Abstract
Accurate radiation dose calculation is essential for successful proton radiotherapy. Monte Carlo (MC) simulation is considered to be the most accurate method. However, the long computation time limits it from routine clinical applications. Recently, graphics processing units (GPUs) have been widely used to accelerate computationally intensive tasks in radiotherapy. We have developed a fast MC dose calculation package, gPMC, for proton dose calculation on a GPU. In gPMC, proton transport is modeled by the class II condensed history simulation scheme with a continuous slowing down approximation. Ionization, elastic and inelastic proton nucleus interactions are considered. Energy straggling and multiple scattering are modeled. Secondary electrons are not transported and their energies are locally deposited. After an inelastic nuclear interaction event, a variety of products are generated using an empirical model. Among them, charged nuclear fragments are terminated with energy locally deposited. Secondary protons are stored in a stack and transported after finishing transport of the primary protons, while secondary neutral particles are neglected. gPMC is implemented on the GPU under the CUDA platform. We have validated gPMC using the TOPAS/Geant4 MC code as the gold standard. For various cases including homogeneous and inhomogeneous phantoms as well as a patient case, good agreements between gPMC and TOPAS/Geant4 are observed. The gamma passing rate for the 2%/2 mm criterion is over 98.7% in the region with dose greater than 10% maximum dose in all cases, excluding low-density air regions. With gPMC it takes only 6–22 s to simulate 10 million source protons to achieve ~1% relative statistical uncertainty, depending on the phantoms and energy. This is an extremely high efficiency compared to the computational time of tens of CPU hours for TOPAS/Geant4. Our fast GPU-based code can thus facilitate the routine use of MC dose calculation in proton therapy.
1. Introduction
Proton therapy allows higher dose conformality compared to conventional radiation therapy. At the same time, uncertainties in dose calculation and delivery can potentially have a bigger impact on the desired dose distribution. An accurate and efficient dose calculation method is of vital importance for the success of a treatment. Consequently, Monte Carlo (MC) dose calculation is desirable and it has been demonstrated that the use of MC in proton therapy could lead to a significant reduction in treatment planning margins (Paganetti 2012). Nevertheless, among all the available methods, pencil-beam-based algorithms are widely used in clinical practice (Hong et al 1996, Schaffner et al 1999) mainly because of their high computational efficiency. Yet, the accuracy of pencil-beam algorithms is not satisfactory. In particular for those cases with a large degree of tissue heterogeneity, a small amount of inaccuracy in dose calculation in pencil-beam algorithms may lead to a significant shift of dose distributions, possibly resulting in underdosage to the tumor and/or overdosage to the critical structures.
MC dose calculation has been introduced into proton clinics but it is applied only for recalculating existing treatment plans for research studies, because it is still too computationally inefficient for routine applications for all patients (Paganetti et al 2008). As a statistical method, the total number of particles simulated determines the accuracy of an MC dose calculation and an enormously large number of particles are usually necessary to yield a desired level of precision. Over the years, despite the great efforts devoted to accelerating the MC dose calculation process, such as using large-scale computational hardware and developing simplified algorithms (Kohno et al 2003, Fippel and Soukup 2004, Li et al 2005, Yepes et al 2009), the available proton MC dose calculation methods still cannot meet the clinically acceptable efficiency. The unsatisfactory efficiency also prohibits the development of advanced treatment techniques for proton therapy, e.g. MC-based treatment planning and adaptive radiotherapy.
Recently, computer graphics processing units (GPUs) have drawn great attention due to their tremendous ability of accelerating a variety of computationally intensive tasks in radiation therapy (Xu and Mueller 2005, Sharp et al 2007, Jacques et al 2008, Samant et al 2008, Yan et al 2008, Gu et al 2009, 2010, Hissoiny et al 2009, Men et al 2009, 2010a, 2010b, Jia et al 2010b). In particular, a set of MC packages has been successfully developed for mega-voltage photon dose calculations and high acceleration factors have been reported (Jia et al 2010a, 2011, Hissoiny et al 2011, Jahnke et al 2012). Kilo-voltage photon dose calculation packages also become available to assess CT dose to patients (Badal and Badano 2009, Jia et al 2012). For the purpose of proton dose calculations, a track-repeating algorithm has been implemented on a GPU platform (Yepes et al 2010). Very recently, a simplified MC method has been developed on a GPU and then used clinically (Kohno et al 2011). Because of these efforts, the calculation time of MC-based proton dose calculation has been greatly shortened. Nonetheless, these packages developed on a GPU utilize, to a certain extent, simplifications and approximations in proton transport physics. A dose calculation engine on a GPU with a full MC simulation is still highly desirable.
It is challenging to develop a full MC dose calculation package for proton therapy on a GPU to achieve both satisfactory accuracy and efficiency. First, protons interact with human tissue through a variety of electromagnetic and nuclear interactions. However, not all of the interactions and transport processes are necessary for dose calculations. It requires a series of investigations how much detail one should include in the simulations to balance accuracy and efficiency. Meanwhile, from the computational perspective, it is not straightforward to achieve high performance in MC particle transport on a GPU platform (Hissoiny et al 2011, Jia et al 2011, Pratx and Xing 2011) because of the inherent conflict between the GPU’s SIMD (single instruction multiple data) processing scheme and the stochastic nature of an MC process. Moreover, GPU-based proton MC dose calculation encounters its own difficulties such as memory writing conflict, as will be discussed later in this paper.
We report here our recent progress toward the development of an MC package, gPMC, for proton dose calculation on a GPU platform. The roadmap of this paper is as follows. In section 2, we describe the physics employed in gPMC and the algorithm structure. Section 3 presents experimental results of our dose calculation in various cases. Finally, we conclude our paper in section 4 and present some further discussions.
2. Methods and materials
2.1. Proton transport in gPMC
The physics employed by gPMC combines those reported in various publications (Kawrakow 2000, Fippel and Soukup 2004, Salvat et al 2009, Geant4 Collaboration 2011). In this subsection, we will first present the data used in gPMC and then the proton transport physics.
