ABSTRACT
Replication of the integrated HIV-1 genome is tightly regulated by a series of cellular factors. In previous work we showed that transactivation of the HIV-1 promoter is regulated by the cellular splicing factor SRSF1. Here we report that SRSF1 can downregulate the replication of B, C, and D subtype viruses by >200-fold in a cell culture system. We show that viral transcription and splicing are inhibited by SRSF1 expression. Furthermore, SRSF1 deletion mutants containing the protein RNA-binding domains but not the arginine serine-rich activator domain can downregulate viral replication by >2,000-fold with minimal impact on cell viability and apoptosis. These data suggest a therapeutic potential for SRSF1 and its RNA-binding domains.
IMPORTANCE Most drugs utilized to treat the HIV-1 infection are based on compounds that directly target proteins encoded by the virus. However, given the high viral mutation rate, the appearance of novel drug-resistant viral strains is common. Thus, there is a need for novel therapeutics with diverse mechanisms of action. In this study, we show that the cellular protein SRSF1 is a strong inhibitor of viral replication. Furthermore, expression of the SRSF1 RNA-binding domains alone can inhibit viral replication by >2,000-fold in multiple viral strains without impacting cell viability. Given the strong antiviral properties of this protein, the RNA-binding domains, and the minimal effects observed on cell metabolism, further studies are warranted to assess the therapeutic potential of peptides derived from these sequences.
INTRODUCTION
Replication of the integrated HIV-1 genome is tightly regulated by a combination of host and viral factors. Interactions between viral sequences and cellular and viral proteins are required to express the viral genome, and alteration of the mechanisms regulating transcription, splicing, and export of the viral transcripts can dramatically affect HIV-1 infectivity and pathogenesis (1–3).
The integrated provirus is transcribed into a single pre-mRNA from a promoter located within the 5′ long terminal repeat (LTR) of the viral genome through RNA polymerase II (RNAP II) and a combination of basal and promoter specific factors (1). The viral protein, Tat, stimulates transcription elongation by binding to a structured RNA element (transactivation responsive region [TAR]), located at the 5′ ends of all nascent HIV-1 transcripts (4, 5) and triggering the recruitment of the P-TEFb complex. The P-TEFb complex is composed of cellular cyclin T1 and the cyclin-dependent kinase 9. P-TEFb activates viral transcription by promoting the release of the NELF and DRB sensitivity-inducing factor transcriptional pausing complex (6, 7) and phosphorylation of the C-terminal domain of RNAP II to facilitate elongation of the viral transcript (8, 9).
The single viral transcript undergoes a complex series of splicing events to generate over 40 mRNA isoforms; thus, the same viral protein is encoded by multiple mRNAs that vary for their 5′ and 3′ untranslated regions (10). Spliced viral mRNAs can be classified in a group of ∼4 kb in length, coding for the Env, Vpu, Vpr, and Vif proteins, and a group of ∼2 kb in length, coding for the Tat, Rev, and Nef proteins (10). Furthermore, ca. 50% of the viral pre-mRNAs leave the nucleus without being spliced. The unspliced 9-kb mRNA encodes the Gag and Gag-Pol polyprotein and is packaged within the nascent virions as viral genome. The complex splicing regulation of the viral transcripts is carried out by several cellular factors, which interact with partially characterized cis-acting elements distributed throughout the genome and selectively enhance or inhibit the use of specific splice sites (1).
We have recently showed that SRSF1, an RNA binding protein (RBP) member of the serine/arginine (SR) proteins family, can inhibit Tat transactivation by directly competing for its binding onto TAR (11). SR proteins are key regulators of gene expression, highly conserved, and widely expressed in eukaryotes. SRSF1 and other members of this protein family regulate the assembly of the splicing machinery (12), integrate multiple steps in RNA metabolism (13), and have been shown to modulate RNAP II activity (14, 15). SRSF1 binds to splicing regulatory sequences within the viral transcript and modulates the splicing of subgenomic clones in vitro and ex vivo (3, 16, 17). Nevertheless, given the presence of multiple putative binding sites for SRSF1 throughout the viral genome and the high mutation rate in the primary sequences among different viral isolates, the overall contribution of this protein to viral replication, in the contest of the full-length virus, is unclear.
Previous studies on the role of SRSF1 in HIV-1 replication have focused on its activity as a splicing regulator and have been carried out utilizing minimal subgenomic substrates and molecular clones derived from the LAV(LAI) viral isolate (10, 18). We sought to analyze the role of SRSF1 in viral gene expression and replication utilizing molecular clones from different viral subtypes. We show that the overexpression of SRSF1 strongly inhibits the viral transcription, splicing, and replication of viruses from the B, C, and D subtypes. Furthermore, we found that the SRSF1 RNA-binding domains (RBDs) alone can downregulate viral replication by >2,000-fold without altering cell viability and apoptosis. Taken together, our results demonstrate the therapeutic potential of SRSF1 and it RBDs.
MATERIALS AND METHODS
Plasmids and cells.
The SRSF1 deletion mutants have been previously described (19). The enhanced green fluorescent protein (EGFP)-tagged SRSF1 clones were obtained by cloning the SRSF1 coding sequences upstream the EGFP gene in the pEGFP-C1 vector (Clontech). The HIV-1 molecular clones—pNL4.3 contributed by Malcolm Martin (18), pMtat(−) contributed by Reza Sadaie (20), pLAI.2 contributed by K. Peden (21), pMJ4 contributed by T. Ndung'u (22), and p94UG114.1.6 contributed by B. H. Hahn (23)—were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Disease (NIAID), National Institutes of Health (NIH).
HEK293 cells were maintained at below 80% confluence. Cells were transfected in 24-well plates with Lipofectamine 2000 (Life Technologies), 0.2 μg of virus coding plasmid, and 0.2 μg of the SRSF1 or control expression plasmids. Proteins expressed were analyzed utilizing the antibodies anti-SRSF1 (provided by A. R. Krainer, Cold Spring Harbor Laboratories), Tat (obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH), and antiserum to HIV-1 Tat from Bryan Cullen), GFP (B-2; Santa Cruz Biotech), β-tubulin (Sigma), and T7-Tag (Sigma). HLM1 was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH; HLM30 cells (obtained from Reza Sadaie) were transfected by electroporation by utilizing a GenePulser II (Bio-Rad).
In vitro infection assay.
