Abstract
When Francis Crick first proposed the central dogma, he predicted that genetic information flows from DNA to RNA and finally to proteins. By this classical concept, the sole purpose of mRNA is to serve as a template for translation. Recent work has expanded our understanding of the function of mRNA well beyond this singular definition. A paper published in this issue sheds more light on the myriad roles mRNAs can play in genetic regulation. Miyakoshi et al (2015a) report an intriguing scenario in Salmonella where a small RNA molecule derived from a larger polycistronic mRNA promotes cross talk between physically unlinked mRNAs via controlling turnover of a global small RNA repressor.
See also: M Miyakoshi et al (June 2015) and D Lalaouna et al (May 2015)
Small RNA regulators were first discovered in bacteria some 35 years ago (Tomizawa et al, 1981), but remained under the radar for an additional two decades until we began to discover their numbers and diverse regulatory roles. Early work on bacterial small RNAs (sRNAs) revealed their roles as post-transcriptional regulators of gene expression, primarily as repressors of translation. The majority of sRNAs characterized to date are produced under specific conditions and go on to base pair with mRNA targets (often with the aid of an RNA chaperone, Hfq) to inhibit translation and promote mRNA degradation. There are, however, a growing number of examples of sRNAs that regulate gene expression by more complicated regulatory mechanisms, acting at all levels encompassed by the central dogma.
Over the last decade, pioneering global screens have identified novel bacterial sRNAs (Zhang et al, 2003; Sittka et al, 2008), mostly located in non-coding intergenic regions of bacterial genomes. These sRNAs are typically between 50- and 250-nt in length and are often conserved among closely related organisms. Recent studies utilizing the power of next generation sequencing have identified a surprisingly large number of Hfq-binding sRNAs located in the 3′ regions of adjacent protein-coding genes (Chao et al, 2012; Kroger et al, 2012). These sRNAs localized 3′ relative to mRNAs have been classified as Type I, expressed from an independent promoter in the upstream coding sequence or 3′ untranslated region (UTR), or Type II, originating from processing of the parental mRNA. Both types share a terminator with the adjacent gene, so that the sRNA and mRNA have the same 3′ end (Miyakoshi et al, 2015b). In this issue of The EMBO Journal, Miyakoshi et al (2015a) characterize a fascinating Type II sRNA (SroC) that is generated by processing of the gltIJKL operon mRNA, which encodes a glutamate/aspartate ABC transporter. SroC is unique because unlike the majority of sRNA regulators that base pair with mRNAs to regulate stability and translation, SroC acts as a ‘sponge,’ base pairing with and regulating activity of the sRNA GcvB, a global regulator of amino acid transport and biosynthesis genes (Fig1).
Figure 1. Functionally related RNAs cross talk through a sponge RNA in bacteria.
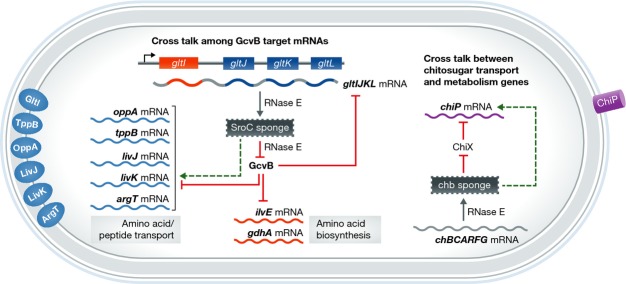
The sRNA GcvB targetome contains amino acid/peptide transporters and amino acid biosynthesis genes. A sponge RNA SroC, originated from one of the targets, base pairs with sRNA GcvB and recruits RNase E. Degradation of GcvB causes derepression of its targetome, including the parental mRNA of SroC (cross talk is indicated by dashed arrows in this figure). In this regulatory circuit, the sponge interacts with multiple mRNAs that are physically unlinked but functionally works in the same pathway. Another example is the chb sponge that degrades ChiX sRNA in a similar RNase E-dependent manner leading to expression of an outer membrane protein ChiP.
