ABSTRACT
The E1 helicase from anogenital human papillomavirus (HPV) types interacts with the cellular WD repeat-containing protein UAF1 in complex with the deubiquitinating enzyme USP1, USP12, or USP46. This interaction stimulates viral DNA replication and is required for maintenance of the viral episome in keratinocytes. E1 associates with UAF1 through a short UAF1-binding site (UBS) located within the N-terminal 40 residues of the protein. Here, we investigated if the E1 UBS could be replaced by the analogous domain from an unrelated protein, the pleckstrin homology domain and leucine-rich repeat protein phosphatase 1 (PHLPP1). We found that PHLPP1 and E1 interact with UAF1 in a mutually exclusive manner and mapped the minimal PHLPP1 UBS (PUBS) to a 100-amino-acid region sufficient for assembly into UAF1-USP complexes. Similarly to the E1 UBS, overexpression of PUBS in trans inhibited HPV DNA replication, albeit less efficiently. Characterization of a PHLPP1-E1 chimeric helicase revealed that PUBS could partially substitute for the E1 UBS in enhancing viral DNA replication and that the stimulatory effect of PUBS likely involves recruitment of UAF1-USP complexes, as it was abolished by mutations that weaken UAF1-binding and by overexpression of catalytically inactive USPs. Although functionally similar to the E1 UBS, PUBS is larger in size and requires both the WD repeat region and C-terminal ubiquitin-like domain of UAF1 for interaction, in contrast to E1, which does not contact the latter. Overall, this comparison of two heterologous UBSs indicates that these domains function as transferable protein interaction modules and provide further evidence that the association of E1 with UAF1-containing deubiquitinating complexes stimulates HPV DNA replication.
IMPORTANCE The E1 protein from anogenital HPV types interacts with the UAF1-associated deubiquitinating enzymes USP1, USP12, and USP46 to stimulate replication of the viral genome. Little is known about the molecular nature of the E1-UAF1 interaction and, more generally, how UAF1-USP complexes recognize their substrate proteins. To address this question, we characterized the UAF1-binding site (UBS) of PHLPP1, a protein unrelated to E1. Using a PHLPP1-E1 chimeric helicase, we show that the PHLPP1 UBS (PUBS) can partially substitute for the E1 UBS in stimulating HPV DNA replication. This stimulation required conserved sequences in PUBS that meditate its interaction with UAF1, including a motif common to the E1 UBS. These results indicate that UAF1-binding sequences function as transferable protein interaction modules and provide further evidence that UAF1-USP complexes stimulate HPV DNA replication.
INTRODUCTION
Human papillomaviruses (HPVs) are small DNA tumor viruses that infect squamous epithelia of the skin and mucosa. Infections by these viruses can lead to the development of benign and malignant tumors, depending on the viral types. Several HPV types are sexually transmitted and the causative agents of cervical cancer, the second most common cancer in women worldwide. These so-called “high-risk” types predominate in high-grade precancerous and cancerous lesions of the cervix and sustain their proliferation (1). In recent years, these oncogenic viral types have also been linked to other malignancies of the anogenital area as well as a growing proportion of oropharyngeal cancers (1).
The genome of HPV is a circular double-stranded DNA molecule that is present in multiple copies in the nucleus of infected cells. Replication of the viral episomes during S phase requires the viral proteins E1 and E2, which act in concert with the host DNA replication machinery (reviewed in reference 2). E1 is a DNA helicase that assembles as a double hexamer at the viral origin of replication to unwind DNA and nucleate the formation of a bidirectional replication fork. E1 can be subdivided into three main functional regions (reviewed in references 3 and 4). Two of these, the central DNA-binding domain and the C-terminal helicase domain, mediate the assembly of E1 at the origin and are sufficient to support viral DNA replication in vitro. The N-terminal region, in contrast, is dispensable for replication in cell-free systems but essential in vivo (5, 6). Several regulatory motifs have been identified in this region, including a bipartite nuclear localization signal (NLS) and, for most HPV types, a nuclear export signal (NES) and a cyclin-binding motif (CBM) that promote the nucleocytoplasmic shuttling of E1 in a Cdk2 phosphorylation-dependent manner (4). We previously reported that the first 40 amino acids of E1 from anogenital HPV types contain a binding site for the USP1-associated factor 1 (UAF1) cellular protein that is required for efficient viral DNA replication and for episomal maintenance of the viral genome in immortalized keratinocytes (7, 8). In noninfected cells, UAF1 forms separate complexes with the three deubiquitinating enzymes USP1, USP12, and USP46 and stimulates their activity (9–12). Accordingly, we recently determined that E1 assembles with UAF1 and either USP1, USP12, or USP46 into ternary complexes that are recruited to the origin of replication and whose deubiquitinase activity stimulates viral DNA replication in transient assays (13).
Little is known about the molecular determinants of E1 that mediate its interaction with UAF1, other than our previous observations that a peptide spanning the E1 UAF1-binding site (UBS; amino acids [aa] 1 to 40) is intrinsically disordered in solution and that mutations of conserved residues within this region abrogate binding (7, 8). Our understanding of the E1-binding surface on UAF1 is also limited. Previous studies have suggested that UAF1 contains an N-terminal region comprised of 8 WD repeats and a C-terminal ring-finger and WD40-associated ubiquitin-like (RAWUL) domain (14, 15). RAWUL domains were first identified computationally as ubiquitin-like regions present in UAF1 and in ring-finger proteins of the PRC1 family of polycomb group complexes (14). A subsequent study proposed that the C terminus of UAF1 contains two tandemly repeated domains of the small ubiquitin-like modifier (SUMO) family, named SLD1 and SLD2, with the latter corresponding to the RAWUL domain (16). However, because SLD1 shows only weak sequence similarity to other ubiquitin-like domains, additional evidence is needed to ascertain its function. We previously reported that a fragment of UAF1 encompassing both the WD repeat region and SLD1, but lacking SLD2, is sufficient for interaction with HPV E1, thus raising the possibility that the two SLDs may function independently (8). Clearly, additional studies are needed to elucidate how E1 interacts with UAF1-USP complexes and, more generally, how UAF1-containing deubiquitinating enzymes recognize their substrates.
To help address these questions and to validate the stimulatory function of UAF1-USP complexes in HPV DNA replication, we set out to characterize the UBS from another protein, unrelated to E1, and test if it could replace the E1 UBS in functional assays. We selected to characterize the UBS from the cellular pleckstrin homology domain and leucine-rich repeat protein phosphatase 1 (PHLPP1), because this protein was found to interact with UAF1 in previous proteomic studies (including our own [13]) (data not shown) and to be a substrate of either USP1, USP12, or USP46, depending on the report (17–21). PHLPP1, and its close homologue PHLPP2, have been shown to regulate the proliferation and survival of normal and cancer cells through their ability to dephosphorylate a common set of substrates, which include the kinases Akt and PKC (22, 23). Thus, PHLPP1 and PHLPP2 are not functionally linked to HPV DNA replication in any way, and their only resemblance to E1 is in their ability to bind UAF1 and associated USPs.
In this study, we report our characterization of the minimal PHLPP1 UAF1-binding site (PUBS) and provide evidence that it is functionally similar to the E1 UBS. Specifically, we show that PUBS is sufficient for assembly into UAF1-USP complexes and that it can impede HPV DNA replication when overexpressed as a transdominant inhibitory domain. We also show that PUBS can function in cis within the context of a PUBS-E1 chimeric helicase, as it can partially substitute for the E1 UBS in enhancing HPV DNA replication. Although functionally related to the E1 UBS, PUBS is significantly larger than the latter (100 compared to 40 amino acids), suggesting that it may engage in additional contacts with UAF1. This is supported by the finding that the interaction of either PHLPP1 or PUBS-E1 with UAF1 requires both the WD repeat region of UAF1 and the C-terminal SUMO-like domain (SLD), in contrast to wild-type (WT) E1, which does not require this SLD. Collectively, these results bring novel insights into the molecular nature of UAF1-binding sites and how these domains interact with UAF1-USP complexes. Importantly, they also provide further evidence that the primary function of the E1 N terminus is to recruit UAF1-containing deubiquitinase complexes to stimulate viral DNA replication.
MATERIALS AND METHODS
Plasmids used in this study.
The plasmids used to express UAF1 and its truncated derivatives tagged at their N terminus with a triple-Flag epitope (3F) were previously described (8). The vector expressing UAF1 fused to green fluorescent protein (GFP) was constructed by inserting the UAF1 open reading frame (ORF) between the EcoRV and KpnI sites of plasmid pGFP2-C2 (BioSignal Packard-Perkin-Elmer). The plasmids encoding GFP-E1 and yellow fluorescent protein (YFP)-E1 from HPV31 were previously described (7, 24). Those expressing codon-optimized 3F-E1 (p31E1), 3F-E2 (p31E2), and red fluorescent protein (RFP)-E2 were also described earlier (8, 25). The plasmid used to express PHLPP1 fused to GFP was constructed by inserting the PHLPP1 ORF (obtained from Life Technologies; Mammalian Gene Collection clone 3916044) between the SacI and BamHI sites of plasmid pGFP2-N1 (BioSignal Packard-Perkin-Elmer). Plasmids encoding PHLPP1 and truncated derivatives fused to hemagglutinin (HA) were constructed by inserting the appropriate PCR fragments between the EcoRV and SalI sites of the HA plasmid AB-1896. This HA vector was constructed by substituting the 3F epitope of pCMV-3Tag-1a (Stratagene) by an HA epitope. Expression vectors for RFP-USP1, USP12, and USP46 and their catalytically inactive versions were described previously (13). The plasmid used to express the HPV31 E1 UAF1-binding site (UBS) fused to YFP was previously described (referred to as N40-YFP in reference 8). An identical cloning strategy was used to construct the plasmid expressing the PHLPP1 UBS fused to YFP (PUBS-YFP). The 3F-E1Δ expression plasmid was previously described (E1 C40 truncation) (26). The plasmid encoding the 3F-PUBS-E1Δ chimeric helicase was generated by inserting the PHLPP1 UBS coding region between the BamHI and XmaI sites of the 3F-E1Δ plasmid. Site-directed mutagenesis was performed with the QuikChange site-directed mutagenesis kit (Stratagene). All DNA constructs were verified by sequencing. Sequences of primers and additional details on the construction of these plasmids will be made available upon request.
Antibodies and Western blotting.
Commercially available antibodies were used to detect β-tubulin (monoclonal antibody; Sigma-Aldrich; catalog no. T4026) and fusion proteins fused to either a 3F epitope (M2 monoclonal antibody; Sigma-Aldrich; catalog no. F1804), GFP (mixture of two mouse monoclonal antibodies; Roche; catalog no. 11814460001), RFP (monoclonal antibody; Abcam; catalog no. AB65856), or an HA epitope (HA.11 antibody; Covance; catalog no. MMS-101P). Rabbit polyclonal antibodies against UAF1 were raised by injecting rabbits (Open Biosystems) with a purified C-terminal fragment of UAF1, as described elsewhere (8). PHLPP1 and USP46 antibodies were purchased from Bethyl Laboratories (catalog no. AB65856 A300-660A) and Sigma-Aldrich (catalog no. SAB1407903), respectively. For Western blot analysis, proteins were transferred onto polyvinylidene difluoride membranes and detected using horseradish peroxidase-conjugated secondary antibodies from GE Healthcare, either sheep anti-mouse IgG (catalog no. NA931) or donkey anti-rabbit IgG (NA934V), with an enhanced chemiluminescence detection kit (GE Healthcare).
Cell culture and transfections.
