Abstract
Machine learning methods have been increasingly used to predict the conversion of mild cognitive impairment (MCI) to Alzheimer's disease (AD), by classifying MCI converters (MCI-C) from MCI non-converters (MCI-NC). However, most of existing methods construct classifiers using only data from one particular target domain (e.g., MCI), and ignore data in the other related domains (e.g., AD and normal control (NC)) that could provide valuable information to promote the performance of MCI conversion prediction. To this end, we develop a novel domain transfer learning method for MCI conversion prediction, which can use data from both the target domain (i.e., MCI) and the auxiliary domains (i.e., AD and NC). Specifically, the proposed method consists of three key components: 1) a domain transfer feature selection (DTFS) component that selects the most informative feature-subset from both target domain and auxiliary domains with different imaging modalities, 2) a domain transfer sample selection (DTSS) component that selects the most informative sample-subset from the same target and auxiliary domains with different data modalities, and 3) a domain transfer support vector machine (DTSVM) classification component that fuses the selected features and samples to separate MCI-C and MCI-NC patients. We evaluate our method on 202 subjects from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) with MRI, FDG-PET and CSF data. The experimental results show that the proposed method can classify MCI-C patients from MCI-NC patients with an accuracy of 79.4%, with the aid of additional domain knowledge learned from AD and NC.
Index Terms: Domain Transfer Learning, Feature Selection, Sample Selection, Mild Cognitive Impairment Converters, Alzheimer's Disease
I. Introduction1
Alzheimer's disease (AD) is characterized by the progressive impairment of neurons and their connections, which result in loss of cognitive functions. In 2007, a report published by Ron et al. indicated that there were 26.6 million AD sufferers worldwide, and forecasted that 1 in 85 people will be affected by 2050 [1]. As the prodromal stage of AD, mild cognitive impairment (MCI) can be further categorized into MCI converters (MCI-C) and MCI non-converters (MCI-NC). Specifically, MCI-C patients will likely progress to AD, while MCI-NC patients will not convert to AD. Existing research has suggested that the individuals with amnestic MCI tend to progress to the probable AD at a rate of approximately 10% to 15% per year [1]. Thus, the accurate diagnosis of AD, especially in the early stage (i.e., MCI), is very important for timely therapy, disease modifying drug development, and possible delay of the disease. Nowadays, many machine learning methods have been proposed to recognize AD patients [2–10]. Recently, an increasing number of studies on AD research begin to address classification of MCI conversion (MCI-C) and MCI non-conversion (MCI-NC) patients based on the high-resolution brain imaging data [4, 7, 11–24].
One challenge for MCI-C prediction is that the number of MCI-C and MCI-NC subjects available for training is generally very small, while the dimensionality of data is often very high, which makes it very challenging to train an accurate classifier. Thus, many advanced machine learning methods have been proposed to address this issue [2,3,7,11, 22,23,49–51,57]. For instance, in [3], a multi-task learning method achieved an accuracy of 73.9% on 43 MCI-C and 48 MCI-NC patients using multi-modality data such as MRI, fluorodeoxyglucose positron emission tomography (FDG-PET), and cerebrospinal fluid (CSF). In [7], a manifold harmonic transform method using cortical thickness data achieved a sensitivity of 63% and specificity of 76% on 72 MCI-C and 131 MCI-NC patients, and in [11] a morphological factor method using MRI data achieved an accuracy of 72.3% on 20 MCI-C and 29 MCI-NC patients. In [22], an orthogonal partial least square to latent structures was used to diagnosis of MCI-C and achieved prediction accuracy of 75.4% and 68% for those MCI subjects converting to AD by 24 and 36 months, respectively. This performance was reported on 162 MCI ADNI patients. In [23], a Gaussian process approach that combined several multi-modality data sources (i.e., MRI, PET, CSF and APOE genotype) was used for classification and achieved an accuracy of 74.1% on classification between 47 MCI-C and 96 MCI-NC patients. In all these referenced studies, the size of dataset is small and also only one domain (i.e., MCI-C and MCI-NC) of subjects is used for training the classification models.
It is worth noting that MCI cohorts are heterogeneous, consisting of MCI-C (that will convert to AD) and MCI-NC (that will remain stable). Because of the characteristic of MCI cohorts, several studies proposed a hypothesis that the MCI-NC subjects are more healthy-like, while the MCI-C subjects are more AD-like, which is consistent with the contention that discrete disease states are an approximation to a continuous disease spectrum [23, 40, 56]. In the literature, several studies have treated AD as MCI-C, and NC as MCI-NC, and then used AD and NC subjects to train a support vector machine (SVM) for MCI-C prediction [23–25]. It demonstrates that the task of classifying MCI-C and MCI-NC subjects is related to the task of classifying AD and NC subjects. On the other hand, in machine learning community, transfer learning has been developed to better deal with the problem involving multiple domains (including target domain and auxiliary domain) of data [26–28], where it does not assume that the auxiliary data have exactly the same distribution as the target data. Recently, in our preliminary work [29], transfer learning has been introduced into medical imaging analysis. Specifically, in [29] a domain transfer support vector machine (DTSVM) was used to classify MCI-C and MCI-NC patients (i.e., target data) with the help of AD and NC patients as the auxiliary data, showing a great performance improvement. However, in [29], it did not use a feature selection step to identify the most discriminative features from imaging data. Also, in [29] all the patients in the auxiliary domain were used for training, without a step to select the most informative samples in the auxiliary domain (i.e., AD/NC subjects) for further improving the classification performance between MCI-C and MCI-NC patients.
To address the above-mentioned limitations, we propose a novel domain transfer learning framework for MCI-C prediction. Specifically, we first develop a domain transfer feature selection (DTFS) method by using both the auxiliary (AD/NC) and target (MCI-C/MCI-NC) domains to select a subset of discriminative features, common to both domains. Then, using the instance-transfer approach, a cross-domain kernel is constructed for transferring the auxiliary domain knowledge. To improve the quality of the source data in the cross-domain kernels, a domain transfer sample selection (DTSS) method is further developed using the multi-task least absolute shrinkage selection operator (Lasso) kernel-based method to select the most informative sample subset. Finally, all selected samples are classified by a domain transfer support vector machine (DTSVM) using adaptive SVMs and multi-kernel learning. The proposed method is evaluated using MRI, FDG-PET and CSF data of 202 subjects from the Alzheimer’s Disease Neuroimaging Initiative (ADNI), and experimental results show that the proposed method can recognize MCI-C patients from MCI-NC ones with 79.4% accuracy by using the aid of additional domain knowledge learned from AD and NC.
II. MATERIALS AND METHODS
The data used in our experiments came from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database. The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies, and non-profit organizations, as a $60 million, 5-year public-private partnership. The primary goal of ADNI has been to test whether the serial MRI, PET, other biological markers, and clinical and neuropsychological assessments can be combined to measure the progression of MCI and early AD. Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials.
ADNI is the result of efforts of many co-investigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada. The initial goal of ADNI was to recruit 800 adults, aged 55 to 90, to participate in the research (approximately 200 cognitively normal older individuals to be followed for 3 years, 400 people with MCI to be followed for 3 years, and 200 people with early AD to be followed for 2 years). The research protocol was approved by each local institutional review board and the written informed consent is obtained from each participant.
A. Subjects
In the ADNI database, there are totally 202 subjects with three modality data (i.e., MRI, PET, and CSF), including 51 AD patients, 99 MCI patients (including 43 MCI converters (MCI-C) and 56 MCI non-converters (MCI-NC)), and 52 normal controls. During the 24-month follow-up period, 43 MCI subjects converted to AD and 56 remained stable. It is worth noting the subject size used in our experiments is very similar to that used in many previous studies [2, 3, 29], and thus is sufficient to compare our proposed method with other methods. In general, the inclusion/exclusion criteria are listed below, and Table 1 provides a summary.
MCI subjects: MMSE scores between 24–30, a memory complaint, having objective memory loss measured by education adjusted scores on Wechsler Memory Scale Logical Memory II, a CDR of 0.5, and absence of significant levels of impairment in other cognitive domains, essentially preserved activities of daily living, and an absence of dementia.
Mild AD: MMSE scores between 20–26, CDR of 0.5 or 1.0, and meeting the National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer ’s disease and Related Disorders Association (NINCDS/ADRDA) criteria for probable AD.
Healthy subjects: MMSE scores between 24–30, a Clinical Dementia Rating (CDR) of 0, non-depressed, non MCI, and non-demented.
Table 1.
Subject information
| Characteristics | AD (n=51) |
NC (n=52) |
MCI-C (n=43) |
MCI-NC (n=56) |
|---|---|---|---|---|
| Age (mean ± SD) | 75.2±7.4 | 75.3±5.2 | 75.8±6.8 | 74.7±7.7 |
| Education (mean ± SD) | 14.7±3.6 | 15.8±3.2 | 16.1±2.6 | 16.1±3.0 |
| MMSE (mean ± SD) | 23.8±2.0 | 29.0±1.2 | 26.6±1.7 | 27.5±1.5 |
| CDR (mean ± SD) | 0.7±0.3 | 0.0±0.0 | 0.5±0.0 | 0.5±0.0 |
AD = Alzheimer’s Disease, NC = Normal Control, MCI = Mild Cognitive Impairment, MCI-C = MCI converter, MCI-NC = MCI non-converter, MMSE = Mini-Mental State Examination, CDR = Clinical Dementia Rating.
