Abstract
Importance
The PIK3CA mutation is one of the most common mutations in Head and Neck Squamous Cell Carcinoma (HNSCC). Through this research we attempt to elicit the role of oncogene dependence and effects of targeted therapy on this PIK3CA mutation.
Objectives
1) To determine the role of oncogene dependence on one of the more common and targetable oncogenes in HNSCC – PIK3CA; 2) To evaluate the consequence of this oncogene on the effectiveness of newly developed targeted therapies.
Study Design
In vitro study.
Setting
Academic research laboratory.
Participants
Cell culture based study assessing the viability of PIK3CA mutated head and neck cell lines when treated with targeted therapy.
Exposures
PIK3CA mutated head and neck cell lines were treated with 17-AAG, GDC-0941, trametinib, and BEZ-235.
Main Outcome and Measures
Assessment of cell viability of HNSCC cell lines characterized for PIK3CA mutations or SCC25 cells engineered to express the PIK3CA hotspot mutations E545K or H1047R
Results
Surprisingly, in engineered cell lines, the hotspot E545K and H1047R mutations conferred decreased, rather than increased, sensitivity as measured by IC50 when treated with the respective HSP90, PI3K, and MEK inhibitors, 17-AAG, GDC-0941, and trametinib, compared to the SCC25 control cell lines. When treated with BEZ-235, H1047R-expressing cell lines showed increased sensitivity to inhibition compared to control while those expressing E545K showed slightly increased sensitivity of unclear significance.
Conclusions and Relevance
1) The PIK3CA mutations within our engineered cell model did not lead to enhanced oncogene-dependent cell death when treated with direct inhibition of the PI3K enzyme yet did show increased sensitivity compared to control with dual PI3K/mTOR inhibition. 2) Oncogene addiction to PIK3CA hot spot mutations, if it occurs, is likely to evolve in vivo molecular changes that remain to be identified. Additional study is required to develop new model systems and approaches to determine the role of targeted therapy in the treatment of PI3K-overactive HNSCC tumors.
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common cancer by incidence worldwide.1,2 Approximately two-thirds of patients present with advanced disease and undergo primary surgical treatment with adjuvant radiation/chemoradiation or primary chemoradiation therapy with salvage surgical treatment. Although some advances in therapy have occurred, survival has not markedly improved in recent decades due to locoregional recurrences, distant metastases and second primary tumors.2 The only targeted therapy that is FDA approved for current use in HNSCC is cetuximab, a monoclonal antibody recognizing the epidermal growth factor receptor (EGFR), which gives a modest improvement in survival when combined with chemotherapy.3 Identification of novel targeted therapies for patients would make a large impact on disease that does not respond to traditional therapies.
To date, most targeted therapies are effective in patients that harbor a mutation or other specific genetic alteration in an oncogenic driver. Frequently occurring genetic mutations in HNSCC include: TP53, CDKN2A, CASP8, FAT1, NOTCH1, PTEN, SYNE1, HRAS, and PIK3CA.4,5 Of these, most are inactivating mutations or are otherwise difficult to target. Notably, however, two genetic events target the oncogenic phosphatidylinositol 3-kinase (PI3K) pathway: PTEN and PIK3CA. PTEN is a tumor suppressor that, when inactivated, leads to increased signaling through the PI3K pathway. PIK3CA is the catalytic subunit of Class IA PI3K enzymes and hotspot mutations in this gene lead to PI3K overactivity. When evaluating all subtypes of HNSCC, PTEN mutations occur in up to 7% of tumors and PIK3CA mutations occur in 8-13% of tumors.4,6 Furthermore, Lui et al. has reported that 30.5% of HNSCC tumors carry mutations in some portion of the PI3K pathway.6
The phosphatidylinositol 3-kinase (PI3K) pathway, when constitutively activated, is most commonly mutated through the PIK3CA gene. This gene encodes the catalytic subunit p110α, which has been shown to be one of the most commonly mutated oncogenes in multiple human malignancies.7 These mutations have been shown to be clustered in exon 9 and exon 20 corresponding to the helical domain mutant E545K and the kinase domain mutant H1047R respectively.7 These mutations have been shown to not only occur in up to 20% of HPV-positive oropharyngeal carcinomas but also are associated with advanced stage, vascular invasion, lymph node metastasis, cancer recurrence, and poor prognosis.8-10
Oncogene addiction is the apparent dependence of some cancers on one or a few genes, promoting continued cell proliferation and maintenance of the malignant phenotype.11 Oncogene-addicted cancer cells are more sensitive to their inhibition than normal tissue. With the development of targeted therapy, oncogene addicted cancers and the oncogenes they depend on are prime targets, given the favorable therapeutic index between cancer cell and normal cell susceptibility. However, to date, only a few oncogene-addicted cancers have been identified and successfully targeted. These cancers include chronic myelogenous leukemia, B-Raf-mutated melanoma, and EGFR-mutated non-small cell lung cancers.12 Our goal in this study is to evaluate the role of one of the more common and targetable oncogenes in HNSCC, PIK3CA, in oncogene dependence and the effectiveness of newly developed targeted therapies.
