Abstract
Background
Tumour necrosis factor (TNF) is central to the immune response to Plasmodium infection. Its plasma concentration is influenced by allele variants in the promoter region of TNF. The study’s objectives were to assess TNF allele variants (TNF−1031, TNF−308): (1) modulation of malaria rates in young Tanzanian children; (2) modulation of the severity of malaria as indicated by haemoglobin concentrations at the time of presentation with febrile episodes; and (3) the association between Plasmodium infection and haemoglobin concentration in symptomless parasite carriers.
Methods
Data from a placebo-controlled trial in which 612 Tanzanian children aged 6–60 months with height-for-age z-score in the range −3 SD to 1.5 SD was utilised. Those with Plasmodium infection at baseline were treated with artemether-lumefantrine. An episode of malaria was predefined as current Plasmodium infection with an inflammatory response (axillary temperature ≥37.5°C or whole blood C-reactive protein concentration ≥8 mg/L) in children reported sick. Linkage disequilibrium (LD) pattern assessment as well as haplotype analysis was conducted using HAPLOVIEW. Cox regression models used in the primary analysis accounted for multiple episodes per child.
Results
Genotyping of 94.9% (581/612) children for TNF−1031 (TNF−1031T>C); allele frequency was 0.39. Corresponding values for rs1800629 (TNF−308G>A) were 95.4% (584/612) and 0.17. Compared to the wild type genotype (TT), malaria rates were increased in the TNF−1031CC genotype (hazard ratio, HR [95% CI]: 1.41 [1.01‒1.97] and 1.31 [0.97‒1.76] for crude analysis and adjusting for pre-specified baseline factors, respectively) but decreased in those with the TNF−308AA genotype (corresponding HR: 0.13 [0.02‒0.63] and 0.16 [0.04‒0.67]). These associations were weaker when analysing first episodes of malaria (P value −0.59 and 0.38, respectively). No evidence that allele variants of TNF−1031 and TNF−308 affected haemoglobin concentration at first episode of malaria, or that they modified the association between Plasmodium infection and haemoglobin concentrations at baseline was observed.
Conclusion
In this cohort of Tanzanian children, the TNF−1031CC genotype was associated with increased rates of malarial episodes, whereas the TNF−308AA genotype was associated with decreased rates.
Keywords: Tumour necrosis factor, Malaria, Plasmodium infection
Background
Tumour necrosis factor (TNF) is central to the immune response to Plasmodium infection because of its pyrogenic properties and its key role in triggering the cascade of pro-inflammatory cytokines that regulate immune cells. It is secreted predominantly by activated macrophages following exposure to Plasmodium antigens, and acts to suppress parasitaemia.
Eight variants have been described within the TNF promoter at positions 1031T>C (rs1799964), −863C>A (rs1800630), −857C>T (rs1799724), −575G>A, −376G>A (rs1800750), −308G>A (rs1800629), −244G>A, and −238G>A (rs361525) relative to the transcription start site [1–7]. These genetic variants are investigated due to the possibility that the genetic changes they introduce, in comparison to the wild type form, could affect the binding of transcription factors, thereby controlling the activity of the promoter sites and affecting resulting mRNA and protein concentrations. There are many studies showing association of TNF variants with various infectious diseases, but others failed to show an association [8].
In relation to malaria, the TNF−1031C allele has been associated with decreased risk of severe malaria in a child cohort of Gambian malaria patients but this observation was not made in similar cohorts from Kenya and Malawi [9]. Also observations from a case–control study in India indicate that the TNF−1031C allele is associated with elevated plasma TNF concentrations and increased susceptibility to severe falciparum malaria [10]. TNF−308AA has been associated with severe cerebral malaria in Gambian children, and with a seven-fold increased risk of death and severe cerebral malaria [11]. Furthermore, a study of Gabon school children observed the TNF−308A allele to be associated with shorter intervals of malaria reinfection [12]. A Kenyan study investigating malarial morbidity in young children found this variant to be associated with preterm birth and early childhood mortality and malaria morbidity, with carriers showing a trend towards high density parasitaemia and severe malarial anaemia [13]. Another study from Sri Lanka found that the TNF−308AA was associated with both severe malaria and other infections [14]. Some studies however have failed to observe any association between TNF−308AA and malaria. A study of malaria patients from Thailand found no association between TNF−308AA and disease severity. This finding is similar to that of Ubalee et al. [15], which did not observe a difference in severe symptoms between patients in Myanmar on the basis of TNF−308AA [15]. Another study of Tanzanian infants observed minor clinical parameters to differ in TNF−308GA heterozygotes but did not observe major differences in other malaria related indices [16].
There have been few cohort studies to longitudinally assess the association of TNF allele variants on malaria outcomes. Children with an elevated TNF response to Plasmodium infection are more likely to become sicker than their peers with a lower TNF response capacity, so that the former may experience more frequent episodes of malaria. On the other hand, in children exposed to chronic or repeated Plasmodium infections, an increased TNF response associated with the TNF−308AA genotype may intensify a hepcidin-mediated block in iron absorption and consequently induce or exacerbate iron deficiency [17], which probably protects against malarial episodes [18, 19].
The aim of this study was therefore to assess the associations of TNF−1031 and TNF−308 allele variants with malaria rates in young Tanzanian children. Additionally, the study aimed to investigate the association of these variants on the severity of malaria as indicated by haemoglobin concentrations at the time of presentation with febrile episodes. Lastly, how these variants modulated the association between Plasmodium infection and haemoglobin concentration in symptomless parasite carriers was assessed.
Methods
Study area and population
Data from a randomized trial that aimed to assess the effect of supplementation with zinc and other micronutrients on malaria rates was used. The trial was conducted between February 2008 and March 2009 in four rural villages in Handeni District, northeastern Tanzania. Residents primarily belong to the Wazigua and Wabondei Bantu tribes, but settlement of migrant plantation workers has resulted in a mixture of tribes with different origins and much intermarriage. The area is mainly populated by poor farmer families involved in subsistence farming. Malaria transmission is intense and perennial, with nearly all infections being due to Plasmodium falciparum [20]. Apart from several local traditional healers, the research dispensary was the only health facility in the area. The study was approved by the Ethical Review Committee of Wageningen University, The Netherlands (Approval number: NL: 04/07) and the National Health Research Ethics Review sub-Committee, Dar es Salaam, Tanzania (Approval number: NIMR/HQ/R.8a/Vol. IX/540). Informed consent was obtained from community leaders, local government officials and parents or guardians. Further details of this trial are described elsewhere [21].
