Abstract
Background
HOX transcript antisense intergenic RNA (HOTAIR), a long non-coding RNA transcribed from the antisense strand of the HOXC gene locus, has been shown to be overexpressed in various carcinomas and is thought to be an indicator of poor prognosis. Recently, HOTAIR was found to be an estrogen-responsive gene. We therefore conducted a meta-analysis to systematically summarize and clarify the association between HOTAIR expression and prognosis in the four main estrogen-dependent tumors.
Methods
A systematic search of studies that examined the association and prognostic impact of HOTAIR in four of the main estrogen-dependent tumors was conducted in PubMed and Embase. Hazard ratios (HRs) and 95% confidence intervals (95% CIs) were calculated to pool the effect size.
Results
A total of 1,200 patients from eight eligible studies were included. The current study found an association between HOTAIR expression and overall survival (OS) in four estrogen-dependent tumor types (HR, 1.99; 95% CI: 1.02–3.90; PHeterogeneity=0.001). Subgroup analyses indicated that high HOTAIR expression appeared to be a potential prognostic biomarker in non-breast cancer patients (HR, 2.72; 95% CI: 1.65–4.48). There was also an increased risk in Asian populations (HR, 2.55; 95% CI: 1.62–4.00) compared with Caucasian populations (HR, 1.19; 95% CI: 0.16–8.83) and in patients without preoperative treatment (HR, 2.55; 95% CI: 1.62–4.00) compared with patients with preoperative treatment (HR, 1.19; 95% CI: 0.16–8.83). In addition, the HRs of patients with high HOTAIR expression for metastasis-free survival (MFS), relapse-free survival (RFS), and disease-free survival (DFS) were 2.30 (P=0.120), 1.39 (P=0.000), and 2.53 (P=0.714), respectively, but there were insufficient data to fully confirm these associations.
Conclusion
HOTAIR may be a predictor of poor prognosis in four of the main estrogen-dependent tumors, especially in cervical, ovarian, and endometrial cancer patients without preoperative treatment in Asian populations. It is important to note that the prognostic value of HOTAIR in MFS, RFS, and DFS should be interpreted with caution due to the limited sample size and sample heterogeneity. Well-designed and larger-scale studies are needed to validate our findings.
Keywords: HOTAIR, prognosis, estrogen-dependent tumors, breast cancer, cervical cancer, ovarian cancer, endometrial cancer
Introduction
In the human genome, only 2% of transcripts are protein-coding RNAs, and almost 90% of genomic DNA in eukaryotic genomes do not encode proteins.1 These non-coding RNAs (ncRNAs) have been dismissed as transcriptional “noise” in past decades but are now classified into two categories: housekeeping ncRNAs and regulatory ncRNAs. Housekeeping ncRNAs, including ribosomal, transfer, small nuclear, and small nucleo-lar RNAs, are usually expressed constitutively. MicroRNAs, small interfering RNAs, and piwi-associated RNAs belong to the short regulatory ncRNA group.2 Notably, long non-coding RNAs (lncRNAs), 200–100,000 nt in length, are crucial players in various key biological processes, including chromatin modification, transcription, and post-transcriptional processing.3,4 Emerging evidence indicates that the dysregulation of lncRNAs may contribute to tumor cell proliferation, apoptosis, and invasion, as well as participate in metastasis.5
The lncRNA HOX transcript antisense intergenic RNA (HOTAIR), an lncRNA with regulatory transcription activity, was identified in 2007.6 HOTAIR is transcribed by RNA polymerase II (RNAP II) from the antisense strand. It is co-expressed with the HOXC gene cluster and participates in the trans downregulation of various genes in a genome-wide fashion,6 although some genes are upregulated by HOTAIR. Aberrant HOTAIR expression is associated with various types of cancer, such as breast, hepatocellular, and gastric, among others, and its expression affects the survival and prognosis of cancer patients.5,7,8
Recently, a study reported that HOTAIR is an estrogen-responsive gene.9 Similar to other protein-coding estradiol (E2)-responsive genes, HOTAIR is activated by E2 and is transcribed from the antisense strand under the coordination of E2 receptors (ERs), ER co-regulators, and general transcription factors associated with RNAP II transcription. In addition, HOTAIR is potentially regulated by the RNAP II machinery in the presence of E2.9 Estrogen induction of cell proliferation is a crucial step in breast and gynecological tumors, such as cervical, ovarian, and endometrial tumors.10 Some studies have shown that HOTAIR upregulation contributes to carcinogenesis in these tumors,5,11–18 whereas a few studies have investigated the role of HOTAIR as a tumor-suppressing gene and have yielded mixed and inconclusive results.19
Currently, there is no meta-analysis investigating the association between these factors. We therefore performed a meta-analysis to evaluate the prognostic significance of HOTAIR expression in four of the main estrogen-dependent tumors.
Methods
Identification
This meta-analysis was performed in accordance with preferred reporting items for systematic reviews and meta-analyses.20 PubMed and Embase were searched for studies using both medical subject heading terms and free-text words. The following search terms were used: HOTAIR, “HOX transcript antisense RNA”, breast cancer/neoplasm/tumor/carcinoma, cervix (or cervical) cancer/neoplasm/tumor/carcinoma, ovary (or ovarian) cancer/neoplasm/tumor/carcinoma, endometrium (endometrial) cancer/neoplasm/tumor/carcinoma, prognos*, predict*, outcome, mortality, survival, recurrence, and metastasis. The last search was performed on March 14, 2015. The citation lists of the retrieved articles were manually screened to ensure the sensitivity of the search strategy.
Inclusion and exclusion criteria
Studies in this meta-analysis had to meet the following inclusion criteria: 1) inclusion of patients with breast cancer, cervical cancer, ovarian cancer, or endometrial cancer; 2) evaluation of the association between HOTAIR and clinical prognosis; 3) sufficient data to estimate hazard ratio (HR) and 95% confidence interval (95% CI); and 4) publication as a full paper in English. Studies were excluded based on the following criteria: 1) duplicated studies, reviews, letters, unpublished data, and comments; 2) published in a language other than English; 3) data that could not be extracted or calculated from the original article; and 4) non-human subjects. Study selection was achieved by two investigators independently according to the inclusion and exclusion criteria by screening the title, abstract, and full text.