Materials and Methods
Sanger and CCLE data
All available cell line data from the Sanger and CCLE databases13,14 were used to compare cell lines that contained PI3K or PTEN mutations to those cell lines that contained neither of these mutations. Published IC50 data for GDC-0941 and 17-AAG were plotted.
Cell Lines and Engineering
We utilized the HNSCC cell lines – SCC25, SCC61, FaDu, HSC-2, CAL-33, and Detroit 562. PIK3CA genes WT, H1047R and E545K were PCR-cloned into pENTR and recombined into pLenti6/V5 as previously described into the SCC25 cell line.15 Lentiviral transduction was used to stably express the PIK3CA genes within the SCC25 cell line. We used commercially available antibodies for V5 (Invitrogen/Life Technologies) and actin (Sigma).
Cell Culture and Drug Treatment
Cells were grown in 10% or 20% Fetal Bovine Serum in DMEM/F-12. For IC50 assays, cells were plated in a 96 well plate with a density of 2,500 cells per well in 200 microliters of the same media. After twenty-four hours, the media was changed and serial dilutions of 17-AAG, GDC-0941, trametinib, and BEZ-235 were added. The cells were next incubated for 96 hours.
Cell Viability
Calcein, Hoechst, and propidium iodide were applied to the cell culture at a concentration of 2 μM, 5 μg/mL, and 2 μM respectively followed by incubation for thirty minutes. The cells were imaged using an automated Cellavista© system (Synentec). The cells that were fluorescent with both Calcein and Hoechst were determined as viable while those that fluoresced with propidium iodide were determined as non-viable. Proper thresholding of cells by the software was confirmed by visual inspection. Statistics and IC50 determinations
The Sanger14 and CCLE13 IC50 data were plotted using Box and Whisker plots including the 5th and 95th percentile using GraphPad. Statistical significance was then determined using the Mann-Whitney test with a value of p<.05. For analysis of experimental data, IC50 values were determined by plotting the log of the cell viability versus the log of the drug concentration. This linear graph was next solved for the point at which fifty percent of the cells were viable and fifty percent were non-viable. The inverse log of this drug concentration was calculated. Viability at each data point was calculated as the average number (from three wells on the same plate) of living cells treated with a certain concentration of drug divided by the average number (from three wells on the same plate) of living cells that received no treatment. GraphPad was used to display the data using the mean and standard error of the mean.
