Abstract
We analyzed eight H10N8 viruses isolated from ducks and chickens in live poultry markets from 2009 to 2013 in China. These viruses showed distinct genetic diversity and formed five genotypes: the four duck isolates formed four different genotypes, whereas the four chicken viruses belong to a single genotype. The viruses bound to both human- and avian-type receptors, and four of the viruses caused 12.7% to 22.5% body weight loss in mice.
TEXT
Influenza viruses bearing the H10 subtype hemagglutinin (HA) have been detected in avian species across wide geographic areas. The first H10 isolate, an H10N7 virus, was detected in chickens in Germany in 1949 (1, 2). Since then, viruses bearing H10 HA and different neuraminidase (NA) subtypes have been widely detected in wild birds and domestic poultry around the world (3–19). Moreover, an H10N4 virus caused an outbreak of a respiratory disease in mink in Sweden in 1984 (20), and more recently, several U.S. turkey workers tested seropositive for H10 influenza virus (15). In March 2010, an H10N7 virus caused an outbreak on a chicken farm in Australia; after processing clinically normal birds from the farm, seven abattoir workers reported conjunctivitis and minor upper respiratory tract symptoms and H10 virus infection was detected in two of the seven workers (11). In 2013, H10N8 virus caused three human infections in China, two of which were fatal (21, 22). Although sequence information about the H10 viruses is increasing (23–26), the biologic properties of these viruses remain largely unknown.
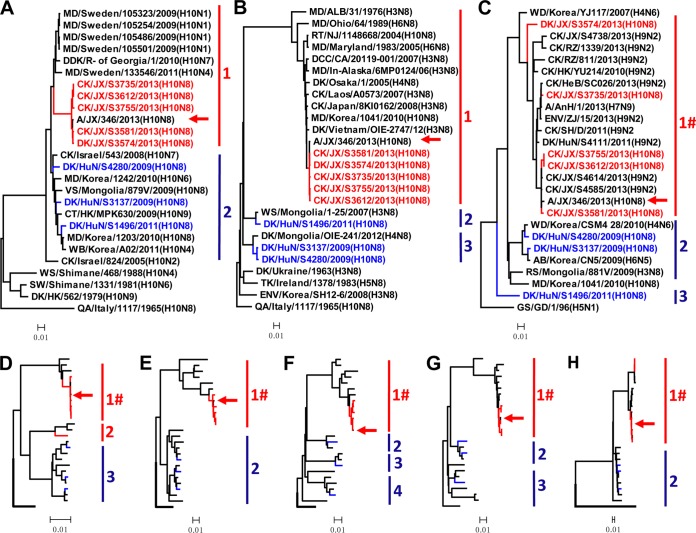
A total of eight H10N8 influenza viruses were isolated from ducks and chickens during our routine surveillance from 2009 to 2013; of these eight viruses, three were isolated in Hunan province (A/duck/Hunan/S4280/2009 [DK/HuN/S4280/09], A/duck/Hunan/S3137/2009 [DK/HuN/S3137/09], and A/duck/Hunan/S1496/2011 [DK/HuN/S1496/11]) (Hunan viruses) and five were isolated in Jiangxi province (A/duck/Jiangxi/S3574/2013 [DK/JX/S3574/13], A/chicken/Jiangxi/S3581/2013 [CK/JX/S3581/13], A/chicken/Jiangxi/S3612/2013 [CK/JX/S3612/13], A/chicken/Jiangxi/S3735/2013 [CK/JX/S3735/13], and A/chicken/Jiangxi/S3755/2013 [CK/JX/S3755/13]) (Jiangxi viruses). To investigate the genetic relationships among these viruses, we sequenced the genomes of all eight viruses. The amino acid motif at the HA cleavage site of these isolates is -R-, which is a characteristic of viruses of low pathogenicity in chickens. The eight genes of the viruses showed distinct diversity, with the HA, NA, PB2, PB1, PA, NP, M, and NS genes of the eight viruses sharing 94.7 to 100, 78.4 to 99.9, 89.9 to 100, 90.1 to 100, 89.7 to 100, 90.0 to 99.9, 90.5 to 100, and 89.3 to 99.9% identity, respectively, at the nucleotide level. The HA, PA, and NS genes each formed two branches in their phylogenetic trees (Fig. 1A, E, and H), whereas the NA, PB2, PB1, and M genes formed three branches each in their phylogenetic trees (Fig. 1B, C, D, and G) and the NP gene formed four branches in its phylogenetic tree (Fig. 1F). Of note, the six internal genes of the H10N8 viruses in branch 1 were clustered with the corresponding genes of H9N2 influenza viruses (Fig. 1C to H), and the NA gene in group 1 belongs to the North American lineage (Fig. 1B).
