Abstract
Background
Chronic bacterial infections occur as a result of the infecting pathogen’s ability to live within a biofilm, hence escaping the detrimental effects of antibiotics and the immune defense system. Burkholderia pseudomallei, a gram-negative facultative pathogen, is distinctive in its ability to survive within phagocytic and non-phagocytic cells, to persist in vivo for many years and subsequently leading to relapse as well as the development of chronic disease. The capacity to persist has been attributed to the pathogen’s ability to form biofilm. However, the underlying biology of B. pseudomallei biofilm development remains unresolved.
Results
We utilised RNA-Sequencing to identify genes that contribute to B. pseudomallei biofilm phenotype. Transcriptome analysis of a high and low biofilm producer identified 563 differentially regulated genes, implying that expression of ~9.5 % of the total B. pseudomallei gene content was altered during biofilm formation. Genes involved in surface-associated motility, surface composition and cell wall biogenesis were over-expressed and probably play a role in the initial attachment of biofilms. Up-regulation of genes related to two component signal transduction systems and a denitrification enzyme pathway suggest that the B. pseudomallei high biofilm producer is able to sense the surrounding environmental conditions and regulate the production of extracellular polymeric substance matrix, a hallmark of microbial biofilm formation.
Conclusions
The transcriptome profile described here provides the first comprehensive view of genes that contribute to the biofilm phenotype in B. pseudomallei.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-015-1692-0) contains supplementary material, which is available to authorized users.
Keywords: B. pseudomallei, Biofilm, Transcriptome
Background
Bacterial cells have evolved a biofilm phenotype over billions of years as part of their successful strategy to colonize biotic and abiotic surfaces when faced with different environmental conditions. Biofilm consists of a biological architecture of aggregated microbes on a surface, enclosed within a mesh of exopolysaccharides, fatty acids, DNA and large surface proteins [1]. Biofilms are closely associated with persistence as the presence of the extracellular matrix surrounding the cells renders the bacteria less susceptible to anti-bacterial agents compared to free-floating cells [2]. As a result, biofilm infections tend to be chronic and are difficult to eradicate. The transition from free-swimming planktonic cells to biofilm producers occurs in response to environmental changes including pH, temperature, nutrient levels and ionic strength. This response involves multiple regulatory networks which translate signals to alter the reorganization of the bacterial cell to survive unfavourable conditions [3]. It is generally believed that quorum sensing contributes to the formation of a functioning biofilm. Human infections involving biofilm have been described in patients with native valve endocarditis, cystic fibrosis, periodontitis as well as chronic bacterial infections such as prostatitis. Biofilm formation in medical devices such as central venous and urinary catheters, prosthetic heart valves, intrauterine devices and contact lenses, is well described [4].
Burkholderia pseudomallei, the causative agent of melioidosis, is known to produce biofilm. A major feature of melioidosis is the difficulty in achieving complete bacterial eradication following an episode of infection and an extended period of antimicrobial treatment is needed for total clearance. Formation of biofilm has been proposed as a contributory factor in the occurrence of persistent infection in the host. Clinical response to antimicrobials is slow and recurrent disease is common [5]. Sawasdidoln et al. [6] demonstrated that B. pseudomallei isolates which were sensitive to doxycycline, ceftazidime, imipenem and trimethoprim/sulfamethoxazole became resistant under conditions that promoted the formation of biofilm.
Levels of humoral antibodies in patients who have had melioidosis remain high and seldom drop to basal level even years after recovery from an acute infection, supporting the notion of persistence [7]. It is possible that B. pseudomallei can adapt to survival in vivo through the formation of biofilm but the mechanism by which this occurs in melioidosis patients is unclear [8]. It has also been reported that B. pseudomallei biofilm does not contribute to the virulence of the organism [9]. Based on studies involving various B. pseudomallei mutants, acapsular mutants may or may not have reduced formation of biofilm [6, 10]. On the other hand, restricted biofilm formation was observed in the fliC flagella mutant [6] and the polyphosphate kinase ppk mutant [11] whilst the role of B. pseudomallei cyclic-di-GMP-phosphodiesterase (CdpA) in biofilm formation and virulence was established with the corresponding cdpA mutant being attenuated in human macrophage cells [12]. A recent report by Lazar-Adler et al. [13] proposed the role of B. pseudomallei Trimeric Autotransporter Adhesins (TAA) in biofilm formation whereby an insertional mutant of the BPSS1439 gene was affected in its ability to form biofilm in addition to being partially attenuated in an acute murine melioidosis model, implying a positive relationship between biofilm formation and bacterial virulence.
A number of studies involving individual mutants of the biofilm-associated genes described above have demonstrated that inactivating these single genes does not completely attenuate biofilm formation. This suggests a more global regulation of multiple B. pseudomallei genes and pathways involved in biofilm formation and may, either directly or indirectly, be related to virulence or persistence in infected hosts. Hence, in this study, a comprehensive transcriptional analysis of representative high and low clinical B. pseudomallei biofilm producers was performed to identify the genes required for biofilm formation in B. pseudomallei. In addition, preliminary virulence studies of these two B. pseudomallei biofilm producers were carried out using the nematode Caenorhabditis elegans and BALB/c mice infection models.
Results
Transcriptome analysis and global transcriptional profile of B. pseudomallei biofilm strains
The sequence based transcriptome approach has been used to study regulatory mechanisms and pathogenicity factors of Pseudomonas syringae [14], Mycobacterium tuberculosis [15], Pseudomonas aeruginosa [16], Listeria monocytogenes and Listeria innocua [17]. We utilised RNA sequencing and comparative transcriptome analysis to identify genes and their respective expression levels that contribute to the B. pseudomallei biofilm phenotype. A total of 84 B. pseudomallei clinical isolates were analysed for biofilm formation (Additional file 1). From this collection, we selected one representative from the high biofilm producers, UM6, and one of the low biofilm producers, UM1 for RNA-Seq analysis. The biofilm formation phenotypes of both these strains is presented in Fig. 1. Both strains were sequenced on the Illumina platform and sequence reads were mapped to the annotated B. pseudomallei strain K96243 genome. The expression analysis demonstrated that approximately 84.5 % of the UM1 and UM6 reads mapping to B. pseudomallei K96243 genes had a calculable ‘fragments per kilobase of million fragments mapped’ (FPKM) value (Additional file 2). The pattern of relative gene expression was similar between the biological replicates with a correlation coefficient of r = 0.86 and r = 0.87 for UM6 and UM1, respectively.
Fig. 1.
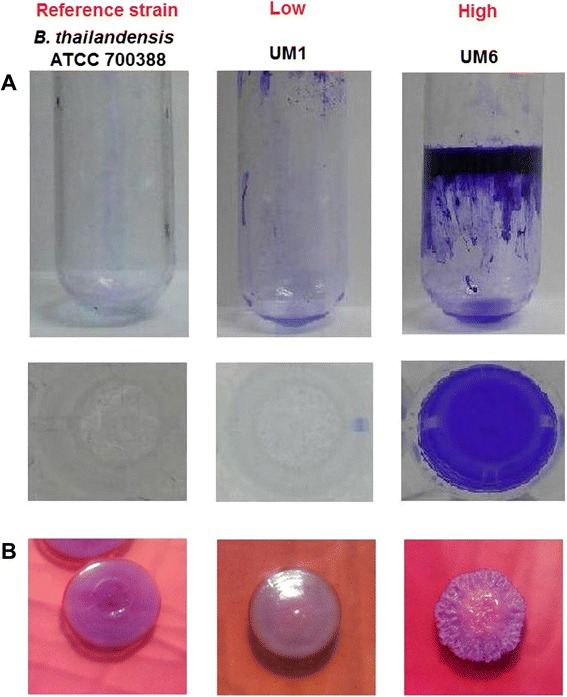
B. pseudomallei biofilm formation phenotypes. a Biofilm formation was measured by crystal violet staining assay using static broth cultures of B. pseudomallei UM1 (low producer) and UM6 (high producer) in a 96-well flat-bottomed microtiter plate and in test tubes. Non-pathogenic B. thailandensis ATCC 700388 was used as the reference strain in this study. b Colony morphology of B. thailandensis ATCC 700388, B. pseudomallei UM1 (low producer) and UM6 (high producer) on Ashdown agar plates after 48 h incubation at 37 °C
We next used the transcriptome data to identify genes that potentially contribute towards the biofilm phenotype in B. pseudomallei as determined by differential transcription analysis between UM6 and UM1. By adopting a q-value of ≤ 0.05 and log2 fold-change above 1 to classify a transcript as being differentially expressed, transcriptional analysis revealed 563 differentially expressed genes (324 up-regulated genes and 239 down-regulated genes) in UM6 relative to UM1. Functional classification of up- and down-regulated genes showed that most of these genes encode core functions such as cell envelope, central intermediary metabolism, energy metabolism, transport, regulatory proteins and cellular processes (Additional file 3). Many genes encoding proteins with unknown function or hypothetical proteins were also modulated in the high biofilm producer, UM6 (Additional file 3). Furthermore, genes predicted to encode proteins that are known to localise as extracellular proteins were observed at a higher percentage in the group of genes with up-regulated expression (Additional file 3). Eleven genes were randomly selected from seven functional categories (Fig. 2a and Additional file 4) for validation by quantitative real-time PCR (qRT-PCR). The expression was verified by qRT-PCR as up- or down-regulated, albeit with magnitudes different from those recorded by RNA-Seq (Fig. 2b and Additional file 5). In lieu of the large number of significantly differentiated genes, only data related to genes that have some functional information are shown and discussed below.