Recruitment
All resident children were eligible for randomization when aged 6–59 months and with a height-for-age z-score in the range −3 SD to 1.5 SD. Children with haemoglobin concentration <70 g/L, signs of chronic illness, and those unlikely to remain permanently resident or comply with the supplementation for the duration of the trial, or whose parents or guardians declined consent, were excluded from the study. Venous blood samples were collected in EDTA tubes and centrifuged immediately. An aliquot of 90 μL erythrocyte sediment with the buffy coat was mixed with 90 μL phosphate-buffered saline and 180 μL of DNA stabilizing buffer (AS1; Qiagen, Hilden, Germany) and stored at 4°C for subsequent genotyping. Plasma samples were stored in liquid nitrogen in the field and at −80°C during transport and subsequent storage until biochemical analysis in The Netherlands. Haemoglobin concentration was measured in an aliquot of whole blood by a haematology analyser (Sysmex KX21, Kobe, Japan). Plasmodium infection was detected in fresh blood by rapid dipstick test (CareStart, Access Bio, Monmouth Jct, USA). Children with a positive test result were treated immediately with artemether-lumefantrine. The location of the child’s homestead was determined using a global positioning system. Further details about recruitment procedures are reported elsewhere [21].
Experimental intervention
Children were randomized within six strata defined by Plasmodium infection (binary) and age class (6–17 months, 18–35 months and 36–60 months) and randomly permuted blocks with size randomly selected of four or eight. They then received daily supplements with either zinc alone (10 mg as gluconate), multi-nutrients without zinc, zinc combined with multi-nutrients or placebo. Supplements, in the form of powder in colour-coded capsules, were contained in blister packs, and administered orally after suspending capsule contents in clean water or breastmilk. Supplementation was performed by local community volunteers, who reported adherence daily to field staff at the research dispensary.
Follow-up and case detection
A clinical officer was on duty at the research clinic day and night. At recruitment, parents or guardians were requested to bring participating children to the dispensary immediately when detecting fever or any other illness during the intervention period. In samples collected at baseline and from sick children, the presence of parasite-specific lactate dehydrogenase (P. falciparum and other Plasmodium species) was detected by rapid tests (CareStart, G0121, Access Bio, Monmouth Jct, NJ). Axillary temperature was measured using an electronic thermometer and dipstick tests were administered for children with guardian-reported fever; for those with positive test results, plasma samples were collected and measured whole-blood C-reactive protein concentrations using a point-of-care test. Plasma was stored as described for the recruitment procedure. Artemether-lumefantrine (Novartis Pharma, Basel, Switzerland) was administered to any child with current Plasmodium infection upon enrolment, or with reported fever and a positive dipstick test result during the follow-up period.
Laboratory procedures
For children who presented with malarial episodes, whole-blood concentrations of haemoglobin and C-reactive protein were measured using point-of-care tests (HemoCue, Ängelholm, Sweden and QuikRead, Orion Diagnostica, Espoo, Finland, respectively). Plasma concentrations of P. falciparum-specific histidine-rich protein-2 (HRP2) in samples collected during the first malaria episode were measured using a commercial enzyme-linked immunosorbent assay kit (Malaria Ag Celisa; Cellabs, Brookvale, NSW, Australia). Plasma concentrations of C-reactive protein were measured (Meander Medical Centre, Amersfoort, The Netherlands) on a Beckman Coulter Unicel DxC880i system according to the manufacturer’s instructions. Genotypes were determined using Illumina’s VeraCode™ GoldenGate Genotyping Assay on a BeadXpress™ platform. TNF variant allele clustering was assessed visually to determine success of genotyping. Further quality control cut-offs were: a GenCall Score of >0.5 and a call rate of ≥0.95 [22]. For the variant TNF−1031, the reference allele was T and the alternate C. Individuals homozygous for the TNF−1031 reference allele (T) are hereon described as wild type while those homozygous for the alternate allele (C) are referred to as homozygote mutant. For the variant TNF−308, the reference allele was G and the alternate was A. Individuals homozygous for the TNF−308 reference allele (G) are hereon described as wild type while those homozygous for the alternate allele (A) are referred to as homozygote mutant.
Statistical analyses
Linkage disequilibrium (LD) pattern assessment as well as haplotype analysis was conducted using HAPLOVIEW [23]. Anthropometric indices were calculated using Epi Info software [24] All analyses were performed using SPSS (v15.0 for Windows, SPSS, Chicago, IL, USA), CIA (v2.1.2) [25] and STATA (v11; College Station, TX, USA). For each TNF genotype, Fisher’s exact test was used to assess whether populations were in Hardy–Weinberg equilibrium. The differences in baseline characteristics were calculated using the homozygote wild-type group as the reference for each variant using CIA (v2.1.2). Normality of variables was assessed by visual inspection of histograms. Because variables of interest (age, haemoglobin concentrations, distance from homestead to dispensary and anthropometric indices) were normally distributed, means, SDs and 95% CIs were reported.
Association of TNF genotype with malaria rates
The primary outcome, an episode of malaria, was predefined as a positive result for either a pLDH or a HRP2 dipstick test with any of the following: (a) confirmed fever (axillary temperature ≥37.5°C as measured by electronic thermometer); or, (b) guardian-reported but unconfirmed 24-h history of fever in the presence of inflammation (whole blood C-reactive protein concentrations >8 mg/L), separated by at least 14 days from a previous malaria episode. Incidence per TNF genotype and incidence ratios based on time to first episodes, with wild-type homozygotes as reference group was calculated. In the primary analysis, group rates were compared using Cox regression with robust estimates of the standard error to account for multiple episodes within children. The extent to which supplementation with either zinc or multi-nutrients including iron modulated the magnitude of the association of TNF genotype on malaria rates was explored. The extent to which adjustment for baseline factors that were a priori expected to be prognostic for malaria (Plasmodium infection status, distance between homestead and clinic [continuous variable], height-for-age z-score [continuous variable], mosquito net use [binary variable]) and experimental intervention modulated the estimated association of genotypes were explored. In this adjusted analysis, experimental intervention as a binary variable indicating pooled groups receiving multi-nutrients (with or without zinc) and pooled groups receiving no multi-nutrients (with or without zinc) was included. Kaplan–Meier analysis with Peto tests were used to assess associations of TNF genotypes with time-to-first episode of malaria.
