Abstract
Quantitative real-time PCR (qRT-PCR) is a powerful technique to quantify gene expression. To standardize gene expression studies and obtain more accurate qRT-PCR analysis, normalization relative to consistently expressed housekeeping genes (HKGs) is required. In this study, ten candidate HKGs including elongation factor 1 α (EF1A), ribosomal protein L11 (RPL11), ribosomal protein L14 (RPL14), ribosomal protein S8 (RPS8), ribosomal protein S23 (RPS23), NADH-ubiquinone oxidoreductase (NADH), vacuolar-type H+-ATPase (ATPase), heat shock protein 70 (HSP70), 18S ribosomal RNA (18S), and 12S ribosomal RNA (12S) from the cowpea aphid, Aphis craccivora Koch were selected. Four algorithms, geNorm, Normfinder, BestKeeper, and the ΔCt method were employed to evaluate the expression profiles of these HKGs as endogenous controls across different developmental stages and temperature regimes. Based on RefFinder, which integrates all four analytical algorithms to compare and rank the candidate HKGs, RPS8, RPL14, and RPL11 were the three most stable HKGs across different developmental stages and temperature conditions. This study is the first step to establish a standardized qRT-PCR analysis in A. craccivora following the MIQE guideline. Results from this study lay a foundation for the genomics and functional genomics research in this sap-sucking insect pest with substantial economic impact.
Introduction
Quantitative real-time PCR (qRT-PCR) is a powerful technique to quantify gene expressions during different biological processes [1]. Although qRT-PCR is one of the premier research tools, limitations still exist, several factors can influence the threshold cycle values including RNA quality, cDNA concentration, and PCR efficiency [2,3]. The most extensively adopted approach in qRT-PCR analysis is to normalize the expressions of target genes through measuring in parallel the expression of a housekeeping gene (HKG). Housekeeping genes, involved in basic cellular functions, are typically believed to possess inherent stable and constitutive expression in different samples under various biotic and abiotic conditions [1].
The cowpea aphid, Aphis craccivora Koch (Hemiptera, Aphidiae), is an important pest of cowpea, Vigna unguiculata (L.), one of the most important food crops in the semiarid tropical regions, including Asia, Africa, southern Europe, and Central and South America. Aphis craccivora typically feeds on several species of legumes (family Fabaceae) worldwide, including alfalfa, beans, chickpea, lentils, lupins, and peanuts. Aphids can infest cowpea through direct feeding on leaves, pods and other aerial tissues of the plant, or indirectly through the transmission of virus diseases [4–6]. A. craccivora can cause great damage even at low population densities because of its ability to transmit at least 14 viruses including the potyviruses, the cowpea aphid-borne mosaic virus and the blackeye cowpea mosaic virus [6,7]. In order to better understanding the molecular basis and facilitate the development of integrated pest management strategies of A. craccivora, Roche 454 pyrosequencing technology was used to generate the transcriptome of A. craccivora [7]. To take advantage of these genomics resources, establishing a standardized qRT-PCR procedure in A. craccivora following the MIQE (Minimum Information for publication of Quantitative real time PCR Experiments) guidelines [8] will be instrumental for the subsequent functional and epi-genomic research.
The objective of this research was to address an important aspect of gene expression studies in A. craccivora. Here, ten candidate HKGs including elongation factor 1 α (EF1A), ribosomal protein L11 (RPL11), ribosomal protein L14 (RPL14), ribosomal protein S8 (RPS8), ribosomal protein S23 (RPS23), NADH-ubiquinone oxidoreductase (NADH), vacuolar-type H+-ATPase (ATPase), heat shock protein 70 (HSP70), 18S ribosomal RNA (18S), and 12S ribosomal RNA (12S) were selected from the publically available A. craccivora transcriptome resources and the sequence obtained from GenBank [7]. The expression profile of these candidate HKGs was investigated across different developmental stages and under various temperature regimes. As a result, a suite of reference genes were recommended for the qRT-PCR analysis in A. craccivora.
Materials and Methods
Ethics statement
The cowpea aphid, Aphis craccivora Koch (Hemiptera, Aphidiae), was collected from a greenhouse on fava bean, Vicia faba (Fabales, Fabaceae), at the University of Kentucky. Aphis craccivora colony was maintained on seedlings of fava bean in a growth chamber at 23°C with a photoperiod of 12: 12 (L: D) and 50% relative humidity. No specific permit was required for the described collection. A. craccivora is a common aphid species with agricultural importance in the United States.