Fig. 2.
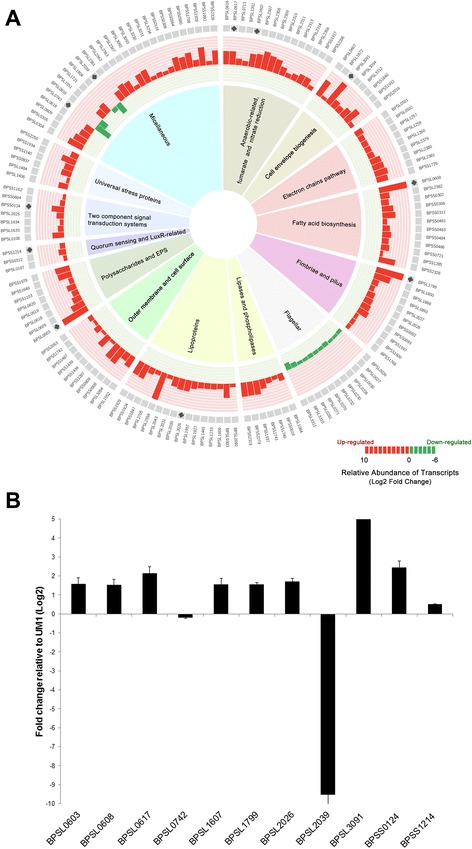
B. pseudomallei biofilm development transcriptome profile. a Expression profiles of modulated-genes according to functional categories. Transcript expression of log2 fold level are depicted by the histogram. The height of the bars correspond to the degree of expression level. Red and green bars represent up-and down-regulation in relative expression levels, respectively. b qRT-PCR analysis of eleven B. pseudomallei genes from seven functional categories differentially expressed as determined by RNA-Seq. The results are from a representative of three reproducible independent experiments
Fimbriae and pilus may be required for initiation of B. pseudomallei biofilm attachment
Motility influences biofilm formation in various pathogens including Enteropathogenic E. coli [18, 19] and P. aeruginosa [20, 21]. A number of fimbriae and pilus-related genes (BPSL1799, BPSL1888, BPSL1893, BPSL2027, BPSL2028, BPSS0092, BPSS1597, BPSS1600 and BPSS1768) were significantly up-regulated in UM6 (Figs. 2a and 3 and Additional file 4) suggesting that these structures may also be important in B. pseudomallei biofilm attachment. To validate the observation of over-expressed pili-related genes, we performed scanning electron microscopy (SEM) on both strains. The micrographs (Fig. 4) demonstrate the presence of pili protruding from UM6 which are not observed in UM1, thus supporting the transcriptional-level analysis (Fig. 4).
Fig. 3.
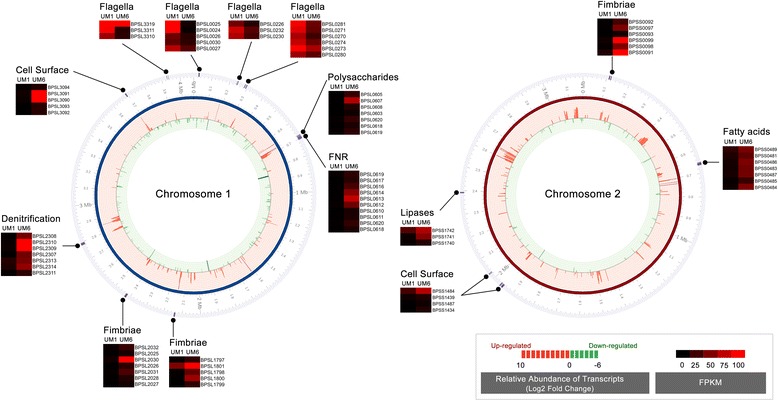
Alteration of surface composition and regulation of anaerobic respiration contribute to B. pseudomallei biofilm phenotype. Transcript expression of log2 fold level are depicted by the histogram in the inner-most ring of the circular maps. The height of the bars correspond to the degree of expression level. Genes uniquely expressed in UM6 and UM1 are coloured in dark red and dark green, respectively. Red and green bars represent up- and down-regulation in relative expression levels, respectively. Hierarchical clustering of B. pseudomallei UM1 (low biofim producer) and UM6 (high biofilm producer) expression profiles according to functional categories. The heat maps indicate the gene transcripts expressed as FPKM. Genes whose expression did not change are coloured in black
Fig. 4.
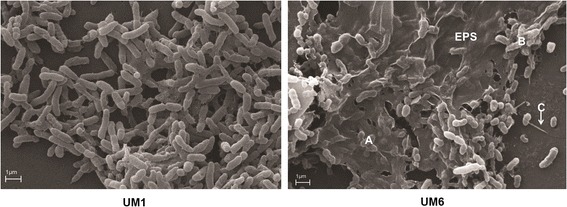
Scanning electron microscope micrograph of biofilm formation by B. pseudomallei on a glass slide. B. pseudomallei UM1demonstrated reduced biofilm formation compared to UM6. a Extracellular polymeric substance (EPS) is clearly visible around the B. pseudomallei UM6 colonies. b The matrix is holding the bacteria together but has not yet been encapsulated. c Pilus protruding from a UM6 colony
To gain more insight into the identified genes, we used the Conserved Domain Database (CDD) to look at protein domains of these genes. BPSL2027 (putative fimbriae-related protein) contains an usher superfamily domain (pfam0057) and PapC C-terminal (pfam13953) and N-terminal conserved domains (pfam13954). Concomitantly, two outer membrane usher proteins, BPSS0093 (4.07 fold) and BPSL1800 (5.99 fold), were over expressed in UM6 and may be involved in assembly of pre-pilins for fimbriae biogenesis. In contrast, a cohort of flagella-related genes encoding the flagella basal body, motor switch and hook proteins (BPSL0026, BPSL0027, BPSL0030, BPSL0226, BPSL0230, BPSL0232, BPSL0270, BPSL0271, BPSL0281, BPSL3310 and BPSL3311) were down-regulated in UM6 (Figs. 2a and 3 and Additional file 4), similar to that previously described for Pseudomonas aeruginosa [22] and Escherichia coli biofilm formation [22]. The down-regulation of flagella-related genes was validated with motility assays performed on solid agar, which demonstrated that the UM6 strain has reduced swimming and swarming motility compared to the UM1 strain (Table 1).
Table 1.
Swimming and swarming analysis of UM1 and UM6. Data are mean ± SD of two independent experiments
| Phenotype | UM1 | UM6 |
|---|---|---|
| Swimming (mm in diameter) | 69.55 ± 7.14* | 53.4 ± 1.98 |
| Swarming (mm in diameter) | 74.55 ± 3.62* | 55.82 ± 2.36 |
*p < 0.05 (Students’ t test)
Over expression of extracellular polymeric substance (EPS) matrix components for development of B. pseudomallei biofilm architecture
Genes encoding for polysaccharides (BPSL0603, BPSL0605, BPSL0618, BPSL0619, BPSL0620, BPSS1649) and EPS (BPSS1978) were over-expressed in UM6 (Fig. 2a and Additional file 4). Observations based on SEM micrographs support the expression profile whereby the presence of EPS matrix encapsulating the bacteria was only seen in the high biofilm producer strain UM6 but not in the low biofilm producer strain UM1 (Fig. 4). Attachment of Gram-negative bacteria to a surface via outer membrane proteins is the first step in biofilm formation, followed by replication to form micro colonies and production of a mature biofilm [23]. Several outer membrane and cell-surface encoded proteins were also over-expressed in UM6, including BPSS0908, BPSS0909, BPSS1287, BPSS1487, BPSS1742, BPSS1434, BPSS1439, BPSS2053, BPSL1552 and BPSL3094 (Figs. 2 and 3 and Additional file 4).
Alteration of B. pseudomallei surface composition in a high biofilm producer
Expression levels of 13 fatty acid biosynthesis-related genes (BPSL0608, BPSL0618, BPSL2382, BPSS0302, BPSS0306, BPSS0311, BPSS0481, BPSS0483, BPSS0484, BPSS0486, BPSS0712, BPSS1285 and BPSS2328), seven phospholipases and lipase-related genes (BPSL1064, BPSS0016, BPSS1740, BPSS1741, BPSS1937, BPSS2279 and BPSS2319) as well as seven cell envelope biogenesis-related genes (BPSL0607, BPSL1872, BPSL3094, BPSL3312, BPSS1840, BPSS1932 and BPSS2016) were up-regulated in the high biofilm producer (Figs. 2 and 3 and Additional file 4). Furthermore, 13 lipoprotein-encoding genes (BPSL0092, BPSL0303, BPSL1233, BPSL1445, BPSL1927, BPSL1957, BPSL2026, BPSL2043, BPSL2359, BPSL2705, BPSS1847, BPSS1924 and BPSS1929) were also over-expressed in UM6. One of the putative lipoproteins (BPSL2026) contained a spore coat protein U domain, SCPU (cl02253), which is generally present in the bacterial family of secreted pili proteins involved in motility and biofilm formation. Concomitantly, three putative SCPU domain containing export protein genes, BPSL1009 (1.95 fold), BPSL2030 (4.01 fold) and BPSL2031 (3.51 fold), were also identified as over-expressed in UM6 (Fig. 3 and Additional file 4).