TNF genotype modulation of the association between Plasmodium falciparum infection and haemoglobin concentration at baseline
Since the additive genetic model was considered, for each genotype, two dummy variables were used to indicate the three classes, resulting in two interaction terms per variant. Regression models were then used to assess the modulation of genotypes on the associations between P. falciparum and haemoglobin concentration. Main terms for P. falciparum infection and genotype dummies were included, and adjusted as pre planned for age class, mosquito net use, height-for-age z-score, and distance between the child’s homestead and the dispensary in regression analyses.
TNF genotype modulation of the association between Plasmodium falciparum infection and haemoglobin concentration at first episode of malaria
The additive genetic model was considered so for each genotype, two dummy variables were used to indicate the three classes. TNF genotype modulation of the association between P. falciparum infection and haemoglobin concentration at first episode of malaria was then assessed by analysis of variance (ANOVA) and adjusted for age class, height-for-age z-score and distance between the child’s homestead and the dispensary and treatment group.
Results
Of 612 children recruited in the original study, 581 (94.9%) and 584 (95.4%) had DNA typed for TNF−1031 and TNF−308, respectively. Only 3% (20/612) of children failed to complete the trial. Minor allele frequencies were 0.39 (TNF−1031) and 0.17 (TNF−308), respectively. Neither of the TNF variants showed a departure from Hardy–Weinberg equilibrium (P = 0.90 and 0.57 for TNF−1031 and TNF−308, respectively). Additionally, the variants TNF−1031 is not in linkage disequilibrium with TNF−308 (r2 = 0.03). No haplotype blocks were identified.
Baseline factors associated with TNF genotypes
Children with TNF−1031C allele seemed to have larger weight-for-age z-scores than their peers with the wild type variant (0.14 SD and 0.41 SD in heterozygotes and homozygotes, respectively, compared to −0.18 SD in those with the wild type genotype) (Table 1). The TNF−308A allele was marginally significantly associated with increased Plasmodium infection, but the evidence was weak (p-value: 0.09). No evidence of other factors being associated with the TNF genotypes investigated was observed.
Table 1.
Factors associated with rs1799964 and rs1800629 allele variants of the TNF gene
| Allele variant/factor | Wild type | Heterozygote | Difference (95% CI) | Homozygote mutant | Difference (95% CI) | p-value |
|---|---|---|---|---|---|---|
| rs1799964 | −1031TT | −1031TC | −1031CC | |||
| n | 356 | 200 | 25 | |||
| Age (months) | 33.4 ± 15.5 | 31.9 ± 16.1 | −1.5 (−4.2,1.2) | 28.06 ± 14.9 | −5.3 (−11.6, 0.99) | 0.18 |
| Sex (%male) | 48.0% (171) | 48.0% (96) | 0.0% (−8.6%, 8.6%) | 44.0% (11) | −4.0% (−22.1%, 15.6%) | 0.93 |
| Haemoglobin concentration, g/L | 102.7 ± 12.3 | 103.4 ± 13.8 | 0.0 (−2.1,2.1) | 101.5 ± 10.4 | −1.2 (−6.2, 3.8) | 0.71 |
| Anaemia, % (n)a | 69.1% (246) | 66.0% (132) | −3.1% (−11.3%, 4.9%) | 76.0% (19) | 6.9% (−13.1%, 20.4%) | 0.50 |
| Plasmodium infection, % (n) | 43.8% (156) | 43.0% (86) | −0.8% (−9.3%, 7.8%) | 36.0% (9) | −7.8% (−24.4%, 12.3%) | 0.75 |
| Inflammation, % (n)b | 32.3% (114) | 32.0% (64) | 0.0% (−7.9%, 8.2%) | 32.0% (8) | −0.3% (−15.6%, 19.8%) | 0.58 |
| Mosquito net use, % (n)c | 32.9% (117) | 32.0% (64) | −0.1% (−8.8%, 7.3%) | 32.0% (8) | −0.9% (−16.5%, 19.3%) | 0.99 |
| Distance from homestead to dispensary, kmd | 3.5 ± 2.1 | 3.5 ± 2.2 | 0.0 (−0.4, 0.4) | 3.8 ± 2.6 | 0.3 (−0.6, 1.2) | 0.90 |
| Anthropometric indices | ||||||
| Height-for-age z-score | −2.45 ± 0.70 | −2.38 ± 0.69 | 0.07 (−0.05, 0.19) | −2.48 ± 0.78 | −0.03 (−0.32, 0.26) | 0.48 |
| Weight-for-height z-score | −0.18 ± 0.81 | −0.04 ± 0.85 | 0.14 (0.00, 0.28) | 0.23 ± 1.00 | 0.41 (0.08, 0.75) | 0.02 |
| Weight-for-age z-score | −1.63 ± 0.74 | −1.52 ± 0.75 | 0.11 (−0.02, 0.24) | −1.42 ± 0.76 | 0.21 (−0.09, 0.51) | 0.13 |
| rs1800629 | −308GG | −308GA | −308AA | |||
| n | 477 | 104 | 3 | |||
| Age | 32.1 ± 15.7 | 34.8 ± 15.3 | 2.7 (−0.6, 6.0) | 47.08 ± 11.7 | 15.0 (−2.9, 32.8) | 0.08 |
| Sex (% male) | 46.8% (223) | 53.8% (56) | 7% (−4%, 17%) | 66.7% (2) | 20% (−26%, 48%) | 0.34 |
| Haemoglobin concentration, g/L | 102.3 ± 12.8 | 105.2 ± 12.2 | 2.9 (0.2, 5.6) | 105.0 ± 19.2 | 2.7 (−11.9, 17.3) | 0.10 |
| Anaemia, % (n)a | 69.0% (329) | 64.4% (67) | −4.5% (−14.9%, 5.0%) | 66.7% (2) | −2.3% (−48.4%, 25.2%) | 0.54 |
| Plasmodium infection, % (n) | 41.1% (196) | 51.9% (54) | 10.8% (0.3%, 21.1%) | 66.7% (2) | 25.6% (−20.5%, 53.1%) | 0.09 |
| Inflammation, % (n)b | 31.0% (148) | 33.7% (35) | 2.6% (−6.8%, 12.9%) | 33.3% (1) | 2.3% (−25.2%, 48.4%) | 0.23 |
| Mosquito net use, % (n)c | 32.9% (157) | 30.8% (32) | −2.1% (−11.3%, 8.1%) | 33.3% (1) | 0.4% (−27.1%, 46.5%) | 0.95 |
| Distance from homestead to dispensary, kmd | 3.6 ± 2.2 | 3.6 ± 2.2 | 0.0 (−0.5, 0.5) | 3.5 ± 1.0 | −0.10 (−2.6, 2.4) | 1.00 |
| Anthropometric indices | ||||||
| Height-for-age z-score | −2.42 ± 0.69 | −2.47 ± 0.74 | −0.05 (−0.20, 0.10) | −2.69 ± 0.72 | −0.27 (−1.06, 0.52) | 0.61 |
| Weight-for-height z-score | −0.11 ± 0.84 | −0.17 ± 0.85 | −0.06 (−0.24, 0.12) | 0.07 ± 0.78 | 0.04 (−0.92, 1.00) | 0.73 |
| Weight-for-age z-score | −1.57 ± 0.75 | −1.64 ± 0.75 | −0.07 (−0.23, 0.09) | −1.56 ± 0.92 | 0.01 (−0.84, 0.86) | 0.72 |
Reference/wild type allele for TNF−1031 is T and TNF−308 is G. For both genotypes the reference/wild type allele is the common allele. p-values for the associations between each factor and the allele variants investigated. Mean ± SD or % [n] unless indicated otherwise % figures represent % with feature/% without feature (number with feature/number without feature). Difference is relative to the wild type group. The difference columns are for each variable a comparison of the heterozygotes and the homozygote mutants to the wild type.
