was combined with crystal packing analysis to determine the position and orientation of the particle. Each individual structure was then determined by the molecular replacement method using the program package PHENIX (16). To avoid model bias, the crystal structure of the empty particle of type 1 poliovirus (Protein Data Bank [PDB] identifier [ID] 1POV) was taken as the search model and a single solution was obtained. After obtaining the initial density map, model building and refinement were carried out iteratively using COOT (17) and PHENIX (16) programs, respectively. The final refinement statistics are summarized in Table 1. Figures were drawn and rendered with PyMOL (version 1.3r1; Schrödinger, LLC) and UCSF Chimera (18).
TABLE 1.
Data collection and refinement statistics
Proteolytic digestion of EV71 VLP.
The proteolytic sensitivity of VP1 in EV71 VLP was assessed by adding 1 μg of trypsin into 1 μg of the particles in a 10-μl system. Trypsin digestion was performed at 16°C and 37°C for 1 h in parallel. The digestion was stopped by dilution of the samples with 1/5 volume of the 5× SDS loading buffer followed by boiling of the mixture for 10 min. The samples were then subjected to SDS-PAGE analysis, and VP1 was visualized by Western blotting using a VP1 antibody (Abnova).
The proteolytic sensitivity of VP0 in EV71 VLP was assessed in parallel. The samples were then subjected to SDS-PAGE analysis, and VP4 was visualized by Western blotting using a VP4 antibody (Biorbyt).
Calculation of electronic potential.
The electrostatic potentials were calculated by PyMOL (version 1.3r1; Schrödinger, LLC). X-ray coordinates (PDB IDs 2PLV, 1EAH, and 1PVC) were used for electrostatic potential calculation for types 1, 2, and 3 of poliovirus, respectively.
Fitting of the X-ray coordinates into the EM density map of EV71 VLPs.
We used UCSF Chimera (18) to visualize the cryo-EM density map of EV71 VLP (EMD-2607) (19), to segment the corresponding density maps, and to compare the structural similarities between electron density and crystal structure of EV71 VLP. After docking the crystal structure of EV71 VLP (PDB ID 4YVS) into the EV71 VLP density map (σ = 1.00), the correlation coefficient (cc) values from comparisons of the cryo-EM density map and crystal structure were calculated using “Fit in Map” in UCSF Chimera (18).
Protein structure accession numbers.
The coordinates and structure factors of EV71 VLP and EV71/CVA16 chimeric VLP have been deposited in the Protein Data Bank (PDB IDs 4YVS and 4YVW, respectively).
RESULTS
Crystal structure of EV71 VLP.
The EV71 VLPs were produced from Saccharomyces cerevisiae by coexpression of EV71-P1 and 3CD. The self-assembled VLPs were then purified by sucrose gradient ultracentrifugation. The quality of purification was deemed good as capsid proteins of VP0, VP1, and VP3 were the only major bands resolved on the SDS-PAGE gels.
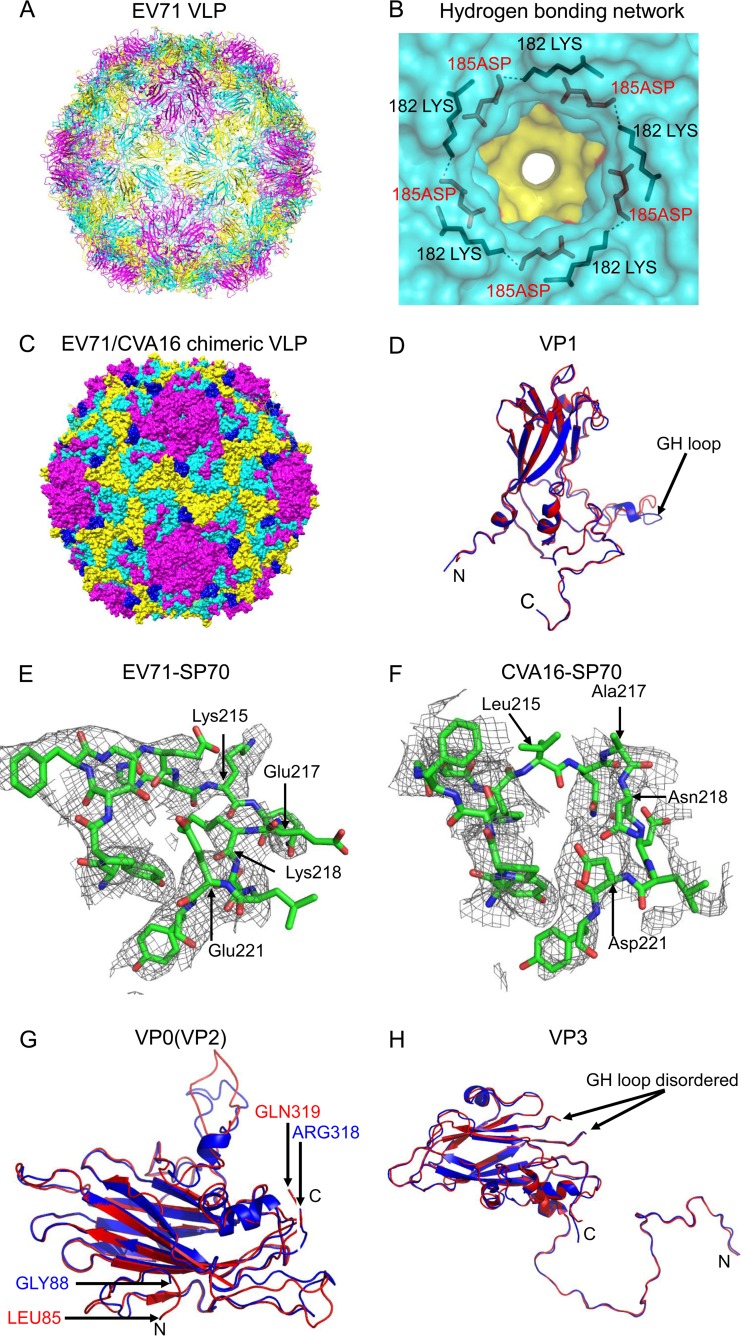
The crystal structure of EV71 VLP was determined at 3.65-Å resolution (Fig. 1A). After iterative model building and structure refinement, the electron density was visible for residues 72 to 297 of VP1, 88 to 318 of VP0 (corresponding to residues 19 to 249 of VP2), and 1 to 176 and 190 to 237 of VP3 (Fig. 1D, G, and H). The VP4 portion of VP0, as well as the N-terminal extensions of VP1, was disordered. Similar to the empty particle, the VLP contained an empty pocket inside VP1. The calculated root mean square deviation (RMSD) from the main chain atoms in VP1, VP0, and VP3 between VLP and empty particle (PDB ID 4RQP) (14) was 0.78 Å (218 Cα), 1.18 Å (231 Cα), and 0.98 Å (224 Cα), respectively. Our structural analysis also revealed a hydrogen-bonding network, contributed by 182K and 185D in VP1 and situated at the 5-fold axis channels. These two residues, 182K and 185D, are conserved in members of species Enterovirus A and D, suggesting that such a hydrogen-bonding network is likely conserved among members of these two species (Fig. 1B).