There are 25 materials predefined in gPMC’s database, which are relevant to proton therapy (Schneider et al 2000). For each material, its mass stopping power ratio with respect to water, fs(E, i), is extracted from TOPAS/Geant4 (Perl et al 2012) and is tabulated, where E is the proton kinetic energy and i labels the material type. Ionization differential cross sections are calculated analytically (Geant4 Collaboration 2011), while the functional forms for nuclear interaction cross sections are implemented according to Fippel et al (Fippel and Soukup 2004). Moreover, water-restricted stopping power, Lw(E), as a function of the energy E is calculated using the Bethe–Bloch equation (Geant4 Collaboration 2011) with a user-specified cut-off energy Ee,min for δ-electron production e.g. Ee,min = 100 keV. gPMC supports proton transport in a voxelized patient geometry. During its initialization stage, the material type i, density ρ and electron density ρe at each voxel are determined based on its CT number according to calibrated conversion curves.
gPMC transports protons in a kinetic energy range of [0.5, 350.0] MeV using the class II condensed history simulation scheme with a continuous slowing down approximation. Specifically, the proton is transported in a step-by-step fashion until its energy is below the cut-off energy Ep, min = 0.5 MeV or it exits the phantom region. For each step, the length is chosen to be d = min (dvox, dhard, dmax), where dvox is the distance to the next voxel boundary. dhard is the distance to the next discrete interaction point, sampled according to the mean free path of all the interactions considered using the fictitious interaction method developed in PENELOPE (Salvat et al 2009). dmax is the maximum distance allowed by the algorithm, which is chosen such that each step length is less than a user-specified distance and the fractional energy decrease per step is less than a certain value. By default, these two criteria are set to be 0.2 cm and 25%, respectively. Once a step length is determined, its equivalent length in water is calculated based on the mass stopping power ratio as
| (1) |
where ρw is the water density. Moreover, the mean energy decrease in this step is calculated by solving an equation
| (2) |
which is numerically carried out using the scheme developed by Kawrakow (2000). The actual energy decrease is further calculated as , where ζ is a Gaussian random number with a zero mean and a certain variance calculated for this step (Geant4 Collaboration 2011). Such a treatment accounts for the energy fluctuations of the proton in this step due to the secondary electron production in ionization with energy lower than the cut-off energy Ee,min. We have also considered multiple scattering due to elastic Coulomb interactions, which is modeled as a random deflection angle of the proton trajectory at each step following a Gaussian distribution (Hagiwara et al 2002, Fippel and Soukup 2004). Lateral deflection is not modeled in gPMC, as this has been shown to be unnecessary for proton beam therapy dose calculations (Fippel and Soukup 2004).
For discrete interactions, ionization events with a δ-electron with energy above Ee,min are considered. The scattered electron energy is sampled using a typical rejection method (Geant4 Collaboration 2011), while its polar scattering angle is determined by the kinematics and an azimuthal angle generated uniformly in the range of [0, 2π]. The generated δ-electrons are not transported in gPMC for simplicity. Instead, their energies are locally deposited. The maximum energy of the δ-electron generated is about 480 keV for a 200 MeV proton beam. Neglecting the electron transport will lead to negligible error in most clinical cases due to the small electron range in human tissue in this energy range. However, electron transport may be necessary in some cases such as lung, where the electron range is not small, as will be demonstrated in section 3.
As for nuclear interactions, gPMC follows an empirical strategy developed previously by Fippel and Soukup (2004). Only proton–proton elastic interactions, proton–oxygen elastic and inelastic interactions are considered. The secondary protons generated in the proton–proton elastic interactions and in the proton–oxygen inelastic interactions are tracked by the same proton transport physics as mentioned above. All other heavy particles are not followed and their energies are locally deposited. Charge-neutral particles produced in the proton–oxygen inelastic events are neglected. Such a simplified modeling of nuclear interactions has been shown to have adequate accuracy for proton therapy dose calculation previously (Fippel and Soukup 2004), as well as in the studies in this paper. However, in some extreme cases, e.g. in high-Z materials, this simplification will lead to errors in dose calculation.
2.2. CUDA implementation
gPMC is developed under the compute unified device architecture (CUDA) platform supported by NVIDIA (2011), which enables us to extend C language to program an NVIDIA GPU. In this section, we discuss the code structure of gPMC, as well as a few relevant issues in the implementation.
2.2.1. Code structure
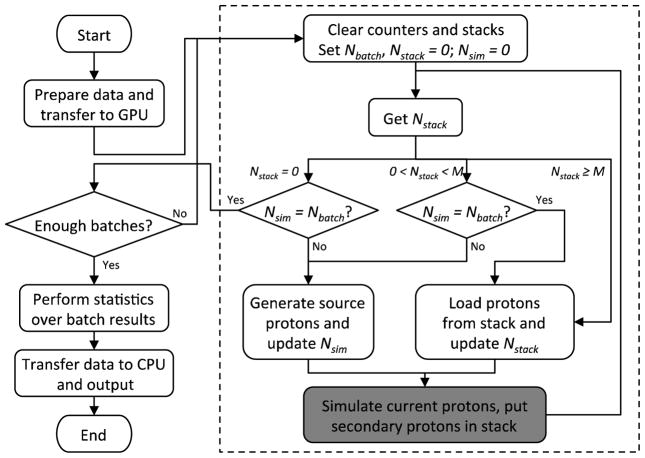
The structure of gPMC is shown in figure 1. Once it is launched, gPMC prepares all the necessary data at the initialization step such as the voxelized geometry, material properties, all the cross section data and random number seeds. All of these data are transferred from the CPU memory to the global memory of a GPU. After the initialization stage, simulation is performed in a batched fashion. In each batch, a certain number of source protons and the generated secondary protons are transported and dose depositions are recorded. These steps are indicated by the dashed box in figure 1 and will be further discussed later. The dose distribution after each batch is stored on a GPU. Finally, a GPU function is called to perform statistical analysis over those results from different batches to obtain the average dose to each voxel and the corresponding uncertainties. The program transfers data from the GPU to the CPU and outputs results, before it exits.
Figure 1.
Flow chart of our gPMC MC simulation. Steps inside the dashed box correspond to the simulation of a batch. The shaded box is the proton transport kernel.