FIG 1.
Phylogenetic analyses of H10N8 viruses isolated between 2009 and 2013 in China. The phylogenetic trees were generated with the PHYLIP program of the CLUSTALX software package (version 1.81). The trees were generated based on the following sequences: HA nucleotides 22 to 1704, NA nucleotides 19 to 1416, PB2 nucleotides 28 to 2307, PB1 nucleotides 25 to 2298, PA nucleotides 25 to 2175, NP nucleotides 46 to 1542, M nucleotides 26 to 1007, and NS nucleotides 27 to 864. The phylogenetic trees of HA (A) and NA (B) were rooted to A/Quail/Italy/1965(H10N8), and those of PB2 (C), PB1 (D), PA (E), NP (F), M (G), and NS (H) were rooted to A/Goose/Guangdong/1/1996(H5N1). The viruses with names in colors were characterized in this study (the viruses isolated in Jiangxi province are colored red, and the viruses isolated in Hunan province are colored blue). Sequences of viruses with names in black were downloaded from available databases. Abbreviations are as follows: AB, aquatic bird; CK, chicken; CT, common teal; DCC, double-crested cormorant; DDK, domestic duck; DK, duck; ENV, environment; GS, goose; MD, mallard; QA, quail; RS, ruddy shelduck; RT, ruddy turnstone; TK, turkey; VS, velvet scoter; WB, wild bird; WS, whistling swan; SW, swan; WD, wild duck; AnH, Anhui; ALB, Alberta; CA, California; JX, Jiangxi; HeB, Hebei; HuN, Hunan; HK, Hong Kong; In-Alaska, interior Alaska; NJ, New Jersey; R- of Georgia, Republic of Georgia; RZ, Ri Zhao; ZJ, Zhejiang; SH, Shanghai; GD, Guangdong. Groups labeled with a red “#” in the phylogenetic trees of the six internal genes contain viruses of only the H10N8, H7N9, and H9N2 subtypes, and the human H10N8 isolate is indicated with a red arrow; 96% sequence identity cutoffs were used to categorize each gene segment in the phylogenetic trees.
On the basis of this genomic diversity, we divided the viruses into five genotypes (Table 1). Of note, the four duck viruses belong to four different genotypes, whereas the four chicken viruses belong to a single genotype, and the eight gene segments of the CK/JX/S3581/13 virus shared 99.2% to 99.9% identity with the human H10N8 virus A/Jiangxi-Donghu/346/2013.
TABLE 1.
Genotypes of H10N8 viruses
| Virus | Group of each gene segment in phylogenetic tree (% similarity to human H10N8 virus)a |
Genotype | |||||||
|---|---|---|---|---|---|---|---|---|---|
| HA | NA | PB2 | PB1 | PA | NP | M | NS | ||
| CK/JX/S3581/13 | 1 (99.8) | 1 (99.9) | 1 (99.4) | 1 (99.8) | 1 (99.5) | 1 (99.5) | 1 (99.2) | 1 (99.8) | 1 |
| CK/JX/S3612/13 | 1 (99.4) | 1 (99.8) | 1 (99.3) | 1 (99.9) | 1 (99.6) | 1 (99.9) | 1 (99.6) | 1 (96.3) | 1 |
| CK/JX/S3735/13 | 1 (99.4) | 1 (99.6) | 1 (97.8) | 1 (99.9) | 1 (99.5) | 1 (99.5) | 1 (100) | 1 (99.8) | 1 |
| CK/JX/S3755/13 | 1 (99.6) | 1 (99.7) | 1 (99.3) | 1 (99.9) | 1 (99.6) | 1 (99.9) | 1 (99.5) | 1 (96.2) | 1 |
| DK/JX/S3574/13 | 1 (99.9) | 1 (99.8) | 1 (96.8) | 2 (91.3) | 1 (99.5) | 1 (99.3) | 1 (99.3) | 1 (99.8) | 2 |
| DK/HN/S1496/11 | 2 (94.8) | 2 (78.5) | 3 (89.9) | 3 (90.1) | 2 (90.2) | 4 (91.2) | 2 (91.3) | 2 (90.9) | 3 |
| DK/HN/S3137/09 | 2 (95.6) | 3 (78.7) | 2 (91.5) | 3 (90.4) | 2 (90.1) | 2 (91.1) | 3 (90.8) | 2 (90.7) | 4 |
| DK/HN/S4280/09 | 2 (95.6) | 3 (78.5) | 2 (91.8) | 3 (90.4) | 2 (90.2) | 3 (90.2) | 2 (91.1) | 2 (90.7) | 5 |
Genome similarity was compared with the H10N8 human isolate A/Jiangxi-Donghu/346/2013. The data were generated based on the following sequences: HA nucleotides 22 to 1704, NA nucleotides 19 to 1416, PB2 nucleotides 28 to 2307, PB1 nucleotides 25 to 2298, PA nucleotides 25 to 2175, NP nucleotides 46 to 1542, M nucleotides 26 to 1007, and NS nucleotides 27 to 864.
