Abstract
Serine protease (SP) and serine protease homolog (SPH) genes in insects encode a large family of proteins involved in digestion, development, immunity, and other processes. While 68 digestive SPs and their close homologs are reported in a companion paper (Kuwar et al., 2015), we have identified 125 other SPs/SPHs in Manduca sexta and studied their structure, evolution, and expression. Fifty-two of them contain cystine-stabilized structures for molecular recognition, including clip, LDLa, Sushi, Wonton, TSP, CUB, Frizzle, and SR domains. There are nineteen groups of genes evolved from relatively recent gene duplication and sequence divergence. Thirty-five SPs and seven SPHs contain 1, 2 or 5 clip domains. Multiple sequence alignment and molecular modeling of the 54 clip domains have revealed structural diversity of these regulatory modules. Sequence comparison with their homologs in Drosophila melanogaster, Anopheles gambiae and Tribolium castaneum allows us to classify them into five subfamilies: A are SPHs with 1 or 5 group-3 clip domains, B are SPs with 1 or 2 group-2 clip domains, C, D1 and D2 are SPs with a single clip domain in group-1a, 1b and 1c, respectively. We have classified into six categories the 125 expression profiles of SP-related proteins in fat body, brain, midgut, Malpighian tubule, testis, and ovary at different stages, suggesting that they participate in various physiological processes. Through RNA-Seq-based gene annotation and expression profiling, as well as intragenomic sequence comparisons, we have established a framework of information for future biochemical research of nondigestive SPs and SPHs in this model species.
Keywords: phylogenetic analysis, insect immunity, hemolymph protein, clip domain, RNA-Seq
1. Introduction
S1A serine proteases (SPs) are a large family of hydrolytic enzymes that cleave peptide bonds at different levels of specificity (Rawlings and Barrett, 1993). For instance, chymotrypsin cuts efficiently next to any accessible aromatic residues in dietary proteins; thrombin hydrolyzes a few protein substrates next to certain Arg residues at a lower rate (kcat); elastase cleaves after small nonpolar residues (e.g. Ala). According to Schechter and Berger (1967), the S1 pocket in a protease, which directly interacts with the P1 side chain in a substrate, determines the enzyme’s primary specificity. Protein interactions via regulatory domains in many nondigestive SPs localize transient responses that are regulated by activation, inactivation, or inhibition (O’Brien and McVey, 1993; Krem and Di Cera, 2002; Jiang and Kanost, 2000). The active site of S1A SPs consists of His, Asp and Ser residues, responsible for the acyl transfer mechanism of catalysis. Substrate binding clefts near the active site largely determine enzyme specificity. Led by a secretion signal peptide, SPs are transported to extracellular, vesicular, or granular locations to execute functions. They are commonly synthesized as inactive zymogens and activated by proteolytic cleavage at a particular peptide bond. Through specific molecular recognition, several SP zymogens can form a cascade pathway in which one SP activates the zymogen of another in each step and, thus, amplify the initial signal in a short period of time. The human blood coagulation is a classic example of such SP cascade systems. In addition to SPs, many serine protease homolog (SPHs) genes have been identified in animal genome projects. These proteins, lacking one or more of the catalytic residues, are not active as enzymes but involved in regulating specific SPs (Park et al., 2010; Jiang et al., 2010). However, molecular mechanism of the SPH regulation remains unclear.
