Abstract
There is an expanding list of examples by which one mRNA can posttranscriptionally influence the expression of others. This can involve RNA sponges that sequester regulatory RNAs of mRNAs in the same regulon, but the underlying molecular mechanism of such mRNA cross talk remains little understood. Here, we report sponge-mediated mRNA cross talk in the posttranscriptional network of GcvB, a conserved Hfq-dependent small RNA with one of the largest regulons known in bacteria. We show that mRNA decay from the gltIJKL locus encoding an amino acid ABC transporter generates a stable fragment (SroC) that base-pairs with GcvB. This interaction triggers the degradation of GcvB by RNase E, alleviating the GcvB-mediated mRNA repression of other amino acid-related transport and metabolic genes. Intriguingly, since the gltIJKL mRNA itself is a target of GcvB, the SroC sponge seems to enable both an internal feed-forward loop to activate its parental mRNA in cis and activation of many trans-encoded mRNAs in the same pathway. Disabling this mRNA cross talk affects bacterial growth when peptides are the sole carbon and nitrogen sources.
Keywords: GcvB, Hfq, noncoding RNA, RNase E, SroC
See also: MS Azam & CK Vanderpool (June 2015)
Introduction
Messenger RNA (mRNA) has long been seen solely as an intermediate in the transport of genetic information from DNA to the translation apparatus for decoding. In this classical scenario of molecular biology, only the end product of the process—the translated protein—may impact the expression of other genes. Recent work, however, has revealed intriguing cases in which mRNAs cross talk with other mRNAs, and thereby, themselves influence physically unlinked genes.
In eukaryotes, one such type of cross talk is mediated by mRNAs that contain multiple base-pairing sites to ‘sponge’ microRNAs with complementary sequence, thus minimizing the microRNA-mediated repression of other target mRNAs in the same regulon (Seitz, 2009; Ebert & Sharp, 2010; Rubio-Somoza et al, 2011). This mRNA cross-activation mechanism has been generally conceptualized in terms of competitive endogenous mRNAs (ceRNAs) which titrate microRNAs from competing mRNAs by changing their relative concentrations (Salmena et al, 2011). However, microRNAs may also be sequestered by other RNAs including a pseudogene transcript (Franco-Zorrilla et al, 2007), long noncoding RNAs (Cesana et al, 2011), or circular RNAs (Hansen et al, 2013; Memczak et al, 2013). Moreover, some viruses use noncoding RNA sponges to inactivate host microRNAs (Cazalla et al, 2010; Marcinowski et al, 2012).
In prokaryotes, mRNA cross talk through competition for small regulatory RNAs (sRNAs) was first reported in the chitosugar utilization pathway of Salmonella enterica and Escherichia coli (Figueroa-Bossi et al, 2009; Overgaard et al, 2009). These enteric model bacteria encode the chitosugar-specific porin ChiP whose mRNA is translationally repressed by an abundant Hfq-dependent sRNA, ChiX, under regular growth conditions. However, chitosugars in the environment trigger transcription of a polycistronic chitobiose mRNA (encoding enzymes for chitosugar breakdown) which contains a short complementarity region for the ChiX sRNA (Plumbridge et al, 2014). As the ChiX sRNA base-pairs with this site, it gets degraded and its cellular concentration decreases, which boosts the synthesis of the relevant porin for chitosugar uptake.
The sRNAs that associate with Hfq often target dozens of mRNAs (Vogel & Luisi, 2011; Sobrero & Valverde, 2012; De Lay et al, 2013) and so constitute nodes in larger posttranscriptional networks that help control many different aspects of bacterial physiology, virulence, and behavior (Storz et al, 2011; Papenfort & Vogel, 2014). However, sponges for such global regulators, which would result in cross talk between multiple functionally related bacterial mRNAs as observed in eukaryotic microRNA regulons, have not been described. Here, we report a conserved RNA sponge in the large GcvB-controlled regulatory circuit of bacterial amino acid synthesis and transport.
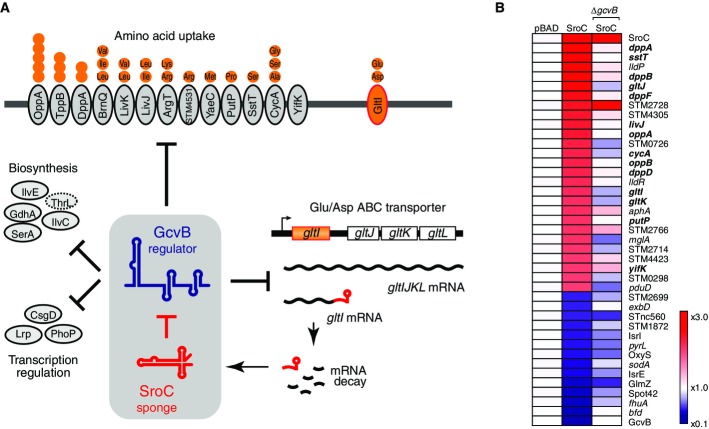
GcvB is a widely conserved, ∽200 nt Hfq-dependent sRNA and one of the most globally acting posttranscriptional regulators in bacteria, potentially regulating ∽1% of all mRNAs in Salmonella and E. coli (Urbanowski et al, 2000; Sharma et al, 2007, 2011; Pulvermacher et al, 2009; Busi et al, 2010; Vanderpool, 2011; Stauffer & Stauffer, 2012; Wright et al, 2013; Yang et al, 2014). Its regulon (Fig1A) is highly enriched with transporters of amino acids and short peptides, including the major ABC transporters Dpp and Opp, but also contains amino acid biosynthesis proteins and major transcription factors such as Lrp, PhoP, and CsgD (Modi et al, 2011; Sharma et al, 2011; Jørgensen et al, 2012; Coornaert et al, 2013). Since GcvB is highly abundant when bacteria grow rapidly in nutrient-rich media, its major role may be to optimize the energy-expensive import and biosynthesis of amino acids (Sharma et al, 2007).
Figure 1.
- Schematic description of GcvB regulation by SroC as shown in this study. GcvB represses mRNAs of amino acid transporters with different substrate specificity. Orange particles and strands indicate environmental amino acids and peptides, respectively. The GcvB regulon also includes mRNAs of amino acid biosynthetic enzymes and transcriptional regulators. The ThrL leader peptide of the threonine synthesis operon is shown by a dotted oval. One of the GcvB targets, Glu/Asp ABC transporter operon gltIJKL, is transcribed in two mRNAs, and the shorter mRNA generates the majority of SroC by processing. Repression of GcvB by SroC results in the derepression of GcvB regulon, including the parental mRNA of SroC.
- Microarray analysis of SroC pulse expression. The Salmonella WT (JVS-1574) or ΔgcvB (JVS-1044) strains harboring pBAD-ctrl (pKP8-35) or pBAD-SroC (pYC6-4) were induced with 0.2% l-arabinose for 10 min. Gene expression in the WT strain containing pBAD-ctrl was normalized to 1 (left column), and genes that showed > twofold change (P-value < 0.1) by SroC overexpression are indicated on the middle column. SroC-induced changes of the genes in ΔgcvB background were analyzed by comparing the expression levels in ΔgcvB (pBAD-SroC) to those in ΔgcvB (pBAD-ctrl) and are shown on the right column. Known GcvB targets are set in boldface. A list of differentially expressed genes and fold changes is included in Supplementary Table S1.
As expected for a regulator of this scope, the cellular level of GcvB is tightly controlled at the level of synthesis; the two transcription factors, GcvA and GcvR, of the glycine cleavage system activate the gcvB gene in response to endogenous glycine (Urbanowski et al, 2000; Ghrist et al, 2001; Heil et al, 2002; Stauffer & Stauffer, 2005). However, GcvB also exhibits an exceptionally low cellular RNA half-life of < 2 min (Vogel et al, 2003), hinting at additional control on the level of RNA stability. We have now discovered that this cellular lability of GcvB is caused by a conserved RNA sponge that is released from one of GcvB's own target mRNAs.
The new GcvB sponge originates from the polycistronic gltIJKL mRNA of a glutamate/aspartate ABC transporter (Willis & Furlong, 1975; Schellenberg & Furlong, 1977; Reitzer & Schneider, 2001), which is a well-studied GcvB target in Salmonella. Aided by the Hfq protein, GcvB binds to the 5′ UTR of gltI to repress translational initiation on this mRNA; of note, the C/A-rich target site of GcvB also acts as a translation enhancer element in this mRNA (Sharma et al, 2007). We show here that this same operon mRNA carries a second GcvB site between gltI and gltJ; however, this region produces a stable RNA that sponges GcvB and causes mRNA cross talk in the amino acid pathway.
Results
Global mRNA cross talk through depletion of GcvB sRNA
We initially sought to identify a function for SroC, which is a stable ∽160 nt RNA fragment encountered in numerous studies of E. coli (Vogel et al, 2003; Tree et al, 2014), Salmonella (Sittka et al, 2008; Kröger et al, 2012, 2013; Ortega et al, 2012), and Klebsiella pneumoniae (Kim et al, 2012). SroC maps to an internal region of the gltIJKL operon mRNA, consistent with it being an mRNA breakdown fragment (Vogel et al, 2003). However, since SroC associates with Hfq protein in vivo (Sittka et al, 2008; Chao et al, 2012; Tree et al, 2014), we speculated that it may regulate other cellular RNAs by base-pairing. Indeed, as shown in Fig1B, a brief overexpression of SroC in a Salmonella wild-type strain altered the levels of 33 mRNAs and seven sRNAs (> twofold change; < 0.1 of P-value; Supplementary Table S1).
Whereas computer predictions of potential SroC sites in these regulated genes returned no significant results, we noted that 14 of the 26 SroC-activated mRNAs (Fig1B) were known targets of GcvB. For example, the dpp and opp operon mRNAs are repressed by GcvB (Urbanowski et al, 2000; Sharma et al, 2007) but activated by SroC. Moreover, since GcvB itself was down-regulated (∽5.4-fold) by SroC, the observed activation of mRNAs may result from a depletion of their repressor GcvB. Indeed, most of these mRNAs were insensitive to SroC pulse expression in a ΔgcvB strain (Fig1B). In contrast, the mild down-regulation of some sRNAs following SroC pulse expression in the wild-type strain was still observed in the ΔgcvB background, suggesting that overexpression of SroC titrates the pool of available Hfq in vivo (Papenfort et al, 2009; Hussein & Lim, 2011; Moon & Gottesman, 2011), which would destabilize other Hfq-dependent sRNAs.