LuxR-like domain is likely to be involved in B. pseudomallei biofilm formation
Quorum sensing (QS) is a form of cell to cell communication that bacteria adopt to coordinate group behaviour in a cell density dependent manner. QS relies on N-acyl homoserine lactones (AHLs) to regulate gene expression in response to cell density dependent cues and is related to biofilm formation and exopolysaccharide production [1, 20, 24]. In addition, QS influences the expression profile of diverse genes including antibiotic tolerance and virulence determinants [2]. The QS system plays a major role in the control of bacterial biofilm formation in many known pathogens including P. aeruginosa [20, 25], Streptococcus pneumoniae [26] and E. coli [27, 28]. In this study, the expression levels of homoserine O-acetyltransferase (BPSL0197) and the LuxR-family transcriptional regulator (BPSS0312), which, together mediate gene expression following association with the cognate AHL (s), were up-regulated in UM6 (Fig. 2a and Additional file 4).
Up-regulation of two component signal transduction systems and stress proteins in the B. pseudomallei high biofilm producer
The two component signal (TCS) transduction system related proteins, a sensor histidine kinase protein and response regulator, are responsible in regulating biofilm formation in a number of bacteria. Several genes related to the two-component signal transduction systems (BPSL0106, BPSL1633, BPSL1634, BPSL2025, BPSL2314, BPSS0124, BPSS0664 and BPSS1162) were up-regulated in UM6 (Fig. 2a and Additional file 4). Interestingly, two putative sensor kinases (BPSL2025 and BPSL1634) demonstrated considerable similarity to the E. coli RcsC sensor protein, particularly at the conserved domains (Additional file 6).
Four genes encoding response regulators (BPSL1633, BPSL2314, BPSS0124 and BPSS1214) that contained a LuxR-like domain (cd06170) were identified as up-regulated in UM6 (Fig. 2a and Additional file 4). Amongst the identified genes, a hypothetical protein (BPSL0106) containing the CpxP component domain (cl01482), was up-regulated by 4.1 fold. Proteins containing the Cpx component domain are known modulators of cell-envelope stress in Gram-negative bacteria including E. coli biofilm-producing cells [29]. In addition, genes encoding two universal stress proteins (BPSS1140, BPSS1934) and one hypothetical protein (BPSS0837) with a universal stress protein family domain (cd00293) as well as genes of three stress–related proteins (BPSS2250, BPSL1484 and BPSL1406) were also up-regulated in UM6.
Modulation of the denitrification enzyme pathway in the B. pseudomallei high biofilm producer
Two anaerobic-related genes (BPSL2309 and BPSL2356), three reductase genes involved in nitrate metabolism (BPSL2351, BPSL1607, BPSS1487) and several genes encoding fumarate and the nitrate reduction (FNR) subfamily were over expressed in UM6 (Figs. 2a and 3 and Additional file 4). The majority of these genes encode for proteins involved in nitrate regulation and dissimilation including nitrate reductases (BPSL2309, BPSL2310, BPSL2311), nitrate-oxide reductase (BPSL1607), nitrate sensor protein (BPSL2313) and nitrate extrusion proteins (BPSL2307, BPSL2308) (Fig. 3 and Additional file 4). Of interest, two nitrite extrusion proteins and a transport-related membrane protein (BPSS2206) contain the major facilitator superfamily (MFS) domain (cd06174), which is involved in the symport, antiport or uniport pumping of various substrates such as sugars, oligosaccharides and antibiotics [30]. Moreover, one of the crp-family transcriptional regulators (BPSS1917) contained the effector domain of the CAP family transcription factor (cd00038) whilst two hypothetical proteins (BPSL0616 and BPSL0617) that also contained the same domain, were up regulated (4.72 fold and 4.74 fold, respectively) in UM6 compared to UM1. Up-regulation of MFS-containing genes has recently been associated with the development of biofilm by P. aeruginosa [31] as well as adherence and biofilm formation for Acinetobacter baumannii [30].
Potential correlation between B. pseudomallei biofilm formation and virulence in nematode and mice models
Biofilm formation has been implicated as a virulence factor in C. elegans infection models for Yersinia pseudotuberculosis [32] and staphylococcal infections [33]. Hence, we used the C. elegans host model to evaluate virulence of the different B. pseudomallei biofilm producers and determine the contribution of biofilm in B. pseudomallei-mediated killing of C. elegans. Nematodes were fed with B. pseudomallei UM1 (low biofilm producer) and UM6 (high producer), respectively, and the non-pathogenic B. thailandensis ATCC 700388 [34]. As shown in Fig. 5a, worms exposed to the laboratory food source E. coli OP50 remained completely viable over the course of the experiment. Worms exposed to UM6 died significantly faster (Logrank (Mantel-Cox) test p < 0.0001) with a mean time to death (TDmean) of 13.897 ± 0.401 h compared to worms exposed to B. thailandensis (TDmean = 77.631 ± 1.638 h) and UM1 (TDmean = 77.876 ± 1.183 h). The preliminary survival/virulence assay demonstrates that B. pseudomallei biofilm production could be a contributing virulence factor in the pathogenesis of this bacterium.
Fig. 5.
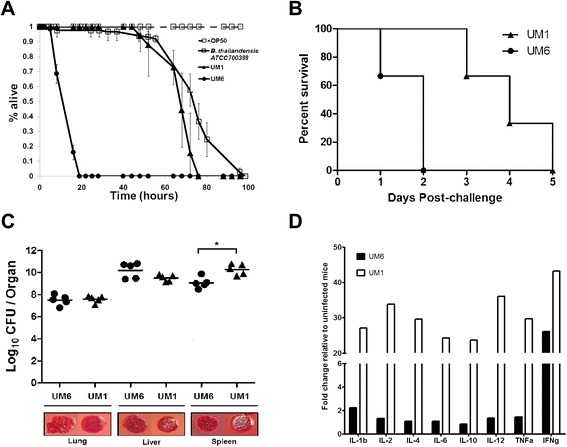
B. pseudomallei biofilm may contribute to bacterial pathogenesis. a B. pseudomallei biofilm contributes towards lethality in C. elegans. One-day old Glp worms were transferred to individual B. pseudomallei isolates UM1 (black line, solid triangles), UM6 (black line, solid circles), B. thailandensis ATCC 700388 (black line, open squares) and E. coli OP50 (black dashed line, open squares). The graph shows the mean ± SD of three replicates (30 worms/replicate) from a representative of two independent experiments. b Mice (n = 5) were challenged intraperitonealy with a lethal dose of B. pseudomallei UM6 (circle) or UM1 (triangle) and their survival was monitored. Mice challenged with UM6 succumbed to disease significantly faster (within 24 h) than those challenged with UM1 [Logrank (Mantel-Cox) test, p-value = 0.0084]. c The bacterial loads in the spleen, liver and lung of B. pseudomallei-infected mice are shown. Each symbol represents one mouse. The horizontal line indicates the geometry mean for each group. Significance was determined using the Mann–Whitney test (*p < 0.05). Representative colony morphologies of B. pseudomallei UM1 and UM6 harvested from the infected mice organs are shown. d B. pseudomallei UM6 attenuates production of various cytokines. Lungs of BALB/c mice challenged with B. pseudomallei UM1 or UM6 were harvested and the levels of 12 pro- and anti-inflammatory cytokines were measured by the mouse cytokine Multi-Analyte ELISArray Kit (Qiagen). Cytokine expression is shown as fold increase compared to the control unchallenged mice
BALB/c mice serve as a well-established animal model for melioidosis. To confirm the findings in C. elegans, mice were challenged intraperitoneally with a lethal dose of UM6 or UM1 and mice survival was monitored. As observed in C. elegans, mice infected with UM6 died significantly faster than those infected with UM1 (Fig. 5b). All mice infected with UM6 succumbed to disease within 24 h with a median survival of 1 day while only 1 mouse infected with UM1 died on day one and the remaining 4 mice succumbed to disease on day 3 with a median survival of 3 days. Although UM6 appeared to be more virulent, bacterial loads in the lungs and livers of UM6 and UM1-infected mice were similar, and the spleens of UM6 infected mice displayed a significantly lower bacterial count compared to UM1 infected mice (Fig. 5c).