The human cervical carcinoma cell line C33A was grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 50 IU/ml of penicillin, and 50 μg/ml streptomycin (Wisent Bioproducts). Transfections were performed using the Lipofectamine 2000 reagent (Life Technologies) according to the manufacturer's protocol.
Coimmunoprecipitation assays.
C33A cells were grown on 100-mm plates and transfected with the indicated plasmid DNA. Cells were harvested 48 h posttransfection in lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 10 μg/ml antipain, 2 μg/ml leupeptin, 2 μg/ml aprotinin, 1 μg/ml pepstatin A, and 1 mM phenylmethylsulfonyl fluoride). Cleared cellular extracts were then immunoprecipitated for 3 h with 40 μl of protein G Sepharose (GE Healthcare) conjugated to 1 μg of anti-Flag or anti-GFP antibodies. The resin was washed 3 times with TBS (50 mM Tris-HCl [pH 7.4], 150 mM NaCl), and the bound proteins were eluted in 5× Laemmli buffer prior to Western blotting.
Luciferase-based HPV31 DNA replication assays.
The HPV31 DNA replication assay was performed as described previously (25, 27). Briefly, C33A cells were seeded at a density of 25,000 cells/well in white, flat-bottom, 96-well plates and transfected 24 h later with a mix of four plasmids: an origin-containing plasmid with a firefly luciferase reporter in cis (pFLORI31), a Renilla luciferase plasmid as an internal control (pRL), and the indicated quantities of E1 and E2 expression vectors. For all experiments, the total quantity of plasmid DNA transfected was adjusted to 100 ng with the YFP or RFP vector as carrier DNA. Firefly and Renilla luciferase activities were measured using the Dual-Glo luciferase assay system (Promega) 48 h posttransfection unless indicated otherwise. The GraphPad Prism 6 software was used to fit the DNA replication data to a sigmoidal dose-response curve described by the following equation: Y = Bottom + (Top − Bottom)/[1 + 10(LogEC50 − X) × HillSlope], where Y is the DNA replication value, Bottom is the DNA replication value obtained in the absence of E2, Top is the maximal level of DNA replication, X is the amount of E2 expression plasmid (in grams), and EC50 is the 50% effective concentration. The DNA replication inhibitor aphidicolin was purchased from EMD Millipore (catalog no. 178273), while ara-C, hydroxyurea, and mimosine were obtained from Sigma-Aldrich (catalog no. C3350000, H8627, and M0253, respectively).
Confocal fluorescence microscopy.
C33A cells were plated at a density of 6 × 105 cells/well on coverslips and were transfected 24 h later with the indicated plasmids. Twenty-four hours posttransfection, cells were fixed with 4% formaldehyde and permeabilized with 0.2% Triton X-100, and their DNA was stained with 1 μg/ml 4,6-diamidino-2-phenylindole (DAPI; Life Technologies; catalog no. D1306). Slides were mounted using Vectashield mounting medium (Vector Laboratories). Images were collected with a Zeiss LSM710 laser scanning confocal microscope and analyzed using the Zen 2009 LE software.
Cell cycle analysis.
Cell cycle profiles were obtained by staining live cells 48 h posttransfection with 6.3 μg/ml Hoechst and 50 μM verapamil. Acquisitions were done using a BD LSR flow cytometer, gated on the GFP-positive population. Data analysis was performed using the FlowJo (v.8.1) software.
Colony formation assay.
C33A cells (∼1.2 × 106) were transfected with 1.5 μg of the indicated plasmids in a 6-well plate. Twenty-four hours posttransfection, cells were trypsinized and seeded on a new plate at a 1/15 dilution in fresh medium. Twenty-four hours later, medium containing G418 (500 μg/ml) or puromycin (2 μg/ml) was added and changed every 3 to 4 days for a period of about 3 weeks or until fully resistant cells were selected. Colonies were fixed in cold methanol for 10 min and stained for 2 min at room temperature with methylene blue (1% [wt/vol] in 60% methanol-H2O).
Bioinformatic tools.
Amino acid sequence alignments were generated with Clustal Omega (28). Sequence logos were generated with WebLogo (29) using an alignment of the E1 UBS from 50 different anogenital HPV types or with an alignment of PUBS from 32 PHLPP1 and PHLPP2 sequences from diverse organisms. The structural model of human UAF1 SLD2 was generated using the Phyre2 server (30) and refined with the ModRefiner algorithm (31). A similar model was obtained with the RaptorX server (not shown) (32). Structure representations were generated with the PyMOL Molecular Graphics System, Schrödinger, LLC.
RESULTS
PHLPP1 and HPV E1 interact with UAF1 in a mutually exclusive manner.
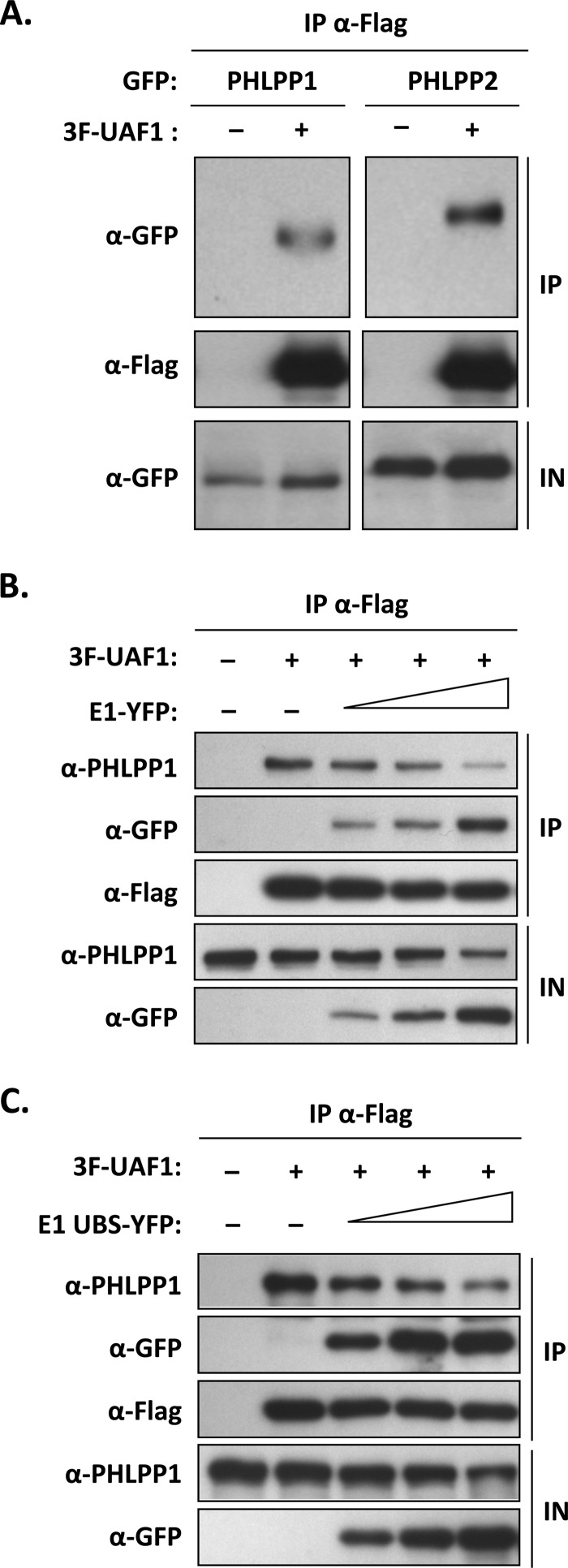
It was previously reported that the cellular PHLPP1 phosphatase interacts with UAF1 and is a substrate of either USP1, USP12, or USP46, depending on the study (17–21). To confirm that PHLPP1 interacts with UAF1, coimmunoprecipitation experiments were performed in transfected C33A cells transiently expressing UAF1 fused to a triple-Flag epitope (3F-UAF1) and PHLPP1 tagged with GFP (PHLPP1-GFP), essentially as described previously (13). These experiments confirmed that PHLPP1, and its close homolog PHLPP2, do interact with UAF1 in vivo (Fig. 1A). We then set out to determine if PHLPP1 competes with HPV E1 for binding to UAF1 or, alternatively, if all three proteins can assemble into a ternary complex. To begin with, we tested if PHLPP1 and E1 could interact with each other in coimmunoprecipitation experiments but found no evidence of their association (data not shown). This result suggested that E1 and PHLPP1 may compete with each other for binding to UAF1 rather than being part of the same complex. This prompted us to test if overexpression of E1 (tagged with YFP [E1-YFP]) could antagonize the interaction of endogenous PHLPP1 with 3F-UAF1. The results presented in Fig. 1B showed that overexpression of E1-YFP, but not of YFP alone, could indeed reduce the amount of PHLPP1 coprecipitated with UAF1 (Fig. 1B). Although this finding is consistent with the notion that E1 competes with PHLPP1 for binding to UAF1, its interpretation has been complicated by the observation that E1 also downregulates the expression of endogenous PHLPP1 (see Western blot of input cell extracts in Fig. 1B). It was previously reported that UAF1, in complex with USP12, increases the stability of PHLPP1 by promoting its deubiquitination (18). This led us to hypothesize that the decrease in PHLPP1 levels caused by overexpression of E1 might be related to its ability to bind UAF1 and, as a result, to compete the interaction of PHLPP1 with UAF1-containing deubiquitinase complexes. To address this possibility, we repeated the coimmunoprecipitation studies presented above using the E1 UBS only, instead of the full-length protein. We previously found that the E1 UBS is contained within the first 40 amino acids of the protein and that this domain, when overexpressed in trans, efficiently competes the interaction of full-length E1 with UAF1 (7, 8). The results presented in Fig. 1C revealed that overexpression of the E1 UBS (fused to YFP as done previously [8]) can also reduce the interaction of endogenous PHLPP1 with UAF1. The E1 UBS was also sufficient to lower the steady-state levels of PHLPP1 in the input cell extracts (Fig. 1C). These results argued that the binding of E1 to UAF1 underlies its ability to reduce the interaction of PHLPP1 with UAF1 and to lower the steady-state levels of PHLPP1. As such, these findings provided the first indication that PHLPP1 and E1 interact with UAF1 in a mutually exclusive manner, a notion that is substantiated by additional experiments presented below.
FIG 1.
PHLPP1 and PHLPP2 interact with UAF1 in a mutually exclusive manner with E1. (A) Coimmunoprecipitation of GFP-tagged PHLPP1 and PHLPP2 with 3F-UAF1. C33A cells were cotransfected with a 3F-UAF1 expression plasmid together with an expression vector for either PHLPP1-GFP or PHLPP2-GFP. UAF1-containing protein complexes were immunoprecipitated with an anti-Flag antibody, and the presence of PHLPP1 and PHLPP12 in the immunoprecipitates (IP) and input cell extracts (IN) was determined by Western blotting with anti-GFP antibodies. (B and C) Inhibitory effect of full-length E1 and of the E1 UBS on the interaction of UAF1 with endogenous PHLPP1. C33A cells were transfected with a 3F-UAF1 expression vector and increasing amounts of E1 expression plasmid encoding either the full-length protein (B) or the UBS alone (C), fused to YFP. UAF1-containing protein complexes were immunoprecipitated with an anti-Flag antibody and the presence of endogenous PHLPP1 in the immunoprecipitates determined by Western blotting with an anti-PHLPP1 antibody.