Samples preparation
For the developmental stage treatment, 10 A. craccivora adults (only unwinged individuals) and 20 nymphs (mixed nymphal stages) were, respectively, placed on fava bean leaves resting on a wet filter paper in a petri dish (9 cm diameter) for 2 d at 22°C. There are six replicates for the adult and nymph stages, respectively; therefore, there were 12 biological samples in total. For the temperature treatment, 10 A. craccivora adults and 20 nymphs (mixed nymphal stages) were, respectively, exposed to 10°C, 22°C, and 30°C, respectively, for 2 d. Each treatment was repeated three times independently, therefore, there were 18 biological samples for the temperature experiment. All the experiments were conducted in a growth chamber with a photoperiod of 14: 10 (L: D) and 50% relative humidity. After treatments, aphids were initially snap frozen in liquid nitrogen in a 1.5 ml microcentrifuge tube and then stored at -80°C for the subsequent total RNA extraction.
Total RNA extraction and cDNA synthesis
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA) according to previously described methods [9,10]. First strand cDNA was synthesized from 1 μg of total RNA with M-MLV reverse transcription kit (Invitrogen, USA) using a random N primer according the manufacturer’s recommendations.
Reference gene selection and primer design and quantitative real-time PCR
A total of 10 housekeeping genes that are commonly used in qRT-PCR analysis were selected as the candidate (Table 1). Majority of these genes have been previously used as the reference genes in other insect species [10–25]. Primers for EF1A was designed based on the sequences obtained from GenBank, and the others were obtained from the transcriptome of A. craccivora [7]. Primers for the qRT-PCR analysis were designed online, https://www.idtdna.com/Primerquest/Home/Index. The information of qRT-PCR amplifications and programs were described in detail in our previous study [9,10]. The standard curve and PCR efficiency of each candidate were constructed and calculated according to previously described methods [9,10].
Table 1. Summary of the ten housekeeping genes tested in this study.
| Gene | Description | Accession No. | Primer sequences (5’-3’) | Length (bp) | Efficiency (%) | Regression coefficient |
|---|---|---|---|---|---|---|
| EF1A | elongation factor 1 α | KC897473 | F: CCAGTAGGTCGTGTTGAAACT | 100 | 102.6 | 0.9997 |
| R: GGTGCATCTCCACGGATTTA | ||||||
| NADH | NADH-ubiquinone oxidoreductase | GAJW01000104 | F: CCTCAGCCTATTGAACGAGAAG | 101 | 109.6 | 0.9976 |
| R: CCTGCCAGTTCCAGTACTAATC | ||||||
| HSP70 | 70 kilodalton heat shock proteins | GAJW01000112 | F: AGTACCATGGAACCCGTAGA | 91 | 99.7 | 0.9992 |
| R: GGGTAGAACCTCCAACCAATAC | ||||||
| 18S | 18S ribosomal RNA | GAJW01000254 | CCTACCGTCGACAGTTGATAAG | 100 | 95.8 | 0.9992 |
| CAAAGACCTGGTGACTCTGAATA | ||||||
| 12S | 12S ribosomal RNA | GAJW01000011 | AGAAACCAACCTGGCTTACAC | 121 | 102.3 | 0.9992 |
| TTGCGACCTCGATGTTGAATTA | ||||||
| RPS23 | ribosomal protein S23 | GAJW01000179 | TACTGCCCGTAAACACGTAAA | 110 | 95.5 | 0.9983 |
| AAGCTCCTCCGAAAGGATTG | ||||||
| RPS8 | ribosomal protein S8 | GAJW01000269 | GTCGTCCGAGCCATTCTTT | 105 | 94.8 | 0.9977 |
| TCCTGTCTTCCTGCGTTTATG | ||||||
| RPL14 | ribosomal protein L14 | GAJW01000046 | CGAGTGGTCTACGTTGTTGAT | 106 | 93.9 | 0.9993 |
| GTACTCCAGTTTCTGGTCCATC | ||||||
| RPL11 | ribosomal protein L11 | GAJW01000099 | GGAACCACTTCATTGCATCTTC | 104 | 106.3 | 0.9991 |
| TGTCTTAGGACGTCAAGGTTTC | ||||||
| ATPase | vacuolar type H+-ATPase | GAJW01000023 | AGAGTGTCCACCATAGTTAGTTG | 95 | 101.3 | 0.9951 |
| ATCTCGGTAGTGGGTAGTTAGA |
Stability of gene expression
All biological replicates were used to calculate the average C t value. The stability of the ten HKGs was evaluated by algorithms geNorm [1], NormFinder [26], BestKeeper [27], and the comparative ΔC t method [28]. Finally, we compared and ranked the tested candidate HKGs based on a web-based analysis tool RefFinder (http://www.leonxie.com/referencegene.php).