We have been studying SP-related proteins from arthropods, because they mediate defense responses such as hemolymph clotting, melanotic encapsulation, antimicrobial peptide induction, and cytokine activation (Jiang et al., 2010). Analogous to some human clotting and complement factors, insect SPs also constitute complex enzyme networks to prevent bleeding and infection. In each insect species with a known genome, SPs and SPHs constitute a large protein family with 50 to 300 members (Christophides et al., 2002; Ross et al., 2003; Zou et al., 2006 and 2007; Waterhouse et al., 2007; Zhao et al., 2010). The corresponding genes are annotated to different accuracies depending on the quality of genome sequences and coverage of expressed sequence tags. Their roles in food digestion, embryo development, and immune responses have been tested in Drosophila melanogaster, Anopheles gambiae, Aedes aegypti, Manduca sexta, Tenebrio molitor, and other insects (Jang et al., 2008; Barillas-Mury, 2007; Zou et al., 2010; Jiang et al., 2010; Park et al., 2010). SP pathways and their regulators (e.g. SPHs, SP inhibitors) are being revealed by genetic and biochemical analyses. Some of the SPs and SPHs contain one or more disulfide-bridged structures named clip domains (Jiang and Kanost, 2000). These proteins were designated CLIPs (Christophides et al., 2002), even though clip-domain SPHs do not clip peptide bond. Constituting the largest group of regulatory domains in the insect SP-related proteins, clip domains in the SPs were classified into group-1a, 1b, and 2 (Ross et al., 2003), which associate with CLIP subfamilies C, D, and B (Waterhouse et al., 2007), respectively. Clip domains in the SPHs (CLIP subfamily A) belong to group-3. While 3D-structures of three clip domains were available (Piao et al., 2005; Huang et al., 2007), little is known about their functions in relation to the structures.
We have investigated an SP-SPH network for proteolytic activation of phenoloxidase (PO), spätzle, and plasmatocyte spreading peptide (PSP) precursors in M. sexta. To date, we have not yet elucidated the entire system and would like to annotate all SP and SPH genes in the genome as a step toward achieving that goal. We also hope, through phylogenetic analysis, to establish a platform for comparing SP/SPH sequences from important insect species, combined with functional data from genetic and biochemical analyses. Furthermore, we want to explore the expression patterns of these genes and provide pertinent guidance for future biochemical studies in M. sexta.
2. Materials and methods
2.1. Identification of M. sexta SPs and SPHs
Hemolymph (serine) proteases (HPs) 1 through 4 were isolated from 5th instar larval hemocyte cDNA library (Jiang et al., 1999). HP5 through HP23 cDNAs were cloned from 5th instar larval fat body and hemocyte libraries (Jiang et al., 2005). Prophenoloxidase activating proteases (PAPs) 1, 2 and 3 were cloned based on amino acid sequences of the purified enzymes (Jiang et al., 1998, 2003a, and 2003b). HP23 and HP24 genomic clones were isolated during the PAP2 gene analysis (Wang et al., 2006). HP25 through HP29 cDNA fragments were identified in the RNA-Seq data of M. sexta hemocytes, fat body, and other tissues (Zou et al., 2008; Zhang et al., 2011; Gunaratna and Jiang, 2013). Gut (serine) proteases (GPs) and their homologs (GPHs) 1 through 70 were uncovered by BLAST search of M. sexta larval midgut EST database (Pauchet et al., 2010) using the PAP1 catalytic domain sequence as a query. The EST pairs (GP8–GP20, GP11–GP65, GP12–GP14, GP18–GP19, GP22–GP49, and GP26–GP48) were encoded by GP8, GP11, GP12, GP18, GP22, and GP26 genes, respectively. Probably due to different locations on cDNAs or sequence variations, the six EST pairs were not assembled into single contigs. The GP62 EST contig was a hybrid of GP61 and GP63 genes. Scolexins A and B were cloned from larval epidermis (Finnerty et al., 1999). SPH1 and SPH2, which form a high Mr cofactor of the PAPs, were cloned from larval fat body cDNA library (Yu et al., 2003). These published protein sequences were used as queries to search Manduca Cufflinks Assembly 1.0 (http://agripestbase.org/manduca/) using the TBLASTN algorithm with default settings. Hits with aligned regions longer than 30 residues and identity over 40% were retained for retrieving corresponding cDNA sequences. Correct open reading frames (ORFs) in the retrieved sequences were identified based on their lengths and domains structure predicted using ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) and SMART (http://smart.embl-heidelberg.de/smart/set_mode.cgi). Errors resulting from misassembled regions in the Manduca Genome Assembly 1.0 (http://agripestbase.org/manduca/) were fixed by BLASTN search of Manduca Oases and Trinity Assemblies 3.0 of RNA-Seq data (http://darwin.biochem.okstate.edu/blast/blast_links.html). These two genome-independent assemblies (Cao and Jiang, 2015) were developed to cross gaps among genome scaffolds or contigs, to detect errors in the genome assembly and gene models, and to profile expression of genes bearing the imperfections. The improved sequences were in many cases verified with Manduca Official Gene Set (OGS2.0, http://agripestbase.org/manduca/) and Manduca Cufflinks Assembly 1.0b (http://darwin.biochem.okstate.edu/blast/blast_links.html) based on all 52 cDNA libraries sequenced by Illumina technology (Cufflinks 1.0 was based on only 33 of the libraries). To uncover all members of SP/SPH gene clusters, which are too similar to distinguish by Cufflinks 1.0/1.0b, the relevant genome contigs were manually examined to identify exons based on the GT-AG rule and sequence alignment, as the exon-intron junctions are absolutely conserved among these clustered genes. One of the sequence pairs (e.g. GP11–GP65) or triplets (e.g. GP8-GP20-SP133) encoded by the same gene was kept, whereas the incomplete genes were excluded from the final gene list.