SroC has been regarded as a stable RNA decay intermediate of the much longer gltIJKL operon mRNA (Fig1A): It carries the characteristic 5′ monophosphate (5′P) of a processed species (Vogel et al, 2003) and its 5′ end within the gltI coding sequence (Supplementary Fig S1A) is not associated with a transcription start site (Kröger et al, 2012). The 3′ end of SroC is likely generated by a conserved ρ-independent transcription terminator in the gltI-gltJ intergenic region (Supplementary Fig S1A). Therefore, it may be the expression of the glt mRNA of which SroC is a secondary product that normally causes the activation of GcvB targets.
To test this, we constructed a series of mutants in the chromosomal gltIJKL locus and evaluated expression changes of GcvB and its representative target OppA. Deletion of gltIJKL increased GcvB levels by eightfold and concomitantly reduced the level of OppA by twofold, whereas the deletion of the downstream gltJKL genes had no effect (Fig2A, lanes 1–3). Importantly, the effects of the gltI deletion on GcvB and OppA were phenocopied by disrupting only the sroC region (lane 4), confirming SroC as the trans-activator region of the gltI mRNA. Again, SroC acted through GcvB since a ΔsroC mutation had no effect on OppA levels in a ΔgcvB background (lanes 5–6).
Figure 2.
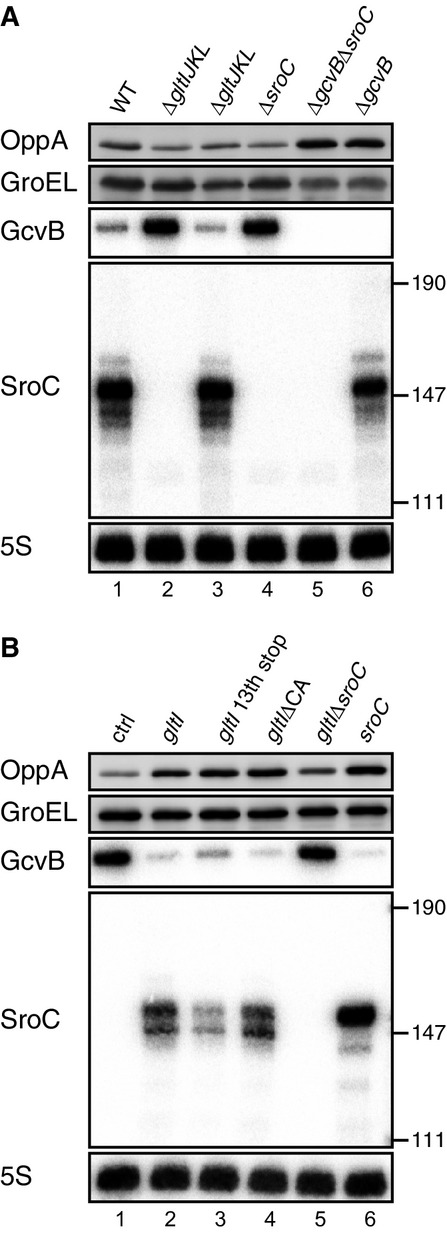
- Expression changes of OppA and GcvB upon deletion of gltIJKL locus. WT (JVS-1574), ΔgltIJKL (JVS-5823), ΔgltJKL (JVS-10795), ΔsroC (JVS-5821), ΔgcvBΔsroC (JVS-5822), and ΔgcvB (JVS-0236) strains were grown to early stationary phase (OD600 of 2.0) in LB medium.
- Expression changes of OppA and GcvB by overexpression of gltI-sroC. ΔgltIJKL strain (JVS-5823) harboring pBAD-ctrl (lane 1), pBAD-gltI (pMM36) derivatives (lanes 2-5), or pBAD-SroC (lane 6) was grown to early stationary phase (OD600 of 2.0) in LB medium supplemented with 0.02% l-arabinose.
Next, we expressed variants of the gltI-sroC region from a plasmid-borne, arabinose-inducible promoter. As in the chromosome, expression of the gltI-sroC mRNA decreased GcvB and increased OppA levels (Fig2B, lane 2). To test whether the GltI protein is involved in the cross-activation of OppA synthesis, we terminated translation of gltI at codon 13. Although the levels of SroC from this construct were somewhat lower, we still observed regulation of GcvB and OppA (Fig2B, lane 3), confirming our hypothesis of RNA-mediated cross talk.
Interestingly, the gltI mRNA itself is a target of GcvB (Fig1A), raising the possibility that base-pairing at the CA-rich target site in the gltI 5′ UTR might contribute to GcvB depletion. However, the same regulation of GcvB and OppA occurred in the presence of a ΔCA mutant plasmid as with the full-length gltI-sroC transcript (Fig2B, lane 4). Lastly, the expression of SroC as a primary transcript was sufficient for both GcvB repression and OppA activation (Fig2B, lane 6), which independently validated the original microarray results.
A gltI mRNA fragment destabilizes the GcvB sRNA
The above results pinpointed SroC as the actual repressor of GcvB but did not reveal on which level GcvB was regulated. Using a transcriptional reporter fusion, we found no significant effect of SroC on gcvB transcription (Supplementary Fig S2). To test regulation at the RNA level, we used a two-plasmid system, wherein GcvB was constitutively produced from one plasmid (pPL-GcvB), and SroC was pulse-expressed from another (pBAD-SroC). Under the conditions used, GcvB levels were ∽sixfold higher as compared to chromosomal expression (Fig3A, lanes 1 and 5). A 10-min pulse of SroC expression depleted the constitutively transcribed GcvB sRNA (Fig3A, lanes 7 versus 8), indicating that SroC—directly or indirectly—destabilized the GcvB transcript. The Hfq protein which associates with both SroC and GcvB in vivo (Sittka et al, 2008) is required for this regulation; that is, SroC failed to deplete GcvB in a Δhfq strain (Fig3A, lane 12).
Figure 3.
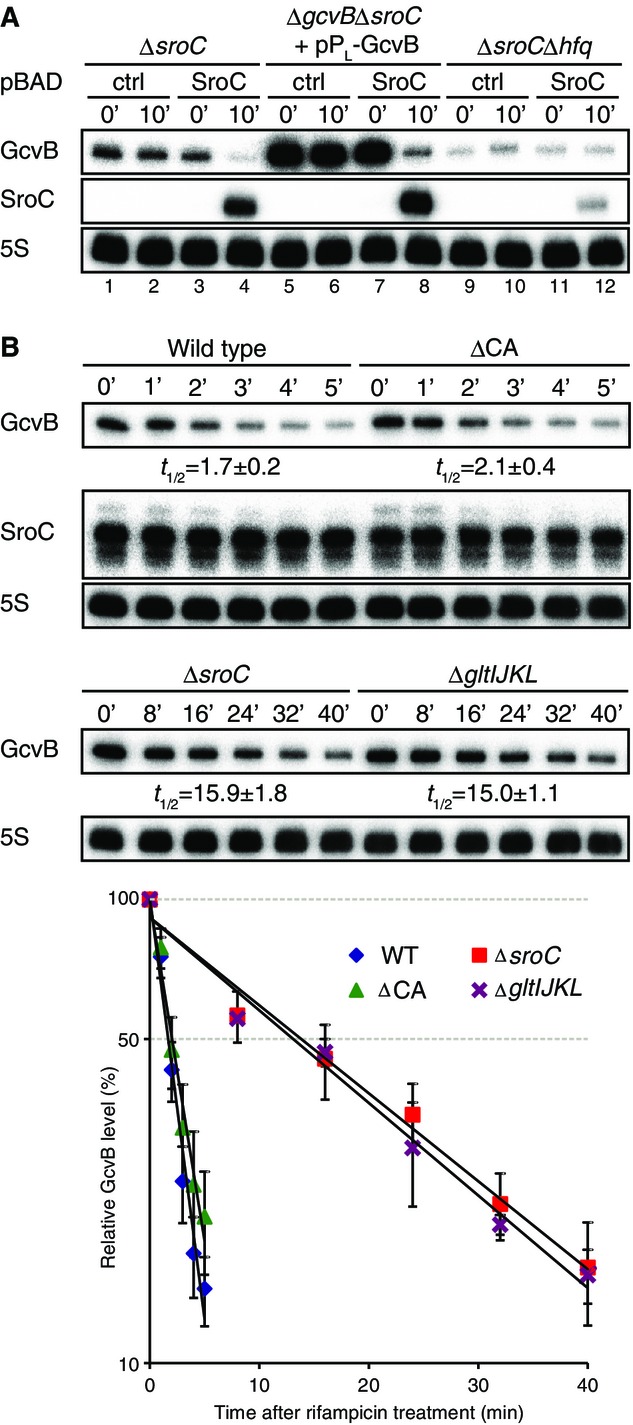
- Salmonella ΔsroC (JVS-5821), ΔgcvBΔsroC (JVS-5822), and ΔsroCΔhfq (JVS-9031) strains were transformed with pBAD-ctrl (pKP8-35) or pBAD-SroC (pYC6-4). ΔgcvBΔsroC was cotransformed with pPL-GcvB (pMM03). Each strain was grown to OD600 of 0.5 (0 min) and was further incubated for 10 min in the presence of 0.2% l-arabinose.
- Salmonella WT (JVS-1574), ΔgltIJKL (JVS-5823), ΔCA (JVS-10741), and ΔsroC (JVS-5821) strains were grown to OD600 of 2.0 prior to the addition of rifampicin. Total RNA was analyzed by Northern blot to determine decay rates of GcvB. The half-lives were determined from three independent experiments; the standard deviation is indicated.
To prove that SroC targets GcvB on the level of RNA stability, we determined changes in RNA half-life in rifampicin treatment experiments. Figure3B shows that deletion of either the gltIJKL locus or the sroC region alone dramatically increased the half-life of GcvB, from ∽1.7 min to > 15 min, whereas the GcvB target site in the 5′ UTR of gltI had little if any effect on the half-life of GcvB (see ΔCA mutant). Thus, ABC transporter mRNA cross talk is largely determined by SroC acting as a negative regulator of GcvB stability.
RNase E catalyzes both GcvB degradation and SroC biogenesis
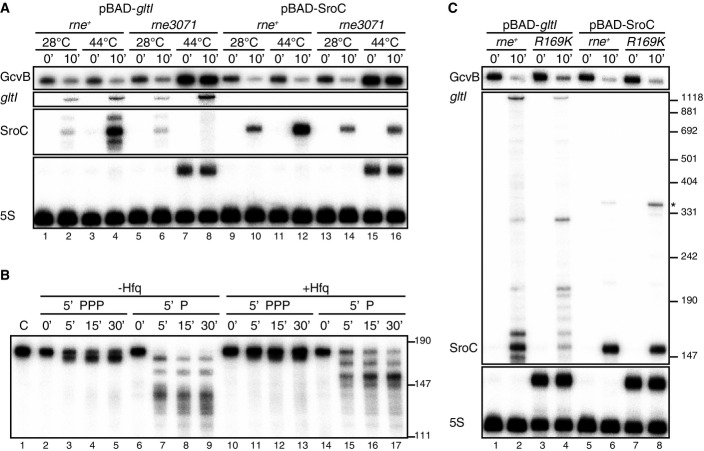
Endoribonuclease RNase E is the key enzyme for transcript destabilization in the regulatory network of Hfq-associated RNAs (Massé et al, 2003; Morita et al, 2005; Saramago et al, 2014). To test its involvement in GcvB depletion, we pulse-expressed the gltI-sroC region after heat inactivation of the essential RNase E protein using the temperature-sensitive rne3071 mutant strain (Fig4A). As expected, SroC depleted GcvB under all conditions in the presence of active RNase E (Fig4A, lanes 1–6), but GcvB levels remained unchanged upon inactivation of RNase E (lanes 7 versus 8; rne3071 at 44°C).