Results
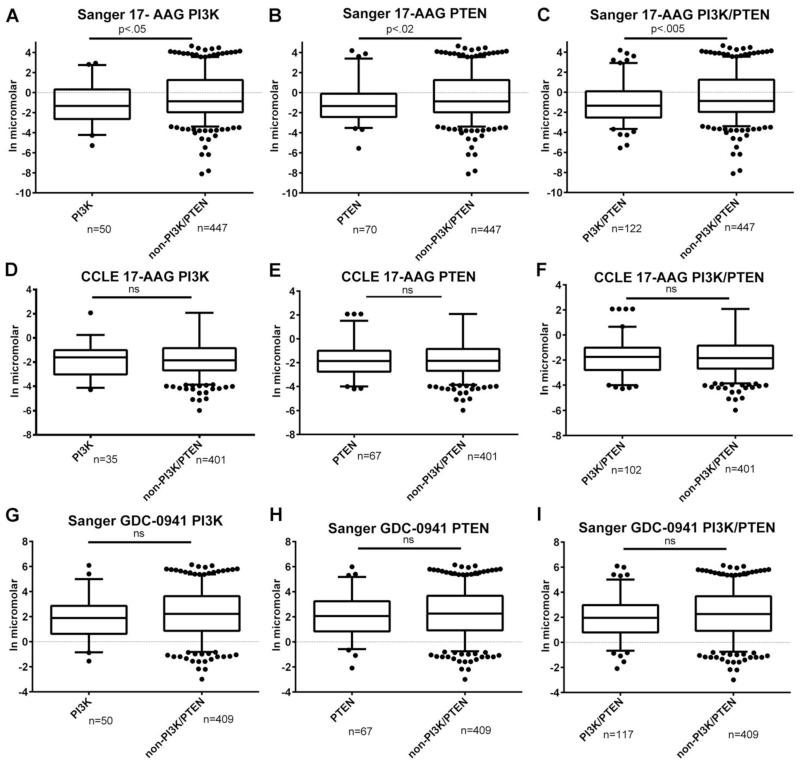
The goal of our study was to investigate whether the commonly mutated PI3K pathway confers oncogene dependence or alters the response to targeted therapies in HNSCC. We began by analyzing publicly available drug response data from two recent studies13,14 that molecularly characterized a large number of cell lines across a diverse set of cancer types. In order to achieve an adequate sample size, it was necessary to include all cancer cell types. IC50 proliferation response data from the Sanger database14 (Figures 1A-C and 1G-I) were analyzed comparing the response of cell lines that contained a mutation in either the catalytic subunit of class Ia PI3K (PIK3CA) or in the opposing enzyme PTEN to those that did not contain these mutations. Only the data from Garnett et al.14, from the Sanger center, included drug data from treatments with a direct PI3K inhibitor, the selective class I PI3K inhibitor GDC-0941. Using the Mann-Whitney test, we found no statistically significant difference in the IC50 response to GDC-0941 between those containing PI3K or PTEN mutations and non-mutated cells (Figure 1G-I), although there was a trend towards greater sensitivity. We also found a trend towards greater sensitivity when comparing solely the PI3K mutated cells with the Mann-Whitney test that were treated with GDC-0941 compared to non-mutated cells but statistical significance was not present. Of note, only 22 of the 526 cell lines tested were HNSCC cells. Of these, three had PI3K mutations and zero had PTEN mutations.
Figure 1. Analysis of drug response data to 17-AAG or GDC-0941 from the Sanger14 and CCLE13 datasets.
Box and Whisker Plots displaying published drug response data A) Sanger data comparing cell lines with PI3K mutations compared to non-PI3K/PTEN mutations when treated with 17-AAG B) Sanger data comparing cell lines with PTEN mutations compared to non-PI3K/PTEN mutations when treated with 17-AAG C) Sanger data comparing cell lines with PI3K or PTEN mutations compared to non-PI3K/PTEN mutations when treated with 17-AAG D) CCLE data comparing cell lines with PI3K mutations compared to non-PI3K/PTEN mutations when treated with 17-AAG E) CCLE data comparing cell lines with PTEN mutations compared to non-PI3K/PTEN mutations when treated with 17-AAG F) CCLE data comparing cell lines with PI3K or PTEN mutations compared to non-PI3K/PTEN mutations when treated with 17-AAG G) Sanger data comparing cell lines with PI3K mutations compared to non-PI3K/PTEN mutations when treated with GDC-0941 H) Sanger data comparing cell lines with PTEN mutations compared to non-PI3K/PTEN mutations when treated with GDC-0941 I) Sanger data comparing cell lines with PI3K or PTEN mutations compared to non-PI3K/PTEN mutations when treated with GDC-0941. Number of cell lines in each group indicated on graph. Statistical significance expressed in p-values on graphs. ns = not significant.
