Abstract
The ability of HIV to establish latent infection in CD4+ lymphocytes represents a major barrier to the eradication of HIV. It is not clear what mechanisms favor latent over productive infection, but prior studies have suggested a role for the viral transcription factor Tat or its RNA target, TAR. Using samples from five individuals who were started on ART within 6 months of infection and achieved a viral load <50 (suppressed), we isolated one- and two-exon tat RNA from HIV propagated ex vivo from baseline plasma and from co-cultures of CD4+ T cells obtained at baseline and suppressed time points. Compared to virus from the baseline plasma (mostly from productively-infected CD4+ T cells), virus from the baseline and suppressed co-cultures (mostly from latently-infected cells) had more Tat variants with impaired transactivation activity. These findings suggest that impaired activity in the Tat-TAR axis may contribute to the establishment of latent infection in CD4+ T cells.
Keywords: HIV, latency, Tat, Transcription, Transactivation, TAR, CD4, Early infection
INTRODUCTION
HIV-1 can establish a latent infection in resting CD4+ T cells (Chun et al., 1997; Chun et al., 1995), and these latently infected cells are a major obstacle to the eradication of HIV (Finzi et al., 1999; Ramratnam et al., 2000; Siliciano et al., 2003; Strain et al., 2003; Wong et al., 1997; Zhang et al., 1999). These cells harbor integrated HIV provirus and do not produce virus constitutively, but can be induced to produce replication-competent virus upon activation. Latently-infected CD4+ T cells have been found in low numbers in all HIV+ patients (Chun et al., 1997). The prolonged half-life of the latent reservoir led to the suggestion that HIV cannot be eradicated by conventional antiretroviral therapy (ART) alone (Finzi et al., 1999; Ramratnam et al., 2000; Siliciano et al., 2003; Strain et al., 2003; Wong et al., 1997; Zhang et al., 1999). Latency also results in archiving of HIV variants with drug resistance mutations, compromising the effectiveness of ART (Martinez-Picado et al., 2000).
It is not clear what determines whether an infected cell will progress to productive or latent infection. Though multiple different mechanisms may contribute to latency (reviewed in (Marcello, 2006)), considerable research has focused on the viral transcription factor Tat (the Trans-activator of transcription) (Arya et al., 1985; Sodroski et al., 1985) and its RNA target, TAR (the Trans-Activation Response region) (Berkhout, Silverman, and Jeang, 1989). Tat, a viral protein encoded by two exons, has been shown to stimulate transcription in a trans fashion. During HIV replication, Tat binds to a stem-loop region of the nascent RNA (TAR) and the cellular cofactor PTEF-b (Positive Transcription Elongation Factor B), leading to phosphorylation of RNA polymerase II and reversing a block to transcriptional elongation (Isel and Karn, 1999; Kao et al., 1987). In the absence of Tat, the arrest in transcriptional elongation results in short, prematurely terminated transcripts (Kao et al., 1987).
Indirect evidence suggests that latency could be due to impaired activity in the Tat-TAR axis. This evidence includes: 1) the presence of latently-infected cell lines with mutations in Tat or TAR (Emiliani et al., 1998; Emiliani et al., 1996); 2) the presence of short, prematurely-terminated RNA transcripts in latently-infected patient cells (Adams et al., 1994; Kao et al., 1987; Lassen, Bailey, and Siliciano, 2004; Lin et al., 2003); and 3) the ability of exogenous Tat to rescue viral expression from latently-infected cells (Adams et al., 1994; Lin et al., 2003; Sonza et al., 2002). However, few if any studies have sought to systematically measure the range of activities in Tat/TAR cloned directly from patient samples, especially from latently-infected cells. In both the SIV macaque model of pathogenic lentiviral infection and in some human studies, it has been noted that regulatory and accessory genes, including tat and nef, are among the earliest to diversify under host immune selection (Addo et al., 2001; Allen et al., 2000; Cao et al., 2003). We hypothesized that natural variation in tat sequence would accumulate soon after primary HIV infection, that sequence variation would lead to a corresponding variation in Tat-mediated transcriptional activity (transactivation), and that latent HIV would be enriched for Tat/TAR variants with impaired transactivation activity.
Despite circumstantial and in vitro data pointing to the role of attenuated Tat activity in latency, testing this hypothesis in patient samples is complicated by the large proportion of replication-incompetent genomes that would be represented by direct PCR amplification from clinical samples (Meyerhans et al., 1989) and by the potential for selection against impaired Tat activity during cell culture procedures that are necessary to demonstrate replication-competence. To minimize sampling the many replication-incompetent forms that collect in peripheral blood mononuclear cells (PBMC) over longer periods of infection (Sanchez et al., 1997), we chose to study individuals initiating treatment soon after infection and to isolate tat RNA from replication-competent virus that was successfully propagating in culture.
Samples were obtained from five individuals with acute or early infection who were started on combined ART and subsequently had decrease in VL to <50 copies/ml (herein referred to as suppressed). Plasma was obtained before therapy (baseline). CD4+ T cells were isolated at both baseline and suppressed time points. Since >95% of the plasma virus in an untreated patient comes from productively-infected cells (Ho et al., 1995; Perelson et al., 1997; Perelson, Essunger, and Ho, 1997; Perelson et al., 1996; Wei et al., 1995; Wodarz and Nowak, 2002), virus from the baseline plasma should be predominantly non-latent virus. Replication-competent, latent virus was obtained by activating CD4+ T cells from suppressed time points and co-culturing them with activated CD8-depleted peripheral blood lymphocytes (PBL) from healthy donors (“suppressed co-cultures”). Given that the plasma virus at these time points is undetectable, most virus recovered from these co-cultures should represent latent virus that has been “rescued” by activation and provision of susceptible donor target cells. Virus from nonsuppressed (baseline) co-cultures should also come mostly from latently-infected cells, as suggested by previous in situ hybridization studies on activated and non-activated PBMC from untreated patients (Derdeyn et al., 1999).
We generated one and two exon tat cDNA from RNA extracted from baseline plasma, from baseline plasma spinoculated onto donor CD8-depleted PBL, and from co-cultures obtained from CD4+ T cells at baseline and suppressed time points. We also isolated the transactivation response region (TAR) from co-cultures obtained at baseline and after suppression. Tat-mediated transcriptional activity (transactivation) was measured by transfection of equal amounts of DNA into reporter TZM-bl or LuSIV cells expressing luciferase under control of the HIV or SIV LTR.
Tat/TAR sequences and Tat function from the baseline plasma (representing predominantly non-latent virus) were compared to those from the baseline and suppressed co-cultures (predominantly latent virus). We found that genotypic variation in tat and TAR sequence accumulated early in infection. The Tat variants exhibited a range of transactivation activities, both within and between individuals, including a number with reduced transactivation activities. There were more Tat alleles with impaired activity in the co-cultures than in the baseline plasma, and the Tat alleles from the co-cultures were on average more impaired than those from the baseline plasma, suggesting that variability in Tat function can contribute to the establishment of latent infection with HIV-1.
RESULTS
Clinical Data
The baseline (pretherapy) samples from the five study participants (Strain et al., 2005) showed a mean CD4 cell count of 469 cells/µl (range: 304 to 542) and a mean VL of 271,570 copies/ml (range: 15,976 to 907,400) (Table 1). After initiation of ART, all subjects had a decrease in VL to <50 copies/ml. When blood was drawn for the suppressed time points (after 1 to 3.5 months of suppression), the mean CD4 count had increased to 548 cells/µl (range: 349 to 632).
Table 1.
Clinical characteristics of the study subjects.
| individual | peak VL | baseline CD4/% | baseline VL | suppressed CD4/% | suppressed VL | regimen |
|---|---|---|---|---|---|---|
| A046 | 268,145 | 304/NA | 268,145 | 349/NA | <50 | AZT, 3TC, ABC |
| A047 | 907,400 | 497/40.9 | 907,400 | 738/47.6 | <50 | AZT, 3TC, ABC |
| A048 | 156,852 | 474/30 | 15,976 | 538/30 | <50 | D4T, 3TC, ABC, APV/RTV |
| A049 | 2,464,790 | 528/22 | 47,438 | 620/43 | <50 | D4T, 3TC, ABC, APV/RTV |
| A050 | 195,568 | 542/16 | 118,891 | 497/29 | <50 | AZT, 3TC, IDV/RTV |
Spinoculation and culture of baseline plasma virus
Spinoculation of the baseline plasma was successful for 3 of 5 individuals (A046, A047, and A049). For A047, A049, and positive control, rising p24 was detected at day 3–6. Virus from A046 grew more slowly, with rise in p24 detected at day 11–14 and a lower peak p24 (data not shown).
Tat and TAR Clones
229 tat clones were isolated (range: 21 to 62 per individual), including 143 two-exon tat clones and 86 one-exon tat clones. Of these 229 clones, there were 58 from baseline plasma (27 from direct extraction and 31 from spinoculation), 73 from baseline co-cultures, and 98 from suppressed co-cultures. We also isolated a total of 116 TAR clones from baseline or suppressed co-cultures.
Tat sequences showed patient specific phylogenetic clustering and variable sequence diversity
Maximum likelihood phylogenetic trees were constructed using the first exon tat sequences (Supplementary Figure 1) and TAR sequences (Supplementary Figure 2). Separate phylogenies were created for two-exon tat sequences (Figure 1) and for one exon tat sequences from each individual. Figure 2 shows a tree for one representative individual, A047. Both tat and TAR sequences from all 5 individuals formed patient-specific clusters that were distinct from each other and from the lab strains HXB2 and NL4-3. For a given individual, there was intermingling of taxa representing clones from baseline plasmas, baseline co-cultures, and suppressed co-cultures.
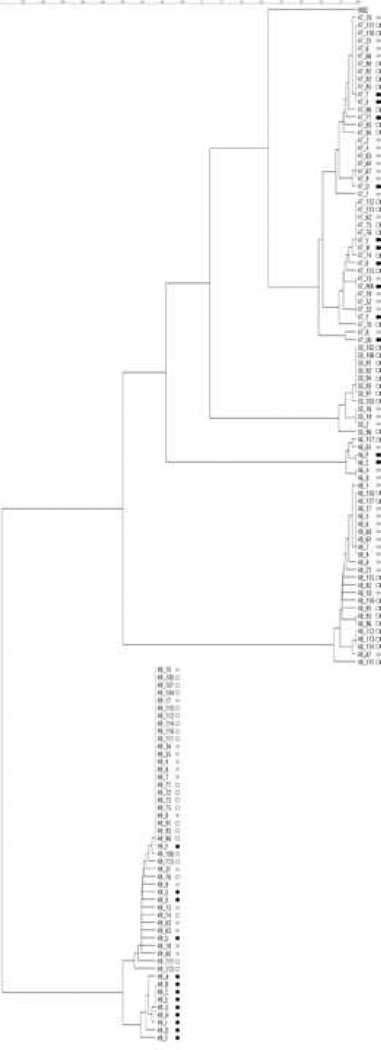
Figure 1.