Data extraction
The data of eligible studies were extracted in duplicate by two investigators independently (Li and Wen). The following details were extracted: name of the first author, year of publication, country of origin, ethnicity of the study population, type of specimen, cancer type, number of patients included in the analysis, detection method of HOTAIR, and cut-off values, as well as the HR and the corresponding 95% CI for overall survival (OS), metastasis-free survival (MFS), recurrence-free survival (RFS), and disease-free survival (DFS). Two authors checked the extracted data and reached a consensus considering all data. Disputes were resolved by discussion. If dissent remained, then the third investigator (Zhang) was consulted.
Statistical analyses
HRs and their associated 95% CIs were used to evaluate the strength of the association between HOTAIR and its prognostic value. For each study, the HRs were extracted from the published data.21 If the HRs could not be obtained directly from the publication or by estimation from the O-E statistic and variance, we calculated the HRs from the total number of events and the P-value in the articles.22 However, if the abovementioned methods were ineffective, then we calculated the HRs and CIs from the Kaplan–Meier survival curves using the HR digitizer software Engauge 4.1 as described previously.22 In this meta-analysis, we adopted a random-effects model that accounted for the additional sources of inter-study variation when heterogeneity existed. This model is more conservative than a fixed-effects model because the latter assumes similar true genetic effects between studies, whereas the former assumes normally distributed effects and parameterizes the inter-study variation.23 Estimates of the HRs with their corresponding 95% CIs were weighted and pooled using the inverse-variance random-effects model. Subgroup analyses by factor for cancer type and ethnicity were also performed, as well as sensitivity analyses to test the effect of each study on the overall pooled results. Because of the limited number (less than 10) of studies included in each analysis, publication bias was not assessed. The data were combined into a meta-analysis using Stata 12.0 analysis software (Stata Corporation, College Station, TX, USA). All statistical texts were two-sided, and significance was defined as a P-value of <0.05.
Results
Description of studies
A total of eight papers were acquired from electronic searches of PubMed and Embase. The literature selection process is shown in Figure 1. We identified eight studies based on the inclusion criteria.5,12–14,16,17,19,24 Table 1 summarizes the main characteristics of the included studies. All studies were published in 2010 or later. A total of 1,200 patients were included in those studies, with a maximum sample size of 336 participants and a minimum sample size of 30 participants. Tissue specimens were evaluated in eight studies: four of these studies evaluated breast cancer;5,17,19,24 two studies evaluated cervical cancer;13,14 and one study each evaluated endometrial cancer12 and ovarian cancer.16 All studies included a high-HOTAIR expression arm and a low-HOTAIR expression arm, while the cut-off values considered in the studies were inconsistent due to variations in the detection methods or discrepancies in the cut-off definitions. A total of eight HRs were analyzed. There were seven studies that assessed OS,5,12–14,16,19,24 two that assessed MFS,5,17 two that assessed RFS,14,19 and two that assessed DFS13,16 in the meta-analysis. HRs with their corresponding 95% CIs could be obtained directly from six studies; HRs and their corresponding 95% CIs were extracted from three studies from their Kaplan–Meier curves.
Figure 1.
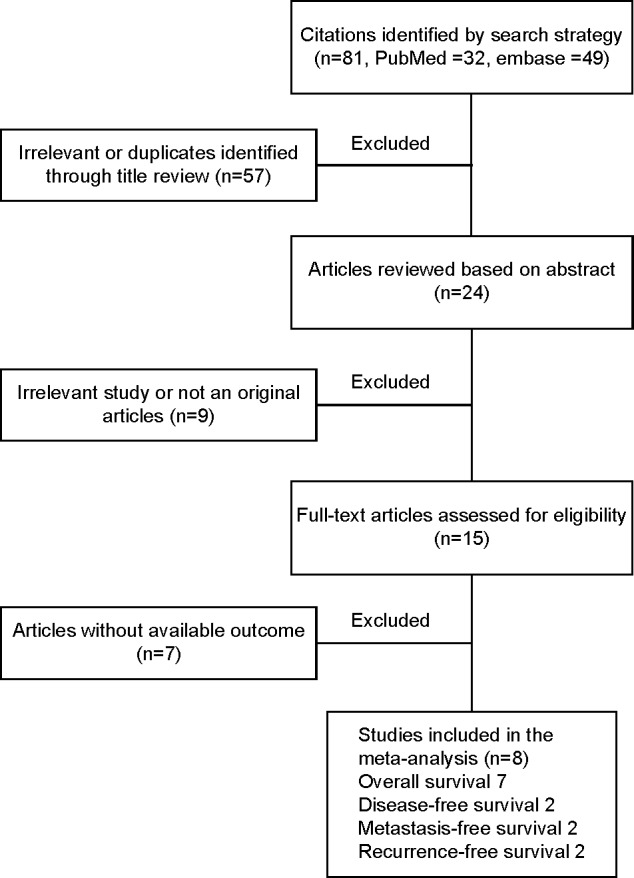
Flow chart of the different stages of the meta-analysis.
Table 1.
Characteristics of studies included in the meta-analysis
| First author | Year | Country | Ethnicity | Specimens | Cancer type | Number | Method | Cut-off | Analysis | Survival* |
|---|---|---|---|---|---|---|---|---|---|---|
| Gupta5 | 2010 | USA | Caucasian | Tissue | Breast cancer | 132 | qRT-PCR | 125-fold compared with normal tissue | Multivariate | OS,a MFSa |
| Lu19 | 2012 | Italy | Caucasian | Tissue | Breast cancer | 336 | qRT-PCR | 10.01 expression index | Multivariate | OS,a RFSa |
| Li24 | 2014 | People’s Republic of China | Asian | Tissue | Breast cancer | 30 | qRT-PCR | .270-fold increased levels | Univariate | OSb |
| Sorensen17 | 2013 | Denmark | Caucasian | Tissue | Breast cancer | 164 | Microarray | Density plot of gene expression of 0.6 | Multivariate | MFSa |
| Kim14 | 2015 | Korea | Asian | Tissue | Cervical cancer | 111 | qRT-PCR | 30-fold compared with normal tissues | Univariate/multivariate | OS,b RFSa |
| He12 | 2014 | People’s Republic of China | Asian | Tissue | Endometrial cancer | 145 | ISH | An staining index score of 6 | Univariate | OSa |
| Huang13 | 2014 | People’s Republic of China | Asian | Tissue | Cervical cancer | 218 | qRT-PCR | Median expression value of HOTAIR | Multivariate/univariate | OS,a DFSb |
| Qiu16 | 2014 | People’s Republic of China | Asian | Tissue | Ovarian cancer | 64 | qRT-PCR | Median value of HOTAIR expression levels | Univariate | OS,b DFSb |
Notes:
Denoted as obtaining HRs directly from publications;
denoted as extracting HRs from Kaplan–Meier curves.