The high biofilm producer was able to kill both mice and nematode relatively quickly suggesting an imbalance between the host proinflammatory and anti-inflammatory responses towards infection. Hence, we asked if the presence of the biofilm deregulated this equilibrium by limiting the cytokine response to infection. To address this question, the mouse cytokine Multi-Analyte ELISArray Kit (Qiagen) was utilised to simultaneously measure 12 cytokines i.e., IL-1α, IL-1β, IL-2, IL-4, IL-6, IL-10, IL-12, IL-17A, Interferon-γ (IFN-γ), Tumour necrosis factor-α (TNF-α), Granulocyte- Colony Stimulating Factor (G-CSF) and Granulocyte-Macrophage Colony Stimulating Factor (GM-CSF). We observed a significant attenuation in the levels of all 12 cytokines within the lungs of mice infected with UM6 compared to lungs from mice infected with UM1 (Fig. 5d). In summary, we propose that high levels of biofilm production attenuate the cytokine response which may explain the increased virulence of B. pseudomallei UM6.
A similar gene expression profile is observed in other B. pseudomallei high and low biofilm producing strains
The transcriptional data presented here have identified genes that most likely contribute towards the biofilm phenotype in B. pseudomallei. To confirm that the observed gene expression pattern is not restricted to UM6 and UM1, we selected a second high biofilm producing strain (UM5) as well as a low B. pseudomallei biofilm producing strain, UM2 (Fig. 6a) to analyse the expression profile. Seven of 11 genes that were modulated in UM6 (Fig. 2b) were analysed, including genes associated with the denitrification pathway, cell envelope and EPS production. Six of the selected genes were also over-expressed in UM5 compared to UM2 (Fig. 6b). Both isolates were also analysed in C. elegans (Fig. 6c) and mice (Fig. 6d) infection assays and we observed that the high biofilm B. pseudomallei strain UM5 contributed to higher killing kinetics in both animal models.
Fig. 6.
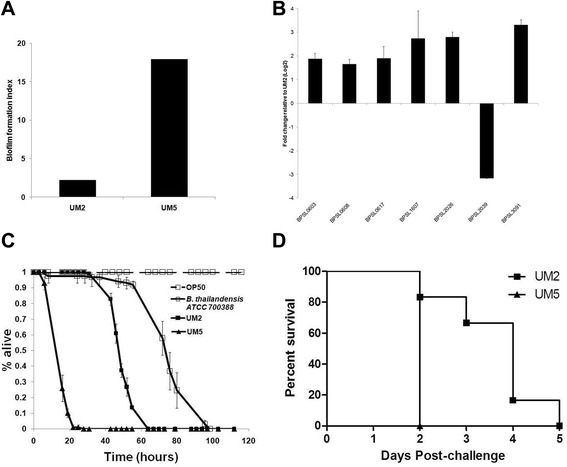
The B. pseudomallei high biofilm producer, UM5, also results in faster killing kinetics and over-expression of biofilm-associated genes compared to a second low biofilm producing isolate, UM2. a Biofilm index of UM2 and UM5. b qRT-PCR analysis of B. pseudomallei genes from seven functional categories differentially expressed by RNA-Seq analysis. The results are from a representative of three reproducible independent experiments. c Killing assay of C. elegans infected with UM2 (black line, closed square), UM5 (black line, closed triangle) and B. thailandensis (black line, open square). The graph shows the mean ± SD of three replicates (30 worms/replicate) from a representative of two independent experiments. d Mice (n = 5) were challenged intraperitonealy with a lethal dose of B. pseudomallei UM5 (triangle) or UM2 (square) and their survival was monitored. Mice challenged with UM5 succumbed to disease significantly faster (within 24 h) than those challenged with UM2 [Logrank (Mantel-Cox) test, p-value = 0.0084]
Discussion
B. pseudomallei biofilm formation may contribute to intracellular survival, dormancy and antibiotic resistance [35] but the mechanism by which this occurs in humans is yet to be demonstrated [8]. To date, only a handful of B. pseudomallei biofilm-associated genes have been studied and have mainly focused on single-gene phenotypes. In this study, we demonstrate that biofilm production is a complex process that involves the differential expression of several genes. RNA sequencing analysis performed on low and high biofilm-producing B. pseudomallei strains identified genes that contribute to biofilm formation. We identified 563 differentially expressed genes during the formation and growth of biofilm, accounting for about 9.5 % of the total B. pseudomallei gene content. The transciptome analysis of biofilm related genes was performed on mid-log bacterial cultures, the pre-biofilm state, to conform to standard sequencing protocols. Keeping in mind that our analysis may not necessarily reflect a true biofilm environment, we subsequently analysed a subset of genes from 7 representative functional groups on UM1 and UM6 cells grown to the stationary phase (mature biofilm state). qRT-PCR analysis demonstrated comparable magnitudes and patterns of gene expression between RNA samples extracted from cells at different growth phases (data not shown). Thus, our findings offer new insights into the different transcriptional landscapes observed between clinical B. pseudomallei high and low biofilm producing isolates.
Biofilm producing pathogens sense environmental signals via the TCS transduction system and adapt to these changes by transcribing genes that planktonic organisms do not [21, 36]. Based on the expression profile of the high biofilm producer, we hypothesise that B. pseudomallei also responds to varied environmental signals, for instance pH, temperature, osmotic pressure and oxygen concentration via activation of various TCS (Fig. 7). Upon encountering the environmental cues that stress the cell membranes, B. pseudomallei most likely activates the RcsB-RcsC TCS and subsequently regulates genes that encode proteins involved in the alteration of surface components, including capsular polysaccharides, cell envelop biogenesis, lipoproteins, phospolipases and fatty acid biosynthesis that are pivotal for survival of B. pseudomallei within the host (Figs. 2a and 7). RcsC sensor kinase is required for biofilm formation in E. coli and regulates genes encoding for proteins that are either localised to the envelope or have activities that affect the structure/properties of the bacterial surface [21, 36]. In addition, the cell-to-cell communication small fatty acid signal molecule, diffusible signal factor (DSF), regulates the expression of factors contributing to virulence, antibiotic tolerance and biofilm formation [37, 38]. DSF is synthesized by putative enoyl-CoA hydratase and putative acyl-coA ligase in Xanthomonas campestris and Burkholderia cecocepacia [39–41]. Three fatty acid biosynthesis genes that encode for Co-A hydratase and ligase are up-regulated in UM6. This, in turn, most likely regulates EPS-associated genes, the core component for maintenance of biofilm architecture and pilus biogenesis-related genes to initiate the attachment of planktonic cells for microcolony formation.
Fig. 7.
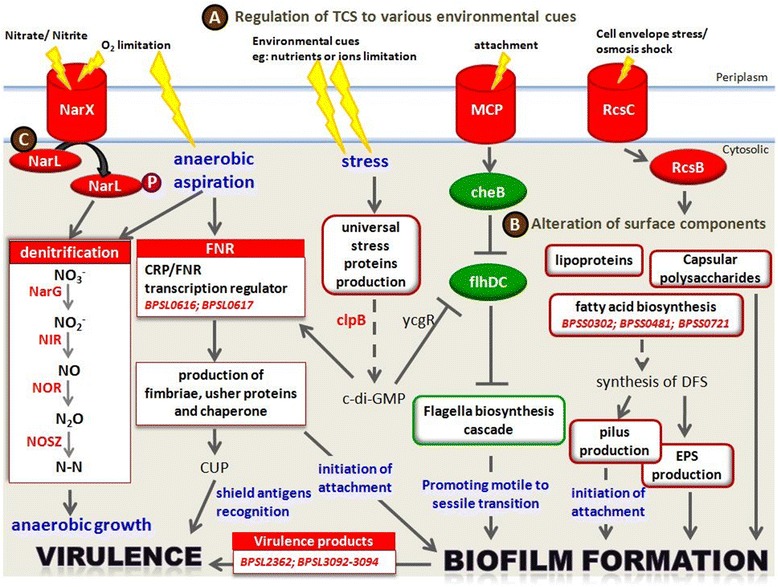
Summary of B. pseudomallei genes that contribute to biofilm development. a In response to varied environmental signals, B. pseudomallei regulates the two-component signal (TCS) transduction system to adapt to environmental changes, by (b) transcribing genes that encode proteins involved in the alteration of surface components and components crucial to initiate attachment of planktonic bacterial cells to solid surfaces for microcolony formation. As the biofilm grows, an oxygen-limited environment forms. c The high biofilm producing B. pseudomallei strain utilises nitrate for anaerobic growth through the denitrification enzyme pathway. Arrow represents activator connection; solid lines represent direct activation; dashed lines represent indirect activation. The boxes and fonts highlighted in red depict the up-regulated biological processes and genes, respectively, while the box highlighted in green depicts the down-regulated biological processes
During the growth of micro colonies, methyl- accepting chemotaxis protein MCP (BPSL2367), a sensor protein in TCS, is up-regulated in UM6 and subsequently suppresses the regulation of CheB which is responsible for flagella switch (Fig. 7). Down regulation of the flagella biosynthesis cascade seen in the high biofilm producer (Figs. 2a and 3) suggests sessile transition of motile B. pseudomallei for biofilm development [28]. Bacterial biofilm formation is known to affect colony morphotype (mucoid vs non-mucoid) [20] and bacterial attachment [42]. This is also true for B. pseudomallei biofilm, whereby the low biofilm producer UM1 that exhibits a mucoid colony morphotype, has lower expression levels of pilus, fimbriae, lipoprotein, polysaccharide and EPS-related genes when compared to the wrinkled colony morphotype observed in UM6 (Fig. 1). This observation is similar to a P. aeruginosa mutant with a wrinkled colony phenotype capable of forming pronounced pellicles that exhibited increased production of exopolysaccharide and EPS [20].