B. MRI, CSF, and PET biomarkers
A detailed description on how the MRI, CSF and PET datasets were acquired can be found in the public ADNI website. In general, structural MR scans were acquired from 1.5T or 3.0T scanners. Raw DICOM MRI scans were downloaded from the public ADNI website (http://adni.loni.usc.edu/) reviewed for quality, and automatically corrected for spatial distortion caused by gradient nonlinearity and B1 field inhomogeneity. PET images were acquired 30–60 minutes post-injection, averaged, spatially aligned, interpolated to a standard voxel size, intensity normalized, and smoothed to a common resolution of 8mm full width at half maximum. CSF data were collected in the morning after an overnight fast using a 20- or 24-gauge spinal needle, frozen within 1 hour of collection, and transported on dry ice to the ADNI Biomarker Core laboratory at the University of Pennsylvania Medical Center. In this study, amyloid β (Aβ42), CSF total tau (t-tau) and tau hyperphosphorylated at threonine 181 (p-tau) are used as features.
C. Image analysis
The MRI and PET images are pre-processed to extract ROI-based features, by following the pipeline in [2]. Specifically, we first perform an anterior commissure (AC)-posterior commissure (PC) correction on all MRI and PET images using MIPAV software [30]. The AC-PC corrected images are resampled to 256×256×256, and the N3 algorithm [31] is then used to correct the intensity inhomogeneity. For the MR images, the skull is stripped using the method described in [32], followed by manual editing and cerebellum removal. Next, we use FAST in the FSL package [33] to segment the human brain into three different types of tissues: gray matter (GM), white matter (WM) and cerebrospinal fluid (CSF). For a given brain image with three segmented tissues (i.e., GM, WM and CSF), we then non-linearly register it to a template with 93 manually labeled regions-of-interests (ROIs) [35], by using a widely-used high-dimensional elastic warping tool (i.e., HAMMER [34]). Finally, we use the volumes of GM tissue of the 93 ROIs, which were normalized by the total intracranial volume (which is estimated by the summation of GM, WM and CSF volumes from all ROIs), as features for a given subject. For PET image, we first align it to its corresponding MRI image of the same subject through affine transformation, and then compute the average intensity of each ROI in the PET image as features. In addition, three CSF biomarkers are also used in this study, namely CSF Aβ42, CSF t-tau and CSF p-tau. As a result, for each subject, we have 93 features derived from the MRI image, 93 features generated from the PET image, and 3 features obtained from the CSF biomarkers.
D. Domain transfer learning for MCI-C prediction
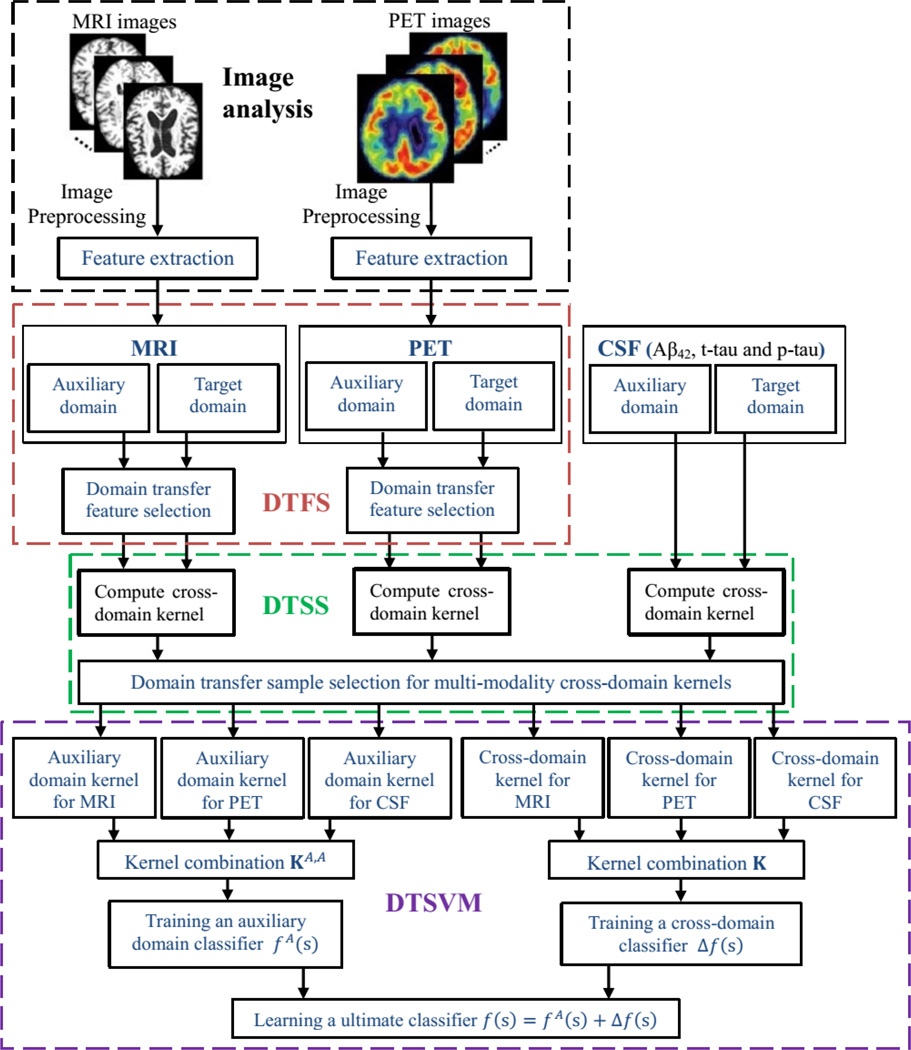
The system diagram illustrated in Fig. 1 outlines the steps and components used in our proposed classification framework. In general, it contains four components, i.e., image pre-processing, domain transfer feature selection (DTFS), domain transfer sample selection (DTSS), and domain transfer support vector machine (DTSVM). At first, all MRI and PET images are pre-processed to get the extracted features as described in Section II.C. Then, for both MRI and PET features, the DTFS component identifies a subset of features (corresponding to brain regions) that are relevant to the disease under study. Next, the DTSS component uses the multi-modality features found by the DTFS component, as well as the CSF features, to compute cross-domain kernels that simultaneously select the most informative cross-domain sample subset. Finally, in the DTSVM component, the auxiliary domain and cross-domain kernels are combined to train the ultimate classifier for classifying MCI-C and MCI-NC patients. In what follows, we will provide the technical details of these three components (i.e., DTFS, DTSS and DTSVM).
Fig. 1.
The system diagram of our proposed classification framework.
1) Domain transfer feature selection (DTFS)
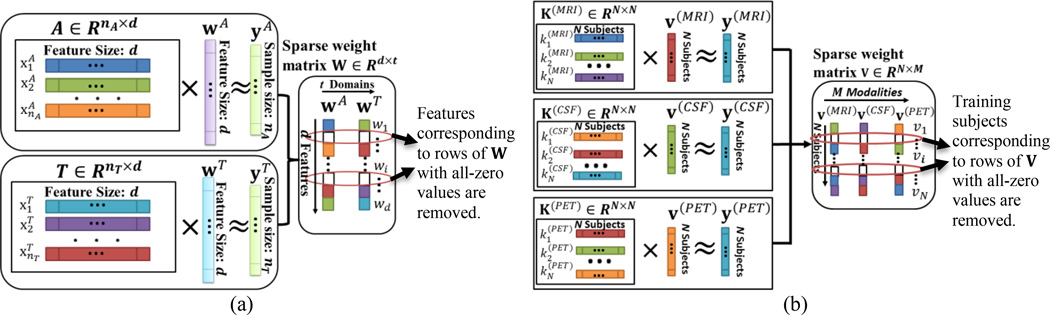
In traditional single-domain learning, we can employ sparse logistic regression [43] for learning weight vectors wA and wT from the auxiliary and target domains, respectively. Then, we can use weight vectors wA and wT for feature selection. Although using single-domain sparse learning can achieve better performance for MCI-C prediction [44], it cannot acquire knowledge from cross-domain. Since the task of classifying MCI-C and MCI-NC patients is related to the task of classifying NC and AD patients, we combine these two learning domains for learning a common weight matrix W that can select a common feature subset, as done in our proposed domain transfer feature selection (DTFS) model, which will be explained below.
Specifically, the proposed DTFS model jointly considers two learning domains: 1) AD and NC classification as the auxiliary domain, denoted as , where is a sample with d features, NA is the number of samples in the auxiliary domain, and is the corresponding class label (with the AD labeled as +1 and the NC labeled as −1); 2) MCI-C/MCI-NC classifications as the target domain, denoted as where is a sample with d features, NT is the number of samples in the target domain, and is the class label (with the MCI-C labeled as +1 and the MCI-NC labeled as −1). The DTFS model combines auxiliary domain A and target domain T for learning a common weight matrix W = [wA, wT]. Fig. 2(a) gives the illustration of DTFS, which optimizes the following objective function with the ‘group sparsity’ (i.e., L2/L1-norm) regularization:
| (1) |
where W ∈ Rd×t is the weight matrix whose row vector wi is the coefficient vector associated with i-th feature across two different learning domains, and b = [bA, bT] ∈ R1×t is the intercept for the classification task both in the auxiliary (t = A) and target (t = T) domains. Also, β > 0 is a regularization parameter that controls the relative contributions of the two terms, and the symbol ′ denotes the transpose of a matrix. According to the optimization algorithm of literature [36] for solving the optimization problem of Eq.1, we can get the sparse matrix W, where all non-zero row vectors wi correspond to features that will be selected, indicating that they are essential for classification both on auxiliary and target domains.
Fig. 2.
Illustration on the proposed DTFS and DTSS models. (a) Using DTFS to select discriminative brain regions. (b) Using DTSS to select informative subjects.