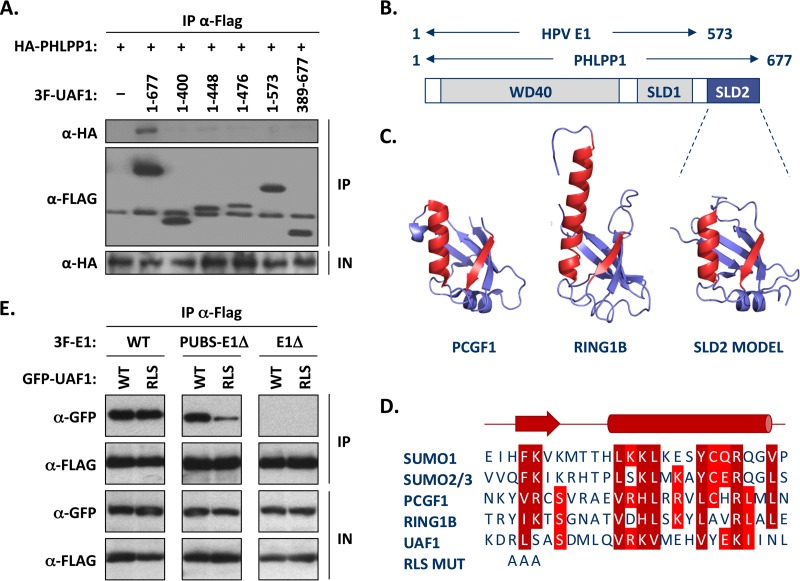
PHLPP1 interacts with UAF1 through a short UBS located in the C-terminal portion of the protein.
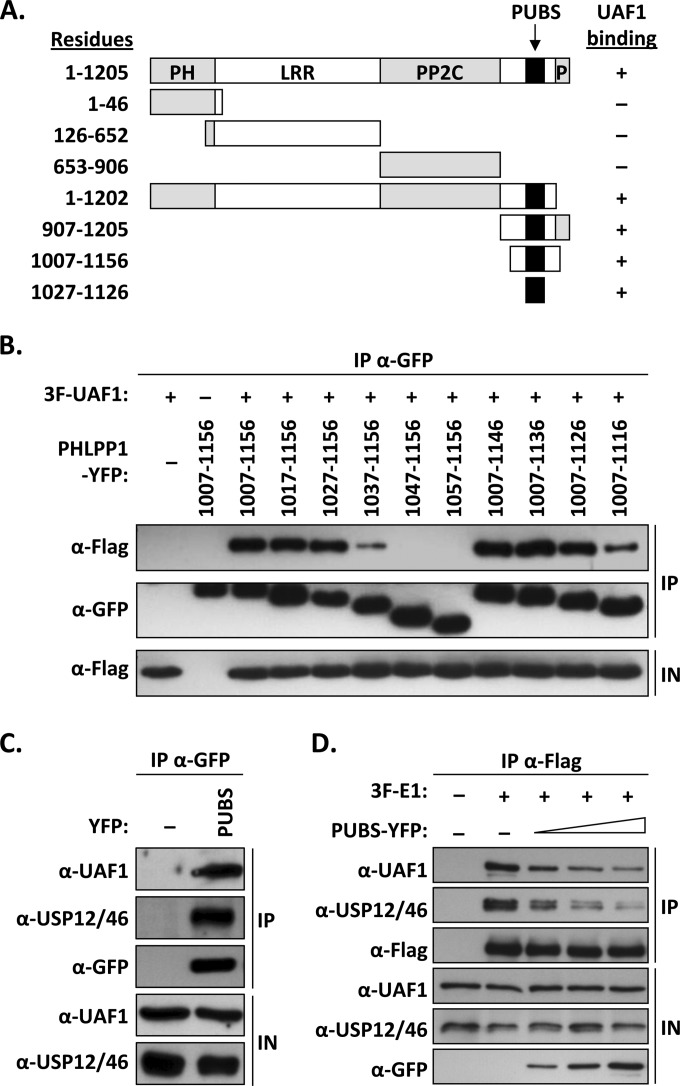
PHLPP1 can be subdivided into four main regions: a pleckstrin homology (PH) domain, a leucine-rich repeat (LRR) region, a PP2C phosphatase domain, and a short PDZ-binding domain at its C terminus (22). To determine which region of PHLPP1 is required for UAF1 interaction, we tested the truncated proteins shown in Fig. 2A for their capacity to associate with UAF1 in coimmunoprecipitation experiments. These studies, which were performed with the 1,205-amino-acid-long isoform of PHLPP1 (GenBank accession number AAI26278.1), indicated that the UBS of PHLPP1 was contained within residues 1007 to 1156, outside the known functional domains of the protein (summarized in Fig. 2A). Smaller deletions, in 10-amino-acid increments, were then used to precisely map the boundaries of the UBS (Fig. 2B). Truncations lacking amino acids downstream of residue 1027, or upstream of residue 1126, showed reduced binding to UAF1 (Fig. 2B). These results suggested that the minimal UBS is encoded within amino acids 1027 to 1126 of PHLPP1. This was confirmed using a fusion of this 100-amino-acid-long domain to YFP; for simplicity, we will refer to this short PHLPP1 UBS as PUBS. As expected, coimmunoprecipitation experiments confirmed that YFP-PUBS could interact with endogenous UAF1, while YFP alone was unable to do so (Fig. 2C). To determine if PUBS could form a ternary complex with UAF1 and its associated USPs, we analyzed the YFP-PUBS immunoprecipitate with an antibody that recognizes both USP12 and USP46. The positive signal shown in Fig. 2C revealed that PUBS assembles into a ternary complex with endogenous UAF12/46. Thus, like the E1 UBS, PUBS can bind to UAF1-associated deubiquitinase complexes.
FIG 2.
Mapping of the UAF1-binding site of PHLPP1. (A) Schematic representation of PHLPP1 and truncated derivatives. Functional regions of the protein are indicated: pleckstrin homology domain (PH), leucine-rich region (LRR), PP2C phosphatase domain (PP2C), and C-terminal PDZ protein-binding domain (P). The position of the PHLPP1 UBS (PUBS; amino acids 1027 to 1126) is indicated by a black box. Amino acid boundaries are indicated on the left. Results from coimmunoprecipitation experiments are summarized on the right (+, binding; −, no binding). (B) Fine mapping of PUBS. C33A cells were cotransfected with an expression vector for 3F-UAF1 together with a plasmid encoding one of the indicated PHLPP1 fragments fused to YFP. YFP-PHLPP1 proteins were immunoprecipitated with anti-GFP antibodies and the presence of 3F-UAF1 in the immunoprecipitates (IP) and input cell extracts (IN) tested by Western blotting using an anti-Flag antibody. (C) Coimmunoprecipitation of endogenous UAF1 and USP12/46 with PUBS. C33A cells were transfected with a plasmid encoding PUBS-YFP or YFP alone (−) as a negative control. YFP proteins were immunoprecipitated as described above, and the immunoprecipitates were probed for the presence of UAF1 and USP12/46 by Western blotting using antibodies against these proteins. (D) Inhibitory effect of PUBS-YFP on the interaction of 3F-E1 with endogenous UAF1 and USP12/46. C33A cells were transfected with 4 μg of 3F-E1 expression vector and increasing amounts of PUBS-YFP expression plasmid (1, 2, and 4 μg). 3F-E1 was immunoprecipitated with an anti-Flag antibody and the presence of coprecipitated UAF1 and USP12/46 analyzed by Western blotting using antibodies against these proteins.
A prediction from the findings presented above is that overproduction of PUBS should prevent the interaction of E1 with UAF1. This was tested by coimmunoprecipitation experiments similar to those presented in Fig. 1. The results presented in Fig. 2D showed that overexpression of PUBS-YFP could, indeed, inhibit the interaction of 3F-E1 with endogenous UAF1, in a dose-dependent manner. As expected, this inhibition was accompanied by a reduction in the levels of USP46 coprecipitated with E1. Thus, conversely to the E1 UBS, which competes with PHLPP1 for binding to UAF1, PUBS competes with E1 for binding to UAF1. These results confirm that the mutually exclusive binding of PHLPP1 and E1 to UAF1 is mediated by their respective UBS.
Overexpression of PUBS inhibits HPV DNA replication.
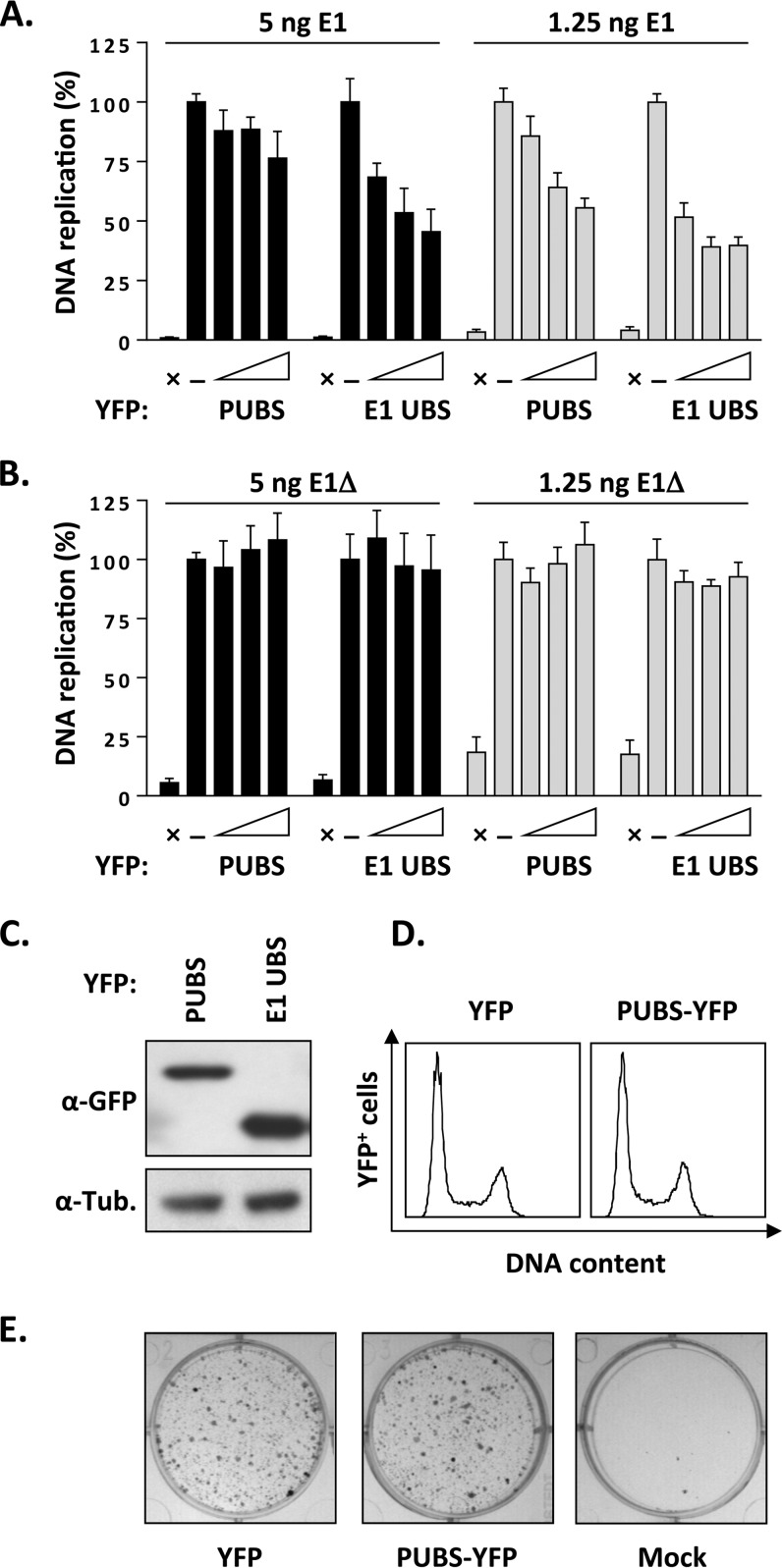
We previously reported that overexpression of the E1 UBS, fused to YFP, inhibits HPV DNA replication by competing with full-length E1 for binding to UAF1 (6). This prompted us to test if PUBS could also inhibit viral DNA replication, as measured using our luciferase-based assay (25, 27). Results presented in Fig. 3A show that PUBS-YFP could reduce HPV DNA replication only weakly, by approximately 20%. This is in contrast to the E1 UBS-YFP protein which inhibited DNA replication by about 50% at the largest amount of expression vector tested (Fig. 3A). We reasoned that the effect of PUBS-YFP might become more apparent at lower concentrations of wild-type E1, if indeed PUBS-YFP competes with full-length E1 for binding to UAF1. To test this possibility, we determined the effect of PUBS in DNA replication assays performed with 4-fold less E1 expression vector (1.25 ng instead of 5 ng as normally used). Under these conditions, the inhibitory activity of PUBS became more obvious (Fig. 3A). As expected, the effect of the E1 UBS was stronger than that of PUBS in these assays. Thus, PUBS-YFP can inhibit HPV DNA replication, although less efficiently than the E1 UBS-YFP. The weaker effect of PUBS might be due, at least in part, to the fact that it is expressed at approximately 3-fold lower levels than the E1 UBS, as determined from the Western blot presented in Fig. 3C.