aHaemoglobin concentration <110 g/L.
bPlasma C-reactive protein concentration ≥8 mg/L.
cData missing for 11 children.
dAs the crow flies, based measurements by global positioning system.
Associations of TNF genotype with malaria rates
Overall, there were 1,511 malaria episodes recorded in 400 child-years of observation (incidence: 3.8/child-year). Of 581 children, 489 children (84%) experienced at least one malaria episode while recurrent episodes occurred in 406 (69%) children (Table 2). Compared to the wild type genotype, malaria rates (all episodes) in homozygotes for the TNF−1031C allele (TNF−1031CC) were elevated by 41% (hazard ratio 1.41; crude analysis). In the analysis of first episodes, malaria rates also seemed elevated for this genotype (hazard ratio 1.17, Table 2), but the statistical evidence for such an association was much weaker (P = 0.59). Only one episode of malaria occurred in homozygotes for the TNF−308A allele (TNF−308AA). This genotype was nonetheless associated with a decrease in malaria rates by 87% (hazard ratio 0.13; all episodes). Malaria rates were similarly decreased in the analysis of first episodes, but the statistical evidence was weak (P = 0.38). Kaplan–Meier analysis did not indicate marked group-specific differences in malaria rates for specific time periods of follow-up for either of the two TNF genotypes investigated (Figure 1).
Table 2.
Associations between TNF genotype and rates of first and all episodes of malaria
| Homozygotes | Heterozygotes | Homozygote mutants | ||||
|---|---|---|---|---|---|---|
| rs1799964 | −1031TT | −1031TC | −1031CC | |||
| All episodes of malaria | ||||||
| n | 356 | 200 | 25 | |||
| Incidence | 2.93 | (907/309.1) | 2.92 | (502/171.9) | 4.15 | (90/21.7) |
| Hazard ratio, crude | 1.00 | Reference | 0.99 | [0.85–1.14] | 1.41 | [1.01–1.97] |
| Hazard ratio, adjusteda | 1.00 | Reference | 0.95 | [0.82–1.09] | 1.35 | [1.01–1.80] |
| First episode of malaria | ||||||
| Incidence | 2.85 | (296/104.0) | 3.09 | (168/54.4) | 3.29 | (20/6.07) |
| Incidence ratio | 1.00 | Reference | 1.08 | [0.89–1.31] | 1.15 | [0.68–1.76] |
| Hazard ratio, crude | 1.00 | Reference | 1.08 | [0.90–1.31] | 1.17 | [0.90–1.31] |
| Hazard ratio, adjusteda | 1.00 | Reference | 1.07 | [0.89–1.31] | 1.11 | [0.66–1.86] |
| rs1800629 | −308GG | −308GA | −308AA | |||
| All episodes of malaria | ||||||
| n | 477 | 104 | 3 | |||
| Incidence | 2.97 | (1216/409.2) | 3.08 | (282/91.5) | 0.36 | (1/2.8) |
| Hazard ratio, crude | 1.00 | Reference | 1.04 | [0.87–1.24] | 0.13 | [0.02–0.63] |
| Hazard ratio, adjusteda | 1.00 | Reference | 1.10 | [0.93–1.29] | 0.14 | [0.04–0.48] |
| First episode of malaria | ||||||
| Incidence | 2.89 | (392/135.6) | 3.27 | (91/27.8) | 0.53 | (1/1.9) |
| Incidence ratio | 1.00 | Reference | 1.13 | [0.90–1.44] | 0.18 | [0.00–0.98] |
| Hazard ratio, crude | 1.00 | Reference | 1.07 | [0.86–1.33] | 0.27 | [0.03–2.86] |
| Hazard ratio, adjusteda | 1.00 | Reference | 1.14 | [0.91–1.44] | 0.33 | [0.03–3.59] |
Values between brackets indicate (episodes/child-years observed) or [95% CIs].
aEstimates adjusted for baseline Plasmodium infection status, distance between homestead and clinic (continuous variable), height-for-age z-score (continuous variable), mosquito net use and experimental intervention. There was no evidence of interactions between genotype and experimental intervention.
Figure 1.
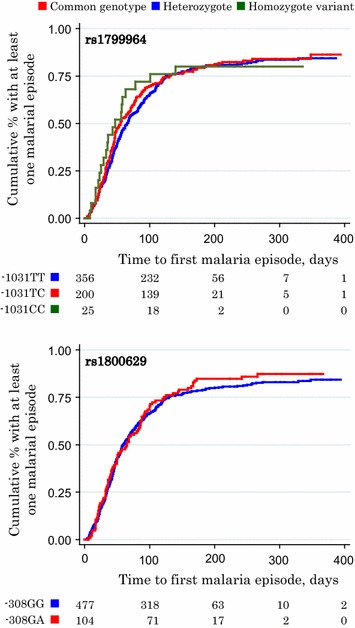
Associations of TNF genotype with time to first episode of malaria. Values indicate the number of children, by TNF genotype group, who had remained free of malaria at 0, 100, 200, 300, and 400 days of follow-up. For TNF −1031 (rs1799964) allele variant (top panel), pairwise group comparison with the wild type genotype as the reference group yielded p-values of 0.78 and 0.74 for heterozygotes rs1799964 TC and homozygotes (rs1799964 CC), respectively; for the TNF −308 allele variant (bottom panel), corresponding p-values for heterozygotes (TNF −308GA) and homozygotes (TNF −308AA) were 0.54 and 0.31, respectively (Peto tests). For homozygotes of the TNF −308A allele (TNF −308AA), the curve is not shown because only one episode occurred at 400 days of follow-up.