2) Domain transfer sample selection (DTSS)
After performing DTFS, we have obtained the most informative features in both auxiliary and target domains. In what follows, we will compute the cross-domain kernel matrix K for implementing the knowledge fusion both on auxiliary and target domains. Here, the instance-transfer approach [37, 38] is used to join the auxiliary domain data (i.e., AD and NC subjects which are more separable than MCI-C and MCI-NC subjects in the reduced feature space via DTFS), to the target domain data (i.e., MCI-C and MCI-NC subjects).
To be specific, we first define the kernel matrices from the auxiliary domain and the target domain as: and , respectively. Here, and are samples in the auxiliary and target domains, respectively, in the reduced feature space via DTFS. Also, as defined before, NA and NT are the numbers of samples in the auxiliary and target domains, respectively. Then, we define the cross-domain kernel matrices from the auxiliary domain to the target domain and also from the target domain to the auxiliary domain as and , respectively. Then the cross-domain kernel matrix K can be computed as:
| (2) |
where N = NA + NT.
In our study, from Eq. (2), three cross-domain kernel matrices are obtained, which correspond to three modalities (i.e., MRI, PET and CSF), denoted as K(MRI), K(PET), and K(CSF), respectively. Moreover, to remove the noisy samples and seek out the most informative samples from multimodal cross-domain, we present a sample selection framework via a multi-task Lasso based kernel learning, namely Domain Transfer Sample Selection (DTSS). Specifically, in our proposed DTSS model, we first employ the kernel learning technique to map sample set from the original space to the kernel space, where multi-task Lasso is then performed for sample selection. Fig. 2(b) gives the illustration of the proposed DTSS method. From Fig. 2(b), DTSS learns a common weight matrix V = [v(MRI), v(PET), v(CSF)], where v(MRI), v(PET), and v(CSF) are the learning weight vectors corresponding to MRI, PET and CSF modalities, respectively, and is to solve the following objective function:
| (3) |
Where , V ∈ RN×M is the weight matrix whose row vector νi is the vector of coefficients associated with the i-th training sample across three different modalities, and M is the number of modalities (M =3 in this study); λ > 0 is a regularization parameter that controls the relative contributions of the two terms; and y(m) ∈ RN×1 is the corresponding class label of cross-domain for the m-th modality. In addition, K(m) is the cross-domain kernel matrix for the m-th modality, i.e., K(MRI), K(PET), and K(CSF), respectively, which can be seen as the similarity between pairwise samples in the cross-domain for the m-th modality. In our study, the widely used Gaussian kernel function is used, which is defined as follows:
| (4) |
where σ is the bandwidth. Due to the use of ‘group sparsity’ (i.e., L2/L1-norm) regularization, many of the rows of V will be zeros, and thus all samples corresponding to non-zero row vectors will be selected. In this paper, the SLEP package [39] is used for solving the optimization problems in DTFS and DTSS.
3) Domain transfer support vector machines (DTSVM)
After performing DTFS and DTSS, we can obtain the most discriminative features and informative samples, upon which we will build a domain transfer support vector machines (DTSVM) classifier for final classification.
Denote and as the new auxiliary and target domains, with the corresponding labels denoted as and , respectively. Here, and , respectively, denote the new auxiliary and target data after feature selection (via DTFS) and sample selection (via DTSS), and and are the numbers of selected samples. Then, we employ multi-kernel learning for multimodal kernel combination [2, 3, 6], which can be used to compute the ultimate auxiliary domain kernel matrix KA,A and the cross-domain kernel matrix K. Here, kernel combination is used to define a new integrated kernel function for the m-th modality data and as follows:
| (5) |
where k(m) denotes the kernel function over the m-th modality, and cm denotes the weight on the m-th modality. From Eq. 5, we can achieve the ultimate auxiliary domain kernel matrix KA,A and cross-domain kernel matrix K, i.e., , . To find the optimal values for weights cm, we constrain them so that Σm cm = 1, 0 ≤ cm ≤ 1 and then adopt a coarse-grid search through cross-validation on the training samples, which has been shown effective in our previous work [2, 3, 6].
Then, we adopt the adaptive SVMs method in [26] to learn the ultimate classifier f(s). This ultimate classifier f(s) is first learned from the auxiliary domain classifier fA(s), which is implemented by adding a delta function for cross-domain in the form of Δf(s) = h′ Φ(s) on the basis of fA(s):
| (6) |
where Φ(s) is a kernel-based nonlinear mapping function, and h is the weight vector of cross-domain classifier. Also, h′ denotes the transpose of h.
To learn the weight vector h in Eq. 6, we use the following objective function, similar to the SVM:
| (7) |
where l is the l-th sample in the cross-domain training subset, , represents the total number of samples in, and ζl is the slack variable that represents the prediction error of objective function of Eq. 7, thus it can be used for nonlinear classification. The parameter C balances contributions between auxiliary classifier and cross-domain training examples. According to [26], we can solve this objective function (i.e., Eq. 7) to obtain the solution for the weight vector h. Then, we can obtain the final solution for f(s). In this study, fA(s) is trained by SVM using the ultimate auxiliary domain kernel matrix KA,A, and Δf(s) is solved by Eq.6 using the ultimate cross-domain kernel matrix K = [k(s, s′)], and k(s, s′) = 〈Φ(s), Φ(s′)〉.
III. RESULTS
A. Experimental Settings
In this study, we use a 10-fold cross-validation strategy to evaluate the performances of different methods. Specifically, the set of subjects in the target domain (i.e., 99 MCI subjects) are partitioned into 10 subsets (each subset with a roughly equal size), and then one subset is successively selected as the test dataset and the remaining subsets are used to train the classifiers. This process is repeated 10 times, and the classification performance is evaluated by the average sensitivity, specificity, accuracy, and area under curve (AUC) measures.
In our experiments, traditional SVM (denoted as SVM) and other methods with the SVM algorithm for classification (i.e., DTSVM and our proposed methods) are implemented using the LIBSVM toolbox [41] with a linear kernel. For DTFS and DTSS components, sparse learning is performed using the SLEP package [39], and regularization parameters β and λ are learned using another 10-fold cross-validation strategy on the training data. In DTSS component, the similarity function bandwidth parameter σ is also learned using the 10-fold cross-validation strategy on the training subsets. Specifically, a hierarchical optimization based grid search is employed for searching the optimal parameters. In our experiments, we first optimize the regularization parameters β and λ simultaneously through grid search with the range of {0.00001, 0.0001, 0.0005, 0.001, 0.004, 0.007, 0.01, 0.02, 0.03, 0.05, 0.06, 0.08, 0.1, 0.2, 0.4, 0.6, 0.8}, with the fixed default values for other parameters (i.e., σ=2, weights cm: cMRI=0.4, cCSF=0.3, cPET=0.3). Then, we adopt a grid search with the range of {2, 4, 8, 16, 32, 64, 128} to optimize the bandwidth parameter σ of similarity function. Finally, we use the grid search to find the optimal weights cm in multi-kernel learning. The performance of the proposed method was compared with the Laplacian SVM (LapSVM) [42]. The LapSVM classifier uses a linear kernel function and its graph Laplacian is constructed by using the k nearest neighbor algorithm, where k (1 ≤ k ≤ 10) is learned through 10-fold cross-validation on training data.
In addition, we use another similar hierarchical optimization based grid search to search the optimal parameters (including k and cm). Specifically, we first find the optimal value for the nearest neighbor k with fixed values for weights cm (i.e., cMRI=0.4, cCSF=0.3, cPET=0.3), and then determine the optimal weights cm in multi-kernel learning through cross-validation. Multi-modality and single-modality biomarkers are used for testing the classification performance of the proposed method, and a multi-kernel learning method is used to combine multi-modality biomarkers (i.e., MRI, CSF and PET) for all classification methods. In particular, multi-kernel learning implement a grid search with range from 0 to 1 and step size 0.1 on the training subsets. It is worth noting that, for optimization of all parameters, it is performed on the training subset by an inner 10-fold cross-validation. In addition, the same feature normalization scheme as in [2] is used in our experiments.