Phylogenetic tree for two-exon tat sequences. Prematurely-truncated clones were excluded from analysis. The first two digits of each clone name identify the subject from which that clone was isolated. Scale bar shows genetic distance. The dot after each clone name indicates the type of sample from which that Tat was cloned. Black dots indicate clones from spinoculated baseline plasma (note that spinoculation was not successful for subjects A048 and A050). Grey dots indicate clones from baseline co-cultures, while white dots indicate clones from suppressed co-cultures.
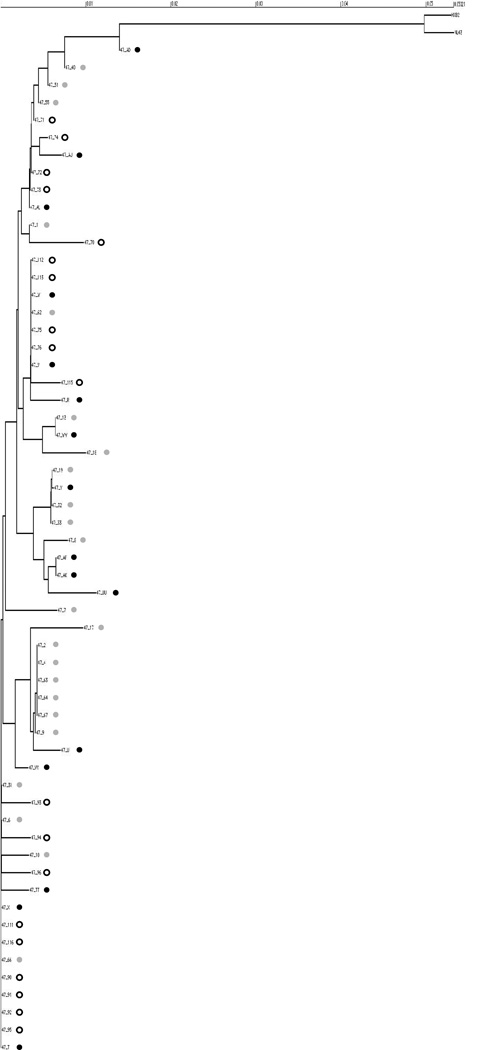
Figure 2.
Phylogenetic tree for one-exon tat from subject A047. Prematurely truncated clones and second exon sequences were excluded from analysis. Scale bar shows genetic distance. Black dots indicate clones from baseline plasma. Grey dots indicate clones from baseline co-cultures, while white dots indicate clones from suppressed co-cultures.
For the five individuals included in this study, mean genetic diversity of Tat during primary infection ranged from 0.2% to 1.0%. 38.2% of Tat clones from the five study subjects had one or more amino acid change(s) relative to the corresponding consensus sequence. The percentage of mutants was 34.5% in the baseline plasma, 45.8% in the baseline co-cultures, and 34.7% in the suppressed co-cultures.
Cloning of NL4-3 tat
Of 22 different clones of NL4-3, there were a total of five amino acid substitutions (4 missense and 1 stop codon) in four clones (18.2% mutants). Two of the four mutants, corresponding to two of twenty evaluable NL4-3 clones (10%), had impaired transactivation activity.
Assays for transactivation activity with both TZM-bl cells and LuSIV cells demonstrated a good dose response relationship
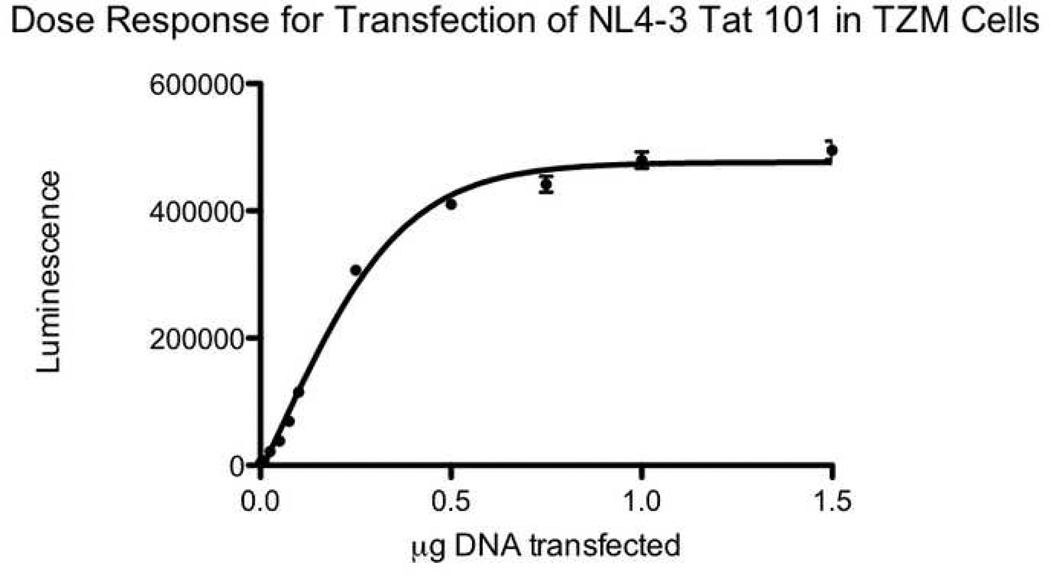
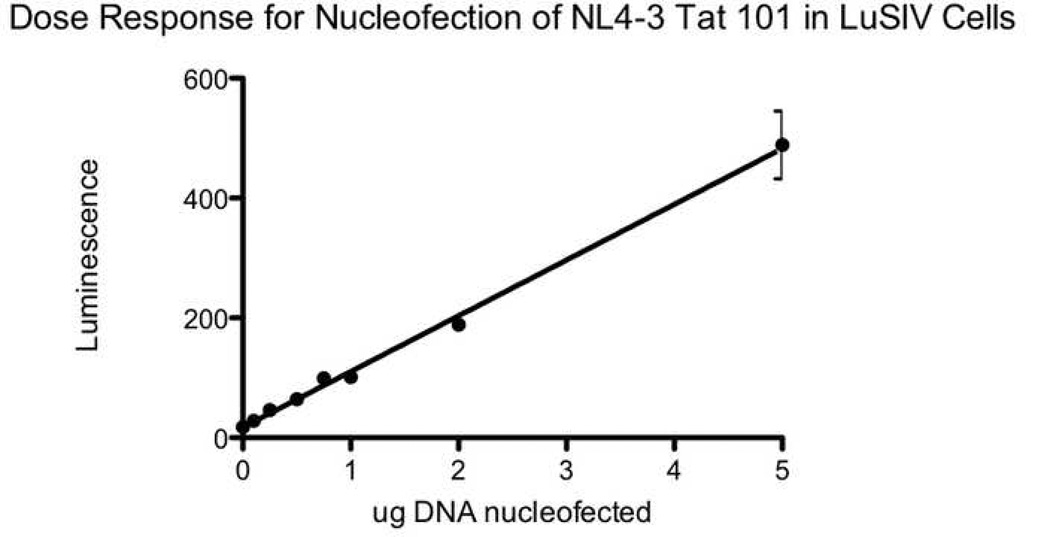
For all expressible tat variants (those cloned into the expression vector in the correct orientation), transactivation activity was measured by transfection of equal amounts of DNA into reporter TZM-bl cells. GFP was used to control for transfection efficiency. The number of GFP positive cells was similar between wells (data not shown). Figure 3A shows a dose response curve for transfection of two-exon NL4-3 Tat into TZM-bl cells. The curve is sigmoidal, with a linear portion at around 0.1µg and saturation at higher doses. The detection limit is about 0.01 to 0.02µg, with a log linear range from 0.075 to 0.5µg. Dose response studies using a luciferin substrate showed similarly shaped curves, albeit with much lower luminescence (data not shown). For a select number of alleles, transactivation activity was also measured by nucleofection of equal amounts of DNA into the T cell line LuSIV. Figure 3B shows a dose response curve for nucleofection of two-exon NL4-3 Tat into LuSIV cells. The lower limit of detection is 0.1–0.25µg, and the curve is linear over a wide range of DNA concentrations from 0.1 to 5µg.
Figure 3.
Dose response curves for transfection of two-exon NL4-3 Tat. 3A) Dose response curve for transfection of NL4-3 Tat in TZM-bl cells. Reporter TZM-bl cells containing an integrated LTR linked to beta-galactosidase and luciferase were transfected with empty vector or 0.001 to 1.5 µg of Tat from NL4-3. After 3 days, cells were lysed with Beta-Glo (luciferin-galactoside) and the luminescence was read at 30 minutes. The x axis shows the dose transfected. The y axis shows absolute luminescence. Error bars show the standard error of measurement (SEM) of two replicate wells. The data are representative of 2–3 separate experiments.
3B) Dose response curve for transfection of LuSIV cells. LuSIV cells containing an integrated LTR-luciferase were nucleofected with empty vector or 0.1 to 5µg of NL4-3 Tat 101. After 3 days, cells were pelleted and lysed with Bright-Glo (luciferin) and the luminescence was read at 5 minutes. The x axis shows the dose transfected. The y axis shows luminescence. Error bars show the SEM.