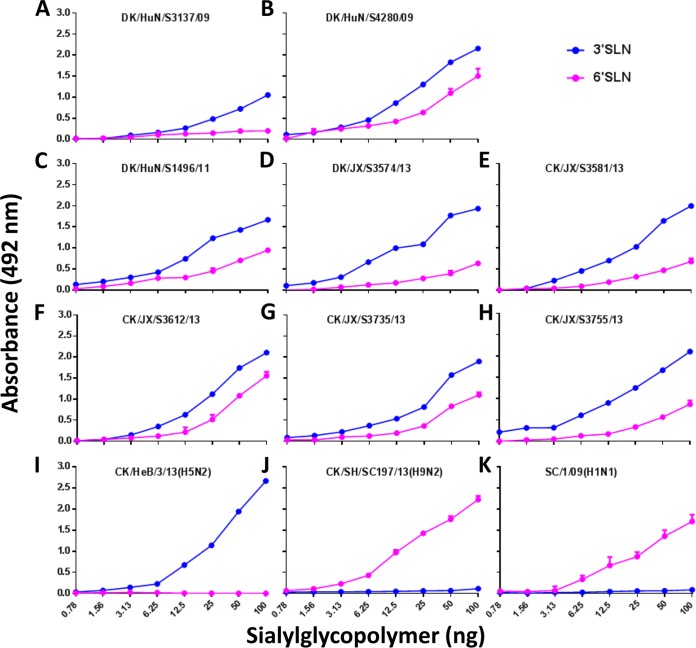
A change in receptor-binding preference from α2,3-linked sialic acids (Sias) (avian-type receptors) to α2,6-linked Sias (human-type receptors) is important for an avian influenza virus to transmit among humans. Previous studies reported that over 30% of H6 influenza viruses bind to human-type receptors (27) and H7N9 viruses bind to both receptor types (28) but that the recent H9N2 viruses bind to only human-type receptors (29). In this study, we found that the HAs of the Jiangxi viruses share over 99.4% identity with each other but that the HAs of the three Hunan viruses are quite diverse. We therefore tested their receptor-binding specificity by using a solid-phase binding assay as described previously (29). All eight H10N8 viruses bound to both α2,3-linked Sias and α2,6-linked Sias, although their affinity for the α2,3-linked Sias was higher than that for the α2,6-linked Sias (Fig. 2A to H). The control H5N2 virus bound to only the α2,3-linked Sias (Fig. 2I), whereas the H9N2 virus and the human influenza virus A/Sichuan/1/2009 (SC/1/09) bound to only the α2,6-linked Sias (Fig. 2J and K).
FIG 2.
Characterization of the receptor-binding properties of H10N8 viruses. The binding of the viruses to two different biotinylated glycans (α2,3 glycan, blue; α2,6 glycan, pink) was tested. The data shown are the means of three repeats from one experiment; the error bars indicate standard deviations.