FIG 1.
Crystal structures of EV71 VLP and EV71/CVA16 chimeric VLP. (A) Cartoon representation of EV71 VLP viewed along the 2-fold axis with VP1, VP0, and VP3 colored in magenta, yellow, and cyan, respectively. (B) Hydrogen-bonding network at the 5-fold axis channel. The amino group in the side chain of Lys182 (colored in black) of VP1 interacted with Asp185 (colored in red) of a neighboring VP1 through hydrogen bonds. (C) Surface representation of EV71/CVA16 chimeric VLP viewed along the 2-fold axis. The capsid proteins VP1, VP0, and VP3 are colored in magenta, yellow, and cyan, respectively. The CVA16-SP70 epitope is colored in blue. (D) Superimposition of VP1 from EV71 VLP and EV71/CVA16 chimeric VLP. Residues 72 to 297 in EV71 VLP (colored in blue) and residues 72 to 296 in EV71/CVA16 chimeric VLP (colored in red) are modeled. (E) Electron densities corresponding to the EV71-SP70 epitope (amino acids 208 to 222 in VP1) in EV71 VLP. These residues are displayed as sticks with Lys215, Glu217, Lys218, and Glu221 labeled. (F) Electron densities corresponding to the CVA16-SP70 region (amino acids 208 to 222 in VP1) in the chimeric VLP. These residues are displayed as sticks with Leu215, Ala217, Asn218, and Asp221 labeled. (G) Superimposition of VP0 from EV71 VLP and EV71/CVA16 chimeric VLP. The color scheme is the same as in panel D. Residues 88 to 318 in EV71 VLP and residues 85 to 319 in EV71/CVA16 chimeric VLP are modeled. (H) Superimposition of VP3 from EV71 VLP and EV71/CVA16 chimeric VLP. The color scheme is the same as in panel D. Residues 1 to 176 and 190 to 237 in EV71 VLP and residues 1 to 176 and 189 to 236 in EV71/CVA16 chimeric VLP are modeled.
Crystal structure of the EV71/CVA16 chimeric VLP.
The crystal structure of the EV71/CVA16 chimeric VLP was determined at 3.8-Å resolution (Fig. 1C). The CVA16-SP70 epitope was well exposed on the particle surface (Fig. 1C). Despite relatively weak electron density, most of the CVA16-SP70 main chain atoms could still be traced (Fig. 1F), which shows structural variations from the SP70 epitope in EV71-VLP (Fig. 1E). After iterative model building and structure refinement, the electron density was visible for residues 72 to 296 of VP1, 85 to 319 of VP0 (corresponding to residues 16 to 250 of VP2), and 1 to 176 and 189 to 236 of VP3 (Fig. 1D, G, and H). The structure of the chimeric VLP was rather similar to that of EV71 VLP, including the lack of ordered structure in the VP4 portion of VP0 and the N-terminal extensions of VP1 and the loss of pocket factors.
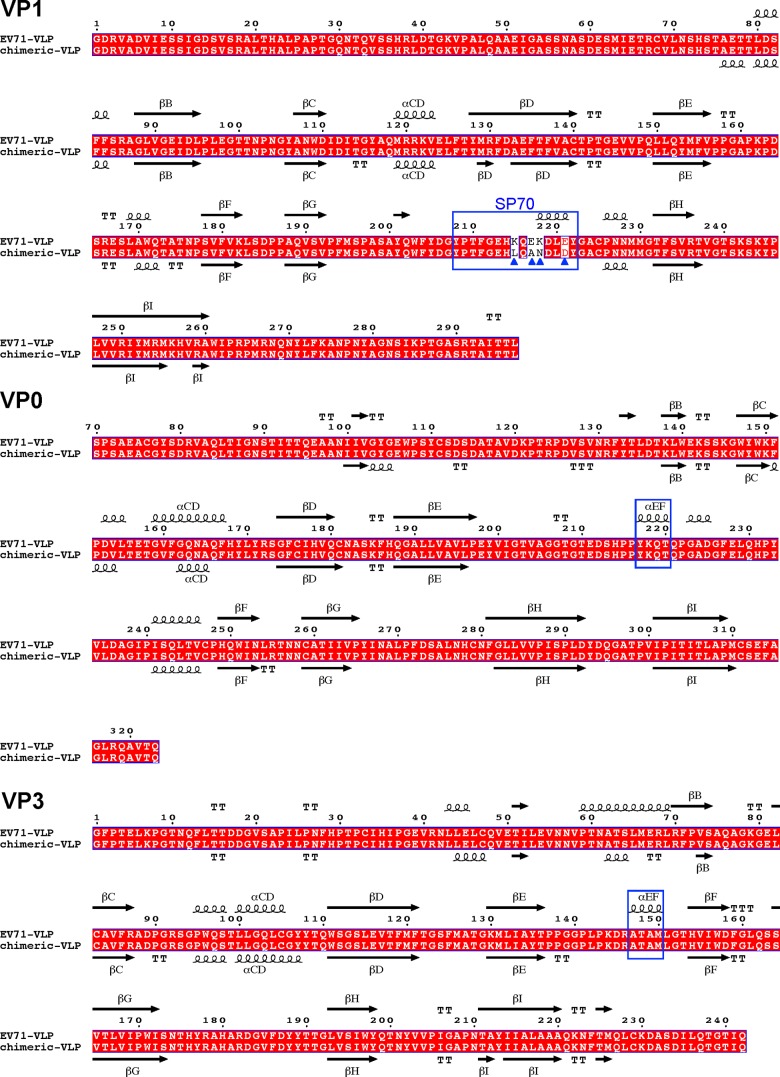
Structure-based sequence alignment between EV71 VLP and the chimeric VLP showed that there were some secondary structure changes in the capsid proteins (Fig. 2). In VP1, residues 218 to 221 in the GH loop located near the junction changed from a 310-helix conformation in EV71 VLP into a loop structure in the chimeric VLP (Fig. 2; see also Fig. 6B). In VP0, residues 217 to 220 located near the junction switched from a helical structure (the EF helix) in EV71 VLP to a loop conformation in the chimeric VLP (Fig. 2). In VP3, residues 147 to 150 located near the 2-fold channel also changed from a helical structure (the EF helix) in EV71 VLP into a loop conformation in the chimeric VLP (Fig. 2).
FIG 2.
Structure-based sequence alignments of EV71 VLP and EV71/CVA16 chimeric VLP. Capsid protein sequences used for the alignment include EV71 VLP and the chimeric VLP whose capsid proteins have been structurally determined. The secondary structure elements for EV71 VLP and chimeric VLP are shown at the top and bottom of the sequence alignment, respectively. The residue numbers correspond to those in EV71 VLP. Conserved residues are shown in white with a red background. Helices and strands are labeled according to the standard picornavirus nomenclature and are represented by coils and arrows, respectively. The blue triangles indicate the residues that are variable between EV71 VLP and the chimeric VLP. VP1 amino acids 208 to 222 corresponding to the SP70 region in EV71 VLP are boxed with a blue rectangle. VP0 residues 217 to 220, boxed with a blue rectangle, switched from a helical structure (the EF helix) in EV71 VLP to a loop conformation in the chimeric VLP. VP3 residues 147 to 150, boxed with a blue rectangle, changed from a helical structure (the EF helix) in EV71 VLP into a loop conformation in the chimeric VLP. This figure was produced using ESPript (33).