FIG 3.
Overexpression of PUBS inhibits HPV DNA replication. (A) HPV DNA replication levels in C33A cells transfected with increasing amounts of PUBS-YFP or E1 UBS-YFP expression vector (25, 50, and 75 ng). The empty YFP vector (75 ng) was used as a negative control (−). Assays were performed with two different amounts of E1 expression vector (5 ng and 1.25 ng), as indicated, and without E1 as a baseline (i.e., no DNA replication) control (×). DNA replication activity is reported as a percentage of the signal obtained with the YFP-negative control. Each value represents the average from two independent experiments, each performed in triplicates, with the standard deviation indicated by an error bar. (B) Same as panel A but using the E1Δ protein instead of wild-type E1. (C) Western blot showing the relative expression of PUBS-YFP and E1 UBS-YFP in transfected C33A cells. Proteins were detected with anti-GFP antibodies. (D) Cell cycle analysis. C33A cells were transfected with a plasmid encoding PUBS-YFP or YFP alone as a control, and their DNA was stained with Hoechst 48 h posttransfection. The cell cycle distribution of the YFP-positive cell population was analyzed by flow cytometry. (E) Colony formation assay. C33A cells were either mock transfected or transfected with an expression vector for PUBS-YFP or E1 UBS-YFP and selected for approximately 3 weeks in bleomycin-containing medium. Drug-resistant colonies were fixed in methanol and stained with methylene blue.
To provide additional evidence that the inhibitory action of PUBS involves a competition with full-length E1 for binding to UAF1, we repeated the experiments presented above using a truncated E1 protein that lacks the first 40 amino acids comprising the UBS (E1Δ protein). We previously reported that the DNA replication activity of the E1Δ protein is approximately 25% that of wild-type E1 and, importantly, that this residual activity is independent of UAF1 and associated USPs. As expected, overexpression of PUBS did not affect the levels of DNA replication catalyzed by the E1Δ protein, similarly to what was observed with the E1 UBS (Fig. 3B). These results provide further support that PUBS inhibits wild-type HPV DNA replication by targeting UAF1. They also suggest, albeit indirectly, that overexpression of PUBS does not significantly alter cell cycle progression, as any delay outside S phase would have resulted in an inhibition of viral DNA replication. To further validate this point, we confirmed that the cell cycle distribution of C33A cells transiently overexpressing PUBS-YFP was similar to that of cells expressing YFP alone (Fig. 3D). We further verified that PUBS-YFP had no effect on the proliferation of C33A cells in a colony formation assay (Fig. 3E). These control experiments indicated that overexpression of PUBS does not affect cellular proliferation, cell cycle progression, or host DNA synthesis. Collectively, the results presented in this section indicate that PUBS inhibits HPV DNA replication by specifically targeting UAF1.
Characterization of a PUBS-E1Δ chimeric helicase.
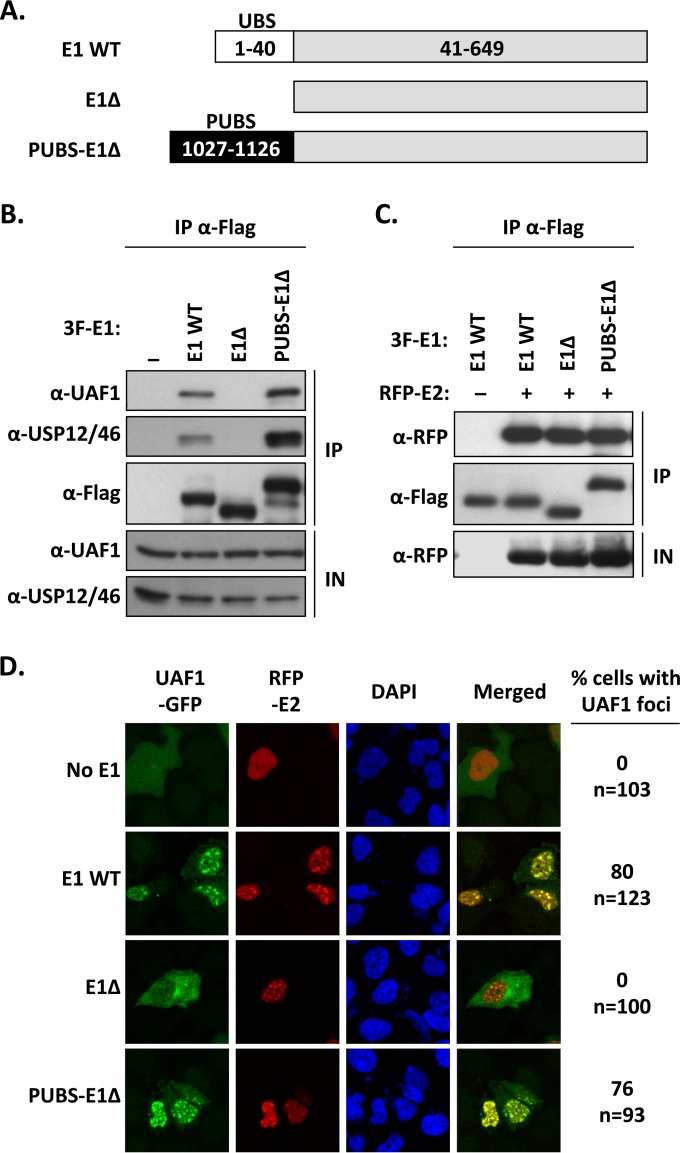
The results presented above suggest that PUBS engages in interactions with UAF1 and its associated USPs similar to interactions with the E1 UBS. This prompted us to test if PUBS could functionally substitute for the E1 UBS in viral DNA replication assays. To do so, we constructed a chimeric helicase in which PUBS was fused to the truncated E1 fragment lacking the N-terminal 40-aa UBS (E1Δ protein, diagrammed in Fig. 4A). The resulting PUBS-E1 chimeric protein was then characterized, first for its ability to interact with UAF1 in coimmunoprecipitation experiments. To do so, the PUBS-E1Δ protein, WT E1, and the E1Δ protein, all tagged with a triple-Flag epitope, were transiently expressed in C33A cells and immunoprecipitated using an anti-Flag antibody. The immunoprecipitates were then probed for the presence of UAF1 and USP46. The results presented in Fig. 4B confirmed that WT E1 interacts with endogenous UAF1 and USP46 but that the truncated E1Δ protein is unable to do so. More importantly, they indicated that the PUBS-E1Δ protein was capable of interacting with both UAF1 and USP46, as anticipated. Similar coimmunoprecipitation experiments were conducted to show that the PUBS-E1Δ protein was as competent as WT E1 and the E1Δ protein at forming a complex with RFP-tagged E2 (RFP-E2) (Fig. 4C). These results confirmed that the fusion of PUBS with the E1Δ protein did not grossly alter the structure or function of either protein fragment.
FIG 4.
The chimeric PUBS-E1Δ helicase interacts with UAF1 and HPV E2 in nuclear foci. (A) Schematic representation of the different E1 proteins. The wild-type protein (WT E1) is represented by a gray bar with its N-terminal UBS indicated by a white box. The E1Δ protein lacks the first 40 amino acids encompassing the E1 UBS. In the PUBS-E1Δ chimeric helicase, the E1 UBS has been replaced by the analogous domain from PHLPP1 (PUBS, black box). Amino acid boundaries of each protein fragment are indicated. All three E1 proteins are tagged with a 3F epitope at their N terminus (not shown). (B) Coimmunoprecipitation of endogenous UAF1 and USP46 with the PUBS-E1Δ protein. C33A cells were transfected with an expression plasmid for 3F-tagged PUBS-E1Δ protein or with 3F-WT E1 as a positive control or 3F-E1Δ protein as a negative control. E1 proteins were immunoprecipitated with an anti-Flag antibody, and the presence of UAF1 and USP12/46 in the immunoprecipitates (IP) and input cell extracts (IN) was analyzed by Western blotting with antibodies against these two proteins. (C) Coimmunoprecipitation of HPV E2. Coimmunoprecipitation experiments were performed as described above but using C33A cells cotransfected with an RFP-E2 expression vector. The ability of the indicated E1 proteins to interact with E2 was determined by probing the immunoprecipitates with an anti-RFP antibody. (D) Fluorescence confocal microscopy showing the intracellular localization of UAF1-GFP and RFP-E2 in cells expressing one of the indicated E1 proteins. Nuclei were stained with DAPI. For each condition, the percentage of cells in which UAF1-GFP is present within E2-containing nuclear foci is indicated on the left. The total number of transfected cells analyzed (n=) is specified underneath the percentage.
When expressed in transfected cells, E1 and E2 accumulate in nuclear foci that are thought to be the precursors of viral DNA replication centers (33). We previously demonstrated that E1, through its UBS, can relocalize UAF1 from the cytoplasm to these foci (8). Thus, as an additional measure of the functionality of the PUBS-E1Δ chimera, we investigated if it could relocalize UAF1-GFP to nuclear foci containing RFP-E2, using fluorescence confocal microscopy. These experiments revealed that the PUBS-E1Δ protein was almost as efficient as WT E1 at recruiting UAF1 to E2-containing foci (Fig. 4D). The E1Δ protein, which was used as a control, was unable to do so. All together, these results provided additional evidence that the PUBS-E1Δ chimeric helicase has the ability to interact with UAF1 and E2, a prerequisite for testing its activity in viral DNA replication assays.
Fusion of PUBS to the E1Δ protein enhances its DNA replication activity.