There have been several reports of human infections caused by H10 viruses (11, 15, 21), but only the H10N8 virus has thus far caused fatal outcomes (21). To understand the virulence of the H10N8 viruses in mammals, we tested the replication and lethality of the eight viruses in mice. Groups of eight 6-week-old female BALB/c mice (Beijing Vital River Laboratories, Beijing, China) were anesthetized with CO2 and inoculated intranasally (i.n.) with 106.0 50% egg infective doses (EID50) of each test virus in a volume of 50 μl. Three mice were euthanized on day 3 postinoculation (p.i.), and the nasal turbinates, lungs, kidneys, spleens, and brains were collected for virus titration in eggs. The remaining five mice in each group were monitored daily for 14 days for weight loss and survival. (The protocol for this animal study was approved by the Committee on the Ethics of Animal Experiments of the Harbin Veterinary Research Institute of the Chinese Academy of Agricultural Sciences.)
Seven of the eight viruses replicated well in the lungs of mice, with mean titers ranging from 3.1 to 5.8 log10 EID50; the eighth virus, DK/HuN/S1496/11, was detected from only two of three mice (Fig. 3A). The five Jiangxi viruses were detected in the turbinates of all of the inoculated mice, with mean titers ranging from 2.8 to 4.0 log10 EID50, whereas virus was detected in the turbinate of only one of three mice inoculated with each of the three Hunan viruses (Fig. 3A). The titers in the Jiangxi virus-infected mice were significantly higher than that of the Hunan viruses (Fig. 3A). Virus was not detected in the spleen, kidneys, or brain of any mice. Mice infected with these viruses showed diverse body weight changes during the observation period: four viruses caused 12.7% to 22.5% body weight loss in mice, whereas the other four viruses did not cause apparent body weight loss (Fig. 3B). All of the mice survived during the observation period.
FIG 3.
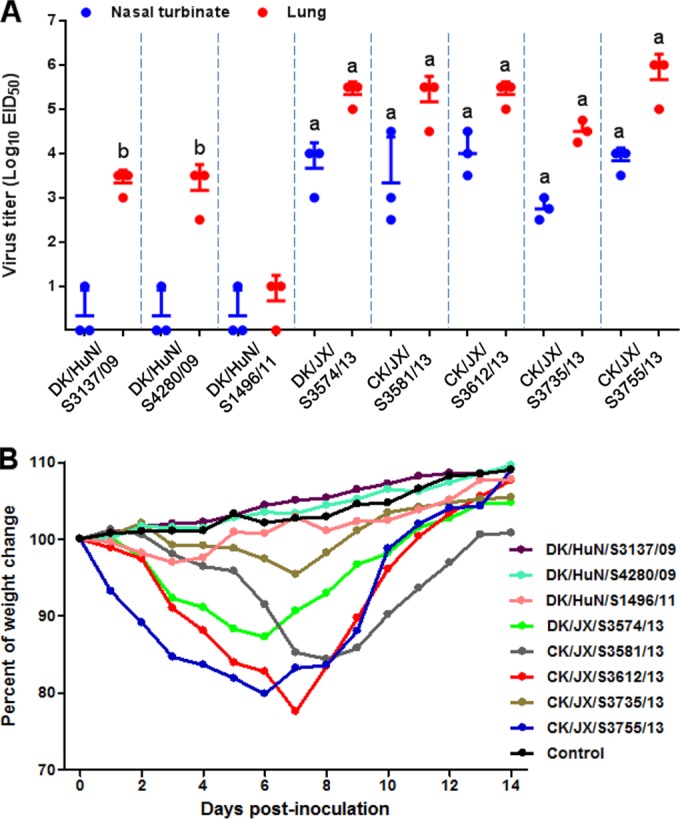
Replication and virulence of H10N8 viruses in mice. (A) Virus titers in organs of mice. The data shown are the means ± standard deviations for each group. Because virus was not detected from spleen, kidney, or brain of any mouse, data for these organs are not shown. a, P < 0.01 compared with the corresponding value for the three Hunan H10N8 virus-inoculated groups; b, P < 0.01 compared with the corresponding value for the DK/HuN/S1496/11-inoculated group. (B) Body weight changes of mice.