Within each batch, a set of protons is transported in parallel on the GPU, each by a GPU thread. Before discussing the details, let us denote the targeted number of source protons to be simulated in a batch by Nbatch, the number of already simulated source protons by Nsim and the number of protons currently in stack by Nstack. Let us also denote by M the maximum number of protons that can be transported by the GPU simultaneously, which is usually limited by the available GPU computation and memory resources. At the beginning of each batch, we first allocate a particle array of length M to store information, e.g. locations and velocities, of all the particles currently being transported. One more array is allocated as a stack to store secondary protons generated during the simulation. The length of the stack is empirically chosen as 64M to provide a large enough space.
The flow of our simulations within a batch is shown in the dashed box in figure 1. Here, we provide a brief discussion for each key step. The simulation starts by clearing all relevant counters and the stack and setting the targeted number of protons Nbatch. It then loops over the following steps. First, gPMC queries for the current number of protons in the stack Nstack and depending on its value, the execution branches out as follows. (1) Nstack = 0: in this case, it checks if the number of simulated protons equals to the targeted number and if so, the current batch finishes. Otherwise, gPMC generates a certain number of protons in the particle array awaiting to be simulated. The number of protons generated is usually M, except at the very end of a batch where this number is adjusted so that the total number of generated protons is Nbatch. (2) 0 < Nstack < M: in this case, although there are protons in the stack, the number of protons is less than M and it is inefficient to simulate them due to the not fully loaded GPU. Therefore, gPMC first checks if Nsim = Nbatch. If so, it loads those protons from stack to the particle array, as no further source protons are needed. Otherwise, gPMC generates a certain number of protons in the particle array. (3) Nstack ≥ M: there are enough protons in the stack, so gPMC simply loads M protons from the stack to the particle array. The particle array is now filled with a set of protons. A GPU kernel is then launched to transport all of them, as indicated by a step shown in the shaded box in figure 1. A number of copies of such a kernel will be executed on the GPU. The number of copies equals the number of protons in the particle array and all of these copies are executed by the processors available on a GPU. In this process, dose deposition to each voxel is recorded and any secondary particles created are put into the stack. After this, gPMC loops back to the query of Nstack.
2.2.2. Dose scoring
We allocate a dose counter in the GPU’s global memory, which is of the same size as the voxel array to hold the dose values to each voxel. During the proton transport, all GPU threads deposit doses to the corresponding locations in this counter. One practical issue is the memory writing conflict in dose deposition. Specifically, when two threads happen to deposit dose information to the same voxel at the same time, a memory writing conflict occurs and the energy deposition has to be serialized in order to obtain correct results. In practice, we have adopted an atomic float addition function developed by Lensch et al (Lensch and Strzodka 2008) to resolve this problem. This function is called atomic in that, once a GPU thread starts writing to a memory address, it has the highest priority and no other threads can interfere with this process. Yet, this serialization apparently compromises the GPU parallel processing capability. Its impacts on the efficiency will be discussed in section 3.
2.2.3. Other issues
There are a few other issues we would like to mention briefly. First, a high-performance pseudo-random number generator CURAND (NVIDIA 2010) developed by NVIDIA is used in gPMC, which offers simple and efficient generation of high-quality pseudo-random numbers using the XORWOW algorithm (Marsaglia 2003). The quality of the random numbers has been tested using the TestU01 ‘Crush’ framework of tests (L’Ecuyer and Simard 2007). Second, the material data, such as stopping power in water, mass stopping power ratio and various cross sections, are stored in the GPU memory at a set of discrete energy values. Hence, interpolation is a frequently required operation to obtain material data at other energy levels. In gPMC, we store all of these data in a linear energy grid with a 0.5 MeV increment in the energy range of interest, and linear interpolation is performed. This linear interpolation can be achieved by the GPU hardware via the so-called texture memory, which ensures the efficiency of this operation.
2.3. Test cases
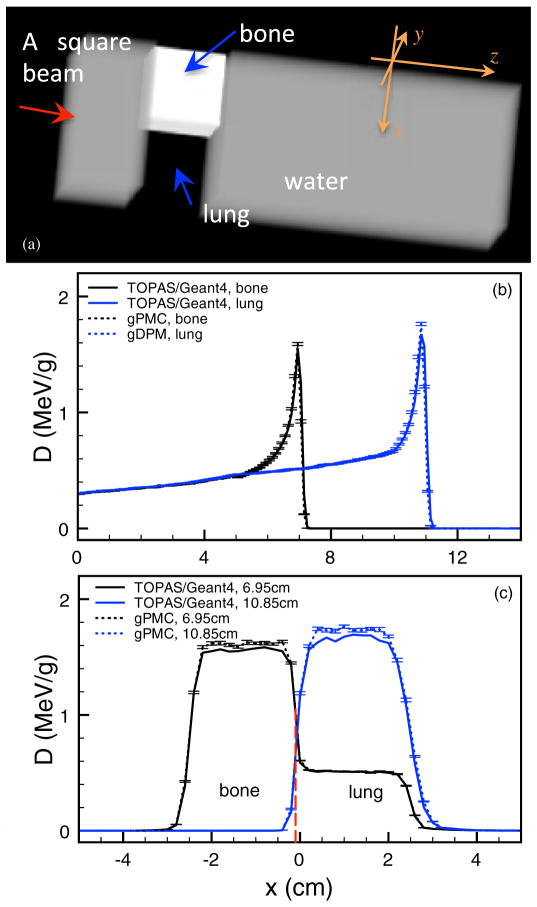
To test gPMC, we have conduced a series of dose calculations in a variety of phantom cases and a patient case. In all the phantom cases we studied, the phantom dimensions are 10.2 × 10.2 × 30 cm3 with a voxel size of 0.2 × 0.2 × 0.1 cm3. We considered four phantom configurations: (1) a homogeneous water phantom, (2) a homogeneous bone phantom with 500 HU, (3) a homogeneous tissue phantom with 200 HU and (4) a water phantom with a lung slab of 5.0 cm thickness at z = 7.5 cm, in which a 5.0×5.0×5.0 cm3 box of bone is inserted at x = −2.6 cm and y = 0 cm, as shown in figure 5(a). The HU number of lung is −700, while that of the bone slab is 500. In these phantom cases, a mono-energetic mono-directional square proton beam of size 5.0 × 5.0 cm2 impinges normally on the phantom surface. We have also selected a head and neck CT scan for the patient study using the same square beam placed on the right side of the patient positioned such that it experiences a very inhomogeneous material setup. The patient is defined by a CT grid with a resolution of 256×256×84 and a voxel size of 1.084 × 1.084 × 2.5 mm3. In all of these cases, the voxel material properties, e.g. density, electron density and material type, are inferred based on conversion curves that are calibrated against our CT scanner. 107 source protons are used in all the cases in gPMC.