Tat variants with impaired activity were present in all sample types but were significantly enriched in those corresponding to latent virus (co-culture samples)
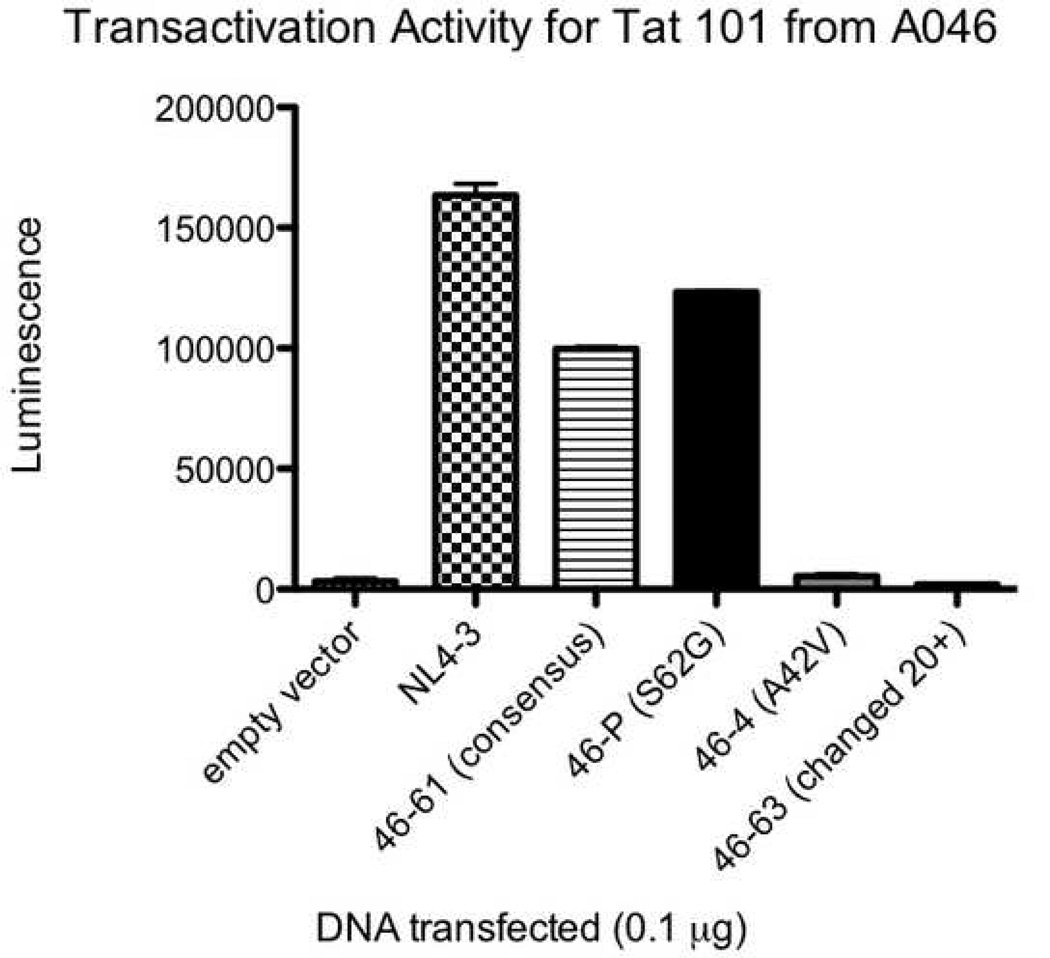
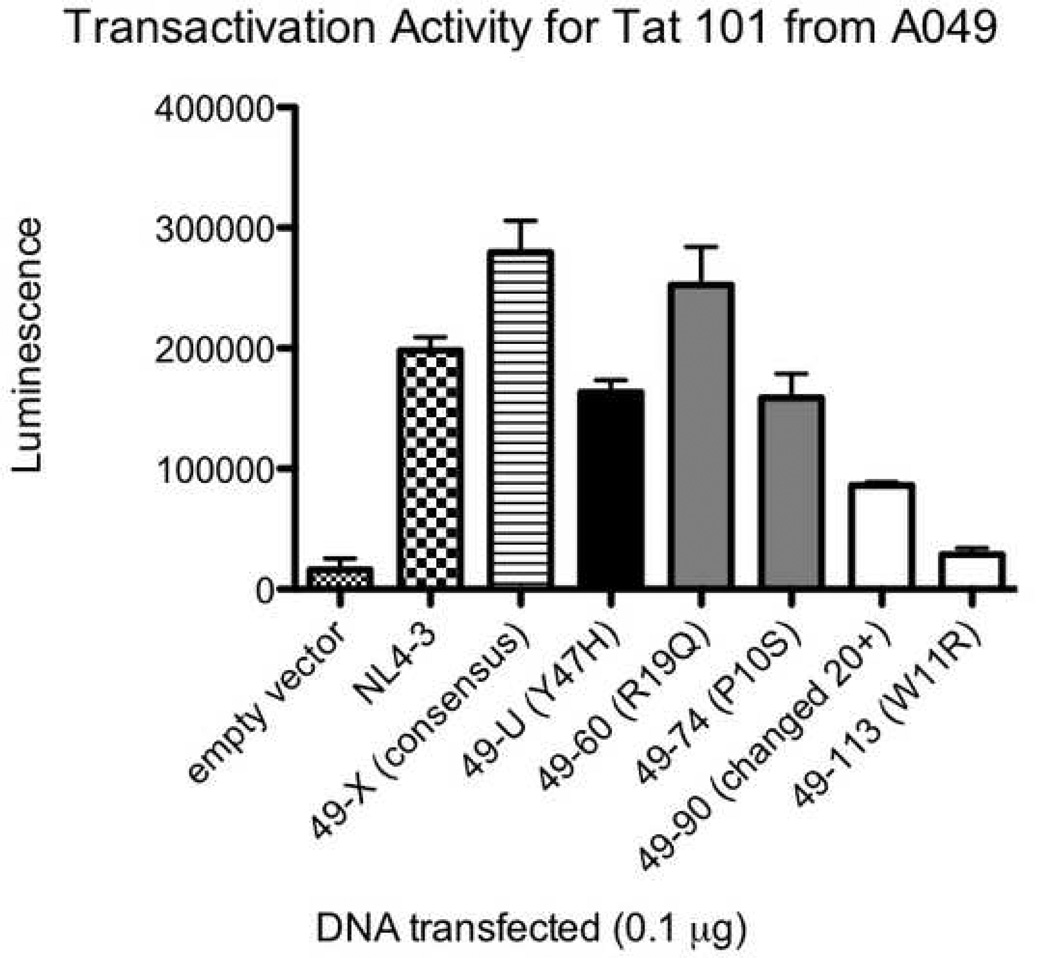
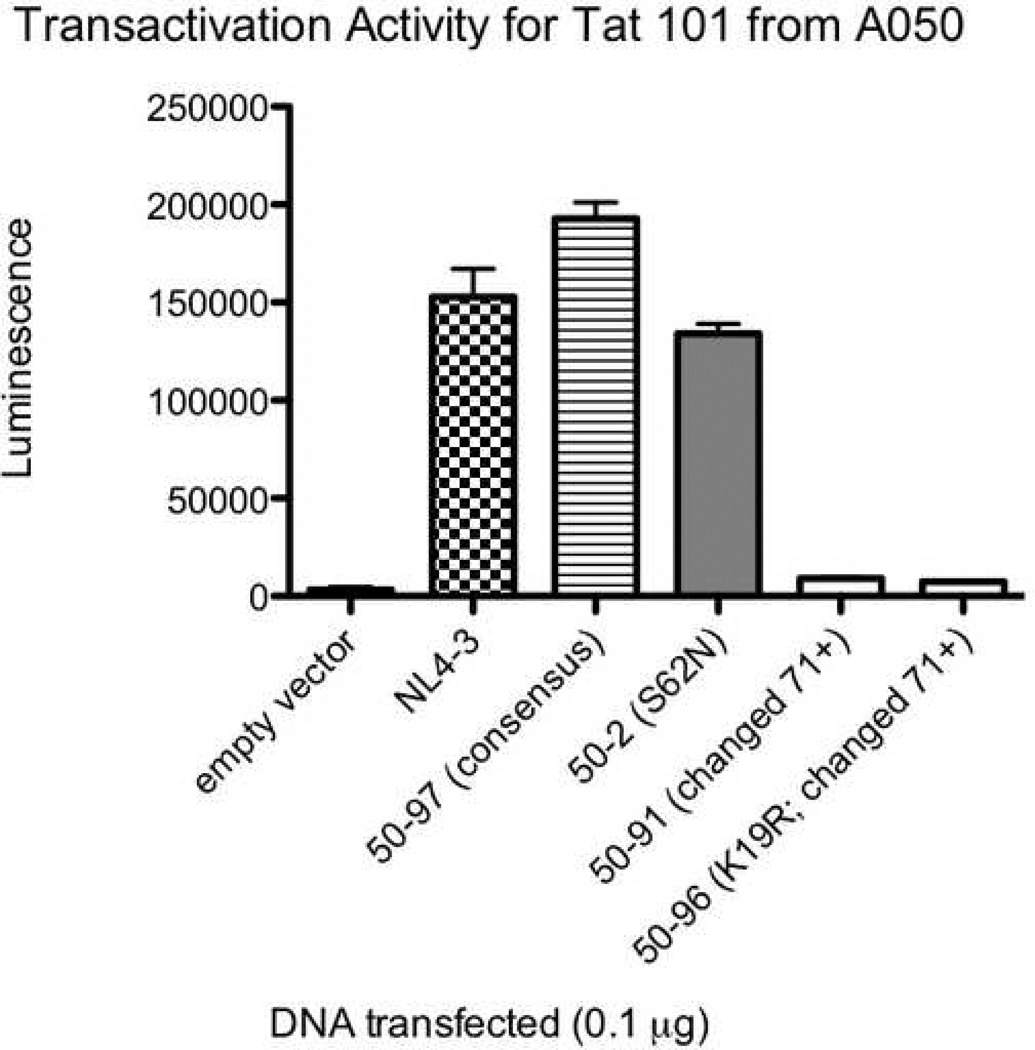
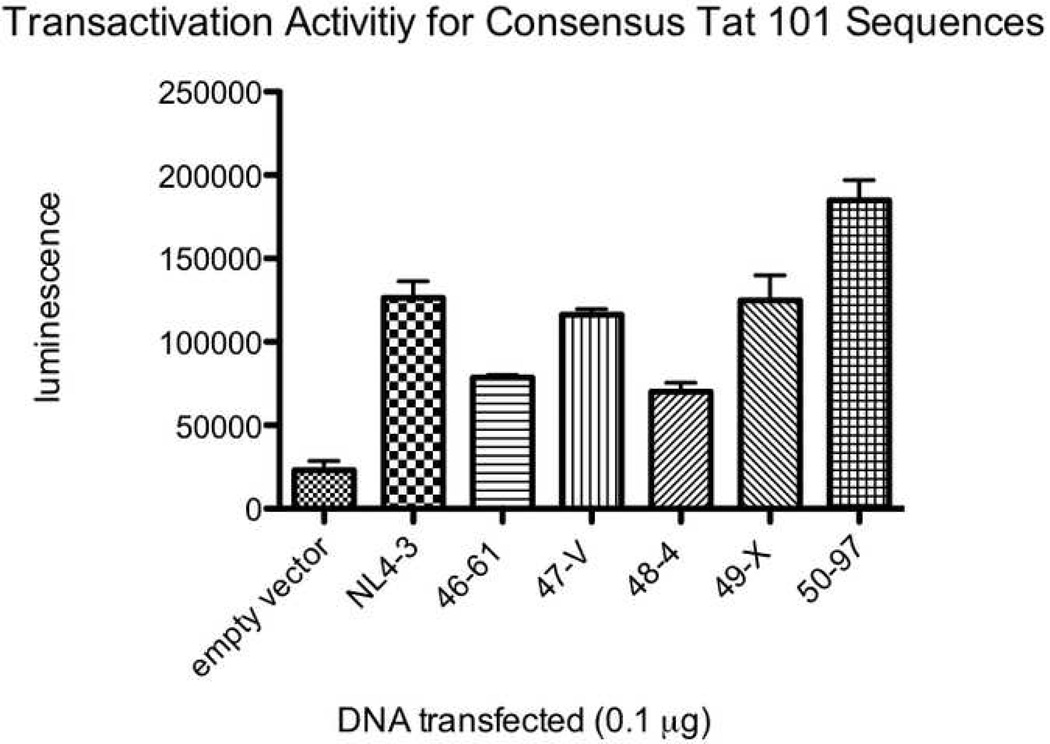
All five study participants had Tat variants with impaired functional activity. The average percentage of clones with reduced transactivation activity was 13.9% in the baseline plasma, 43.3% in the baseline co-cultures (p=0.023 for comparison with baseline plasma), and 21.5% in the suppressed co-cultures (p=NS). Many of these were missense mutations, though there were a number of variants that appeared to have arisen from abnormal splicing and/or deletion. Figure 4 A, B, and C show the transactivation activities for NL4-3 Tat and selected two-exon Tat variants (all obtained using the same primers) from each of three different individuals. The Tat variants exhibited a range of activities relative to the consensus sequence (the most common sequence for that individual) and relative to NL4-3. A separate comparison of consensus one and two-exon Tats from different individuals revealed considerable variability in Tat activities between individuals (Figure 5).
