Summary
Hepatocyte growth factor (HGF) is a mesenchyme-derived pleiotropic factor that regulates cell growth, motility, mitogenesis, and morphogenesis in a variety of cells, and increased serum levels of HGF have been linked to a number of clinical and subclinical cardiovascular disease phenotypes. However, little is currently known regarding what genetic factors influence HGF levels, despite evidence of substantial genetic contributions to HGF variation. Based upon ethnicity-stratified single-variant association analysis and trans-ethnic meta-analysis of 6201 participants of the Multi-Ethnic Study of Atherosclerosis (MESA), we discovered five statistically significant common and low-frequency variants: HGF missense polymorphism rs5745687 (p.E299K) as well as four variants (rs16844364, rs4690098, rs114303452, rs3748034) within or in proximity to HGFAC. We also identified two significant ethnicity-specific gene-level associations (A1BG in African Americans; FASN in Chinese Americans) based upon low-frequency/rare variants, while meta-analysis of gene-level results identified a significant association for HGFAC. However, identified single-variant associations explained modest proportions of the total trait variation and were not significantly associated with coronary artery calcium or coronary heart disease. Our findings indicate genetic factors influencing circulating HGF levels may be complex and ethnically diverse.
Keywords: protein, genetic association, genome-wide analysis, circulation
INTRODUCTION
Hepatocyte growth factor (HGF) is a mesenchyme-derived pleiotropic factor that regulates cell growth, motility, mitogenesis, and morphogenesis in a variety of cells (Zarnegar & Michalopoulos, 1995). It is secreted as a single polypeptide that is subsequently cleaved to form an active heterodimer (Nakamura & Mizuno, 2010). HGF initiates a tyrosine kinase signaling cascade as the sole ligand to c-Met, an epithelial cell membrane receptor, which in turn induces a variety of biological effects related to tissue regeneration and repair. Originally discovered as a mitogen for hepatocytes (Nakamura et al., 1987, Nakamura et al., 1984), several in vitro studies (Matsumoto & Nakamura, 1992, Nakamura, 1991) have shown HGF to have a regenerative effect on multiple tissue types and elevated circulating HGF levels have been observed in organ injury patients (Funakoshi & Nakamura, 2003). Despite increased circulation of HGF resulting in a systemic exposure of the protein, accumulation of HGF remains localized to the injured tissue (Tajima et al., 1992).
While increased circulation of HGF is a biological response to acute tissue injury, elevated levels have also been associated with a wide variety of cardiovascular disease (CVD) states. In humans, serum HGF has been associated with advanced age, current smoking and diabetes (Miller et al., 2014), systolic blood pressure (Aspinall & Pockros, 2006, Billich, 2002), intima medial thickness (Miller et al., 2014, Billich, 2002), and aorto-iliac artery atherosclerosis (Nieva et al., 2012). Obesity has been associated with higher levels of serum HGF(Rehman et al., 2003) with concomitant decreases following weight loss(Swierczynski et al., 2005). Higher serum HGF has also been associated with presence of coronary atherosclerosis (Mamelak et al., 2012) and chronic renal disease (Tsien et al., 2009). Furthermore, higher serum concentrations have been associated with myocardial infarction, unstable angina, acute aortic dissection, and pulmonary thromboembolism compared to patients with stable angina (Solivetti et al., 1998).
Although a familial aggregation study (Vistoropsky et al., 2008) demonstrated circulating HGF to be highly heritable (h2 = 48.4%), little is currently known regarding what genetic factors influence HGF levels. Previous genetic association analyses with single nucleotide polymorphisms (SNPs) have been limited to two narrowly-focused studies: a single candidate-gene study, which implicated CYP19A1 haplotypes with plasma HGF levels in post-menopausal women (Lin et al., 2012); and a follow-up analysis of a keratoconus risk variant (HGF SNP rs3735520), which was associated with increased serum HGF levels in an Australian control population (Burdon et al., 2011). Genetic variants within the protein encoding gene HGF have also been associated with a number of complex diseases, including hypertension (Motone et al., 2004), myopia (Yanovitch et al., 2009), and ovarian cancer (Goode et al., 2011), although the connection between these variants and circulating HGF levels is unknown. Characterization of genetic factors that influence HGF levels may provide additional insight into biological mechanisms behind HGF regulation and identify new biomarkers for disease risk.
In this study, we present the first extensive investigation of common, low-frequency, and rare-variant genetic associations with serum HGF levels in a large, multi-ethnic cohort. In addition to ethnicity-stratified analyses, we also employ meta-analytic methods to combine results across sub-cohorts, gaining insight into genetic effects common across ethnicities.
METHODS
Study participants
The Multi-Ethnic Study of Atherosclerosis (MESA), described in greater detail elsewhere (Nieva et al., 2012), enrolled 6814 participants aged 45-84 from 2000-2002 without existing clinical CVD. This parent study population included 38% non-Hispanic White American (EUR), 28% African American (AFA), 22% Hispanic American (HIS), and 12% Chinese American (CHN) subjects. MESA participants were examined at six field centers located in Baltimore, MD; Chicago, IL; Forsyth County, NC; Los Angeles County, CA; northern Manhattan, NY; and Saint Paul, MN. For this study, Exam 1 serum was available for HGF measurement on 6769 participants. MESA and its ancillary studies were approved by the Institutional Review Board at participating centers and all participants gave written informed consent.
At each visit, information on demographics, cardiovascular risk factors, past medical history and co-morbidities, social history, family history, and medications was collected through a combination of self-administered questionnaires and interview-administered questionnaires. Height was measured while participants were standing without shoes, heels together against a vertical mounted ruler. Body mass index (BMI) was calculated as weight (kg)/height2 (m2). Resting seated blood pressure was measured three times using an automated oscillometric method (Dinamap), and the average of the second and third readings are used in analyses. Hypertension was defined according to the Seventh Report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure (JNC 7) guidelines as systolic blood pressure ≥140 mmHg, diastolic blood pressure ≥90 mmHg, or use of anti-hypertensive medications (Chobanian et al., 2003). Diabetes was defined as any participant who self-reported a physician diagnosis, used diabetic medication, had a fasting glucose ≥126 mg/dL, or a non-fasting glucose of ≥200 mg/dL. Serum glucose was assayed by a hexokinase/glucose-6-phosphate dehydrogenase method. Triglycerides were measured in plasma by a glycerol blanked enzymatic method, and cholesterol was measured in plasma using a cholesterol oxidase method. HDL cholesterol was measured by the cholesterol oxidase method after precipitation of non-HDL-cholesterol with magnesium/dextran. LDL-cholesterol was calculated in specimens having a triglyceride <400 mg/dL via the Friedewald equation.