In summary, our genetic studies indicate that the four duck viruses belong to four different genotypes, suggesting that they were introduced into ducks independently; the four chicken viruses belong to one genotype and appear to be hybrids of a duck virus and the local H9N2 viruses (Table 1). The ability of H10N8 viruses to bind to human-type receptors facilitates their infection of humans, as occurred with the H7N9 viruses (28). The more efficient replication in mice of the viruses isolated in Jiangxi province than of the three duck viruses isolated in Hunan province suggests that the internal genes of the H9N2 viruses may have further increased the replicative ability and virulence of H10N8 viruses in mammals; of course, the surface proteins may have also contributed to the difference of the virulence. Although the viruses in our studies were all isolated from healthy birds, two H10 influenza viruses, A/turkey/England/384/79 and A/mandarin duck/Singapore/805/F-72/7/93, were reported to be highly pathogenic in chickens (3, 30). Therefore, it is important to continue monitoring the evolution of H10N8 influenza viruses and to evaluate their potential to cause disease in poultry and pandemics in humans.
Nucleotide sequence accession numbers.
The nucleotide sequences of the eight viruses determined in this study have been deposited in GenBank under accession numbers KP861987 to KP862050.
ACKNOWLEDGMENTS
We thank S. Watson for editing the manuscript and Yamasa Corporation Co. Ltd. for synthesizing the sialylglycopolymer.
This work was supported by the National Science and Technology Major Project (2012ZX10004214).
REFERENCES
- 1.Feldmann H, Kretzschmar E, Klingeborn B, Rott R, Klenk HD, Garten W. 1988. The structure of serotype H10 hemagglutinin of influenza A virus: comparison of an apathogenic avian and a mammalian strain pathogenic for mink. Virology 165:428–437. doi: 10.1016/0042-6822(88)90586-7. [DOI] [PubMed] [Google Scholar]
- 2.Englund L, Hard ASC. 1998. Two avian H10 influenza A virus strains with different pathogenicity for mink (Mustela vison). Arch Virol 143:653–666. doi: 10.1007/s007050050321. [DOI] [PubMed] [Google Scholar]
- 3.Kim HR, Lee YJ, Oem JK, Bae YC, Kang MS, Kang HM, Choi JG, Park CK, Kwon YK. 2012. Characterization of H10 subtype avian influenza viruses isolated from wild birds in South Korea. Vet Microbiol 161:222–228. doi: 10.1016/j.vetmic.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 4.Vijaykrishna D, Deng YM, Su YC, Fourment M, Iannello P, Arzey GG, Hansbro PM, Arzey KE, Kirkland PD, Warner S, O'Riley K, Barr IG, Smith GJ, Hurt AC. 2013. The recent establishment of North American H10 lineage influenza viruses in Australian wild waterfowl and the evolution of Australian avian influenza viruses. J Virol 87:10182–10189. doi: 10.1128/JVI.03437-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonfante F, Fusaro A, Zanardello C, Patrono LV, De Nardi R, Maniero S, Terregino C. 2014. Lethal nephrotropism of an H10N1 avian influenza virus stands out as an atypical pathotype. Vet Microbiol 173:189–200. doi: 10.1016/j.vetmic.2014.07.023. [DOI] [PubMed] [Google Scholar]
- 6.Fries AC, Nolting JM, Bowman AS, Killian ML, Wentworth DE, Slemons RD. 2014. Genomic analyses detect Eurasian-lineage H10 and additional H14 influenza A viruses recovered from waterfowl in the Central United States. Influenza Other Respir Viruses 8:493–498. doi: 10.1111/irv.12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okamatsu M, Nishi T, Nomura N, Yamamoto N, Sakoda Y, Sakurai K, Chu HD, Thanh LP, Van Nguyen L, Van Hoang N, Tien TN, Yoshida R, Takada A, Kida H. 2013. The genetic and antigenic diversity of avian influenza viruses isolated from domestic ducks, Muscovy ducks, and chickens in northern and southern Vietnam, 2010–2012. Virus Genes 47:317–329. doi: 10.1007/s11262-013-0954-7. [DOI] [PubMed] [Google Scholar]