Figure 4.
Transactivation activities for NL4-3 Tat and selected two-exon Tat variants from individuals A046 (4A), A049 (4B), and A050 (4C). TZM-bl cells were transfected with 0.1µg of either empty vector, NL4-3 Tat, a clone containing the consensus Tat sequence (the most common sequence for that individual), or one of several Tat variants. All Tat clones were obtained using the same primers. The x axis shows the clone transfected. The y axis shows luminescence (minus the luminescence for cells alone). Error bars show the SEM of two replicate wells for that experiment. Black bars represent clones from the baseline plasma, grey bars represent clones from the baseline co-cultures, and white bars represent clones from the suppressed co-cultures. The data are representative of 2–4 different experiments.
Figure 5.
Consensus two-exon Tat activities from NL4-3 and the five subjects. TZM-bl cells were transfected with 0.1µg of either empty vector, NL4-3 Tat, or a clone containing the consensus Tat sequence (the most common sequence for that individual) for each of the five individuals (A046, A047, A048, A049, and A050). All Tat clones were obtained using the same primers. The x axis shows the clone transfected. The first two digits of the clone name indicate the patient from which it was derived. The y axis shows luminescence (minus the luminescence for cells alone). Error bars show the SEM of two replicate wells for that experiment.
The vast majority of mutations resulted in impaired Tat activity, though a few mutations (found exclusively in the baseline plasma) resulted in activities greater than that of the consensus sequence. Table 2 shows a list of some of the variants with one or two missense mutations and their fractional activities relative to that of the corresponding consensus sequence when tested in TZM-bl cells. When a subset of these alleles was tested in LuSIV cells, the order of Tat activities was the same, although the actual magnitudes differed. In general, Tat variants that had impaired activity in TZM-bl cells showed even greater impairment in LuSIV cells. For example, the mutations P10S, W11R, and Y47H from A049 had relative tat activities of 51.4%, 12.0%, and 62.1% (respectively) in TZM cells, compared to 2.2%, 1.7%, and 5.9% (respectively) in LuSIV cells.
Table 2.
Transactivation activities relative to consensus. For clone names (far left column), the first two digits of each clone name identify the subject from which that clone was isolated. The mutations (second column from left) are listed in order of amino acid position within the protein. Letters in parentheses indicate the amino acid in NL4-3 (in cases where that differed from the subject’s consensus sequence). The relative transactivation activity of each mutant Tat (third column) was calculated by taking the average luminescence (from two replicates), subtracting out the average background luminescence (from empty vector), and dividing by the average background-subtracted luminescence of the Tat allele with the consensus sequence (most common sequence for that individual) obtained using the same forward and reverse primers. Results were then averaged over 2–4 separate experiments. The far right column shows the standard deviation of the averages from the 2–4 separate experiments.
| clone | mutation | % consensus | std dev |
|---|---|---|---|
| 49-102 | V4A; K71* | 33.8 | 23.7 |
| 46-AH | R7G | 93.9 | 6.5 |
| 50-84 | E9G; K19R; R52Q | 7.6 | 3.1 |
| 49-74 | P10S | 51.4 | 8.3 |
| 49-113 | W11R | 12.0 | 3.4 |
| 48-115 | (K)Q12R | 81.4 | 6.2 |
| 50-BH | G15R; K19R | 2.5 | 1.9 |
| 49-60 | (K)R19Q | 89.9 | 27.3 |
| 50-80 | K19R | 64.7 | 9.0 |
| 47-70 | K29E | 50.1 | 12.4 |
| 49-9 | K29R | 69.2 | 14.7 |
| 48-36 | (F)L32P | 2.7 | 1.2 |
| 48-110 | C34R | 4.9 | 5.4 |
| 49-K | F38L | 9.8 | 11.1 |
| 47-2 | 39I | 92.1 | 4.4 |
| 47-U | 39I; S75P | 125.3 | 12.8 |
| 46-4 | A42V | 2.1 | 0.1 |
| 47-13 | I45T | 52.0 | 1.5 |
| 49-U | Y47H | 62.1 | 5.0 |
| 49-13 | A58V | 65.3 | 16.4 |
| 46-P | S62G | 123.6 | 1.1 |
| 50-16 | S62N | 68.3 | 10.2 |
| 47-96 | (T)N64D | 80.1 | 19.0 |
| 47-R | (S)P70S | 94.4 | 5.1 |
| 47-TT | Q72R | 92.3 | 5.6 |
| 48-1 | P73H | 96.3 | 29.5 |
| 47-8 | E86K | 65.0 | 6.1 |
| 49-106 | E92G | 80.2 | 11.1 |
| 47-7 | (T)A97T | 88.7 | 5.9 |
Most clones from the baseline plasma had normal (consensus) or greater activity, and relatively few (8 clones in 4 subjects) had impaired activity. No clones from the co-cultures (baseline or suppressed) had activity greater than that of the consensus sequence, and while many had consensus sequence and activity, a number of clones from both co-cultures showed impaired Tat activity. For one individual (A046), all of the Tat clones from the suppressed co-culture had consensus sequence, although the majority (6 of 7) of evaluable clones from the baseline co-culture from this same individual had substantially impaired activity.
For all five individuals, the mean Tat activities from the suppressed co-cultures were less than the mean Tat activities from the baseline plasmas (p=0.019 by the t test for correlated samples; p=0.05 by the paired Wilcoxon rank sum test, which is as high as can be achieved with this test using five paired samples). The mean Tat activities from the baseline co-cultures were also less than the mean Tat activities from the baseline plasmas, though the results were not statistically significant.
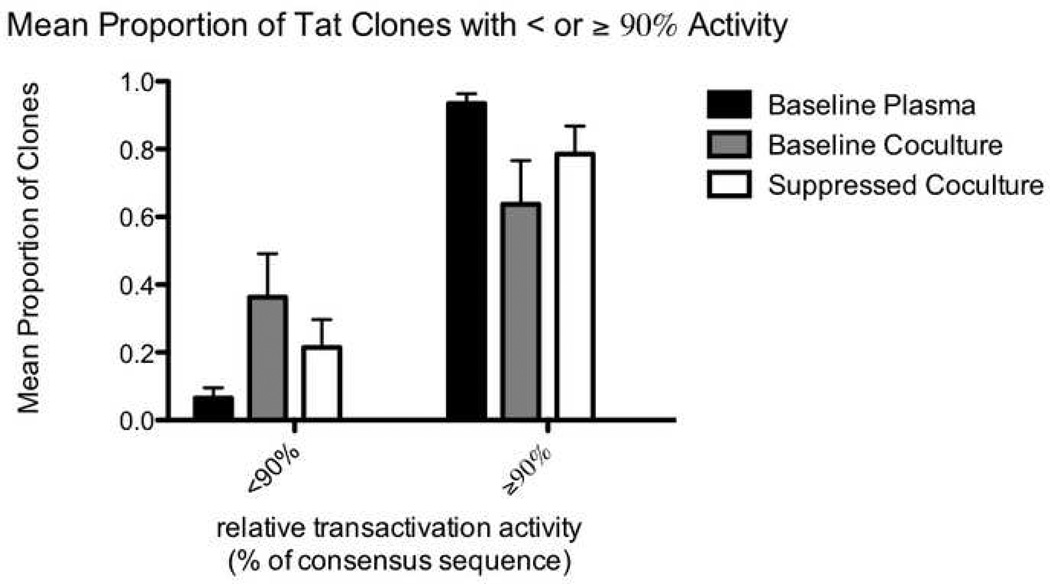
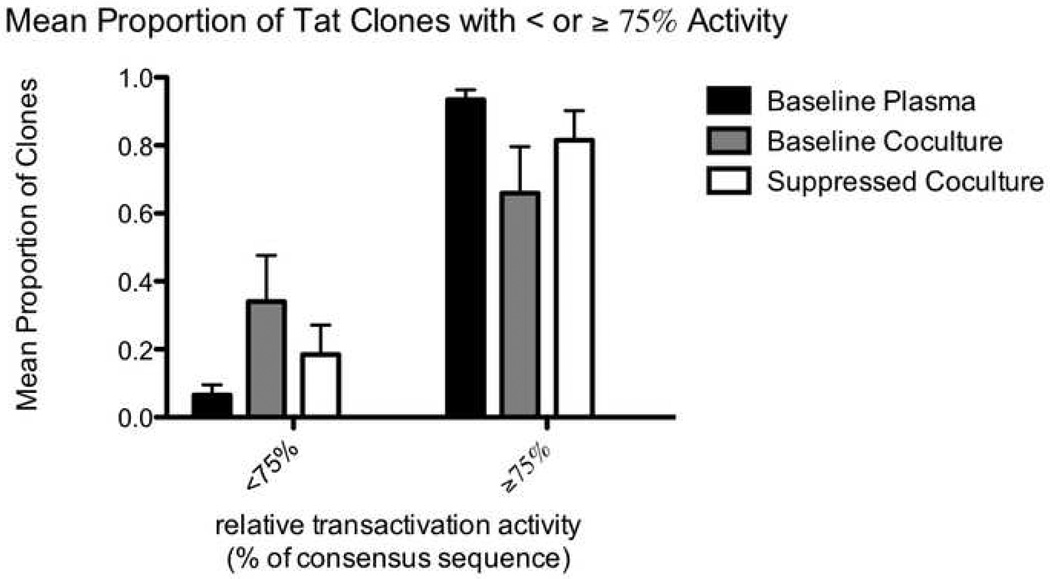
For each of the three types of samples (baseline plasma, baseline co-culture, and suppressed co-culture), we also calculated the mean (across five subjects) proportion of Tat sequences with less than or greater than 90%, 75%, 50%, and 25% of consensus Tat activity (figure 6). Compared to the baseline plasma (black bars), the suppressed co-cultures (white bars) had a larger proportion of clones with <90% activity (p=0.039 using the t test for correlated samples; figure 6A) and a trend towards more clones with <75% activity (p=0.077; figure 6B). The baseline co-cultures (grey bars) appeared to have an even larger proportion of clones with impaired activity, though comparison to the baseline plasma yielded p values of 0.052 (for <90% activity) and 0.070 (for <75% activity). Compared to the baseline plasma, both co-cultures also had a trend towards more clones with <50% and <25% activity.