Sample preparation
Blood samples were obtained from fasting participants at MESA Exam 1 and processed within 30 minutes from phlebotomy. Serum was obtained allowing blood samples to clot at room temperature for 40 minutes. Samples were centrifuged at 4°C at 2000 g × 15 minutes or 3000 g × 10 minutes for a total of 30,000 g-minutes and subsequently stored at −70°C. DNA extraction was performed for genotyping assays.
Measurement of HGF
Circulating levels of HGF protein were measured at MESA Exam 1 in serum by a quantitative sandwich enzyme-linked immunosorbent assay (ELISA) using the Human soluble HGF/CD62P Immunoassay kit (R&D Systems, Minneapolis, MN), with a lower limit of detection of 40 pg/mL. The interassay laboratory coefficients of variation for the HGF method were 12.0%, 8.0%, and 7.4% at respective mean concentrations of 686.6, 2039.1, and 4079.5 pg/mL for lyophilized manufacturer’s controls; and 10.4% at a mean concentration of 687.7 pg/mL for an in-house pooled serum control.
Genotyping and annotation
The assayed genotype data consisted of three individual genotype panels on all MESA participants who consented for genetic studies: the Illumina Exome BeadChip (Huyghe et al., 2013), the Illumina Cardio-MetaboChip (Mamelak et al., 2012), and the Illumina iSelect ITMAT/Broad/CARe (IBC) Chip (Keating et al., 2008). Each of the three panels had quality control measures individually performed on the genotype data prior to merging them together using Plink v1.07 (Purcell et al., 2007) under genome build NCBI build 37. This produced a genotype dataset for all MESA samples that passed quality control procedures, with a total sample size of N = 6323 (AFA = 1635; CHN = 766; EUR = 2491; HIS = 1431) for 377,173 variants with minor allele frequency (MAF) >0% in at least one subcohort. Population stratification was assessed using STRUCTURE (Pritchard et al., 2000) and EIGENSTRAT (Price et al., 2006) for participants with genome-wide SNP data. All variants in the final merged dataset were annotated using BioR (Kocher et al., 2014). All reported calculations of linkage disequilibrium are based on the 1000 Genomes EUR Phase 1 population unless otherwise noted.
Statistical analysis
Single-variant association analysis
SNP-trait associations for HGF serum levels were tested by linear regression under an assumed additive genetic model with PLINK v1.07 (Purcell et al., 2007), adjusting for age, sex, BMI, smoking status, and the first three ancestry-informative principal components (PCs). Analyses were conducted stratified by ethnicity on any autosomal SNP in the merged genotype data, and SNPs with a subcohort-specific empirical MAF <0.5% and/or call rate <90% were excluded from corresponding single-variant analysis. To combine results across ethnicity, trans-ethnic meta-analysis of association results on all SNPs observed in ≥2 subcohorts was performed using METASOFT (Han & Eskin, 2011), which provides association results under a fixed-effect model. Cochran’s Q statistics and accompanying p-values were calculated to evaluate effect heterogeneity across ethnicities.
Gene-based association analysis
For low-frequency and rare variants, single-variant association testing may be less powerful than gene-level aggregation tests, which combine the effects of multiple variants within a given gene (Lee et al., 2014). Ethnicity-stratified gene-based tests were performed such that variants were included in a gene set if they corresponded to an ethnicity-specific MAF ≤5% as well as a maximum MAF across ethnicities ≤10%. Furthermore, we selected variants that were potentially functional based upon the BioR annotation, defined as being either non-synonymous, start- or stop-codon altering, or a splice-site variant for the given gene. To avoid Type I error inflation due to unstable parameter estimation, gene-level associations were evaluated if at least two such variants were present and if the total minor allele count (MAC) across all included variants was ≥20. Testing was conducted using the optimally unified rare-variant association method SKAT-O (Lee et al., 2012), which combines gene-burden testing with the kernel machine method SKAT (Wu et al., 2011). These analyses were adjusted for the same set of covariates used in the single-variant analyses. We additionally conducted a meta-analysis of the ethnicity-specific test results using MetaSKAT (Lee et al., 2013), assuming fixed effects across subcohorts under the SKAT-O model. All testing was completed under the linear-weighted kernel using the default Beta distribution weighting scheme, along with ethnicity-specific MAFs for MetaSKAT.
Statistical significance
To adjust for multiple testing, a single-SNP association was declared to be statistically significant if the nominal P-value was below a Bonferroni-corrected alpha level of 0.05, with the adjustment factor based upon the total number of SNPs evaluated across any subcohort (255,930 variants, P < 1.95E-07). Similarly defined thresholds were applied for the gene-level testing based upon the total number of evaluated genes (12,492 genes, P < 4.00e-06).
Association with clinical and sub-clinical disease
Follow-up analysis for all significant single-variant meta-analysis results was performed for marginal association with subclinical and clinical CVD by examining their association with coronary artery calcium (CAC) and coronary heart disease (CHD) across the entire MESA cohort with available genetic data (Table S1), stratified by ethnicity. We evaluated CAC Agatston score associations using a Tobit model(Fornage et al., 2004) to account for inflated zero measurements, while incident CHD was evaluated under Cox proportional hazards model. All associations were considered under both a minimally adjusted (age and sex) and fully adjusted model (age, sex, BMI, smoking and alcohol use status, LDL and HDL cholesterol, triglycerides, and hypertension and diabetes status).