B. Classification results for recognizing MCI-C and MCI-NC patients
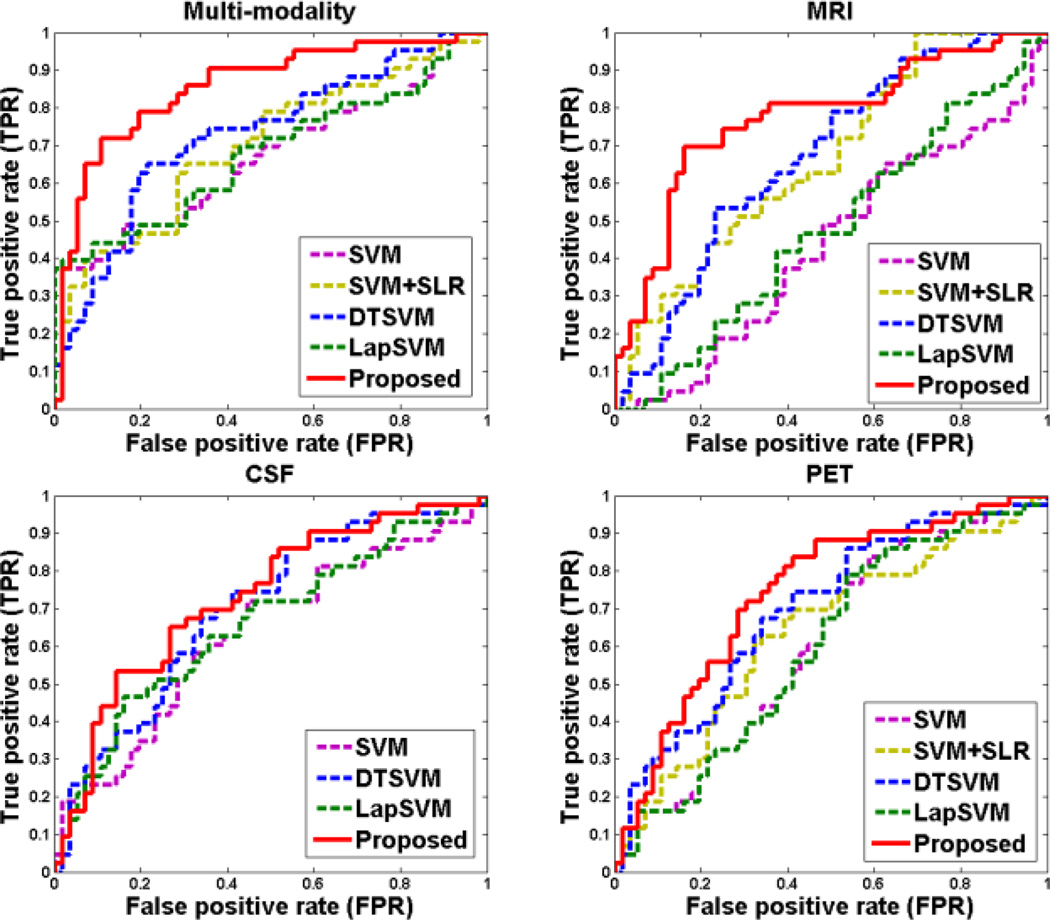
To investigate the effectiveness of the proposed method, we first evaluate the classification performance of our method for recognizing MCI-C and MCI-NC patients using both multi-modality and single-modality biomarkers. Table 2 shows the classification performance of five different methods, including SVM (traditional SVM), SVM+SLR, DTSVM [29], LapSVM [42], and the proposed method (i.e., DTFS+DTSS+DTSVM). We also perform DeLong's method [53] on the AUC between the proposed method and each of other four comparison methods, with the corresponding p-values shown in Table 2. Note that DeLong's test is a nonparametric statistical test for comparing AUC between two ROC curves, which can be employed to assess statistical significance via computing z-scores for the AUC estimate [54, 55]. Here, SLR denotes a sparse feature selection method with logistic regression loss function [43], and ‘SVM+SLR’ represents the method that first applies SLR for feature selection and then adopts SVM for classification. Note that each value in Table 2 is the averaged result of the 10-fold cross-validation, which was performed 10 different times. In addition, we plot the ROC curves achieved by different methods in Fig. 3.
Table 2.
Comparison of performance measures of five different methods for MCI-C/MCI-NC classification.
| Modality | Method | ACC % |
SEN % |
SPE % |
AUC | p-value |
|---|---|---|---|---|---|---|
| Multi-modality (MRI+CSF+PET) | SVM | 63.8 | 58.8 | 67.7 | 0.683 | <0.0001 |
| SVM+SLR | 67.3 | 73.9 | 58.8 | 0.728 | <0.0001 | |
| DTSVM | 69.4 | 64.3 | 73.5 | 0.736 | <0.0001 | |
| LapSVM | 64.6 | 55.9 | 71.1 | 0.689 | <0.0001 | |
| Proposed | 79.4 | 84.5 | 72.7 | 0.848 | - | |
| MRI | SVM | 53.9 | 47.6 | 57.7 | 0.554 | <0.0001 |
| SVM+SLR | 63.6 | 67.5 | 58.3 | 0.696 | <0.0001 | |
| DTSVM | 63.3 | 59.8 | 66.0 | 0.700 | <0.0001 | |
| LapSVM | 58.3 | 52.4 | 62.9 | 0.593 | <0.0001 | |
| Proposed | 73.4 | 74.3 | 72.1 | 0.764 | - | |
| CSF | SVM | 60.8 | 55.2 | 65.0 | 0.647 | <0.0010 |
| DTSVM | 66.2 | 60.3 | 70.8 | 0.701 | <0.0500 | |
| LapSVM | 61.1 | 55.6 | 65.3 | 0.651 | <0.0010 | |
| Proposed | 67.6 | 74.6 | 61.5 | 0.715 | - | |
| PET | SVM | 58.0 | 52.1 | 62.5 | 0.612 | <0.0001 |
| SVM+SLR | 60.9 | 65.1 | 55.4 | 0.662 | <0.0001 | |
| DTSVM | 67.0 | 59.6 | 72.7 | 0.732 | <0.0100 | |
| LapSVM | 59.1 | 53.3 | 63.5 | 0.607 | <0.0001 | |
| Proposed | 71.6 | 76.4 | 67.9 | 0.741 | - |
ACC=Accuracy, SEN=Sensitivity, SPE=Specificity.
Fig. 3.
ROC curves of different methods for MCI-C/MCI-NC classification with multi-modality and single-modality data, respectively.
As can be seen in Table 2 and Fig. 3, the proposed method consistently outperforms SVM, SVM+SLR, DTSVM and LapSVM in terms of the classification accuracy, sensitivity and AUC measures. Also, Table 2 indicates that both the proposed method and DTSVM significantly outperform SVM and SVM+SLR methods, while LapSVM is only slightly better than SVM. Moreover, statistical test measures via DeLong's method [53] (i.e., p-value) demonstrate the superiority of the proposed method over other comparison methods. These results imply that using knowledge learned from auxiliary domain (i.e., AD/NC classification) can effectively improve the performance of MCI-C/MCI-NC classification. These results also suggest that transfer learning is more suitable than semi-supervised learning for the case of data coming from different domains.
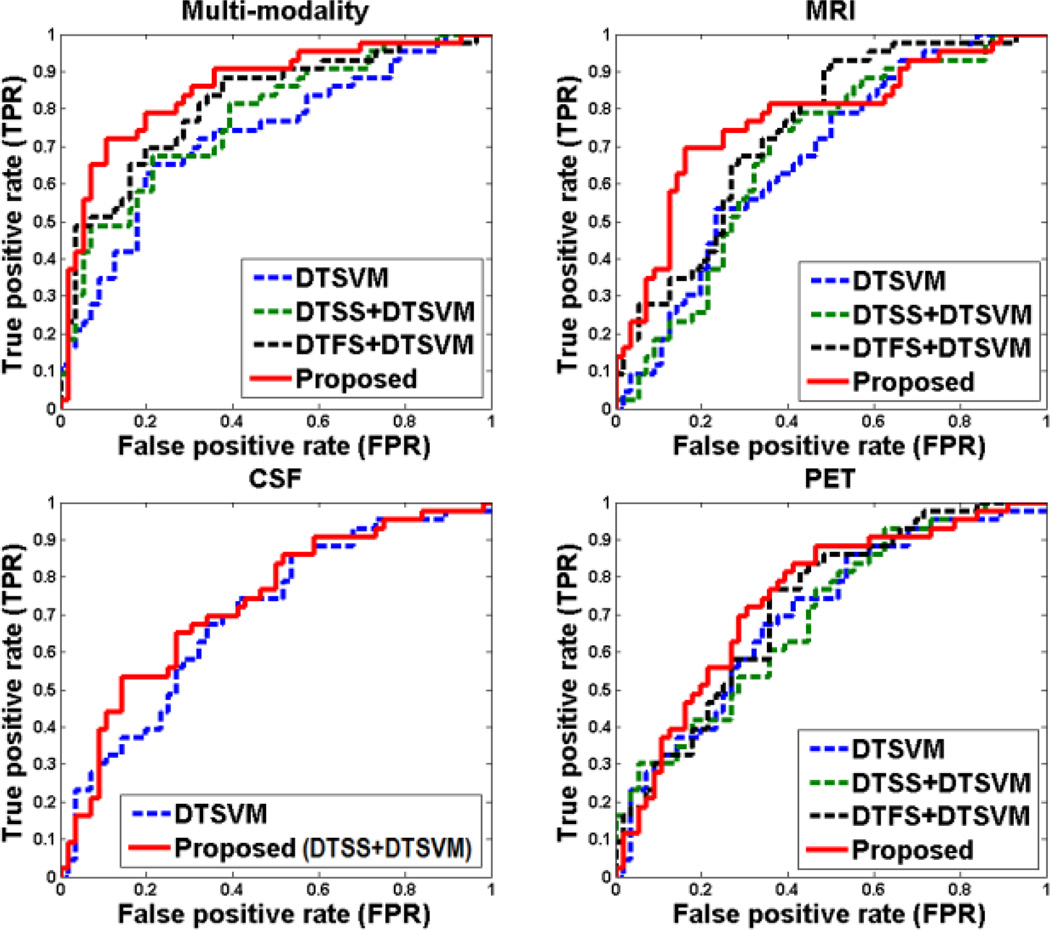
On the other hand, to investigate the relative contributions of the three components (i.e., DTFS, DTSS, and DTSVM) in our proposed method, we compare our method with its three variants, i.e., DTSVM, DTSS+DTSVM, and DTFS+DTSVM, with experimental results shown in Table 3. It is worth noting that, DTSS+DTSVM is based on DTSVM classifier using only the DTSS sample selection method, and DTFS+DTSVM is also based on DTSVM classifier using only the DTFS feature selection method. For using CSF biomarkers, feature selection was not performed because there are only three features in CSF. In Fig. 4, we also plot the ROC curves achieved by different methods using both multi-modality and single-modality biomarkers, respectively. In addition, we also report the p-values, which are computed by DeLong's method [53] on the AUC between the proposed method and its other three variants methods in Table 3.