H9 cells (obtained from Robert Gallo through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH) were infected at high multiplicity of infection (MOI; ≤100) with virus generated by collecting the supernatant from HEK293 cells transfected with the proviral clone pNL4-3. At 4 days postinfection, cells were transfected with the EGFP-tagged clones by electroporation utilizing a GenePulser II (Bio-Rad). At 24 h posttransfection, EGFP-expressing cells were sorted using a BD FACSAria II cell sorter. Cells that displayed a peak EGFP signal were gated and collected. After sorting, the cells were collected and plated in 96-well plates at a concentration of 50,000 cells/well in triplicates. Virus was collected every 24 h for the following 4 days and quantified utilizing TZM-bl cells.
RNA extraction and transcripts analysis.
Total RNA was extracted 48 h after transfection with a total RNA isolation kit (Agilent) and DNase treated with Turbo DNase (Ambion). RNA was reverse transcribed utilizing a random pd(N)6 primer and Superscript II RT (Life Technologies). Quantitative PCR (qPCR) analysis of the viral transcripts was performed as previously described (3) utilizing the primers described in Table SA1 in the supplemental material. Each sample was normalized for the relative content in the housekeeping genes GAPDH and Tubulin. qPCR was performed utilizing a Stratagene Mx3005P real-time PCR system and SYBR green dye and analyzed with MxPro V3.0 software. Each assay was carried out with a minimum of three independent transfections, while qPCR assays were carried out in duplicates. The data are represented as means ± the standard errors of the mean (SEM). Alternative splicing events in cellular genes were quantified by semiquantitative reverse transcription-PCR (RT-PCR) assays utilizing the primer sets described in Table SA1 in the supplemental material. To avoid amplification bias, 28 cycles were utilized for each PCR assay. PCR products were analyzed and quantified utilizing a 2100 Bioanalyzer (Agilent Technologies) and 2100 Expert Software. The data are represented as means ± the SEM.
Cellular assays.
The viral replication assay was carried out utilizing TZM-bl cells contributed by J. C. Kappes (24) obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, and seeded 24 h before infection in 96-well plates at 50% confluence in 200 μl of Dulbecco modified Eagle medium supplemented with 8% fetal calf serum and gentamicin. Supernatant collected from HEK293 cells 72 h after the transfection carried out with the proviral constructs was utilized to infect the TZM-bl cells. At 48 h postinfection, the cells were lysed, and the luciferase expression was assayed and quantified utilizing a BMG PolarStar Omega reader. Each assay was carried out with a minimum of three independent transfections. TZM-bl infections from each independent assay were carried out in triplicates. Luciferase data were analyzed utilizing the MARS data analysis software. Cell viability was measured in HEK293 cells at 72 h posttransfection utilizing the CellTiter-Glo (Promega) ATP production assay. The assay was performed in HEK293 cells at 72 h posttransfection and in H9 cells at 5 days posttransfection. Apoptotic events were detected utilizing a FAM FLICA caspase 9 apoptosis detection kit (Marker Gene Technologies). The data are represented as means ± the SEM. The assay was performed in HEK293 cells at 72 h posttransfection.
RESULTS
SRSF1 inhibits Tat transactivation.
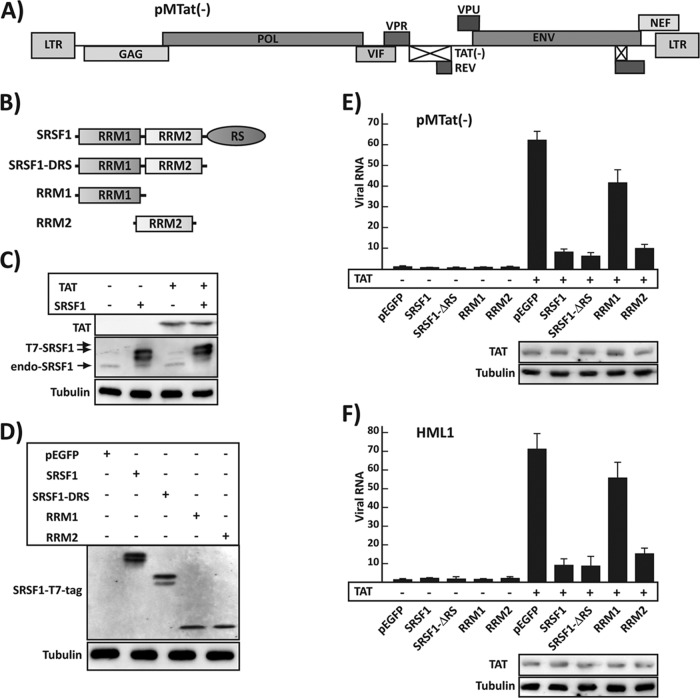
The cellular machineries regulating the transcription and processing of eukaryotic RNAs are intimately coupled. Components of the RNAP II transcription complex modulate the processing of the nascent pre-mRNA and, in some cases, the processing feeds back to regulate transcription (25, 26); therefore, it is important to study the regulation of viral gene expression within the contest of the full-length virus. In our previous work we utilized a series of reporter minigenes and biochemical assays to show that SRSF1 inhibits viral transactivation by recognizing a sequence within TAR that overlaps the Tat binding site. Here, we set out to study the role of SRSF1 in HIV transcription in the context of the full-length virus. We used the proviral clone pMtat(−), which contains a full-length viral genome but lacks a functional copy of the Tat gene (Fig. 1A) (20). HEK293 cells were cotransfected with the proviral vector pMtat(−) and expression clones for Tat, SRSF1, and SRSF1 deletion mutants or the control EGFP. Tat increased the amount of viral transcripts by >60-fold, whereas SRSF1 overexpression reduced transactivation to 8-fold (Fig. 1E), confirming our previous observations, obtained utilizing a subgenomic reporter construct (11).
FIG 1.
SRSF1 inhibition of HIV-1 transcription. (A) Schematic representation of the pMtat(−) proviral clone. (B) Schematic representation of the SRSF1 deletion clones containing the RNA recognition motifs (RRMs) and the Arg-Ser-rich domain (RS). (C) Tat and SRSF1 expression in HEK293 cells transfected with SRSF1 and Tat coding plasmids. Proteins are detected with the anti-Tat, anti-SRSF1, and anti-tubulin antibodies. The SRSF1 expressed from the pSRSF1 vector is tagged with a T7 epitope (T7-SRSF1) and migrates more slowly than the endogenous protein (endo-SRSF1). (D) The T7-tagged SRSF1 proteins was detected utilizing an anti-T7 tag. (E) Quantification of viral mRNA in a transient-transfection system. HEK293. Cells were transfected with the proviral clone pMtat(−) and the indicated vector in the presence (+) or absence (−) of the Tat expression construct. Viral mRNA was quantified by qPCR (primers P1 and P2), and data were normalized for the α-tubulin mRNA content of each sample and expressed as the fold increase versus the pEGFP, Tat− transfection control. The Tat expression level is shown for the cells transfected with pTat. (F) Quantification of viral mRNA in the integrated provirus. HLM1 cells were transfected with the indicated expression vector in the presence (+) or absence (−) of Tat, and viral mRNA was quantified by qPCR and expressed as the fold increase versus the pEGFP, Tat− transfection control. The Tat expression level is shown for the cells transfected with pTat. All data are represented as means ± the SEM.