RESULTS
A total of 6201 participants (AFA=1550; CHN=762; EUR=2477; HIS=1412) with genotype information (subject call rate >90%) were successfully phenotyped for association analysis in our study. Distributional characteristics of the genotyped variants per ethnicity can be found in Table S2 and Figure S1, while summaries of the HGF levels and their association with traditional CVD risk factors are presented in Table 1 and Table S3, respectively.
Table 1. Summary statistics for serum HGF levels by race/ethnicity and in total.
| AFA (N=1550) |
CHN (N=762) |
EUR (N=2477) |
HIS (N=1412) |
Total (N=6201) |
|
|---|---|---|---|---|---|
| N | 1550 | 762 | 2477 | 1412 | 6201 |
| Mean (SD) | 934.4 (247.9) | 840.8 (217.5) | 915.6 (254.6) | 1035.8 (268.9) | 938.5 (258.9) |
| Median | 892.4 | 813.5 | 884.7 | 993.7 | 903.3 |
| Q1, Q3 | 766.6, 1064.8 | 698.4, 951.5 | 733.6, 1065.2 | 848.4, 1181.6 | 757.4, 1085.1 |
| Range | (403.2-2151.8) | (292.4-2135.9) | (313.6-2117.0) | (317.7-2094.1) | (292.4-2151.8) |
AFA, African American; CHN, Chinese American; EUR, European American; HIS, Hispanic American
Single-variant associations
Manhattan plots for the stratified single-SNP association analyses are presented in Figure S2. The top SNPs per ethnicity are reported in Table 2. Only one SNP, HGF missense variant rs5745687 (PEUR = 5.374E-11), exceeded the statistical significance threshold, while rs200231675 was borderline significant (PCHN = 2.68E-07). The variant allele for rs5745687 showed strong evidence of association with reduced HGF levels in the EUR, with each additional copy corresponding to a decrease in serum HGF of 85.7 pg/ml.
Table 2. Top Ethnicity-Stratified Single-SNP Association Results, Adjusted for Age, Sex, BMI, Smoking, and First Three PCs.
| SNP | Gene | Chr | Position | Ethnicity | MAF | Ref/Alt | Protein | Beta | SE | P |
|---|---|---|---|---|---|---|---|---|---|---|
| rs5745687 | HGF | 7 | 81359051 | EUR | 0.073 | A/G | p.E299K | −85.7 | 12.5 | 7.83E-12 |
| rs200231675* | DPH1 | 17 | 1944781 | CHN | 0.007 | G/C | p.V370L | 323.9 | 62.4 | 2.68E-07 |
| rs57257204* | NOTCH2 | 1 | 120500640 | HIS | 0.064 | G/A | Intron | 93.2 | 18.6 | 5.77E-07 |
| rs7163763* | CHRNA7 | 15 | 31822215 | AFA | 0.017 | T/A | Intron | 157.7 | 32.2 | 1.04E-06 |
AFA, African American; CHN, Chinese American; EUR, European American; HIS, Hispanic American; MAF, minor allele frequency; SE, standard error
Not statistically significant at α = 1.95E-07
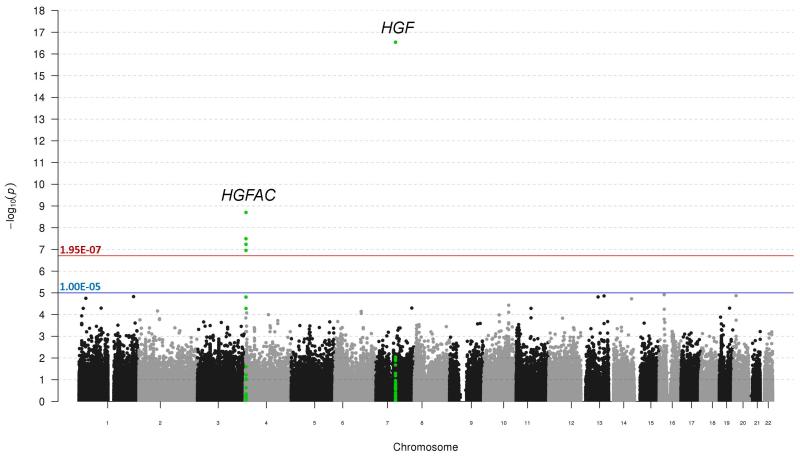
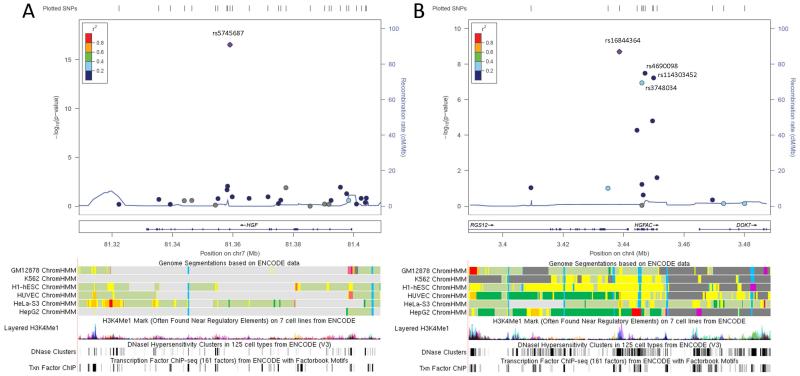
A Manhattan plot of the single-variant trans-ethnic meta-analysis results is presented in Figure 1, with significant findings highlighted in Table 3. Statistically significant results were localized to two loci: EUR-associated SNP rs5745687 (PMETA = 2.88E-17) as well as a cluster of four SNPs (rs16844364, rs4690098, rs114303452, rs3748034) within or in proximity to gene HGFAC. Regional association plots (Pruim et al., 2010) for these loci are presented in Figure 2. Although Cochran’s Q statistic is known to be conservative (Higgins et al., 2003), results for these five SNPs indicated little to no evidence of variation in true effect size across subcohorts. Analyses assuming effect heterogeneity (Han & Eskin, 2011) did not result in any changes to significance status. A QQ-plot of all single-SNP analyses (meta- and stratified) is displayed in Figure S3, while the proportion of explained variation of HGF by clinical characteristics and significant meta-analysis SNPs is reported in Table 4. Overall, the explained variability by the associated SNPs was very modest, ranging from 0.9-2.6%. Empirical evaluation of pair-wise LD with the four significant SNPs at the HGFAC locus across ethnicity indicated moderate LD between rs16844364 and rs3748034 (r2 from 0.13-0.47), with all other calculations <0.05.