Another molecule that might modify PI3K signaling is HSP90. HSP90 is a chaperone that controls the levels of many oncogenic proteins, including the downstream target of PI3K, the kinase Akt.13,14,16 Both the Barretina et al.13 and Garnett14 et al. studies tested the response of cancer cell lines to the HSP90 inhibitor 17-AAG.13,14 Our analysis of the Garnett et al.14 study indicates that the presence of PI3K or PTEN mutations is associated with greater sensitivity to 17-AAG (Fig 1A-B). Cell lines with PI3K or PTEN mutations analyzed together further increased the statistical significance of the difference between the response of non-mutated and mutated cell line IC50 response (Fig 1C). However, our analysis of data from Barretina et al.13 yielded no difference in response to 17-AAG (Fig 1D-1F). The reasons for these differences are unclear but could include cancer cell types tested or testing conditions.
The Barretina et al.13 and Garnett et al.14 genomic studies included some HNSCC cell lines, but most of them were not tested for drug responsiveness. To investigate whether the presence of PI3K pathway activating mutations might confer sensitivity to targeted therapies in HNSCC, we tested a panel of HPV-negative HNSCC cell lines with 17-AAG of which FaDu and SCC25 parental were the only cell lines that did not carry PI3K mutations. These data yielded IC50 results of 3 nM (FaDu), 10 nM (SCC25), 22 nM (Detroit 562), 13 nM (HSC-2), 50 nM (CAL-33), and 31 nM (SCC-61). In these cell lines, there was no clear difference to treatment with 17-AAG although there appeared to be a trend towards increased resistance of those cells with PI3K mutations compared to those cells that did not contain a PI3K mutation (Figure 2A). These same cell lines were treated with GDC-0941 yielding IC50 results of 86 nM (SCC25), 36 nM (Detroit 562), 75 (FaDu), 231 nM (SCC-61), 111 nM (HSC-2), and 158 nM (CAL-33). There was no clear difference in sensitivity to GDC-0941 between cell lines that contained a PI3K mutation compared to those that did not (Figure 2B).
Figure 2. Cell viability of HNSCC cell lines treated with 17-AAG, GDC-0941 or BEZ-235.
Cell viability data for HPV-negative HNSCC cell lines when treated with A) 17-AAG (nM), B) GDC-0941 (nM), or C) BEZ-235 (nM). Mean cell viability plotted with error bars denoting standard error of the mean. Stars denote cell lines containing PIK3CA hotspot mutations (E545K or H1047R). A) Experiments were performed with triplicate measurements and, with four independent experiments except the 200 nM concentration of SCC25 parental was performed in triplicate measurements with three independent experiments; 5 nM concentration of Detroit 562 was performed in triplicate measurements in two independent experiments; 50, 100, and 200 nM concentrations of CAL-33; 1, 5, and 10 nM concentrations of SCC-61; 1 and 10 nM concentrations of HSC-2 were performed in triplicate measurements with one independent experiment. B) Experiments were performed with triplicate measurements and, with three independent experiments except for the HSC-2 and CAL-33 cell lines, which were performed in, triplicate measurements and two independent experiments. C) Experiments were performed with triplicate measurements and, with three independent experiments except for CAL-33 that was performed in two independent experiments.
The Lui et al study6 evaluated HNSCC cell lines that contained PIK3CA hotspot mutations or WT expression and their sensitivity to BEZ-235, a combined PI3K/mTOR inhibitor, and found those cell lines with PIK3CA mutations were more sensitive to PI3K/mTOR inhibition compared to those with WT mutations. We evaluated the response of a panel of HPV-negative cell lines – SCC25, FaDu, Detroit 562, HSC-2, and CAL-33 – of which SCC25 and FaDu were the only cell lines that did not carry PI3K mutations to BEZ-235. These data yielded IC50 results of 31 nM (SCC25), 24 nM (FaDu), 5 nM (Detroit 562), 2 nM (HSC-2), and 34 nM (Cal-33). Since two out of the three cell lines with hotspot PIK3CA mutations were more sensitive than control cells to BEZ-235, these data suggest a potential increased sensitivity to BEZ-235 in cell lines carrying PIK3CA mutations (Figure 2C).