SRSF1 is composed of two RNA recognition motif (RRM) RNA-binding domains (RBDs), which interact with specific RNA sequences, and an RS (arginine/serine-rich) carboxy-terminal domain, which is required for protein-protein interaction but does not appear to affect the RNA binding specificity of the protein (27). Our previous results indicated that the ability of SRSF1 to inhibit transactivation was solely dependent on its binding to TAR. To determine whether this was also true in the context of the full-length virus, we expressed a series of SRSF1 deletion mutants (Fig. 1B and D) in the absence or presence of Tat.
The mutant carrying a deletion of the protein-interacting RS domain reduced transactivation to only 6-fold, which is lower than the one observed utilizing the wild-type protein (8-fold). Expression of a mutant clone carrying only RRM2 decreased transactivation with efficiency similar to the wild-type protein (10-fold), whereas RRM1 reduced transactivation by <50%. These data are consistent with our previous observations suggesting an inhibition mechanism solely dependent on the RNA binding specificity of SRSF1 for a sequence overlapping the Tat binding site onto TAR and with findings suggesting that the RNA binding activity of SRSF1 is mostly dependent on RRM2 (28).
One of the limitations of utilizing a transient-transfection system to study the mechanism of viral replication is the presence of multiple nonintegrated copies of the viral genome. Although the transcription machinery appears to similarly regulate the integrated and nonintegrated viral genome, it is conceivable that the transcriptional effect of SRSF1 might differ in the course of the natural infection, with cells carrying a single copy of the genome integrated in the chromosomal DNA. The results obtained using HLM1 cells, which carry a single copy of the integrated pMtat(−) (Fig. 1F), validate the data obtained with the transfection of pMtat(−) and indicate that the transient-transfection system utilized can be adopted to test the effects of SRSF1 on viral production.
SRSF1 RBDs inhibit viral production.
SRSF1 exerts a key role in the regulation of viral splicing. Previous work carried out by us and others characterized splicing regulatory sequences bound by SRSF1 that modulate selection of specific splice sites by the cellular splicing machinery (3, 16, 17). Furthermore, in silico analysis of the viral transcript performed using the ESEfinder 3.0 (29), a SR protein functional binding site prediction matrix, indicates that several putative high-affinity SRSF1 binding sites are present throughout the viral genome (Fig. 2B), revealing a pervasive role for this splicing factor in the regulation of the viral pre-mRNA.
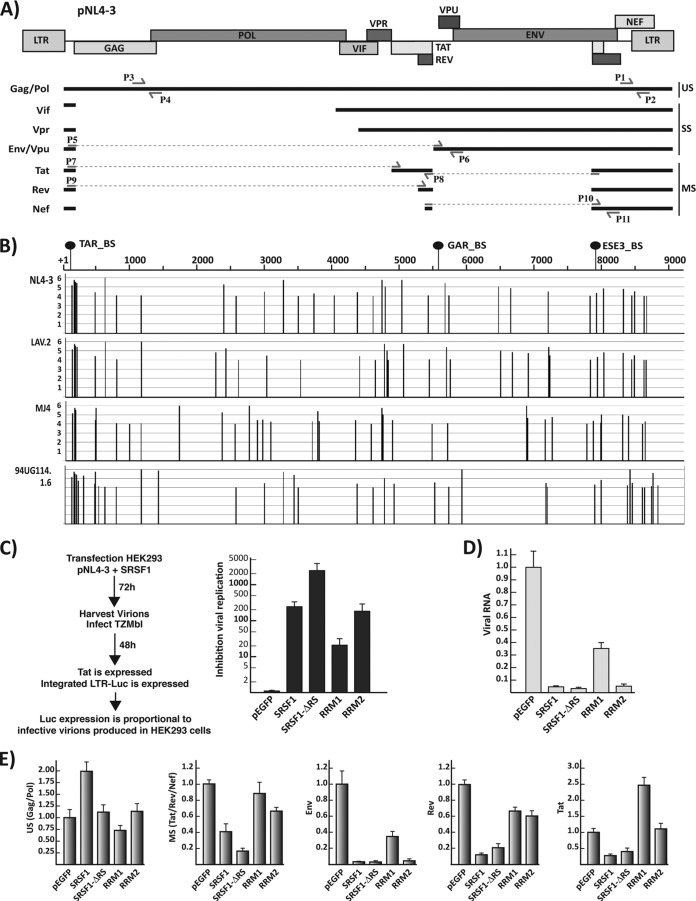
FIG 2.
SRSF1 inhibition of HIV-1 replication. (A) Schematic representation of the pNL4-3 proviral clone. The relative position of the viral genes is indicated on the map on top. The main mRNAs—classified as unspliced (US), singly spliced (SS), and multiply spliced (MS)—are indicated. Multiple mRNAs coding for each viral gene are generated by the choice of several alternative 5′ and 3′ splice sites (10). Locations of the primers utilized in the qPCR assays to amplify the total viral mRNA (P1 and P2) and specific for the gag/pol (P3 and P4), env/vpu (P5, P6), rev (P9 and P8), tat (P7 and P8), and the multiple spliced mRNAs (tat, rev, and nef) (P10 and P11) are indicated. (B) SRSF1 binding sites within the viral transcript. The sequences of four molecular clones were analyzed for consensus SRSF1 binding sites utilizing the ESEfinder 3.0 (29). A high-threshold stringency of 4 was chosen for the SRSF1 matrix analysis to reduce the number false positives and the significance of the output data. The position and score of each predicted SRSF1 binding site is plotted for each viral molecular clone in relation to the standard NL4-3 genome map. The location of the SRSF1 binding sites that have been experimentally characterized is indicated in relation to the NL4-3 genome (top). (C) Viral replication assay. HEK293 cells were cotransfected with the viral expression clone pNL4-3 and the indicated SRSF1 plasmid. Viral production was assayed as indicated in the schematics on the left. The amount of supernatant utilized for the assay was normalized for the amount of cells present in the plate. Inhibition of viral replication was defined as the: the luciferase counts from TZM-bl infected with the supernatant of HEK293 transfected with pNL4-3 and the control pEGFP/the luciferase counts from TZM-bl infected with the supernatant of HEK293 transfected with pNL4-3 and the indicated SRSF1 clone. (D) Quantification of total viral mRNA from the transfected HEK293 cells by qPCR (primers P1 and P2). The results are visualized as the fold increase or decrease versus the control pEGFP transfection. (E) Quantification of the single viral transcripts. Expression of the gag/pol, env, tat, rev, and multiply spliced mRNAs relative to the total amount of viral transcript was calculated by normalizing the amount of specific transcripts for the total amount of viral mRNA in each sample, and each value is expressed as the fold increase or decrease versus control pEGFP transfection. All data are represented as means ± the SEM.