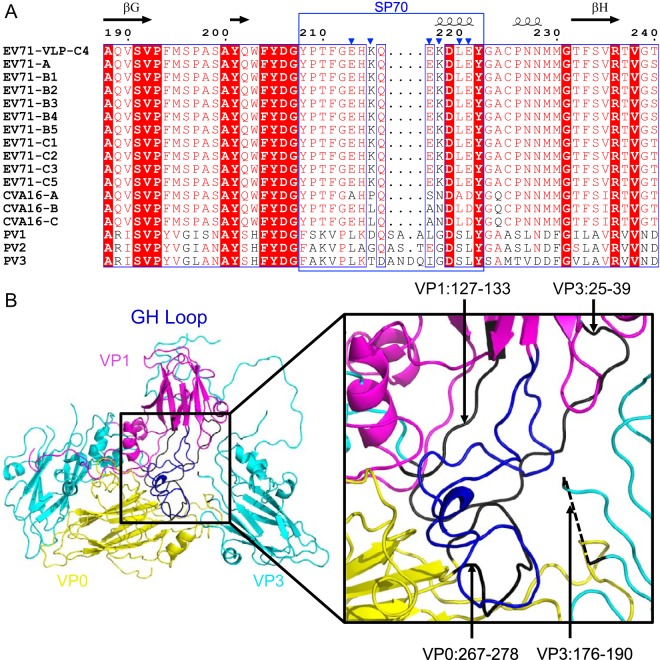
FIG 6.
Amino acid sequence variations present in the corresponding SP70 region of different enteroviruses and extensive interactions that the VP1 GH loop made with other regions in EV71 VLP. (A) Multiple-sequence alignment of VP1 from different strains of EV71, CVA16, and poliovirus (PV) showing the amino acid sequence variations in the corresponding SP70 region. VP1 sequences used for the alignment include strains A to C5 of EV71, strains A to C of CVA16, and types 1 to 3 of poliovirus. The residue numbers correspond to those in EV71. Conserved residues are shown in white with a red background. The secondary structure elements for EV71 VLP are shown at the top of the sequence alignment. Alpha-helix and beta strands are labeled according to standard picornavirus nomenclature and are represented by coils and arrows, respectively. The blue triangles indicate the VP1 residues that are variable between EV71 and CVA16. The corresponding SP70 regions (residues 208 to 222 in VP1) are boxed with a blue rectangle. This figure was produced using ESPript (33). (B) Shown on the left are cartoon representations of capsid proteins VP1 (colored in magenta), VP0 (colored in yellow), and VP3 (colored in cyan). The VP1 SP70 region (colored in blue), located near the pseudo-3-fold junction, makes interactions with amino acids 127 to 133 of VP1 (colored in black), amino acids 267 to 278 of VP0 (colored in black), and amino acids 25 to 39 and 176 to 190 of VP3 (colored in black).
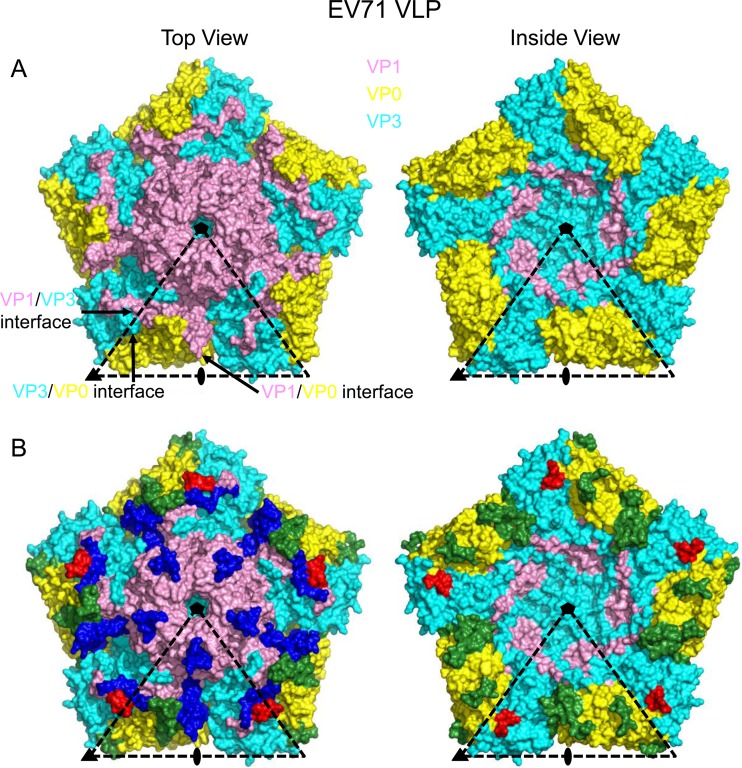
Superimposition of the individual VP1, VP0, and VP3 proteins in the chimeric VLP with those of EV71 VLP resulted in calculated RMSD values of 1.58 Å (225 Cα), 1.86 Å (231 Cα), and 1.01 Å (223 Cα) for equivalent Cα atoms, respectively. Thus, VP1 and VP0 undergo more substantial conformational changes than VP3 upon amino acid replacement in the VP1 GH loop. Structural variations could be observed in all three capsid proteins, including amino acids 95 to 105 (capsid surface near the 5-fold axis), 208 to 229 (near the pseudo-3-fold junction), and 280 to 292 (C terminus, on the capsid surface) of VP1; amino acids 93 to 101 (VP0/VP3 interface), 110 to 130 (near the pseudo-3-fold junction), 201 to 227 (capsid surface), 292 to 298 (VP0/VP3 interface), and 313 to 318 (near the pseudo-3-fold junction) of VP0; and amino acids 56 to 65 (VP0/VP3 interface) and 159 to 165 (VP0/VP3 interface) of VP3. These regions with structural changes were mapped onto the pentameric structure of EV71 VLP (Fig. 3), showing that the majority of these structural variations occurred at the particle surface or interfaces between the capsid proteins. In summary, the overall structure of the particle and the disposition of all three capsid proteins, VP1, VP0, and VP3, were largely maintained upon the 4-amino-acid replacement in the VP1 GH loop.
FIG 3.
Structural changes between EV71 VLP and chimeric VLP mapped onto a pentameric structure of EV71 VLP (left, top view; right, inside view). (A) Surface representation of a pentameric structure of EV71 VLP viewed along the 5-fold symmetry axis. VP1, VP0, and VP3 are colored in pink, yellow, and cyan, respectively. Positions of 5-, 3-, and 2-fold symmetry axes and interfaces between pairs of capsid proteins are indicated. (B) Significant structural variations occur at all three capsid proteins, including residues 95 to 105 (near the 5-fold axis), 208 to 229 (near the pseudo-3-fold junction), and 280 to 292 (C terminus, on capsid surface) of VP1 (colored in blue); residues 24 to 32 (VP0-VP3 interface), 41 to 61 (pseudo-3-fold junction), 132 to 158 (capsid surface), 223 to 229 (VP0-VP3 interface), and 244 to 249 (near the pseudo-3-fold junction) of VP0 (colored in green); and residues 56 to 65 (VP0-VP3 interface) and 159 to 165 (VP0-VP3 interface) of VP3 (colored in red).