Associations of TNF genotype with haemoglobin concentration at first malarial episode
No evidence that haemoglobin concentrations at first episode of malaria varied by variants of TNF−1031 was observed (Figure 2).
Figure 2.
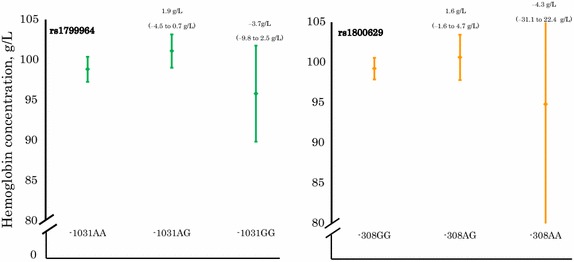
Haemoglobin concentration at first episode of malaria by TNF genotype. Left panel TNF −1031 (rs1799964) genotype; right panel TNF −308 (rs1800629) genotype. Genotypes are homozygous wild type, heterozygous and homozygous mutant from left to right. The data shown are means and their corresponding 95% CI. Values on top of bars indicate differences in group means with corresponding 95% CIs as obtained by multivariate regression analysis. All values are adjusted for Plasmodium infection status, distance between homestead and clinic, height-for-age z-score, mosquito net use, and experimental intervention. The 95% CI for haemoglobin concentration of the TNF −308AA genotype is outside the indicated range; this 95% CI is: 68.0–121.6 g/L.
TNF genotype modulation of the association between Plasmodium infection and haemoglobin concentration at baseline
No evidence that heterozygosity for either TNF−1031 or TNF−308 modified the association between Plasmodium infection at baseline and haemoglobin concentrations at baseline (p-values: 0.47 and 0.57, respectively) was observed. Similarly, no evidence that this association was modified by homozygosity for TNF−1031C (p-value = 0.56) was found (Figure 3).
Figure 3.
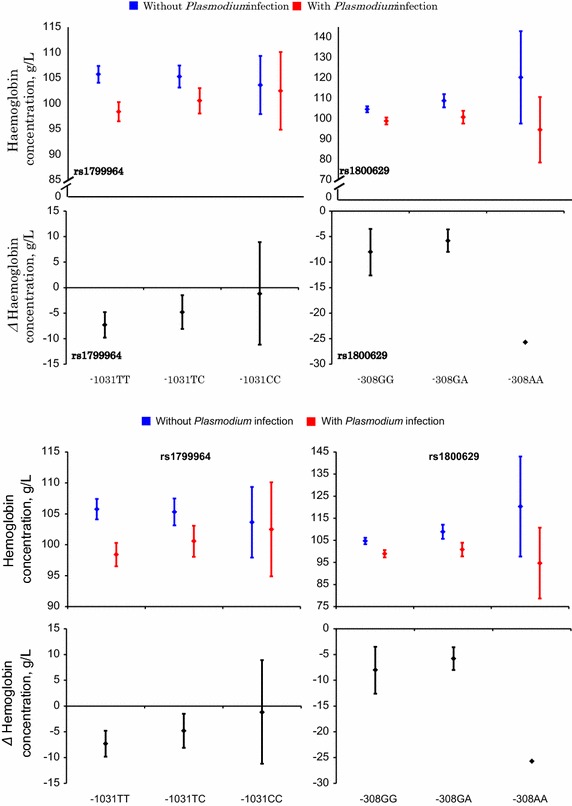
Plasmodium infection and haemoglobin concentrations at baseline, by TNF genotype. The data shown are means and their corresponding 95% CI. No confidence interval indicated for TNF −308AA because there was only one case of Plasmodium infection. All values are adjusted for age class, mosquito net use, height-for-age z-score, and distance from homestead. Red panels indicate haemoglobin concentration means and their corresponding 95% CI for those with Plasmodium infection while blue indicate haemoglobin concentration means and their corresponding 95% CI for those without Plasmodium infection. Black panels are for each genotype, difference in haemoglobin concentrations (means and 95% CI) between those with Plasmodium infection and those without Plasmodium infection.
Discussion
In this cohort of Tanzanian children, malaria rates due to P. falciparum were increased in homozygotes for the TNF−1031C allele (TNF−1031CC) and decreased in homozygotes for the TNF−308A allele (TNF−308AA). No evidence that allele variants of TNF−1031 and TNF−308 were associated with haemoglobin concentration at first episode of malaria, or that they modified the association between Plasmodium infection and haemoglobin concentrations at baseline was found.
The associations between TNF allele variants and malaria rates were generally more pronounced in the analysis of all episodes than in the analysis of first episodes (Table 2). The question arises how such differences should be interpreted. Some children experience malaria more frequently than others. This between-individual variation may be due to differences in exposure to infectious mosquito bites, immunity (and thus age) and therapy-seeking behaviour. In the presence of such heterogeneity in susceptibility to malaria, ‘high risk’ individuals tend to become sick more rapidly than ‘low risk’ individuals, and once they have experienced an episode they are no longer considered in a time-to-first-event analysis. With the passing of follow-up time, only the ‘low risk’ individuals remain in the risk set and, accordingly, the event rate observed will decrease. In cohort studies such as the one described herein, this decrease is generally more pronounced in groups with high overall event rates, which have ‘high-risk’ individuals dropping out more efficiently. In a time-to-first-event analysis, group contrasts can become underestimated and associations with malaria may appear decrease with time. For this reason, the analysis of all episodes reflects with more validity associations between TNF allele variants and malaria rates than analysis of first episodes. In addition, the former better reflects associations with the total population burden of disease.
There was no evidence that the association between TNF−1031CC genotype and malaria rates were confounded by Plasmodium infection status, distance between homestead and clinic, height-for-age z-score and mosquito net use factors assessed at baseline, or by the experimental intervention. Similarly, no evidence for such confounding on the association between TNF−308AA and malaria rates was found. These associations should nonetheless be interpreted with caution, because the statistical evidence was weak in the analysis of first episodes, even though the direction of these associations was similar.