Figure 5.
(a) Illustration of the configuration for the inhomogeneous phantom. (b) Depth dose curves through the centers of the bone insert and the lung insert for the inhomogeneous phantom. (c) Lateral profiles at the depths of 6.95 and 10.85 cm.
Our test focuses on both accuracy and efficiency. For the accuracy test, we have performed simulations with the same configurations using TOPAS (Perl et al 2012), a MC system based on Geant4 (Agostinelli et al 2003) for proton therapy dose calculations which has been validated extensively by the proton therapy community. For the homogeneous phantom, a pencil beam is used in the TOPAS simulation instead of a square broad beam, and the result is integrated to obtain the dose distribution corresponding to the broad beam. This integration effectively reduces the resulting dose uncertainty. As for the gPMC simulations, we calculated dose from the broad beam directly due to a severe slowing down in the pencil-beam cases, as will be discussed in section 3. The results are quantitatively measured using a γ -test (Low et al 1998). As for the efficiency, we have recorded the computation time of each case to demonstrate the gain in computational efficiency achieved by gPMC.
As for the hardware, we use an NVIDIA Tesla C2050 card for the gPMC dose calculations. It has a total of 448 processor cores, each with a clock speed of 1.15 GHz. It is also equipped with a 3 GB GDDR5 memory shared by all processor cores. Such a GPU card is manufactured by NVIDIA for the purpose of scientific computing. It supports error correction codes to protect data from random errors occurring in data transfer and manipulation. As for the CPUs, we execute TOPAS on a single dedicated node on a CPU cluster. The node consists of a few 3 GHz CPU processors with 2 GB of RAM.
3. Results
3.1. Dose distributions
We first present the dose calculation results in the water phantom case. This is of particular interest, as in those cases with materials other than water, all the proton step lengths are first scaled to the equivalent lengths in water using the mass stopping power ratio and the dose depositions are computed in water. Hence, the accuracy of dose calculations in water serves as the foundation for the accuracy in all other cases. To study the water case, we generate a phantom with physical properties identical to pure water, namely ρ = 1.0 g cm−3, ρe = 3.34 × 1023 cm−3 and fs (E) = 1 irrespective of the energy.
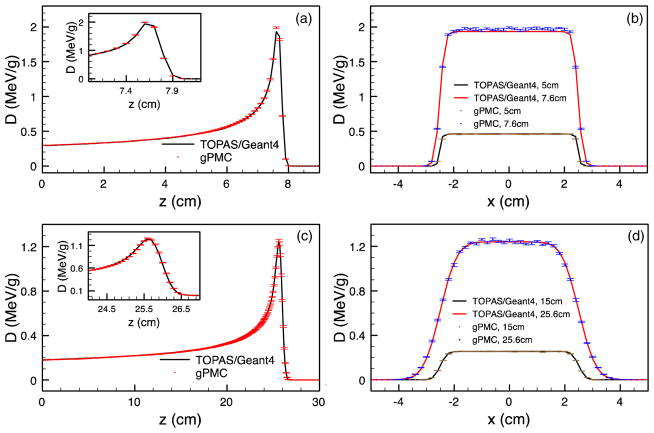
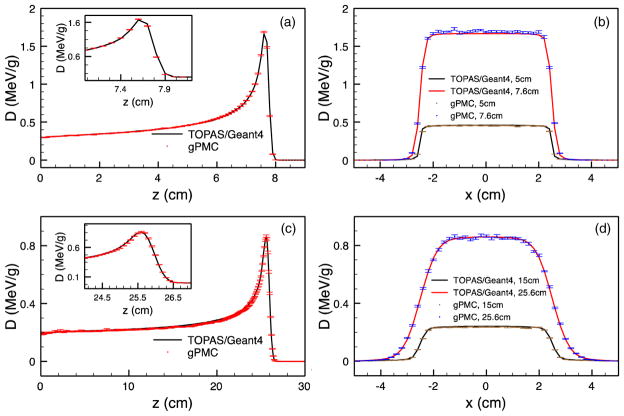
Figure 2 illustrates the dose calculation results in water when only simulating electromagnetic interactions. As proton deposits energy mainly through this channel, it is necessary and desirable to first verify the dose calculation accuracy with only electromagnetic interactions. Figure 3 further demonstrates the dose calculation results with all interactions. In both figures, the top and the bottom rows correspond to the cases with 100 and 200 MeV source protons, while the left and the right columns are the depth dose curves and the lateral profiles, respectively. Between the two lateral profiles presented for each case, one of them is taken at the Bragg peak, while the other is approximately at the half way of the peak depth. The error bars in gPMC results correspond to one standard deviation of the dose and those for the TOPAS results are not drawn for clarity, which are much smaller due to the integration of the pencil-beam results. The dose distributions in the two codes match well, especially the ranges, which can be clearly verified by the zoom-in view of those depth dose curves. A certain number of discrepancies in the peak heights can be seen, especially when comparing the profiles. These can be possibly ascribed to a small difference (less than a voxel) between the peak locations in gPMC and TOPAS results. Plotting profiles at this high dose gradient region exaggerates the difference.
Figure 2.
Depth dose curves (left) and lateral profiles (right) for a water phantom with only electromagnetic interactions. The top and bottom rows are for 100 and 200 MeV sources, respectively. Insets are zoomed-in views of the depth curves near the Bragg peak.
Figure 3.
Depth dose curves (left) and lateral profiles (right) for a water phantom with all interactions. The top and bottom rows are for 100 and 200 MeV sources, respectively. Insets are zoomed-in views of the depth curves near the Bragg peak.