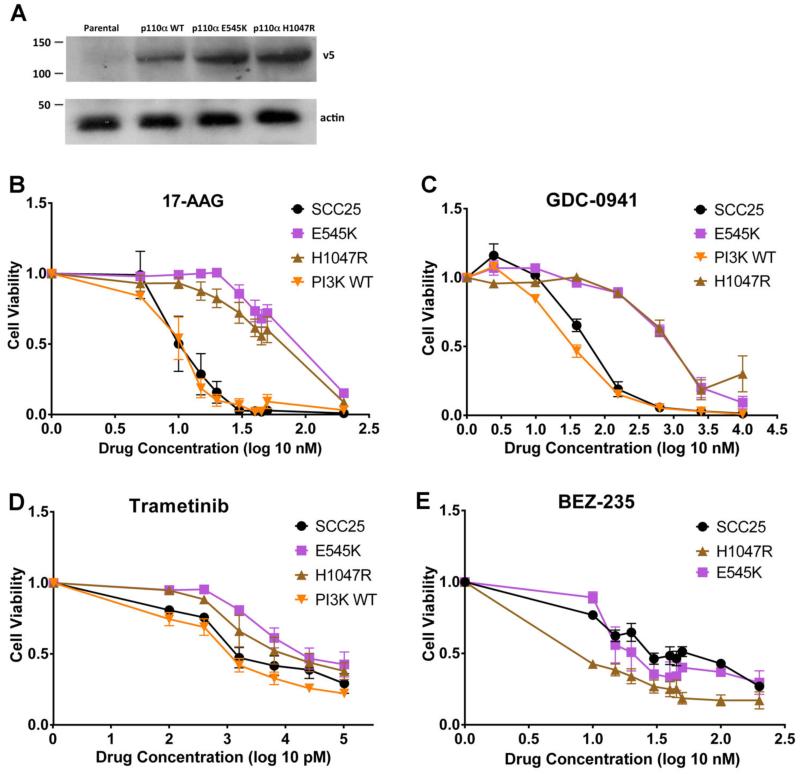
Due to the variability of drug response in PI3K-mutated and non-mutated cells in our panel, we sought to create a cleaner system in which the only difference between the cell lines was the presence of PI3K mutations. Therefore, we used lentiviral transduction to express wild-type PI3K or PI3K hotspot mutants E545K or H1047R in SCC25 HNSCC cells (Figure 3A). The resulting cell lines were then tested for response to candidate-targeted therapies by growing them for 96 hours in the presence of the selected drugs.
Figure 3. Response of PIK3CA-engineered SCC25 cell lines to 17-AAG, GDC-0941, Trametinib, or BEZ-235.
A) Western blot showing expression of the V5-tagged PIK3CA protein in parental, p110α wild type (WT)-, p110a E545K-, and p110a H1047R-expressing SCC25 cell lines (V5 probe, as indicated). Actin was used as a oading control. (B-E) Cell viability plots of SCC25 Parental, SCC25 wild-type, and PIK3CA hotspot mutants (E545K and H1047R) when treated with B) 17-AAG, C) GDC-0941, D) Trametinib, and E) BEZ-235. Mean cell viability plotted with error bars denoting standard error of the mean. Figure 3B was performed in triplicate measurements and with four independent experiments except for PI3K WT being performed in two independent experiments for the concentrations of 5, 10, and 200 nM. Figure 3C was performed in triplicate measurements and with three independent experiments except for SCC25 Parental being performed in triplicate measurements with four independent experiments for all concentrations. Figure 3D was performed in triplicate measurements and with three independent experiments except for the 400 nM concentration was performed with triplicate measurements and one independent experiment for all cell lines. Figure 3E was performed in triplicate measurements and with three independent experiments except for the SCC25 parental 200 nM concentration, which was only performed with triplicate measurements, and one independent experiment.