Table 3.
Comparison of performances of different variants of our proposed method.
| Modality | Method | ACC % |
SEN % |
SPE % |
AUC | p-value |
|---|---|---|---|---|---|---|
| Multi-modality (MRI+CSF+PET) | DTSVM | 69.4 | 64.3 | 73.5 | 0.736 | <0.0001 |
| DTSS+DTSVM | 71.3 | 84.0 | 61.4 | 0.755 | <0.0001 | |
| DTFS+DTSVM | 76.5 | 81.2 | 71.9 | 0.836 | <0.0010 | |
| Proposed | 79.4 | 84.5 | 72.7 | 0.848 | - | |
| MRI | DTSVM | 63.3 | 59.8 | 66.0 | 0.700 | <0.0001 |
| DTSS+DTSVM | 65.6 | 66.2 | 65.3 | 0.686 | <0.0001 | |
| DTFS+DTSVM | 69.4 | 72.0 | 67.3 | 0.743 | <0.0010 | |
| Proposed | 73.4 | 74.3 | 72.1 | 0.764 | - | |
| CSF | DTSVM | 66.2 | 60.3 | 70.8 | 0.701 | <0.0500 |
| Proposed (DTSS+DTSVM) | 67.6 | 74.6 | 61.5 | 0.715 | - | |
| PET | DTSVM | 67.0 | 59.6 | 72.7 | 0.732 | <0.0100 |
| DTSS+DTSVM | 68.1 | 72.9 | 60.8 | 0.726 | <0.0010 | |
| DTFS+DTSVM | 68.7 | 81.0 | 59.0 | 0.733 | <0.0500 | |
| Proposed | 71.6 | 76.4 | 67.9 | 0.741 | - |
Fig. 4.
ROC curves of different variants of our proposed method.
From Table 3 and Fig. 4, each component can boost the classification performance compared with SVM. It is worth noting that, according to Fig. 4 and statistical significance assessment in Table 3, the MRI features (as opposed to PET and CSF biomarkers) most benefit from the proposed method as compared with other individual components (DTSVM, DTSS, and DTFS). This observation shows that using MRI features in the auxiliary domain can more efficiently capture those discriminative features between MCI-C and MCI-NC patients, than using PET or CSF features. Also, it indicates that the structural changes in a brain from MRI scans are more distinctive than the changes in a brain reflected by PET and CSF biomarkers. In general, our proposed method that integrates all the three components together achieves the best performance.
C. Discriminative brain regions detection
To evaluate the efficacy of our proposed domain transfer feature selection (DTFS) method for detecting the discriminative brain regions, we compare our proposed DTFS method with the commonly used single-domain based feature selection methods including paired t-test and sparse logistic regression (SLR) [43]. It is worth noting that in the feature selection step, both paired t-test and SLR methods only use data from target domain (i.e., MCI-C/MCI-NC) while DTFS uses data from both target and auxiliary (i.e., AD/NC) domains. In addition, we also compare our method with a baseline method where all features are used (i.e., no feature selection). For evaluating the performances of different feature selection methods including DTFS, paired t-test, SLR and Baseline, the classifier DTSVM is used for subsequent classification after feature selection. Table 4 shows results achieved by different feature selection methods. Here, each value in Table 4 is the averaged result of 10-fold cross-validation strategy in 10 independent runs. In addition, we also compute the p-value on the AUC between the DTFS method and other three methods via DeLong's method [53], which is shown in Table 4. As can be seen from Table 4, for MRI biomarker, there is no statistical significance between the DTFS and SLR (P=0.054), and also for PET biomarker, there is no statistical significance between the DTFS and the Baseline method (P=0.063). In general, in most cases our proposed DTFS method outperforms the other three methods, which suggests that the combination of auxiliary and target domains may provide complementary information for seeking out the most discriminative brain regions.
Table 4.
Performance comparison of four different feature selection methods: Baseline (no feature selection), paired t-test, SLR, and DTFS.
| Modality | Method | ACC % |
SEN % |
SPE % |
AUC | p-value |
|---|---|---|---|---|---|---|
| Multi-modality (MRI+CSF+PET) | Baseline | 69.4 | 64.3 | 73.5 | 0.736 | <0.0001 |
| t-test | 72.3 | 73.1 | 71.7 | 0.760 | <0.0010 | |
| SLR | 73.1 | 79.3 | 68.3 | 0.824 | <0.0010 | |
| DTFS | 76.5 | 81.2 | 71.9 | 0.836 | - | |
| MRI | Baseline | 63.3 | 59.8 | 66.0 | 0.700 | <0.0001 |
| t-test | 65.3 | 63.1 | 66.9 | 0.699 | <0.0010 | |
| SLR | 67.2 | 63.4 | 70.2 | 0.741 | >0.0500 | |
| DTFS | 69.4 | 72.0 | 67.3 | 0.743 | - | |
| PET | Baseline | 67.0 | 59.6 | 72.7 | 0.732 | >0.0500 |
| t-test | 66.6 | 69.2 | 64.5 | 0.718 | <0.0500 | |
| SLR | 67.7 | 67.9 | 67.6 | 0.714 | <0.0500 | |
| DTFS | 68.7 | 76.4 | 59.1 | 0.733 | - |
Furthermore, we list all selected brain regions with the highest frequency of occurrence by DTFS on MRI and PET images in Table 5. Here, to get these features (i.e., brain regions), we count the frequency of each feature and selected across all folds and all runs (i.e., a total of 100 times for 10-fold cross-validation with 10 independent runs), and then regarded those features with frequency of 100 (i.e., always selected in all folds and all runs) as selected stable features. On the other hand, in the supplementary Tables S1 and S2, we also listed all selected stable brain regions by paired t-test and SLR methods on MRI and PET images, respectively.
Table 5.
Selected brain regions by the DTFS method on MRI and PET images, respectively.
| MRI (Brain regions) | PET (Brain regions) |
|---|---|
| amygdala right | angular gyrus left |
| anterior limb of internal capsule left | amygdala right |
| entorhinal cortex left | cuneus right |
| inferior frontal gyrus right | inferior occipital gyrus left |
| temporal lobe WM left | inferior temporal gyrus left |
| inferior temporal gyrus right | hippocampal formation right |
| insula left | lingual gyrus left |
| fornix left | middle frontal gyrus left |
| hippocampal formation left | medial frontal gyrus right |
| perirhinal cortex left | middle occipital gyrus left |
| precuneus left | middle temporal gyrus left |
| precentral gyrus left | precuneus left |
| superior parietal lobule right | postcentral gyrus right |
| supramarginal gyrus left | superior frontal gyrus right |
| superior parietal lobule left temporal pole right |
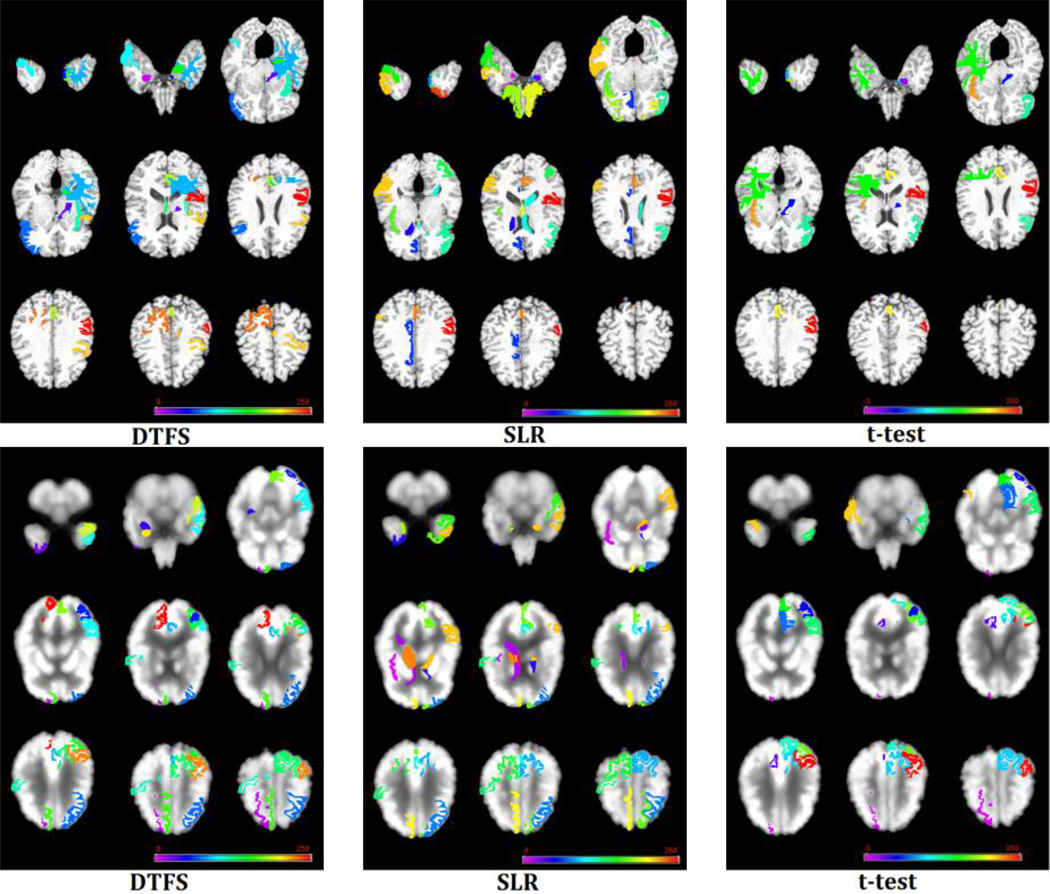
Finally, Fig. 5 visually shows the selected brain regions with the highest frequency of occurrence by DTFS, paired t-test and SLR on MRI and PET images, respectively. As can be seen from Fig. 5, our proposed DTFS method successfully finds out the most discriminative brain regions (e.g., hippocampal, amygdala, temporal lobe, precuneus, and insula) that are known to be related to Alzheimer’s disease [3, 4, 44, 45].