Figure 4.
- Salmonella ΔgltIJKL rne+ (JVS-9257) and ΔgltIJKL rne3071 (JVS-9258) strains transformed with pBAD-gltI or pBAD-SroC were grown to OD600 of 0.3 at 28°C and further incubated at 44°C for 30 min (OD600 of ˜0.5, indicated as time point 0). SroC expression was then induced for 10 min in the presence of 0.2% of l-arabinose. Total RNA was analyzed by Northern blots.
- 100 nM of 5′PPP or 5′P in vitro-transcribed preSroC was incubated with 100 nM of purified RNase E (1–529) at 30°C for the indicated time in the presence (lanes 10–17) or absence (lanes 2–9) of 100 nM Hfq. The same amount of preSroC was loaded in lane 1 as a control. SroC transcripts were detected using 5′-end-labeled oligonucleotide JVO-2907.
- Salmonella ΔgltIJKL rne+ (JVS-9257) and ΔgltIJKL rneR169K (JVS-11001) strains transformed with pBAD-gltI or pBAD-SroC were grown to OD600 of 0.5 (0 min) at 37°C and was further incubated for 10 min in the presence of 0.2% l-arabinose. The asterisk indicates transcriptional read-through to the rrnB terminator located downstream on the plasmid. Total RNA was prepared from the Salmonella strains and subjected to Northern blot analysis. 9S rRNA accumulated in the rneR169K strain. Estimated size from pUC8 marker is indicated on the right.
A caveat of this experiment was that RNase E also seemed essential to process SroC from the gltI-sroC precursor (Fig4A, lanes 7–8). To overcome this problem, we expressed SroC as a primary transcript, which permitted sufficient SroC accumulation under all RNase E +/− conditions tested (lanes 10, 12, 14, and 16) and helped confirm that RNase E was required for the targeted degradation of GcvB (lanes 16).
The observed requirement of RNase E for the biogenesis of SroC (Fig4A, lanes 7–8) is consistent with a model in which SroC is an mRNA processing fragment with 5′P (Vogel et al, 2003). To confirm this, we subjected a potential SroC precursor RNA to in vitro cleavage with purified RNase E (Fig4B). Preliminary experiments with serial 5′ truncations of the gltI-sroC region in vivo (Supplementary Fig S3) helped define SroC as requiring a 20-nucleotide leader as a minimal processing-proficient precursor (preSroC) for the in vitro cleavage reactions. Figure4B shows that in vitro-transcribed preSroC containing a 5′P after tobacco acid pyrophosphatase treatment was readily converted to the mature form of SroC by treatment with a catalytic domain (1–529) containing variant of RNase E (lanes 14–17). Correct maturation required the presence of the Hfq protein; without Hfq in the reaction, RNase E generated SroC variants with aberrant 5′ ends (lanes 6–9). Similarly important was the 5′ group of the substrate: preSroC with a 5′ triphosphate (5′PPP), incubated with or without Hfq, was a poor substrate for RNase E (lanes 2–5 and 10–13), suggesting that SroC maturation occurs by 5′-end-dependent mRNA decay initiated in the upstream gltI region.
We further explored the observed 5′P dependence in SroC biogenesis by constructing a chromosomal rneR169K mutant allele which expresses a variant of RNase E that is defective in 5′P sensing (Callaghan et al, 2005; Jourdan & McDowall, 2008; Bandyra et al, 2012). The 5′P sensing defect of this Salmonella mutant was apparent from the accumulation of the 5S rRNA precursor (Fig4C), as previously reported in E. coli (Garrey et al, 2009; Anupama et al, 2011). In line with our in vitro results, SroC biogenesis from the gltI-sroC transcript was dramatically altered in the rneR169K strain (Fig4C, lanes 1–4): Instead of mature SroC, longer processing intermediates accumulated. Intriguingly, however, the lower levels of 5′P-SroC in the rneR169K background still permitted GcvB regulation, and this was independently confirmed by the expression of SroC as a primary (5′PPP) transcript in this background (lanes 5–8; note that this construct would also yield 5′P SroC due to enzymatic 5′ pyrophosphate removal in vivo (Hui et al, 2014)). These results suggest the involvement of two different mechanisms of RNase E catalysis; that is, SroC is processed by the 5′-end-dependent pathway and GcvB is degraded by the 5′-end-independent pathway of the enzyme. While the molecular details of RNase E action on the GcvB-SroC complex are being investigated, we already note a striking difference to the inactivation of ChiX sRNA on the chbBC mRNA where base-pairing changes the 3′ terminator region of ChiX, rendering it susceptible to PNPase (Figueroa-Bossi et al, 2009; Plumbridge et al, 2014).
Direct interaction between GcvB and SroC
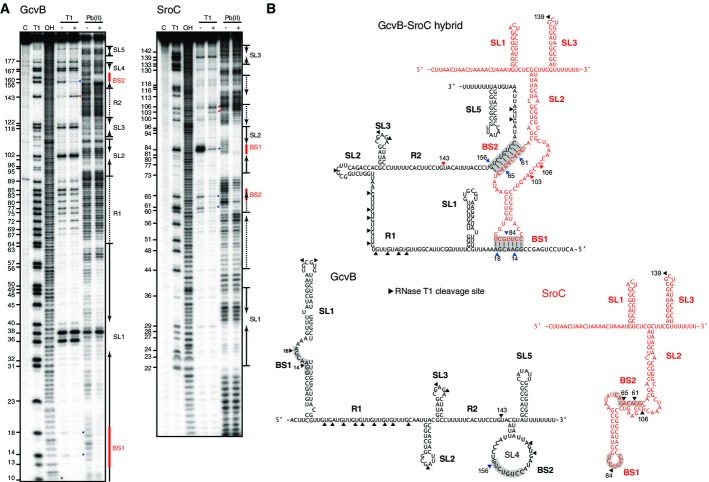
SroC-mediated depletion of GcvB requires the Hfq protein (Fig3A) whose primary role in the cell is to facilitate RNA interactions (Vogel & Luisi, 2011; Sobrero & Valverde, 2012; De Lay et al, 2013). To identify base-pairing regions, we probed the structures of GcvB, SroC, and their hybrid in vitro with lead(II) and RNase T1 (Fig5A). While the cleavage pattern of GcvB alone agreed with previous results (Sharma et al, 2007, 2011), the presence of a fivefold excess of SroC suppressed T1 cleavages at guanosine (G) positions 14, 18, and 156 in GcvB; concomitantly, G143 became more susceptible (Fig5B). SroC also protected stem loop 1 (SL1) of GcvB from backbone cleavage by lead(II). Conversely, GcvB protected SroC from T1 attack at G84, located in a rare accessible region at the tip of SL2 in the otherwise tightly folded SroC RNA. GcvB also protected SL2 of SroC during lead(II) probing, and suppressed T1 cleavage sites at G61 and G65 in the internal bulge of SroC. These results indicate that GcvB and SroC interact by base-pairing at two short complementary regions of 7 or 8 nucleotides. These predicted binding sites, hereafter referred to as BS1 and BS2, are only 14 nucleotides apart in SroC, but located in very distant regions of GcvB (Fig5B). While the BS1 region of GcvB has not been implicated in base-pairing with other targets before, the BS2 site partially overlaps with the GcvB target region for the phoP mRNA in E. coli (Coornaert et al, 2013).
Figure 5.
- 20 nM of 5′-end-labeled GcvB and SroC were subjected to RNase T1 and lead(II) cleavage in the absence and presence of 100 nM cold sRNAs, SroC, and GcvB, respectively. Lane C: untreated RNA; lane T1: RNase T1 ladder of denatured RNA; lane OH: alkaline ladder. The position of G residues cleaved by RNase T1 is indicated at the left of picture.
- Secondary structures of GcvB and SroC, paired or alone. RNase T1 cleavage sites are indicated by arrowheads; color indicates those that appeared (red) or disappeared (blue) upon base-pairing. The base pairing sites BS1 and BS2 are shadowed.
Alignment of SroC sequences reveals that the GcvB interaction sites are conserved in numerous enteric bacteria that also encode GcvB (Supplementary Fig S1A); the interaction sites in GcvB are also conserved (Supplementary Fig S1B). Using our two-plasmid approach, we confirmed that the GcvB and SroC homologues of E. coli are equally functional (Supplementary Fig S1C). In Yersinia pestis, however, the BS2 site is not conserved and SL2 containing the other GcvB binding regions is followed by a uridine stretch; as expected, these mutations generate a truncated SroC homologue which is unable to regulate GcvB (Supplementary Fig S1D).
Base-pairing with SroC destabilizes the regulator GcvB
To address the importance of the predicted RNA interaction for GcvB destabilization in vivo, point mutations were introduced in the BS1 and BS2 regions of the plasmid-expressed sRNAs (Fig6A). Either one of two C->G changes at SroC positions 63 (BS1) and 87 (BS2) weakened the repression of GcvB, while the individual compensatory mutations in GcvB (G14C, G160C) rendered GcvB a little less sensitive to SroC (Fig6B, lanes 4, 6, 10, 12). However, combining the two mutations in GcvB or SroC almost eliminated regulation when either sRNA was paired with its wild-type partner sRNA, but regulation was fully rescued by combination of the double compensatory mutations (GcvB G14C/G160C and SroC C87G/C63G). These in vivo results provide strong evidence that SroC destabilizes GcvB through two independent additive RNA interactions. The additive function may be explained by the relatively weak binding strength of each of the two interactions: Their predicted changes in minimal free energy of −13.6 kcal/mol (BS1) and −16.7 kcal/mol (BS2) are well above the range of, for example, the many mRNA interactions of the RybB sRNA (Papenfort et al, 2010).
Figure 6.

- GcvB-SroC base-pairing regions. The compensatory base pair changes in GcvB and SroC are indicated in red.