Recently, the CRP/FNR superfamily protein Bcam1349 of Burkholderia cenocepacia (Bcc) was reported as a cyclic diguanosine monophosphate (c-di-GMP) effector in regulating biofilm formation and is involved in virulence [43]. This protein regulates the increased production of cellulose and fimbriae [43]. Two hypothetical proteins (BPSL0616 and BPSL0617) with the CRP/FNR superfamily conserved domain were significantly up-regulated in UM6 (Fig. 2a and Additional file 4). Protein sequence analysis revealed that BPSL0617 has 69 % identity to Bcam1349, suggesting that it is the ortholog of the Bcc Bcam1349 (Additional file 7). Of note, the neighboring genes of these two hypothetical proteins were also up- regulated and a number of these genes are involved in surface protein modification (Additional files 4 and 8). Another member of the CRP/FNR family protein, the c-di-GMP response regulator ClpB heat-shock protein (BPSL1484) is also up-regulated in UM6. Furthermore, a cohort of fimbriae related genes, including three chaperone-usher gene clusters (BPSL1799–BPSL1801; BPSL2026–BPSL2028; BPSS0091–BPSS0093) which make up the chaperone-usher fimbriae pathway (CUP) were also over expressed in UM6 (Additional files 4 and 7). Up-regulation of fimbriae genes in biofilm formation seen in this study are similar to that in Klebsiella pneumoniae [44] and E. coli [45] which promotes adhesion to abiotic surfaces. In addition, P. aeruginosa fimbrial CupE/Csu proteins that contain the SCPU sub-domain are reportedly involved in structuring of biofilm [46]. Concomitantly, three hypothetical proteins with the conserved SCPU domain (BPSL1009, BPSL2030 and BPSL2031) up-regulated in the high biofilm producer may likely play a role in B. pseudomallei biofilm formation.
As the micro colonies mature into a progressively thick biofilm, a nutrient and oxygen-limited environment forms [21, 25] and the anaerobic fitness of the pathogen is pivotal for survival in the biofilm [47]. P. aeruginosa uses nitrate as an alternate electron acceptor through a denitrification enzyme pathway during the anaerobic growth of biofilm [48, 49] and this is regulated by ANR, an ortholog of the E. coli FNR [48, 49]. This transcriptional profiling suggests that the high B. pseudomallei biofilm producer may sense oxygen limitation through the NarX/NarL TCS and subsequently activate the nitrate reductase operon (BPSL2307–BPSL2314) and several types of nitrate reductases. This suggests that the facultative anaerobe B. pseudomallei is also likely to utilise nitrate for anaerobic growth in biofilm through the denitrification pathway regulated by BPSL0617 (Fig. 7). To our knowledge, this is the first report that describes the involvement of the TCS and denitrification enzyme pathways within the anaerobic environment in B. pseudomallei biofilm development.
Biofilm formation in other pathogenic bacteria such as Staphylococcus aureus and Streptococcus pneumoniae has been reported to be associated with altered host immune responses [50, 51]. Our preliminary study suggests that B. pseudomallei biofilm is probably a contributing factor towards virulence in both C. elegans and BALB/c mice models (Fig. 5a and b). Furthermore, UM6, the high biofilm producer strain failed to elicit the expected cytokine response even though the number of recoverable CFU was similar for the two strains (Fig. 5d). Many inflammatory cytokines referred to as the “core host immune response” molecules commonly seen in general inflammation infections including melioidosis [52], were not over expressed in UM6 infected-mice. This attenuation of in vivo inflammation suggests that intracellular B. pseudomallei are camouflaged from the host immune defense response by the biofilm causing the host to succumb to the infection. Although our findings challenge the previous report by Taiweechaisupapong et al. [9], both studies are limited by the small number (n = 2) of isolates to conclude a positive or negative association between B. pseudomallei biofilm formation and virulence and the analysis of a larger B. pseudomallei strain cohort should be undertaken.
Conclusions
In summary, this is the first report of the complete transcriptome profile of a B. pseudomallei biofilm producer. We have identified genes that are likely involved in the development of the B. pseudomallei biofilm phenotype, including quorum sensing, motility and surface composition-related genes (Fig. 2a). Interestingly, many of these genes are clustered together in the genome and may be regulated as an operon (Additional file 8). We postulate that the ability to sense various environmental cues and adapt to anaerobic conditions via the denitrification enzyme pathway is pivotal for the formation of B. pseudomallei biofilm in the infected host which subsequently allows for persistent infection in chronic melioidosis. B. pseudomallei is particularly recalcitrant to antibiotic treatment and this is most likely attributable to biofilm formation. Thus, novel strategies designed to thwart B. pseudomallei biofilm formation or to block a specific biofilm developmental stage, such as the use of anti-adhesion agents and inhibitors which interfere with signal transduction, are exciting avenues for the development of potent and bioavailable treatment strategies.
Methods
Bacteria
The four clinical B. pseudomallei isolates (UM1, UM2, UM5 and UM6) as well as two reference strains, B. thailandensis ATCC 700388 [53], and B. pseudomallei K96243 [54] used in this study are listed in Additional file 9.
Sample cultivation, RNA isolation and sequencing
Overnight cultures of B. pseudomallei (K96243, UM1 and UM6) were diluted 1:100 in 50 mL BHI broth and were grown at 37 °C until mid- logarithmic phase (OD600 = 0.5). Total RNA was isolated from two biological replicates of B. pseudomallei UM6 and UM1 harvested at mid- logarithmic growth phase using TRIzol (Invitrogen Life Technologies, CA, USA) according to the manufacturer’s instructions. Residual DNA was completely removed using QIAGEN’s RNase-Free DNase Set and complete DNA removal was validated by performing PCR with the B. pseudomallei recA gene primers. The integrity of the total RNA was assessed on the Agilent 2100 Bioanalyzer. Total RNA (10 μg) was subjected to 23 s and 16 s ribosomal RNA removal using the MicrobExpress kit (Ambion, CA, USA). Ribosomal depleted RNA was resuspended in 5 μL elution buffer (Qiagen, GmbH, Germany). A total of 15.5 μL Elute Prime Fragment Mix from the (non-stranded) TruSeq RNA Sample Prep kit (Illumina, CA, USA) was mixed with 4 μL of ribosomal depleted RNA and used for RNA fragmentation followed by cDNA synthesis, end-repair, TruSeq indexed-adapter ligation and PCR enrichment as per the TruSeq RNA sample preparation protocol (Illumina, CA, USA). A total of 6 libraries (2 biological replicates of each bacterial sample), each labelled with a unique index, were multiplexed in one flow cell lane and the sequencing run was performed on the Illumina HiSeq2000 sequencing platform.
Mapping and analysis of Illumina reads
Sequence reads from each sample were quality pre-processed using the FASTX-toolkit fastq_quality_filter. Trimming was based on the minimum accepted lllumina quality value of 20 and minimum accepted read size of 30 bp. The pre-processed reads were separated between paired and orphan reads using the Python script. Only the paired reads were used in the analysis while orphan reads were discarded. After pre-processing, an average of 7.5 million reads, corresponding to 95 % of the total reads, were mapped to chromosomes 1 and 2 of the B. pseudomallei strain K96243 genome sequence (GenBank Accession numbers NC006350 and NC006351). Due to the absence of genome sequences for both UM1 and UM6, this approach may be biased against the accessory genome of B. pseudomallei, however, transcripts that mapped to the core genes were the main interest of this study. Mapping generating four total transcriptome profiles (Additional file 2) using the alignment tool TopHat version 2.02 [55] integrated with Bowtie version 0.12.7 [56]. The TopHat default settings were used: 20 alignments per read were allowed with up to 1 mismatch per alignment. To determine differential expression of known transcripts, the resulting aligned reads were analysed by Cuffdiff, a part of the Cufflinks package version 2.02 [55] and expression of those transcripts was reported as fragments per kilobase of transcript per million mapped reads (FPKM). Overall, ≥ 87 % of the generated transcriptome reads were mapped to the B. pseudomallei K96243 reference genome. Transcripts with a q-value of ≤ 0.05 and log2 fold-change above 1 were considered as differentially expressed transcripts. Sequence reads were deposited in the database of the European Nucleotide Archive with accession number PRJEB6085 and are accessible via http://www.ebi.ac.uk/ena/data/view/PRJEB6085. The sample accession numbers are ERR475457 (UM1;1st replicate), ERR475458 (UM1; 2nd replicate), ERR475459 (UM6;1st replicate) and ERR475460 (UM6; 2nd replicate).
Hierarchical clustering
Selected data were organized by a hierarchical clustering with the web-based software Cluster 3.0. The clustering algorithm is based on an uncentered correlation metric, with average linkage clustering and visualized using Java Treeview V1.1.3. [52]
PSORT
The cellular localization of each differentially expressed gene was predicted using PSORTb version 3.0.2 (http://www.psort.org/psortb/). For the run, the following parameters were used: Organism type: Bacteria; Gram stain: Negative. BPSLt38 was excluded from the analysis as it is a tRNA.