Mapping of the neutralization epitopes.
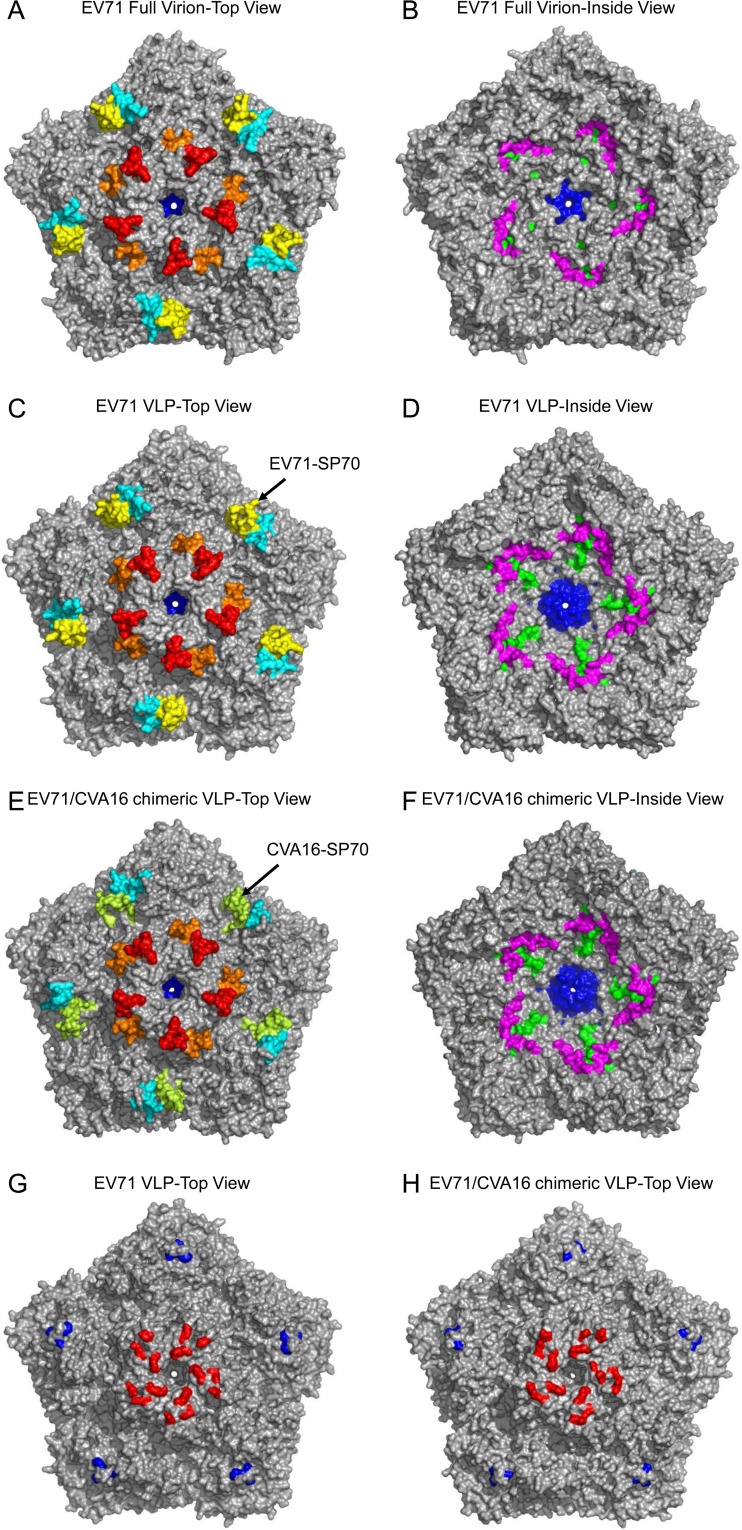
The preservation of neutralization epitopes is critical for ensuring high efficacy of good vaccine candidates. For EV71, these regions were mapped to the BC, EF, and GH loops and C terminus of VP1; the EF loop of VP2; and the N terminus of VP3 (Fig. 2), respectively. These linear neutralization epitopes of EV71 were mapped onto the structures of EV71 VLP and EV71/CVA16 chimeric VLP (Fig. 4C to F). Instead of the SP70 epitope of EV71, the corresponding SP70 region of CVA16 (CVA16-SP70) was mapped onto the structure of the chimeric VLP (Fig. 4E). Compared to the full virion (Fig. 4A and B), the linear neutralization epitopes of EV71 were well preserved on EV71 VLP and the chimeric VLP. In addition, the known conformational neutralization epitopes were also mapped onto both VLP structures and shown to be maintained (Fig. 4G and H). Thus, both the linear and conformational neutralization epitopes are well preserved on EV71 VLP and EV71/CVA16 chimeric VLP.
FIG 4.
Linear and conformational neutralization epitopes located on EV71 VLP and EV71/CVA16 chimeric VLP. For linear neutralization epitopes, a top view (A, C, and E) and an inside view (B, D, and F) were each provided for a pentameric structure of EV71 full virion, EV71 VLP, and EV71/CVA16 chimeric VLP, respectively. Residues 97 to 105, 163 to 177, 208 to 222, and 253 to 267 of VP1 are colored in red, orange, yellow, and green, respectively; residues 141 to 150 of VP2 (210 to 219 of VP0) are colored in cyan; residues 1 to 15 and 28 to 42 of VP3 are colored in blue and magenta, respectively. The CVA16-SP70 region (residues 208 to 222 of VP1) is colored in light yellow-green. For the conformational neutralization epitope, a top view was provided for a pentameric structure of EV71 VLP (G) and EV71/CVA16 chimeric VLP (H). Residues 98, 145, 242, and 244 of VP1 are colored in red; residues 59, 62, and 67 of VP3 are colored in blue.
Change of the local surface charge potential resulted from the amino acid replacement in the VP1 GH loop.