Abbreviations: DFS, disease-free survival; HOTAIR, HOX transcript antisense intergenic RNA; HRs, hazard ratios; ISH, in situ hybridization; MFS, metastasis-free survival; OS, overall survival; qRT-PCR, quantitative reverse transcription-polymerase chain reaction; RFS, recurrence-free survival.
Overall survival
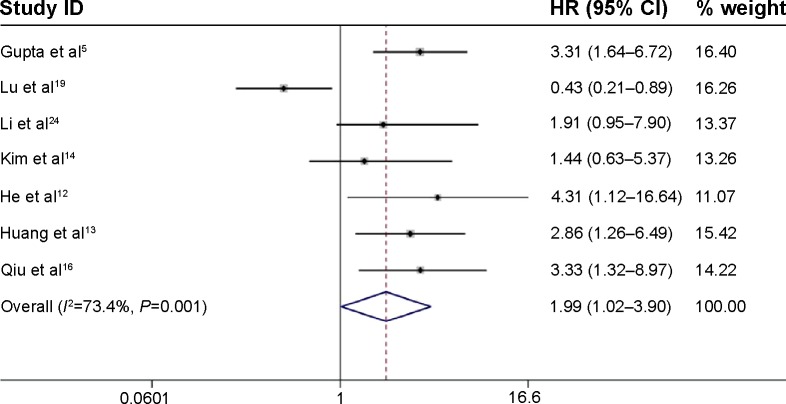
The main results of this meta-analysis are shown in Figure 2. Based on variations in the levels of HOTAIR expression in a total of 1,036 patients with four types of cancer, seven studies reported HRs for the OS. The combined analysis showed that HOTAIR expression was associated with poorer OS (pooled HR, 1.99; 95% CI: 1.02–3.90) with obvious heterogeneity (χ2=22.55, df=6, P=0.001, I2=73.4%).
Figure 2.
Forest plots of studies evaluating the association between HOTAIR expression and overall survival.
Note: Weights are from random-effects analysis.
Abbreviations: CI, confidence interval; HOTAIR, HOX transcript antisense intergenic RNA; HR, hazard ratio.
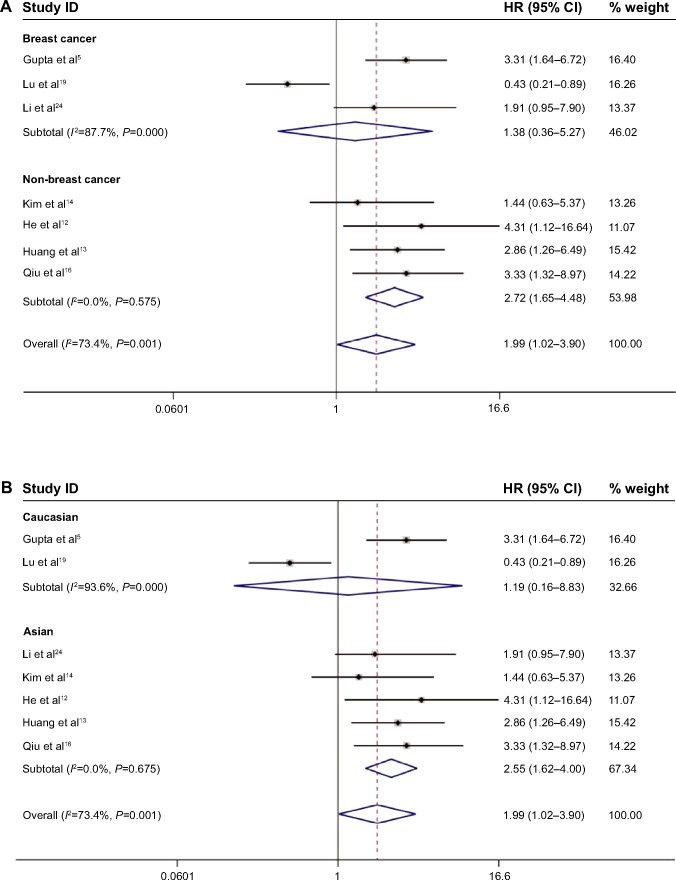
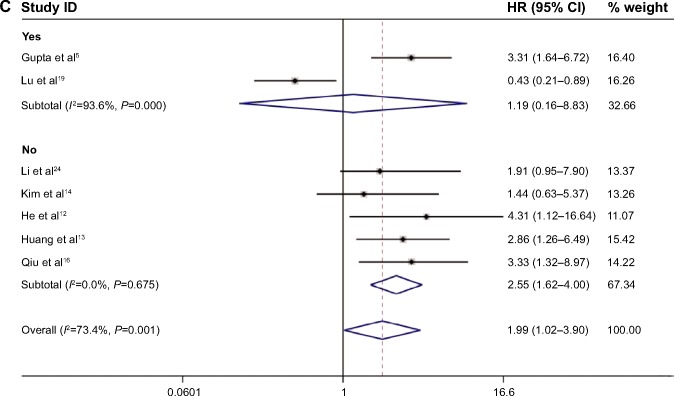
Due to the presence of heterogeneity, subgroup analyses based on the cancer type, ethnicity, and preoperative treatment status were performed (Table 2). Subgroup analysis by cancer type indicated that high HOTAIR expression was significantly associated with a poorer outcome in three non-breast cancer patients (HR, 2.72; 95% CI: 1.65–4.48) but not in breast cancer patients (HR, 1.38; 95% CI: 0.36–5.27; Figure 3A). We found a significant association between HOTAIR expression and OS in Asian cancer patients (HR, 2.55; 95% CI: 1.62–4.00) but not in Caucasians (HR, 1.19; 95% CI: 0.16–8.83; Figure 3B). Among non-preoperative treatment patients, we found an accordant role of higher HOTAIR expression in OS with similar HRs (2.55), whereas there was no significant association between HOTAIR and OS in those patients who received preoperative treatment (Figure 3C). Significant heterogeneity existed across the studies in the subgroups of Caucasian breast cancer patients and preoperative treatment patients, but there was no significant heterogeneity in the subgroups of Asian patients, patients with cervical and two other estrogen-dependent tumors, or patients without preoperative treatment. To further explore the source of the heterogeneity, a meta-regression was performed using covariates including sample size, detection method, and HRs from univariate/multivariate analyses to analyze the heterogeneity (Table 3). However, none of the examined factors accounted for the inter-study heterogeneity in the meta-regression.