Results
Transcriptional profiling of candidate reference genes
The entire candidate HKGs were visualized as a single amplicon with expected size on a 1.5% agarose gel (S1 Fig). Furthermore, gene-specific amplification was confirmed by a single peak in real-time melting-curve analysis (S2 Fig). Standard curves were created for all the candidate HKGs, and the PCR efficiency and correlation coefficient for each standard curve were shown in Table 1.
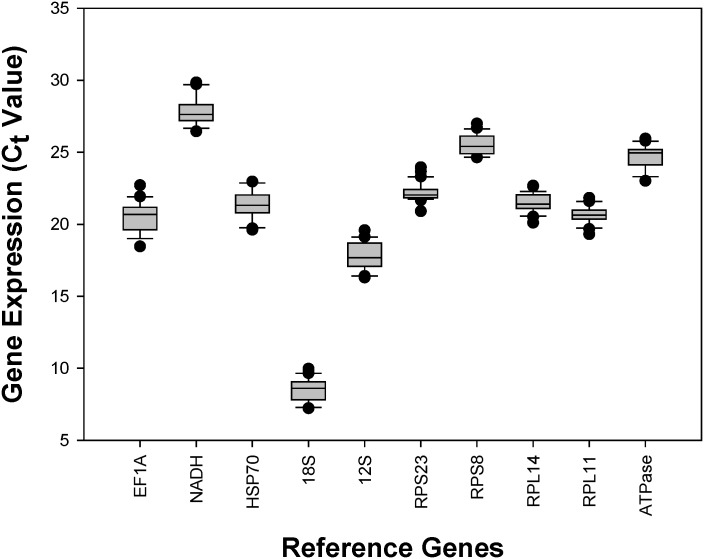
The mean and the standard deviation (SD) of the C t values were calculated for all the samples (S1 Table). RPL11 (SD = 0.61) had the least variable expression level and it was reflected in its low SD values. On the contrary, EF1A (SD = 1.09) had the most variable expression levels, and it was shown in its high SD values. Additionally, 18S had the lowest C t values (C tavg = 8.50), suggesting that it had the highest expression level, whereas, NADH was the least expressed gene among the candidates (C tavg = 27.82) (Fig 1, S1 Table).
Fig 1. Expression profiles of candidate housekeeping genes in Aphis craccivora.
The expression level of candidate housekeeping genes in 30 tested samples are documented in C t value. The median is represented by the line in the box. The interquartile rang is bordered by the upper and lower edges, which indicate the 75th and 25th percentiles, respectively.
Selection of the best candidate reference genes
Based on geNorm, under the impact of temperature, RPL14 and RPS8 were co-ranked as the most stable genes. The overall order from the most stable to the least stable reference genes was: RPL14 = RPS8, RPL11, RPS23, ATPase, 12S, HSP70, NADH, EF1A, 18S (Table 2). Under the impact of development, RPL14 and RPS8 were co-ranked as the most stable genes. The overall order from the most stable to the least stable reference genes was: RPL14 = RPS8, RPL11, ATPase, RPS23, 12S, HSP70, NADH, EF1A, 18S (Table 3).