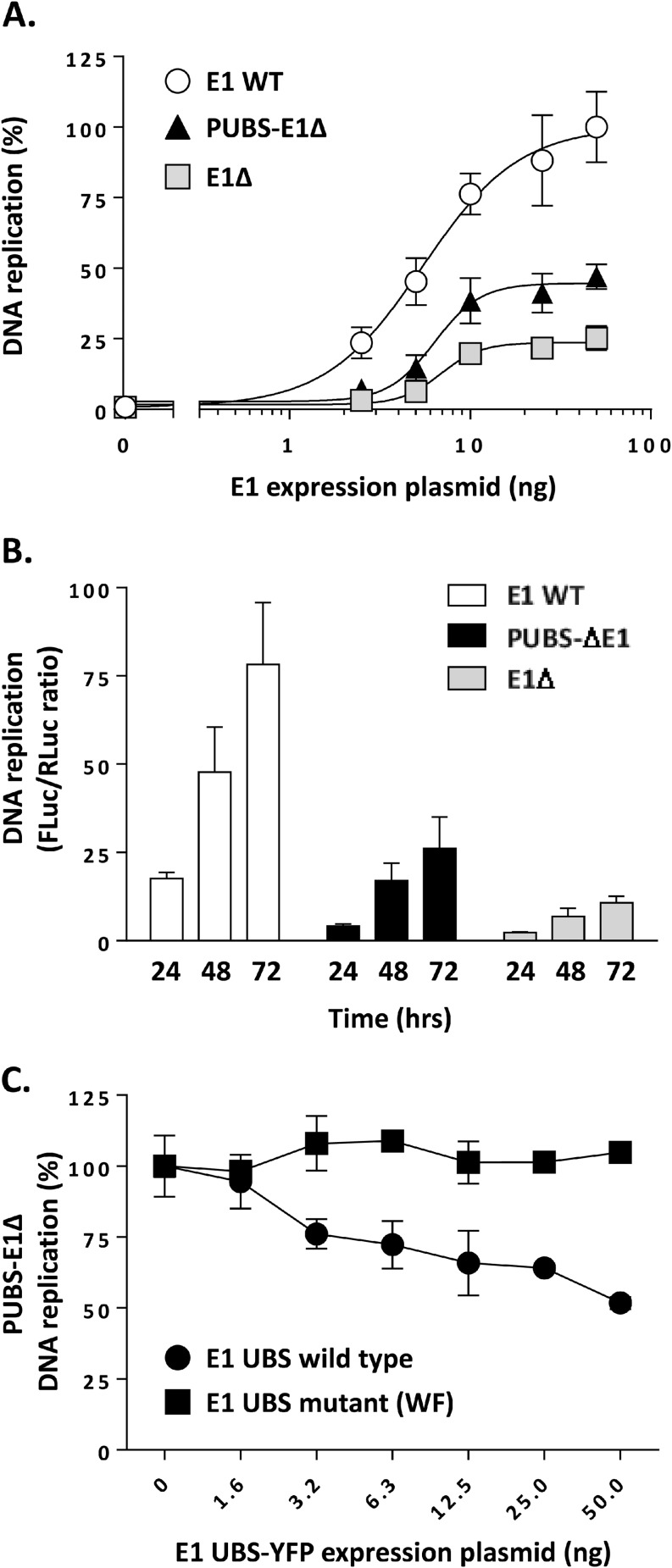
The DNA replication activity of the PUBS-E1Δ protein was then compared to that of WT E1 and of the E1Δ protein, using increasing amounts of E1 expression vector in our luciferase-based HPV31 DNA replication assay. As we observed previously, the levels of replication obtained with the E1Δ protein increased in a dose-dependent manner and reached a plateau at approximately 25% of those achieved with wild-type E1 (Fig. 5A). The replication levels measured for the PUBS-E1Δ protein were intermediary, reaching about 45% those of wild-type E1. The quality of each dose-response curve was confirmed by the goodness-of-fit value obtained by fitting the data to a sigmoid: R2 = 0.94 for WT E1, R2 = 0.92 for PUBS-E1Δ protein, and R2 = 0.92 for E1Δ protein. Thus, the fusion of PUBS to the E1Δ protein enhances the replication activity of the protein, although not up to the levels measured with wild-type E1. This stimulatory effect of PUBS was observed at different E1 concentrations (Fig. 5A) and irrespective of the length of the assay (24, 48, 72 h; Fig. 5B). Next, we tested the activity of E1, E1Δ protein, and PUBS-E1Δ protein under conditions of DNA replication stress to investigate if PUBS and/or the E1 UBS modulates the capacity of E1 to deal with such an impediment. To do so, we titrated the known DNA replication inhibitors aphidicolin, ara-C, and hydroxyurea into the luciferase assay. Mimosine, which inhibits HPV DNA replication indirectly by blocking cell cycle progression at the G1-S boundary, was used as a control inhibitor, as its potency should be unaffected by the E1 protein used in the assay. As shown in Table 1, the potency of each inhibitor (EC50) was not markedly changed by the type of E1 used in the DNA replication assay, indicating that neither PUBS nor the E1 UBS has a significant influence on the ability of E1 to cope with DNA replication stress. Finally, we began to investigate if the stimulatory effect of PUBS was indeed due to its ability to interact with UAF1-USP complexes. As a first experiment, we tested if the DNA replication levels measured with the PUBS-E1Δ protein could be reduced by competition with the E1 UBS peptide (E1 UBS-YFP). Results, presented in Fig. 5C, showed that the E1 UBS reduced PUBS-E1Δ DNA replication by approximately 50% at the largest amount tested, in contrast to the UAF1-binding-defective UBS (WF mutant) (8), which was used as a negative control. Therefore, these results demonstrate that PUBS can partially replace the E1 UBS in enhancing HPV DNA replication and suggest that this effect is mediated through the interaction of PUBS with UAF1.
FIG 5.
PUBS stimulates the DNA replication activity of the E1Δ protein. (A) DNA replication activities of the indicated E1 proteins measured in cells transfected with increasing amounts of E1 expression vector (2.5, 5, 20, 25, and 50 ng). DNA replication activity is reported as a percentage of the maximal signal obtained with WT E1, which was set at 100%. Each value represents the average from two independent experiments, each performed in triplicates, with the standard deviation indicated by an error bar. (B) DNA replication activities of the indicated E1 proteins measured at different times posttransfection (24, 48, and 72 h). (C) Overexpression of the E1 UBS inhibits DNA replication supported by the PUBS-E1Δ protein. Levels of DNA replication, catalyzed by the PUBS-E1Δ protein, in C33A cells transfected with increasing amounts of E1 UBS-YFP expression vector (1.6 to 50 ng in 2-fold increments). A mutant UBS defective for UAF1 binding (WF mutant) was used as a specificity control. DNA replication activity is reported as a percentage of the signal obtained with YFP alone as a negative control. Each value is the average from two independent experiments, each performed in triplicates, with the standard deviation indicated by an error bar.
TABLE 1.
EC50 of pharmacological inhibitors in HPV31 DNA replication assays supported by different E1 proteins
| Inhibitor | EC50 |
||
|---|---|---|---|
| WT E1 | E1Δ protein | PUBS-E1Δ protein | |
| Aphidicolin | 760 ± 86 nM | 1,368 ± 380 nM | 1,105 ± 23 nM |
| Ara-C | 380 ± 81 nM | 558 ± 165 nM | 651 ± 18 nM |
| Hydroxyurea | 312 ± 33 μM | 466 ± 106 μM | 302 ± 17 μM |
| Mimosine | 193 ± 5 μM | 144 ± 19 μM | 177 ± 5 μM |
Conserved sequences in PUBS are needed for interaction with UAF1 and to stimulate HPV DNA replication.
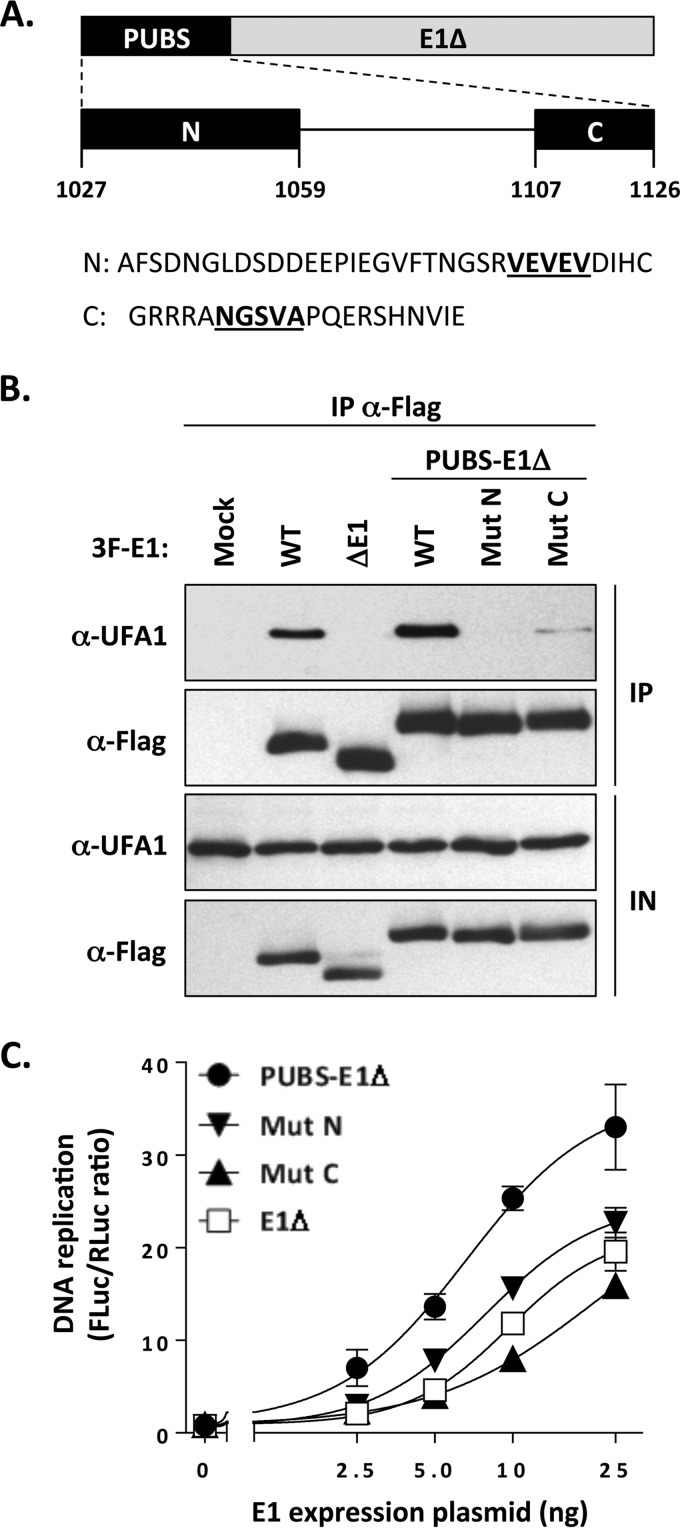
To gain additional evidence that the stimulatory effect of PUBS described above was due to the recruitment of UAF1-USP complexes, we set out to create mutations in the PUBS-E1Δ protein that disrupt its interaction with UAF1. Alignment of the amino acid sequences of PUBS from PHLPP1 and PHLPP2 of diverse species revealed that the N and C termini of PUBS are the most highly conserved (data not shown). Based on this analysis, we created one mutation within the N-terminal region that deletes a conserved VEVEV motif (MutN; Fig. 6A). We focused on this motif because of its similarity to a portion of the E1 UBS, as discussed below and shown in Fig. 9B. We also created a mutation in the C-terminal region of PUBS that deletes a conserved NGSVA motif that has the potential to be part of a β-turn and hence may be important for the structure and/or function of PUBS (MutC; Fig. 6A). As can be seen in Fig. 6B, both MutN and MutC reduced the interaction of the PUBS-E1Δ protein in coimmunoprecipitation experiments. Importantly, these two mutations also significantly reduced the ability of PUBS to stimulate HPV DNA replication, as the two mutant PUBS-E1Δ enzymes displayed activity similar to that of the E1Δ protein (Fig. 6C). All together, these results indicate that conserved regions in PUBS mediate its interaction with UAF1 and its ability to enhance HPV DNA replication.
FIG 6.