In figure 4 we present the dose calculation results for the cases with other materials. In this figure, we studied the two different phantom materials, namely bone and tissue, and two different source energies of 100 and 200 MeV. In all of these cases, the results from gPMC and TOPAS are in good agreement.
Figure 4.
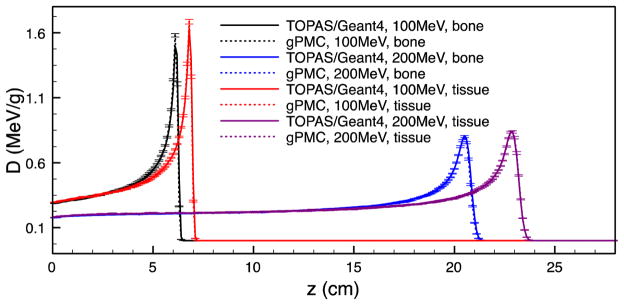
Depth dose curves in a bone phantom and in a tissue phantom for 100 and 200 MeV sources.
Finally, we show the dose calculation results in an inhomogeneous phantom, schematically shown in figure 5(a). Figure 5(b) depicts the depth dose curves on two straight lines that are parallel to the z axis and are through the bone insert and the lung insert, respectively. The two Bragg peaks are located at different depths due to the different insert materials along the beam path. The calculated dose values, in particular the peak locations, given by gPMC agree well with those given by TOPAS. Figure 5(c) presents two lateral dose profiles taken at z = 6.95 cm and z = 10.85 cm, respectively. The calculation results from gPMC and from TOPAS are in excellent agreement.
Additionally, we have studied the dose calculations in a patient case. The results are shown in figure 6. A square monogenetic beam impinges on the patient from the right side. The resulting dose distribution calculated by gPMC at one transverse slice is demonstrated in figure 6(a) in a color wash format, which is further overlaid on the patient CT image. Bragg peaks are clearly observed at the distal end of the beam and the peak locations vary due to the non-flat patient surface, as well as the nasal cavity on the beam path. To demonstrate the accuracy of our calculations, we plot the dose profiles along two straight lines as indicated by the dashed lines in figure 6(a). The result are shown in figures 6(b) and (c). Again, the gPMC and the TOPAS results agree well within statistical uncertainty.
Figure 6.
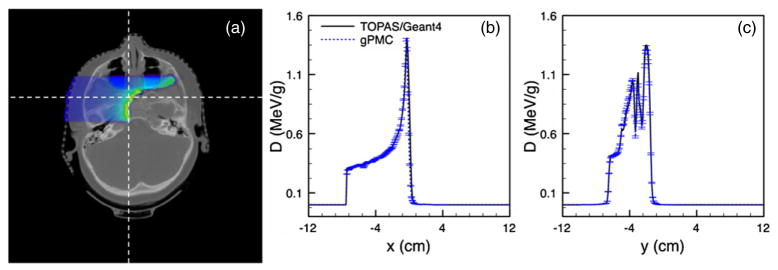
(a) Dose distribution in a patient case. (b), (c) Dose profiles along a horizontal and a vertical line as indicated by the dashed lines in (a).
3.2. Quantitative analysis
First, we quantify the precision of our simulation by calculating the relative uncertainty at each voxel σ/D, where σ is the uncertainty at the voxel estimated by the dose results in all batches in our simulation and D is the dose at the voxel. We further average the relative uncertainty σ/D over a high dose region where the local dose D exceeds 10% of its maximum value Dmax inside the entire phantom. The quantity indicates the achieved simulation precision. The results are summarized in table 1. In all the cases studied, it is found that is less than 1% in gPMC with 107 source protons simulated.
Table 1.
Average relative uncertainty ( ), γ-test passing rates (Pt ) with different criteria and computation time (T) for all different test cases.
| Phantom | Source energy (MeV) | (%) | Pγ (1 mm/1%) (%) | Pγ (2 mm/2%) (%) | T (s) |
|---|---|---|---|---|---|
| Water, EM | 100 | 0.8 | 99.2 | 99.6 | 6.66 |
| 200 | 0.8 | 99.9 | 99.9 | 19.12 | |
| Water | 100 | 0.9 | 99.3 | 99.7 | 6.76 |
| 200 | 1.1 | 97.3 | 99.9 | 21.14 | |
| Bone | 100 | 0.9 | 98.6 | 98.7 | 6.68 |
| 200 | 1.1 | 98.1 | 99.9 | 20.98 | |
| Tissue | 100 | 0.9 | 99.0 | 99.4 | 7.08 |
| 200 | 1.1 | 97.6 | 99.9 | 22.29 | |
| Inhomogeneous phantom | 100 | 0.9 | 99.9 | 99.9 | 9.52 |
| Patient | 100 | 1.0 | 95.1 | 99.9 | 10.08 |
We then measure the agreements between the gPMC result and the TOPAS result using a γ-test. Specifically, for each case, a 3D γ index distribution is calculated using a GPU-based program (Gu et al 2011) and a voxel is said to pass the test, if its γ value is found to be lower than 1. The passing rate Pγ is calculated over the high dose region where D > 10%Dmax to quantify the overall test result, which is simply the quotient of the number of voxels inside the high dose region which pass the test and the total number of voxels inside this region. Low-density regions defined to be the HU number lower than −900 are excluded in the patient case from the test, where the dose discrepancy may be large due to the lack of electron transport. These regions correspond to the nasal cavity in the patient, where dose values are of no clinical importance. In practice, we use two different γ-test criteria, namely 2 mm/2% and 1 mm/1%. The former is a widely used criterion in clinical practice, while the latter is a stricter one that is used in our study to further demonstrate the accuracy of gPMC. In table 1, we list all the passing rates Pγ. We found that Pγ of 2 mm/2% is above 98.7% and for the majority of the testing cases it is above 99%. When using the strict criteria of 1 mm/1%, gPMC results still pass the test with a high rate of Pγ. These numbers clearly demonstrate the achieved accuracy of gPMC in both the phantom and the patient cases. We would like to remark that, although only test cases with 100 or 200 MeV source energies are presented here, we have validated gPMC in other cases with different energies. The agreements are found to be of the same level as those presented here.