Table 2. A summary of ranking for reference gene candidates using five different statistical approaches.
| RefFinder | geNorm | NormFider | ΔCt | BestKeeper | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genes | GM | Genes | SV | Genes | SV | Genes | SV | Genes [r] | Genes SD | ||
| RPS8 | 1.19 | RPL14 | 1.035 | RPS8 | 0.792 | RPS8 | 1.45 | RPL14 | 0.860 | HSP70 | 0.88 |
| RPL14 | 2.00 | RPS8 | 1.035 | RPL14 | 0.805 | RPL14 | 1.46 | RPL11 | 0.748 | RPS8 | 0.95 |
| RPL11 | 3.00 | RPL11 | 1.092 | RPL11 | 0.955 | RPL11 | 1.54 | 18S | 0.700 | RPL11 | 0.97 |
| HSP70 | 3.64 | ATPase | 1.167 | ATPase | 1.14 | ATPase | 1.62 | RPS8 | 0.670 | RPL14 | 1.01 |
| ATPase | 4.47 | RPS23 | 1.227 | HSP70 | 1.238 | HSP70 | 1.69 | ATPase | 0.376 | ATPase | 1.03 |
| RPS23 | 6.26 | 12S | 1.276 | NADH | 1.367 | NADH | 1.77 | EF1A | 0.299 | RPS23 | 1.04 |
| 12S | 6.74 | HSP70 | 1.432 | 12S | 1.388 | 12S | 1.78 | HSP70 | 0.276 | 12S | 1.04 |
| NADH | 6.93 | NADH | 1.52 | RPS23 | 1.394 | RPS23 | 1.78 | NADH | 0.231 | NADH | 1.05 |
| EF1A | 9.24 | EF1A | 1.578 | 18S | 1.488 | EF1A | 1.86 | 12S | 0.183 | EF1A | 1.26 |
| 18S | 9.74 | 18S | 1.639 | EF1A | 1.512 | 18S | 1.89 | RPS23 | 0.001 | 18S | 1.40 |
12 samples were from developmental stage group as input.
Geometric mean (GM); Stability Value (SV); Pearson’s correlation coefficient ([r]); Standard Deviation (SD).
Table 3. A summary of ranking for reference gene candidates using five different statistical approaches.
| RefFinder | geNorm | NormFider | ΔCt | BestKeeper | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genes | GM | Genes | SV | Genes | SV | Genes | SV | Genes [r] | Genes SD | ||
| RPS8 | 1.41 | RPL14 | 0.775 | RPS8 | 0.813 | RPS8 | 1.44 | RPL11 | 0.692 | RPL14 | 0.86 |
| RPL14 | 1.73 | RPS8 | 0.775 | RPL11 | 0.883 | RPL14 | 1.48 | RPS8 | 0.624 | ATPase | 0.89 |
| RPL11 | 2.45 | RPL11 | 0.912 | RPL14 | 0.976 | RPL11 | 1.51 | RPL14 | 0.605 | RPL11 | 0.90 |
| ATPase | 3.36 | RPS23 | 1.142 | ATPase | 1.009 | ATPase | 1.52 | EF1A | 0.477 | RPS8 | 0.94 |
| RPS23 | 5.23 | ATPase | 1.225 | 12S | 1.204 | HSP70 | 1.64 | 18S | 0.462 | RPS23 | 1.00 |
| 12S | 5.96 | 12S | 1.284 | RPS23 | 1.206 | NADH | 1.65 | HSP70 | 0.402 | HSP70 | 1.01 |
| HSP70 | 6.74 | HSP70 | 1.469 | HSP70 | 1.242 | 12S | 1.68 | ATPase | 0.375 | 12S | 1.04 |
| NADH | 8.00 | NADH | 1.569 | NADH | 1.364 | RPS23 | 1.77 | 12S | 0.359 | NADH | 1.14 |
| EF1A | 9.24 | EF1A | 1.632 | EF1A | 1.439 | EF1A | 1.81 | NADH | 0.249 | 18S | 1.16 |
| 18S | 9.74 | 18S | 1.684 | 18S | 1.522 | 18S | 1.88 | RPS23 | 0.177 | EF1A | 1.30 |
18 samples were from temperature group as input.
Geometric mean (GM); Stability Value (SV); Pearson’s correlation coefficient ([r]); Standard Deviation (SD).
According to the ΔC t method, under the impact of temperature, RPS8 was the top-ranked gene. The overall order from the most stable to the least stable reference genes was: RPS8, RPL14, RPL11, ATPase, HSP70, NADH, 12S, RPS23, EF1A, 18S (Table 2, S2 Table). Under the impact of development, RPS8 was also the top-ranked gene. The overall order from the most stable to the least stable reference genes was: RPS8, RPL14, RPL11, ATPase, HSP70, NADH, 12S, RPS23, EF1A, 18S (Table 3, S3 Table).