SroC was initially thought to represent a decay intermediate derived from gltIJKL mRNA. However, since SroC associates with Hfq in vivo, Miyakoshi et al postulated that it might regulate other mRNAs via base pairing. Using an elegant combination of genetic and biochemical experiments, the authors demonstrated that SroC base pairs not with other mRNAs, but instead with GcvB. SroC-GcvB base pairing induces RNase E-mediated degradation of GcvB. The target regulon of GcvB includes dozens of genes encoding transporters for amino acids and short peptides, amino acid biosynthesis functions and transcription factors (Miyakoshi et al, 2015a,b), and all of these targets are repressed at the level of translation or mRNA stability through base pairing interactions with GcvB. Thus, the result of SroC-mediated repression of the repressor is activation of the genes in the GcvB regulon. Since the parental mRNA of SroC, the gltIJKL operon, is also a target of GcvB sRNA, SroC-mediated antagonism of GcvB constitutes a feed-forward loop that derepresses gltIJKL mRNA, which in turn allows more SroC production. Interestingly, RNase E plays two separate roles in this complex regulatory network. First, RNase E processes a suboperonic transcript that terminates after gltI to produce mature SroC. Second, enhanced turnover of GcvB mediated by SroC also depends on RNase E (Fig1). All steps of the regulatory process, from SroC biogenesis to GcvB turnover, additionally require the chaperone protein Hfq.
RNA sponges have also been identified in eukaryotes, where some mRNAs containing multiple microRNA (miRNA) binding sites competitively titrate miRNA regulators away from their natural targets (Bak & Mikkelsen, 2014). Viruses of eukaryotes (Bak & Mikkelsen, 2014) and bacteria (Tree et al, 2014) have been reported to make non-coding RNA sponges that similarly bind to and inhibit activities of cellular miRNAs or sRNAs. The activity of ChiX sRNA in Salmonella is likewise controlled by a sponge RNA. ChiX represses synthesis of an outer membrane transporter for chitosugars (chitoporin, encoded by chiP) (Figueroa-Bossi et al, 2009) when chitosugars are absent. When chitosugars are present, the chb operon, encoding chitosugar metabolism functions, is transcribed and processed, yielding a chb mRNA-derived sponge RNA. The chb RNA sponge base pairs with ChiX and promotes its degradation. Reduced levels of ChiX allow derepression of chiP mRNA and increased synthesis of the chitoporin (Fig1).
More intriguing RNA sponges have just been discovered by Lalaouna et al (2015). These new sponges share many characteristics with SroC. They are derived from larger RNA precursors, namely the internal transcribed spacers (ITS) and external transcribed spacers (ETS) of tRNA operons. The 3′ ETS of the glyW-cysT-leuZ mRNA (3′ETSleuZ) is produced via RNase E-mediated processing and base pairs with at least two distinct sRNA regulators, RyhB and RybB (Lalaouna et al, 2015). 3′ETSleuZ base pairing with RyhB and RybB prevents their pairing with mRNA targets, which lowers their effective concentrations and sets a threshold for their regulatory activities. In the absence of the 3′ETSleuZ sponge, RyhB can regulate its target mRNAs more efficiently. In other words, without the sponge, lower concentrations of RyhB are required to observe significant regulation of mRNA targets.
The three sponge RNAs described here all provide a mechanism for cross talk between mRNAs. The chb RNA sponge appears to have the most limited regulatory scope in that it only coordinates functions for chitosugar transport (chiP) and metabolism (chb). SroC promotes a more extensive cross talk among the many mRNAs within the GcvB regulon. The 3′ETSleuZ RNA sponge has potentially the most expansive regulatory role in that it can mediate communication between the mRNAs comprising the regulons of two independent sRNAs. Functionally, this implies a physiological link between iron homeostasis (controlled by RyhB) and membrane stress (regulated by RybB).