Figure 1. Manhattan plot for meta-analysis results for genetic associations with serum HGF levels across all four sub-cohorts.
The –log10 p-value for each genetic variant evaluated are plotted against its respective chromosomal location. Bonferroni adjusted significance and suggestive significance are indicated by red and blue lines, respectively. Results in the region of HGF and HGFAC are highlighted in green.
Table 3. Significant single-SNP trans-ethnic meta-analysis results.
| Stratified Analysis P (MAF) | Meta-Analysis | Cochran’s Q | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||||
| SNP | Gene | Chr | Position | AFA | CHN | EUR | HIS | Ref/Alt | Protein | Beta | SE | P | Stat | P |
| rs5745687 | HGF | 7 | 81359051 | 3.1E-04 (0.014) |
NA* (0.001) |
7.8E-12 (0.073) |
3.0E-04 (0.042) |
A/G | p.E299K | −89.1 | 10.5 | 2.88E-17 | 1.43 | 0.49 |
| rs16844364 | RGS12 | 4 | 3438643 | 0.014 (0.129) |
0.001 (0.451) |
0.045 (0.225) |
2.7E-06 (0.275) |
G/C | Intron | 29.7 | 4.9 | 1.98E-09 | 6.58 | 0.09 |
| rs4690098 | HGFAC | 4 | 3447156 | 0.771 (0.056) |
0.007 (0.445) |
5.3E-05 (0.226) |
8.4E-04 (0.230) |
T/C | Intron | −28.8 | 5.2 | 3.22E-08 | 4.16 | 0.24 |
| rs114303452 | HGFAC | 4 | 3449915 | NA* (0.002) |
NA* (0.000) |
2.9E-06 (0.017) |
3.4E-03 (0.005) |
G/A | p.K566R | 125.5 | 23.1 | 5.85E-08 | 1.07 | 0.30 |
| rs3748034 | HGFAC | 4 | 3446091 | 0.346 (0.046) |
0.007 (0.308) |
0.005 (0.144) |
9.7E-05 (0.127) |
C/A | p.A218S | 32.2 | 6.1 | 1.11E-07 | 3.20 | 0.36 |
AFA, African American; CHN, Chinese American; EUR, European American; HIS, Hispanic American; MAF, minor allele frequency; SE, standard error
Stratified analysis results not available for meta-analysis due to low empirical MAF.
Figure 2. Regional association plots of meta-analysis results near rs5745687 and rs16844364.
LocusZoom association plots of the meta-analysis results in the neighborhood (+/− 50 Kb) of (A) rs5745687 and (B) rs16844364. Associated variant rs4690098 in proximity to rs16844364 is also labeled. LD patterns used for plotting are based upon 1000 Genomes European (March 2012) data. Corresponding regulatory annotation tracks from UCSC Genome browser include layered ENCODE H3K27Ac histone modifications, DNaseI hypersensitivity clusters, transcription factor ChIP-seq data, and Chromatin HMM state segmentations. The orange color for the chromatin segments indicates strong enhancer regions.
Table 4. Explained variation by clinical characteristics and significant meta-analysis single SNPs.
| *Proportion of Explained Variation (R2) | ||||
|---|---|---|---|---|
|
| ||||
| Source | AFA | CHN | EUR | HIS |
| Clinical Characteristics | 0.117 | 0.160 | 0.171 | 0.120 |
| SNPs | 0.009 | 0.015 | 0.026 | 0.021 |
| Clinical + SNPs | 0.124 | 0.175 | 0.197 | 0.144 |
AFA, African American; CHN, Chinese American; EUR, European American; HIS, Hispanic American
Based on linear regression for subjects with complete data
Clinical characteristics: Age, sex, BMI, and smoking status
Although the previously reported associated SNP rs3735520, located in the promoter region of HGF, was included in our genotype data, we did not replicate the finding in any of our analyses (min. P-value: PAFA=0.018). We similarly found no significant associations with any variants in the aromatase gene CYP19A1, although those findings corresponded to a highly specific study population. HGF variants in our data previously associated with disease phenotypes (rs2074725 and hypertension (Motone et al., 2004), rs2214825 and ovarian cancer mortality (Goode et al., 2011)) also did not achieve statistical significance in our study.
Gene-based associations
A QQ-plot of the complete gene-level association results is presented in Figure S4, with significant and suggestive (P<1E-04) findings reported in Table S4. Two genes were significantly associated in our stratified analyses: A1BG (PAFA = 6.93E-07; Table S5) and FASN (PCHN = 4.34E-07; Table S6), while no significant gene-based findings were observed for the remaining subcohorts. Results from the MetaSKAT analysis revealed HGFAC to be significant (P = 5.09E-07; Table S7) when combining results across ethnicities. Examination of the stratified analyses for HGFAC suggests this meta-analysis result to be driven largely by variants present in the EUR and HIS subcohorts (PAFA = 0.706, PCHN = 0.77, PEUR = 1.12E-05, PHIS = 0.013).
Associations with CVD
Table S8 reports the ethnicity-specific association p-values of serum HGF with CAC and CHD under minimally and fully adjusted models. These findings indicate strong associations with HGF and clinical and subclinical disease in both European and African Americans, with some attenuation after adjustment for risk factors in the latter. Association analysis results for the five significant meta-analysis SNPs with CAC and CHD among all subjects with available genetic data are reported by ethnicity in Table S9. No findings were significant after adjusting for multiple testing (5 SNPs by 4 subcohorts by 2 phenotypes; α = 0.05/40 = 0.00125).