Based on NormFinder, under the impact of temperature, RPS8 was the most reliable and stable reference gene. The overall order from the most stable to the least stable reference genes was: RPS8, RPL11, RPL14, ATPase, 12S, RPS23, HSP70, NADH, EF1A, 18S (Table 2). Under the impact of development, RPS8 was also the top-ranked gene. The overall order from the most stable to the least stable reference genes was: RPS8, RPL14, RPL11, ATPase, HSP70, NADH, 12S, RPS23, 18S, EF1A (Table 3).
According to BestKeeper, the stability of a gene is directly proportional to the [r] value, while it is inversely proportional to the SD value. Under the impact of temperature, RPL11 had the highest [r] value, and RPL14 had the lowest SD value across all the samples (Table 2, S4 Table). Under the impact of development, RPL14 had the highest [r] value, and HSP70 had the least variable expression levels across all the samples (Table 3, S5 Table)
Comprehensive ranking of best reference genes using RefFinder
Under the impact of temperature, according to RefFinder, the comprehensive ranking of candidate reference genes from the most to the least stable was: RPS8, RPL14, RPL11, ATPase, RPS23, 12S, HSP70, NADH, EF1A, 18S (Table 2). Under the impact of development, the comprehensive ranking of candidate reference genes from the most to the least stable was: RPS8, RPL14, RPL11, HSP70, ATPase, RPS23, 12S, NADH, EF1A, 18S (Table 3). Interestingly, RPL11, RPS8, and RPL14 were the three most stable HKGs throughout different developmental stages and temperature conditions.
Quantitative analysis of candidate reference genes based on geNorm
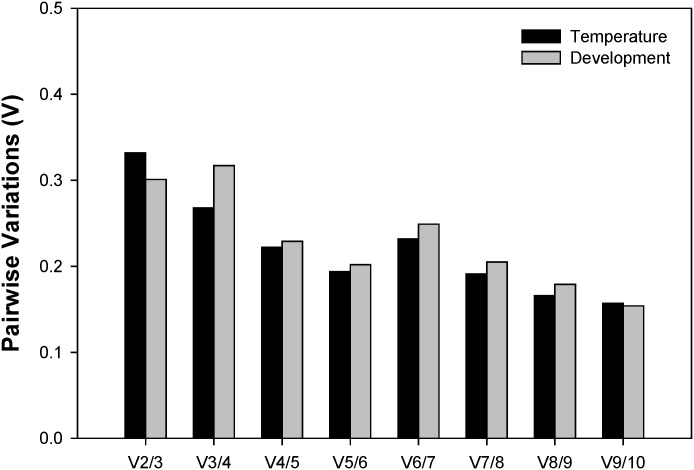
To decide the minimal number of genes mandatory for normalization, the V-value was computed by geNorm. geNorm analysis revealed that the pair-wise variation value V6/7 is higher than V5/6 (Fig 2). Increasing variation in this ratio corresponds to decreasing expression stability, due to the inclusion of a relatively unstable sixth gene. Therefore, five genes (PRL14, RPS8, RPL11, ATPase, and RPS23) are necessary for accurate normalization. Including a sixth reference gene has no significant effect on the normalization factor (Fig 2).
Fig 2. Pairwise variation (V) analysis of the candidate reference genes based on geNorm.
The pair-wise variation (Vn/Vn+1) was analyzed to determine the best number of references genes demanded for qRT-PCR data normalization [1]. The value V6/7 is higher than V5/6; this is due to the inclusion of a relative unstable sixth gene. Increasing variation in this ratio corresponds to decreasing expression stability.
Discussion
qRT-PCR quantification demands a comprehensive normalization by housekeeping genes to counteract confounding variations in experimental data. Housekeeping genes have been considered to be expressed in all cell types of the organism at a constant level to maintain basic cellular functions. However, there are no "universal" reference genes that are stably expressed and appropriate for the entire cell and tissue, and all kinds of test conditions [1]. Most gene expression studies in the literature use one single housekeeping gene; this will deeply influence the outcome of the statistical analysis and may bring about inaccurate data interpretation [29]. Therefore, customized reference gene selection under specific experimental conditions is highly recommended [11].