Two mechanisms have been proposed to explain SRSF1's role in splicing regulation (27). In one model, SRSF1 recruits, via protein-protein interactions with the RS domain, an essential component of the basal splicing machinery onto 5′ and 3′ splice sites. In the second model, SRSF1 activates splicing by competing with and inhibiting the binding of splicing repressors in the proximity of the regulated splice site. Furthermore, this protein has also been shown to inhibit the splicing of a number of cellular exons (30–32), although the mechanism of action is unclear.
Given the multiple SRSF1 binding sites present throughout the viral genome, the complex alternative splicing patterns required to generate multiple mRNAs encoding the viral gene products and the diverse mechanisms by which this protein exerts its functions in splicing and transcription, it is conceivable that expression of the SRSF1 full-length sequence or deletion mutants, which retain the RNA binding functions, might considerably alter viral replication. We sought to determine how the wild type and its single-domain deletion mutants might affect the biogenesis of the viral mRNAs and viral replication at large. To this end, we utilized a viral production assay. HEK293 cells were cotransfected with the proviral vector pNL4-3, which codes for a replication competent copy of the viral genome, and the SRSF1 full-length or RBD domain constructs. Transfection of HEK293 cells is highly efficient, thus ensuring coexpression of the proviral and SRSF1 constructs. Viral production was analyzed by collecting the supernatant from the transfected HEK293 cells and infecting TZM-bl cells (24), which contain a copy of the luciferase gene under the control of the HIV-1 LTR promoter. Infected cells express the viral protein Tat, which in turn activates the viral LTR promoter. Thus, the number of infective viral copies is reliably quantified by the activity of the luciferase gene product. Overexpression of SRSF1 decreased viral production by >200-fold. Surprisingly, a mutant lacking the RS domain exhibited decreased production by >2,000-fold, while the result obtained following expression of the RRM2 alone was comparable to the wild type (Fig. 2C). These data are in agreement with our previous results, which showed that downregulation of SRSF1 by small interfering RNA induces an increase in viral production (3).
Next, we determined the effect that the SRSF1 mutants have on the levels of viral mRNA and on the splicing of specific viral mRNA isoforms. Messengers were quantified utilizing a series of qPCR assays with primers sets designed to anneal either to a region common to all viral mRNAs or to specific spliced mRNAs species as previously described (3). The wild-type SRSF1 downregulated total viral RNA production by >20-fold; comparable results were achieved with the clone carrying the deletion of the RS domain or the RRM2 alone, while the RRM1 alone decreased viral RNA levels to roughly one-third of the control (Fig. 2D). Each SRSF1 mutant caused a deregulation of the relative levels of the single viral mRNAs analyzed (Fig. 2E), which differs from the pattern obtained upon overexpression of the wild-type protein. This was expected given the diverse mechanisms by which SRSF1 exerts its splicing functions and the multiple SRSF1 binding sites present within the viral genome.
Our data indicate that SRSF1 exerts pleiotropic effects on the regulation of the viral genome and that its RBDs can inhibit viral replication by presumably competing with other cellular and viral modulators of RNA biogenesis.
SRSF1 RBDs inhibit divergent viral isolates.
The primary sequence of HIV-1 is highly variable among single isolates. Multiple viral strains with different geographical distributions have been identified. Based on genetic similarities, the numerous virus strains are classified into types, groups, and subtypes or clades. More than 90% of HIV-1 infections belong to HIV-1 group M. Within group M there are known to be at least nine genetically distinct subtypes. Subtype B is the most common subtype in Europe, the Americas, and Japan, subtype C is predominant in Southern and East Africa and India and subtype D is limited to East and Central Africa.
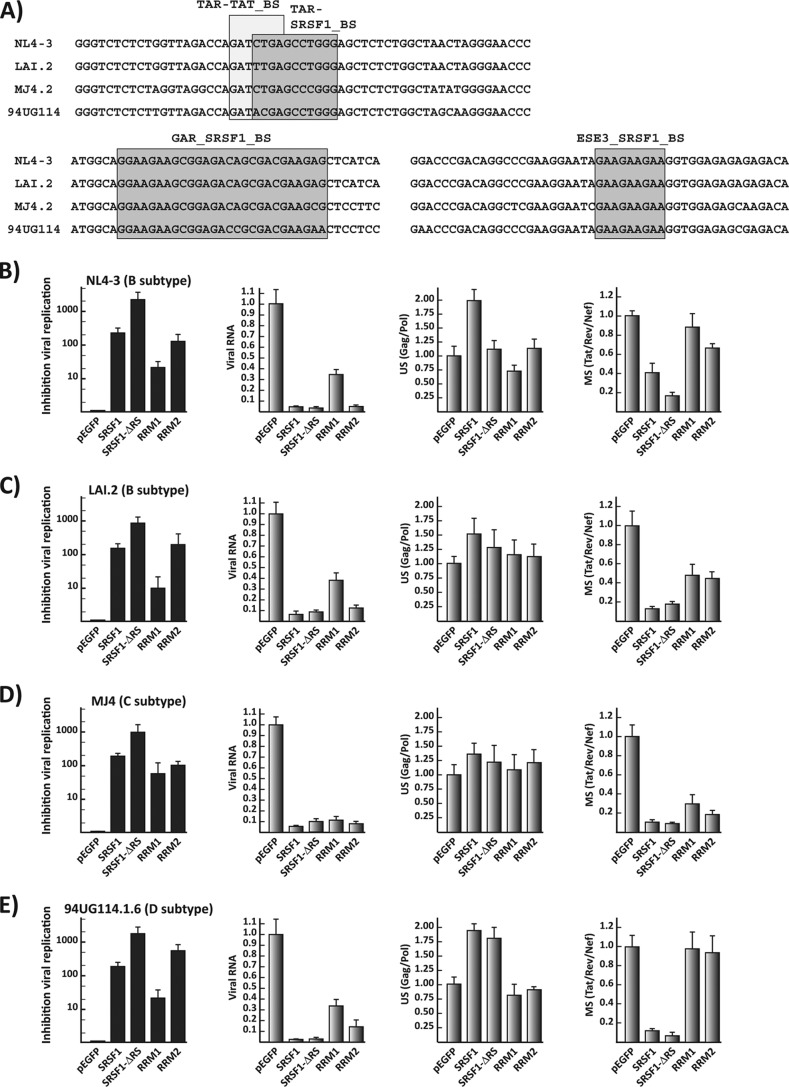
Our studies utilize molecular clones derived from early isolates of the B subtype. Since SRSF1 inhibition of HIV replication is dependent on its binding onto multiple sequences within the viral transcript, its activity could greatly vary among different viral strains. Computational analysis of the putative SRSF1 binding sites in the four viral isolates showed multiple potential targets for this cellular factor (Fig. 2B). Furthermore, comparison of the binding sites, which have been previously experimentally characterized, revealed a high degree of homology among the viral isolates (Fig. 3A).