3.3. Computation time
Finally we report the computation time in the last column of table 1. It is found that the computation time ranges from 6 to 22 s depending on the case. In particular, the beam energy has dominant impacts on the computation time, as a high-energy proton in general travels longer than a low-energy one, which inevitably requires more computation time. It is also found that phantom complexity impacts the computation efficiency. Note that the time reported here is purely for the particle transport time, which does not contain other components such as loading the data from a hard drive and converting the CT number to the material properties. These extra components add up to approximately 3 s to the computation time depending on the phantom size.
The TOPAS simulations are all conducted on a CPU cluster where, depending on the geometrical complexity and beam energy, between 2 and 80 CPU hours are typically required to complete the simulations. If we were to compare the computation time in gPMC and in TOPAS, enormously large speed-up factors would be concluded. Yet, this comparison is not fair, as TOPAS utilizes much more detailed simulation schemes to handle proton transport and secondary particle generations and transport. It can thus predict properties other than just dose in a voxel. However, for pure proton dose calculation gPMC reaches a satisfactory level of accuracy at a much improved computation efficiency compared to what has been currently achieved by using other general purposed CPU-based MC packages.
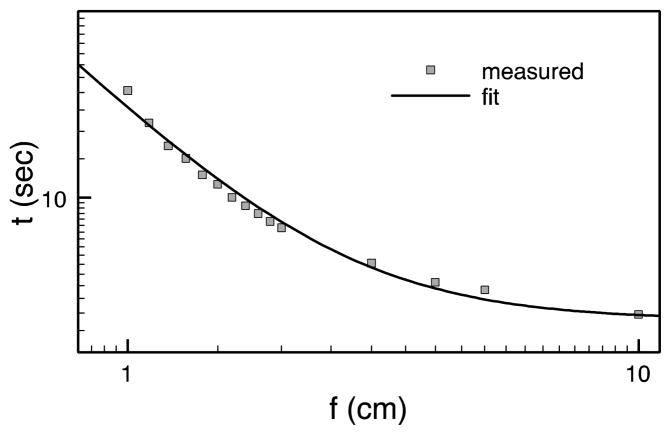
The dose calculation time reported in table 1 are for a square beam of size 5×5 cm2. We would like to point out that the proton dose calculation time in gPMC also depends on the beam size due to a memory conflict problem. In fact, while two GPU threads happen to update the same dose counter, a memory writing conflict occurs and these updates have to be serialized. This serialization apparently counteracts the available parallel processing power of a GPU. A higher frequency of conflict occurrences leads to a lower computational efficiency. Although this memory writing conflict also occurs in photon beam dose calculations (Jia et al 2010a, 2011), it is, however, exacerbated in the context of proton beams. This is because protons travel almost in a straight line, and a parading column of protons in a beam, especially in a small-size beam, marches in almost locked steps, which leads to a high frequency of memory writing conflicts. To test this, we have performed dose calculations in the water phantom with a fixed proton beam energy of 200 MeV but of different square field size f. We have to first turn off the dose deposition and obtain the pure particle transport time, denoted as t0. For each field size, we record the dose calculation time including the dose deposition ttot and calculate the time for dose deposition only as t = ttot − t0. The dependence of t on the field size f is shown in figure 7. As the field size decrease, the deposition time increases dramatically. In particular, when it comes to the cases with a field size of 1 cm or less, the dose deposition time is comparable to or even more than the particle transport time.
Figure 7.
Dose deposition time as a function of the field size.
To further understand this effect, let us denote the dose deposition time per event as Δt and suppose there are N events occurring in the simulation. Among them, a portion of p < 1 encounters the memory conflict and the depositions are serialized, which leads to a time of NpΔt. The rest of (1 − p) are deposited in parallel by all GPU threads, and hence the time is NΔt (1− p)/Nthread. As a simple argument, the probability of this memory conflict is inversely proportional to the field size f2, namely p ≈ α/f2. Hence, the total dose deposition time is
| (3) |
A simple fit of the data in this form leads to the solid curve in figure 7. The successful data fit validates this argument.
4. Discussion and conclusions
We have successfully developed a proton MC dose calculation code, gPMC. It supports proton transport in the energy range of 0.5–300 MeV. Proton transport is modeled by the class II condensed history simulation scheme with a continuous slowing down approximation. Ionization, elastic and inelastic proton nucleus interactions are considered. Energy straggling and multiple scattering are modeled. gPMC is developed on a GPU architecture under the NVIDIA CUDA platform to achieve a high computational efficiency. Simulations in various phantom cases and in a patient case indicate that gPMC leads to dose calculation results that are in good agreement with that of TOPAS, which is based on the popular Geant4 code. We have conducted a γ-test to quantify the agreement. It is found that gPMC achieves a passing rate of over 98% with a 2 mm/2% criterion and a passing rate of over 95% with a 1 mm/1% criterion. With a powerful yet affordable NVIDIA Tesla C2050 GPU, the simulation time of 107 source protons ranges from 6 to 22 s depending on the source proton energy and the phantom complexity, corresponding to relative statistical uncertainties around 1%.
Despite the success, there are a few issues to be addressed in future studies. First, the memory writing conflict in dose deposition highly limits the dose calculation efficiency, especially when it comes to a small field size. There are a few approaches potentially alleviating this issue. For example, one could allocate more than one dose counters, and assign each counter to a subset of all GPU threads. Each thread deposits dose to its own counter during the simulation and only at the end of the dose calculation will the dose results be accumulated. While this method removes the memory conflict to a certain extent, it adds an overhead of dose addition. This strategy, and other potential solutions to this issue, will be our future research topic. On the other hand, in real clinical contexts, the problem may not be as severe as it looks like. First of all, we usually calculate the dose distribution from a broad beam of a few centimeters in size, where the memory conflict problem is not quite severe, as indicated in figure 7. Even in intensity modulated proton therapy where dose distributions from pencil beams are needed for treatment plan optimization, it is still possible to calculate doses for a few pencil beams simultaneously and store them in different counters. This also effectively reduces the probability of memory conflict.