Gene ontology
Functional classifications were carried out based on Comprehensive Microbial Resources (CMR) annotations (www.cmr.jcvi.org) as previously described by Chieng et al. [57].
Quantitative real-time PCR (qRT-PCR)
qRT-PCRs were performed with total DNase-treated RNA on the Bio-RadiCycler (BioRad Laboratories, USA) to quantify the expression of eleven genes from seven functional categories. Briefly, 20 μL reactions were performed using the iScript™ One-Step RT-PCR kit with SYBR Green according to the manufacturer’s instructions (BioRad Laboratories, USA), primers at a final concentration of 1 μM and a data acquisition temperature of 76 °C. In order to control for variation in RNA concentration, cycle threshold (Ct) values were normalized to B. pseudomallei 16 s rRNA that does not change with infection [58]. Primer sets used in this study are shown in Additional file 10.
Scanning electron microscopy analysis of biofilm formation
Bacteria were cultured as previously described [34]. Briefly, overnight cultures of B. pseudomallei (UM1 and UM6) were diluted 1:100 into 50 mL of fresh BHI broth and grown overnight in a shaking incubator at 37 °C. At the end of the incubation, the bacterial density was adjusted to OD600 = 1 using a spectrophotometer. For each isolate, 2 mL of bacterial suspension was added to a 12-well plate with 10 mm × 10 mm glass slides placed inside each well. Biofilms were allowed to form on the slides at 37 °C for 48 h following which, the samples were fixed in 4 % (v/v) glutaraldehyde in 0.05 M phosphate buffer (pH 7.0) at 4 °C for 12 h. Subsequently, the samples were washed three times in phosphate buffer, dehydrated through a graded ethanol series, dried in a critical-point drying apparatus with liquid carbon dioxide, sputter coated with gold and viewed using a LEO 1450VP (Electron Microscopy Unit, Universiti Kebangsaan Malaysia).
Motility assays
Motility assays on solid agar were performed using B. pseudomallei that had been cultured on Ashdown’s agar at 37 °C in air for 48 h. Swim agar plates were composed of 1 % tryptone, 0.5 % NaCl, 0.3 % agar whilst 0.5 % agar plates were used to observe swarming. Bacterial cells from an isolated colony was point inoculated into the centre of a swim plate or on the surface of a swarm plate using a sterile toothpick. Plates were incubated at 37 °C in air for up to 72 h, after which the widest colony diameter was measured represented by the circular turbid zone (swim plates) or migratory growth pattern (swarm plates) formed by the bacterial cells migrating away from the point of inoculation [59].
C. elegans survival assays
The wild type C. elegans N2 strain used in this study was obtained from the Tan Laboratory at Stanford University. The nematode was propagated on nematode growth medium (NGM) and fed on the normal food source, E. coli OP50 [60], at 16 °C.
C. elegans survival assays were performed as previously described [61, 62] with minor modifications. B. pseudomallei isolates (UM1, UM2, UM5 and UM6), B. thailandensis ATCC 700388 and E. coli OP50 were grown overnight in 1 mL Brain Heart Infusion (BHI) broth or LB broth at 37 °C. Ten μL of an overnight culture was spread over a small area on 3.5-cm NGM plates and incubated at 37 °C for 24 h. Plates were then allowed to equilibrate to room temperature for 12–24 h before use. Glp worms were prepared as previously described [63] and thirty age-matched Glp worms were transferred to NGM plates seeded with individual Burkholderia isolates and incubated at 25 °C. The number of live and dead worms was scored at 4–6 h intervals. For all the assays, E. coli OP50 was used in place of B. pseudomallei as the negative control.
Ethics statement
All animal experiments were performed in accordance with the Universiti Kebangsaan Malaysia animal ethics guideline formulated in accordance to the guidelines of the National Health and Medical Research Council of Australia. The experiments were approved by the Universiti Kebangsaan Malaysia Animal Ethics Committee (UKMAEC) under approval number FST/SBB/2010/SHEILA/24-AUGUST/320.
Mice survival assay
Female BALB/c mice, aged 8–10 weeks old, were obtained from the Animal House Facility, Universiti Kebangsaan Malaysia (UKM). Mice were maintained under specific-pathogen-free conditions in a positive pressure environment at 20–25 °C, subjected to a 12 h light/dark cycle and fed with a protein-enriched diet and water ad libitum. B. pseudomallei isolates, UM1, UM2, UM5 and UM6 were cultured as described previously. Mice were challenged intraperitoneally with ~1 × 106 CFU of B. pseudomallei UM1 or UM6 and their survival was monitored. The lung, liver and spleen were aseptically removed from mice that succumbed to disease and individually homogenized in 5 mL of PBS. Organ homogenates were serially diluted with PBS and the dilution was plated on Ashdown agar. The bacterial load in each organ was determined as CFU per organ. The remaining homogenates were centrifuged and supernatants were used for cytokine analysis. Statistical analysis on the difference in organ bacterial load was performed using the Mann–Whitney test within the GraphPad Prism version 4.0 (GraphPad Software) software package.
Measurement of proinflammatory cytokine levels
Mouse cytokine Multi-Analyte ELISArray Kit (Qiagen) was used to measure levels of IL-1α, IL-1β, IL-2, IL-4, IL-6, IL-10, IL-12, IL-17A, Interferon-γ (IFN-γ), Tumour necrosis factor-α (TNF-α), Granulocyte-Colony Stimulating Factor (G-CSF) and Granulocyte-Macrophage Colony Stimulating Factor (GM-CSF) in the organ homogenate supernatants from mice infected with B. pseudomallei strains UM1 or UM6. The arrays were performed according to the manufacturer’s instructions. The absorbance was measured at 450 nm with an automated Sunrise ELISA reader (Tecan, Switzerland).
Acknowledgements
We thank Jia-Shiun Khoo (Codon Genomics) and Yung-Chie Tan (Codon Genomics) for data annotation and Mei-Perng Lim (UKM) for technical assistance. This study was supported by the Malaysia Genome Institute-Stanford University International Research Grant (06-05-15-MB003) awarded to SN by the Ministry of Science, Technology and Innovation, Malaysia.
Additional files
Biofilm production by clinical isolates of B. pseudomallei . Diagram shows the relative comparison of biofilm formation by 87 B. pseudomallei clinical isolates (black bars). B. thailandensis ATCC 700388 was used as the reference for calculation of biofilm-forming capacity (red bar). Biofilm formation of P. aeruginosa ATCC 27852 (blue bar) and three other B. pseudomallei reference strains (K96243, ATCC 23343 and NCTC 13178) (yellow bars) were also included.
Analysis of transcriptome sequencing reads mapped to the K96243 genome.
Summary of significant differentially expressed genes. Classification of (A) biological function and (B) predicted cellular localization as analysed by PSORT. Pie charts indicate the percentage of up- and down-regulated genes that were significantly regulated in UM6 (high biofilm producer) compared to UM1 (low biofilm producer). Genes were divided into functional categories based on Comprehensive Microbial Resources (CMR) annotations.
Transcriptional changes of genes involved in B. pseudomallei biofilm formation. Shown is the expression profile for significantly regulated genes of UM6 (high biofilm producer) compared to UM1 (low biofilm producer). The genes transcripts are expressed as fragments per kilobase of transcript per million mapped reads (FPKM).
qRT-PCR analysis of genes found to be differentially regulated by RNASeq.
Two putative B. pseudomallei sensor kinases, BPSL2025 and BPSL1634 are members of RcsC. The amino sequences of two putative B. pseudomallei sensor kinases, BPSL2026 (YP_108622.1) and BPSL1634 (YP_108248.1) were aligned with E. coli sensor protein RcsC (NP_416722.2) using ClustalW. Conserved residues are highlighted in red and the conserved domains are underlined.
B. pseudomallei hypothetical protein BPSL0617 is a member of the CRP/FNR family protein. The amino sequence of BPSL0617 (YP_107246.1) was aligned with BCAM1349 (YP_002233964.1) using ClustalW. Conserved residues are highlighted in red.
B. pseudomallei genes clusters that contribute to biofilm development. Genomic organization of the B. pseudomallei gene clusters that contribute to biofilm development as identified in this study. Arrows indicate the direction of transcription and colours depict the expression profile. Up and down-regulated genes are coloured in red and green, respectively.
Details and biofilm formation index of B. pseudomallei clinical isolates and reference bacterial strains used in this study.
Primer sequences used for quantitative RT-PCR.
Footnotes
Chui-Yoke Chin, Yuka Hara and Ahmad-Kamal Ghazali contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AKG performed bioinformatics analysis and data interpretation and drafted the manuscript. SJY performed bioinformatics analysis and data interpretation. CCH participated in the RNA-Seq, bioinformatics analysis and data interpretation. YH performed the real time PCR, electron microscopy, mice experiments and drafting of the manuscript. CK performed the real time PCR, YCW isolated and prepared the RNA samples. NR performed the C. elegans survival experiments. SFK performed the biofilm production experiments. CYC performed all other experiments, analysed the transcriptome data and wrote the manuscript. SDP participated in the experimental design, data analysis and drafting of the manuscript. SN participated in the experimental design, data analysis and wrote the manuscript. All the authors have read and approved the final manuscript.