Similar to our results in the unmanipulated HNSCC cell lines (Figure 2A), E545K- and H1047R-expressing cells were found to have decreased sensitivity to 17-AAG compared to the SCC25 parental and wild-type cell lines (Figure 3B). Thus, E545K- and H1047R-expressing SCC25 cells had an IC50 of 83 nM and 53 nM respectively while the SCC25 wild-type PI3K-expressing and parental cell lines had an IC50 of 9 nM and 12 nM respectively. In contrast to our results with unmanipulated cell lines (Figure 2B), treatment with GDC-0941 also revealed a relative resistance in PI3K-mutant carrying cells (Figure 3C). Thus, E545K- and H1047R-SCC25 cells had IC50 values of 935 nM and 1316 nM respectively while the SCC25 parental and wild-type cell lines had IC50 values of 44 nM and 38 nM respectively.
Because of its use in clinical trials and potential for interfacing cell survival pathways,17 we also tested the response of our engineered cell lines to trametinib, which is a selective inhibitor of MEK 1/MEK2 activation and kinase activity. Similar to the trends with 17-AAG and GDC-0941, E545K- and H1047R-SCC25 cells exhibited decreased sensitivity to trametinib (Figure 3D). However, the differences were small, with respective IC50 values of 39 nM and 29 nM while the SCC25 parental and wild type cell lines had IC50 values of 7 nM and 2 nM respectively.
We next tested the response of our engineered cell lines to combined PI3K/mTOR inhibition using BEZ-235. Similar to the trends we had seen in our unmanipulated cell line panel (Figure 2C), expression of the H1047R hot spot mutation led to increased sensitivity compared to the SCC25 parental cells with respective IC50 values of 6 nM and 31 nM respectively (Figure 3E). By contrast, expression of the E545K mutation led to a very small increase in sensitivity of unclear significance (27 nM compared to 31 nM for control). This may be due to the decreased activity of E545K compared to H1047R.18
Discussion
In this study, we investigated the role of PI3K pathway mutations in mediating sensitivity to targeted therapies. Analysis of publically available data from large scale cancer cell line studies13,14 yielded a mixed result, with sensitivity of PI3K and PTEN mutant cells to the HSP90 inhibitor 17-AAG in one study but not in another. Only one study tested a class I PI3K inhibitor and the results suggested sensitivity but were not statistically significant. In a small-scale panel of genetically unrelated HNSCC cell lines, the presence of PI3K mutations also yielded an unclear result, with some cell lines more sensitive than others to PI3K, HSP90, and combined PI3K/mTOR inhibitors. Finally, we created a set of isogenically related HNSCC cell lines and tested them for sensitivity to PI3K, HSP90, MEK inhibitors, and combination PI3K/mTOR inhibitors. Surprisingly, in the latter system, PI3K mutations conferred decreased sensitivity to the class I PI3K inhibitor GDC-0941, the HSP-90 inhibitor 17-AAG, and the MEK inhibitor trametinib. However, the H1047R-expressing cell line exhibited increased responsiveness to the dual PI3K/mTOR inhibitor BEZ-235. These data suggest that PI3K mutations do not universally confer oncogene dependence, at least as measured by IC50. They also suggest that more sophisticated model systems are needed to understand the role of PI3K mutations in targeted therapy of HNSCC.
Our data do provide further support for the findings of Lui et al6 that cells expressing PI3K mutations may exhibit increased sensitivity to dual PI3K/mTOR inhibition. However, our data suggest that it is not universally applicable to all tumors and may not be applicable to all hotspot mutations. Nonetheless, our findings suggest that oncogene dependence in cells expressing PI3K mutations may be due to dependence on mTOR survival signaling and that proper targeting may depend on shutting down that pathway.
Another finding that was similar between the unmanipulated HNSCC cell lines and genetically engineered ones was relative resistance of PI3K mutant-carrying cell lines to 17-AAG. This is in contrast to data from the analysis of data from Garnett et al.14 in which PI3K-activated cell lines overall exhibited greater sensitivity to 17-AAG. One possible explanation for the discrepancy with the Garnett et al.14 data is that tumor type may influence the response to 17-AAG even in the context of oncogenic mutations. For example in Fig 1A, only 3 of the 50 cell lines with PIK3CA mutations tested in that study were HNSCC cell lines.