Fig. 5.
Selected stable brain regions by three different methods on (Top) MRI and (Bottom) PET images. Note that different colors indicate different brain regions.
IV. DISCUSSION
In this paper, we propose a domain transfer learning framework to recognize MCI-C and MCI-NC patients by using AD and NC subjects as auxiliary domain. We have evaluated the performance of our method on 202 baseline subjects from ADNI database, and the experimental results show that our proposed method can consistently and substantially improve the classification performance, with an overall classification accuracy of 79.4% for recognizing MCI-C and MCI-NC patients.
A. Domain transfer learning
Domain transfer learning is a recently-developed machine learning technique, which is able to learn a set of related models from the target domain and its related auxiliary domain for improving the classification performance in target domain [26–28, 38, 46]. Different from conventional machine learning methods, domain transfer learning does not require target and auxiliary domains having the same data distribution [38, 46], and it can effectively use data from auxiliary domain for improving the performance in the target domain [26–28, 38, 46]. However, to the best of our knowledge, there are few studies using domain transfer learning for neuroimaging based diagnosis of brain diseases [29, 47]. It is worth noting that in our preliminary study [29], domain transfer learning is performed only on the classification stage (i.e., DTSVM). To further improve the performance, we implement domain transfer learning throughout the whole process including domain transfer feature selection (i.e., DTFS), domain transfer sample selection (i.e., DTSS) and domain transfer classification (i.e., DTSVM). Our experimental results have validated the efficacy of the proposed domain transfer learning method for recognizing MCI-C and MCI-NC patients.
Recently, many studies in early diagnosis of AD focus on predicting the conversion of MCI to AD, i.e., identifying the MCI-C from MCI-NC [3, 4, 7, 13, 15, 17, 20, 23–25, 45, 48–51]. Several studies adopted the correlated domain knowledge to design classifiers, or feature selection methods for predicting the conversion from MCI to AD [3, 49, 50]. The regression based biomarkers and multiple clinical variables (MMSE and ADAS-Cog scores) were used as auxiliary domain knowledge for feature selection in the literatures [3, 49]. And researchers in [50] proposed an improved multi-task feature selection method for finding discriminative brain regions using multi-modality data (MRI and PET). Different from the above studies, our method adopts more informative auxiliary domain knowledge (i.e., AD/NC learning task) for feature selection. For the validity of using AD/NC learning task as auxiliary domain, we applied the method of [3, 49] on our used dataset (i.e., 99 MCI, and their corresponding MMSE and ADAS-Cog scores), and achieved an accuracy of 71.7% and an AUC of 0.766 using the three modalities data (i.e., MRI, PET, and CSF). Also, we apply the method of [50] on the dataset used in this study, and achieve an accuracy of 67.8% and an AUC of 0.696 with two modalities data (i.e., MRI and PET). Especially, our method achieve an accuracy of 79.4% and an AUC of 0.848 with three modalities data (i.e., MRI, PET and CSF), and achieve an accuracy of 77.3% and an AUC of 0.842 with two modalities data (i.e., MRI and PET). This result further validates our assumption that using AD and NC subjects as auxiliary domain can significantly improve the performance of MCI-C/MCI-NC classification.
In addition, our current study uses only AD and NC subjects as auxiliary domain. However, according to the principle of domain transfer learning, multiple auxiliary domain knowledge can also be utilized, as long as these multi-domain learning tasks are related to the target-domain learning task. Therefore, other relevant learning tasks, e.g., data of other dementia type, could be utilized to further promote the learning performances of our proposed methods.
B. Validation and Optimizing
To evaluate the performance of our proposed domain transfer learning method, the 10-fold cross-validation strategy was adopted. Besides the outer 10-fold cross-validation, we further perform an inner 10-fold cross-validation on the training data, to find the optimal parameters. Our proposed method is evaluated on 43 MCI-C and 56 MCI-NC subjects, which include 93 MRI features and 93 PET features extracted from the original MR and PET images, as well as 3 CSF features. To overcome the potential over-fitting problem caused by the issue of small sample size (i.e., the sample size is considerably smaller than the feature dimensionality), we propose a domain transfer feature selection method to select the most discriminative features. Similar to several studies in early diagnosis of AD [3, 13, 44, 49–51, 58] where many machine learning methods were developed for selecting discriminative brain regions, our proposed method can also be used for detecting discriminative brain regions from a larger number of brain images on the clinical application. In addition, according to our experimental results, we found that the over-fitting issue rarely occurs, because of the following two possible reasons. First, the number of samples is comparable to the number of features after feature selection (DTFS). Second, instead of simply concatenating multimodal features into a long vector, the multi-kernel SVM [2] we adopted can compute kernel matrices by using feature subsets from individual modalities, which helps avoid the over-fitting problem.
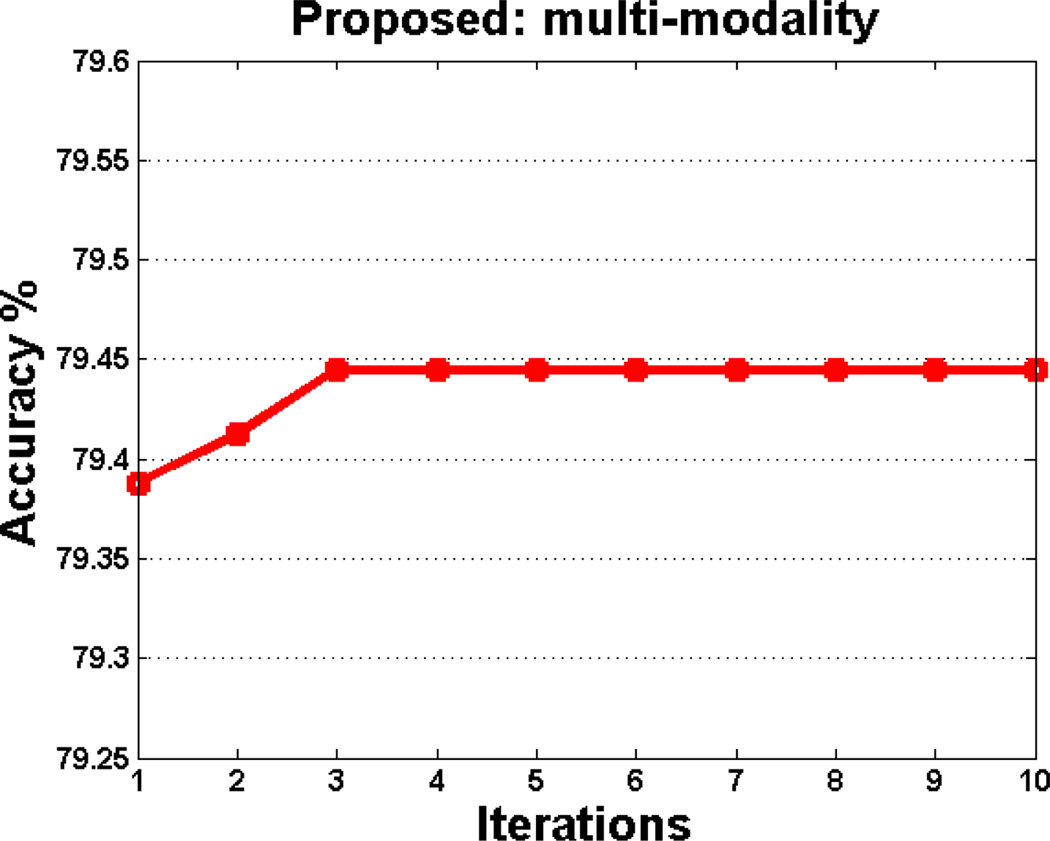
In this paper, a hierarchical optimization based coarse-grid search is employed for searching optimal parameters (i.e., β, λ, σ, and cm). Using this optimization strategy, it cannot guarantee the joint optimality of parameter values. For addressing this problem, we further design an iterative optimization algorithm to find the optimal parameters jointly. Specifically, we first perform a grid search to find the optimal parameters β and λ. With determination of these parameters, we then optimize other parameters (i.e., σ and cm). Afterwards, the above procedures are reported iteratively. In Fig. 6, we plot the change of the classification accuracy with respect to different iterations using the iterative optimization algorithm.
Fig. 6.
Classification accuracy of our proposed method in multimodal case with respect to different iterations, achieved by iterative optimization algorithm.
As shown in Fig. 6, the accuracy first rises with the increase of iteration number, and then keep stable when the iteration number is larger than 3. However, it is very time-consuming to perform such iterative optimization algorithm. We plan to adopt such iterative optimization strategy to find the optimal parameters in the future for further boosting the classification performance.
C. Predicting the conversion of MCI to AD
As mentioned above, many studies in early diagnosis of AD focus on predicting the conversion of MCI to AD using multi-modality data [3, 4, 10, 22, 23, 50, 52]. Accordingly, we report some representative results as follows.