The observation that TNF−1031CC genotype was associated with increased malaria rates whilst the TNF−308AA genotype was associated with a decrease in these rates may be due to the fact that these variants tag different casual variants. This is indicated by the fact the two variants are not in linkage disequilibrium as well as no haplotype blocks were identified. A haplotype analysis previously conducted observed that many variants in TNF are inept markers of each other to the point that if one variant in TNF was a true disease susceptibility locus, most of the other variants in TNF would appear neutral [26]. This suggests that the disease associations with TNF variants are independent of each other. It may be reasonable to expect that TNF variants, in weak linkage disequilibrium with each other, will also be in weak linkage disequilibrium with more distant variants in the central major histocompatibility class. This is because the strength of linkage disequilibrium is determined by among others the physical proximity of two mutations and proximity in time and lineage [27]. These factors are further modified by the effects of genetic drift, migration, and natural selection [28, 29]. So it is plausible that TNF variants, while inefficient markers of each other, may be good markers of variants in more distant genes that have different disease associations.
Findings described here add to the growing but conflicting evidence that allele variants of the TNF gene are associated with susceptibility to malaria [9, 30, 31]. Several factors can explain apparent discrepancies in results obtained thus far in various studies. First, studies conducted have varied in design. Most studies compared hospitalized cases of severe malaria with hospital- or community-based controls [9, 30, 32, 33]. Such studies can be implemented quickly and at a relatively low cost, but they are inherently vulnerable to selection bias if the distribution of TNF allele variants differs between cases and controls in the absence of a true association between these allele variants and malaria. As a consequence, for studies that recruited controls from a hospital setting, the strength of the association between TNF allele variants and malaria will be underestimated if these controls suffered from a condition that is associated with TNF genotype. This is plausible, because TNF genotypes have also been associated with a variety of disorders other than malaria [34–37]. A better approach is to select controls that are representative of the population that produced the cases. Clark et al. [9] selected as controls cord blood samples obtained from birth clinics in the same hospital from which they recruited cases of severe malaria. Even so, bias may still have occurred in that study if TNF genotypes varied in frequency of delivery in hospital, or in the frequency of care-seeking behaviour and hospital admission for severe malaria. Such differentials may have occurred, for example, if the geographical distribution of TNF genotypes in relation to the hospital is uneven, or if TNF genotypes vary in the risk of birth complications. In addition, it is difficult to rule out that the results of TNF genotyping performed by Clark et al. in cord blood may have been affected by maternal DNA originating from contamination of cord blood at birth by maternal cells or by contact with maternal blood or tissue [38]. In this cohort study, bias may have occurred if there was differential loss to follow-up such that the risk of being lost to follow-up was related both to malaria and TNF genotype, or if there were group differentials in the detection of malarial episodes. However, loss to follow-up was very low (3%), genotyping was successful for almost all (95%) children, and the study is believed to have captured virtually all malaria cases [39].
Secondly, both false positives (type I errors) and false negatives (type II errors) may occur especially among studies including relatively small numbers of subjects. Indeed our study had small numbers compared to studies that have previously been conducted to investigate these kind of associations. Due to this deficiency, this study may have had reduced chance of detecting a true effect due to low power. Furthermore, the likelihood that the statistically significant results reflect a true effect may be in question. To try and reduce the possibility of sample size affecting study conclusions we have avoided other possible sources of bias by having rigorous genotyping quality measures as well as diagnosis of P. falciparum malaria.
Thirdly, the relationship between TNF genotype and susceptibility to malaria may depend on population-specific factors. For example, as noted in the introduction, TNF allele variants that are associated with an enhanced TNF response to chronic or repeated infections can possibly protect against newly acquired Plasmodium infections through an altered iron metabolism. Such protection is conceivably more effective in children who already have marginal iron status than in their iron-replete peers, and in children who are frequently exposed to infection. There may also be population-specific masking of gene variant effects (epistasis) or gene–environment interactions specific to certain populations. Additionally, haplotype heterogeneity between populations studied may exist. It has been proposed that the causal allele variants regulating TNF responses may actually be located downstream of the TNF gene and that TNF allele variants considered are in linkage disequilibrium with the causal variant in some populations, but not in others [9]. Much of the literature investigating the issue of whether TNF promoter polymorphisms have any functional effect on TNF transcription or influence disease susceptibility appears to report negative results, emphasizing the conclusion that polymorphisms at this locus are functionally silent and exist only due to linkage disequilibrium with selectable human leukocyte antigen (HLA) alleles. Due to this, recent studies have increasingly included more markers in the HLA classes. The relevance of HLA antigens to disease is well recognized [40, 41] and may account for many of those studies that find positive evidence of association to TNF. This emphasizes the role of TNF variants as markers for HLA or other disease loci located in the major histocompatibility class (MHC) [42, 43]. Several studies conducted within the African population have found associations between HLA alleles and P.falciparum malaria [44–46]. It is important to note that differences in the distribution of HLA alleles could be a cause of variation in associations between TNF polymorphisms and P.falciparum malaria in different geographical areas. With the increasing evidence of HLA associations with malaria, it may well be that TNF−1031 and TNF−308 are not functional variants but markers of causal variants located elsewhere.
Conclusion
In pre-school Tanzanian children living in an area of intense transmission, the TNF−1031CC genotype was associated with increased rates of malarial episodes, whereas the TNF−308AA genotype was associated with decreased rates. Further studies are needed to confirm these findings. Additionally, further studies considering associations with the wider MHC should be conducted so as to identify candidate functional variants associated with severity of malaria.
Authors’ contributions
HV, JV and AYD designed and conducted research, WNGW analysed data and HV, WNGW, AMB, and EJF wrote the manuscript. WNGW, HV and AMB had primary responsibility for final content. All authors read and approved the final manuscript.
Acknowledgements
We thank the mothers/guardians for allowing their children to participate in the study, and Richard Crooijmans, Animal Breeding Group of Wageningen University, The Netherlands, for genotyping.
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interests.
Contributor Information
Wanjiku N Gichohi-Wainaina, Email: wngichohi@gmail.com.
Alida Melse-Boonstra, Email: Alida.melse@wur.nl.
Edith J Feskens, Email: Edith.feskens@wur.nl.
Ayse Y Demir, Email: AY.Demir@meandermc.nl.
Jacobien Veenemans, Email: Jacobien.Veenemans@gmail.com.
Hans Verhoef, Email: Hans.Verhoef@icloud.com.