DISCUSSION
In this study, we evaluated common and rare genetic associations with circulating levels of serum HGF in four ethnically distinct populations within the MESA cohort, constituting the first large-scale genetic association analysis for this trait. HGF is highly expressed in vascular smooth muscle cells (Zarnegar & Michalopoulos, 1995) and plays a critical part in the regulation of cellular adhesion. Moreover, HGF is present in atherosclerotic lesions but not in normal vessels (Solivetti et al., 1998) and c-Met is expressed on vascular smooth muscle cells isolated from the intima of atherosclerotic plaque of carotid arteries (Fischer et al., 2014). Consequently, identification of genetic associations with circulating HGF may elucidate heritability of CVD and provide further insight into the molecular mechanisms of atherosclerosis.
Overall, we identified five single variants to be significantly associated with serum HGF levels based on stratified and meta-analysis findings. HGF polymorphism rs5745687 was highly significant in both our EUR analysis and our trans-ethnic meta-analyses, with consistent effect size observed across ethnicities. The SNP rs5745687 is a missense variant (p.E299K) located in exon 8 of HGF, the protein-coding gene of HGF. Annotation analysis using HaploReg V2 (Ward & Kellis, 2012) with the 1000 Genomes EUR Phase 1 population data did not indicate any other variants in high LD (r2 > 0.6) with rs5745687, while functional predictions from SIFT (Kumar et al., 2009) and PolyPhen (Adzhubei et al., 2013) classified the variant allele effect to be tolerated and benign, respectively. Despite evidence that r5745687 is unlikely to be tagging an underlying causal SNP, the functional role of the variant allele in relation to circulating HGF is unclear.
We also identified four single-SNP associations within or in proximity to gene HGFAC based upon our trans-ethnic meta-analyses, and HGFAC was a significant gene-level finding in our MetaSKAT rare-variant meta-analysis. HGFAC encodes the serine protease HGFA that activates HGF by cleaving the single chain pro-HGF to form the active heterodimeric protein (Kataoka et al., 2003). Common SNP rs16844364 is an intronic RGS12 polymorphism that is also within 5 kb of the transcription start site of HGFAC and 8.5 kb of associated HGFAC intronic variant rs4690098. Despite their relative proximity, the two SNPs are independent of one another (r2 < 0.2). HaploReg annotation for rs16844364 indicated overlap with enhancer-associated histone modifications across multiple cell types, indicating strong evidence for potential regulatory function. Query of the FANTOM5 Transcribed Enhancer Atlas (Andersson et al., 2014) resulted in an identified enhancer site 226 bp from rs16844364 (chr4:3438789-3438989) preferentially expressed in blood vessel cells. However, we are unaware of any published eQTL findings that associate rs16844364 with regulation of HGFAC expression and suppositions about the functional relationship between rs16844364 and HGFAC are conjectural. Variant rs16844401 within exon 12 of HGFAC was recently found to be associated with circulating fibrinogen levels in large meta-analysis of European Americans (Sabater-Lleal et al., 2013). Although this SNP did not demonstrate any evidence of association with HGF in our single-SNP analyses, it was included in our gene-based testing for HGFAC.
We also identified two significant gene-based results in our stratified analyses: A1BG (α-1-B glycoprotein) in the AFA subcohort and FASN (fatty acid synthase, FASN) in the CHN subcohort. A1BG is a protein coding gene for a plasma glycoprotein of unknown function, and is primarily expressed in the liver (Lonsdale et al., 2013). A pharmacogenomic association with the nonsynonymous A1BG SNP rs893184 was previously reported for treatment-related outcomes in high risk CVD white and Hispanic subjects with hypertension (McDonough et al., 2013). However, this particular SNP was not associated with HGF levels in our study, and our gene-based association may be due to a different biological mechanism. FASN is a homodimer multi-enzyme that solely catalyzes fatty acid synthesis in mammals. Upregulation of FASN has been observed in multiple cancers, and inhibition of FASN has been linked to concomitant dysregulation of c-Met protein activation by HGF (Coleman et al., 2009, Zaytseva et al., 2012). It is hypothesized that this may be due to disruption of lipid rafts, which are key factors in compartmentalization of growth factor receptors like c-Met, and aberrant FASN activity could play a role in circulating levels of HGF through this biological mechanism.
There are some notable limitations to our analyses. While our merged genotype dataset is highly enriched for plausibly functional coding variation, it does not provide complete coverage of all functional variants nor were the merged chips designed to tag genome-wide variation. Moreover, much of the array design of the genotype panels used in our analyses is based upon European ancestry. This may limit the ability to detect functional variants in the other ethnicities evaluated in our study. There were also multiple suggestive common variant associations (P < 1E-05, data not shown) in our stratified analyses, indicating that larger ethnicity-specific sample sizes may be needed to identify polymorphisms of modest effect. Subcohort sample sizes for gene-based rare-variant association testing were smaller than generally recommended (Wu et al., 2011), and true associations may have gone undetected. Finally, although our meta-analyses provide a type of internal validation of our single variant findings through consistent effect estimates across ethnicities with diverse phenotype distributions and LD patterns, additional external validation and functional evaluation is necessary to confirm our findings.
In summary, our study identified multiple biologically plausible single-SNP and gene-based associations with serum HGF. In addition to a variant within the protein coding gene HGF, common and rare variants related to HGFAC, a gene coding an activating protein for HGF, were also identified to be associated with protein levels. Our gene-based results indicate rare-variation in A1BG and FASN may also play an important role in regulating circulating HGF in African and Chinese Americans, respectively. Although we did not identify any associations with these variants and CVD, our findings may be relevant to other traits where HGF levels are of consequence, including cancer. These results also suggest genetic factors regulating HGF may be diverse and differ across ethnicity, and that future work evaluating the functional consequences of rare variation within HGFAC, A1BG, and FASN is warranted.
Supplementary Material
Acknowledgements
The authors thank the other investigators, the staff, and the participants of the MESA study for their valuable contributions. A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org. MESA is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support is provided by grants and contracts N01 HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169 and RR-024156. Funding for adhesion protein levels was provided by NHLBI by grant R01HL98077. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR000124, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. NIH/NIEHS P50 ES015915 “The Multi-Ethnic Study of Atherosclerosis and Air Pollution (MESA Air) is supported by the U.S. Environmental Protection Agency (EPA) under Science to Achieve Results (STAR) Program Grant # RD831697 and NIH/NIEHS P50 ES015915 award. Although the research described in this presentation has been funded wholly or in part by the United States Environmental Protection Agency through RD831697 to the University of Washington, it has not been subjected to the Agency’s required peer and policy review and therefore does not necessarily reflect the views of the Agency and no official endorsement should be inferred.”