Moreover, it may be necessary to further improve the accuracy of gPMC in low-density regions in some clinical cases. For the current gPMC version, although the dose in lung with −700 HU is found to be acceptable, we do have observed discrepancies in some cases with even lower HU values due to the lack of electron transport. Since the dose in lung is important and should be calculated accurately, electron transport should be included in gPMC when necessary. In fact, such a module is already available in our previously developed dose calculation package for photons, gDPM (Jia et al 2010a, 2011). By integrating that module into gPMC, it is expected that the dose calculation accuracy in the extremely low-density area will be improved. It is also our research topic to assess the necessity of including this electron transport module for real clinical problems.
Another context requiring further improvement in accuracy is when a high-Z material presents. In fact, we have performed dose calculations in gold, but the results do not agree with TOPAS/Geant4. The discrepancy in the high-Z materials can be mainly ascribed to that only nuclear interactions with H and O are considered in the current gPMC. To have an accurate dose calculation result in high-Z materials, the nuclear interactions will have to be modified. As a proton dose calculation package for proton therapy, the 25 materials included currently should be sufficient for most clinical cases. Those high-Z materials will be supported in the future release of gPMC.
For dose calculations in real clinical cases, a phase space file is usually used to provide information regarding the source particles. Currently, gPMC does not support the function of loading particles from a phase space file. But a module for this purpose is under development. One potential issue in this approach is that the extra computation time due to loading particles may not be negligible because of the already very short simulation time achieved so far and the usually very large size of a phase space file. It hence requires further investigations to quantify this computational burden and the practicality of this method.
Acknowledgments
This work is supported in part by the University of California Lab Fees Research Program and in part by R01 CA140735 (‘PBeam: Fast and Easy Monte Carlo System for Proton Therapy’).
Contributor Information
Xun Jia, Email: xunjia@ucsd.edu.
Jan Schümann, Email: jschuemann@partners.org.
Harald Paganetti, Email: hpaganetti@partners.org.
Steve B Jiang, Email: sbjiang@ucsd.edu.
References
- Agostinelli S, et al. GEANT4—a simulation toolkit. Nucl Instrum Methods Phys Res. 2003;506:250–303. [Google Scholar]
- Badal A, Badano A. Accelerating Monte Carlo simulations of photon transport in a voxelized geometry using a massively parallel graphics processing unit. Med Phys. 2009;36:4878–80. doi: 10.1118/1.3231824. [DOI] [PubMed] [Google Scholar]
- Fippel M, Soukup M. A Monte Carlo dose calculation algorithm for proton therapy. Med Phys. 2004;31:2263–73. doi: 10.1118/1.1769631. [DOI] [PubMed] [Google Scholar]
- Geant4 Collaboration. Geant4 Physics Reference Manual Journal. 2011 http://geant4.web.cern.ch/geant4/UserDocumentation/UsersGuides/PhysicsReferenceManual/fo/PhysicsReferenceManual.pdf.
- Gu X, et al. GPU-based ultra fast dose calculation using a finite size pencil beam model. Phys Med Biol. 2009;54:6287–97. doi: 10.1088/0031-9155/54/20/017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu XJ, Jia X, Jiang SB. GPU-based fast gamma index calculation. Phys Med Biol. 2011;56:1431–41. doi: 10.1088/0031-9155/56/5/014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, et al. Implementation and evaluation of various demons deformable image registration algorithms on a GPU. Phys Med Biol. 2010;55:207–19. doi: 10.1088/0031-9155/55/1/012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara K, et al. Review of particle physics. Phys Rev D. 2002;66:010001. [Google Scholar]
- Hissoiny S, Ozell B, Bouchard H, Despres P. GPUMCD: a new GPU-oriented Monte Carlo dose calculation platform. Med Phys. 2011;38:754–64. doi: 10.1118/1.3539725. [DOI] [PubMed] [Google Scholar]
- Hissoiny S, Ozell B, Després P. Fast convolution-superposition dose calculation on graphics hardware. Med Phys. 2009;36:1998–2005. doi: 10.1118/1.3120286. [DOI] [PubMed] [Google Scholar]
- Hong L, et al. A pencil beam algorithm for proton dose calculations. Phys Med Biol. 1996;41:1305–30. doi: 10.1088/0031-9155/41/8/005. [DOI] [PubMed] [Google Scholar]
- Jacques R, Taylor R, Wong J, McNutt T. Towards real-time radiation therapy: GPU accelerated superposition/convolution. Comput Methods Programs Biomed. 2010;98:285–92. doi: 10.1016/j.cmpb.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Jahnke L, Fleckenstein J, Wenz F, Hesser J. GMC: a GPU implementation of a Monte Carlo dose calculation based on Geant4. Phys Med Biol. 2012;57:1217–29. doi: 10.1088/0031-9155/57/5/1217. [DOI] [PubMed] [Google Scholar]
- Jia X, Gu X, Graves YJ, Folkerts M, Jiang SB. GPU-based fast Monte Carlo simulation for radiotherapy dose calculation. Phys Med Biol. 2011;56:7017–31. doi: 10.1088/0031-9155/56/22/002. [DOI] [PubMed] [Google Scholar]
- Jia X, et al. Development of a GPU-based Monte Carlo dose calculation code for coupled electron–photon transport. Phys Med Biol. 2010a;55:3077–86. doi: 10.1088/0031-9155/55/11/006. [DOI] [PubMed] [Google Scholar]
- Jia X, Lou Y, Li R, Song WY, Jiang SB. GPU-based fast cone beam CT reconstruction from undersampled and noisy projection data via total variation. Med Phys. 2010b;37:1757–60. doi: 10.1118/1.3371691. [DOI] [PubMed] [Google Scholar]