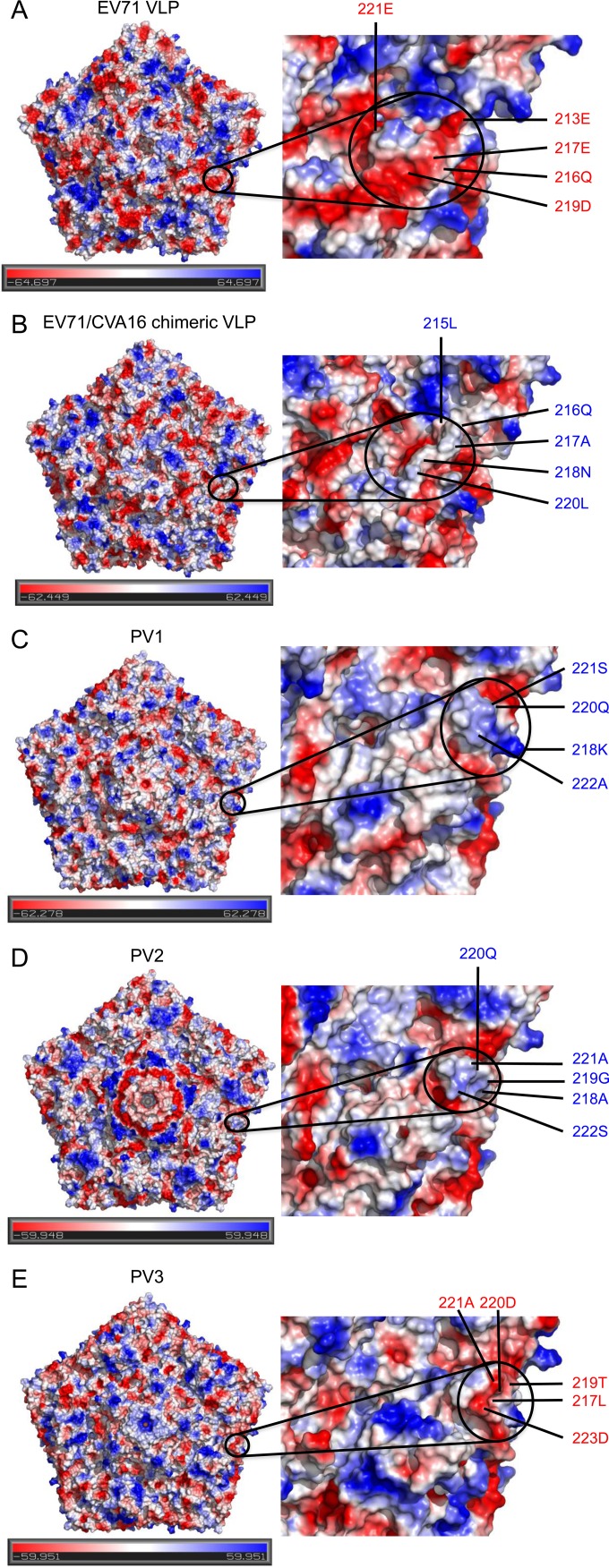
The SP70 epitope in EV71 VLP and the corresponding region in the chimeric VLP were both exposed on the particle surface. All of the 4 amino acid mutations occurred in the capsid protein VP1: K215L, E217A, K218N, and E221D. The electrostatic surface of a pentamer from EV71 VLP showed strongly negatively charged surface patches (colored in red) contributed by 213E, 216Q, 217E, 219D, and 221E in the VP1 GH loop (Fig. 5A). Such clustered negatively charged surface patches, however, disappeared in the chimeric VLP and were replaced with a relatively neutral surface (colored in white) contributed by 215L, 216Q, 217A, 218N, and 220L (Fig. 5B). Such amino acid sequence variations and the dramatic change of local surface charge potential were likely responsible for the additional capability of the chimeric VLP to elicit neutralizing responses against CVA16 infections.
FIG 5.
Electrostatic potential calculated for pentameric structures from EV71 VLP, EV71/CVA16 chimeric VLP, and poliovirus types 1 to 3. (A) Electrostatic surface of a pentamer from EV71 VLP, showing strongly negatively charged patches in EV71-SP70 (colored in red, contributed by 213E, 216Q, 217E, 219D, and 221E, all from VP1). Note that the electrostatic potentials calculated by PyMOL (version 1.3r1; Schrödinger, LLC) are crude approximations. (B) A neutral surface is shown in the corresponding SP70 region (colored in white, contributed by 215L, 216Q, 217A, 218N, and 220L) of EV71/CVA16 chimeric VLP. (C) A positively charged surface is shown in the corresponding SP70 region (colored in blue, contributed by 218K, 220Q, 221S, and 222A) of type 1 poliovirus. (D) A rather weakly positively charged surface is shown in the corresponding SP70 region (colored in blue, contributed by 218L, 219G, 220Q, 221A, and 222S) of type 2 poliovirus. (E) A negatively charged surface is shown in the corresponding SP70 region (colored in red, contributed by 217L, 219T, 220D, 221A, and 223D) of type 3 poliovirus.
To examine if such local charge differences also existed in different types of polioviruses, electrostatic surface potentials of a pentamer from all three types of polioviruses were each calculated for comparison (Fig. 5C to E). Residues 217 to 223 of VP1 in poliovirus are part of neutralization complex site 2 (20) and map to the VP1 GH loop exposed on the virus surface. This region displayed a positively charged surface patch in type 1 poliovirus, in contrast to a weakly positive charged surface or a rather neutral surface patch in type 2 poliovirus and a negatively charged surface patch in type 3 poliovirus (Fig. 5C to E), suggesting that the surface charge potential contributed by the VP1 GH loop was involved in conferring differences in immunogenicity.
The VP1 GH loop is involved in extensive interactions with other capsid regions.
A multiple-sequence alignment of VP1 amino acid sequences from different strains of EV71, CVA16, and poliovirus was performed to examine the sequence variation in the corresponding SP70 region (Fig. 6A). Amino acid sequences in the corresponding SP70 region from different strains of EV71 were rather conserved. Such sequence conservation was also observed in the corresponding SP70 region from different strains of CVA16, although not as significant as in EV71. In contrast, different types of polioviruses exhibited significant sequence variations at the corresponding SP70 region.
The SP70 region, mapped to the VP1 GH loop and located near the pseudo-3-fold junction, participated in interactions with multiple capsid protein regions, including residues 127 to 133 of VP1, residues 267 to 278 (GH loop) of VP0, and residues 25 to 39 (the N-terminal regions) and 176 to 190 (GH loop) of VP3 (Fig. 6B). Replacement of residues in the VP1 GH loop could lead to a rearrangement of the local interaction network, which may be propagated into other capsid regions and lead to both local and remote conformational changes.
Portions of VP1 and VP0 are externalized from EV71 VLP.
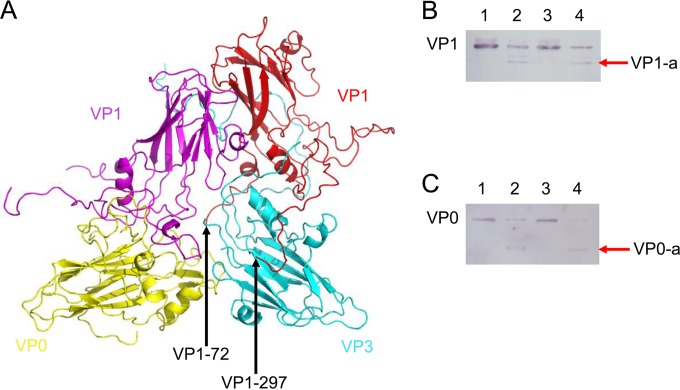
We observed a relative structural flexibility near the pseudo-3-fold junction in EV71 VLP, with the N-terminal regions of VP1 starting to become disordered near this junction (Fig. 7A). To test whether portions of VP1 and VP0 were transiently exposed, purified EV71 VLPs were digested by trypsin at 16°C and 37°C, respectively. A truncated product (VP1-a) was obtained when probed with VP1 antibodies (Fig. 7B). A VP0 truncated product (VP0-a) was also detected with VP4 antibodies (Fig. 7C). The presence of these truncated products suggested that portions of VP1 and VP0 were externalized at least transiently.