References
- 1.D’Alfonso S, Richiardi PM. A polymorphic variation in a putative regulation box of the TNFA promoter region. Immunogenetics. 1994;39:150–154. doi: 10.1007/BF00188619. [DOI] [PubMed] [Google Scholar]
- 2.Higuchi T, Seki N, Kamizono S, Yamada A, Kimura A, Kato H, et al. Polymorphism of the 5′-flanking region of the human tumor necrosis factor (TNF)-alpha gene in Japanese. Tissue Antigens. 1998;51:605–612. doi: 10.1111/j.1399-0039.1998.tb03002.x. [DOI] [PubMed] [Google Scholar]
- 3.Uglialoro AM, Turbay D, Pesavento PA, Delgado JC, McKenzie FE, Gribben JG, et al. Identification of three new single nucleotide polymorphisms in the human tumor necrosis factor-alpha gene promoter. Tissue Antigens. 1998;52:359–367. doi: 10.1111/j.1399-0039.1998.tb03056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brinkman BM, Giphart MJ, Verhoef A, Kaijzel EL, Naipal AM, Daha MR, et al. Tumor necrosis factor alpha-308 gene variants in relation to major histocompatibility complex alleles and Felty’s syndrome. Hum Immunol. 1994;41:259–266. doi: 10.1016/0198-8859(94)90044-2. [DOI] [PubMed] [Google Scholar]
- 5.Wilson AG, di Giovine FS, Blakemore AI, Duff GW. Single base polymorphism in the human tumour necrosis factor alpha (TNF alpha) gene detectable by NcoI restriction of PCR product. Hum Mol Genet. 1992;1:353. doi: 10.1093/hmg/1.5.353. [DOI] [PubMed] [Google Scholar]
- 6.Brinkman BM, Kaijzel EL, Huizinga TW, Giphart MJ, Breedveld FC, Verweij CL. Detection of a C-insertion polymorphism within the human tumor necrosis factor alpha (TNFA) gene. Hum Genet. 1995;96:493. doi: 10.1007/BF00191815. [DOI] [PubMed] [Google Scholar]
- 7.Hamann A, Mantzoros C, Vidal-Puig A, Flier JS. Genetic variability in the TNF-alpha promoter is not associated with type II diabetes mellitus (NIDDM) Biochem Biophys Res Commun. 1995;211:833–839. doi: 10.1006/bbrc.1995.1887. [DOI] [PubMed] [Google Scholar]
- 8.Bayley JP, Ottenhoff TH, Verweij CL. Is there a future for TNF promoter polymorphisms? Genes Immun. 2004;5:315–329. doi: 10.1038/sj.gene.6364055. [DOI] [PubMed] [Google Scholar]
- 9.Clark TG, Diakite M, Auburn S, Campino S, Fry AE, Green A, et al. Tumor necrosis factor and lymphotoxin-alpha polymorphisms and severe malaria in African populations. J Infect Dis. 2009;199:569–575. doi: 10.1086/596320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinha S, Mishra SK, Sharma S, Patibandla PK, Mallick PK, Sharma SK, et al. Polymorphisms of TNF-enhancer and gene for FcgammaRIIa correlate with the severity of falciparum malaria in the ethnically diverse Indian population. Malar J. 2008;7:13. doi: 10.1186/1475-2875-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGuire W, Hill AV, Allsopp CE, Greenwood BM, Kwiatkowski D. Variation in the TNF-alpha promoter region associated with susceptibility to cerebral malaria. Nature. 1994;371:508–510. doi: 10.1038/371508a0. [DOI] [PubMed] [Google Scholar]
- 12.Meyer CG, May J, Luty AJ, Lell B, Kremsner PG. TNFalpha-308A associated with shorter intervals of Plasmodium falciparum reinfections. Tissue Antigens. 2002;59:287–292. doi: 10.1034/j.1399-0039.2002.590406.x. [DOI] [PubMed] [Google Scholar]
- 13.Aidoo M, McElroy PD, Kolczak MS, Terlouw DJ, ter Kuile FO, Nahlen B, et al. Tumor necrosis factor-alpha promoter variant 2 (TNF2) is associated with pre-term delivery, infant mortality, and malaria morbidity in western Kenya: Asembo Bay Cohort Project IX. Genet Epidemiol. 2001;21:201–211. doi: 10.1002/gepi.1029. [DOI] [PubMed] [Google Scholar]
- 14.Wattavidanage J, Carter R, Perera KL, Munasingha A, Bandara S, McGuinness D, et al. TNFalpha*2 marks high risk of severe disease during Plasmodium falciparum malaria and other infections in Sri Lankans. Clin Exp Immunol. 1999;115:350–355. doi: 10.1046/j.1365-2249.1999.00804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ubalee R, Suzuki F, Kikuchi M, Tasanor O, Wattanagoon Y, Ruangweerayut R, et al. Strong association of a tumor necrosis factor-alpha promoter allele with cerebral malaria in Myanmar. Tissue Antigens. 2001;58:407–410. doi: 10.1034/j.1399-0039.2001.580610.x. [DOI] [PubMed] [Google Scholar]
- 16.Stirnadel HA, Stockle M, Felger I, Smith T, Tanner M, Beck HP. Malaria infection and morbidity in infants in relation to genetic polymorphisms in Tanzania. Trop Med Int Health. 1999;4:187–193. doi: 10.1046/j.1365-3156.1999.43381.x. [DOI] [PubMed] [Google Scholar]
- 17.Atkinson SH, Rockett KA, Morgan G, Bejon PA, Sirugo G, O’Connell MA, et al. Tumor necrosis factor SNP haplotypes are associated with iron deficiency anemia in West African children. Blood. 2008;112:4276–4283. doi: 10.1182/blood-2008-06-162008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gwamaka M, Kurtis JD, Sorensen BE, Holte S, Morrison R, Mutabingwa TK, et al. Iron deficiency protects against severe Plasmodium falciparum malaria and death in young children. Clin Infect Dis. 2012;54:1137–1144. doi: 10.1093/cid/cis010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prentice AM, Cox SE. Iron and malaria interactions: research needs from basic science to global policy. Adv Nutr. 2012;3:583–591. doi: 10.3945/an.111.001230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellman R, Maxwell C, Finch R, Shayo D. Malaria and anaemia at different altitudes in the Muheza district of Tanzania: childhood morbidity in relation to level of exposure to infection. Ann Trop Med Parasitol. 1998;92:741–753. doi: 10.1080/00034989858989. [DOI] [PubMed] [Google Scholar]