References
- 1.Lazar V. Quorum sensing in biofilms–how to destroy the bacterial citadels or their cohesion/power? Anaerobe. 2011;17(6):280–285. doi: 10.1016/j.anaerobe.2011.03.023. [DOI] [PubMed] [Google Scholar]
- 2.Drenkard E. Antimicrobial resistance of Pseudomonas aeruginosa biofilms. Microbes Infect. 2003;5(13):1213–1219. doi: 10.1016/j.micinf.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 3.Kostakioti M, Hadjifrangiskou M, Hultgren SJ. Bacterial biofilms: development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb Perspect Med. 2013;3(4):a010306. doi: 10.1101/cshperspect.a010306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15(2):167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puthucheary SD, Sam IC. Why is the response rate slow in ceftazidime therapy for melioidosis? Expert Rev Anti Infect Ther. 2012;10(1):5–7. doi: 10.1586/eri.11.158. [DOI] [PubMed] [Google Scholar]
- 6.Sawasdidoln C, Taweechaisupapong S, Sermswan RW, Tattawasart U, Tungpradabkul S, Wongratanacheewin S. Growing Burkholderia pseudomallei in biofilm stimulating conditions significantly induces antimicrobial resistance. PLoS One. 2010;5(2):e9196. doi: 10.1371/journal.pone.0009196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vasu C, Vadivelu J, Puthucheary SD. The humoral immune response in melioidosis patients during therapy. Infection. 2003;31(1):24–30. doi: 10.1007/s15010-002-3020-2. [DOI] [PubMed] [Google Scholar]
- 8.Chantratita N, Wuthiekanun V, Boonbumrung K, Tiyawisutsri R, Vesaratchavest M, Limmathurotsakul D, Chierakul W, Wongratanacheewin S, Pukritiyakamee S, White NJ, et al. Biological relevance of colony morphology and phenotypic switching by Burkholderia pseudomallei. J Bacteriol. 2007;189(3):807–817. doi: 10.1128/JB.01258-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taweechaisupapong S, Kaewpa C, Arunyanart C, Kanla P, Homchampa P, Sirisinha S, Proungvitaya T, Wongratanacheewin S. Virulence of Burkholderia pseudomallei does not correlate with biofilm formation. Microb Pathog. 2005;39(3):77–85. doi: 10.1016/j.micpath.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Yuen CW, Ong EB, Mohamad S, Manaf UA, Najimudin N. Construction and characterization of a Burkholderia pseudomallei wzm deletion mutant. J Microbiol Biotechnol. 2012;22(10):1336–1342. doi: 10.4014/jmb.1203.03059. [DOI] [PubMed] [Google Scholar]
- 11.Tunpiboonsak S, Mongkolrob R, Kitudomsub K, Thanwatanaying P, Kiettipirodom W, Tungboontina Y, Tungpradabkul S. Role of a Burkholderia pseudomallei polyphosphate kinase in an oxidative stress response, motilities, and biofilm formation. J Microbiol. 2010;48(1):63–70. doi: 10.1007/s12275-010-9138-5. [DOI] [PubMed] [Google Scholar]
- 12.Lee HS, Gu F, Ching SM, Lam Y, Chua KL. CdpA is a Burkholderia pseudomallei cyclic di-GMP phosphodiesterase involved in autoaggregation, flagellum synthesis, motility, biofilm formation, cell invasion, and cytotoxicity. Infect Immun. 2010;78(5):1832–1840. doi: 10.1128/IAI.00446-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazar Adler NR, Dean RE, Saint RJ, Stevens MP, Prior JL, Atkins TP, Galyov EE. Identification of a predicted trimeric autotransporter adhesin required for biofilm formation of Burkholderia pseudomallei. PLoS One. 2013;8(11):e79461. doi: 10.1371/journal.pone.0079461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Filiatrault MJ, Stodghill PV, Bronstein PA, Moll S, Lindeberg M, Grills G, Schweitzer P, Wang W, Schroth GP, Luo S, et al. Transcriptome analysis of Pseudomonas syringae identifies new genes, noncoding RNAs, and antisense activity. J Bacteriol. 2010;192(9):2359–2372. doi: 10.1128/JB.01445-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arnvig KB, Comas I, Thomson NR, Houghton J, Boshoff HI, Croucher NJ, Rose G, Perkins TT, Parkhill J, Dougan G, et al. Sequence-based analysis uncovers an abundance of non-coding RNA in the total transcriptome of Mycobacterium tuberculosis. PLoS Pathog. 2011;7(11):e1002342. doi: 10.1371/journal.ppat.1002342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dotsch A, Eckweiler D, Schniederjans M, Zimmermann A, Jensen V, Scharfe M, Geffers R, Haussler S. The Pseudomonas aeruginosa transcriptome in planktonic cultures and static biofilms using RNA sequencing. PLoS One. 2012;7(2):e31092. doi: 10.1371/journal.pone.0031092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wurtzel O, Sesto N, Mellin JR, Karunker I, Edelheit S, Becavin C, Archambaud C, Cossart P, Sorek R. Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol Syst Biol. 2012;8:583. doi: 10.1038/msb.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pruss BM, Besemann C, Denton A, Wolfe AJ. A complex transcription network controls the early stages of biofilm development by Escherichia coli. J Bacteriol. 2006;188(11):3731–3739. doi: 10.1128/JB.01780-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wood TK, Gonzalez Barrios AF, Herzberg M, Lee J. Motility influences biofilm architecture in Escherichia coli. Appl Microbiol Biotechnol. 2006;72(2):361–367. doi: 10.1007/s00253-005-0263-8. [DOI] [PubMed] [Google Scholar]
- 20.Ueda A, Wood TK. Connecting quorum sensing, c-di-GMP, pel polysaccharide, and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885) PLoS Pathog. 2009;5(6):e1000483. doi: 10.1371/journal.ppat.1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harmsen M, Yang L, Pamp SJ, Tolker-Nielsen T. An update on Pseudomonas aeruginosa biofilm formation, tolerance, and dispersal. FEMS Immunol Med Microbiol. 2010;59(3):253–268. doi: 10.1111/j.1574-695X.2010.00690.x. [DOI] [PubMed] [Google Scholar]
- 22.Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S, Greenberg EP. Gene expression in Pseudomonas aeruginosa biofilms. Nature. 2001;413(6858):860–864. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]
- 23.Felek S, Jeong JJ, Runco LM, Murray S, Thanassi DG, Krukonis ES. Contributions of chaperone/usher systems to cell binding, biofilm formation and Yersinia pestis virulence. Microbiology. 2011;157(Pt 3):805–818. doi: 10.1099/mic.0.044826-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gamage AM, Shui G, Wenk MR, Chua KL. N-Octanoylhomoserine lactone signalling mediated by the BpsI-BpsR quorum sensing system plays a major role in biofilm formation of Burkholderia pseudomallei. Microbiology. 2011;157(Pt 4):1176–1186. doi: 10.1099/mic.0.046540-0. [DOI] [PubMed] [Google Scholar]
- 25.Hassett DJ, Cuppoletti J, Trapnell B, Lymar SV, Rowe JJ, Yoon SS, Hilliard GM, Parvatiyar K, Kamani MC, Wozniak DJ, et al. Anaerobic metabolism and quorum sensing by Pseudomonas aeruginosa biofilms in chronically infected cystic fibrosis airways: rethinking antibiotic treatment strategies and drug targets. Adv Drug Deliv Rev. 2002;54(11):1425–1443. doi: 10.1016/S0169-409X(02)00152-7. [DOI] [PubMed] [Google Scholar]
- 26.Vidal JE, Ludewick HP, Kunkel RM, Zahner D, Klugman KP. The LuxS-dependent quorum-sensing system regulates early biofilm formation by Streptococcus pneumoniae strain D39. Infect Immun. 2011;79(10):4050–4060. doi: 10.1128/IAI.05186-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ren D, Bedzyk LA, Setlow P, England DF, Kjelleberg S, Thomas SM, Ye RW, Wood TK. Differential gene expression to investigate the effect of (5Z)-4-bromo- 5-(bromomethylene)-3-butyl-2 (5H)-furanone on Bacillus subtilis. Appl Environ Microbiol. 2004;70(8):4941–4949. doi: 10.1128/AEM.70.8.4941-4949.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wood TK. Insights on Escherichia coli biofilm formation and inhibition from whole-transcriptome profiling. Environ Microbiol. 2009;11(1):1–15. doi: 10.1111/j.1462-2920.2008.01768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X, Ma Q, Wood TK. The R1 conjugative plasmid increases Escherichia coli biofilm formation through an envelope stress response. Appl Environ Microbiol. 2008;74(9):2690–2699. doi: 10.1128/AEM.02809-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sahu PK, Iyer PS, Gaikwad MB, Talreja SC, Pardesi KR, Chopade BA. An MFS transporter-like ORF from MDR Acinetobacter baumannii AIIMS 7 is associated with adherence and biofilm formation on biotic/abiotic surface. Int J Microbiol. 2012;2012:490647. doi: 10.1155/2012/490647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waite RD, Papakonstantinopoulou A, Littler E, Curtis MA. Transcriptome analysis of Pseudomonas aeruginosa growth: comparison of gene expression in planktonic cultures and developing and mature biofilms. J Bacteriol. 2005;187(18):6571–6576. doi: 10.1128/JB.187.18.6571-6576.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joshua GW, Karlyshev AV, Smith MP, Isherwood KE, Titball RW, Wren BW. A Caenorhabditis elegans model of Yersinia infection: biofilm formation on a biotic surface. Microbiology. 2003;149(Pt 11):3221–3229. doi: 10.1099/mic.0.26475-0. [DOI] [PubMed] [Google Scholar]