As representatives of an emerging group of RNAs that act as sponges to control the activities of base pairing-dependent sRNA regulators, SroC, chb RNA and 3′ETSleuZ provide new experimental challenges and opportunities. Many existing computational algorithms for sRNA identification have focused on conservation (of sequence or structure) found in intergenic regions separating protein-coding genes. Findings with sponge RNAs suggest that we must now expand our concept of where and how to find novel sRNAs as well as what types of functional activities can be expected. Lalaouna et al note strong conservation of ITS and ETS of many tRNA operons, suggesting that there are potentially many additional tRNA-derived RNA regulators, possibly more sponges, to be discovered. Another important implication of the work with SroC and other sponge RNAs is related to the ‘repression of a repressor’ mechanism, which yields activation of the target genes of the sRNA regulator (GcvB, in the case of SroC). While there are a number of examples of direct activation of mRNA targets by sRNA regulators, sponge RNAs like SroC and 3′ETSleuZ may be responsible for pervasive indirect activation of mRNA targets belonging to sRNA regulons. With the sponge RNAs essentially acting as competitors for base pairing with sRNAs, the outcome of competition will depend on the relative levels of sponge, sRNA and mRNA targets as well as the kinetics and binding affinities that define each interaction. Thus, control of sponge RNA levels by different regulatory inputs provides another parameter that allows cells to sensitively tune expression of genes post-transcriptionally.
References
- Bak RO, Mikkelsen JG. miRNA sponges: soaking up miRNAs for regulation of gene expression. Wiley Interdiscip Rev RNA. 2014;5:317–333. doi: 10.1002/wrna.1213. [DOI] [PubMed] [Google Scholar]
- Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J. An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 2012;31:4005–4019. doi: 10.1038/emboj.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa-Bossi N, Valentini M, Malleret L, Bossi L. Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev. 2009;23:2004–2015. doi: 10.1101/gad.541609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroger C, Dillon SC, Cameron AD, Papenfort K, Sivasankaran SK, Hokamp K, Chao Y, Sittka A, Hebrard M, Handler K, Colgan A, Leekitcharoenphon P, Langridge GC, Lohan AJ, Loftus B, Lucchini S, Ussery DW, Dorman CJ, Thomson NR, Vogel J, et al. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc Natl Acad Sci USA. 2012;109:E1277–E1286. doi: 10.1073/pnas.1201061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalaouna D, Carrier MC, Semsey S, Brouard JS, Wang J, Wade JT, Massé E. A 3′ external transcribed spacer in a tRNA transcript acts as a sponge for small RNAs to prevent transcriptional noise. Mol Cell. 2015;58:393–405. doi: 10.1016/j.molcel.2015.03.013. [DOI] [PubMed] [Google Scholar]
- Miyakoshi M, Chao Y, Vogel J. Cross talk between ABC transporter mRNAs via a target mRNA-derived sponge of the GcvB small RNA. EMBO J. 2015a;34:1478–1492. doi: 10.15252/embj.201490546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakoshi M, Chao Y, Vogel J. Regulatory small RNAs from the 3′ regions of bacterial mRNAs. Curr Opin Microbiol. 2015b;24:132–139. doi: 10.1016/j.mib.2015.01.013. [DOI] [PubMed] [Google Scholar]
- Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JC, Vogel J. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J, Itoh T, Selzer G, Som T. Inhibition of ColE1 RNA primer formation by a plasmid-specified small RNA. Proc Natl Acad Sci USA. 1981;78:1421–1425. doi: 10.1073/pnas.78.3.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tree JJ, Granneman S, McAteer SP, Tollervey D, Gally DL. Identification of bacteriophage-encoded anti-sRNAs in pathogenic Escherichia coli. Molecular cell. 2014;55:199–213. doi: 10.1016/j.molcel.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]