- Salmonella ΔgcvBΔsroC (JVS-5822) strain was cotransformed with pPL-GcvB and pBAD-SroC derivatives (see Supplementary Table S4). Each transformant was grown to OD600 of 0.5 (0 min) and incubated for another 10 min in the presence of 0.2% l-arabinose. Total RNA was prepared from the cells and subjected to Northern blot analysis. The fold repression of GcvB was determined from three independent replicates; standard deviation is shown.
- The chromosomally modified wild-type sroC strain (JVS-10108), sroC** mutant (JVS-10111) and sroC deletion mutant (JVS-5821) were grown to early stationary phase in LB medium (OD600 of 2.0) prior to the addition of rifampicin. Cultures were harvested at the indicated time points after rifampicin treatment. RNA was isolated and analyzed by Northern blot as in Fig3B.
To quantitatively assess the effect of this bipartite interaction with physiological concentrations of SroC and GcvB, we introduced scar-less point mutations of C63G/C87G (sroC**) in the chromosomal gltI-sroC locus and determined the half-life of GcvB. While SroC itself remained stable, the sroC** strain exhibited the same stabilization of GcvB as observed in ΔsroC (Fig6C), validating that base-pairing is required for SroC-mediated GcvB destabilization in vivo. Of note, the disruption of GcvB-SroC pairing by point mutations affects only the stability of GcvB, suggesting that SroC does not undergo coupled degradation with its partner RNA by RNase E as previously seen with Hfq-dependent sRNA-mRNA pairs (Massé et al, 2003).
SroC ensures expression of its parental mRNA along with other GcvB targets
To investigate how SroC affects the expression of GcvB targets in a chromosomal context, we constructed chromosomal translational superfolder GFP (sfGFP) fusions for oppA and gltI; the design of the gltI-sfgfp fusion maintains wild-type levels of SroC (see Materials and Methods). Using bacterial fluorescence as a proxy, we monitored GcvB- or SroC-dependent expression changes of OppA-sfGFP and GltI-sfGFP during growth in a minimal medium supplemented with 0.4% casamino acids as sole carbon and nitrogen sources (Fig7A and B). A comparison of wild-type and sroC** backgrounds revealed that both fusions require SroC for full expression. However, this difference between the wild-type and sroC** strains is eliminated upon the introduction of a ΔgcvB mutation. Single-cell measurements revealed largely homogenous expression patterns for each fusion (Fig7C and D), ruling out the possibility that SroC affects variability of GcvB target expression in the population. These experiments under physiological conditions confirm that mRNA cross talk via the SroC-GcvB axis affects ABC transporter expression both in cis and trans.
Figure 7.

- A–D Salmonella strains with oppA (A and C) and gltI (B and D) translational fusions to a sfgfp reporter gene in WT, sroC**, ΔgcvB, and ΔgcvB sroC** background were grown in minimal medium supplemented with 0.4% casamino acids. Fluorescence of bulk culture (A and B) normalized by OD595 was measured in 100 μl culture per well on 96-well plate over growth by TECAN plate reader. Fluorescence of 50,000 cells grown to exponential phase (C and D) was monitored by FACS. See also Supplementary Fig S4.
Salmonella and E. coli have three major peptide permeases, oligo-/di-peptide ABC transporters OppABCDF/DppABCDF and tripeptide symporter TppB (Hiles & Higgins, 1986), all of which are under the posttranscriptional control of GcvB (Urbanowski et al, 2000; Sharma et al, 2007, 2011; Pulvermacher et al, 2008). Given that SroC alleviates the repression of these multiple ABC transporters, we expected this sponge to promote growth under peptide-limiting conditions. To address this, we compared the growth of the sroC base-pairing mutant strain in two media—tryptone and casamino acids—that both offer carbon and nitrogen for growth but differ in their content of importable peptides (Supplementary Fig S4). As expected, inactivation of SroC increased the doubling time of Salmonella during growth on tryptone which is enzymatically digested from cow milk casein and thus abundant in peptides (Table1). By contrast, we observed no difference between the wild-type and ΔsroC strains in casamino acids medium. This latter medium is composed of 19 single amino acids which can be taken up by multiple amino acid transporters (Reitzer, 2005), most of which are not regulated by GcvB. This result indicates that one of the physiological roles of SroC is to fine-tune the expression of GcvB targets according to nutrient availability.
Table 1.
SroC promotes growth on peptides
| Strain | 0.4% tryptonea | 0.4% casamino acidsb |
|---|---|---|
| Wild-type | 181 ± 6 min | 140 ± 1 min |
| sroC** | 235 ± 2 min | 145 ± 1 min |
| ΔgcvB | 172 ± 2 min | 134 ± 5 min |
| ΔgcvB sroC** | 173 ± 4 min | 135 ± 6 min |
Doubling time for strain grown in the M9 minimal medium supplemented with 0.4% tryptone, calculated from triplicate cultures. “±” denote standard deviation. The associated growth curves are shown in Supplementary Fig S4A and B.
As above but culture in media supplemented with 0.4% casamino acids.
Discussion
By investigating a possible cellular function of a stable mRNA breakdown fragment, we have discovered a mechanism of mRNA cross talk in one of the most global RNA regulons currently known in bacteria. We show that the synthesis of gltI mRNA provides both, the template for protein synthesis of a Glu/Asp ABC transporter, and the precursor of an RNA antagonist of the GcvB small RNA which controls a large mRNA regulon of amino acid-related proteins. Through licensing GcvB for degradation by RNase E, the SroC sponge relieves its own parental mRNA from GcvB-mediated repression, resulting in a dual RNA containing feed-forward loop that might facilitate GltIJKL protein synthesis under certain conditions. Concomitantly, the SroC-mediated depletion of GcvB has the potential to cross-activate dozens of Salmonella mRNAs with amino acid-related transport and biosynthesis functions. Nucleotide conservation patterns in gltI and GcvB argue that this unusual RNA interaction has been maintained by selection and that the gltI-SroC-GcvB axis is functional in many enterobacterial species (Supplementary Fig S1). As will be discussed below, SroC may also be seen as a new subtype of the emerging class of 3′ UTR-derived sRNAs whose roles in bacterial physiology are just beginning to be appreciated.
Bacterial mRNA cross talk via antagonists of small RNAs
The GcvB regulon is the first bacterial RNA regulon in which a sponge-mediated cross talk would affect many physically unlinked mRNAs. Nonetheless, although studies of the paradigmatic chitosugar utilization pathway have focused on cross-activation of a single porin mRNA upon the degradation of ChiX sRNA (Figueroa-Bossi et al, 2009; Overgaard et al, 2009), we speculate that other ChiX targets such as the DpiA/B two-component system (Mandin & Gottesman, 2009b) are also likely to be affected. Likewise, it has been speculated that the RprA sRNA is trapped by the abundant csgD mRNA under certain growth conditions; this should affect the synthesis of additional RprA targets, foremost the rpoS mRNA (Mika et al, 2012).
In both the above scenarios, ChiX or RprA would be sequestered by intact mRNAs, whereas the primary GcvB antagonist is SroC and not its parental mRNA species. That is, SroC exceeds the glt transcripts with respect to both, cellular abundance and enrichment by Hfq (Chao et al, 2012). Nonetheless, the outcomes of this competition for mRNA cross talk will depend on how good a competitor SroC is compared to other targets; this will be additionally influenced by the transcription rate of the gltI operon synthesis, the net SroC levels, and the affinity and binding kinetics of GcvB for SroC as compared to the binding of other GcvB targets. While these parameters remain to be determined, we speculate that this novel mRNA-sRNA-sRNA-mRNA scheme of cross talk may be advantageous for two reasons: (i) Unlike the high molecular weight glt mRNAs, SroC is not expected to be diffusion-limited, increasing its chance to interact with GcvB in the cell; (ii) the activity of SroC may be subject to independent further posttranscriptional control, allowing for the integration of extra input signals in this complex RNA-based circuit.
Enriched stable mRNA fragments such as SroC have been observed in Hfq coIP experiments in diverse organisms (Zhang et al, 2003; Sonnleitner et al, 2008; Berghoff et al, 2011; Chao et al, 2012; Möller et al, 2014). Given that the concentration of Hfq is limiting in vivo (Fender et al, 2010; Moon & Gottesman, 2011), we predict that some of these stable mRNA fragments function as sRNA sponges akin to SroC.
A lesson for experimental target prediction
Examples of positive regulation by sRNAs were for many years considered to be rare exceptions to target repression, but they have recently received increased attention following reports of new direct and indirect mechanisms of mRNA activation (Sonnleitner et al, 2011; Fröhlich et al, 2013; Göpel et al, 2013; Jørgensen et al, 2013; Papenfort et al, 2013; Salvail et al, 2013). Nonetheless, the activation of a large number of apparent targets by SroC (Fig1B) was unprecedented. We can now explain this by a high rate of false positives, caused by titration of another sRNA. Thus, SroC holds an important lesson for future experimental target predictions as we enter the era of systematic screening of sRNA functions (Papenfort et al, 2008; Mandin & Gottesman, 2009a; Nichols et al, 2011; De Lay & Gottesman, 2012; Brochado & Typas, 2013). Curated sRNA target databases (Cao et al, 2010) will be needed to identify obvious false positives among target activation patterns.
SroC as a new type of 3′ UTR-derived small RNA
RNA species such as SroC pose a challenge to functional transcript annotation. From a genetic point of view that may ignore the underlying mechanism, it is the gltI mRNA that cross talks with other ABC transporter mRNAs via GcvB. On the molecular level, however, SroC is an Hfq-associated sRNA produced from the 3′ UTR of the gltI mRNA. While there has been much effort to predict and characterize conserved freestanding sRNA genes, there is increasing evidence for widespread functions of conserved 3′ UTR-embedded sRNAs. Such transcripts usually share the terminator with their ‘host’ mRNA and fall into two classes according to their biogenesis (Chao et al, 2012). The first class includes the Hfq-dependent DapZ or MicL sRNAs, which are independently expressed from mRNA gene internal promoters; both DapZ and MicL act in pathways unrelated to their respective mRNA loci (Chao et al, 2012; Guo et al, 2014). Members of the other class, for example, s-SodF sRNA of Streptomyces coelicolor (Kim et al, 2014), are strictly generated by mRNA processing, for lack of a gene internal promoter. Interestingly, similar to SroC regulating (indirectly) amino acid-related mRNAs, the s-SodF sRNA mediates cross talk of two superoxide dismutase-encoding mRNAs (Kim et al, 2014). Clearly, more examples are needed to address whether biogenesis type and target choice of 3′ UTR-embedded sRNAs are correlated.