- Jia X, Yan H, Gu X, Jiang SB. Fast Monte Carlo simulation for patient-specific CT/CBCT imaging dose calculation. Phys Med Biol. 2012;57:577–90. doi: 10.1088/0031-9155/57/3/577. [DOI] [PubMed] [Google Scholar]
- Kawrakow I. Accurate condensed history Monte Carlo simulation of electron transport: I. EGSnrc, the new EGS4 version. Med Phys. 2000;27:485–98. doi: 10.1118/1.598917. [DOI] [PubMed] [Google Scholar]
- Kohno R, et al. Clinical implementation of a GPU-based simplified Monte Carlo method for a treatment planning system of proton beam therapy. Phys Med Biol. 2011;56:N287. doi: 10.1088/0031-9155/56/22/N03. [DOI] [PubMed] [Google Scholar]
- Kohno R, et al. Experimental evaluation of validity of simplified Monte Carlo method in proton dose calculations. Phys Med Biol. 2003;48:1277–88. doi: 10.1088/0031-9155/48/10/303. [DOI] [PubMed] [Google Scholar]
- L’Ecuyer P, Simard R. TestU01: a C library for empirical testing of random number generators. ACM Trans Math Softw. 2007;33:22. [Google Scholar]
- Lensch H, Strzodka R. Massively parallel computing with CUDA. 2008 http://www.mpi-inf.mpg.de/~strzodka/lectures/ParCo08/
- Li JS, Shahine B, Fourkal E, Ma CM. A particle track-repeating algorithm for proton beam dose calculation. Phys Med Biol. 2005;50:1001–10. doi: 10.1088/0031-9155/50/5/022. [DOI] [PubMed] [Google Scholar]
- Low DA, Harms WB, Mutic S, Purdy JA. A technique for the quantitative evaluation of dose distributions. Med Phys. 1998;25:656–61. doi: 10.1118/1.598248. [DOI] [PubMed] [Google Scholar]
- Marsaglia G. Xorshift RNGs. J Stat Softw. 2003;8(14) available at http://www.jstatsoft.org/v08/i14. [Google Scholar]
- Men C, et al. GPU-based ultra fast IMRT plan optimization. Phys Med Biol. 2009;54:6565–73. doi: 10.1088/0031-9155/54/21/008. [DOI] [PubMed] [Google Scholar]
- Men CH, Jia X, Jiang SB. GPU-based ultra-fast direct aperture optimization for online adaptive radiation therapy. Phys Med Biol. 2010a;55:4309–19. doi: 10.1088/0031-9155/55/15/008. [DOI] [PubMed] [Google Scholar]
- Men CH, Romeijn HE, Jia X, Jiang SB. Ultrafast treatment plan optimization for volumetric modulated arc therapy (VMAT) Med Phys. 2010b;37:5787–91. doi: 10.1118/1.3491675. [DOI] [PubMed] [Google Scholar]
- NVIDIA. CUDA CURAND Library. 2010 (available at http://docs.nvidia.com/curand/index.html)
- NVIDIA. NVIDIA CUDA Compute Unified Device Architecture, Programming Guide, 4.0. 2011 (available from http://developer.download.nvidia.com/compute/DevZone/docs/html/C/doc/CUDA_C_Programming_Guide.pdf)
- Paganetti H. Range uncertainties in proton therapy and the role of Monte Carlo simulations. Phys Med Biol. 2012;57:R99–117. doi: 10.1088/0031-9155/57/11/R99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paganetti H, Jiang H, Parodi K, Slopsema R, Engelsman M. Clinical implementation of full Monte Carlo dose calculation in proton beam therapy. Phys Med Biol. 2008;53:4825–53. doi: 10.1088/0031-9155/53/17/023. [DOI] [PubMed] [Google Scholar]
- Perl J, Shin J, Schumann J, Faddegon B, Paganetti H. TOPAS: an innovative proton Monte Carlo platform for research and clinical applications. Med Phys. 2012 doi: 10.1118/1.4758060. at press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratx G, Xing L. GPU computing in medical physics: a review. Med Phys. 2011;38:2685–97. doi: 10.1118/1.3578605. [DOI] [PubMed] [Google Scholar]
- Salvat F, Fernández-Varea JM, Sempau J. PENELOPE-2008: A Code System for Monte Carlo Simulation of Electron and Photon Transport. Issy-les-Moulineaux, France: OECD-NEA; 2009. [Google Scholar]
- Samant SS, Xia JY, Muyan-Ozcelilk P, Owens JD. High performance computing for deformable image registration: towards a new paradigm in adaptive radiotherapy. Med Phys. 2008;35:3546–53. doi: 10.1118/1.2948318. [DOI] [PubMed] [Google Scholar]
- Schaffner B, Pedroni E, Lomax A. Dose calculation models for proton treatment planning using a dynamic beam delivery system: an attempt to include density heterogeneity effects in the analytical dose calculation. Phys Med Biol. 1999;44:27–41. doi: 10.1088/0031-9155/44/1/004. [DOI] [PubMed] [Google Scholar]
- Schneider W, Bortfeld T, Schlegel W. Correlation between CT numbers and tissue parameters needed for Monte Carlo simulations of clinical dose distributions. Phys Med Biol. 2000;45:459–78. doi: 10.1088/0031-9155/45/2/314. [DOI] [PubMed] [Google Scholar]
- Sharp GC, Kandasamy N, Singh H, Folkert M. GPU-based streaming architectures for fast cone-beam CT image reconstruction and demons deformable registration. Phys Med Biol. 2007;52:5771–83. doi: 10.1088/0031-9155/52/19/003. [DOI] [PubMed] [Google Scholar]
- Xu F, Mueller K. Accelerating popular tomographic reconstruction algorithms on commodity PC graphics hardware. IEEE Trans Nucl Sci. 2005;52:654–63. [Google Scholar]
- Yan GR, Tian J, Zhu SP, Dai YK, Qin CH. Fast cone-beam CT image reconstruction using GPU hardware. J X-Ray Sci Technol. 2008;16:225–34. [Google Scholar]
- Yepes PP, Mirkovic D, Taddei PJ. A GPU implementation of a track-repeating algorithm for proton radiotherapy dose calculations. Phys Med Biol. 2010;55:7107–20. doi: 10.1088/0031-9155/55/23/S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yepes P, Randeniya S, Taddei PJ, Newhauser WD. Monte Carlo fast dose calculator for proton radiotherapy: application to a voxelized geometry representing a patient with prostate cancer. Phys Med Biol. 2009;54:N21–8. doi: 10.1088/0031-9155/54/1/N03. [DOI] [PMC free article] [PubMed] [Google Scholar]