- 8.Slavec B, Krapez U, Racnik AJ, Hari A, Wernig JM, Dovc A, Zadravec M, Lindtner-Knific R, Marhold C, Zorman-Rojs O. 2012. Surveillance of influenza A viruses in wild birds in Slovenia from 2006 to 2010. Avian Dis 56:999–1005. doi: 10.1637/10175-041012-ResNote.1. [DOI] [PubMed] [Google Scholar]
- 9.Pasick J, Pedersen J, Hernandez MS. 2012. Avian influenza in North America, 2009–2011. Avian Dis 56:845–848. doi: 10.1637/10206-041512-Reg.1. [DOI] [PubMed] [Google Scholar]
- 10.Vittecoq M, Grandhomme V, Champagnon J, Guillemain M, Crescenzo-Chaigne B, Renaud F, Thomas F, Gauthier-Clerc M, van der Werf S. 2012. High influenza a virus infection rates in mallards bred for hunting in the Camargue, south of France. PLoS One 7:e43974. doi: 10.1371/journal.pone.0043974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arzey GG, Kirkland PD, Arzey KE, Frost M, Maywood P, Conaty S, Hurt AC, Deng YM, Iannello P, Barr I, Dwyer DE, Ratnamohan M, McPhie K, Selleck P. 2012. Influenza virus A (H10N7) in chickens and poultry abattoir workers, Australia. Emerg Infect Dis 18:814–816. doi: 10.3201/eid1805.111852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raleigh PJ, Flynn O, O'Connor M, Minihan D, Connell J, Sammin DJ, Markey BK. 2011. Phylogenetic analysis of H and N2 genes of avian influenza viruses detected in Ireland between 2003 and 2007. Epidemiol Infect 139:1191–1201. doi: 10.1017/S0950268810002372. [DOI] [PubMed] [Google Scholar]
- 13.Ferro PJ, Budke CM, Peterson MJ, Cox D, Roltsch E, Merendino T, Nelson M, Lupiani B. 2010. Multiyear surveillance for avian influenza virus in waterfowl from wintering grounds, Texas coast, USA. Emerg Infect Dis 16:1224–1230. doi: 10.3201/eid1608.091864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langstaff IG, McKenzie JS, Stanislawek WL, Reed CE, Poland R, Cork SC. 2009. Surveillance for highly pathogenic avian influenza in migratory shorebirds at the terminus of the East Asian-Australasian Flyway. N Z Vet J 57:160–165. doi: 10.1080/00480169.2009.36896. [DOI] [PubMed] [Google Scholar]
- 15.Kayali G, Ortiz EJ, Chorazy ML, Gray GC. 2010. Evidence of previous avian influenza infection among US turkey workers. Zoonoses Public Health 57:265–272. doi: 10.1111/j.1863-2378.2009.01231.x. [DOI] [PubMed] [Google Scholar]
- 16.De Marco MA, Campitelli L, Foni E, Raffini E, Barigazzi G, Delogu M, Guberti V, Di Trani L, Tollis M, Donatelli I. 2004. Influenza surveillance in birds in Italian wetlands (1992–1998): is there a host restricted circulation of influenza viruses in sympatric ducks and coots? Vet Microbiol 98:197–208. doi: 10.1016/j.vetmic.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 17.Senne DA. 2003. Avian influenza in the Western Hemisphere including the Pacific Islands and Australia. Avian Dis 47:798–805. doi: 10.1637/0005-2086-47.s3.798. [DOI] [PubMed] [Google Scholar]
- 18.Alexander DJ. 2003. Report on avian influenza in the Eastern Hemisphere during 1997–2002. Avian Dis 47:792–797. doi: 10.1637/0005-2086-47.s3.792. [DOI] [PubMed] [Google Scholar]
- 19.Pfitzer S, Verwoerd DJ, Gerdes GH, Labuschagne AE, Erasmus A, Manvell RJ, Grund C. 2000. Newcastle disease and avian influenza A virus in wild waterfowl in South Africa. Avian Dis 44:655–660. doi: 10.2307/1593107. [DOI] [PubMed] [Google Scholar]