Figure 6.
Mean proportion of clones with relative Tat activity greater than or less than 90% (6A) and 75% (6B). For each of the three types of samples (baseline plasma, baseline co-culture, and suppressed co-culture) from each individual, we calculated the proportion of tat sequences in that sample with less than or greater than a given activity level (90% or 75%) relative to the appropriate consensus sequence. The means of the proportions for the five individuals are displayed on the y axis. Black bars represent clones from the baseline plasma, grey bars represent clones from the baseline co-cultures, and white bars represent clones from the suppressed co-cultures.
For each of the three types of samples, we also examined the relationship between genetic variation in tat (as measured by mean pairwise genetic distance) and frequency of impaired alleles. There was no correlation between the two variables (R2 = 0.0147; p=0.49, Spearman’s Rank Test), suggesting that the increased prevalence of impaired Tats in the co-cultures (especially baseline co-cultures) is not merely a reflection of increased genetic diversity in those samples, but instead represents the specific enrichment of attenuated alleles in the latent reservoir.
DISCUSSION
In this study, we surveyed the range of Tat/TAR sequences in early infection to investigate whether impaired activity in the Tat-TAR axis contributes to the establishment of latent infection in vivo. In five individuals with acute or early infection, we found evidence of considerable variation in Tat sequence and in Tat-mediated transcriptional activity. These findings concur with previously published observations that Tat is among the earliest of genes to diversify under immune selection (Addo et al., 2001; Allen et al., 2000; Cao et al., 2003; Guillon et al., 2006). In functional assays using reporter cell lines with wild-type TAR, we found that all subjects had a number of Tat variants with impaired activity (ranging from 0 to 99%), while two subjects had Tat variants with increased activity (the latter was seen in the baseline plasma but not in the co-cultures). Compared to virus from the baseline plasma (mostly from productively-infected cells), virus from the suppressed co-cultures (representing mostly latent virus) had more Tat alleles with impaired (<90%) activity, and the average Tat activity from the suppressed co-cultures was lower than that from the baseline plasma. These findings suggest that naturally occurring variation in Tat transactivation activity affects the complex host/viral interactions that control the balance between productive and latent infection, and that attenuated Tat alleles may favor latent infection.
Virus from baseline co-cultures also had more impaired Tat alleles than virus from baseline plasma. These baseline co-cultures could represent a mixture of productively-infected and latently-infected cells. However, in situ hybridization studies (Derdeyn et al., 1999) of PBMC from untreated patients show that the proportion of cells that can be induced by activation to transcribe HIV RNA greatly exceeds the proportion that are HIV RNA+ in the absence of activation, suggesting that even in the untreated patient, the majority of the virus in PBMC is actually in the form of latent, inducible virus. This conclusion is also in keeping with the observation that activated lymphocytes which support productive HIV infection express homing and adhesion molecules that cause their recruitment and retention in lymphoid tissues and effector sites. Taken together, these observations suggest that while the virus from both baseline and suppressed co-cultures could represent a mixture of latent and non-latent virus, latent virus is likely to predominate in both cases.
The finding of impaired Tat alleles in the baseline co-cultures may seem discordant from that of Guillon et al, who sequenced Tat from PBMC-associated RNA from serial samples of four individuals with untreated early HIV-1 infection (Guillon et al., 2006). They found evidence of Tat and Rev mutations that appeared to arise from mutational escape of MHC-1 epitopes, but most of these mutations did not result in significant changes in Tat activity. The apparent discrepancy is likely explained by differences in methodology. For example, their PBMC had not been activated, so most of the cell-associated tat RNA would be expected to come from the small number of cells that are constitutively expressing full length HIV-1 RNA transcripts, which are likely to be productively-infected rather than latently-infected cells.
One limitation of our study is the small number of subjects studied (dictated solely by the availability of stored samples). These findings will have to be confirmed in larger studies with more participants. There are other caveats that are intrinsic to studies of this kind. All studies of viral genetics are complicated by the dilemma of whether sequences amplified directly from patient material (plasma or PBL) are preferable to those obtained from virus isolated in co-culture. While direct amplification obviates some ex vivo selection artifacts, it cannot distinguish viral variants of greater or lesser importance based on phenotypic traits. In the current study, we felt it was important to utilize an experimental design that could distinguish replication-defective from replication-competent virus by the criteria of spreading infection based on increasing p24 levels.
Given the time required to culture virus until the p24 is detectable (to demonstrate replication competence), contamination with even small numbers of productively-infected cells might allow non-latent virus to outcompete and outgrow latent virus with impaired Tat activity. The use of a three NRTI regimen (in two patients) and the short duration of suppression may have increased the likelihood of such “contamination” with productively infected cells (though the half life of these cells on ART is extremely short). We chose not to perform additional ex vivo manipulation to exclude activated, productively-infected cells since, as discussed earlier, these cells are likely in the minority and we cannot exclude the possibility that a population of latently infected cells become activated during isolation and ex vivo processing. It should be noted that when patient CD4+ T cells were cultured without activation, they never resulted in spreading infection and positive p24, which seems to suggest that there were not too many productively-infected cells. Even if the suppressed co-cultures were “contaminated” with productively-infected cells, these cells would be expected to have wild type Tat, and the presence of these cells would cause our methods to underestimate the true contribution of impaired Tat to latency.
It is also possible that some of the cells going into the co-cultures (especially the baseline co-culture) had unintegrated but replication-competent genomes (pre-integration latency). However, multiple studies have shown that, at least in resting CD4+ T cells, unintegrated HIV DNA has a very short half life (on the order of days). Thus, in CD4+ T cells, most unintegrated HIV (including “pre-integration latency”) is likely the product of very recent infection. In the untreated patient (baseline co-cultures), where >95% of the plasma virus comes from productively-infected cells, one would expect that most of the new infection of peripheral CD4+ T cells (and hence, the unintegrated HIV DNA) is also from productively-infected cells, which would be expected to have wild-type Tat.
Even if all of the virus came from truly latently-infected cells, if a small number of latently-infected cells had normal Tat sequence and function, these would be expected to rapidly outcompete those with impaired Tat activity. Also, the 1–3 week duration of culture (necessary to detect +p24) allows time for reversion or back-mutation of impaired Tat alleles back to a more wild-type phenotype. As a result of all of these factors, our experimental design may underestimate both the fraction of viruses with impaired Tat activity and the magnitude of impairment.
We tried to choose a duration of co-culture that was long enough to get spreading infection and detectable, increasing p24 (thus ensuring that most of the virus is replication competent), but not so long as to facilitate back-mutation or overgrowth by virus with normal Tat activity. For this reason, we chose the earliest time point with positive p24, which was 7 days for most of the baseline co-cultures. However, the co-cultures from suppressed time points grew more slowly (which may be due to lower inoculums or impairment in a viral factor such as Tat), so that p24 was usually not detectable until 14–21 days. If the data seem to suggest that in some cases the baseline co-cultures have more impaired Tats than the suppressed co-cultures (though the numbers are not statistically significant), it could be that the longer duration of the suppressed co-culture allowed more time for back-mutation or overgrowth from wild-type Tat.
In both baseline and suppressed co-cultures, we were able to detect mutant Tats with impaired activity. Although mutations can arise in culture or in the cloning process (from PCR to sequencing), the use of proofreading enzymes and non-nested PCR protocols should reduce the latter risk. The increased frequency of mutations and impaired Tats in the patient samples compared to the NL4-3-infected PBL suggests that many of these mutations did in fact arise in vivo. To the extent possible (for example, by using the baseline plasma to infect donor PBL), we tried to ensure similar culture conditions in the spinoculated baseline plasma, baseline co-culture, and suppressed co-culture. Mutations introduced by cloning should be equally likely for all types of samples (baseline plasma, baseline co-culture, or suppressed co-culture). Also, for many mutations, the same mutation was seen from different PCR reactions or different co-culture wells, suggesting that they were not introduced by PCR or cloning. Some wells had several Tat variants, including some that were totally defective and should not be replication competent. It is likely that these wells had a mix of viruses (at least one of which was replication competent), and/or that there was some trans-complementation.