Table 2.
Results of subgroup analysis of the independent role of HOTAIR in OS/MFS/RFS/DFS
| Subgroup analysis | No of studies | No of patients | Pooled HR (95% CI) | Heterogeneity
|
|
|---|---|---|---|---|---|
| I2 (%) | P-value | ||||
| Overall survival | |||||
| Cancer type | |||||
| Breast cancer | 3 | 498 | 1.38 (0.36–5.27) | 87.7 | 0.000 |
| Non-breast cancer | 4 | 538 | 2.72 (1.65–4.48) | 0.0 | 0.575 |
| Ethnicity | |||||
| Caucasian | 2 | 468 | 2.55 (1.62–4.00) | 0.0 | 0.675 |
| Asian | 5 | 568 | 1.19 (0.16–8.83) | 93.6 | 0.000 |
| Preoperative treatment | |||||
| No | 5 | 568 | 1.19 (0.16–8.83) | 93.6 | 0.000 |
| Yes | 2 | 468 | 2.55 (1.62–4.00) | 0.0 | 0.675 |
| Metastasis | 2 | 296 | 2.30 (1.19–4.44) | 58.5 | 0.120 |
| Recurrence | 2 | 447 | 1.39 (0.13–14.64) | 86.2 | 0.007 |
| Disease-free | 2 | 282 | 2.53 (1.27–5.03) | 0.0 | 0.714 |
Abbreviations: CI, confidence interval; DFS, disease-free survival; HOTAIR, HOX transcript antisense intergenic RNA; HR, hazard ratio; MFS, metastasis-free survival; OS, overall survival; RFS, recurrence-free survival.
Figure 3.
Meta-analysis of the pooled HRs of OS with increased HOTAIR expression.
Notes: (A) Subgroup analysis of HRs of OS by factor of cancer types. (B) Subgroup analysis of HRs of OS by factor of ethnicity. (C) Subgroup analysis of HRs of OS by factor of preoperative treatment status. Weights are from random-effects analysis.
Abbreviations: CI, confidence interval; HOTAIR, HOX transcript antisense intergenic RNA; HRs, hazard ratios; OS, overall survival.
Table 3.
Results of meta-regression of the independent value of HOTAIRin overall survival
| Covariate | No of study | No of patients | exp(b) | SE | P-value |
|---|---|---|---|---|---|
| Sample size | |||||
| ≤132 | 4 | 337 | 0.6355 | 0.4206 | 0.524 |
| >132 | 3 | 699 | |||
| Detection method | |||||
| qRT-PCR | 6 | 891 | 0.4192 | 0.4415 | 0.447 |
| Non-qRT-PCR | 1 | 145 | |||
| Univariate/multivariate | |||||
| Univariate | 4 | 350 | 0.6436 | 0.4334 | 0.542 |
| Multivariate | 3 | 686 | |||
Abbreviations: HOTAIR, HOX transcript antisense intergenic RNA; qRT-PCR, quantitative reverse transcription-polymerase chain reaction; SE, standard error.
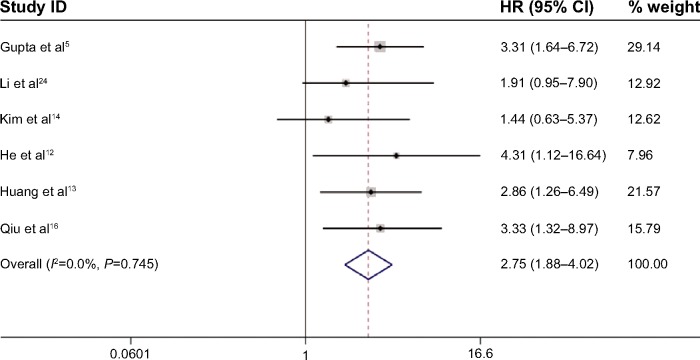
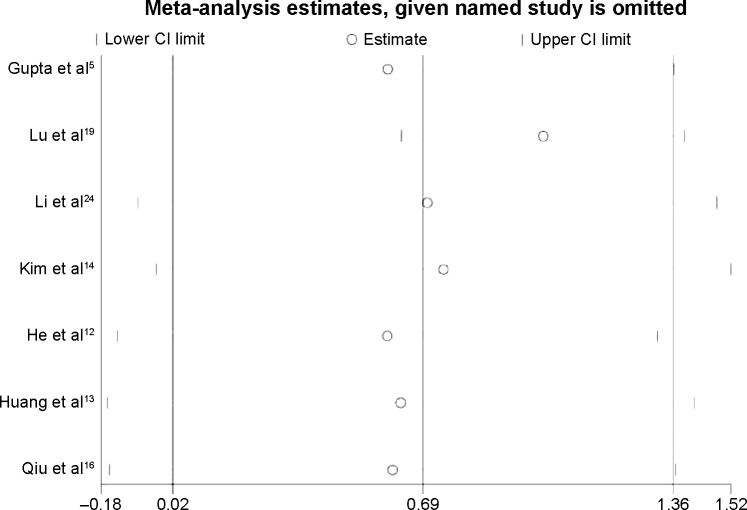
To explore the source of the heterogeneity, we used the “hatred” macro command in STATA to determine which study resulted in the heterogeneity in the meta-analysis. The heterogeneity decreased from 73.4% to 0.0% if we excluded the study by Lu et al19 from the overall pooled analysis. After removing this study, we again pooled the other six included studies, and the heterogeneity was eliminated (Figure 4). Sensitivity analysis confirmed the stability of our conclusions, and we deleted single study from the overall pooled analysis one at a time to determine the influence of the removed data set on the overall HR (Figure 5). The association between HOTAIR and OS remained stable after the exclusion of the Lu et al study19 and any other study from the sensitivity analysis. Table 4 summarizes the sensitivity analysis outcome. Because only seven studies were included, publication bias was not assessed.