FIG 3.
SRSF1 inhibition of replication in divergent viral subtypes. (A) Primary sequences alignments of the characterized SRSF1 binding sites in molecular clones from the B (NL4.3 and LAI.2), C (MJ4), and D (94UG114.1.6) viral subtypes. (B to E) Quantification of viral replication and viral mRNAs for the four viral molecular clones after expression of the SRSF1 constructs. Replication for each viral clone was assayed as described in Fig. 2C. The relative amount of viral RNA was quantified as described in Fig. 2D. The relative amounts of gag/pol and multiple spliced mRNAs were quantified as described in Fig. 2E, normalized for the total viral mRNA content of each sample, and expressed as the fold increase or decrease versus the control pEGFP transfection. All data are represented as means ± the SEM.
To study the effect of SRSF1 on isolates from the B, C, and D subtypes, we analyzed the production of these viruses in HEK293 cells cotransfected with the molecular clones of each viral isolate and plasmids coding for SRSF1 and its deletion mutants. Overexpression of SRSF1 inhibited viral production by at least 100-fold in all of the viral isolates (Fig. 3B to E). Consistent with the results obtained with the NL4-3 isolate, the strongest inhibitor of viral production was the mutant carrying the RS deletion (>1,000-fold in all isolates), while the inhibition achieved by the clone expressing the RRM2 alone was comparable to the one achieved by the full-length protein. Analysis of the viral transcripts among the different isolates indicated that the intracellular level of all viral messengers were similarly impacted by the SRSF1 wild-type sequence and the RS deletion mutant. Expression of the single RRMs leads to more distinctive differences in the relative amounts of unspliced and multiple spliced messengers among the isolates (Fig. 3B to E).
Given the heterogeneity of the sequences recognized by SRSF1 and the different roles played by the single domains in determining its RNA-binding specificity (28, 33), we expected substantial differences in the total and relative amounts of viral mRNAs generated by divergent viruses upon expression of each SRSF1 mutant. Nevertheless, the synthesis and processing of viral mRNAs was severely disrupted in all of the molecular clones tested, and the production of all three viral subtypes analyzed was strongly inhibited by SRSF1 and its single RBD.
The SRSF1 RBDs have minimal impact on cell viability and apoptosis.
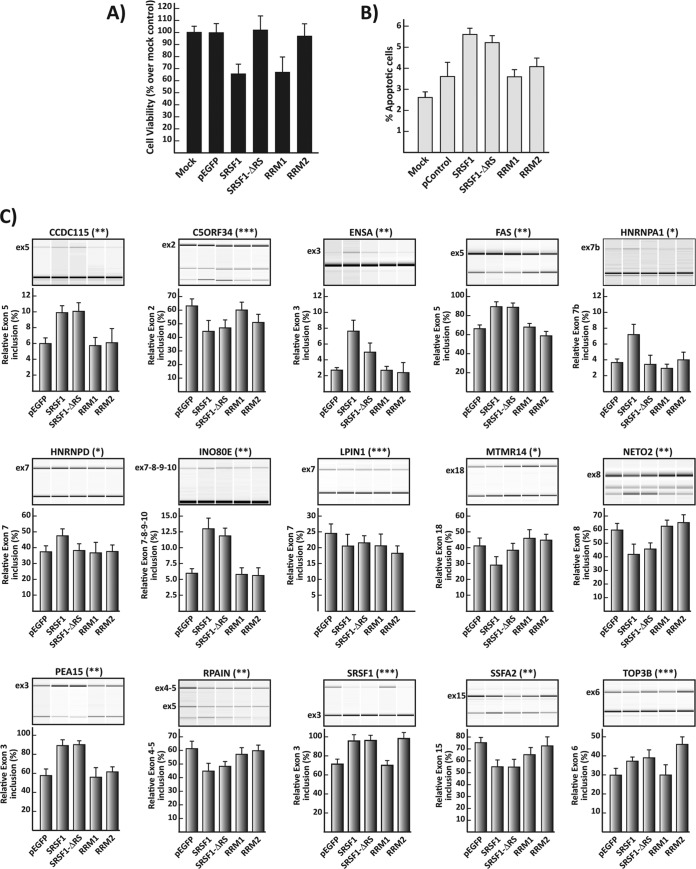
The strong inhibition of viral production observed upon overexpression of SRSF1 suggests a therapeutic potential for this protein. Nevertheless, SRSF1 is an essential gene, which is associated with disease and neoplastic transformation. Thus, it is plausible that its overexpression will induce major changes in cell metabolism, which can result in altered cell viability, proliferation, morphology, and apoptosis, making it a less likely therapeutic candidate. Our data also show that the SRSF1 RRMs can inhibit viral production with an activity comparable (RRM2) or higher (RRM1+2) to the full-length protein. Given the multiple mechanisms utilized by this protein to exert its cellular functions, it is conceivable that the single RRM might impact cell metabolism less severely than the wild-type sequence. To explore the effects that these deletion mutants have on cell viability and apoptosis, we analyzed both parameters in HEK293 cells transduced with SRSF1 and its deletion mutants. Cell viability was reduced by >30% upon overexpression of the wild-type SRSF1 and single RRM1 (Fig. 4A). Surprisingly, deletion mutants carrying RRM1+2 (ΔRS) or RRM2 alone did not significantly impact cell viability. Apoptotic events increased upon transfection of the wild-type and RS domain deletion clones but not the ones expressing the single RRM1 and RRM2 compared to the control EGFP vector (Fig. 4B).