Conserved regions in PUBS mediate its interaction with UAF1 and its DNA replication stimulatory activity. (A) Schematic representation of the PUBS-E1Δ chimeric helicase with the PUBS region enlarged underneath. The highly conserved N- and C-terminal regions of PUBS are diagrammed as black boxes labeled N and C, respectively. Amino acid boundaries are indicated. Also shown is the amino acid sequence of the N and C regions in which the motifs that were deleted to create the MutN and MutC mutations are underlined. (B) Coimmunoprecipitation of endogenous UAF1 with the WT and mutant PUBS-E1Δ proteins. C33A cells were transfected with the indicated 3F-E1 expression plasmids, and the encoded protein was immunoprecipitated with an anti-Flag antibody. The presence of UAF1 in the immunoprecipitates (IP) and input cell extracts (IN) was determined by Western blotting with an anti-UAF1 antibody. Wild-type E1 and the E1Δ protein were used as a positive and negative control, respectively. (C) DNA replication activities of wild-type and mutant PUBS-E1Δ proteins. DNA replication activities, presented as RLuc/FLuc ratios, were measured in cells transfected with increasing amounts of the indicated E1 expression vector (2.5, 5, 10, and 25 ng). The E1Δ protein was used as a negative control. Each value represents the average from two independent experiments, each performed in triplicates, with the standard deviation indicated by an error bar.
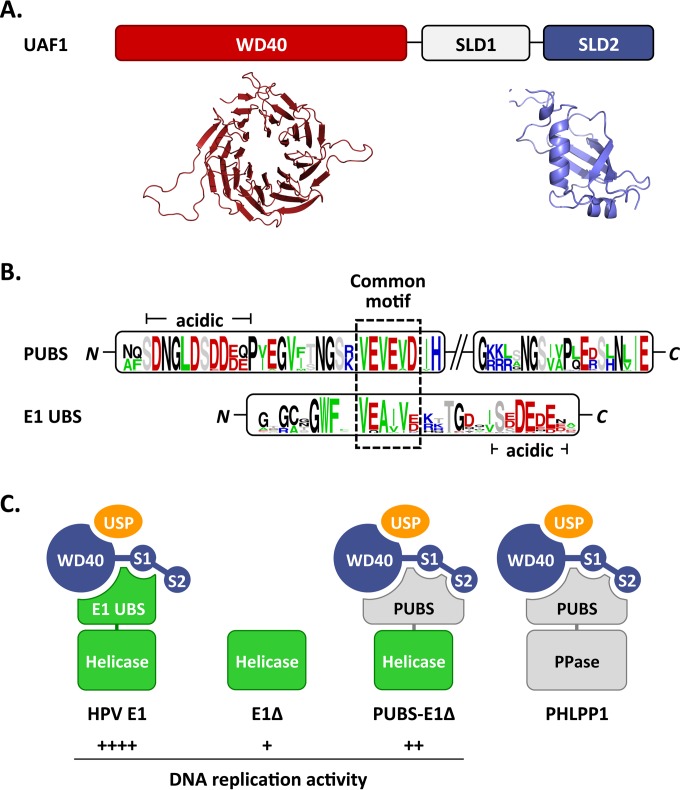
FIG 9.
Proposed interactions between HPV E1, PHLPP1, and UAF1-containing deubiquitinating enzymes. (A) Schematic representation of UAF1 showing the WD repeat region and two SUMO-like domains (SLD1 and SLD2). A structural model of the WD repeat region of human UAF1 obtained from the WDSP database (40) is presented underneath (colored in red), together with the model of SLD2 generated in this study (colored in blue). (B) Sequence logos of the PHLPP1 UBS (PUBS) and E1 UBS. Only the conserved N- and C-terminal regions of PUBS are shown. A sequence motif conserved between the N-terminal region of PUBS and the E1 UBS is boxed. A stretch of negatively charged amino acids found in both UBSs is also indicated (acidic). (C) Cartoon representation of the proposed complexes formed by E1 and PHLPP1 with UAF1-containing deubiquitinating enzymes. UAF1 is represented as a tripartite protein (blue) comprised of a WD40 repeat region and of two SUMO-like domains, SLD1 (S1) and SLD2 (S2). USP1, USP12, and USP46, which form mutually exclusive complexes with the WD repeat region of UAF1, are represented by a yellow oval. HPV E1 is diagrammed as a bipartite protein (green) comprised of a helicase domain linked to the E1 UBS, which contacts UAF1 independently of SLD2. This UBS is missing in the E1Δ protein and replaced by the PHLPP1 UBS in the PUBS-E1Δ chimeric enzyme. PHLPP1 is also shown as a bipartite protein (gray) made of a phosphatase domain (PPase) attached to PUBS, which requires all three subdomains of UAF1 for interaction. The relative levels of HPV DNA replication supported by the proposed E1-containing complexes are indicated by plus signs.
The stimulatory effect of PUBS on DNA replication is dependent on USP activity.
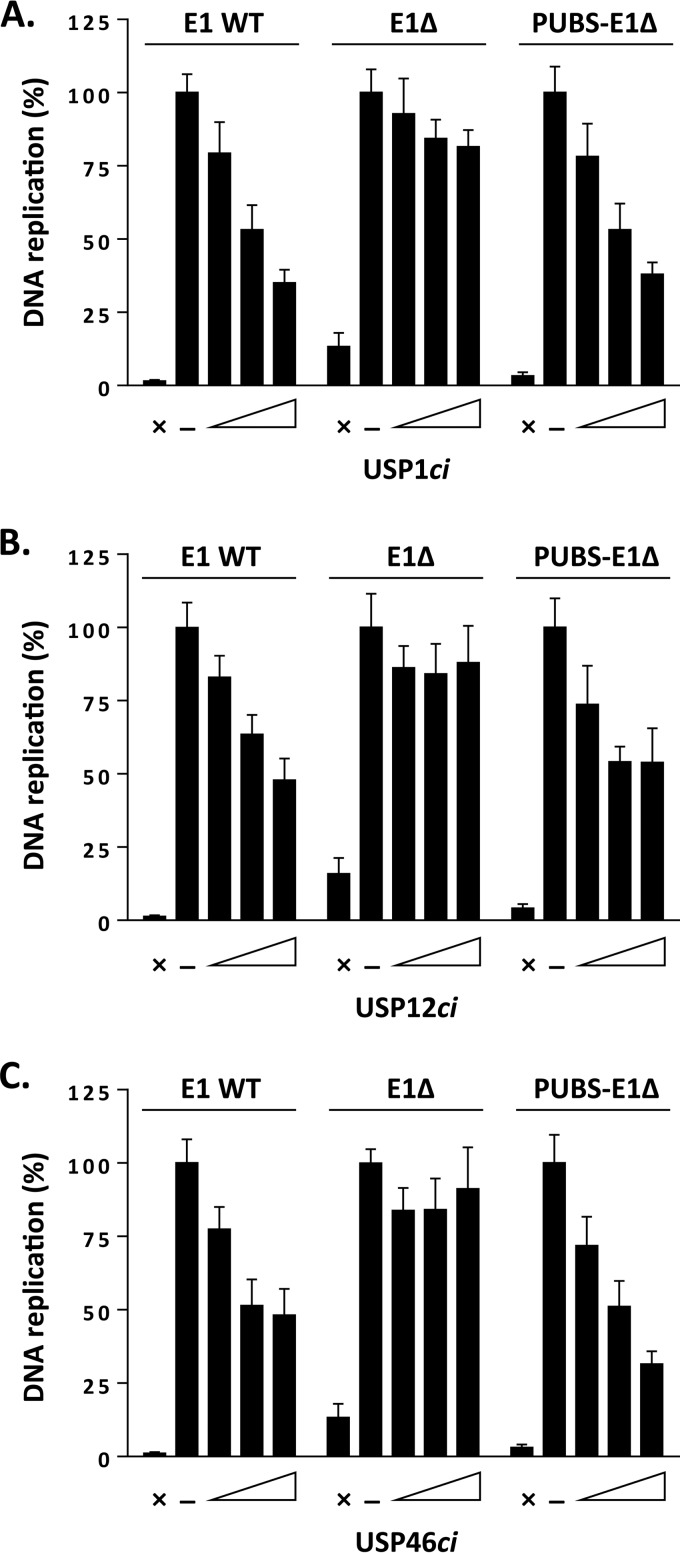
To test if the stimulatory effect of PUBS described above was dependent on the enzymatic activity of UAF1-containing deubiquitinase complexes, we tested if it could be abrogated by overexpression of catalytically inactive USP1 (USP1ci), USP12ci, and USP46ci. We previously reported that these USPci enzymes inhibit the ability of wild-type E1 to support viral DNA replication by 40 to 70%, while having much less of an effect on the E1Δ protein (13). The results presented in Fig. 7A to C and quantified in Table 2 confirmed these findings. Importantly, they also revealed that the activity of the PUBS-E1Δ chimera was sensitive to dominant negative inhibition by USP1ci, USP12ci, and USP46ci or by a triple combination of these inactive enzymes. From these results, we conclude that the stimulatory effect of PUBS is dependent on the enzymatic activity of USP1, USP12, and/or USP46.
FIG 7.
The DNA replication stimulatory effect of PUBS is antagonized by catalytically inactive USPs. (A to C) Levels of HPV DNA replication in cells expressing the indicated E1 proteins and increasing amounts of RFP-tagged USP1ci (A), USP12ci (B), or USP46ci (C). Cells cotransfected with the empty RFP vector were used as a negative control (−). DNA replication activity is reported as a percentage of the signal obtained with the negative control, which was set at 100%. Assays performed in the absence of E1 were used as baseline (i.e., no DNA replication) controls (×). Each value represents the average from three independent experiments, each performed in triplicates, with the standard deviation indicated by an error bar.
TABLE 2.
EC50 of catalytically inactive USPs in HPV31 DNA replication assays supported by different E1 proteinsa
| USP1ci | WT E1 |
E1Δ protein |
PUBS-E1Δ protein |
|||
|---|---|---|---|---|---|---|
| EC50 (ng) | Imax (%) | EC50 (ng) | Imax (%) | EC50 (ng) | Imax (%) | |
| 1 | 11 | 87 | 32 | 19 | 11 | 73 |
| 12 | 34 | 72 | NA | NA | 27 | 40 |
| 46 | 38 | 70 | NA | NA | 75 | 50 |
| 1 + 12 + 46 | 20 | 75 | NA | NA | 15 | 58 |
EC50, amount of USPci plasmid needed to inhibit HPV DNA replication by 50%; Imax, maximal level of inhibition achievable with high amounts of USPci; NA, not applicable.
PUBS-E1 requires both the WD repeat region and C-terminal SUMO-like domain of UAF1 for interaction.
Although the studies presented so far indicate that PUBS is functionally similar to the E1 UBS, it shows very limited sequence similarity to the E1 UBS and is larger in size (100 compared to 40 aa). This prompted us to investigate if PHLPP1 interacts with UAF1 in a different manner than E1. As mentioned in the introduction, UAF1 contains eight WD repeats at its N terminus and two SUMO-like domains (SLD1 and SLD2) at its C terminus (Fig. 8B) (14–16). We previously reported that the interaction of E1 with UAF1 requires the first 573 amino acids of the protein, which encompass both the WD repeat region and SLD1 (8). To determine if these two domains are also sufficient for interaction with PHLPP1, we performed coimmunoprecipitation experiments using HA-tagged PHLPP1 and the same set of truncated 3F-UAF1 proteins that we previously used to map the E1-binding region (8). Interestingly, we found that the interaction of UAF1 with PHLPP1 requires not only the WD repeat region and SLD1, as E1 does, but also SLD2 (Fig. 8A). Similar results were obtained for PHLPP2 (data not shown).