SroC is a previously unknown variation of the theme of 3′ UTR-derived sRNAs whose biogenesis warrants deeper investigation. The single gltIJKL promoter produces both, the full-length gltIJKL operon mRNA to make the entire Glu/Asp ABC transport system and the shorter mRNA of the periplasmic Glu/Asp-binding protein, GltI, alone (Kröger et al, 2013). The periplasmic high-affinity binding protein recaptures endogenous compounds leaking from the cell (Stirling et al, 1989) and is therefore made in much higher stoichiometry than the other components (Boos & Lucht, 1996). This is commonly achieved by gene order (first position in ABC transporter operons) and selective stabilization of a monocistronic mRNA from 3′ to 5′ exonucleolytic decay. By contrast, the monocistronic gltI mRNA and its SroC derivative are generated by a leaky Rho-independent terminator. We note that the SroC region harbors additional conserved nucleotides outside the GcvB contact regions, which raises the possibility that a cis-acting element or a trans-acting factor regulates transcription termination in the gltIJ intergenic region and, thus, the release of SroC and with it the levels of GcvB.
Regulating the regulator GcvB
The cellular level of GcvB has been known to be tightly controlled at the level of transcription, through the activity of the GcvA/GcvR transcription factors that respond to glycine availability (Urbanowski et al, 2000; Ghrist et al, 2001; Heil et al, 2002; Stauffer & Stauffer, 2005). Such tight transcriptional control reflects the situation of many other sRNAs, some of which possess the most highly regulated promoters in their respective regulons (Pfeiffer et al, 2007; Mutalik et al, 2009). However, GcvB is also a paradigm for a very unstable sRNA, displaying the shortest (< 2 min) half-life of twenty sRNAs in the first systematic evaluation of sRNA stability (Vogel et al, 2003). The molecular cause of its lability remained elusive; GcvB possesses terminal structures (Sharma et al, 2007) that should protect it from rapid degradation.
At first, coupled degradation whereby sRNAs are degraded as they base-pair with targets (Massé et al, 2003) seemed a straightforward explanation for this lability, given the extensive GcvB target regulon (Pulvermacher et al, 2009; Modi et al, 2011; Sharma et al, 2011; Coornaert et al, 2013; Wright et al, 2013). Our study, however, now reveals a singular factor—the SroC sponge—to account for much of the instability of GcvB (Figs3B and 6C) demonstrating that GcvB is regulated at both the level of synthesis and decay, with consequences for the whole GcvB regulon (Fig1B). Comparison of intracellular copy numbers suggests that SroC targets GcvB in a near-stoichiometric manner. Stationary-phase Salmonella grown in rich medium expresses ∽50 copies of SroC per cell (Supplementary Fig S5); if SroC is genomically depleted, the steady-state level of GcvB increases by ∽fourfold and the half-life by ∽eightfold (Fig3B). When the level of SroC is reduced by half, GcvB levels increase by only ∽threefold (Supplementary Fig S5). We consider SroC itself as a true sponge as under no condition, GcvB affects its steady-state levels.
Others recently reported the discovery of a prophage-specific anti-GcvB sRNA, named AgvB, in a strain of enterohemorrhagic E. coli (Tree et al, 2014). Physiological levels of AgvB when it acts remain unknown, but an overexpression experiment suggests that it can antagonize GcvB without affecting sRNA stability. Importantly, unlike SroC which binds GcvB outside the established mRNA binding sites, the AgvB sRNA may directly compete with mRNA regulation by mimicking a C/A-rich target site complementary to the R1 seed region of GcvB. Clearly, although these two GcvB antagonists differ in their molecular mechanism, their existence argues that control of the global regulator GcvB at the RNA level equally benefits endogenous gene expression and horizontally acquired genetic elements. It will also be interesting to see whether functionally analogous riboregulators of amino acid-related mRNAs, for example, the enterobacterial DapZ sRNA, the Staphylococcus RsaE sRNA, or the α-proteobacterial AbcR sRNAs (Geissmann et al, 2009; Bohn et al, 2010; Caswell et al, 2012; Chao et al, 2012; Torres-Quesada et al, 2013; Overlöper et al, 2014), are kept in check by similar types of RNA sponges.
Outlook
An important physiological question to answer in the future is why the gltIJKL locus evolved to express such a potent GcvB sponge. This operon encodes a Glu/Asp ABC transporter, which together with the GltP and GltS proteins supplies bacteria with Glu or Asp (Schellenberg & Furlong, 1977; Reitzer, 2004). This could be relevant for Salmonella infection of epithelial cells, for glutamate is limiting in the Salmonella-containing vacuole (Bowden et al, 2010). However, a review of published Salmonella mutagenesis data sets (Supplementary Table S2) has so far failed to suggest a suitable system to study SroC effects in the nutrient-poor environment of bacterial hosts. Under some conditions, the SroC-GcvB axis may also affect the mRNAs of the global transcriptional regulators, Lrp, CsgD, and PhoP (Modi et al, 2011; Sharma et al, 2011; Coornaert et al, 2013; Wright et al, 2013). Lrp alone affects the expression of ∽10% of all genes (Cho et al, 2008, 2011), which include prominent GcvB targets such as oppABCDF, dppABCDF, tppB, livJ-livKHMGF, brnQ-proY, cycA, thrLABC, and gdhA genes. This would suggest that a plethora of coherent or incoherent feed-forward loops in response to leucine and glycine levels alters gene expression dynamics (Beisel & Storz, 2010).
A combination of approaches profiling both gene expression and metabolic changes at high resolution will be needed to address how global mRNA cross talk through SroC affects the expression kinetics and hierarchy of GcvB targets. In addition, we do not rule out the possibility that SroC regulates other mRNAs independent of GcvB, for example, the phage-derived STM2728 mRNA. Last but not least, its exceptional intracellular stability (Vogel et al, 2003) asks the question whether SroC itself is turned over in a controlled manner, perhaps through adaptor-mediated recruitment of RNase E as recently shown for other sRNAs (Suzuki et al, 2006; Göpel et al, 2013).
Materials and Methods
Bacterial strains and growth conditions
Salmonella enterica serovar Typhimurium strain SL1344 (JVS-1574) was used as a wild-type strain. The strains used in this study are listed in Supplementary Table S3. Bacterial cells were grown at 37°C with shaking at 220 rpm in LB broth (Lennox) medium or minimal medium (the same as M9 but without NH4Cl; 12.8 g Na2HPO4 7H2O, 3 g KH2PO4, 0.5 g NaCl per liter). Minimal medium was supplemented with tryptone or casamino acids at a final concentration of 0.4% as a sole carbon and nitrogen source. Solutions of tryptone and casamino acids (BD Biosciences) were sterilized by filtration. Where appropriate, media were supplemented with antibiotics at the following concentrations: 100 μg/ml ampicillin (Ap), 50 μg/ml kanamycin (Km), 20 μg/ml chloramphenicol (Cm) and 500 μg/ml rifampicin.
Strain construction
Deletion strains were constructed by the lambda Red system (Datsenko & Wanner, 2000). sroC, gltIJKL, and gltJLK were deleted using pKD4 as a template and primer pairs, JVO-0303/JVO-0304, JVO-5007/JVO-5008, and JVO-11569/JVO-5008, respectively. The resulting Km-resistant strains were confirmed by PCR, and the mutant loci were transduced into appropriate genetic backgrounds by P22 phage. To eliminate the resistance genes from the chromosome, strains were transformed with the temperature-sensitive plasmid pCP20 expressing FLP recombinase (Datsenko & Wanner, 2000).
Chromosomal mutant strains, sroC (JVS-10108, JVS-10111) and gltIΔCA (JVS-10741), were constructed by scar-less mutagenesis using a two-step lambda Red system (Blank et al, 2011). DNA fragments containing a CmR resistance marker and a I-SceI recognition site amplified with primer pairs JVO-0303/JVO-0304 and JVO-7448/JVO-11078 from the template plasmid pWRG100 were integrated into the chromosomal sroC and gltIp region, respectively, by lambda Red recombinase expressed from pKD46 (Datsenko & Wanner, 2000). The resultant mutant was transformed by pWRG99, and the mutant allele amplified from pBAD-SroC or pBAD-gltI derivatives (Supplementary Table S4) with JVO-9194/JVO-9195 or JVO-7451/JVO-9611 was integrated by the lambda Red recombinase expressed from pWRG99, which subsequently expressed I-SceI endonuclease by supplementation of 2 μg/ml of anhydrotetracyclin to select the resultant recombinant in which the CmR I-SceI allele was eliminated. Successful recombinants were confirmed by Cm sensitivity, PCR, and sequencing.
To integrate oppA and gltI translational fusions into Salmonella chromosome, oppA16aa-sfgfp and gltI16aa-sfgfp were amplified with primer pairs JVO-10629/JVO-9762 and JVO-10631/JVO-10688 using pDP124 (pCBP derivative replaced with sfgfp) (Wahl et al, 2009) as a template and were recombined by lambda Red system (Datsenko & Wanner, 2000). To omit the downstream terminator sequence and Km resistance gene, the C-terminal 23 aa of the gltI sequence was fused with sfgfp by scar-less mutagenesis (Blank et al, 2011). A PCR fragment amplified with JVO-10927/JVO-10668 from pWRG100 was first integrated into the gltI-sfgfp KmR strain, and next, the CmR I-SceI allele was replaced by an 80-mer dsDNA of JVO-10966/JVO-10967 using pWRG99.
Salmonella rne3071 mutant (JVS-7000) and its parental strain (JVS-6999) were kindly provided by L. Bossi. Mutation in the 5′-end sensor of RNase E (R169K) was introduced into the chromosomal rne gene by a previously described procedure in Figueroa-Bossi et al (2009). A DNA fragment was amplified with the JVO-10059 (the same as ppC66) and JVO-11002 primers from genomic DNA of strain JVS-6999 (contains the Cm resistance gene in the IGR between rluC and rne) and integrated into the wild-type strain by lambda Red recombinase expressed from pKD46 (Datsenko & Wanner, 2000). The mutant (JVS-10999) was selected for Cm resistance and small colony formation and was confirmed by sequencing; it also showed the expected accumulation of 9S rRNA.
Plasmid construction
A complete list of all plasmids used in this study can be found in Supplementary Table S4. GcvB expression plasmid pMM03 and its control pMM01 were constructed by replacing the XhoI-AvrII fragment of pTP11 and pTP09 (Sharma et al, 2007) with that containing a Cm resistance marker and a p15A ori from pZA31-luc (Lutz & Bujard, 1997), respectively. To overexpress SroC and gltI-sroC mRNA under the control of arabinose-inducible promoter, sroC and gltI-sroC fragments were amplified with JVO-4625/JVO-4626 and JVO-7775/JVO-0306 and cloned into a pBAD backbone by the procedure described previously in Papenfort et al (2006), to yield pYC6-4 and pMM36, respectively. pMM44 and pMM45 expressing gltIΔCA and preSroC were generated by PCR with JVO-1973/JVO-1974 and JVO-7997/JVO-4531 using pMM36 as a template and self-ligation of the PCR fragments. The plasmid-borne gcvB and sroC were mutated by site-directed mutagenesis using overlapping PCR with primer pairs, JVO-9090/JVO-9091 (gcvB G14C), JVO-9214/JVO-9215 (gcvB G160C), JVO-7578/JVO-9020 (sroC C87G), and JVO-9216/JVO-9217 (sroC C63G), respectively.