FIG 4.
Effects on cellular viability, apoptosis, and splicing of SRSF1 deletion mutants. (A) Cell viability was measured by quantifying cellular ATP production at 72 h posttransfection. The viability of the cells transfected with the SRSF1 clones is given relative to that of the mock-transfected control. (B) Apoptotic events in transfected cells. Apoptosis in HEK293 cells transfected with the control and SRSF1 vectors was detected by visualizing the active apoptotic marker caspase 9 at 72 h posttransfection. (C) RT-PCR analysis of SRSF1 target genes. Primers located in the exons flanking the alternatively spliced exon allow amplification of isoforms either including or excluding the regulated exon. For each gene, we show the graphic output of the analysis (top) and the relative level of exon inclusion (bottom) in cells expressing the indicated SRSF1 or control plasmid 48 h after transfection. The splicing of three genes is responsive solely for the expression of the full-length SRSF1 (*). Eight genes respond to both the wild type and the ΔRS clone (**); four genes respond to the wild type, the ΔRS clone, and the RRM2 clone (***). All data are represented as means ± the SEM.
The relevance of this protein's role as a global regulator of transcription, mRNA stability, nuclear export, and translation is still unclear. On the contrary, its pervasive role in mRNA splicing regulation has been widely studied, and it is crucial for cell survival, development, and replication. To determine whether the deletion clones were differently affecting the splicing of cellular genes, we selected a panel of 15 genes, whose splicing is regulated by SRSF1 (28, 34), and we measured the inclusion of specific exons. Only 4 of the genes analyzed were similarly regulated by the wild type and the deletion mutants (Fig. 4C). Three of the genes surveyed were regulated solely by the wild-type SRSF1, whereas eight genes were regulated by both the wild type and the RS deletion mutant. Similar results, indicating marked differences in the splicing pattern generated by each SRSF1 mutant, had been previously obtained surveying a series of genes involved in mammary epithelial cell transformation (34). Taken together, these data indicate that the SRSF1 RRM domains can inhibit viral replication with a reduced impact on cellular metabolism.
SRSF1 effects on viral replication, cell viability, and apoptosis are dose dependent.
Next, we sought to determine the minimal amount of wild-type or truncated mutant SRSF1 plasmid that could be transfected to maintain the inhibition of HIV-1 replication and minimize the effects on cellular viability and apoptosis. We analyzed the dose-dependent response on the inhibition of HIV-1 virion production (Fig. 5A), cell viability (Fig. 5B), and apoptotic events (Fig. 5C) upon transfection of increasing amounts of the SRSF1 plasmids. A reduction by 50% of the input plasmids (Fig. 5D) abolished the effects on cell viability and apoptosis for the RS deletion and single RRM2 clones, while maintaining the maximum inhibition of viral production. Cell viability and apoptosis were reduced to control levels when the input of the plasmid coding for the wild-type SRSF1 was decreased by 8-fold, this reduced the inhibition of viral production to roughly 20-fold compared to the >200-fold reduction observed at the higher DNA input.
FIG 5.
Dose-dependent response to SRSF1 expression. (A) Inhibition of viral replication was assayed as described in Fig. 2C. The proviral clone pNL4-3 was cotransfected with the indicated amount of each SRSF1 plasmid. Cell viability (B) and apoptotic events (C) in cells transfected with increasing amounts of SRSF1, deletion mutants, and control plasmid DNA was measured at 72 h after transfection. All data are represented as means ± the SEM.
SRSF1 inhibits HIV replication in an in vitro infection system.
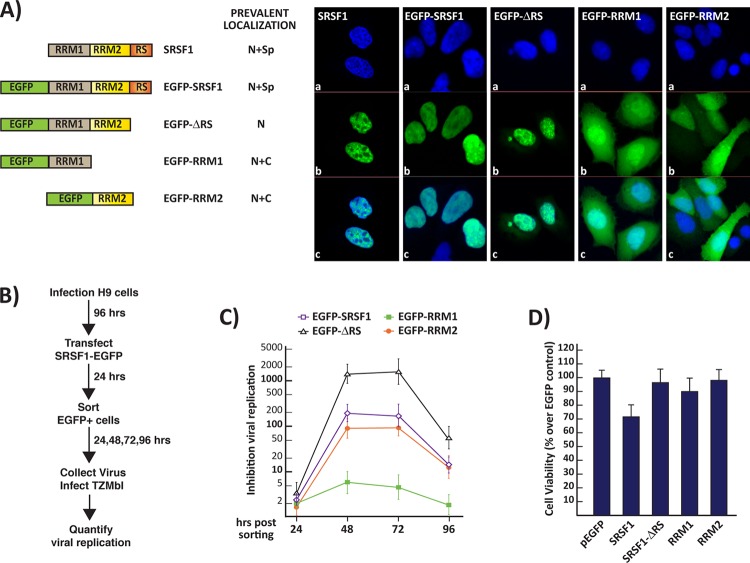
To show that SRSF1 can inhibit viral replication in infected cells that carry an integrated copy of the viral genome, we utilized the leukocyte-derived H9 cell line, which can be easily infected with a number of viral strains and allows for efficient viral replication. H9 cells were infected with viral particles (NL4-3 clone) at a high MOI (≥100) and grown for 4 days to ensure the homogenous infection of the cell population. The infected cells were transiently transfected utilizing EGFP-tagged SRSF1 clones. The EGFP-tagged proteins exhibited cellular localization similar to the untagged ones, as previously reported (19) (Fig. 6A). Fluorescence-activated cell sorting was utilized to select EGFP-expressing cells.
FIG 6.
(A) Localization of endogenous SRSF1 and EGFP-tagged clones. (a) Nuclear DAPI (4′,6′-diamidino-2-phenylindole) staining. (b) Endogenous SRSF1 was detected by immunofluorescence while tagged SRSF1-EGFP clones were detected by fluorescence microscopy. (c) Merged panel a and b images show localization of the endogenous and tagged proteins within the nuclei and cytosol. (B) Viral replication assay. H9 cells were infected with NL4-3 virus at a high MOI (≥100). The infected cells were then transfected with the SRSF1-EGFP clones, sorted for EGFP expression, and assayed for viral replication in the following days, as summarized in the schematics on the left. Inhibition of viral replication was measured as described as described in Fig. 2C for four consecutive days following cell sorting. (C) Cell viability of the infected cells transfected with the EGFP-tagged SRSF1 clones was measured by quantifying cellular ATP production 5 days posttransfection. (D) Viability of the cells transfected with the SRSF1-EGFP clones is expressed relative to that of the EGFP-transfected control. All data are represented as means ± the SEM.