Recently, there is an influx of reference gene selection studies in insects, including convergent lady beetle, Hippodamia convergens; sweet potato whitefly, Bemisia tabaci; diamondback moth, Plutella xylostella; brown planthopper, Nilaparvata lugens; beet armyworm, Spodoptera exigua; oriental leafworm moth, Spodoptera litura; oriental fruit fly, Bactrocera dorsalis; Colorado potato beetle, Leptinotarsa decemlineata; soybean aphid, Aphis glycines; Russian wheat aphid, Diuraphis noxia; bird cherry-oat aphid, Rhopalosiphum padi; pea aphid, Acyrthosiphon pisum; bumblebees, Bombus terrestris and Bombus lucorum; western flower thrips, Frankliniella occidentalis; and honeybee, Apis mellifera [10–25]. Here, the expression profiles of ten HKGs from A. craccivora were evaluated across different developmental stages and temperature conditions. Our results are largely consistent with previous studies. For example, RPS8 (the component of the 40S ribosomal subunit) and RPL14 (60S ribosomal subunit) were the most stable HKGs across different developmental stage and temperature conditions, whereas the expression of 18S varied under the two conditions [14,16]. Not surprisingly, the comprehensive rankings (RefFinder) of these candidate reference genes under the two experimental conditions were, in principal, comparable to the rankings complied by the four algorithms, geNorm, Normfinder, BestKeeper, and the ΔC t method, respectively (Tables 2 and 3). Based on the comprehensive analyses, RPS8, RPL14, and RPL11 were the most stable A. craccivora HKGs under different developmental stages and temperature conditions.
There has been ongoing discussion about the optimal number of reference genes warrant for qRT-PCR analysis [9,14]. To prevent biased normalization, multiple instead of a single reference gene have been gradually adopted to normalize the expression of target genes under test conditions [30]. Our results showed that the pair-wise variation value of V6/7 is higher than that of V5/6 (Fig 2), suggesting that five reference genes are warranted for the accurate normalization in A. craccivora under different developmental stages and temperature conditions.
A phloem-feeding cowpea aphid, A. craccivora, is one of the key pests of cowpea, a major protein source for people in West Africa. Most recently, Roche 454-based pyrosequencing generated 176,262 raw reads from an A. craccivora transcriptome, and de novo assembly produced 7,647 transcripts [7]. Building on this newly developed genomic resource, we carried out the first reference gene selection study in one of the major pest species of cowpea. Although studies involving different developmental stages and /or temperature regimes have been limited [31–34], the advent of the Genomics Era will facilitate our understanding of A. craccivora, and eventually will lead to the development of integrated pest management strategies. Therefore, this study not only sheds light on establishing a standardized qRT-PCR procedure for quantification of gene expression in A. craccivora, but also lays a solid foundation for the genomics and functional genomics research in this sap-sucking insect pest.
Supporting Information
M, EZ Load 100 bp Molecular Ruler; Templates in the PCR reactions were as follows: 1) EF1A; 2) NADH; 3) HSP70; 4) 18S; 5)12S; 6) RPS23; 7) RPS8; 8) RPL14; 9) RPL11; 10) ATPase.
(TIFF)
(TIFF)
(DOCX)
(DOCX)
(DOCX)
18 samples were from temperature group as input.
(DOCX)
12 samples were from developmental stage group as input.