Abbreviations
- HGF
hepatocyte growth factor
- CVD
cardiovascular disease
- SNPs
single nucleotide polymorphisms
- MESA
Multi-Ethnic Study of Atherosclerosis
- EUR
non-Hispanic White American
- AFA
African American
- HIS
Hispanic American
- CHN
Chinese American
- ELISA
enzyme-linked immunosorbent assay
- MAF
minor allele frequency
- PCs
principal component
- MAC
minor allele count
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
REFERENCES
- Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013 doi: 10.1002/0471142905.hg0720s76. Chapter 7, Unit7 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, Ntini E, Arner E, Valen E, Li K, Schwarzfischer L, Glatz D, Raithel J, Lilje B, Rapin N, Bagger FO, Jorgensen M, Andersen PR, Bertin N, Rackham O, Burroughs AM, Baillie JK, Ishizu Y, Shimizu Y, Furuhata E, Maeda S, Negishi Y, Mungall CJ, Meehan TF, Lassmann T, Itoh M, Kawaji H, Kondo N, Kawai J, Lennartsson A, Daub CO, Heutink P, Hume DA, Jensen TH, Suzuki H, Hayashizaki Y, Muller F, Forrest AR, Carninci P, Rehli M, Sandelin A. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–61. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspinall RJ, Pockros PJ. SCV-07 (SciClone Pharmaceuticals/Verta) Curr Opin Investig Drugs. 2006;7:180–5. [PubMed] [Google Scholar]
- Billich A, SciClone Pharmaceuticals Thymosin alpha1. Curr Opin Investig Drugs. 2002;3:698–707. [PubMed] [Google Scholar]
- Burdon KP, Macgregor S, Bykhovskaya Y, Javadiyan S, Li X, Laurie KJ, Muszynska D, Lindsay R, Lechner J, Haritunians T, Henders AK, Dash D, Siscovick D, Anand S, Aldave A, Coster DJ, Szczotka-Flynn L, Mills RA, Iyengar SK, Taylor KD, Phillips T, Montgomery GW, Rotter JI, Hewitt AW, Sharma S, Rabinowitz YS, Willoughby C, Craig JE. Association of polymorphisms in the hepatocyte growth factor gene promoter with keratoconus. Invest Ophthalmol Vis Sci. 2011;52:8514–9. doi: 10.1167/iovs.11-8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chobanian AV, Bakris GL, Black HR, Cushman WC, Green LA, Izzo JL, Jr., Jones DW, Materson BJ, Oparil S, Wright JT, Jr., Roccella EJ. Seventh report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure. Hypertension. 2003;42:1206–52. doi: 10.1161/01.HYP.0000107251.49515.c2. [DOI] [PubMed] [Google Scholar]
- Coleman DT, Bigelow R, Cardelli JA. Inhibition of fatty acid synthase by luteolin post-transcriptionally down-regulates c-Met expression independent of proteosomal/lysosomal degradation. Mol Cancer Ther. 2009;8:214–24. doi: 10.1158/1535-7163.MCT-08-0722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Vazquez-Garcia I, Illingworth CJ, Mustonen V. High-definition reconstruction of clonal composition in cancer. Cell Rep. 2014;7:1740–52. doi: 10.1016/j.celrep.2014.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornage M, Boerwinkle E, Doris PA, Jacobs D, Liu K, Wong ND. Polymorphism of the soluble epoxide hydrolase is associated with coronary artery calcification in African-American subjects: The Coronary Artery Risk Development in Young Adults (CARDIA) study. Circulation. 2004;109:335–9. doi: 10.1161/01.CIR.0000109487.46725.02. [DOI] [PubMed] [Google Scholar]
- Funakoshi H, Nakamura T. Hepatocyte growth factor: from diagnosis to clinical applications. Clin Chim Acta. 2003;327:1–23. doi: 10.1016/s0009-8981(02)00302-9. [DOI] [PubMed] [Google Scholar]
- Goode EL, Chenevix-Trench G, Hartmann LC, Fridley BL, Kalli KR, Vierkant RA, Larson MC, White KL, Keeney GL, Oberg TN, Cunningham JM, Beesley J, Johnatty SE, Chen X, Goodman KE, Armasu SM, Rider DN, Sicotte H, Schmidt MM, Elliott EA, Hogdall E, Kjaer SK, Fasching PA, Ekici AB, Lambrechts D, Despierre E, Hogdall C, Lundvall L, Karlan BY, Gross J, Brown R, Chien J, Duggan DJ, Tsai YY, Phelan CM, Kelemen LE, Peethambaram PP, Schildkraut JM, Shridhar V, Sutphen R, Couch FJ, Sellers TA. Assessment of hepatocyte growth factor in ovarian cancer mortality. Cancer Epidemiol Biomarkers Prev. 2011;20:1638–48. doi: 10.1158/1055-9965.EPI-11-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B, Eskin E. Random-Effects Model Aimed at Discovering Associations in Meta-Analysis of Genome-wide Association Studies. Am J Hum Genet. 2011;88:586–598. doi: 10.1016/j.ajhg.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Bmj. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyghe JR, Jackson AU, Fogarty MP, Buchkovich ML, Stancakova A, Stringham HM, Sim X, Yang L, Fuchsberger C, Cederberg H, Chines PS, Teslovich TM, Romm JM, Ling H, Mcmullen I, Ingersoll R, Pugh EW, Doheny KF, Neale BM, Daly MJ, Kuusisto J, Scott LJ, Kang HM, Collins FS, Abecasis GR, Watanabe RM, Boehnke M, Laakso M, Mohlke KL. Exome array analysis identifies new loci and low-frequency variants influencing insulin processing and secretion. Nat Genet. 2013;45:197–201. doi: 10.1038/ng.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka H, Miyata S, Uchinokura S, Itoh H. Roles of hepatocyte growth factor (HGF) activator and HGF activator inhibitor in the pericellular activation of HGF/scatter factor. Cancer Metastasis Rev. 2003;22:223–36. doi: 10.1023/a:1023051500010. [DOI] [PubMed] [Google Scholar]