Figure 4.
Forest plots of studies evaluating the hazard ratios (HRs) of HOTAIR for overall survival, excluding the study generating heterogeneity (Lu et al19).
Note: Weights are from random-effects analysis.
Abbreviations: CI, confidence interval; HOTAIR, HOX transcript antisense intergenic RNA.
Figure 5.
Sensitivity analysis of the effect of individual studies on the pooled HRs for increased HOTAIR expression and overall survival of patients.
Abbreviations: CI, confidence interval; HOTAIR, HOX transcript antisense intergenic RNA; HRs, hazard ratios.
Table 4.
Sensitivity analysis
| Sensitivity analysis | Heterogeneity test
|
Pooled effect size
|
||
|---|---|---|---|---|
| I2 (%) | Tau2 | HR (95% CI) | P-value | |
| Lu et al19 included | 73.4 | 0.5865 | 1.99 (1.02–3.90) | 0.001 |
| Lu et al19 excluded | 0.0 | 0.0000 | 2.75 (1.88–4.02) | 0.745 |
Abbreviations: CI, confidence interval; HR, hazard ratio.
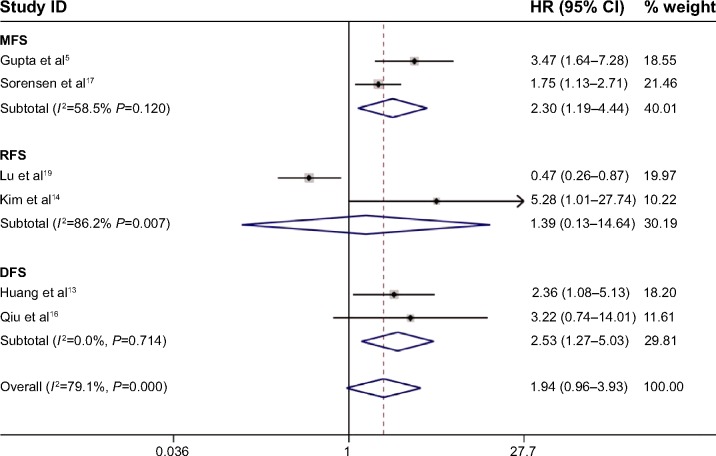
MFS, RFS, and DFS
Meta-analyses showed the prognostic significance of HOTAIR for MFS, RFS, and DFS in cancer patients (Figure 6). Two studies comprising 296 patients reported HRs for MFS and showed that HOTAIR expression exceeding the cut-off value was more likely to be associated with an obviously shorter MFS (HR, 2.30; 95% CI: 1.19–4.44) with heterogeneity (I2=58.5%, P=0.120). The prognostic value of HOTAIR for RFS was evaluated in two studies comprising 447 patients. In those studies, HOTAIR expression was not significantly associated with RFS (HR, 1.39; 95% CI: 0.13–14.64) with obvious heterogeneity (I2=86.2%, P=0.007). Two studies comprising 282 patients reported HRs for the DFS, suggesting that HOTAIR overexpression predicted a worse outcome for DFS (HR, 2.53; 95% CI: 1.27–5.03) without heterogeneity (I2=0%, P=0.714). The sensitivity analysis changed after deleting any of the included studies with respect to the MFS and the RFS but remained stable with respect to the DFS. Subgroup analysis, meta-regression, and publication bias were not applicable in the analyses of MFS, RFS, and DFS due to the limited number of included studies (less than 10; Table 4).
Figure 6.
Meta-analysis of the independent role of HOTAIR in the metastasis/recurrence/disease-free survival of four estrogen-dependent cancers.
Note: Weights are from random-effects analysis.
Abbreviations: CI, confidence interval; DFS, disease-free survival; HOTAIR, HOX transcript antisense intergenic RNA; HR, hazard ratio; MFS, metastasis-free survival; RFS, recurrence-free survival.
Discussion
HOTAIR is an lncRNA that is transcribed from the anti-sense strand of the HOXC gene locus on chromosome 12.6 According to a genome-wide analysis, HOTAIR silencing affects the expression of various types of genes associated with carcinogenesis, tumor suppression, apoptosis, cell differentiation, and development.6 HOTAIR has also been found to be overexpressed in various non-estrogen-dependent cancers and to contribute to cancer development, mainly via chromosome remodeling and transcription and post-transcriptional processing.25 Targeting the polycomb repressive complex 2 and lysine-specific demethylase 1 complexes to chromatin for coupled histone methylation and demethylation processes of histone H3 lysine 27, HOTAIR then binds various target genes, such as the HOXC cluster.6 HOTAIR was recently reported as a hallmark of cancer, especially in predicting the OS of Asian patients or patients with digestive system carcinoma.26 The promotion of cell proliferation by estrogen is a significant step in the tumorigenesis of breast and gynecological target tissues, and the mitogenic actions of estrogens are critical in the etiology and progression of human breast and gynecological cancers.27 Breast and gynecological cancers are hormone-dependent tumors, and endogenous (reproductive factors) and exogenous (primarily hormonal contraceptives and hormone replacement therapy) exposure to estrogens are the major environmental risk factors for those malignant tumors.28–30 The effects of estrogens are mediated by ERα and ERβ, which are typically thought to act via ligand-dependent binding to the estrogen-response elements (EREs) of target genes, resulting in transcriptional regulation. Bhan et al9 recently reported that HOTAIR is an estrogen-responsive gene. HOTAIR is transcriptionally induced upon treatment with E2 and suppressed upon exposure to the anti-estrogen agent tamoxifen. In the same study, sequence analysis showed that the HOTAIR promoter contained multiple functional EREs located near transcription start sites, especially ERE2 and ERE3, which exhibited the highest responses to E2 treatment in a luciferase-based reporter assay.9 Moreover, ERα and ERβ bound to the ERE regions in the HOTAIR promoter in the presence of E2, emphasizing the crucial roles of ERs during E2-induced HOTAIR expression. ERs combined with various ER co-regulators; for example, the histone methylases MLL1 and MLL3 and CBP/p300 bound the HOTAIR promoter in an E2-dependent manner. The level of histone H3K4 trimethylation, histone acetylation, and RNAP II recruitment was enhanced at the HOTAIR promoter in the presence of E2.9 This mechanism of HOTAIR expression may potentially contribute to cancer progression. It is important to note that the HOTAIR gene might be dysregulated by estrogenic endocrine-disrupting chemicals and synthetic estrogens associated with major health risks, specifically, bisphenol A (BPA), a well-known estrogenic endocrine disruptor, and diethylstilbestrol (DES), a synthetic estrogen. The epigenetic mechanism of the transcriptional activation of HOTAIR produced by BPA and DES was similar to that of E2-mediated HOTAIR activation. Exposure to BPA or DES might induce the expression of HOTAIR, even without estrogen, and this expression, in turn, might cause adverse health effects, including cancer and other hormonally regulated diseases.31