Microarray analysis
Microarray analysis of SroC overexpression from PBAD promoter was performed as described (Papenfort et al, 2006; Sharma et al, 2011). The wild-type (JVS-1574) or ΔgcvB (JVS-1044) strains were transformed with pKP8-35 (pBAD-ctrl) or pYC6-4 (pBAD-SroC) and were grown in LB medium. At optical density (OD600) of 1.5, l-arabinose was added to cultures at a final concentration of 0.2% to induced SroC expression for 10 min, and total RNA was prepared with SV total RNA isolation system (Promega). The gene expression in WT strain containing pBAD-ctrl was normalized (to 1) and used as a standard for differential gene expression analysis. SroC-induced changes in ΔgcvB were analyzed by comparing the expression levels in ΔgcvB (pBAD-SroC) to those in ΔgcvB (pBAD-ctrl) and plotted in Fig1B. A list of differentially expressed genes and fold changes can be found in Supplementary Table S1.
Northern blot analysis
Bacterial culture was immediately frozen at an appropriate condition by the addition of 0.2 vol/vol of stop solution (95% ethanol and 5% phenol). Total RNA was isolated using the TRIzol reagent (Invitrogen). Five μg of total RNA was denatured at 95°C for 5 min in RNA loading buffer (95% v/v formamide, 10 mM EDTA, 0.1% w/v xylene cyanole, 0.1% w/v bromophenol blue) and separated by gel electrophoresis on 4% polyacrylamide/7 M urea gels in 1× TBE buffer. RNA was transferred from the gel onto Hybond-XL nylon membrane (GE Healthcare) by electroblotting. The membrane was cross-linked by UV light, and after prehybridization in Roti-Hybri-Quick buffer (Roth), a [32P]-labeled probe was hybridized at 42°C. Membrane was washed in three subsequent 15-min steps in 2× SSC/0.1% SDS, 1× SSC/0.1% SDS, and 0.5× SSC/0.1% SDS buffers at 42°C. To detect GcvB, SroC, and 5S rRNA, oligonucleotides JVO-0750, JVO-2907, and JVO-0322 were 5′-end-labeled with [32P]-γ-ATP by T4 polynucleotide kinase (Fermentas) and purified over G25 columns (GE Healthcare). Signals were visualized on Typhoon FLA 7000 (GE Healthcare) and quantified using AIDA software (Raytest).
Western blot analysis
Western blot was performed following a previously published protocol (Urban & Vogel, 2007). Briefly, 1 OD of bacteria culture was collected by centrifugation for 2 min at 16,100 g at 4°C, and the pellet was dissolved in 100 μl of 1× protein loading buffer. After heating for 5 min at 95°C, 2 μl of samples were separated on 10% SDS–PAGE. OppA and GroEL were detected as described previously in Sharma et al (2007).
In vitro structure probing
In vitro transcripts of Salmonella GcvB and SroC were generated with the T7 Megascript Kit (Ambion) using DNA templates amplified with oligonucleotides JVO-0941/JVO-8375 and JVO-7588/JVO-8374, respectively. A total of 20 pmol of RNA was 5′-end-labeled and purified as described previously in Papenfort et al (2006). Structure probing was performed using 0.1 pmol of RNA in 10 μl reactions as previously described in Sharma et al (2007). 5′-end-labeled RNA was denatured for 1 min at 95°C followed by incubation on ice for 5 min and hybridized with fivefold excess of cold RNA for 10 min at 37°C in the presence of 1 μg of yeast tRNA and 1× structure buffer (10 mM Tris, pH 7.0, 100 mM KCl, 10 mM MgCl2; Ambion). The RNA mixture was digested with a final concentration of 5 mM lead(II) (Fluka) or 0.005 units of RNase T1 (Ambion) for 1.5 or 3 min at 37°C. RNase III cleavage reaction was conducted with 2 units of ShortCut RNase III (NEB) in the presence of 1 mM DTT for 5 min at 37°C. RNase T1 ladder was generated by incubating 0.2 pmol of denatured RNA with 0.1 units of RNase T1 in 1× sequencing buffer (Ambion) for 5 min at 37°C. OH ladder was obtained by incubating 0.2 pmol of RNA in alkaline hydrolysis buffer (Ambion) for 5 min at 95°C. Reactions were stopped by adding 12 μl loading buffer II (95% v/v formamide, 18 mM EDTA, 0.025% SDS, xylene cyanole, bromophenol blue; Ambion). Samples were denatured for 3 min at 95°C and run on 6% polyacrylamide/7 M urea sequencing gels in 1× TBE buffer at 40 W for 90 min. Gels were dried and analyzed using Typhoon FLA 7000 (GE Healthcare) and AIDA software (Raytest).
In vitro RNase E cleavage assay
In vitro transcripts of preSroC were generated with the T7 Megascript Kit (Ambion) using a DNA template generated with oligonucleotides JVO-8373/JVO-8374 and were hydrolyzed by tobacco acid pyrophosphatase (Epicentre). In a 10 μl reaction buffer (25 mM Tris–HCl, pH 7.5, 50 mM NaCl, 50 mM KCl, 10 mM MgCl2, 1 mM DDT), 100 nM of 5′PPP or 5′P preSroC were bound with 100 nM Hfq for 10 min at 30°C and incubated with 100 nM of RNase E (1–529). Reactions were stopped by the addition of equal volume of RNA loading buffer (95% v/v formamide, 10 mM EDTA, 0.1% w/v xylene cyanole, 0.1% w/v bromophenol blue) and heating for 5 min at 95°C. Processing products were separated by gel electrophoresis on 4% polyacrylamide/7 M urea gels and were visualized by Northern blot as described above.
Fluorescence monitoring during growth
Salmonella strains were precultured in 2 ml of 0.4% casamino acids medium overnight. On a 96-well plate, 100 μl of 0.4% tryptone or casamino acids medium was inoculated by 1/100 of preculture. The plate was incubated at 37°C with shaking at 3-mm amplitude with monitoring GFP fluorescence (excitation at 485 ± 20 nm, emission at 535 ± 25 nm) and optical density (absorbance at 595 ± 10 nm) every 15 min using an Infinite 200 Pro machine (Tecan). GFP fluorescence was normalized by OD595, and that of GFP-negative cells grown under the same condition was subtracted.
Flow cytometry
Salmonella translational fusion strains were grown in 0.4% casamino acids medium to exponential phase, and cells were fixed with 4% paraformaldehyde and resuspended in 1× PBS buffer (pH 7.4). GFP fluorescense intensity was quantified for 50,000 events by flow cytometry using FACSCalibur (BD Biosciences). Data were analyzed by Cyflogic software (CyFlo Ltd).
Acknowledgments
We thank Kai Papenfort and Cynthia Sharma for comments, ideas, and stimulating discussions; Stan Gorski for editing the manuscript; K. Papenfort and H. J. Mollenkopf for microarrays analysis; Lionello Bossi for Salmonella rne and rne3071 strains; Roman Gerlach for providing pWRG99 and pWRG100 plasmids; Kazuei Igarashi for OppA antiserum; and Ben Luisi for purified RNase E proteins. M.M. is supported by JSPS Postdoctoral Fellowships for Research Abroad. The Vogel laboratory received funds from DFG Priority Program SPP1258 (Vo875/3-2), the Bavarian BioSysNet program, and BMBF eBio programme RNASys.