In [3], a multi-task feature selection method achieved an accuracy of 73.9% and an AUC of 0.797 on 43 MCI-C and 48 MCI-NC ADNI subjects by using multi-modality data (MRI, FDG-PET and CSF). The method proposed in [4], which combines statistical analysis and pattern classification methods, achieved an accuracy of 62% and an AUC of 0.734 on 69 MCI-C and 170 MCI-NC ADNI subjects by using multi-modality data (MRI and CSF). Also, researchers in [10] proposed a multi-kernel pattern classification method, achieving an AUC of 0.791 on 119 MCI ADNI subjects with multi-modality data (including MRI, FDG-PET, CSF, and APOE). Researchers in [22] proposed a multivariate data analysis method and achieved an accuracy of 68.5% and an AUC of 0.76 on 81 MCI-C and 81 MCI-NC ADNI subjects with multi-modality data (i.e., MRI and CSF). In [23], a Gaussian process classification method was proposed and achieved an accuracy of 74.1% and an AUC of 0.795 on 47 MCI-C and 96 MCI-NC ADNI subjects with multi-modality data (i.e., MRI, FDG-PET, CSF, and APOE). By using the data employed in this study, we apply the Gaussian process classification method proposed in [23], and achieve an accuracy of 72.1% and an AUC of 0.797. In [50], an inter-modality relationship constrained multi-task feature selection method was proposed, which achieved an accuracy of 67.8% and an AUC of 0.696 on 43 MCI-C and 56 MCI-NC ADNI subjects with MRI and FDG-PET data. Trzepacz et al. [52] used a statistical analyses method and achieved an accuracy of 76% on 20 MCI-C and 30 MCI-NC ADNI subjects by using MRI, FDG-PET, and PIB-PET data. As shown in Section IV, our proposed domain transfer learning classification method achieves an accuracy of 79.4% and an AUC of 0.848 with three modalities (i.e., MRI, FDG-PET, and CSF).
D. Extension for recognizing MCI and AD/NC
To further investigate the efficacy of our proposed domain transfer learning method, besides recognizing MCI-C and MCI-NC, we also apply our method for recognizing MCI and AD/NC. Specifically, we have two new classification tasks, i.e., MCI vs. NC classification and MCI vs. AD classification. It is worth noting that, AD subjects are regarded as the auxiliary data for MCI vs. NC classification, and NC subjects are regarded as the auxiliary data for MCI vs. AD classification. In the Appendix, we show the experimental results by comparing different methods with both multi-modality and sing-modality biomarkers for MCI vs. NC classification and MCI vs. AD classification in Table S3 and Table S4, respectively.
As we can see from Table S3 and Table S4, our proposed method achieves consistently better performance than other methods in terms of four performance evaluation measures. Specifically, for multi-modality case, our proposed method achieves the classification accuracy of 86.4%, sensitivity of 92.8%, specificity of 73.8%, and AUC of 0.924 in MCI vs. NC classification; and the classification accuracy of 82.7%, sensitivity of 89.2%, specificity of 69.6%, and AUC of 0.906 in MCI vs. AD classification.
In addition, we also list the results achieved by different variants of our proposed method in Table S5 and Table S6 in the Appendix. Table S5 and S6 indicate that our method that integrates all the three components together achieves the best performance. These results further validate the efficacy of our proposed domain transfer learning method that uses data from auxiliary domain for aiding the classification in the target domain.
E. Limitations
The current study is limited by several factors. First, our proposed method is based on the single auxiliary domain data (i.e., AD and NC) from the ADNI database. In the future work, we will investigate whether using data from more auxiliary domains (e.g., fMRI or PIB-PET data, or even the related disease data, e.g., vascular dementia) can further improve the performance. Second, our current method cannot deal with subjects with incomplete multi-modality of data (i.e., missing of some modalities of data) and thus used only 202 subjects with all three modalities of data in ADNI. It is interesting to extend our current method for dealing with subjects with missing modalities of data for further improvement of performance, which is also one of our future works. Finally, in the current study we computed the total PET signal from each ROI, but did not perform any partial volume correction based on the segmented tissues from MR image. For further performance improvement, in our future work we will address the issues of partial volume effect and different point spread functions between modalities.
V. CONCLUSION
In this paper, we propose a general domain transfer learning based framework that consists of domain transfer feature selection (DTFS), domain transfer sample selection (DTSS) and domain transfer support vector machine (DTSVM), for MCI conversion prediction. Here, the main idea of our domain transfer learning based method is to exploit the auxiliary domain data (AD/NC subjects) to improve the classification between MCI-C and MCI-NC in the target domain. To the best of our knowledge, our study is among the first in neuroimaging to use and transfer the knowledge learned from the auxiliary task with multimodal data (i.e., AD vs. NC classification) for guiding the target task (i.e., MCI-C vs. MCI-NC classification). We have validated the efficacy of our proposed method using 202 subjects from the ADNI database with multi-modality data (including MRI, FDG-PET and CSF). The experimental results show that our proposed method achieves significantly better performance than the traditional methods for MCI conversion prediction, by effectively adopting the extra domain knowledge learned from AD and NC.
Acknowledgment
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott, AstraZeneca AB, Bayer Schering Pharma AG, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan Corporation, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson and Johnson, Eli Lilly and Co., Medpace, Inc., Merck and Co., Inc., Novartis AG, Pfizer Inc, F. Hoffman-La Roche, Schering-Plough, Synarc, Inc., as well as non-profit partners the Alzheimer's Association and Alzheimer's Drug Discovery Foundation, with participation from the U.S. Food and Drug Administration. Private sector contributions to ADNI are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuron Imaging at the University of California, Los Angeles. This work was supported in part by NIH grants EB006733, EB008374, EB009634, MH100217, AG041721, and AG042599, and by the National Natural Science Foundation of China (Nos. 61422204, 61473149, and 61473190), the Jiangsu Natural Science Foundation for Distinguished Young Scholar (No. BK20130034), the Specialized Research Fund for the Doctoral Program of Higher Education (No. 20123218110009), the NUAA Fundamental Research Funds (No. NE2013105), and the Scientific and Technological Research Program of Chongqing Municipal Education Commission (No. KJ131108).
Footnotes
Data used in this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative database (http://adni.loni.usc.edu/).
Contributor Information
Bo Cheng, School of Computer Science and Technology, Nanjing University of Aeronautics and Astronautics, Nanjing 210016, China; School of Computer Science and Engineering, Chongqing Three Gorges University, Chongqing 404000, China.
Mingxia Liu, School of Computer Science and Technology, Nanjing University of Aeronautics and Astronautics, Nanjing 210016, China; School of Information Science and Technology, Taishan University, Taian 271021, China.
Daoqiang Zhang, School of Computer Science and Technology, Nanjing University of Aeronautics and Astronautics, Nanjing 210016, China.
Brent C. Munsell, Department of Computer Science at the College of Charleston, Charleston, SC 29424, USA
Dinggang Shen, Department of Radiology and BRIC, University of North Carolina at Chapel Hill, NC 27599, USA, and also with Department of Brain and Cognitive Engineering, Korea University, Seoul, Republic of Korea.