FIG 7.
Portions of VP1 and VP0 in EV71 VLP are at least transiently exposed. (A) The pseudo-3-fold junction is surrounded by VP1, VP0, and VP3, colored in magenta, yellow, and cyan, respectively. VP1 from a neighboring protomer is colored in red. The visible N terminus of VP1 (residue 72) is located at the base of the junction (from top view). (B) A truncated product (VP1-a) was detected. Lane 1, incubation at 16°C for 1 h; lane 2, digestion by trypsin at 16°C for 1 h; lane 3, incubation at 37°C for 1 h; lane 4, digestion by trypsin at 37°C for 1 h. VP1-a was detected in lanes 2 and 4. (C) A band smaller than VP0 (VP0-a) was detected. Lane 1, incubation at 16°C for 1 h; lane 2, digestion by trypsin at 16°C for 1 h; lane 3, incubation at 37°C for 1 h; lane 4, digestion by trypsin at 37°C for 1 h. VP0-a was detected in lanes 2 and 4.
Fitting of the EV71 VLP crystal structure into the EM density map.
Recently, Gong et al. reported the cryo-EM structure of EV71 VLP produced in insect cells at 5.2-Å resolution (EMBL-EBI identifier EMD-2607) (19). The crystal structure of our EV71 VLP produced in yeast was fitted into the reported cryo-EM density map (σ = 1), which showed that the crystal structure agreed well with the cryo-EM structural features, except for a few protruding loops (cc = 0.81).
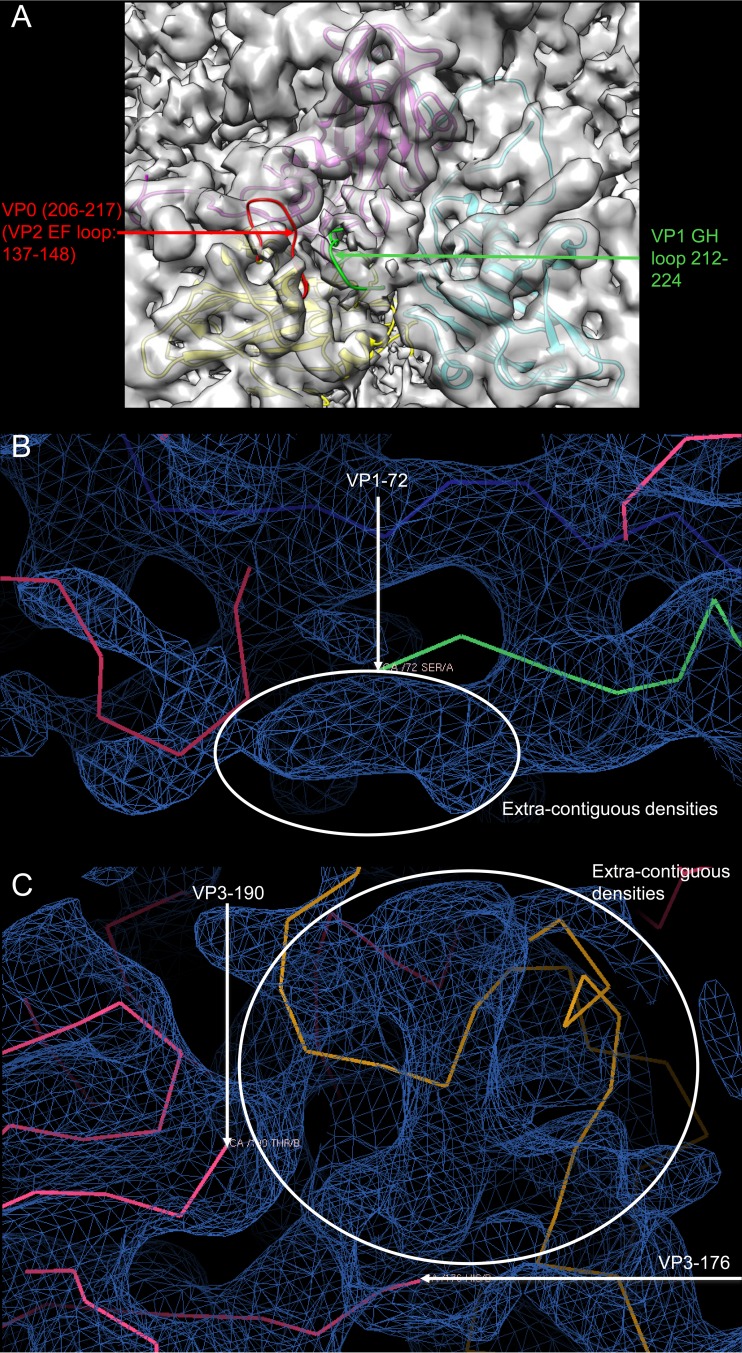
In the crystal structure, residues 212 to 224 of VP1 (located in the GH loop) (Fig. 8A) and residues 206 to 217 of VP0 (located in the EF loop) (Fig. 8A) were ordered. However, in the cryo-EM structure, there were no corresponding electron densities. These two regions are both EV71 neutralizing epitopes. In the crystal structure, the N-terminal residues 1 to 71 were disordered, while in the cryo-EM reconstruction, some extracontiguous densities ahead of residue 72 of VP1 could be observed (Fig. 8B). We speculate that these contiguous densities corresponded to the N-terminal residues of VP1. In the crystal structure, residues 177 to 189 of VP3 were disordered, whereas in the cryo-EM structure, some extracontiguous densities between residues 177 and 189 could be seen (Fig. 8C), which may correspond to residues 177 to 189 in the GH loop of VP3. All these differences were located near the junction region.
FIG 8.
Fitting of the EV71 VLP crystal structure into the EM density map. (A) One asymmetric unit in the EV71 VLP crystal structure was represented as ribbons with VP1, VP0, and VP3 colored in magenta, yellow, and cyan, respectively. Amino acids 206 to 217 of VP0 (colored in red) and 212 to 224 of VP1 (colored in green) are both loops near the pseudo-3-fold junction. (B) The cryo-EM density map is colored in blue. The VLP crystal structure is shown in Cα. Some extracontiguous densities are seen ahead of residue 72 of VP1. (C) The cryo-EM density map is colored in blue. The VLP crystal structure is shown in Cα. There are some extracontiguous densities between residues 176 and 190 of VP3.