FIG 8.
The WD repeat region and C-terminal SUMO-like domain of UAF1 are required for interaction with PHLPP1 and PUBS-E1Δ proteins. (A) Mapping of the UAF1 domain required for interaction with PHLPP1, by coimmunoprecipitation. C33A cells were cotransfected with an expression vector for HA-tagged PHLPP1 together with a plasmid encoding 3F-UAF1 or the indicated truncated derivative. The amino acid boundaries of each UAF1 fragment are indicated. UAF1 proteins were immunoprecipitated with an anti-Flag antibody and the presence of HA-PHLPP1 in each immunoprecipitate (IP) and input cell extract (IN) tested by Western blotting with an anti-HA antibody. Only the full-length UAF1 protein could interact with PHLPP1 in this assay. (B) Schematic representation of UAF1 showing the location of the WD40 repeat region and of the two SUMO-like domains (SLD1 and SLD2). The regions of UAF1 needed for interaction with PHLPP1 (aa 1 to 677) and with E1 (aa 1 to 573) (8) are summarized above. (C) Homology model of SLD2 from human UAF1 based on the RAWUL domains of PCGF1 (34), RING1B (35), and related proteins. The structures of PCGF1 (PDB 4HPM) and RING1B (PDB 3GS2) are shown for comparison. The β-strand and α-helix that make up part of the interaction surface on PCGF1 and other SUMO-like domains are colored in red, and their sequences are aligned below. (D) Amino acid sequence alignment of the β-strand and α-helix of SUMO1 to SUMO3, PCGF1, and RING1B with UAF1 SLD2. RLS MUT refers to the mutation introduced in the β-strand of UAF1 that changes the RLS sequence to three alanines. (E) Coimmunoprecipitation of wild-type and RLS-mutated UAF1 with either wild-type E1 (WT), PUBS-E1Δ, or E1Δ protein. C33A cells were cotransfected with an expression vector for the indicated 3F-E1 protein together with a plasmid encoding either wild-type or RLS mutant GFP-UAF1. Following the immunoprecipitation of E1 with an anti-Flag antibody, the presence of GFP-UAF1 in each immunoprecipitate (IP) and input cell extract (IP) was tested by Western blotting with anti-GFP antibodies.
The aforementioned finding raised the possibility that the interaction of the PUBS-E1 chimeric helicase with UAF1-USP complexes also requires SLD2. To test this possibility, we wished to create a mutation in SLD2 that would affect its interaction with target proteins. To do so, we generated a homology model of SLD2 using the Phyre2 server (30), which predicted a high degree of structural similarity between SLD2 and the RAWUL domain of PCGF1 (34) and RING1B (35). Although the sequence similarity between SLD2 and these two proteins was noted previously (14), their structures were not available at the time for homology modeling. The ubiquitin fold of PCGF1 and RING1B is characterized by a specific arrangement of secondary structure elements, including the presence of an α-helix and of a β-sheet that becomes extended by one or two additional β-strands from an interacting protein partner (34, 35). The α-helix and β-strand are colored in red in the structures of PCGF1, RING1B, and the UAF1 SLD2 model shown in Fig. 8C, and their sequences are aligned with the corresponding regions of SUMO1 to SUMO3 in Fig. 8D. Based on this model of SLD2, we chose to mutate the 3 amino acids RLS in the β-strand by changing them to three alanines (Fig. 8D). When tested in coimmunoprecipitation experiments, the UAF1 RLS mutant protein showed reduced binding to the PUBS-E1Δ protein but not to wild-type E1 (Fig. 8E). As anticipated, no interaction was detected between UAF1 and the E1Δ protein, which was used as a negative control. These results confirmed that the SLD2 domain of UAF1 is needed for optimal interaction with the PUBS-E1Δ protein but not with wild-type E1 and suggest that PUBS and the E1 UBS interact with UAF1 in slightly different manners.
DISCUSSION
In this study, we have shown that the cellular phosphatase PHLPP1 contains a UAF1-binding site (UBS) that is functionally similar to the one present at the N terminus of E1. These two UBSs are both sufficient for complex formation with UAF1 and its associated deubiquitinating enzymes USP1, USP12, and USP46 (Fig. 2C and data not shown). As a result, both UBSs can inhibit HPV DNA replication when overexpressed in trans as competitive inhibitors (Fig. 3A and B). The similarities between the PHLPP1 and E1 UBS also extend to the fact that both are sufficient to compete the interaction of UAF1 with full-length E1 (Fig. 2D) (8), a finding which likely explains the mutually exclusive binding of PHLPP1 and E1 to UAF1 (Fig. 1). These results raise the possibility that both UBSs interact with a common surface, or with overlapping regions, on UAF1 (Fig. 9A), although we cannot rule out an allosteric mechanism of competition at the moment. In support of the notion that these UBSs interact with a common surface on UAF1, we note that the N-terminal portion of PUBS resembles the E1 UBS in its amino acid composition and sequence and, in particular, by the presence of a similar sequence motif flanked by a region rich in acid residues (Fig. 9B). Satisfyingly, deletion of this motif (ΔVEVEV; MutN in Fig. 6) in the context of the PUBS-E1 helicase reduced its interaction with UAF1, similarly to what we observed previously with mutations in the VEA(I/V)V motif of HPV11 and HPV31 E1 (7, 8). Thus, it is tempting to speculate that this common motif in E1 and PHLPP1 is a critical determinant of their interaction with UAF1-USP complexes. PUBS, however, clearly requires additional sequences for interaction with UAF1, as shown by the fact that deletion of 10 residues at its C terminus (Fig. 2B), or deletion of the NGSVA sequence (MutC in Fig. 6), reduced binding to UAF1. These sequences could be needed for the proper three-dimensional structure of PUBS and/or to make additional contacts with UAF1. In support of the latter possibility, we found that PHLPP1 and the PUBS-E1Δ chimeric helicase require SLD2 for interaction with UAF1 (Fig. 8), in contrast to E1, which relies on the WD repeat region and SLD1 only (8). To ascertain this difference, we created a homology model of SLD2 (Fig. 8C) and mutated a conserved β-strand that forms part of the protein interaction surface in PCGF1, RING1B, and SUMO1 to SUMO3 (Fig. 8D). This mutation (RLS changed to AAA) reduced the interaction of UAF1 with the PUBS-E1Δ protein but not with wild-type E1 in coimmunoprecipitation assays, thus confirming that SLD2 is specifically needed for interaction with PUBS (Fig. 8D). A previous study of FANCI and ELG1 revealed that both proteins interact with UAF1 via its SLD2 domain (16). FANCI and ELG1 bind to FANCD2 and PCNA, respectively, and mediate their deubiquitination by UAF1-USP complexes (10, 16, 36). Interestingly, mutagenesis of putative SUMO interaction motifs (SIMs) in FANCI and ELG1 abolished their interaction with UAF1, suggesting that UAF1-associated deubiquitinating enzymes are brought to their substrates through SIM-SLD2 interactions (16). This prompted us to analyze the amino acid sequence of PUBS for the presence of putative SIMs. Of the two motifs identified using the GPS-SUMO server (37), one corresponds to the VEVEV motif described above and needed for interaction with UAF1. The second putative SIM, IVISA, is located between the two conserved regions of PUBS, but mutagenesis of this motif did not significantly affect UAF1 binding (not shown). Thus, it remains a possibility that SLD2 interacts with the SIM-like VEVEV motif in PUBS, although we feel this is unlikely given that SLD2 is dispensable for interaction with the analogous VEA(I/V)V motif in E1. Clearly, detailed structural and mutagenesis studies will be needed to pinpoint the exact contacts that the PHLPP1 and E1 UBS make with UAF1 and the role of the WD repeat β-propeller region and of the two SUMO-like domains in these interactions. Although our results indicate that PHLPP1 and E1 compete with each other for binding to UAF1, we emphasize that this finding was derived from experiments in which one of the two proteins was overexpressed in order to make the amount of UAF1 limiting for interaction. It is currently unknown if E1 accumulates to sufficiently high levels in HPV-infected cells to overcome the binding capacity of UAF1 and sequester UAF1-containing deubiquitinase complexes away from PHLPP1 and/or other UAF1-binding proteins. The low levels of E1 in undifferentiated cells make this scenario unlikely. E1 expression is expected to be much higher in differentiated cells undergoing viral DNA amplification than in undifferentiated cells. However, as the E1 UBS is likely removed from the rest of the protein by caspase-3/7 cleavage to promote genome amplification (38), it is unclear if the free UBS has any function in these differentiated cells. Given these uncertainties, we have refrained from suggesting that E1 reduces the interaction of PHLPP1 with UAF1 under physiological conditions and chose to limit our interpretation of their mutually exclusive binding as an indication that both proteins contact UAF1 in a similar manner.
A major impetus for characterizing the PHLPP1 UBS was to test if it could replace the E1 UBS in stimulating HPV DNA replication. Characterization of a PHLPP1-E1 chimeric helicase (PUBS-E1Δ) showed that PUBS could substitute for the E1 UBS in recruiting UAF1 and its associated USPs, while having little to no effect on E2 binding (Fig. 4B). Accordingly, the PUBS-E1Δ protein induced a relocalization of UAF1 into E1-E2-containing nuclear foci, similarly to wild-type E1 (Fig. 4C). Furthermore, the activity of the PUBS-E1Δ enzyme in HPV DNA replication assays was approximately twice that of the E1Δ protein, but not as high as that of wild-type E1 (Fig. 5). The fact that PUBS could partially substitute for the E1 UBS in stimulating the replication activity of the E1Δ protein provided further evidence that both domains are functionally related and strengthened our hypothesis that the main role of the E1 N-terminal region is to function as a UAF1-USP-recruiting module (summarized in Fig. 9C). This is further supported by the observation that the stimulatory effect of PUBS could be abrogated by overexpression of catalytically inactive USP, thus confirming that a deubiquitinase activity is necessary for increased HPV DNA replication (Fig. 6). As encouraging as these results may be, it is also noteworthy that the fusion of PUBS to the E1Δ protein did not stimulate HPV DNA replication up to wild-type levels, despite the fact that the chimeric helicase interacts efficiently with UAF1-USP complexes. An obvious explanation for this mitigated rescue might be the need of the PUBS-E1Δ protein to engage SLD2 for interaction with UAF1, in contrast to wild-type E1. In addition, we do not exclude the possibility that the N-terminal 40 amino acids of E1 play a structural role and/or contribute to another, UAF1-independent function of the helicase, which would not be restored in the PUBS-E1Δ chimeric enzyme. Along this line, we note that the N-terminal region of E1, although generally more divergent than the rest of the protein, nevertheless contains a few residues that are highly conserved between HPV types, including types whose encoded E1 does not interact with UAF1.
It was previously reported that UAF1, in complex with USP12, can increase the stability of PHLPP1 by promoting its deubiquitination (18). Consistent with this report, we observed that the steady-state levels of PHLPP1 decreased slightly when its interaction with UAF1 was outcompeted by overexpression of E1 or of the E1 UBS (see input cell extracts in Fig. 1B and C). However, we did not observe any significant differences in the steady-state accumulation of the PUBS-E1Δ protein relative to that of the E1Δ protein or compared to mutant versions of this chimeric enzyme that are defective for interaction with UAF1 (MutN and MutC) (Fig. 6B). These results are similar to what we have observed with mutations in the E1 UBS, which affect the steady state of E1 by less than 2-fold (D. Gagnon and M. Archambault, unpublished data). We do not believe that these observations necessarily rule out a role for PUBS and the E1 UBS in controlling the stability of the PUBS-E1Δ protein and wild-type E1, respectively. Rather, we believe that these results simply reflect the fact that E1, the PUBS-E1Δ protein, and the E1Δ protein are stable enzymes when expressed in transfected C33A cells, perhaps because E1-expressing cells are blocked in S phase and undergo a DNA damage response (24, 39). We are currently investigating if E1 becomes destabilized under certain conditions, such as at the end of viral DNA replication, during a specific phase of the cell cycle, or in the absence of a DNA damage response. Thus, a definitive answer as to the role of the E1 UBS in modulating the ubiquitination state of E1 awaits the identification of conditions under which E1 is unstable.