- 21.Veenemans J, Milligan P, Prentice AM, Schouten LR, Inja N, van der Heijden AC, et al. Effect of supplementation with zinc and other micronutrients on malaria in Tanzanian children: a randomised trial. PLoS Med. 2011;8:e1001125. doi: 10.1371/journal.pmed.1001125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.illumina: Analyzing GoldeGate Genotyping Data
- 23.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 24.Epi Info. http://www.cdc.gov/epiinfo. Accessed 1 Aug 2011
- 25.Altman GDM, Bryant TN, Gardner MJ. Statistics with confidence. 2. London: BMJ Books; 2000. [Google Scholar]
- 26.Ackerman H, Usen S, Mott R, Richardson A, Sisay-Joof F, Katundu P, et al. Haplotypic analysis of the TNF locus by association efficiency and entropy. Genome Biol. 2003;4:R24. doi: 10.1186/gb-2003-4-4-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nordborg M, Tavare S. Linkage disequilibrium: what history has to tell us. Trends Genet. 2002;18:83–90. doi: 10.1016/S0168-9525(02)02557-X. [DOI] [PubMed] [Google Scholar]
- 28.Thompson EA, Neel JV. Allelic disequilibrium and allele frequency distribution as a function of social and demographic history. Am J Hum Genet. 1997;60:197–204. [PMC free article] [PubMed] [Google Scholar]
- 29.Reich DE, Cargill M, Bolk S, Ireland J, Sabeti PC, Richter DJ, et al. Linkage disequilibrium in the human genome. Nature. 2001;411:199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- 30.McGuire W, Knight JC, Hill AV, Allsopp CE, Greenwood BM, Kwiatkowski D. Severe malarial anemia and cerebral malaria are associated with different tumor necrosis factor promoter alleles. J Infect Dis. 1999;179:287–290. doi: 10.1086/314533. [DOI] [PubMed] [Google Scholar]
- 31.Gimenez F, Barraud de Lagerie S, Fernandez C, Pino P, Mazier D. Tumor necrosis factor alpha in the pathogenesis of cerebral malaria. Cell Mol Life Sci. 2003;60:1623–1635. doi: 10.1007/s00018-003-2347-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Randall LM, Kenangalem E, Lampah DA, Tjitra E, Mwaikambo ED, Handojo T, et al. A study of the TNF/LTA/LTB locus and susceptibility to severe malaria in highland papuan children and adults. Malar J. 2010;9:302. doi: 10.1186/1475-2875-9-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Church J, Maitland K. Invasive bacterial co-infection in African children with Plasmodium falciparum malaria: a systematic review. BMC Med. 2014;12:31. doi: 10.1186/1741-7015-12-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wajant H. The role of TNF in cancer. Results Probl Cell Differ. 2009;49:1–15. doi: 10.1007/400_2008_26. [DOI] [PubMed] [Google Scholar]
- 35.Daniele G, Guardado Mendoza R, Winnier D, Fiorentino TV, Pengou Z, Cornell J, et al. The inflammatory status score including IL-6, TNF-alpha, osteopontin, fractalkine, MCP-1 and adiponectin underlies whole-body insulin resistance and hyperglycemia in type 2 diabetes mellitus. Acta Diabetol. 2014;51:123–131. doi: 10.1007/s00592-013-0543-1. [DOI] [PubMed] [Google Scholar]
- 36.Mootoo A, Stylianou E, Arias MA, Reljic R. TNF-alpha in tuberculosis: a cytokine with a split personality. Inflamm Allergy Drug Targets. 2009;8:53–62. doi: 10.2174/187152809787582543. [DOI] [PubMed] [Google Scholar]
- 37.Elahi MM, Asotra K, Matata BM, Mastana SS. Tumor necrosis factor alpha −308 gene locus promoter polymorphism: an analysis of association with health and disease. Biochim Biophys Acta. 2009;1792:163–172. doi: 10.1016/j.bbadis.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 38.Hall JM, Lingenfelter P, Adams SL, Lasser D, Hansen JA, Bean MA. Detection of maternal cells in human umbilical cord blood using fluorescence in situ hybridization. Blood. 1995;86:2829–2832. [PubMed] [Google Scholar]
- 39.Veenemans J, Schouten LR, Ottenhof MJ, Mank TG, Uges DR, Mbugi EV, et al. Effect of preventive supplementation with zinc and other micronutrients on non-malarial morbidity in Tanzanian pre-school children: a randomized trial. PLoS One. 2012;7:e41630. doi: 10.1371/journal.pone.0041630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Vries RR. HLA and disease: past, present and future. Neth J Med. 1994;45:302–308. [PubMed] [Google Scholar]
- 41.Tomlinson IP, Bodmer WF. The HLA system and the analysis of multifactorial genetic disease. Trends Genet. 1995;11:493–498. doi: 10.1016/S0168-9525(00)89159-3. [DOI] [PubMed] [Google Scholar]
- 42.Jawaheer D, Li W, Graham RR, Chen W, Damle A, Xiao X, et al. Dissecting the genetic complexity of the association between human leukocyte antigens and rheumatoid arthritis. Am J Hum Genet. 2002;71:585–594. doi: 10.1086/342407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newton J, Brown MA, Milicic A, Ackerman H, Darke C, Wilson JN, et al. The effect of HLA-DR on susceptibility to rheumatoid arthritis is influenced by the associated lymphotoxin alpha-tumor necrosis factor haplotype. Arthritis Rheum. 2003;48:90–96. doi: 10.1002/art.10719. [DOI] [PubMed] [Google Scholar]
- 44.Lyke KE, Fernandez-Vina MA, Cao K, Hollenbach J, Coulibaly D, Kone AK, et al. Association of HLA alleles with Plasmodium falciparum severity in Malian children. Tissue Antigens. 2011;77:562–571. doi: 10.1111/j.1399-0039.2011.01661.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamazaki A, Yasunami M, Ofori M, Horie H, Kikuchi M, Helegbe G, et al. Human leukocyte antigen class I polymorphisms influence the mild clinical manifestation of Plasmodium falciparum infection in Ghanaian children. Hum Immunol. 2011;72:881–888. doi: 10.1016/j.humimm.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 46.Osafo-Addo AD, Koram KA, Oduro AR, Wilson M, Hodgson A, Rogers WO. HLA-DRB1*04 allele is associated with severe malaria in northern Ghana. Am J Trop Med Hyg. 2008;78:251–255. [PubMed] [Google Scholar]