DISCUSSION
In this study, we presented the crystal structures of EV71 VLP and EV71/CVA16 chimeric VLP. Both structures were similar to that of the naturally occurring empty particle produced from virus-infected cells. Mapping studies demonstrated that the major neutralization epitopes of EV71 were all preserved on both VLPs, in agreement with the observation that a potent neutralization response against EV71 could be elicited. The structure of chimeric VLP revealed that the corresponding SP70 epitope in CVA16 was well displayed on the particle surface. In addition, upon replacement of the 4 amino acid residues in the SP70 epitope, the strongly negatively charged surface patches presented on the EV71 VLP surface changed into a neutral surface on the chimeric VLP. Together, these structural observations provided explanations why the chimeric VLP can elicit neutralization responses against both EV71 and CVA16 infections. Interestingly, structural analysis showed that the corresponding region (the VP1 GH loop or part of the complex site 2) in polioviruses also revealed differences in the local charge potential (Fig. 5C to E). It is unclear whether such differences in the amino acid sequences and the local surface charge potential in different types of polioviruses were also related to variations in immunogenicity.
An effective HFMD vaccine should elicit strong cross-neutralizing responses against both EV71 and CVA16 infections. Structure-based design of chimeric antigens could be used to develop vaccines against multiple pathogens. The VLP system could be used as an efficient platform to screen the inserted epitope for the introduced neutralization capability. Such structure-based vaccine design was also illustrated by the production of stabilized FMDV VLPs (21). In that study, a disulfide bond was rationally engineered by replacing a single histidine residue at position 93 of VP2, located at the icosahedral 2-fold axis between adjacent pentamers with a cysteine residue. This engineered disulfide bond across the 2-fold axis relating two pentamers enhanced the stability of FMDV VLPs, whereas there were no significant differences in antibody titers between the cattle vaccinated with wild-type VLPs and those vaccinated with engineered VLPs. In the present study, based on the structural analysis on EV71 and CVA16, a replacement in the VP1 GH loop was performed with the replacement of four amino acids, and the resulting chimeric VLP showed potential as a bivalent vaccine against both EV71 and CVA16 infections.
Given these observations, it would be interesting to conduct more in-depth immunological analyses of the responses induced by the chimeric particles. How the responses elicited by EV71 VLP compare qualitatively and quantitatively with those induced by the chimeric VLP and whether antigenic variant viruses are selected more readily by anti-chimeric VLP antibodies should be investigated.
To our knowledge, this study is the first report of a chimeric enterovirus VLP resulting from the replacement in the GH loop of VP1. Previously, the VP1 BC loop of enteroviruses was used as the site for either replacement or insertion to produce recombinant viruses (22–27). Structural studies have been performed to examine the conformational changes in these recombinant viruses. The two polioviruses (types 1 and 2 or types 1 and 3) contributing to the poliovirus chimeras were closely related, and relatively small conformational alterations were found in the exchanged loop when it was placed in a different context (22, 23, 28). In this study, replacement in the VP1 GH loop was performed, and the resulting chimeric VLP exhibited conformational changes compared with the parent VLP. Structural analysis indicated that the VP1 GH loop was involved in the extensive interactions with the other regions of the capsid, which might be the reason why the replacement of the amino acid residues in the VP1 GH loop led to structural rearrangements, both locally and remotely.
Recently, our structure study on virus particles produced from cells infected with EV71 recombinant viruses showed that the VP1 BC loop is an ideal insertion site for foreign epitopes, thus providing insights into multivalent vaccine design (14). Instead of the VP1 BC loop, this study discussed the impact of the residue replacement in the VP1 GH loop and its implication for novel vaccine design.
In the poliovirus study, a cryo-EM study showed that during the transition of virion into an A-particle, gaps were produced in the junction area to allow for the exit of VP4 and N-terminal regions of VP1 (29, 30). In our study on EV71 uncoating, capsid regions near the pseudo-3-fold junction experienced a significant structural flexibility, demonstrated by the drastic conformational changes in all capsid proteins nearby (12). In addition, mass spectrometry analysis demonstrated that the intact virions of poliovirus and rhinovirus could experience “breathing” and transiently expose VP4 and the N-terminal regions of VP1, both situated inside the particle in the crystal structure (31, 32). Furthermore, proteolytic digestion of the naturally occurring empty particle of EV71 and N-terminal sequencing analysis revealed that at least 18 residues from the N terminus of VP1 are at least transiently externalized, suggesting that the junction site is an intrinsically dynamic area (14). Here, we demonstrated that such structural flexibility also existed in the yeast-produced VLP, revealed by the observation that portions of VP1 and VP0 get externalized at least transiently. Taken together, such structural flexibility in the capsid proteins may be an inherent property for enterovirus particles, including VLPs, which may be attributed to the types of amino acid residues surrounding the junction region and the interaction network nearby. It is thus imaginable that amino acid mutations introduced into the junction region, such as the VP1 GH loop, may confer immunogenicity changes and structural variations, as reported in this study with only the replacement of 4 amino acids in the VP1 GH loop.
ACKNOWLEDGMENTS
This study was supported by the 100 Talents Program of the Chinese Academy of Sciences, a Shanghai Pu-Jiang Career Development Award (grant no. 09PJ1411400), the 973 Project (grant no. 2010CB912403), and the National Natural Science Foundation of China (31370730 and 31070144).
We thank Cheng-Feng Qin for helpful discussions. We also thank the staff at the Shanghai Synchrotron Radiation Facility (beamline BL17U1) for onsite assistance and all other lab members for their helpful discussions. We are also grateful to the referees who kindly helped to make the paper much clearer in many places by providing many stimulating thoughts.