Author contributions
MM and JV designed the study; MM and YC performed the experiments; MM, YC, and JV analyzed the data; MM and JV wrote the paper with input from YC.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary Information
Review Process File
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
References
- Anupama K, Leela JK, Gowrishankar J. Two pathways for RNase E action in Escherichia coli in vivo and bypass of its essentiality in mutants defective for Rho-dependent transcription termination. Mol Microbiol. 2011;82:1330–1348. doi: 10.1111/j.1365-2958.2011.07895.x. [DOI] [PubMed] [Google Scholar]
- Bandyra KJ, Said N, Pfeiffer V, Gorna MW, Vogel J, Luisi BF. The seed region of a small RNA drives the controlled destruction of the target mRNA by the endoribonuclease RNase E. Mol Cell. 2012;47:943–953. doi: 10.1016/j.molcel.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel CL, Storz G. Base pairing small RNAs and their roles in global regulatory networks. FEMS Microbiol Rev. 2010;34:866–882. doi: 10.1111/j.1574-6976.2010.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghoff BA, Glaeser J, Sharma CM, Zobawa M, Lottspeich F, Vogel J, Klug G. Contribution of Hfq to photooxidative stress resistance and global regulation in Rhodobacter sphaeroides. Mol Microbiol. 2011;80:1479–1495. doi: 10.1111/j.1365-2958.2011.07658.x. [DOI] [PubMed] [Google Scholar]
- Blank K, Hensel M, Gerlach RG. Rapid and highly efficient method for scarless mutagenesis within the Salmonella enterica chromosome. PLoS One. 2011;6:e15763. doi: 10.1371/journal.pone.0015763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohn C, Rigoulay C, Chabelskaya S, Sharma CM, Marchais A, Skorski P, Borezée-Durant E, Barbet R, Jacquet E, Jacq A, Gautheret D, Felden B, Vogel J, Bouloc P. Experimental discovery of small RNAs in Staphylococcus aureus reveals a riboregulator of central metabolism. Nucleic Acids Res. 2010;38:6620–6636. doi: 10.1093/nar/gkq462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boos W, Lucht JM. Periplasmic binding protein-dependent ABC transporters. In: Neidhardt FC, Curtiss R III, Ingraham JL, Lin CC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, DC: ASM Press; 1996. pp. 1175–1209. [Google Scholar]
- Bowden SD, Ramachandran VK, Knudsen GM, Hinton JC, Thompson A. An incomplete TCA cycle increases survival of Salmonella Typhimurium during infection of resting and activated murine macrophages. PLoS One. 2010;5:e13871. doi: 10.1371/journal.pone.0013871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochado AR, Typas A. High-throughput approaches to understanding gene function and mapping network architecture in bacteria. Curr Opin Microbiol. 2013;16:199–206. doi: 10.1016/j.mib.2013.01.008. [DOI] [PubMed] [Google Scholar]
- Busi F, Le Derout J, Cerciat M, Régnier P, Hajnsdorf E. Is the secondary putative RNA-RNA interaction site relevant to GcvB mediated regulation of oppA mRNA in Escherichia coli. Biochimie. 2010;92:1458–1461. doi: 10.1016/j.biochi.2010.06.020. [DOI] [PubMed] [Google Scholar]
- Callaghan AJ, Marcaida MJ, Stead JA, McDowall KJ, Scott WG, Luisi BF. Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature. 2005;437:1187–1191. doi: 10.1038/nature04084. [DOI] [PubMed] [Google Scholar]
- Cao Y, Wu J, Liu Q, Zhao Y, Ying X, Cha L, Wang L, Li W. sRNATarBase: a comprehensive database of bacterial sRNA targets verified by experiments. RNA. 2010;16:2051–2057. doi: 10.1261/rna.2193110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell CC, Gaines JM, Ciborowski P, Smith D, Borchers CH, Roux CM, Sayood K, Dunman PM, Roop II RM. Identification of two small regulatory RNAs linked to virulence in Brucella abortus 2308. Mol Microbiol. 2012;85:345–360. doi: 10.1111/j.1365-2958.2012.08117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D, Yario T, Steitz JA. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science. 2010;328:1563–1566. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J. An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 2012;31:4005–4019. doi: 10.1038/emboj.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho BK, Barrett CL, Knight EM, Park YS, Palsson BØ. Genome-scale reconstruction of the Lrp regulatory network in Escherichia coli. Proc Natl Acad Sci USA. 2008;105:19462–19467. doi: 10.1073/pnas.0807227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho BK, Federowicz S, Park YS, Zengler K, Palsson BØ. Deciphering the transcriptional regulatory logic of amino acid metabolism. Nat Chem Biol. 2011;8:65–71. doi: 10.1038/nchembio.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coornaert A, Chiaruttini C, Springer M, Guillier M. Post-transcriptional control of the Escherichia coli PhoQ-PhoP two-component system by multiple sRNAs involves a novel pairing region of GcvB. PLoS Genet. 2013;9:e1003156. doi: 10.1371/journal.pgen.1003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lay N, Gottesman S. A complex network of small non-coding RNAs regulate motility in Escherichia coli. Mol Microbiol. 2012;86:524–538. doi: 10.1111/j.1365-2958.2012.08209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lay N, Schu DJ, Gottesman S. Bacterial small RNA-based negative regulation: Hfq and its accomplices. J Biol Chem. 2013;288:7996–8003. doi: 10.1074/jbc.R112.441386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. Emerging roles for natural microRNA sponges. Curr Biol. 2010;20:R858–R861. doi: 10.1016/j.cub.2010.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fender A, Elf J, Hampel K, Zimmermann B, Wagner EG. RNAs actively cycle on the Sm-like protein Hfq. Genes Dev. 2010;24:2621–2626. doi: 10.1101/gad.591310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa-Bossi N, Valentini M, Malleret L, Fiorini F, Bossi L. Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev. 2009;23:2004–2015. doi: 10.1101/gad.541609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- Fröhlich KS, Papenfort K, Fekete A, Vogel J. A small RNA activates CFA synthase by isoform-specific mRNA stabilization. EMBO J. 2013;32:2963–2979. doi: 10.1038/emboj.2013.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrey SM, Blech M, Riffell JL, Hankins JS, Stickney LM, Diver M, Hsu YH, Kunanithy V, Mackie GA. Substrate binding and active site residues in RNases E and G: role of the 5′-sensor. J Biol Chem. 2009;284:31843–31850. doi: 10.1074/jbc.M109.063263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann T, Chevalier C, Cros MJ, Boisset S, Fechter P, Noirot C, Schrenzel J, François P, Vandenesch F, Gaspin C, Romby P. A search for small noncoding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation. Nucleic Acids Res. 2009;37:7239–7257. doi: 10.1093/nar/gkp668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghrist AC, Heil G, Stauffer GV. GcvR interacts with GcvA to inhibit activation of the Escherichia coli glycine cleavage operon. Microbiology. 2001;147:2215–2221. doi: 10.1099/00221287-147-8-2215. [DOI] [PubMed] [Google Scholar]
- Göpel Y, Papenfort K, Reichenbach B, Vogel J, Görke B. Targeted decay of a regulatory small RNA by an adaptor protein for RNase E and counteraction by an anti-adaptor RNA. Genes Dev. 2013;27:552–564. doi: 10.1101/gad.210112.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo MS, Updegrove TB, Gogol EB, Shabalina SA, Gross CA, Storz G. MicL, a new σE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Dev. 2014;28:1620–1634. doi: 10.1101/gad.243485.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- Heil G, Stauffer LT, Stauffer GV. Glycine binds the transcriptional accessory protein GcvR to disrupt a GcvA/GcvR interaction and allow GcvA-mediated activation of the Escherichia coli gcvTHP operon. Microbiology. 2002;148:2203–2214. doi: 10.1099/00221287-148-7-2203. [DOI] [PubMed] [Google Scholar]
- Hiles ID, Higgins CF. Peptide uptake by Salmonella Typhimurium. The periplasmic oligopeptide-binding protein. Eur J Biochem. 1986;158:561–567. doi: 10.1111/j.1432-1033.1986.tb09791.x. [DOI] [PubMed] [Google Scholar]
- Hui MP, Foley PL, Belasco JG. Messenger RNA degradation in bacterial cells. Annu Rev Genet. 2014;48:537–559. doi: 10.1146/annurev-genet-120213-092340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein R, Lim HN. Disruption of small RNA signaling caused by competition for Hfq. Proc Natl Acad Sci USA. 2011;108:1110–1115. doi: 10.1073/pnas.1010082108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen MG, Nielsen JS, Boysen A, Franch T, Møller-Jensen J, Valentin-Hansen P. Small regulatory RNAs control the multi-cellular adhesive lifestyle of Escherichia coli. Mol Microbiol. 2012;84:36–50. doi: 10.1111/j.1365-2958.2012.07976.x. [DOI] [PubMed] [Google Scholar]
- Jørgensen MG, Thomason MK, Havelund J, Valentin-Hansen P, Storz G. Dual function of the McaS small RNA in controlling biofilm formation. Genes Dev. 2013;27:1132–1145. doi: 10.1101/gad.214734.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdan SS, McDowall KJ. Sensing of 5′ monophosphate by Escherichia coli RNase G can significantly enhance association with RNA and stimulate the decay of functional mRNA transcripts in vivo. Mol Microbiol. 2008;67:102–115. doi: 10.1111/j.1365-2958.2007.06028.x. [DOI] [PubMed] [Google Scholar]
- Kim D, Hong JS, Qiu Y, Nagarajan H, Seo JH, Cho BK, Tsai SF, Palsson BØ. Comparative analysis of regulatory elements between Escherichia coli and Klebsiella pneumoniae by genome-wide transcription start site profiling. PLoS Genet. 2012;8:e1002867. doi: 10.1371/journal.pgen.1002867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HM, Shin JH, Cho YB, Roe JH. Inverse regulation of Fe- and Ni-containing SOD genes by a Fur family regulator Nur through small RNA processed from 3′UTR of the sodF mRNA. Nucleic Acids Res. 2014;42:2003–2014. doi: 10.1093/nar/gkt1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröger C, Dillon SC, Cameron AD, Papenfort K, Sivasankaran SK, Hokamp K, Chao Y, Sittka A, Hébrard M, Händler K, Colgan A, Leekitcharoenphon P, Langridge GC, Lohan AJ, Loftus B, Lucchini S, Ussery DW, Dorman CJ, Thomson NR, Vogel J, et al. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc Natl Acad Sci USA. 2012;109:E1277–E1286. doi: 10.1073/pnas.1201061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröger C, Colgan A, Srikumar S, Händler K, Sivasankaran SK, Hammarlöf DL, Canals R, Grissom JE, Conway T, Hokamp K, Hinton JC. An infection-relevant transcriptomic compendium for Salmonella enterica Serovar Typhimurium. Cell Host Microbe. 2013;14:683–695. doi: 10.1016/j.chom.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandin P, Gottesman S. A genetic approach for finding small RNAs regulators of genes of interest identifies RybC as regulating the DpiA/DpiB two-component system. Mol Microbiol. 2009a;72:551–565. doi: 10.1111/j.1365-2958.2009.06665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandin P, Gottesman S. Regulating the regulator: an RNA decoy acts as an OFF switch for the regulation of an sRNA. Genes Dev. 2009b;23:1981–1985. doi: 10.1101/gad.1846609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcinowski L, Tanguy M, Krmpotic A, Rädle B, Lisnić VJ, Tuddenham L, Chane-Woon-Ming B, Ruzsics Z, Erhard F, Benkartek C, Babic M, Zimmer R, Trgovcich J, Koszinowski UH, Jonjic S, Pfeffer S, Dölken L. Degradation of cellular mir-27 by a novel, highly abundant viral transcript is important for efficient virus replication in vivo. PLoS Pathog. 