- Keating BJ, Tischfield S, Murray SS, Bhangale T, Price TS, Glessner JT, Galver L, Barrett JC, Grant SFA, Farlow DN, Chandrupatla HR, Hansen M, Ajmal S, Papanicolaou GJ, Guo YR, Li MY, Derohannessian S, De Bakker PIW, Bailey SD, Montpetit A, Edmondson AC, Taylor K, Gai XW, Wang SS, Fornage M, Shaikh T, Groop L, Boehnke M, Hall AS, Hattersley AT, Frackelton E, Patterson N, Chiang CWK, Kim CE, Fabsitz RR, Ouwehand W, Price AL, Munroe P, Caulfield M, Drake T, Boerwinkle E, Reich D, Whitehead AS, Cappola TP, Samani NJ, Lusis AJ, Schadt E, Wilson JG, Koenig W, Mccarthy MI, Kathiresan S, Gabriel SB, Hakonarson H, Anand SS, Reilly M, Engert JC, Nickerson DA, Rader DJ, Hirschhorn JN, Fitzgerald GA. Concept, Design and Implementation of a Cardiovascular Gene-Centric 50 K SNP Array for Large-Scale Genomic Association Studies. PLoS One. 2008:3. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher JP, Quest DJ, Duffy P, Meiners MA, Moore RM, Rider D, Hossain A, Hart SN, Dinu V. The Biological Reference Repository (BioR): a rapid and flexible system for genomics annotation. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1082. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Lee S, Abecasis GR, Boehnke M, Lin X. Rare-variant association analysis: study designs and statistical tests. Am J Hum Genet. 2014;95:5–23. doi: 10.1016/j.ajhg.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, Christiani DC, Wurfel MM, Lin X. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am J Hum Genet. 2012;91:224–37. doi: 10.1016/j.ajhg.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Teslovich TM, Boehnke M, Lin X. General framework for meta-analysis of rare variants in sequencing association studies. Am J Hum Genet. 2013;93:42–53. doi: 10.1016/j.ajhg.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Gunter MJ, Manson JE, Rexrode KM, Cook NR, Kraft P, Cochrane BB, Chlebowski RT, Ho GYF, Zhang SMM. The Aromatase Gene (CYP19A1) Variants and Circulating Hepatocyte Growth Factor in Postmenopausal Women. PLoS One. 2012:7. doi: 10.1371/journal.pone.0042079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, Hasz R, Walters G, Garcia F, Young N, Foster B, Moser M, Karasik E, Gillard B, Ramsey K, Sullivan S, Bridge J, Magazine H, Syron J, Fleming J, Siminoff L, Traino H, Mosavel M, Barker L, Jewell S, Rohrer D, Maxim D, Filkins D, Harbach P, Cortadillo E, Berghuis B, Turner L, Hudson E, Feenstra K, Sobin L, Robb J, Branton P, Korzeniewski G, Shive C, Tabor D, Qi LQ, Groch K, Nampally S, Buia S, Zimmerman A, Smith A, Burges R, Robinson K, Valentino K, Bradbury D, Cosentino M, Diaz-Mayoral N, Kennedy M, Engel T, Williams P, Erickson K, Ardlie K, Winckler W, Getz G, Deluca D, Macarthur D, Kellis M, Thomson A, Young T, Gelfand E, Donovan M, Meng Y, Grant G, Mash D, Marcus Y, Basile M, Liu J, Zhu J, Tu ZD, Cox NJ, Nicolae DL, Gamazon ER, Im HK, Konkashbaev A, Pritchard J, Stevens M, Flutre T, Wen XQ, Dermitzakis ET, Lappalainen T, Guigo R, Monlong J, Sammeth M, Koller D, Battle A, Mostafavi S, Mccarthy M, Rivas M, Maller J, Rusyn I, Nobel A, Wright F, Shabalin A, Feolo M, Sharopova N, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamelak AN, Drazin D, Shirzadi A, Black KL, Berci G. Infratentorial supracerebellar resection of a pineal tumor using a high definition video exoscope (VITOM(R)) J Clin Neurosci. 2012;19:306–9. doi: 10.1016/j.jocn.2011.07.014. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Nakamura T. Hepatocyte growth factor: molecular structure, roles in liver regeneration, and other biological functions. Crit Rev Oncog. 1992;3:27–54. [PubMed] [Google Scholar]
- Mcdonough CW, Gong Y, Padmanabhan S, Burkley B, Langaee TY, Melander O, Pepine CJ, Dominiczak AF, Cooper-Dehoff RM, Johnson JA. Pharmacogenomic association of nonsynonymous SNPs in SIGLEC12, A1BG, and the selectin region and cardiovascular outcomes. Hypertension. 2013;62:48–54. doi: 10.1161/HYPERTENSIONAHA.111.00823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, White BS, Dees ND, Griffith M, Welch JS, Griffith OL, Vij R, Tomasson MH, Graubert TA, Walter MJ, Ellis MJ, Schierding W, Dipersio JF, Ley TJ, Mardis ER, Wilson RK, Ding L. SciClone: Inferring Clonal Architecture and Tracking the Spatial and Temporal Patterns of Tumor Evolution. PLoS Comput Biol. 2014;10:e1003665. doi: 10.1371/journal.pcbi.1003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motone M, Katsuya T, Ishikawa K, Iwashima Y, Sugimoto K, Yamamoto K, Fu Y, Matsuo A, Ohishi M, Rakugi H, Ogihara T. Association between hepatocyte growth factor gene polymorphism and essential hypertension. Hypertens Res. 2004;27:247–51. doi: 10.1291/hypres.27.247. [DOI] [PubMed] [Google Scholar]