Oncogene dependence is a complex phenomenon and may include diverse mechanisms. In hyperproliferative, early lesions, such as CML, the oncogenic lesion may be an initiating factor that is required for ongoing imbalances in proliferation or differentiation state. In CML, ~95% of patients carry the BCR-ABL translocation and inhibition of that gene fusion product leads to remission in the majority of patients. However, later stage cancers may either be resistant to targeted therapy or have developed more complex molecular dependencies. For example, the presence of modifying molecular expression patterns and mutations may determine whether or not a patient with an “oncogene-addiction” mutation will respond to targeted therapy. In some cases, enhanced sensitivities to targeted therapies may develop over time in cancers expressing mutated oncogenes.
PI3K mutations often develop in late-stage cancers.6 Thus, it is likely that cancers with PI3K mutations are not strictly dependent on this pathway for tumor growth. However, as the most commonly mutated pathway in cancer, it is an attractive target for therapy.6 One possibility that our data and the data from Lui et al.6 suggest is that cancers with PI3K pathway mutations may develop dependence on certain pathways over time by a shift in their signaling state to adjust to the aberrantly active PI3K/mTOR signaling. Another possibility is the PI3K is more important for controlling other hallmarks of cancer that we and the Garnett et al.14 and Barretina et al.13 studies did not test, such as invasion or serum-independent growth that are generally enhanced in late-stage cancers. A final possibility is that PI3K inhibition alone is not sufficient enough of a blockade to cause cell death due to the ability of PI3K mutant-expressing cells to shift to other survival pathways, notably mTOR. Thus, only with dual PI3K/mTOR therapy are we able to see the increased sensitivity conferred by one of the PI3K hotspot mutations.
Our finding that exogenous expression of PI3K mutant molecules led to resistance to PI3K- and HSP90-targeted with sensitivity only to PI3K/mTOR dual inhibition therapy suggests several future directions. First, the advent of genome editing tools allows introduction of mutations into the endogenous gene. This might represent a better model system than the exogenous expression system we created, because it should separate the effects of overexpressing proteins from the mutation status. Second, primary cancer cells or cell lines in which the mutation naturally occurred may be superior to engineered cell lines if the oncogene dependence requires evolution of the tumor in the presence of the oncogene. Third, it is possible that IC50 analysis of cell proliferation is not sufficient to analyze the effect of PI3K mutations, despite its utility in other oncogene-addiction models. Lastly, investigation of how the signaling state of PI3K mutant-expressing cells shifts upon exposure to PI3K inhibitors may further identify optimal targeted therapy approaches.
Conclusion
This study demonstrates that PI3K plays a complex role in oncogene dependence. New model systems and approaches will likely need to be developed to determine the role of targeted therapy in the treatment of PI3K-overactive HNSCC tumors.
Acknowledgements
We thank Vito Quaranta and members of his laboratory for use of and help with the CellaVista microscope, Dr. Jim Netterville, and the Weaver laboratory for advice and assistance with this project. We thank Dr. Gerard Milano for providing the CAL-33 cell line and Dr. Harvey Babich for providing the HSC-2 cell line. Funding was provided by NIH-NCI R01-CA163592 to AMW and Pilot funding from the Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation via the Vanderbilt-Ingram Cancer Center to AMW and EDW (Weaver/Wirtz).
Footnotes
Eric D Wirtz – Participated in design, data acquisition, analysis, and writing of the manuscript. No conflicts of interest or financial disclosures.
Daisuke Hoshino – Generated and validated the engineered cell lines and participated in analysis, interpretation of the data and editing of the manuscript. No conflicts of interest or financial disclosures.
Anthony T Maldonado – Participated in analysis and interpretation of the data and editing the manuscript. No conflicts of interest or financial disclosures.
Darren R Tyson – Participated in conceptualization and design, analysis and interpretation of the data and editing of the manuscript. No conflicts of interest or financial disclosures.
Alissa M Weaver – Participated in conceptualization and design, analysis and interpretation of the ata, and writing of the manuscript. No conflicts of interest or financial disclosures.
The views expressed in this manuscript are those of the authors and do not reflect the official policy or position of the Department of the Army, Department of Defense, or the U.S. Government.