In this study, we performed a meta-analysis of the published data on the expression of HOTAIR in four of the main estrogen-dependent tumors (breast, cervical, ovarian, and endometrial cancers) and the association of HOTAIR with survival in each study. The eight eligible studies included 1,200 patients, and HOTAIR expression levels were analyzed. First, we performed a meta-analysis to assess the current evidence of the association between HOTAIR expression and OS in a total of 1,036 patients. An association between HOTAIR expression and OS was found in the global results with obvious heterogeneity. There were many factors that might have been responsible for the heterogeneity, for example, variations in the cancer types, discrepancies in the cut-off definitions, or diverse detection methods. In addition, there were differences in the genetic heterogeneity based on the ethnic backgrounds. Finally, environmental factors also had a strong influence.
Subgroup analysis, meta-regression, and sensitivity analysis were performed to demonstrate the inter-study heterogeneity concerning the independent prognostic role of HOTAIR in OS. Subgroup analyses according to cancer type, sample ethnicities, and preoperative status were performed using a random-effects model. Among the cancer types evaluated, non-breast cancer (cervical cancer, endometrial cancer, and ovarian cancer) was significantly linked with a poorer outcome for patients who expressed high levels of HOTAIR without heterogeneity, suggesting that HOTAIR may play an important role in these cancers that commonly overexpress HOTAIR. These results were consistent with the finding that HOTAIR contributes to the development of cervical cancer and is considered as a cancer biomarker.14 Moreover, HOTAIR expression was higher in ovarian and endometrial cancers and played an important role in cancer progression and metastasis. HOTAIR may serve as a biomarker for the malignancy of those cancers and may represent a novel therapeutic target.16,32 However, we failed to find a significant association between HOTAIR and OS in breast cancer patients. This finding can be explained by the observation that HOTAIR induces a repressed chromatin state by promoting the formation of H3 lysine 27, indicating that HOTAIR may function as a tumor suppressor gene by inhibiting the proliferation of breast cancer stem cells.5 In line with this study, another study reported that patients with high HOTAIR expression levels in their breast cancer samples had better survival than patients with low expression due to variations in the levels of intergenic DNA methylation.19 Additionally, different biological types of breast cancer might also be somewhat responsible for the negative outcome, which was not analyzed in our meta-analysis due to limited data. A large proportion of patients with breast cancer were ER positive or progesterone receptor positive, and the prognostic value of HOTAIR expression in ER- and progesterone receptor-negative breast cancer might be different. It has been reported that HOTAIR expression is not E2 responsive in the ER-negative human breast cancer cell line MDAMB231, demonstrating the involvement of ERs in E2-induced HOTAIR expression.9 This observation indicates that HOTAIR is tightly regulated by ER in breast cancer cells. However, the results were inconclusive. It has recently been reported that HOTAIR expression is lower in ductal carcinomas but higher in ER-negative tumors. In addition, in this previous study, a high level of HOTAIR expression was not involved in nodal metastases or prognosis in ER-positive patients. High HOTAIR expression was only associated with poor prognosis in ER-negative patients who were node positive.33 Thus, further independent investigations are needed to identify whether HOTAIR is an independent prognostic biomarker in breast cancer.
It is also possible that HOTAIR may be regarded as a risk factor in Asian populations because we observed a strong association between Asian ethnicity and OS in cancer patients without heterogeneity on ethnicity subgroup analysis. However, when we only considered Caucasian populations, no obvious association was found on subgroup analysis. In line with a previous study,34 the current study further validated that HOTAIR is an independent prognostic factor for OS in Asian patients but not in Caucasian patients. This lack of association may be attributed to variations in life styles, ethnic genetic heterogeneity, etc. Notably, HOTAIR expression was a prognostic indicator for OS in patients without preoperative treatment, such as radiotherapy, chemotherapy, or hormonal therapy. However, HOTAIR expression was not a prognostic indicator in patients with preoperative treatment. To some extent, these latter patients showed the true prognostic effect of HOTAIR on survival within a controlled population because the treatment modality modified the prognostic value of HOTAIR. In summary, HOTAIR is an independent prognostic indicator for non-breast cancer patients without preoperative treatment in an Asian population without heterogeneity.