Post-sorting, the virus-containing media from the infected cells expressing the SRSF1 and control EGFP proteins were collected at 24-h intervals and utilized to quantify viral production. At 48 h after transfection of the SRSF1 expression clones, viral replication was reduced to the levels observed in the viral production assay carried out in HEK293 cells (Fig. 6C). At day 4 post-sorting, the inhibitory effect of the transfected SRSF1 clones was considerably diminished; this is likely explained by the progressive loss of the epigenic expression plasmid.
Next, we compared the viability of EGFP control and SRSF1-transfected but not infected H9 cells at day 5 posttransfection. Similarly to the results obtained in HEK293 cells, the wild-type SRSF1 sequence induced a reduction in cell viability, while the deletion clones did not. These data show that viral replication can be inhibited efficiently by expressing the truncated SRSF1 mutants carrying the RRM2 domain either alone (200-fold inhibition) or in combination with the RRM1 domain (>2,000-fold inhibition) without significantly impact cell viability in a stable cell line, suggesting a therapeutic potential for these RBDs.
DISCUSSION
The mammalian splicing factor SRSF1 exerts pleiotropic effects on cellular genes (13, 14). In our previous work, we showed that this RBP plays a role in the regulation of transactivation of the viral promoter by competing with the viral protein Tat (11) and in the splicing regulation of the viral transcripts (3, 16). In the present study, we show that overexpression of SRSF1 can inhibit viral replication by over 200-fold by reducing the activation of the viral promoter and deregulating the splicing of the viral transcripts. We also observed that the SRSF1 RRM1+2 or RRM2 alone can also efficiently downregulate viral replication. Surprisingly, the decrease in viral replication obtained by expressing RRM1+2 was increased by a factor of 10 compared to the one obtained by overexpressing the wild-type protein (>2,000 versus 200-fold, respectively), while the single RRM2 domain was sufficient to reduce viral replication to the level observed with the full-length protein. This may be due to the role played by the RS domain in promoting the association of SRSF1 with nuclear speckles and spliceosomal complexes (19). The proteins lacking the RS domain are unlikely to be sequestered within these nucleoplasmic complexes, making them available to bind key sequences within the viral transcript inhibiting its transcription and splicing.
The current treatment of HIV-1 is mostly based on compounds that directly target the activities of proteins encoded by the virus. However, these proteins mutate rapidly, generating novel drug-resistant viral strains. There is a need for novel therapeutics with mechanisms of action that differ from the ones currently on the market. Transcription and cellular factors regulating HIV RNA processing are expressed in most cell types and regulate a multitude of cellular events, making them less ideal therapeutic target candidates. Nevertheless, small molecules that inhibit the splicing activity of the splicing factors SR proteins have been shown to efficiently repress viral replication in peripheral blood mononuclear cells with little cell toxicity (2).
We have shown that although SRSF1 significantly affected cell viability and apoptosis, RRM1+2 or RRM2 alone had a minimal impact on cell viability while retaining an extremely potent antiviral activity in divergent viral isolates. These data suggest that the SRSF1 RRMs have a strong therapeutic potential.
SRSF1 antiviral activity is a result of the protein's affinity for specific RNA sequences and the inhibition of the transcription and processing of the viral messenger. The RNA-binding specificity of SRSF1 appears to be mostly dependent on RRM2, since it interacts with the target RNA with efficiency similar to the one for RRM1 and RRM2 combined (28). Our data indicate that inhibition of viral replication differs greatly upon the expression of RRM2 and RRM1+2 (∼200-fold inhibition versus >2,000-fold inhibition); thus, factors other than the RNA-binding specificity of these subdomains may account for their antiviral activity.
SRSF1 is a shuttling protein primarily localized within the nucleus and associated with spliceosomal components in subnuclear structures named nuclear speckles (35). The SRSF1 clone containing RRM1+2 (ΔRS) exhibits decreased nuclear and nuclear speckle localization (Fig. 6A). The deletion mutant with the single RRM2 domain shows a diffuse cellular localization with no evident association with speckles. This suggests that the different efficiency in antiviral activity observed upon expression of the RRM1+2 compared to the RRM2 alone is likely due to the differences in intracellular localization of the two deletion mutants.
To analyze the effect of SRSF1 on viral production, we cotransfected plasmids coding for full-length replication competent HIV-1 molecular clones and SRSF1 coding plasmids in HEK293 cells. This cell line lacks the receptor for HIV-1 and cannot be directly infected. Nevertheless, transfection is highly efficient (>98%), thus ensuring that the virus and the SRSF1 coding plasmids are both present in majority of the cell population, yielding extremely robust and reliable data.
Although the viral genome is expressed from the nonintegrated plasmid, it reliably mimics the expression of the viral genes observed in activated CD4+ T cells, the main HIV-1 target. Furthermore, we confirmed the inhibition of viral replication in an in vitro viral infection-replication assay utilizing the leukocyte derived H9 cell line. To evaluate the therapeutic potential that the SRSF1 protein domains might have in a system that more closely mimics the natural course of the viral infection, future studies will be aimed at the delivery of the deletion mutants lacking the RS domain in primary CD4+ T cells.
Supplementary Material
ACKNOWLEDGMENTS
We thank Adrian Krainer for providing the SRSF1 expression clones and Michelle Caputi for the manuscript editing.
This study was supported by NIH/NIAID grant R15 AI093229 to M.C. The BMG FluoStar Omega reader was acquired with funds provided from the Charles E. Schmidt Foundation.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00693-15.