- Nakamura T. Structure and function of hepatocyte growth factor. Prog Growth Factor Res. 1991;3:67–85. doi: 10.1016/0955-2235(91)90014-u. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Mizuno S. The discovery of hepatocyte growth factor (HGF) and its significance for cell biology, life sciences and clinical medicine. Proc Jpn Acad Ser B Phys Biol Sci. 2010;86:588–610. doi: 10.2183/pjab.86.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Nawa K, Ichihara A. Partial purification and characterization of hepatocyte growth factor from serum of hepatectomized rats. Biochem Biophys Res Commun. 1984;122:1450–9. doi: 10.1016/0006-291x(84)91253-1. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Nawa K, Ichihara A, Kaise N, Nishino T. Purification and subunit structure of hepatocyte growth factor from rat platelets. FEBS Lett. 1987;224:311–6. doi: 10.1016/0014-5793(87)80475-1. [DOI] [PubMed] [Google Scholar]
- Nieva J, Wendel M, Luttgen MS, Marrinucci D, Bazhenova L, Kolatkar A, Santala R, Whittenberger B, Burke J, Torrey M, Bethel K, Kuhn P. High-definition imaging of circulating tumor cells and associated cellular events in non-small cell lung cancer patients: a longitudinal analysis. Phys Biol. 2012;9:016004. doi: 10.1088/1478-3975/9/1/016004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, De Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman J, Considine RV, Bovenkerk JE, Li J, Slavens CA, Jones RM, March KL. Obesity is associated with increased levels of circulating hepatocyte growth factor. J Am Coll Cardiol. 2003;41:1408–13. doi: 10.1016/s0735-1097(03)00231-6. [DOI] [PubMed] [Google Scholar]
- Sabater-Lleal M, Huang J, Chasman D, Naitza S, Dehghan A, Johnson AD, Teumer A, Reiner AP, Folkersen L, Basu S, Rudnicka AR, Trompet S, Malarstig A, Baumert J, Bis JC, Guo X, Hottenga JJ, Shin SY, Lopez LM, Lahti J, Tanaka T, Yanek LR, Oudot-Mellakh T, Wilson JF, Navarro P, Huffman JE, Zemunik T, Redline S, Mehra R, Pulanic D, Rudan I, Wright AF, Kolcic I, Polasek O, Wild SH, Campbell H, Curb JD, Wallace R, Liu S, Eaton CB, Becker DM, Becker LC, Bandinelli S, Raikkonen K, Widen E, Palotie A, Fornage M, Green D, Gross M, Davies G, Harris SE, Liewald DC, Starr JM, Williams FM, Grant PJ, Spector TD, Strawbridge RJ, Silveira A, Sennblad B, Rivadeneira F, Uitterlinden AG, Franco OH, Hofman A, Van Dongen J, Willemsen G, Boomsma DI, Yao J, Swords Jenny N, Haritunians T, Mcknight B, Lumley T, Taylor KD, Rotter JI, Psaty BM, Peters A, Gieger C, Illig T, Grotevendt A, Homuth G, Volzke H, Kocher T, Goel A, Franzosi MG, Seedorf U, Clarke R, Steri M, Tarasov KV, Sanna S, Schlessinger D, Stott DJ, Sattar N, Buckley BM, Rumley A, Lowe GD, Mcardle WL, Chen MH, Tofler GH, Song J, Boerwinkle E, Folsom AR, et al. Multiethnic meta-analysis of genome-wide association studies in >100 000 subjects identifies 23 fibrinogen-associated Loci but no strong evidence of a causal association between circulating fibrinogen and cardiovascular disease. Circulation. 2013;128:1310–24. doi: 10.1161/CIRCULATIONAHA.113.002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solivetti FM, Thorel MF, Di Luca Sidozzi A, Bucher S, Donati P, Panichelli V. Role of high-definition and high frequency ultrasonography in determining tumor thickness in cutaneous malignant melanoma. Radiol Med. 1998;96:558–61. [PubMed] [Google Scholar]
- Swierczynski J, Korczynska J, Goyke E, Adrych K, Raczynska S, Sledzinski Z. Serum hepatocyte growth factor concentration in obese women decreases after vertical banded gastroplasty. Obes Surg. 2005;15:803–8. doi: 10.1381/0960892054222678. [DOI] [PubMed] [Google Scholar]
- Tajima H, Higuchi O, Mizuno K, Nakamura T. Tissue distribution of hepatocyte growth factor receptor and its exclusive down-regulation in a regenerating organ after injury. J Biochem. 1992;111:401–6. doi: 10.1093/oxfordjournals.jbchem.a123769. [DOI] [PubMed] [Google Scholar]
- Tsien CI, Cao Y, Lawrence TS. Functional and metabolic magnetic resonance imaging and positron emission tomography for tumor volume definition in high-grade gliomas. Semin Radiat Oncol. 2009;19:155–62. doi: 10.1016/j.semradonc.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Vistoropsky Y, Trofimov S, Malkin I, Kobyliansky E, Livshits G. Genetic and environmental determinants of hepatocyte growth factor levels and their association with obesity and blood pressure. Ann Hum Biol. 2008;35:93–103. doi: 10.1080/03014460701822003. [DOI] [PubMed] [Google Scholar]
- Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Lee S, Cai TX, Li Y, Boehnke M, Lin XH. Rare-Variant Association Testing for Sequencing Data with the Sequence Kernel Association Test. Am J Hum Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovitch T, Li YJ, Metlapally R, Abbott D, Viet KNT, Young TL. Hepatocyte growth factor and myopia: Genetic association analyses in a caucasian population. Mol Vis. 2009;15:1028–1035. [PMC free article] [PubMed] [Google Scholar]
- Zarnegar R, Michalopoulos GK. The many faces of hepatocyte growth factor: from hepatopoiesis to hematopoiesis. J Cell Biol. 1995;129:1177–80. doi: 10.1083/jcb.129.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaytseva YY, Rychahou PG, Gulhati P, Elliott VA, Mustain WC, O’connor K, Morris AJ, Sunkara M, Weiss HL, Lee EY, Evers BM. Inhibition of fatty acid synthase attenuates CD44-associated signaling and reduces metastasis in colorectal cancer. Cancer Res. 2012;72:1504–17. doi: 10.1158/0008-5472.CAN-11-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.