- 33.Begun J, Gaiani JM, Rohde H, Mack D, Calderwood SB, Ausubel FM, Sifri CD. Staphylococcal biofilm exopolysaccharide protects against Caenorhabditis elegans immune defenses. PLoS Pathog. 2007;3(4):e57. doi: 10.1371/journal.ppat.0030057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koh SF, Tay ST, Puthucheary SD. Colonial morphotypes and biofilm forming ability of Burkholderia pseudomallei. Trop Biomed. 2013;30(3):428–433. [PubMed] [Google Scholar]
- 35.Pibalpakdee P, Wongratanacheewin S, Taweechaisupapong S, Niumsup PR. Diffusion and activity of antibiotics against Burkholderia pseudomallei biofilms. Int J Antimicrob Agents. 2012;39(4):356–359. doi: 10.1016/j.ijantimicag.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 36.Ferrieres L, Clarke DJ. The RcsC sensor kinase is required for normal biofilm formation in Escherichia coli K-12 and controls the expression of a regulon in response to growth on a solid surface. Mol Microbiol. 2003;50(5):1665–1682. doi: 10.1046/j.1365-2958.2003.03815.x. [DOI] [PubMed] [Google Scholar]
- 37.Ryan RP, Dow JM. Communication with a growing family: diffusible signal factor (DSF) signaling in bacteria. Trends Microbiol. 2011;19(3):145–152. doi: 10.1016/j.tim.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 38.Boon C, Deng Y, Wang LH, He Y, Xu JL, Fan Y, Pan SQ, Zhang LH. A novel DSF-like signal from Burkholderia cenocepacia interferes with Candida albicans morphological transition. ISME J. 2008;2(1):27–36. doi: 10.1038/ismej.2007.76. [DOI] [PubMed] [Google Scholar]
- 39.Ryan RP, Dow JM. Intermolecular interactions between HD-GYP and GGDEF domain proteins mediate virulence-related signal transduction in Xanthomonas campestris. Virulence. 2010;1(5):404–408. doi: 10.4161/viru.1.5.12704. [DOI] [PubMed] [Google Scholar]
- 40.Deng Y, Wu J, Tao F, Zhang LH. Listening to a new language: DSF-based quorum sensing in Gram-negative bacteria. Chem Rev. 2011;111(1):160–173. doi: 10.1021/cr100354f. [DOI] [PubMed] [Google Scholar]
- 41.Bi H, Christensen QH, Feng Y, Wang H, Cronan JE. The Burkholderia cenocepacia BDSF quorum sensing fatty acid is synthesized by a bifunctional crotonase homologue having both dehydratase and thioesterase activities. Mol Microbiol. 2012;83(4):840–855. doi: 10.1111/j.1365-2958.2012.07968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hay ID, Gatland K, Campisano A, Jordens JZ, Rehm BH. Impact of alginate overproduction on attachment and biofilm architecture of a supermucoid Pseudomonas aeruginosa strain. Appl Environ Microbiol. 2009;75(18):6022–6025. doi: 10.1128/AEM.01078-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fazli M, O'Connell A, Nilsson M, Niehaus K, Dow JM, Givskov M, Ryan RP, Tolker-Nielsen T. The CRP/FNR family protein Bcam1349 is a c-di-GMP effector that regulates biofilm formation in the respiratory pathogen Burkholderia cenocepacia. Mol Microbiol. 2011;82(2):327–341. doi: 10.1111/j.1365-2958.2011.07814.x. [DOI] [PubMed] [Google Scholar]
- 44.Schroll C, Barken KB, Krogfelt KA, Struve C. Role of type 1 and type 3 fimbriae in Klebsiella pneumoniae biofilm formation. BMC Microbiol. 2010;10:179. doi: 10.1186/1471-2180-10-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lasaro MA, Salinger N, Zhang J, Wang Y, Zhong Z, Goulian M, Zhu J. F1C fimbriae play an important role in biofilm formation and intestinal colonization by the Escherichia coli commensal strain Nissle 1917. Appl Environ Microbiol. 2009;75(1):246–251. doi: 10.1128/AEM.01144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giraud C, Bernard CS, Calderon V, Yang L, Filloux A, Molin S, Fichant G, Bordi C, de Bentzmann S. The PprA-PprB two-component system activates CupE, the first non-archetypal Pseudomonas aeruginosa chaperone-usher pathway system assembling fimbriae. Environ Microbiol. 2011;13(3):666–683. doi: 10.1111/j.1462-2920.2010.02372.x. [DOI] [PubMed] [Google Scholar]
- 47.Hamad MA, Austin CR, Stewart AL, Higgins M, Vazquez-Torres A, Voskuil MI. Adaptation and antibiotic tolerance of anaerobic Burkholderia pseudomallei. Antimicrob Agents Chemother. 2011;55(7):3313–3323. doi: 10.1128/AAC.00953-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Alst NE, Picardo KF, Iglewski BH, Haidaris CG. Nitrate sensing and metabolism modulate motility, biofilm formation, and virulence in Pseudomonas aeruginosa. Infect Immun. 2007;75(8):3780–3790. doi: 10.1128/IAI.00201-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Borriello G, Richards L, Ehrlich GD, Stewart PS. Arginine or nitrate enhances antibiotic susceptibility of Pseudomonas aeruginosa in biofilms. Antimicrob Agents Chemother. 2006;50(1):382–384. doi: 10.1128/AAC.50.1.382-384.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blanchette-Cain K, Hinojosa CA, Akula Suresh Babu R, Lizcano A, Gonzalez-Juarbe N, Munoz-Almagro C, Sanchez CJ, Bergman MA, Orihuela CJ. Streptococcus pneumoniae biofilm formation is strain dependent, multifactorial, and associated with reduced invasiveness and immunoreactivity during colonization. MBio. 2013;4(5):e00745–00713. doi: 10.1128/mBio.00745-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thurlow LR, Hanke ML, Fritz T, Angle A, Aldrich A, Williams SH, Engebretsen IL, Bayles KW, Horswill AR, Kielian T. Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J Immunol. 2011;186(11):6585–6596. doi: 10.4049/jimmunol.1002794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chin CY, Monack DM, Nathan S. Genome wide transcriptome profiling of a murine acute melioidosis model reveals new insights into how Burkholderia pseudomallei overcomes host innate immunity. BMC Genomics. 2010;11:672. doi: 10.1186/1471-2164-11-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brett PJ, Deshazer D, Woods DE. Characterization of Burkholderia pseudomallei and Burkholderia pseudomallei-like strains. Epidemiol Infect. 1997;118(2):137–148. doi: 10.1017/S095026889600739X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holden MT, Titball RW, Peacock SJ, Cerdeno-Tarraga AM, Atkins T, Crossman LC, Pitt T, Churcher C, Mungall K, Bentley SD, et al. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc Natl Acad Sci U S A. 2004;101(39):14240–14245. doi: 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chieng S, Carreto L, Nathan S. Burkholderia pseudomallei transcriptional adaptation in macrophages. BMC Genomics. 2012;13:328. doi: 10.1186/1471-2164-13-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen Y, Wong J, Sun GW, Liu Y, Tan GY, Gan YH. Regulation of type VI secretion system during Burkholderia pseudomallei infection. Infect Immun. 2011;79(8):3064–3073. doi: 10.1128/IAI.05148-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boonbumrung K, Wuthiekanun V, Rengpipat S, Day NP, Peacock SJ. In vitro motility of a population of clinical Burkholderia pseudomallei isolates. J Med Assoc Thai. 2006;89(9):1506–1510. [PubMed] [Google Scholar]
- 60.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tan MW, Mahajan-Miklos S, Ausubel FM. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci U S A. 1999;96(2):715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tan MW, Rahme LG, Sternberg JA, Tompkins RG, Ausubel FM. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc Natl Acad Sci U S A. 1999;96(5):2408–2413. doi: 10.1073/pnas.96.5.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee SH, Ooi SK, Mahadi NM, Tan MW, Nathan S. Complete killing of Caenorhabditis elegans by Burkholderia pseudomallei is dependent on prolonged direct association with the viable pathogen. PLoS One. 2011;6(3):e16707. doi: 10.1371/journal.pone.0016707. [DOI] [PMC free article] [PubMed] [Google Scholar]