REFERENCES
- 1.McMinn PC. 2012. Recent advances in the molecular epidemiology and control of human enterovirus 71 infection. Curr Opin Virol 2:199–205. doi: 10.1016/j.coviro.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Foo DG, Ang RX, Alonso S, Chow VT, Quak SH, Poh CL. 2008. Identification of immunodominant VP1 linear epitope of enterovirus 71 (EV71) using synthetic peptides for detecting human anti-EV71 IgG antibodies in Western blots. Clin Microbiol Infect 14:286–288. doi: 10.1111/j.1469-0691.2007.01904.x. [DOI] [PubMed] [Google Scholar]
- 3.Foo DG, Alonso S, Phoon MC, Ramachandran NP, Chow VT, Poh CL. 2007. Identification of neutralizing linear epitopes from the VP1 capsid protein of enterovirus 71 using synthetic peptides. Virus Res 125:61–68. doi: 10.1016/j.virusres.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 4.Kirk K, Poh CL, Fecondo J, Pourianfar H, Shaw J, Grollo L. 2012. Cross-reactive neutralizing antibody epitopes against enterovirus 71 identified by an in silico approach. Vaccine 30:7105–7110. doi: 10.1016/j.vaccine.2012.09.030. [DOI] [PubMed] [Google Scholar]
- 5.Liu CC, Chou AH, Lien SP, Lin HY, Liu SJ, Chang JY, Guo MS, Chow YH, Yang WS, Chang KH, Sia C, Chong P. 2011. Identification and characterization of a cross-neutralization epitope of enterovirus 71. Vaccine 29:4362–4372. doi: 10.1016/j.vaccine.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 6.Lee H, Cifuente JO, Ashley RE, Conway JF, Makhov AM, Tano Y, Shimizu H, Nishimura Y, Hafenstein S. 2013. A strain-specific epitope of enterovirus 71 identified by cryo-electron microscopy of the complex with fab from neutralizing antibody. J Virol 87:11363–11370. doi: 10.1128/JVI.01926-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiener TK, Jia Q, Meng T, Chow VT, Kwang J. 2014. A novel universal neutralizing monoclonal antibody against enterovirus 71 that targets the highly conserved “knob” region of VP3 protein. PLoS Negl Trop Dis 8:e2895. doi: 10.1371/journal.pntd.0002895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi J, Huang X, Liu Q, Huang Z. 2013. Identification of conserved neutralizing linear epitopes within the VP1 protein of coxsackievirus A16. Vaccine 31:2130–2136. doi: 10.1016/j.vaccine.2013.02.051. [DOI] [PubMed] [Google Scholar]
- 9.Li HY, Han JF, Qin CF, Chen R. 2013. Virus-like particles for enterovirus 71 produced from Saccharomyces cerevisiae potently elicits protective immune responses in mice. Vaccine 31:3281–3287. doi: 10.1016/j.vaccine.2013.05.019. [DOI] [PubMed] [Google Scholar]
- 10.Plevka P, Perera R, Cardosa J, Kuhn RJ, Rossmann MG. 2012. Crystal structure of human enterovirus 71. Science 336:1274. doi: 10.1126/science.1218713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Peng W, Ren J, Hu Z, Xu J, Lou Z, Li X, Yin W, Shen X, Porta C, Walter TS, Evans G, Axford D, Owen R, Rowlands DJ, Wang J, Stuart DI, Fry EE, Rao Z. 2012. A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71. Nat Struct Mol Biol 19:424–429. doi: 10.1038/nsmb.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lyu K, Ding J, Han JF, Zhang Y, Wu XY, He YL, Qin CF, Chen R. 2014. Human enterovirus 71 uncoating captured at atomic resolution. J Virol 88:3114–3126. doi: 10.1128/JVI.03029-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao H, Li HY, Han JF, Deng YQ, Zhu SY, Li XF, Yang HQ, Li YX, Zhang Y, Qin ED, Chen R, Qin CF. 2015. Novel recombinant chimeric virus-like particle is immunogenic and protective against both enterovirus 71 and coxsackievirus A16 in mice. Sci Rep 5:7878. doi: 10.1038/srep07878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyu K, Wang GC, He YL, Han JF, Ye Q, Qin CF, Chen R. 2015. Crystal structures of enterovirus 71 (EV71) recombinant virus particles provide insights into vaccine design. J Biol Chem 290:3198–3208. doi: 10.1074/jbc.M114.624536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tong L, Rossmann MG. 1997. Rotation function calculations with GLRF program. Methods Enzymol 276:594–611. doi: 10.1016/S0076-6879(97)76080-4. [DOI] [PubMed] [Google Scholar]
- 16.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 19.Gong M, Zhu H, Zhou J, Yang C, Feng J, Huang X, Ji G, Xu H, Zhu P. 2014. Cryo-electron microscopy study of insect cell-expressed enterovirus 71 and coxsackievirus a16 virus-like particles provides a structural basis for vaccine development. J Virol 88:6444–6452. doi: 10.1128/JVI.00200-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minor PD, Ferguson M, Katrak K, Wood D, John A, Howlett J, Dunn G, Burke K, Almond JW. 1991. Antigenic structure of chimeras of type 1 and type 3 polioviruses involving antigenic sites 2, 3 and 4. J Gen Virol 72:2475–2481. doi: 10.1099/0022-1317-72-10-2475. [DOI] [PubMed] [Google Scholar]
- 21.Porta C, Kotecha A, Burman A, Jackson T, Ren J, Loureiro S, Jones IM, Fry EE, Stuart DI, Charleston B. 2013. Rational engineering of recombinant picornavirus capsids to produce safe, protective vaccine antigen. PLoS Pathog 9:e1003255. doi: 10.1371/journal.ppat.1003255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burke KL, Dunn G, Ferguson M, Minor PD, Almond JW. 1988. Antigen chimaeras of poliovirus as potential new vaccines. Nature 332:81–82. doi: 10.1038/332081a0. [DOI] [PubMed] [Google Scholar]
- 23.Martin A, Wychowski C, Couderc T, Crainic R, Hogle J, Girard M. 1988. Engineering a poliovirus type 2 antigenic site on a type 1 capsid results in a chimaeric virus which is neurovirulent for mice. EMBO J 7:2839–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murray MG, Kuhn RJ, Arita M, Kawamura N, Nomoto A, Wimmer E. 1988. Poliovirus type 1/type 3 antigenic hybrid virus constructed in vitro elicits type 1 and type 3 neutralizing antibodies in rabbits and monkeys. Proc Natl Acad Sci U S A 85:3203–3207. doi: 10.1073/pnas.85.9.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evans DJ, McKeating J, Meredith JM, Burke KL, Katrak K, John A, Ferguson M, Minor PD, Weiss RA, Almond JW. 1989. An engineered poliovirus chimaera elicits broadly reactive HIV-1 neutralizing antibodies. Nature 339:385–388. doi: 10.1038/339385a0. [DOI] [PubMed] [Google Scholar]
- 26.Jenkins O, Cason J, Burke KL, Lunney D, Gillen A, Patel D, McCance DJ, Almond JW. 1990. An antigen chimera of poliovirus induces antibodies against human papillomavirus type 16. J Virol 64:1201–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin T, Porta C, Lomonossoff G, Johnson JE. 1996. Structure-based design of peptide presentation on a viral surface: the crystal structure of a plant/animal virus chimera at 2.8 A resolution. Fold Des 1:179–187. doi: 10.1016/S1359-0278(96)00030-2. [DOI] [PubMed] [Google Scholar]
- 28.Yeates TO, Jacobson DH, Martin A, Wychowski C, Girard M, Filman DJ, Hogle JM. 1991. Three-dimensional structure of a mouse-adapted type 2/type 1 poliovirus chimera. EMBO J 10:2331–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Butan C, Filman DJ, Hogle JM. 2014. Cryo-electron microscopy reconstruction shows poliovirus 135S particles poised for membrane interaction and RNA release. J Virol 88:1758–1770. doi: 10.1128/JVI.01949-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bubeck D, Filman DJ, Cheng N, Steven AC, Hogle JM, Belnap DM. 2005. The structure of the poliovirus 135S cell entry intermediate at 10-angstrom resolution reveals the location of an externalized polypeptide that binds to membranes. J Virol 79:7745–7755. doi: 10.1128/JVI.79.12.7745-7755.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis JK, Bothner B, Smith TJ, Siuzdak G. 1998. Antiviral agent blocks breathing of the common cold virus. Proc Natl Acad Sci U S A 95:6774–6778. doi: 10.1073/pnas.95.12.6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Q, Yafal AG, Lee YM, Hogle J, Chow M. 1994. Poliovirus neutralization by antibodies to internal epitopes of VP4 and VP1 results from reversible exposure of these sequences at physiological temperature. J Virol 68:3965–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]