2012;8:e1002510. doi: 10.1371/journal.ppat.1002510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landhaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Mika F, Busse S, Possling A, Berkholz J, Tschowri N, Sommerfeldt N, Pruteanu M, Hengge R. Targeting of csgD by the small regulatory RNA RprA links stationary phase, biofilm formation and cell envelope stress in Escherichia coli. Mol Microbiol. 2012;84:51–65. doi: 10.1111/j.1365-2958.2012.08002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi SR, Camacho DM, Kohanski MA, Walker GC, Collins JJ. Functional characterization of bacterial sRNAs using a network biology approach. Proc Natl Acad Sci USA. 2011;108:15522–15527. doi: 10.1073/pnas.1104318108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller P, Overlöper A, Förstner KU, Wen TN, Sharma CM, Lai EM, Narberhaus F. Profound Impact of Hfq on Nutrient Acquisition, Metabolism and Motility in the Plant Pathogen Agrobacterium tumefaciens. PLoS One. 2014;9:e110427. doi: 10.1371/journal.pone.0110427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon K, Gottesman S. Competition among Hfq-binding small RNAs in Escherichia coli. Mol Microbiol. 2011;82:1545–1562. doi: 10.1111/j.1365-2958.2011.07907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutalik VK, Nonaka G, Ades SE, Rhodius VA, Gross CA. Promoter strength properties of the complete sigma E regulon of Escherichia coli and Salmonella enterica. J Bacteriol. 2009;191:7279–7287. doi: 10.1128/JB.01047-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols RJ, Sen S, Choo YJ, Beltrao P, Zietek M, Chaba R, Lee S, Kazmierczak KM, Lee KJ, Wong A, Shales M, Lovett S, Winkler ME, Krogan NJ, Typas A, Gross CA. Phenotypic landscape of a bacterial cell. Cell. 2011;144:143–156. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega AD, Gonzalo-Asensio J, García-del Portillo F. Dynamics of Salmonella small RNA expression in non-growing bacteria located inside eukaryotic cells. RNA Biol. 2012;9:469–488. doi: 10.4161/rna.19317. [DOI] [PubMed] [Google Scholar]
- Overgaard M, Johansen J, Møller-Jensen J, Valentin-Hansen P. Switching off small RNA regulation with trap-mRNA. Mol Microbiol. 2009;73:790–800. doi: 10.1111/j.1365-2958.2009.06807.x. [DOI] [PubMed] [Google Scholar]
- Overlöper A, Kraus A, Gurski R, Wright PR, Georg J, Hess WR, Narberhaus F. Two separate modules of the conserved regulatory RNA AbcR1 address multiple target mRNAs in and outside of the translation initiation region. RNA Biol. 2014;11:624–640. doi: 10.4161/rna.29145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Pfeiffer V, Mika F, Lucchini S, Hinton JC, Vogel J. σE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol Microbiol. 2006;62:1674–1688. doi: 10.1111/j.1365-2958.2006.05524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Pfeiffer V, Lucchini S, Sonawane A, Hinton JC, Vogel J. Systematic deletion of Salmonella small RNA genes identifies CyaR, a conserved CRP-dependent riboregulator of OmpX synthesis. Mol Microbiol. 2008;68:890–906. doi: 10.1111/j.1365-2958.2008.06189.x. [DOI] [PubMed] [Google Scholar]
- Papenfort K, Said N, Welsink T, Lucchini S, Hinton JC, Vogel J. Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol Microbiol. 2009;74:139–158. doi: 10.1111/j.1365-2958.2009.06857.x. [DOI] [PubMed] [Google Scholar]
- Papenfort K, Bouvier M, Mika F, Sharma CM, Vogel J. Evidence for an autonomous 5′ target recognition domain in an Hfq-associated small RNA. Proc Natl Acad Sci USA. 2010;107:20435–20440. doi: 10.1073/pnas.1009784107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Sun Y, Miyakoshi M, Vanderpool CK, Vogel J. Small RNA-mediated activation of sugar phosphatase mRNA regulates glucose homeostasis. Cell. 2013;153:426–437. doi: 10.1016/j.cell.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Vogel J. Small RNA functions in carbon metabolism and virulence of enteric pathogens. Front Cell Infect Microbiol. 2014;4:91. doi: 10.3389/fcimb.2014.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer V, Sittka A, Tomer R, Tedin K, Brinkmann V, Vogel J. A small non-coding RNA of the invasion gene island (SPI-1) represses outer membrane protein synthesis from the Salmonella core genome. Mol Microbiol. 2007;66:1174–1191. doi: 10.1111/j.1365-2958.2007.05991.x. [DOI] [PubMed] [Google Scholar]
- Plumbridge J, Bossi L, Oberto J, Wade JT, Figueroa-Bossi N. Interplay of transcriptional and small RNA-dependent control mechanisms regulates chitosugar uptake in Escherichia coli and Salmonella. Mol Microbiol. 2014;92:648–658. doi: 10.1111/mmi.12573. [DOI] [PubMed] [Google Scholar]
- Pulvermacher SC, Stauffer LT, Stauffer GV. The role of the small regulatory RNA GcvB in GcvB/mRNA posttranscriptional regulation of oppA and dppA in Escherichia coli. FEMS Microbiol Lett. 2008;281:42–50. doi: 10.1111/j.1574-6968.2008.01068.x. [DOI] [PubMed] [Google Scholar]
- Pulvermacher SC, Stauffer LT, Stauffer GV. Role of the sRNA GcvB in regulation of cycA in Escherichia coli. Microbiology. 2009;155:106–114. doi: 10.1099/mic.0.023598-0. [DOI] [PubMed] [Google Scholar]
- Reitzer L, Schneider BL. Metabolic context and possible physiological themes of σ54-dependent genes in Escherichia coli. Microbiol Mol Biol Rev. 2001;65:422–444. doi: 10.1128/MMBR.65.3.422-444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzer L. Biosynthesis of glutamate, aspartate, asparagine, L-alanine, and D-alanine. In: Curtiss R, Kaper JB, Karp PD, Neidhardt FC, Slauch JM, editors. EcoSal. Washington, DC: ASM Press; 2004. Plus. [DOI] [PubMed] [Google Scholar]
- Reitzer L. Catabolism of amino acids and related compounds. In: Curtiss R, Kaper JB, Karp PD, Neidhardt FC, Slauch JM, editors. EcoSal. Washington, DC: ASM Press; 2005. Plus. [DOI] [PubMed] [Google Scholar]
- Rubio-Somoza I, Weigel D, Franco-Zorilla JM, García JA, Paz-Ares J. ceRNAs: miRNA target mimic mimics. Cell. 2011;147:1431–1432. doi: 10.1016/j.cell.2011.12.003. [DOI] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvail H, Caron MP, Bélanger J, Massé E. Antagonistic functions between the RNA chaperone Hfq and an sRNA regulate sensitivity to the antibiotic colicin. EMBO J. 2013;32:2764–2778. doi: 10.1038/emboj.2013.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saramago M, Bárria C, Dos Santos RF, Silva IJ, Pobre V, Domingues S, Andrade JM, Viegas SC, Arraiano CM. The role of RNases in the regulation of small RNAs. Curr Opin Microbiol. 2014;18:105–115. doi: 10.1016/j.mib.2014.02.009. [DOI] [PubMed] [Google Scholar]
- Schellenberg GD, Furlong CE. Resolution of the multiplicity of the glutamate and aspartate transport systems of Escherichia coli. J Biol Chem. 1977;252:9055–9064. [PubMed] [Google Scholar]
- Seitz H. Redefining microRNA targets. Curr Biol. 2009;19:870–873. doi: 10.1016/j.cub.2009.03.059. [DOI] [PubMed] [Google Scholar]
- Sharma CM, Darfeuille F, Plantinga TH, Vogel J. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev. 2007;21:2804–2817. doi: 10.1101/gad.447207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CM, Papenfort K, Pernitzsch SR, Mollenkopf HJ, Hinton JC, Vogel J. Pervasive post-transcriptional control of genes involved in amino acid metabolism by the Hfq-dependent GcvB small RNA. Mol Microbiol. 2011;81:1144–1165. doi: 10.1111/j.1365-2958.2011.07751.x. [DOI] [PubMed] [Google Scholar]
- Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JC, Vogel J. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobrero P, Valverde C. The bacterial protein Hfq: much more than a mere RNA-binding factor. Crit Rev Microbiol. 2012;38:276–299. doi: 10.3109/1040841X.2012.664540. [DOI] [PubMed] [Google Scholar]
- Sonnleitner E, Sorger-Domenigg T, Madej MJ, Findeiss S, Hackermüller J, Hüttenhofer A, Stadler PF, Bläsi U, Moll I. Detection of small RNAs in Pseudomonas aeruginosa by RNomics and structure-based bioinformatic tools. Microbiology. 2008;154:3175–3187. doi: 10.1099/mic.0.2008/019703-0. [DOI] [PubMed] [Google Scholar]
- Sonnleitner E, Gonzalez N, Sorger-Domenigg T, Heeb S, Richter AS, Backofen R, Williams P, Hüttenhofer A, Haas D, Bläsi U. The small RNA PhrS stimulates synthesis of the Pseudomonas aeruginosa quinolone signal. Mol Microbiol. 2011;80:868–885. doi: 10.1111/j.1365-2958.2011.07620.x. [DOI] [PubMed] [Google Scholar]
- Stauffer LT, Stauffer GV. GcvA interacts with both the alpha and sigma subunits of RNA polymerase to activate the Escherichia coli gcvB gene and the gcvTHP operon. FEMS Microbiol Lett. 2005;242:333–338. doi: 10.1016/j.femsle.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Stauffer LT, Stauffer GV. Antagonistic Roles for GcvA and GcvB in hdeAB Expression in Escherichia coli. ISRN Microbiol. 2012;2012:697308. doi: 10.5402/2012/697308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling DA, Hulton CS, Waddell L, Park SF, Stewart GS, Booth IR, Higgins CF. Molecular characterization of the proU loci of Salmonella Typhimurium and Escherichia coli encoding osmoregulated glycine betaine transport systems. Mol Microbiol. 1989;3:1025–1038. doi: 10.1111/j.1365-2958.1989.tb00253.x. [DOI] [PubMed] [Google Scholar]
- Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell. 2011;43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Babitzke P, Kushner SR, Romeo T. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev. 2006;20:2605–2617. doi: 10.1101/gad.1461606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Quesada O, Millán V, Nisa-Martínez R, Bardou F, Crespi M, Toro N, Jiménez-Zurdo JI. Independent activity of the homologous small regulatory RNAs AbcR1 and AbcR2 in the legume symbiont Sinorhizobium meliloti. PLoS One. 2013;8:e68147. doi: 10.1371/journal.pone.0068147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tree JJ, Granneman S, McAteer SP, Tollervey D, Gally DL. Identification of bacteriophage-encoded anti-sRNAs in pathogenic Escherichia coli. Mol Cell. 2014;55:199–213. doi: 10.1016/j.molcel.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban JH, Vogel J. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 2007;35:1018–1037. doi: 10.1093/nar/gkl1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowski ML, Stauffer LT, Stauffer GV. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol Microbiol. 2000;37:856–868. doi: 10.1046/j.1365-2958.2000.02051.x. [DOI] [PubMed] [Google Scholar]
- Vanderpool CK. Combined experimental and computational strategies define an expansive regulon for GcvB small RNA. Mol Microbiol. 2011;81:1129–1132. doi: 10.1111/j.1365-2958.2011.07780.x. [DOI] [PubMed] [Google Scholar]
- Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jäger JG, Hüttenhofer A, Wagner EG. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 2003;31:6435–6443. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl A, Hubert P, Sturgis JN, Bouveret E. Tagging of Escherichia coli proteins with new cassettes allowing in vivo systematic fluorescent and luminescent detection, and purification from physiological expression levels. Proteomics. 2009;9:5389–5393. doi: 10.1002/pmic.200900240. [DOI] [PubMed] [Google Scholar]
- Willis RC, Furlong CE. Purification and properties of a periplasmic glutamate-aspartate binding protein from Escherichia coli K12 strain W3092. J Biol Chem. 1975;250:2574–2580. [PubMed] [Google Scholar]
- Wright PR, Richter AS, Papenfort K, Mann M, Vogel J, Hess WR, Backofen R, Georg J. Comparative genomics boosts target prediction for bacterial small RNAs. Proc Natl Acad Sci USA. 2013;110:E3487–E3496. doi: 10.1073/pnas.1303248110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Figueroa-Bossi N, Bossi L. Translation enhancing ACA motifs and their silencing by a bacterial small regulatory RNA. PLoS Genet. 2014;10:e1004026. doi: 10.1371/journal.pgen.1004026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Review Process File
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6