- 20.Berg M, Englund L, Abusugra IA, Klingeborn B, Linne T. 1990. Close relationship between mink influenza (H10N4) and concomitantly circulating avian influenza viruses. Arch Virol 113:61–71. doi: 10.1007/BF01318353. [DOI] [PubMed] [Google Scholar]
- 21.Chen H, Yuan H, Gao R, Zhang J, Wang D, Xiong Y, Fan G, Yang F, Li X, Zhou J, Zou S, Yang L, Chen T, Dong L, Bo H, Zhao X, Zhang Y, Lan Y, Bai T, Dong J, Li Q, Wang S, Zhang Y, Li H, Gong T, Shi Y, Ni X, Li J, Zhou J, Fan J, Wu J, Zhou X, Hu M, Wan J, Yang W, Li D, Wu G, Feng Z, Gao GF, Wang Y, Jin Q, Liu M, Shu Y. 2014. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. Lancet 383:714–721. doi: 10.1016/S0140-6736(14)60111-2. [DOI] [PubMed] [Google Scholar]
- 22.Vachieri SG, Xiong X, Collins PJ, Walker PA, Martin SR, Haire LF, Zhang Y, McCauley JW, Gamblin SJ, Skehel JJ. 2014. Receptor binding by H10 influenza viruses. Nature 511:475–477. doi: 10.1038/nature13443. [DOI] [PubMed] [Google Scholar]
- 23.Zhang T, Bi Y, Tian H, Li X, Liu D, Wu Y, Jin T, Wang Y, Chen Q, Chen Z, Chang J, Gao GF, Xu B. 2014. Human infection with influenza virus A(H10N8) from live poultry markets, China, 2014. Emerg Infect Dis 20:2076–2079. doi: 10.3201/eid2012.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su S, Qi W, Zhou P, Xiao C, Yan Z, Cui J, Jia K, Zhang G, Gray GC, Liao M, Li S. 2014. First evidence of H10N8 avian influenza virus infections among feral dogs in live poultry markets in Guangdong province, China. Clin Infect Dis 59:748–750. doi: 10.1093/cid/ciu345. [DOI] [PubMed] [Google Scholar]
- 25.Qi W, Zhou X, Shi W, Huang L, Xia W, Liu D, Li H, Chen S, Lei F, Cao L, Wu J, He F, Song W, Li Q, Li H, Liao M, Liu M. 2014. Genesis of the novel human-infecting influenza A(H10N8) virus and potential genetic diversity of the virus in poultry, China. Euro Surveill 19(25):pii:20841 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20841. [DOI] [PubMed] [Google Scholar]
- 26.Ni X, He F, Hu M, Zhou X, Wang B, Feng C, Wu Y, Li Y, Tu J, Li H, Liu M, Chen H, Chen S. 2 October 2014. Investigation of avian influenza virus in poultry and wild birds due to novel avian-origin influenza A(H10N8) in Nanchang City, China. Microbes Infect doi: 10.1016/j.micinf.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 27.Wang G, Deng G, Shi J, Luo W, Zhang G, Zhang Q, Liu L, Jiang Y, Li C, Sriwilaijaroen N, Hiramatsu H, Suzuki Y, Kawaoka Y, Chen H. 2014. H6 influenza viruses pose a potential threat to human health. J Virol 88:3953–3964. doi: 10.1128/JVI.03292-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Q, Shi J, Deng G, Guo J, Zeng X, He X, Kong H, Gu C, Li X, Liu J, Wang G, Chen Y, Liu L, Liang L, Li Y, Fan J, Wang J, Li W, Guan L, Li Q, Yang H, Chen P, Jiang L, Guan Y, Xin X, Jiang Y, Tian G, Wang X, Qiao C, Li C, Bu Z, Chen H. 2013. H7N9 influenza viruses are transmissible in ferrets by respiratory droplet. Science 341:410–414. doi: 10.1126/science.1240532. [DOI] [PubMed] [Google Scholar]
- 29.Li X, Shi J, Guo J, Deng G, Zhang Q, Wang J, He X, Wang K, Chen J, Li Y, Fan J, Kong H, Gu C, Guan Y, Suzuki Y, Kawaoka Y, Liu L, Jiang Y, Tian G, Li Y, Bu Z, Chen H. 2014. Genetics, receptor binding property, and transmissibility in mammals of naturally isolated H9N2 avian influenza viruses. PLoS Pathog 10:e1004508. doi: 10.1371/journal.ppat.1004508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wood GW, Banks J, Strong I, Parsons G, Alexander DJ. 1996. An avian influenza virus of H10 subtype that is highly pathogenic for chickens, but lacks multiple basic amino acids at the haemagglutinin cleavage site. Avian Pathol 25:799–806. doi: 10.1080/03079459608419182. [DOI] [PubMed] [Google Scholar]