(DOCX)
Acknowledgments
We would like to express our gratitude to Drs. Xun Zhu (Chinese Academy of Agricultural Sciences) and Zhen Li (China Agricultural University) for their assistance with the data analysis. The granting agencies have no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The information reported in this paper (No.14-08-070) is part of a project of the Kentucky Agricultural Experiment Station and is published with the approval of the Director.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by a start-up fund from the University of Kentucky to XGZ, a grant from USDA BRAG grant (Award Agreement No.: 3048108827) to XGZ, and a Special Fund for Agroscience Research in the Public Interest (Award Agreement No.: 201303028) to YL. The granting agencies have no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Strube C, Buschbaum S, Wolken S, Schnieder T (2008) Evaluation of reference genes for quantitative real-time PCR to investigate protein disulfide isomerase transcription pattern in the bovine lungworm Dictyocaulus viviparus . Gene 425: 36–43. 10.1016/j.gene.2008.08.001 [DOI] [PubMed] [Google Scholar]
- 3. Bustin SA, Benes V, Nolan T, Pfaffl MW (2005) Quantitative real-time RT-PCR-a perspective. J Mol Endocrinol 34: 597–601. [DOI] [PubMed] [Google Scholar]
- 4. Kamphuis LG, Gao L, Singh KB (2012) Identification and characterization of resistance to cowpea aphid (Aphis craccivora Koch) in Medicago truncatula. BMC Plant Biol 12: 101 10.1186/1471-2229-12-101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Annan IB, Tingey WM, Schaefers GA, Tjallingii WF, Backus EA, Saxena KN (2000) Stylet penetration activities by Aphis craccivora (Homoptera: Aphididae) on plants and excised plant parts of resistant and susceptible cultivars of cowpea (Leguminosae). Annu Entomol Soc Am 93: 133–140. [Google Scholar]
- 6. Sainsbury F, Cañizares MC, Lomonossoff GP (2010) Cowpea mosaic virus: the plant virus-based biotechnology workhorse. Annu Rev Phytopathol 48: 437–455. 10.1146/annurev-phyto-073009-114242 [DOI] [PubMed] [Google Scholar]
- 7. Agunbiade TA, Sun WL, Coates BS, Djouaka R, Tamò M, Ba MN, et al. (2013) Development of reference transcriptomes for the major field insect pests of cowpea: a toolbox for insect pest management approaches in West Africa. PLOS ONE 8: e79929 10.1371/journal.pone.0079929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55: 611–622. 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- 9. Yang CX, Pan HP, Liu Y, Zhou XG (2015) Stably expressed housekeeping genes across developmental stages in the two-spotted spider mite, Tetranychus urticae . PLOS ONE 10: e0120833 10.1371/journal.pone.0120833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pan HP, Yang XW, Siegfried BD, Zhou XG (2015) A comprehensive selection of reference genes for RT-qPCR analysis in a predatory lady beetle, Hippodamia convergens (Coleoptera: Coccinellidae). PLOS ONE 10.1371/journal.pone.0125868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liang P, Guo YJ, Zhou XG, Gao XW (2014) Expression Profiling in Bemisia tabaci under insecticide treatment: indicating the necessity for custom reference gene selection. PLOS ONE 9: e87514 10.1371/journal.pone.0087514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li RM, Xie W, Wang SL, Wu QJ, Yang NN, Yang X, et al. (2013) Reference gene selection for qRT-PCR analysis in the sweetpotato whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae). PLOS ONE 8: e53006 10.1371/journal.pone.0053006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhu X, Yuan M, Shakeel M, Zhang YJ, Wang SL, Wang X, et al. (2014) Selection and evaluation of reference genes for expression analysis using qRT-PCR in the beet armyworm Spodoptera exigua (Hübner)(Lepidoptera: Noctuidae). PLOS ONE 9: e84730 10.1371/journal.pone.0084730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fu W, Xie W, Zhang Z, Wang SL, Wu QJ, Liu Y, et al. (2014) Exploring valid reference genes for quantitative real-time PCR analysis in Plutella xylostella (Lepidoptera: Plutellidae). Int J Biol Sci 9: 792–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shi XQ, Guo WC, Wan PJ, Zhou LT, Ren XL, Ahmat T, et al. (2013) Validation of reference genes for expression analysis by quantitative real-time PCR in Leptinotarsa decemlineata (Say). BMC Res Notes 6: 93 10.1186/1756-0500-6-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yuan M, Lu YH, Zhu X, Wan H, Shakeel M, Zhan S, et al. (2014) Selection and evaluation of potential reference genes for gene expression analysis in the Brown Planthopper, Nilaparvata lugens (Hemiptera: Delphacidae) using reverse-transcription quantitative