In summary, the results presented in this study demonstrate that artificial recruitment of UAF1-USP complexes through the fusion of E1 with a heterologous UBS is sufficient to facilitate HPV DNA replication. As such, these findings provide further evidence that a main function of the E1 N-terminal region from anogenital HPV types is to recruit UAF1-associated USP1, USP12, or USP46, whose catalytic activity is required for optimal replication of the viral episome.
ACKNOWLEDGMENTS
We thank Fanny Bergeron-Labrecque, Julien Leconte, and Matthieu Rousseau for technical assistance. We also thank Mireille Cartier and all members of the Archambault laboratory for critical reading of the manuscript and Tom Melendy for helpful discussions.
M.L. was supported by a studentship from the Fonds de la Recherche en Santé du Québec (FRSQ) and a CIHR Frederick Banting and Charles Best doctoral scholarship award. This work was supported by a grant from the Canadian Institutes of Health Research (CIHR).
REFERENCES
- 1.Forman D, de Martel C, Lacey CJ, Soerjomataram I, Lortet-Tieulent J, Bruni L, Vignat J, Ferlay J, Bray F, Plummer M, Franceschi S. 2012. Global burden of human papillomavirus and related diseases. Vaccine 30(Suppl 5):F12–F23. doi: 10.1016/j.vaccine.2012.07.055. [DOI] [PubMed] [Google Scholar]
- 2.Kadaja M, Silla T, Ustav E, Ustav M. 2009. Papillomavirus DNA replication—from initiation to genomic instability. Virology 384:360–368. doi: 10.1016/j.virol.2008.11.032. [DOI] [PubMed] [Google Scholar]
- 3.D'Abramo CM, Fradet-Turcotte A, Archambault J. 2011. Human papillomavirus DNA replication: insights into the structure and regulation of a eukaryotic DNA replisome, p 217–239. In Gaston EK. (ed), Small DNA tumour viruses. Horizon Scientific Press, Poole, United Kingdom. [Google Scholar]
- 4.Bergvall M, Melendy T, Archambault J. 2013. The E1 proteins. Virology 445:35–56. doi: 10.1016/j.virol.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amin AA, Titolo S, Pelletier A, Fink D, Cordingley MG, Archambault J. 2000. Identification of domains of the HPV11 E1 protein required for DNA replication in vitro. Virology 272:137–150. doi: 10.1006/viro.2000.0328. [DOI] [PubMed] [Google Scholar]
- 6.Sun Y, Han H, McCance DJ. 1998. Active domains of human papillomavirus type 11 E1 protein for origin replication. J Gen Virol 79(Part 7):1651–1658. [DOI] [PubMed] [Google Scholar]
- 7.Cote-Martin A, Moody C, Fradet-Turcotte A, D'Abramo CM, Lehoux M, Joubert S, Poirier GG, Coulombe B, Laimins LA, Archambault J. 2008. Human papillomavirus E1 helicase interacts with the WD repeat protein p80 to promote maintenance of the viral genome in keratinocytes. J Virol 82:1271–1283. doi: 10.1128/JVI.01405-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lehoux M, Fradet-Turcotte A, Lussier-Price M, Omichinski JG, Archambault J. 2012. Inhibition of human papillomavirus DNA replication by an E1-derived p80/UAF1-binding peptide. J Virol 86:3486–3500. doi: 10.1128/JVI.07003-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohn MA, Kee Y, Haas W, Gygi SP, D'Andrea AD. 2009. UAF1 is a subunit of multiple deubiquitinating enzyme complexes. J Biol Chem 284:5343–5351. doi: 10.1074/jbc.M808430200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohn MA, Kowal P, Yang K, Haas W, Huang TT, Gygi SP, D'Andrea AD. 2007. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol Cell 28:786–797. doi: 10.1016/j.molcel.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 11.Kee Y, Yang K, Cohn MA, Haas W, Gygi SP, D'Andrea AD. 2010. WDR20 regulates activity of the USP12 x UAF1 deubiquitinating enzyme complex. J Biol Chem 285:11252–11257. doi: 10.1074/jbc.M109.095141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Villamil MA, Chen J, Liang Q, Zhuang Z. 2012. A noncanonical cysteine protease USP1 is activated through active site modulation by USP1-associated factor 1. Biochemistry 51:2829–2839. doi: 10.1021/bi3000512. [DOI] [PubMed] [Google Scholar]
- 13.Lehoux M, Gagnon D, Archambault J. 2014. E1-mediated recruitment of a UAF1-USP deubiquitinase complex facilitates human papillomavirus DNA replication. J Virol 88:8545–8555. doi: 10.1128/JVI.00379-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanchez-Pulido L, Devos D, Sung ZR, Calonje M. 2008. RAWUL: a new ubiquitin-like domain in PRC1 ring finger proteins that unveils putative plant and worm PRC1 orthologs. BMC Genomics 9:308. doi: 10.1186/1471-2164-9-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park J, Lee BS, Choi JK, Means RE, Choe J, Jung JU. 2002. Herpesviral protein targets a cellular WD repeat endosomal protein to downregulate T lymphocyte receptor expression. Immunity 17:221–233. doi: 10.1016/S1074-7613(02)00368-0. [DOI] [PubMed] [Google Scholar]
- 16.Yang K, Moldovan GL, Vinciguerra P, Murai J, Takeda S, D'Andrea AD. 2011. Regulation of the Fanconi anemia pathway by a SUMO-like delivery network. Genes Dev 25:1847–1858. doi: 10.1101/gad.17020911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sowa ME, Bennett EJ, Gygi SP, Harper JW. 2009. Defining the human deubiquitinating enzyme interaction landscape. Cell 138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gangula NR, Maddika S. 2013. WD repeat protein WDR48 in complex with deubiquitinase USP12 suppresses Akt-dependent cell survival signaling by stabilizing PH domain leucine-rich repeat protein phosphatase 1 (PHLPP1). J Biol Chem 288:34545–34554. doi: 10.1074/jbc.M113.503383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X, Stevens PD, Yang H, Gulhati P, Wang W, Evers BM, Gao T. 2013. The deubiquitination enzyme USP46 functions as a tumor suppressor by controlling PHLPP-dependent attenuation of Akt signaling in colon cancer. Oncogene 32:471–478. doi: 10.1038/onc.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McClurg UL, Summerscales EE, Harle VJ, Gaughan L, Robson CN. 2014. Deubiquitinating enzyme Usp12 regulates the interaction between the androgen receptor and the Akt pathway. Oncotarget 5:7081–7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhiqiang Z, Qinghui Y, Yongqiang Z, Jian Z, Xin Z, Haiying M, Yuepeng G. 2012. USP1 regulates AKT phosphorylation by modulating the stability of PHLPP1 in lung cancer cells. J Cancer Res Clin Oncol 138:1231–1238. doi: 10.1007/s00432-012-1193-3. [DOI] [PubMed] [Google Scholar]
- 22.Brognard J, Newton AC. 2008. PHLiPPing the switch on Akt and protein kinase C signaling. Trends Endocrinol Metab 19:223–230. doi: 10.1016/j.tem.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newton AC, Trotman LC. 2014. Turning off AKT: PHLPP as a drug target. Annu Rev Pharmacol Toxicol 54:537–558. doi: 10.1146/annurev-pharmtox-011112-140338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fradet-Turcotte A, Bergeron-Labrecque F, Moody CA, Lehoux M, Laimins LA, Archambault J. 2011. Nuclear accumulation of the papillomavirus E1 helicase blocks S-phase progression and triggers an ATM-dependent DNA damage response. J Virol 85:8996–9012. doi: 10.1128/JVI.00542-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fradet-Turcotte A, Morin G, Lehoux M, Bullock PA, Archambault J. 2010. Development of quantitative and high-throughput assays of polyomavirus and papillomavirus DNA replication. Virology 399:65–76. doi: 10.1016/j.virol.2009.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morin G, Fradet-Turcotte A, Di Lello P, Bergeron-Labrecque F, Omichinski JG, Archambault J. 2011. A conserved amphipathic helix in the N-terminal regulatory region of the papillomavirus E1 helicase is required for efficient viral DNA replication. J Virol 85:5287–5300. doi: 10.1128/JVI.01829-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gagnon D, Fradet-Turcotte A, Archambault J. 2015. A quantitative and high-throughput assay of human papillomavirus DNA replication. Methods Mol Biol 1249:305–316. doi: 10.1007/978-1-4939-2013-6_23. [DOI] [PubMed] [Google Scholar]
- 28.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 31.Xu D, Zhang Y. 2011. Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys J 101:2525–2534. doi: 10.1016/j.bpj.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kallberg M, Wang H, Wang S, Peng J, Wang Z, Lu H, Xu J. 2012. Template-based protein structure modeling using the RaptorX Web server. Nat Protoc 7:1511–1522. doi: 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swindle CS, Zou N, Van Tine BA, Shaw GM, Engler JA, Chow LT. 1999. Human papillomavirus DNA replication compartments in a transient DNA replication system. J Virol 73:1001–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Junco SE, Wang R, Gaipa JC, Taylor AB, Schirf V, Gearhart MD, Bardwell VJ, Demeler B, Hart PJ, Kim CA. 2013. Structure of the polycomb group protein PCGF1 in complex with BCOR reveals basis for binding selectivity of PCGF homologs. Structure 21:665–671. doi: 10.1016/j.str.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang R, Taylor AB, Leal BZ, Chadwell LV, Ilangovan U, Robinson AK, Schirf V, Hart PJ, Lafer EM, Demeler B, Hinck AP, McEwen DG, Kim CA. 2010. Polycomb group targeting through different binding partners of RING1B C-terminal domain. Structure 18:966–975. doi: 10.1016/j.str.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee KY, Yang K, Cohn MA, Sikdar N, D'Andrea AD, Myung K. 2010. Human ELG1 regulates the level of ubiquitinated proliferating cell nuclear antigen (PCNA) through its interactions with PCNA and USP1. J Biol Chem 285:10362–10369. doi: 10.1074/jbc.M109.092544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Q, Xie Y, Zheng Y, Jiang S, Liu W, Mu W, Liu Z, Zhao Y, Xue Y, Ren J. 2014. GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res 42:W325–W330. doi: 10.1093/nar/gku383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moody CA, Fradet-Turcotte A, Archambault J, Laimins LA. 2007. Human papillomaviruses activate caspases upon epithelial differentiation to induce viral genome amplification. Proc Natl Acad Sci U S A 104:19541–19546. doi: 10.1073/pnas.0707947104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakakibara N, Mitra R, McBride AA. 2011. The papillomavirus E1 helicase activates a cellular DNA damage response in viral replication foci. J Virol 85:8981–8995. doi: 10.1128/JVI.00541-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, Hu XJ, Zou XD, Wu XH, Ye ZQ, Wu YD. 2015. WDSPdb: a database for WD40-repeat proteins. Nucleic Acids Res 43:D339–D344. doi: 10.1093/nar/gku1023. [DOI] [PMC free article] [PubMed] [Google Scholar]