Where it is possible to compare our Tat phenotypes with results of previously published genotype/phenotype studies, the results generally agree. For example, we found that the C34R mutant has virtually no Tat activity, in agreement with prior mutagenesis studies that suggest that mutation of any of six key Cys residues (including C34) virtually abolishes Tat activity (Garcia et al., 1988; Kuppuswamy et al., 1989; Sadaie, Benter, and Wong-Staal, 1988). We found that the mutation Y47H reduced activity to 62% that of the wild type 400bp Tat from A049, while Verhoef et al found that two different forms of Y47H mutation reduced activity to 40–50% that of the wild type 3.1kb Tat construct from LAI (Verhoef, Koper, and Berkhout, 1997). In addition, we found that early truncations and missense mutations in the N-terminal part of the first exon (4 to 42) generally resulted in dramatically reduced transactivation activity, while mutations in the distal first exon and second exon had considerably less effect. These results match those of prior mutagenesis studies that implicate the first 48 N-terminal amino acid residues as the key determinants of transactivation activity (Garcia et al., 1988; Kuppuswamy et al., 1989; Seigel et al., 1986).
Aside from initiation and elongation of transcription, other functions have been ascribed to Tat, including both pre-integration and post-transcriptional processes. These other Tat functions were not assessed in this study. It is possible that Tat variants with lesser degrees of impairment in transactivation activity have other effects (for example, on splicing) that may contribute to latency.
While our findings implicate attenuated Tat transactivation as one factor in the establishment or maintenance of latency, wild-type (consensus) Tat was frequently recovered from co-culture samples. Even though our experimental design should underestimate the true proportion with impaired Tat activity, it seems likely that latent infection can occur with wild-type or near wild-type Tat function. Indeed, both in vitro and in vivo model systems for HIV latency have been described utilizing wild-type HIV (Brooks et al., 2001; Jordan, Bisgrove, and Verdin, 2003), suggesting that additional factors contribute to latency. These could include many human cellular factors, perhaps associated with cellular activation or differentiation state, including those that modulate Tat activities and those that are Tat-independent. For example, in resting CD4+ T cells, PTEF-b function is very low and is probably limiting for Tat (Ghose et al., 2001). Variation in viral factors other than Tat, such as Rev or Nef, could also contribute to latency (Pomerantz, Seshamma, and Trono, 1992). Aside from blocks at the level of initiation or elongation of transcription, latency could be due to a number of post-transcriptional processes, including altered splicing of mRNA (Pomerantz, Seshamma, and Trono, 1992), a block in export of mRNA from the nucleus (Lassen et al., 2006), RNA interference (Bennasser et al., 2005; Huang et al., 2007; Omoto et al., 2004; Weinberg and Morris, 2006), or a combination of mechanisms (Lassen et al., 2004; Lassen, Bailey, and Siliciano, 2004). Given that Tat and Rev both play roles in splicing and other post-transcriptional processes, mutations in either or both of these overlapping genes could contribute to altered splicing or a block to nuclear export.
In summary, the current study supports the proposal that impaired Tat activity may contribute to latent infection with HIV. The data presented here suggest that Tat variation develops very early after infection and that attenuated Tat activity may favor the establishment of latency. These findings deserve further study. Even if latent infection is not always the result of impaired Tat activity, it is possible that Tat activators could be used to “flush out” HIV from latently-infected cells, or that inhibitors of Tat could be used to prevent release of virus from latently- or productively-infected cells. To facilitate such strategies, it is important have a better understanding of the range of Tat activities associated with latent infection.
MATERIALS AND METHODS
Subjects
The study subjects were participants in the Los Angeles and San Diego primary infection cohort (Strain et al., 2005). These subjects all had acute or early infection and were started on combined ART within 6 months of seroconversion. Blood was drawn for labs, plasma, and CD4+ T cells before initiation of ART and at various time points after ART. For this study, we looked through the stored samples and chose all subjects who had the following samples: 1) unused plasma dating from before the initiation of ART (baseline); 2) frozen cell pellets from multiple positive co-cultures from baseline; and 3) frozen cell pellets from at least one time point after the viral load had fallen to <50 copes/ml. Five subjects (A046 to A050) met these criteria. All subjects gave written, informed consent in accordance with local IRB guidelines.
Spinoculation of Baseline Plasma
A portion of the baseline plasma was used for direct extraction of viral RNA. To enhance the yield of replication-competent virus, provide a better comparator for the co-cultures, and isolate spliced two-exon tat, the remainder of the baseline plasma was used to inoculate and infect CD8-depleted PBL from healthy donors. This inoculation was done under conditions of centrifugation (spinoculation), as prior studies have shown that spinoculation of plasma results in a high efficiency of infection (Harrington et al., 2000; O'Doherty, Swiggard, and Malim, 2000). PBMC were isolated from buffy coats of two healthy donors using Ficoll centrifugation, depleted of CD8+ cells using Dynabeads CD8 (Dynal/Invitrogen), washed, and resuspended in complete RPMI + 10% FBS at 1 million cells/ml. A portion of the resting CD8-depleted PBMC was activated for 3 days using PHA (3 µg/ml, Roche) and IL-2 (20 IU/ml, Chiron). After 3 days, the activated CD8-depleted PBL from the two donors were washed twice and resuspended in complete RPMI to a final concentration of 20 million cells/ml. 125µl from each donor (a total of 5 million cells) were pipetted into each well of a 48 well plate, followed by either 500µl of baseline plasma or controls. The 48 well plates were spun at 1,200g for 2h at room temperature and incubated for 3 hrs at 37C. The cells were then resuspended, mixed with 4.3ml of complete RPMI (20% FBS) + IL-2 (20 IU/ml), and transferred to a 6 well plate (day 0). 300µl were removed from each well, spun to pellet cells, and saved as separate cell pellet and supernatant. On days 4,6,9,11,14,16,18, and 21, 1ml was removed from each well, spun to pellet cells, saved at −80 (as separate cell pellet and supernatant), and replaced with 1ml of complete RPMI (20%) + IL-2 (20IU/ml). On day 7, 1ml was removed for sampling and replaced with 1ml containing 0.75 million freshly activated CD8-depleted PBL from each of the two healthy donors. The frozen supernatants were thawed and assayed for p24 by ELISA (Perkin Elmer). A separate spinoculation experiment was done using mixed, resting CD8-depleted PBL (five million cells + 1ml of baseline plasma in a 24 well plate) followed by washing, activation with PHA (5 µg/ml for 1 day), washing of PHA, and serial sampling of 2ml per well.
Co-cultures
At baseline and suppressed time points, blood was used to isolate CD4+ T cells and establish co-cultures, as described previously (Strain et al., 2003; Strain et al., 2005; Wong et al., 1997). Briefly, CD4+ T cells were isolated from each subject using RosetteSep (StemCell Technologies), stimulated with anti-CD3/CD28 beads (Dynal/Invitrogen), and incubated with CD8-depleted PBL isolated from two healthy donors. Supernatants were assayed weekly for evidence of increasing p24 production indicative of replication-competent virus. Corresponding cell pellets were frozen at −80.
For both baseline and suppressed time points, at least 2 (and usually 3) co-culture wells were used to isolate virus. By terminal dilution estimates, these wells contained approximately 10 to 33 latently-infected cells. Co-cultures from baseline time points typically had positive p24 by day 7, and cell pellets from this day were used to isolate viral RNA. Co-cultures from suppressed time points grew more slowly, with p24 often detectable only at day 14 (used for 4 subjects) or day 21 (used for one subject). When incubated alone, unstimulated patient CD4+ T cells invariably had negative p24 despite culture for ≥ 21 days.
Preparation of lab virus stocks and cloning of NL4-3 Tat
To get a series of corresponding tat clones from NL4-3, and to provide an estimate for the diversity that can be introduced during cell culture and cloning, tat was cloned from NL4-3-infected donor PBL. Viral stocks were generated by transfecting the plasmid pNL4-3 (obtained from the NIH AIDS Research Reagent program) into 293T cells, using the supernatant (harvested at day 3) to infect pooled donor PBMC, and harvesting the supernatants from the infected PBMC (pooled from days 7 to 19). These viral stocks were used to infect fresh door PBL, and total cellular RNA was isolated after 2 days.
RNA Extraction
Genomic HIV-1 RNA was isolated from aliquots of baseline plasma (thawed from −80) using the QIAmpViral RNA Mini Kit (Qiagen) according to the manufacturer’s instructions. Total RNA was isolated from cell pellets using Trireagent BD (Molecular Research Center) per the manufacturer’s instructions.
cDNA Synthesis
Tat- and TAR-specific cDNA were generated using Transcriptor reverse transcriptase (Roche) per the manufacturer’s protocol with incubation at 51C for 1hr. For Tat-specific cDNA, we used the reverse primer 8776 rev cDNA rev (5’-CCTTTCYAAGCCCTGTCTTATTC-3’, positions 8761-8783 of HXB2, located downstream of the second exon of rev). For TAR-specific cDNA, we used the reverse primer TAR R (5’-GCACTCAAGGCAAGCTTTAT-3’, HXB2 positions 528–547).
PCR for tat and TAR
PCR was done using the high fidelity polymerase Platinum Taq Hifi (Invitrogen) using total reaction volume of 50µl with 5µl of 10x buffer, 1µl of 10mM dNTPs, 4.5µl of 50mM MgSO4, 36µl of H20, 1µl (5–10 pmol) of each primer, and 0.5µl of Platinum Taq Hifi. To amplify one-exon tat from baseline plasma, we used the primers 5TTOP (5’-CACCATGGAGCCAGTAGATCCTAG-3’, positions 5831–5850, with leader sequence for TOPO cloning) and R-vpu-Tat (5’-TCATTGCCACTGTCTTCTGCTCT-3’, positions 6206–6228). PCR conditions included 10pmol of each primers with 95C for 5”, 40 cycles of (95C for 1”, 58C for 1”, and 72C for 1”), and 72C for 10.” To amplify two-exon tat from cell pellets (spinoculated baseline plasma or co-cultures), we used the primers 5TTOP and 3-T-101-rev (5’-AGAGTAAGTCTCTCAAGCGG-3’, positions 8531–8550 of rev) with 10pmol of each primer and annealing T of 57 to 61C. To amplify two-exon tat that also contained the full two-exon rev, a separate PCR was done with the primers 734-xSpl1-4F (5’-GCGGCGACTGAATTGG-3’, spanning the splice site from 743 to 5777) and 8706 rev PCR rev (5’-CTATCTGTCCCCTCAGCTAC-3’, 8688 to 8707) using 5pmol of each primer and an annealing T of 61C. To amplify TAR, we used the primers TAR F (5’-CCTCAGATCCTGCATATAAGC-3’, positions 413–433) and TAR R (as above) with 10pmol of each primer and annealing T of 55C.