Meta-regression analysis showed that no included stratifying factors, such as sample size, detection method, and HRs from univariable or multivariable analysis, contributed to the main heterogeneity across studies. The “hatred” macro command combined with sensitivity analysis explored the source of the heterogeneity. After excluding the study by Lu et al19 the heterogeneity was eliminated, suggesting that the abovementioned study contributed to the source of heterogeneity. Lu et al19 reported that intergenic DNA methylation might be biologically relevant in the regulation of HOTAIR expression and that HOTAIR may not be an independent prognostic biomarker. To explore the differences between the study by Lu et al19 and other studies, we re-analyzed this paper carefully. There are several possible explanations for the discrepancies in this study regarding the heterogeneity of this association. First, different sample sizes and different cut-off criteria for grouping HOTAIR expression may explain the inconsistency between their results and those of other studies. For example, the Lu et al study,19 which included the largest sample size in the meta-analysis, reported the only negative results among all of the included studies; consequently, these results were weighted heavily in the overall results. For the survival analysis, unlike the other five studies that divided HOTAIR expression into two arms (high and low), the levels of HOTAIR expression were grouped into three categories (low, medium, and high) in Lu et al study19 based on tertiles of the distributions to assess the association between HOTAIR and the risk of disease progression or death. Next, there were differences in the clinicopathological features and therapy regimens of the patients. For example, most of the patients had early-stage disease (36.4% stage I, 53.4% stage II, and 10.3% stages III and IV) in the abovementioned study.19 Moreover, some percentage of the patients in the abovementioned study may have received preoperative treatments, and the author did not divide those patients into a no-adjuvant therapy group and an adjuvant therapy group. Ozgur et al35 reported that levels of HOTAIR expression and other lncRNAs (eg, MALAT1) were repressed in bleomycin-treated HeLa and MCF7 cells, suggesting differential regulation of lncRNAs under geno-toxic stress. Moreover, endocrine disruptors could disrupt the ncRNAs and could activate antisense transcripts. HOTAIR gene expression and misregulation might be induced by endocrine-disrupting chemicals, synthetic estrogens, and other environmental toxins/chemicals.31 Thus, it is reasonable to conclude that different therapeutic regimens could modify the effects of HOTAIR expression. Finally, the abovementioned study19 is a large sample cohort study with a rather long follow-up time (median follow-up time, 86 months; range, 8–108 months) among the studies of the meta-analysis. Considering the research type and the long follow-up time, the validity of the cohort study could have been affected by losses during the long follow-up period.
The present study further analyzed the association between HOTAIR expression and cancer progression. The prognostic effects of HOTAIR on MFS, RFS, and DFS were respectively evaluated in two studies with 296 patients, in two studies with 447 patients, and in two studies with 282 patients. We detected that the prognostic significance of HOTAIR for the DFS was strong (HR, 2.53; 95% CI: 1.27–5.03) without heterogeneity regardless of the data source or cut-off definition. This finding suggested that HOTAIR expression might be a meaningful predictor of DFS. Subgroup analysis showed that HOTAIR was an independent prognostic factor for MFS (HR, 2.30; 95% CI: 1.19–4.44), albeit with heterogeneity. It should be emphasized that all included studies in the MFS subgroup analysis involved breast cancer patients. Given previous findings that HOTAIR overexpression could contribute to breast cancer cell invasion and that HOTAIR knock down inhibited cancer invasiveness through Matrigel in vitro,5 our results indicate that HOTAIR might serve as an independent prognostic factor for breast cancer metastasis. However, HOTAIR expression was not significantly associated with a poor RFS (HR, 1.39; 95% CI: 0.13–14.64) with significant heterogeneity. We are unable to draw a definite conclusion regarding the relationships between HOTAIR and MFS, RFS, or DFS owing to the limited number of studies.
This study is the first meta-analysis focused on the association between the prognostic value of HOTAIR and four of the main estrogen-dependent tumors. Compared with the former meta-analysis examining HOTAIR and various cancers,34 more specific and related studies were included according to the E2 responsiveness of the examined cancer. These analyses have limitations. First, it should be noted that there are other risk factors for cervical cancer that are more important than hormone levels. One major risk factor for cervical cancer is having multiple sexual partners, resulting in an increased risk of human papillomavirus infection. Another factor is cigarette smoking, which is strongly related to a lower socioeconomic status and ethnicity.36 Second, the number of included studies that examined HOTAIR and clinical prognosis is limited. However, the sensitivity analysis results remain stable after excluding each of the included studies. Third, there is no accepted and validated method to assess HOTAIR expression. Hence, there may have been considerable heterogeneity that was not able to be fully interpreted using random-effects modeling. Fourth, several HRs could not be directly obtained from the primary studies, requiring extraction of the HRs from Kaplan–Meier curves. Finally, there are differences in paper quality across the studies that might have contributed to the heterogeneity.
In conclusion, our results suggested that HOTAIR might predict a poor prognosis in four of the main estrogen-dependent cancers, especially in cervical, ovarian, and endometrial cancers in Asian populations without preoperative treatment. Nevertheless, there were insufficient data to fully confirm the association between HOTAIR and its prognostic value in MFS, RFS, and DFS, and the results should be interpreted with caution; well-designed studies with larger sample sizes and additional ethnic groups are required to confirm the risks identified in the present meta-analysis. This study is the first to show HOTAIR as a biomarker for predicting the prognosis of patients with estrogen-dependent tumors. However, more comprehensive studies and larger samples are necessary to confirm this association.