PCR. PLOS ONE 9: e86503 10.1371/journal.pone.0086503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shen GM, Jiang HB, Wang XN, Wang JJ (2010) Evaluation of endogenous references for gene expression profiling in different tissues of the oriental fruit fly Bactrocera dorsalis (Diptera: Tephritidae). BMC Mol Biol 11: 76 10.1186/1471-2199-11-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lu YH, Yuan M, Gao XW, Kang TH, Zhan S, Wan H, et al. (2013) Identification and validation of reference genes for gene expression analysis using quantitative PCR in Spodoptera litura (Lepidoptera: Noctuidae). PLOS ONE 8: e68059 10.1371/journal.pone.0068059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bansal R, Mamidala P, Mian MR, Mittapalli O, Michel AP (2012) Validation of reference genes for gene expression studies in Aphis glycines (Hemiptera: Aphididae). J Econ Entomol 105: 1432–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sinha DK, Smith CM (2014) Selection of reference genes for expression analysis in Diuraphis noxia (Hemiptera: Aphididae) fed on resistant and susceptible wheat plants. Sci Rep 4: 5059 10.1038/srep05059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang CX, Pan HP, Liu Y, Zhou XG (2014) Selection of reference genes for expression analysis using quantitative real-time PCR in pea aphid, Acyrthosiphon pisum (Harris) (Hemiptera, Aphidiae). PLOS ONE 9: e110454 10.1371/journal.pone.0110454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu KK, Liu WW, Mar T, Liu Y, Wu YF, Wang XF (2014) Sequencing and validation of reference genes to analyze endogenous gene expression and quantify yellow dwarf viruses using RT-qPCR in viruliferous Rhopalosiphum padi . PLOS ONE 9: e97038 10.1371/journal.pone.0097038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Horňáková D, Matoušková P, Kindl J, Valterová I, Pichová I (2010) Selection of reference genes for real-time polymerase chain reaction analysis in tissues from Bombus terrestris and Bombus lucorum of different ages. Anal Biochem 397: 118–120. 10.1016/j.ab.2009.09.019 [DOI] [PubMed] [Google Scholar]
- 24. Cameron RC, Duncan EJ, Dearden PK (2013) Stable reference genes for the measurement of transcript abundance during larval caste development in the honeybee. Apidologie 44: 357–366. [Google Scholar]
- 25. Yang CX, Li H, Pan HP, Ma YB, Zhang DY, Liu Y, et al. (2014) Validation of reference genes for gene expression studies in non viruliferous and viruliferous Frankliniella occidentalis (Thysanoptera: Thripidae). PeerJ PrePrints, 2: e662v1. [Google Scholar]
- 26. Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64: 5245–5250. [DOI] [PubMed] [Google Scholar]
- 27. Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett 26: 509–515. [DOI] [PubMed] [Google Scholar]
- 28. Silver N, Best S, Jiang J, Thein SL (2006) Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol 7: 33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ferguson BS, Nam H, Hopkins RG, Morrison RF (2010) Impact of reference gene selection for target gene normalization on experimental outcome using real-time qRT-PCR in adipocytes. PLOS ONE 5: e15208 10.1371/journal.pone.0015208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Veazey KJ, Golding MC (2011) Golding selection of stable reference genes for quantitative RT-PCR comparisons of mouse embryonic and extra-embryonic stem cells. PLOS ONE 6: 27592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Berg GN (1984) The effect of temperature and host species on the population growth potential of the cowpea aphid, Aphis craccivora Koch (Homoptera: Aphididae). Aust J Zool 32: 345–352. [Google Scholar]
- 32. Obopile M, Ositile B (2010) Life table and population parameters of cowpea aphid, Aphis craccivora Koch (Homoptera: Aphididae) on five cowpea Vigna unguiculata (L. Walp.) varieties. J Pest Sci 83: 9–14. [Google Scholar]
- 33. Chen CY, Lai CY, Kuo MH (2009) Temperature effect on the growth of Buchnera endosymbiont in Aphis craccivora (Hemiptera: Aphididae). Symbiosis 49: 53–59. 21347170 [Google Scholar]
- 34. Chen CY, Chiu MC, Kuo MH (2013) Effect of warming with temperature oscillations on a low-latitude aphid, Aphis craccivora . B Entomol Res 103: 406–413. 10.1017/S0007485312000867 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
M, EZ Load 100 bp Molecular Ruler; Templates in the PCR reactions were as follows: 1) EF1A; 2) NADH; 3) HSP70; 4) 18S; 5)12S; 6) RPS23; 7) RPS8; 8) RPL14; 9) RPL11; 10) ATPase.
(TIFF)
(TIFF)
(DOCX)
(DOCX)
(DOCX)
18 samples were from temperature group as input.
(DOCX)
12 samples were from developmental stage group as input.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.