References
- 1.Ron B, et al. Forecasting the global burden of Alzheimer's disease. Alzheimer's & dementia : the journal of the Alzheimer's Association. 2007;3:186–191. doi: 10.1016/j.jalz.2007.04.381. [DOI] [PubMed] [Google Scholar]
- 2.Zhang D, et al. Multimodal classification of Alzheimer's disease and mild cognitive impairment. NeuroImage. 2011 Apr 1;55:856–867. doi: 10.1016/j.neuroimage.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang D, et al. Multi-modal multi-task learning for joint prediction of multiple regression and classification variables in Alzheimer's disease. NeuroImage. 2012 Jan 16;59:895–907. doi: 10.1016/j.neuroimage.2011.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davatzikos C, et al. Prediction of MCI to AD conversion, via MRI, CSF biomarkers, and pattern classification. Neurobiology of Aging. 2011 Dec;32:2322.e19–2322.e27. doi: 10.1016/j.neurobiolaging.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Filipovych R, et al. Semi-supervised pattern classification of medical images: application to mild cognitive impairment (MCI) NeuroImage. 2011 Apr 1;55:1109–1119. doi: 10.1016/j.neuroimage.2010.12.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang D, Shen D. Semi-supervised multimodal classification of Alzheimer’s disease. Proceeding of IEEE International Symposium on Biomedical Imaging. 2011:1628–1631. [Google Scholar]
- 7.Cho Y, et al. Individual subject classification for Alzheimer's disease based on incremental learning using a spatial frequency representation of cortical thickness data. NeuroImage. 2012 Feb 1;59:2217–2230. doi: 10.1016/j.neuroimage.2011.09.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Filipovych R, et al. Semi-supervised cluster analysis of imaging data. NeuroImage. 2011 Feb 1;54:2185–2197. doi: 10.1016/j.neuroimage.2010.09.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ritchie LJ, Tuokko H. Patterns of cognitive decline, conversion rates, and predictive validity for 3 models of MCI. Journal of Alzheimer's Disease and Other Dementias. 2010 Nov;25:592–603. doi: 10.1177/1533317510382286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinrichs C, et al. Predictive markers for AD in a multi-modality framework: an analysis of MCI progression in the ADNI population. Neuroimage. 2011 Mar 15;55:574–589. doi: 10.1016/j.neuroimage.2010.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aksu Y, et al. An MRI-derived definition of MCI-to-AD Conversion for long-term, automatic prognosis of MCI patients. Plos One. 2011 Oct 12;6 doi: 10.1371/journal.pone.0025074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borroni B, et al. Combined 99mTc-ECD SPECT and neuropsychological studies in MCI for the assessment of conversion to AD. Neurobiology of Aging. 2006 Jan;27:24–31. doi: 10.1016/j.neurobiolaging.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 13.Duchesne S, Mouiha A. Morphological Factor Estimation via High-Dimensional Reduction: Prediction of MCI Conversion to Probable AD. Int J Alzheimers Dis. 2011;2011:914085. doi: 10.4061/2011/914085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chetelat G, et al. Using voxel-based morphometry to map the structural changes associated with rapid conversion in MCI: a longitudinal MRI study. NeuroImage. 2005 Oct 1;27:934–946. doi: 10.1016/j.neuroimage.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 15.Leung KK, et al. Increasing power to predict mild cognitive impairment conversion to Alzheimer's disease using hippocampal atrophy rate and statistical shape models. Proceeding of International Conference on Medical Image Computing and Computer-Assisted Intervention. 2010;13:125–132. doi: 10.1007/978-3-642-15745-5_16. [DOI] [PubMed] [Google Scholar]
- 16.Risacher SL, Saykin AJ, West JD, et al. Baseline MRI predictors of conversion from MCI to probable AD in the ADNI cohort. Current Alzheimer Research. 2009 Aug;6:347–361. doi: 10.2174/156720509788929273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Misra C, et al. Baseline and longitudinal patterns of brain atrophy in MCI patients, and their use in prediction of short-term conversion to AD: results from ADNI. NeuroImage. 2009 Feb 15;44:1415–1422. doi: 10.1016/j.neuroimage.2008.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mosconi L, et al. MCI conversion to dementia and the APOE genotype: a prediction study with FDG-PET. Neurology. 2004 Dec 28;63:2332–2340. doi: 10.1212/01.wnl.0000147469.18313.3b. [DOI] [PubMed] [Google Scholar]
- 19.Morbelli S, et al. Mapping brain morphological and functional conversion patterns in amnestic MCI: a voxel-based MRI and FDG-PET study. European Journal of Nuclear Medicine and Molecular Imaging. 2010 Jan;37:36–45. doi: 10.1007/s00259-009-1218-6. [DOI] [PubMed] [Google Scholar]
- 20.Cuingnet R, et al. Automatic classification of patients with Alzheimer's disease from structural MRI: a comparison of ten methods using the ADNI database. NeuroImage. 2011 May 15;56:766–781. doi: 10.1016/j.neuroimage.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 21.Wolz R, et al. Multi-Method Analysis of MRI Images in Early Diagnostics of Alzheimer's Disease. PLoS ONE. 2011;6:e25446. doi: 10.1371/journal.pone.0025446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Westman E, et al. Combining MRI and CSF measures for classification of Alzheimer's disease and prediction of mild cognitive impairment conversion. NeuroImage. 2012 Aug 1;62:229–238. doi: 10.1016/j.neuroimage.2012.04.056. [DOI] [PubMed] [Google Scholar]
- 23.Young J, et al. Accurate multimodal probabilistic prediction of conversion to Alzheimer's disease in patients with mild cognitive impairment. NeuroImage: Clinical. 2013;2:735–745. doi: 10.1016/j.nicl.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Da X, et al. Integration and relative value of biomarkers for prediction of MCI to AD progression: Spatial patterns of brain atrophy, cognitive scores, APOE genotype and CSF biomarkers. NeuroImage: Clinical. 2014;4:164–173. doi: 10.1016/j.nicl.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coupé P, et al. Scoring by nonlocal image patch estimator for early detection of Alzheimer's disease. NeuroImage: Clinical. 2012;1:141–152. doi: 10.1016/j.nicl.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J, et al. Cross-domain video concept detection using adaptive SVMs; Proceedings of the 15th international conference on Multimedia; 2007. pp. 188–197. [Google Scholar]
- 27.Duan L, et al. Domain adaptation from multiple sources via auxiliary classifiers; Proceeding of International Conference Machine Learning; 2009. pp. 289–296. [Google Scholar]
- 28.Jiang W, et al. Cross-domain learning methods for high-level visual concept classification; Proceeding of 15th IEEE International Conference on Image Processing; 2008. pp. 161–164. [Google Scholar]
- 29.Cheng B, et al. Domain transfer learning for MCI conversion prediction. Proceeding of International Conference on Medical Image Computing and Computer-Assisted Intervention- MICCAI 2012. 2012;7510:82–90. doi: 10.1007/978-3-642-33415-3_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.CIT. Medical Image Processing, Analysis and Visualization (MIPAV) 2012 http://mipav.cit.nih.gov/clickwrap.php.
- 31.Sled JG, et al. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998 Feb;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, et al. Medical Image Computing and Computer-Assisted Intervention. Toronto, Canada: 2011. Robust Deformable-Surface- Based Skull-Stripping for Large-Scale Studies; pp. 635–642. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, et al. Segmentation of brain MR images through a hidden Markov random field model and the expectation maximization algorithm. IEEE Transactions on Medical Imaging. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- 34.Shen D, Davatzikos C. HAMMER: Hierarchical attribute matching mechanism for elastic registration. IEEE Transactions on Medical Imaging. 2002 Nov;21:1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- 35.Kabani N, et al. A 3D atlas of the human brain. Neuroimage. 1998;7:S717. [Google Scholar]
- 36.Argyriou A, et al. Convex multi-task feature learning. Machine Learning. 2008;73:243–272. [Google Scholar]
- 37.Dai W, et al. Boosting for transfer learning; Proceedings of the 24th international conference on Machine learning; 2007. pp. 193–200. [Google Scholar]
- 38.Pan SJ, Yang Q. A survey on transfer learning. IEEE Transactions on Knowledge and Data Engineering. 2010 Oct;22:1345–1359. [Google Scholar]
- 39.Liu J, et al. SLEP: sparse learning with efficient projections. Arizona State University. 2009 Available: http://www.public.asu.edu/~jye02/Software/SLEP. [Google Scholar]
- 40.Ferrarini L, et al. Morphological hippocampal markers for automated detection of Alzheimer's disease and mild cognitive impairment converters in magnetic resonance images. Journal of Alzheimer's Disease. 2009;17:643–659. doi: 10.3233/JAD-2009-1082. [DOI] [PubMed] [Google Scholar]
- 41.Chang CC, Lin CJ. LIBSVM: a library for support vector machines. 2001 [Google Scholar]
- 42.Belkin M, et al. Manifold regularization: a geometric framework for learning from labeled and unlabeled examples. Journal of Machine Learning Research. 2006 Nov;7:2399–2434. [Google Scholar]
- 43.Liu J, et al. Large-scale sparse logistic regression; Proceeding of the 15th ACM SIGKDD conference on knowledge discovery and data mining; 2009. [Google Scholar]
- 44.Ye J, et al. Sparse learning and stability selection for predicting MCI to AD conversion using baseline ADNI data. Bmc Neurology. 2012 Jun 25;12 doi: 10.1186/1471-2377-12-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eskildsen SF, et al. Prediction of Alzheimer's disease in subjects with mild cognitive impairment from the ADNI cohort using patterns of cortical thinning. NeuroImage. 2013;65:511–521. doi: 10.1016/j.neuroimage.2012.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duan LX, et al. Domain transfer multiple kernel learning. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2012 Mar;34:465–479. doi: 10.1109/TPAMI.2011.114. [DOI] [PubMed] [Google Scholar]
- 47.Schwartz Y, et al. Improving Accuracy and Power with Transfer Learning Using a Meta-analytic Database. Proceeding of International Conference on Medical Image Computing and Computer-Assisted Intervention-MICCAI 2012. 2012;7512:248–255. doi: 10.1007/978-3-642-33454-2_31. [DOI] [PubMed] [Google Scholar]
- 48.Lehmann M, et al. Visual ratings of atrophy in MCI: prediction of conversion and relationship with CSF biomarkers. Neurobiology of Aging. 2012 Apr 17; doi: 10.1016/j.neurobiolaging.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 49.Zhang D, et al. Predicting future clinical changes of MCI patients using longitudinal and multimodal biomarkers. PLoS One. 2012;3:e33182. doi: 10.1371/journal.pone.0033182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu F, et al. Inter-modality relationship constrained multi-modality multi-task feature selection for Alzheimer's Disease and mild cognitive impairment identification. NeuroImage. 2014 Jan 1;84:466–475. doi: 10.1016/j.neuroimage.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H, et al. Hierarchical interactions model for predicting Mild Cognitive Impairment (MCI) to Alzheimer's Disease (AD) conversion. Plos One. 2014;9:e82450. doi: 10.1371/journal.pone.0082450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trzepacz PT, et al. Comparison of neuroimaging modalities for the prediction of conversion from mild cognitive impairment to Alzheimer's dementia. Neurobiology of Aging. 2014 Jan;35:143–151. doi: 10.1016/j.neurobiolaging.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 53.DeLong ER, et al. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 54.Sabuncu MR, et al. Clinical prediction from structural brain MRI scans: a large-scale empirical study. Neuroinformatics. 2015;13:31–46. doi: 10.1007/s12021-014-9238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robin X, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12 doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh N, et al. Genetic, structural and functional imaging biomarkers for early detection of conversion from MCI to AD. Proceeding of International Conference on Medical Image Computing and Computer-Assisted Intervention-MICCAI 2012. 2012;7510:132–140. doi: 10.1007/978-3-642-33415-3_17. [DOI] [PubMed] [Google Scholar]
- 57.Fan Y, et al. Unaffected family members and schizophrenia patients share brain structure patterns: a high-dimensional pattern classification study. Biological psychiatry. 2008;63:118–124. doi: 10.1016/j.biopsych.2007.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Y, et al. Discriminant analysis of longitudinal cortical thickness changes in Alzheimer's disease using dynamic and network features. Neurobiology of aging. 2012;33:427. e15–427. e30. doi: 10.1016/j.neurobiolaging.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]