Cloning and Sequencing
For cloning of both tat and TAR, the expression vector pcDNA3.1/V5-His TOPO TA kit (Invitrogen) was used, as specified by the manufacturer. Plasmid preps were done using QIAprep Spin Miniprep columns (Qiagen) or QIAfilter Plasmid Maxi columns (Qiagen). DNA concentrations were determined using the average of at least two measurements obtained on a ND-1000 Spectrophotometer (NanoDrop). Sequencing was done using a 3130xl Genetic Analyzer (ABI).
Phylogenetic Analysis
Initial multiple sequence alignments were generated using Multalin with default gap parameters and the DNA 5-0 substitution matrix. Subsequent manual aligning was performed using the Se-Al sequence alignment editor. A master phylogeny, including sequence data from all subjects and common lab strains, was created using Dnadist and Neighbor within the PHYLIP 3.6 software suite (Felsenstein, 1993) to check for sequence contamination. Maximum likelihood phylogenies describing sequences from each individual host were built using the HyPhy software package (Pond, Frost, and Muse, 2005) implementing the HKY85 model of sequence evolution and the UPGMA (Unweighted Pair Group Method with Arithmetic Mean) Clustering Method. All diversity and divergence measurements were calculated using pairwise maximum likelihood estimation (MLE). Genbank accession numbers for all sequences in this manuscript will be provided upon acceptance.
Assessment of transactivation activity
Transactivation activity was measured by transfection of equal amounts of DNA into reporter TZM-bl cells (generous gift of Drs. John Kappes and Xiaoyun Wu) obtained through the NIH AIDS Research Reagent program (Derdeyn et al., 2000; Roos et al., 2000; Wei et al., 2002). These cells have an integrated HIV-1 LTR driving expression of β-galactosidase and luciferase.
Each Tat variant was tested in TZM-bl cells in at least two experiments with two replicates. Each well of a 24-well plate was seeded with 300,000 TZM-bl cells in 1ml of DMEM. TZM-bl cells were transfected using Lipofectamine 2000 (Invitrogen) per the manufacturer’s protocol. A GFP-encoding plasmid (pCMV-GFP) was co-transfected to control for transfection efficiency. To each well with 400µl OPTI-MEM media we added 100µl of OPTI-MEM containing 2.5µl of lipofectamine (from mix), 0.3µg of GFP (from mix), and 0.1µg of tat DNA. Medium was changed to DMEM after 5–6 hrs. On the third day post-transfection, fluorescence microscopy was done to look for GFP, medium was removed, and cells were lysed with Beta-Glo (Promega), which contains a luciferin-galactoside substrate. Luminescence was measured at 30 minutes using a Victor 3 luminometer (Perkin Elmer).
For a subset of variants, the transactivation activities were also measured in a T-lymphocyte derived cell line, LuSIV (a derivative of the CEM cell line with a reporter luciferase under control of an integrated LTR), obtained from the NIH AIDS Research Reagent Program (generous gift of Drs. Jason Roos and Janice Clements) (Roos et al., 2000). LuSIV cells were transfected using a Nucleofector II (Amaxa Biosystems) with 1 million LuSIV cells per cuvette, 1µg of DNA, and nucleofection program A-020. After 2 days, cells were pelleted and lysed with Bright-Glo (Promega). Luminescence was measured at 5 minutes using the Victor 3 luminometer.
The relative transactivation activity of each mutant Tat was calculated by taking the average luminescence (from two replicates), subtracting the average background luminescence (from empty vector), and normalizing to the background-subtracted luminescence of the Tat allele with the consensus sequence (most common sequence for that individual) obtained using the same forward and reverse primers. Results were then averaged over at least 2–4 separate experiments. In a separate analysis, the relative activity of each mutant was compared by normalizing to the activity of NL4-3 Tat obtained with the same primers from NL4-3-infected PBL cultures. Given that spinoculation was successful in only three subjects, the Tats from spinoculation of the baseline plasma were grouped with those obtained by direct extraction of the baseline plasma.
Statistics
The mean transactivation activities (normalized to the appropriate consensus sequence for each individual) from the baseline plasma (mostly non-latent virus) were separately compared to those from the baseline and suppressed co-cultures (mostly latent virus) using the paired Wilcoxon ranked sum test and the t test for correlated samples. In addition, we calculated the proportion of Tat alleles with less than a given percentage (25%, 50%, 75%, or 90%) of consensus Tat activity. Results from baseline plasmas were compared to those from baseline or suppressed co-cultures using the paired Wilcoxon ranked sum test and the t test for correlated samples.
Supplementary Material
Phylogenetic tree for one-exon tat sequences. To maximize the amount of usable sequence that could be compared across all patients, prematurely-truncated clones and sequence data from the second exon were excluded from analysis. The first two digits of each clone name identify the subject from which that clone was isolated. Scale bar shows genetic distance.
Phylogenetic tree for TAR sequences. Prematurely-truncated clones were excluded from analysis. The first digit of each clone name identifies the subject from which that clone was isolated. Scale bar shows genetic distance.
ACKNOWLEDGMENTS
We thank Caroline Ignacio, Nancy Keating, and Ruby Lam for technical assistance. TZM-bl cells were obtained from Drs. John Kappes and Xiaoyun Wu through the NIH AIDS Research Reagent program. LuSIV cells were obtained from Drs. Jason Roos and Janice Clements through the NIH AIDS Research Reagent Program. This work was supported in part by the National Institute of Health (NIH; NS051145 (JW/SY), T32 AI60530 (DH/SY), and 1K01DA024654-01 (SP)) and the U.S. Department of Veterans Affairs VA Merit Award (JW/SY).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adams M, Sharmeen L, Kimpton J, Romeo JM, Garcia JV, Peterlin BM, Groudine M, Emerman M. Cellular latency in human immunodeficiency virus-infected individuals with high CD4 levels can be detected by teh presence of promoter proximal transcripts. PNAS. 1994;91:3862–3866. doi: 10.1073/pnas.91.9.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addo MM, Altfeld M, Rosenberg ES, Eldridge RL, Philips MN, Habeeb K, Khatri A, Brander C, Robbins GK, Mazzara GP, Goulder PJ, Walker BD. The HIV-1 regulatory proteins Tat and Rev are frequently 32 targeted by cytotoxic T lymphocytes derived from HIV-1-infected individuals. Proc Natl Acad Sci U S A. 2001;98(4):1781–1786. doi: 10.1073/pnas.98.4.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen TM, O'Connor DH, Jing P, Dzuris JL, Mothe BR, Vogel TU, Dunphy E, Liebl ME, Emerson C, Wilson N, Kunstman KJ, Wang X, Allison DB, Hughes AL, Desrosiers RC, Altman JD, Wolinsky SM, Sette A, Watkins DI. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature. 2000;407(6802):386–390. doi: 10.1038/35030124. [DOI] [PubMed] [Google Scholar]
- Arya SK, Guo C, Josephs SF, Wong-Staal F. Transactivator gene of human T-lymphotropic virus type III (HTLV-III) Science. 1985;229:69–73. doi: 10.1126/science.2990040. [DOI] [PubMed] [Google Scholar]
- Bennasser Y, Le SY, Benkirane M, Jeang KT. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity. 2005;22(5):607–619. doi: 10.1016/j.immuni.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Berkhout B, Silverman RH, Jeang K-T. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell. 1989;59:273–282. doi: 10.1016/0092-8674(89)90289-4. [DOI] [PubMed] [Google Scholar]
- Brooks DG, Kitchen SG, Kitchen CM, Scripture-Adams DD, Zack JA. Generation of HIV latency during thymopoiesis. Nat Med. 2001;7(4):459–464. doi: 10.1038/86531. [DOI] [PubMed] [Google Scholar]
- Cao J, McNevin J, Holte S, Fink L, Corey L, McElrath MJ. Comprehensive analysis of human immunodeficiency virus type 1 (HIV-1)- specific gamma interferon-secreting CD8+ T cells in primary HIV-1 infection. J Virol. 2003;77(12):6867–6878. doi: 10.1128/JVI.77.12.6867-6878.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, Kuo YH, Brookmeyer R, Zieger MA, Barditch-Crovo P, Siliciano RF. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- Chun TW, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF. In vivo fate of HIV-1 infected T cells: Quantitative analysis of the transition to stable latency. Nature Medicine. 1995;1(12):1284–1290. doi: 10.1038/nm1295-1284. [DOI] [PubMed] [Google Scholar]
- Derdeyn CA, Decker JM, Sfakianos JN, Wu X, O'Brien WA, Ratner L, Kappes JC, Shaw GM, Hunter E. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J Virol. 2000;74(18):8358–8367. doi: 10.1128/jvi.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derdeyn CA, Kilby JM, Miralles GD, Li LF, Sfakianos G, Saag MS, Hockett RD, Bucy RP. Evaluation of distinct blood lymphocyte populations in human immunodeficiency virus type 1-infected subjects in the absence or presence of effective therapy. J Infect Dis. 1999;180(6):1851–1862. doi: 10.1086/315117. [DOI] [PubMed] [Google Scholar]
- Emiliani S, Fischle W, Ott M, Van Lint C, Amella CA, Verdin E. Mutations in the tat gene are responsible for human immunodeficiency virus type 1 postintegration latency in the U1 cell line. J Virol. 1998;72(2):1666–1670. doi: 10.1128/jvi.72.2.1666-1670.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emiliani S, Van Lint C, Fischle W, Paras P, Jr, Ott M, Brady J, Verdin E. A point mutation in the HIV-1 Tat responsive element is associated with postintegration latency. Proc Natl Acad Sci U S A. 1996;93(13):6377–6381. doi: 10.1073/pnas.93.13.6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenetic Inference Package (Phylip) 3.5. University of Washington: 1993. [Google Scholar]
- Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, Quinn TC, Chaisson RE, Rosenberg E, Walker B, Gange S, Gallant J, Siliciano RF. Latent infection of CD4 Tcells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nature Medicine. 1999;5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- Garcia JA, Harrich D, Pearson L, Mitsuyasu R, Gaynor RB. Functional domains required for tat-induced transcriptional activation of the HIV- 1 long terminal repeat. EMBO J. 1988;7(10):3143–3147. doi: 10.1002/j.1460-2075.1988.tb03181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghose R, Liou LY, Herrmann CH, Rice AP. Induction of TAK (cyclin T1/P-TEFb) in purified resting CD4(+) T lymphocytes by combination of cytokines. J Virol. 2001;75(23):11336–11343. doi: 10.1128/JVI.75.23.11336-11343.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillon C, Stankovic K, Ataman-Onal Y, Biron F, Verrier B. Evidence for CTL-mediated selection of Tat and Rev mutants after the onset of the asymptomatic period during HIV type 1 infection. AIDS Res Hum Retroviruses. 2006;22(12):1283–1292. doi: 10.1089/aid.2006.22.1283. [DOI] [PubMed] [Google Scholar]
- Harrington R, Wu L, Pullen H, Emerman M. Direct detection of infectious HIV-1 in blood using a centrifugation-indicator cell assay. J Virol Methods. 2000;88(1):111–115. doi: 10.1016/s0166-0934(00)00181-6. [DOI] [PubMed] [Google Scholar]
- Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- Huang J, Wang F, Argyris E, Chen K, Liang Z, Tian H, Huang W, Squires K, Verlinghieri G, Zhang H. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat Med. 2007;13(10):1241–1247. doi: 10.1038/nm1639. [DOI] [PubMed] [Google Scholar]
- Isel C, Karn J. Direct evidence that HIV-1 Tat stimulates RNA polymerase II carboxyl-terminal domain hyperphosphorylation during transcriptional elongation. J Mol Biol. 1999;290(5):929–941. doi: 10.1006/jmbi.1999.2933. [DOI] [PubMed] [Google Scholar]
- Jordan A, Bisgrove D, Verdin E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. Embo J. 2003;22(8):1868–1877. doi: 10.1093/emboj/cdg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao SY, Calma AF, Luciw PA, Peterlin BM. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987;330:489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- Kuppuswamy M, Subramanian T, Srinivasan A, Chinnadurai G. Multiple functional domains of Tat, the trans-activator of HIV-1, defined by mutational analysis. Nucleic Acids Res. 1989;17(9):3551–3561. doi: 10.1093/nar/17.9.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassen K, Han Y, Zhou Y, Siliciano J, Siliciano RF. The multifactorial nature of HIV-1 latency. Trends Mol Med. 2004;10(11):525–531. doi: 10.1016/j.molmed.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Lassen KG, Bailey JR, Siliciano RF. Analysis of human immunodeficiency virus type 1 transcriptional elongation in resting CD4+ T cells in vivo. J Virol. 2004;78(17):9105–9114. doi: 10.1128/JVI.78.17.9105-9114.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassen KG, Ramyar KX, Bailey JR, Zhou Y, Siliciano RF. Nuclear retention of multiply spliced HIV-1 RNA in resting CD4+ T cells. PLoS Pathog. 2006;2(7):e68. doi: 10.1371/journal.ppat.0020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Irwin D, Kanazawa S, Huang L, Romeo J, Yen TS, Peterlin BM. Transcriptional profiles of latent human immunodeficiency virus in infected individuals: effects of Tat on the host and reservoir. J Virol. 2003;77(15):8227–8236. doi: 10.1128/JVI.77.15.8227-8236.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marcello A. Latency: the hidden HIV-1 challenge. Retrovirology. 2006;3:7. doi: 10.1186/1742-4690-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Picado J, DePasquale MP, Kartsonis N, Hanna GJ, Wong J, Finzi D, Rosenberg E, Gunthard HF, Sutton L, Savara A, Petropoulos CJ, Hellmann N, Walker BD, Richman DD, Siliciano R, D'Aquila RT. Antiretroviral resistance during successful therapy of HIV type 1 infection. Proc Natl Acad Sci U S A. 2000;97(20):10948–10953. doi: 10.1073/pnas.97.20.10948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerhans A, Cheynier R, Albert J, Seth M, Kwok S, Sninsky J, Morfeldt-Manson L, Asjo B, Wain-Hobson S. Temporal fluctuations in HIV quasispecies in vivo are not reflected by sequential HIV isolations. Cell. 1989;58:901–910. doi: 10.1016/0092-8674(89)90942-2. [DOI] [PubMed] [Google Scholar]
- O'Doherty U, Swiggard WJ, Malim MH. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J Virol. 2000;74(21):10074–10080. doi: 10.1128/jvi.74.21.10074-10080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoto S, Ito M, Tsutsumi Y, Ichikawa Y, Okuyama H, Brisibe EA, Saksena NK, Fujii YR. HIV-1 nef suppression by virally encoded microRNA. Retrovirology. 2004;1:44. doi: 10.1186/1742-4690-1-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelson AS, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, Markowitz M, Ho DD. Decay characteristics of HIV-1 infected compartments during combination therapy. Nature. 1997;387:188–191. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- Perelson AS, Essunger P, Ho DD. Dynamics of HIV-1 and CD4+ lymphocytes in vivo. Aids. 1997;11(Suppl A):S17–S24. [PubMed] [Google Scholar]
- Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. HIV-1 dynamics in vivo: virion clearance rate, infected cell lifetime, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- Pomerantz RJ, Seshamma T, Trono D. Efficient replication of human immunodeficiency virus type 1 requires a threshold level of rev: potential implications for latency. Journal of Virology. 1992;66:1809–1813. doi: 10.1128/jvi.66.3.1809-1813.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21(5):676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Ramratnam B, Mittler JE, Zhang L, Boden D, Hurley A, Fang F, Macken C, Perelson AS, Markowitz M, Ho DD. The decay of the latent reservoir of replication competent HIV-1 is inversely correlated with the extent of residual viral replication during prolonged anti-retroviral therapy. Nature Medicine. 2000;6:82–85. doi: 10.1038/71577. [DOI] [PubMed] [Google Scholar]
- Roos JW, Maughan MF, Liao Z, Hildreth JE, Clements JE. LuSIV cells: a reporter cell line for the detection and quantitation of a single cycle of HIV and SIV replication. Virology. 2000;273(2):307–315. doi: 10.1006/viro.2000.0431. [DOI] [PubMed] [Google Scholar]
- Sadaie MR, Benter T, Wong-Staal F. Site-directed mutagenesis of two trans-regulatory genes (tat-III,trs) of HIV-1. Science. 1988;239(4842):910–913. doi: 10.1126/science.3277284. [DOI] [PubMed] [Google Scholar]
- Sanchez G, Xu X, Chermann JC, Hirsch I. Accumulation of defective viral genomes in peripheral blood mononuclear cells fo HIV infected individuals. Journal of Virology. 1997;71:2233–2240. doi: 10.1128/jvi.71.3.2233-2240.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seigel LJ, Ratner L, Josephs SF, Derse D, Feinberg MB, Reyes GR, O'Brien SJ, Wong-Staal F. Transactivation induced by human Tlymphotropic virus type III (HTLV III) maps to a viral sequence encoding 58 amino acids and lacks tissue specificity. Virology. 1986;148(1):226–231. doi: 10.1016/0042-6822(86)90419-8. [DOI] [PubMed] [Google Scholar]
- 35.Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ, Siliciano RF. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med. 2003;9(6):727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- Sodroski JG, Patarca R, Rosen CA, Wong-Staal F, Haseltine WA. Location of the trans-activating region on the genome of human T-cell lymphotropic virus type III. Science. 1985;229:74–77. doi: 10.1126/science.2990041. [DOI] [PubMed] [Google Scholar]
- Sonza S, Mutimer HP, O'Brien K, Ellery P, Howard JL, Axelrod JH, Deacon NJ, Crowe SM, Purcell DF. Selectively reduced tat mRNA heralds the decline in productive human immunodeficiency virus type 1 infection in monocyte-derived macrophages. J Virol. 2002;76(24):12611–12621. doi: 10.1128/JVI.76.24.12611-12621.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strain MC, Gunthard HF, Havlir DV, Ignacio CC, Smith DM, Leigh-Brown AJ, Macaranas TR, Lam RY, Daly OA, Fischer M, Opravil M, Levine H, Bacheler L, Spina CA, Richman DD, Wong JK. Heterogeneous clearance rates of long-lived lymphocytes infected with HIV: intrinsic stability predicts lifelong persistence. Proc Natl Acad Sci U S A. 2003;100(8):4819–4824. doi: 10.1073/pnas.0736332100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strain MC, Little SJ, Daar ES, Havlir DV, Gunthard HF, Lam RY, Daly OA, Nguyen J, Ignacio CC, Spina CA, Richman DD, Wong JK. Effect of treatment, during primary infection, on establishment and clearance of cellular reservoirs of HIV-1. J Infect Dis. 2005;191(9):1410–1418. doi: 10.1086/428777. [DOI] [PubMed] [Google Scholar]
- Verhoef K, Koper M, Berkhout B. Determination of the minimal amount of Tat activity required for human immunodeficiency virus type 1 replication. Virology. 1997;237(2):228–236. doi: 10.1006/viro.1997.8786. [DOI] [PubMed] [Google Scholar]
- Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother. 2002;46(6):1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Ghosh SK, Taylor ME, Jonson VA, Emini EA, Deutsch P, Lifson JD, Bonhoeffer S, Nowak MA, Hahn BH, Saag MS, Shaw GM. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- Weinberg MS, Morris KV. Are viral-encoded microRNAs mediating latent HIV-1 infection? DNA Cell Biol. 2006;25(4):223–231. doi: 10.1089/dna.2006.25.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz D, Nowak MA. Mathematical models of HIV pathogenesis and treatment. Bioessays. 2002;24(12):1178–1187. doi: 10.1002/bies.10196. [DOI] [PubMed] [Google Scholar]
- Wong JK, Hezareh M, Gunthard HF, Havlir DV, Ignacio CC, Spina CA, Richman DD. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278(5341):1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- Zhang L, Ramratnam B, Tenner-Racz K, He Y, Vesanen M, Lewin S, Talal A, Racz P, Perelson AS, Korber BT, Markowitz M, Ho DD. Quantifying residual HIV-1 replication in patients receiving combination antiretroviral therapy. New England Journal of Medicine. 1999;340:1605–1613. doi: 10.1056/NEJM199905273402101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic tree for one-exon tat sequences. To maximize the amount of usable sequence that could be compared across all patients, prematurely-truncated clones and sequence data from the second exon were excluded from analysis. The first two digits of each clone name identify the subject from which that clone was isolated. Scale bar shows genetic distance.
Phylogenetic tree for TAR sequences. Prematurely-truncated clones were excluded from analysis. The first digit of each clone name identifies the subject from which that clone was isolated. Scale bar shows genetic distance.