Acknowledgments
This research was supported by grants from the National Science Foundation of China (81172181) and by the Health Department of Heilongjiang province funds (684).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Knauss JL, Sun T. Regulatory mechanisms of long noncoding RNAs in vertebrate central nervous system development and function. Neuroscience. 2013;235:200–214. doi: 10.1016/j.neuroscience.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yao Y, Li J, Wang L. Large intervening non-coding RNA HOTAIR is an indicator of poor prognosis and a therapeutic target in human cancers. Int J Mol Sci. 2014;15(10):18985–18999. doi: 10.3390/ijms151018985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9(6):703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ponting CP, Oliver PL, Reik W. Evolution and functions of long non-coding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kogo R, Shimamura T, Mimori K, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71(20):6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 8.Yang Z, Zhou L, Wu LM, et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18(5):1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 9.Bhan A, Hussain I, Ansari KI, Kasiri S, Bashyal A, Mandal SS. Anti-sense transcript long noncoding RNA (lncRNA) HOTAIR is transcrip-tionally induced by estradiol. J Mol Biol. 2013;425(19):3707–3722. doi: 10.1016/j.jmb.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henderson BE, Ross R, Bernstein L. Estrogens as a cause of human cancer: the Richard and Hinda Rosenthal Foundation award lecture. Cancer Res. 1988;48(2):246–253. [PubMed] [Google Scholar]
- 11.Chisholm KM, Wan Y, Li R, Montgomery KD, Chang HY, West RB. Detection of long non-coding RNA in archival tissue: correlation with polycomb protein expression in primary and metastatic breast carcinoma. PLoS One. 2012;7(10):e47998. doi: 10.1371/journal.pone.0047998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He X, Bao W, Li X, et al. The long non-coding RNA HOTAIR is upregulated in endometrial carcinoma and correlates with poor prognosis. Int J Mol Med. 2014;33(2):325–332. doi: 10.3892/ijmm.2013.1570. [DOI] [PubMed] [Google Scholar]
- 13.Huang L, Liao LM, Liu AW, et al. Overexpression of long noncoding RNA HOTAIR predicts a poor prognosis in patients with cervical cancer. Arch Gynecol Obstet. 2014;290(4):717–723. doi: 10.1007/s00404-014-3236-2. [DOI] [PubMed] [Google Scholar]
- 14.Kim HJ, Lee DW, Yim GW, et al. Long non-coding RNA HOTAIR is associated with human cervical cancer progression. Int J Oncol. 2015;46(2):521–530. doi: 10.3892/ijo.2014.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Wang Y, Yu J, Dong R, Qiu H. A high level of circulating HOTAIR is associated with progression and poor prognosis of cervical cancer. Tumour Biol. 2015;36(3):1661–1665. doi: 10.1007/s13277-014-2765-4. [DOI] [PubMed] [Google Scholar]
- 16.Qiu JJ, Lin YY, Ye LC, et al. Overexpression of long non-coding RNA HOTAIR predicts poor patient prognosis and promotes tumor metastasis in epithelial ovarian cancer. Gynecol Oncol. 2014;134(1):121–128. doi: 10.1016/j.ygyno.2014.03.556. [DOI] [PubMed] [Google Scholar]
- 17.Sorensen KP, Thomassen M, Tan Q, et al. Long non-coding RNA HOTAIR is an independent prognostic marker of metastasis in estro-gen receptor-positive primary breast cancer. Breast Cancer Res Treat. 2013;142(3):529–536. doi: 10.1007/s10549-013-2776-7. [DOI] [PubMed] [Google Scholar]
- 18.Su X, Malouf GG, Chen Y, et al. Comprehensive analysis of long non-coding RNAs in human breast cancer clinical subtypes. Oncotarget. 2014;5(20):9864–9876. doi: 10.18632/oncotarget.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu L, Zhu G, Zhang C, et al. Association of large noncoding RNA HOTAIR expression and its downstream intergenic CpG island methylation with survival in breast cancer. Breast Cancer Res Treat. 2012;136(3):875–883. doi: 10.1007/s10549-012-2314-z. [DOI] [PubMed] [Google Scholar]
- 20.Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Med. 2009;6(7):e1000100. doi: 10.1371/journal.pmed.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17(24):2815–2834. doi: 10.1002/(sici)1097-0258(19981230)17:24<2815::aid-sim110>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 22.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li D, Duan Y, He L. Association study of serotonin 2A receptor (5-HT2A) gene with schizophrenia and suicidal behavior using systematic meta-analysis. Biochem Biophys Res Commun. 2006;340(3):1006–1015. doi: 10.1016/j.bbrc.2005.12.101. [DOI] [PubMed] [Google Scholar]
- 24.Li JT, Wang LF, Zhao YL, et al. Nuclear factor of activated T cells 5 maintained by Hotair suppression of miR-568 upregulates S100 calcium binding protein A4 to promote breast cancer metastasis. Breast Cancer Res. 2014;16(5):454. doi: 10.1186/s13058-014-0454-2. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339(2):159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 26.Zhang S, Chen S, Yang G, et al. Long noncoding RNA HOTAIR as an independent prognostic marker in cancer: a meta-analysis. PLoS One. 2014;9(8):e105538. doi: 10.1371/journal.pone.0105538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bardin A, Boulle N, Lazennec G, Vignon F, Pujol P. Loss of ERbeta expression as a common step in estrogen-dependent tumor progression. Endocr Relat Cancer. 2004;11(3):537–551. doi: 10.1677/erc.1.00800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hulka BS. Epidemiologic analysis of breast and gynecologic cancers. Prog Clin Biol Res. 1997;396:17–29. [PubMed] [Google Scholar]
- 29.Hulka BS, Moorman PG. Breast cancer: hormones and other risk factors. Maturitas. 2008;61(1–2):203–213. doi: 10.1016/j.maturitas.2008.11.016. discussion 213. [DOI] [PubMed] [Google Scholar]
- 30.Senechal C, Cottereau E, de Pauw A, et al. Environmental and genetic risk factors for endometrial carcinoma. Bull Cancer. 2015;102(3):256–269. doi: 10.1016/j.bulcan.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 31.Bhan A, Hussain I, Ansari KI, Bobzean SA, Perrotti LI, Mandal SS. Bisphenol-A and diethylstilbestrol exposure induces the expression of breast cancer associated long noncoding RNA HOTAIR in vitro and in vivo. J Steroid Biochem Mol Biol. 2014;141:160–170. doi: 10.1016/j.jsbmb.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.You QY, Tao H, Ling B. Long noncoding RNA HOX transcript anti-sense intergenic RNA (HOTAIR) as a foe and novel potential therapeutic target for endometrial carcinoma. Int J Gynecol Cancer. 2014;24(9):1536. doi: 10.1097/IGC.0000000000000277. [DOI] [PubMed] [Google Scholar]
- 33.Gokmen-Polar Y, Vladislav IT, Neelamraju Y, Janga SC, Badve S. Prognostic impact of HOTAIR expression is restricted to ER-negative breast cancers. Sci Rep. 2015;5:8765. doi: 10.1038/srep08765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deng Q, Sun H, He B, et al. Prognostic value of long non-coding RNA HOTAIR in various cancers. PLoS One. 2014;9(10):e110059. doi: 10.1371/journal.pone.0110059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ozgur E, Mert U, Isin M, Okutan M, Dalay N, Gezer U. Differential expression of long non-coding RNAs during genotoxic stress-induced apoptosis in HeLa and MCF-7 cells. Clin Exp Med. 2013;13(2):119–126. doi: 10.1007/s10238-012-0181-x. [DOI] [PubMed] [Google Scholar]
- 36.Bowyer HL, Dodd RH, Marlow LA, Waller J. Association between human papillomavirus vaccine status and other cervical cancer risk factors. Vaccine. 2014;32(34):4310–4316. doi: 10.1016/j.vaccine.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]