References
- 1.Rothenberg Michael S, Ellisen Leif W. The molecular pathogenesis of head and neck squamous cell carcinoma. The Journal of Clinical Investigation. 2012;122(6):1951–1957. doi: 10.1172/JCI59889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leemans CR, Braakhuis BJM, Brakenhoff R. The molecular biology of head and neck cancer. Nature Reviews. 2011 Jan;11(1):9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 3.Dorsey K, Agulnik M. Promising New Molecular Targeted Therapies in Head and Neck Cancer. Drugs. 2013 Mar;73(4):315–325. doi: 10.1007/s40265-013-0025-3. [DOI] [PubMed] [Google Scholar]
- 4.Stransky, et al. The Mutational Landscape of Head and Neck Squamous Cell Carcinoma. Science. 2011 Aug;333(6046):1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agrawal N, Frederick MJ, Pickerking CR, et al. Exome Sequencing of Head and Neck Squamous Cell Carcinoma Reveals Inactivating Mutations in NOTCH1. Science. 2011 Aug;333(6046):1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lui, et al. Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer Discovery. 2013 Jul;3(7):761–769. doi: 10.1158/2159-8290.CD-13-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du L, Shen J, Weems A, Lu SL. Role of Phosphatidylinositol-3-Kinase Pathway in Head and Neck Squamous Cell Carcinoma. Journal of Oncology. 2012 May;2012:450179. doi: 10.1155/2012/450179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He Y, Van’t Veer LJ, Mikolajewska-Hanclich I, et al. PIK3CA mutations predict local recurrences in rectal cancer patients. Clin Cancer Res. 2009 Nov;15(22):6956–6962. doi: 10.1158/1078-0432.CCR-09-1165. [DOI] [PubMed] [Google Scholar]
- 9.Saal LH, Holm K, Maurer M, et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Research. 2005 Apr;65(7):2554–2559. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- 10.Akagi I, Miyashita M, Makino H, et al. Overexpression of PIK3CA is associated with lymph node metastasis in esophageal squamous cell carcinoma. International Journal of Oncology. 2009;34(3):767–775. doi: 10.3892/ijo_00000202. [DOI] [PubMed] [Google Scholar]
- 11.Pellicano F, Mukherjee L, Holyoake TL. Cancer Cells Escape from Oncogene Addiction: Understanding the Mechanisms Behind Treatment Failure for More Effective Targeting. Cancer Stem Cells. 2014 Feb;:1678. doi: 10.1002/stem.1678. doi: 10. 1002/stem. [DOI] [PubMed] [Google Scholar]
- 12.Torti D, Trusolino L. Oncogene addiction as a foundational rationale for targeted anti-cancer therapy: Promises and perils. EMBO Mol Med. 2011 Nov;3(11):623–36. doi: 10.1002/emmm.201100176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modeling of anticancer drug sensitivity. Nature. 2012 Mar;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garnett MJ, Edelman EJ, Heidorn SJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012 Mar;483(7391):570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoshino D, Jourquin J, Emmons SW. Network analysis of the focal adhesion to invadopodia transition identifies a PI3K-PKCα invasive signaling axis. Science Signaling. 2012 Sep;5(241):ra66. doi: 10.1126/scisignal.2002964. doi: 10.1126/scisignal.2002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jhaveri K, Modi S. HSP 90 Inhibitors for Cancer Therapy and Overcoming Drug Resistance. Advances in Pharmacology. 2012;65:471–517. doi: 10.1016/B978-0-12-397927-8.00015-4. [DOI] [PubMed] [Google Scholar]
- 17.King JW, Nathan PD. Role of the MEK inhibitor trametinib in the treatment of metastatic melanoma. Future Oncology. 2014;10(9):1559–1570. doi: 10.2217/fon.14.89. [DOI] [PubMed] [Google Scholar]
- 18.Meyer, et al. Expression of PIK3CA mutant E545K in the mammary gland induces heterogeneous tumors but is less potent than mutant H1047R. Oncogenesis. 2013 Sep;2:e74. doi: 10.1038/oncsis.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]