REFERENCES
- 1.Karn J, Stoltzfus CM. 2012. Transcriptional and posttranscriptional regulation of HIV-1 gene expression. Cold Spring Harb Perspect Med 2:a006916. doi: 10.1101/cshperspect.a006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakkour N, Lin YL, Maire S, Ayadi L, Mahuteau-Betzer F, Nguyen CH, Mettling C, Portales P, Grierson D, Chabot B, Jeanteur P, Branlant C, Corbeau P, Tazi J. 2007. Small-molecule inhibition of HIV pre-mRNA splicing as a novel antiretroviral therapy to overcome drug resistance. PLoS Pathog 3:e159. doi: 10.1371/journal.ppat.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jablonski JA, Caputi M. 2009. Role of cellular RNA processing factors in human immunodeficiency virus type 1 mRNA metabolism, replication, and infectivity. J Virol 83:981–992. doi: 10.1128/JVI.01801-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkhout B, Silverman RH, Jeang KT. 1989. Tat transactivates the human immunodeficiency virus through a nascent RNA target. Cell 59:273–282. doi: 10.1016/0092-8674(89)90289-4. [DOI] [PubMed] [Google Scholar]
- 5.Gaynor RB. 1995. Regulation of HIV-1 gene expression by the transactivator protein Tat. Curr Top Microbiol Immunol 193:51–77. [DOI] [PubMed] [Google Scholar]
- 6.Fujinaga K, Irwin D, Huang Y, Taube R, Kurosu T, Peterlin BM. 2004. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol Cell Biol 24:787–795. doi: 10.1128/MCB.24.2.787-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H. 1998. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J 17:7395–7403. doi: 10.1093/emboj/17.24.7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peterlin BM, Price DH. 2006. Controlling the elongation phase of transcription with P-TEFb. Mol Cell 23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 9.Bres V, Yoh SM, Jones KA. 2008. The multi-tasking P-TEFb complex. Curr Opin Cell Biol 20:334–340. doi: 10.1016/j.ceb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Purcell DFJ, Martin MA. 1993. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication and infectivity. J Virol 67:6365–6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paz S, Krainer AR, Caputi M. 2014. HIV-1 transcription is regulated by splicing factor SRSF1. Nucleic Acids Res 42:13812–13823. doi: 10.1093/nar/gku1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin S, Fu XD. 2007. SR proteins and related factors in alternative splicing. Adv Exp Med Biol 623:107–122. doi: 10.1007/978-0-387-77374-2_7. [DOI] [PubMed] [Google Scholar]
- 13.Zhong XY, Wang P, Han J, Rosenfeld MG, Fu XD. 2009. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol Cell 35:1–10. doi: 10.1016/j.molcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ji X, Zhou Y, Pandit S, Huang J, Li H, Lin CY, Xiao R, Burge CB, Fu XD. 2013. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell 153:855–868. doi: 10.1016/j.cell.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. 2008. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol 15:819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caputi M, Freund M, Kammler S, Asang C, Schaal H. 2004. A bidirectional SF2/ASF- and SRp40-dependent splicing enhancer regulates human immunodeficiency virus type 1 rev, env, vpu, and nef gene expression. J Virol 78:6517–6526. doi: 10.1128/JVI.78.12.6517-6526.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tange TO, Kjems J. 2001. SF2/ASF binds to a splicing enhancer in the third HIV-1 tat exon and stimulates U2AF binding independently of the RS domain. J Mol Biol 312:649–662. doi: 10.1006/jmbi.2001.4971. [DOI] [PubMed] [Google Scholar]
- 18.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol 59:284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caceres JF, Misteli T, Screaton GR, Spector DL, Krainer AR. 1997. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J Cell Biol 138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sadaie MR, Benter T, Wong-Staal F. 1988. Site-directed mutagenesis of two trans-regulatory genes (tat-III,trs) of HIV-1. Science 239:910–913. doi: 10.1126/science.3277284. [DOI] [PubMed] [Google Scholar]
- 21.Peden K, Emerman M, Montagnier L. 1991. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology 185:661–672. doi: 10.1016/0042-6822(91)90537-L. [DOI] [PubMed] [Google Scholar]
- 22.Ndung'u T, Renjifo B, Essex M. 2001. Construction and analysis of an infectious human immunodeficiency virus type 1 subtype C molecular clone. J Virol 75:4964–4972. doi: 10.1128/JVI.75.11.4964-4972.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao F, Robertson DL, Carruthers CD, Li Y, Bailes E, Kostrikis LG, Salminen MO, Bibollet-Ruche F, Peeters M, Ho DD, Shaw GM, Sharp PM, Hahn BH. 1998. An isolate of human immunodeficiency virus type 1 originally classified as subtype I represents a complex mosaic comprising three different group M subtypes (A, G, and I). J Virol 72:10234–10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother 46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fong N, Bentley DL. 2001. Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: different functions for different segments of the CTD. Genes Dev 15:1783–1795. doi: 10.1101/gad.889101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pandit RA, Svasti S, Sripichai O, Munkongdee T, Triwitayakorn K, Winichagoon P, Fucharoen S, Peerapittayamongkol C. 2008. Association of SNP in exon 1 of HBS1L with hemoglobin F level in β0-thalassemia/hemoglobin E. Int J Hematol 88:357–361. doi: 10.1007/s12185-008-0167-3. [DOI] [PubMed] [Google Scholar]
- 27.Long JC, Caceres JF. 2009. The SR protein family of splicing factors: master regulators of gene expression. Biochem J 417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 28.Clery A, Sinha R, Anczukow O, Corrionero A, Moursy A, Daubner GM, Valcarcel J, Krainer AR, Allain FH. 2013. Isolated pseudo-RNA-recognition motifs of SR proteins can regulate splicing using a noncanonical mode of RNA recognition. Proc Natl Acad Sci U S A 110:E2802–E2811. doi: 10.1073/pnas.1303445110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. 2003. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res 31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemaire R, Winne A, Sarkissian M, Lafyatis R. 1999. SF2 and SRp55 regulation of CD45 exon 4 skipping during T cell activation. Eur J Immunol 29:823–837. doi:. [DOI] [PubMed] [Google Scholar]
- 31.Jiang ZH, Zhang WJ, Rao Y, Wu JY. 1998. Regulation of Ich-1 pre-mRNA alternative splicing and apoptosis by mammalian splicing factors. Proc Natl Acad Sci U S A 95:9155–9160. doi: 10.1073/pnas.95.16.9155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu X, Yang D, Ding JH, Wang W, Chu PH, Dalton ND, Wang HY, Bermingham JR Jr, Ye Z, Liu F, Rosenfeld MG, Manley JL, Ross J Jr, Chen J, Xiao RP, Cheng H, Fu XD. 2005. ASF/SF2-regulated CaMKIIδ alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell 120:59–72. doi: 10.1016/j.cell.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 33.Chandler SD, Mayeda A, Yeakley JM, Krainer AR, Fu XD. 1997. RNA splicing specificity determined by the coordinated action of RNA recognition motifs in SR proteins. Proc Natl Acad Sci U S A 94:3596–3601. doi: 10.1073/pnas.94.8.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anczukow O, Rosenberg AZ, Akerman M, Das S, Zhan LX, Karni R, Muthuswamy SK, Krainer AR. 2012. The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat Struct Mol Biol 19:220–228. doi: 10.1038/nsmb.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Misteli T, Caceres JF, Spector DL. 1997. The dynamics of a pre-mRNA splicing factor in living cells. Nature 387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.