2.2. Sequence properties of M. sexta SPs and SPHs
Sequences were categorized as SPs or SPHs by examining the presence of a His-Asp-Ser catalytic triad. If all three residues were found in the conserved TAAHC, DIAL, and GDSGGP regions, the proteins were considered to be SPs. Sequences lacking one or more of the key residues were designated SPHs. The signal peptide was predicted by SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/) (Petersen et al., 2011) and Signal-3L (http://www.csbio.sjtu.edu.cn/bioinf/Signal-3L/) (Shen and Chou, 2007). Domain structure of each protein was predicted using InterProScan (http://www.ebi.ac.uk/Tools/pfa/iprscan/). While some clip domains were found by prediction, others were discovered by manual inspection of the sequences for a cysteine doublet in the region amino-terminal to the protease or protease-like domain (PD or PLD). SPs and SPHs containing four additional cysteine residues upstream of the doublet were designated cSPs and cSPHs, respectively, to indicate the presence of a clip domain (Jiang and Kanost, 2000). For predicting the substrate specificities of SPs, residues 190, 216 and 226 (chymotrypsin numbering) (Perona and Craik, 1995), which determine the primary substrate-binding pocket, were identified from the aligned PD/PLD sequences. SPs containing Asp190, Gly216, and Gly/Ala/Ser226 are predicted to have trypsin-like specificity; SPs with Ser/Thr190, Gly216, and Gly/Ala/Ser226 are classified as chymotrypsin-like. When position 216 or 226 is occupied by a larger, usually nonpolar residue, these SPs are presumably elastase-like.
2.3. Multiple sequence alignment and phylogenetic analysis
Multiple sequence alignments of SP catalytic domains and SPH protease-like domains were performed using MUSCLE, one module of MEGA 6.0 (http://www.megasoftware.net). The following parameters were used: refining alignment, gap opening penalty = −2.9, gap extension penalty = 0, hydrophobicity multiplier = 1.2, maximum iterations = 100, clustering method (for iterations 1 and 2) = UPGMB, and maximum diagonal length = 24. In order to compare equivalent regions in all these sequences, the PD and PLD domain sequences with four extra residues preceding them were aligned. The aligned sequences were used to construct neighbour-joining phylogenetic trees using MEGA 6.0 with bootstrap method for the phylogeny test (1000 replications, Poisson model, uniform rates, and complete deletion of gaps or missing data). For multiple sequence alignment of the clip domains, the sequences from one residue before Cys-1 to one after Cys-6 were analyzed.
2.4. Protein structure modeling
Amino acid sequences of the 54 clip domains were submitted to the I-TASSER server (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) for 3D-structure prediction (Zhang, 2008). Models were built based on multiple-threading alignments by LOMETS and iterative TASSER simulations (Roy et al., 2010). The best model was automatically selected for the production of molecular graphics using PyMol (DeLano Scientific, Palo Alto, CA). Based on the phylogenetic tree of PD/PLD domains, groups or subgroups of the models were separately aligned and displayed in groups. For pairs with high sequence identity, the “align” command of Pymol was used to perform sequence and then structural alignment. For pairs with low sequence identity, the “cealign” command of Pymol, which performs a structure-based alignment was used. In each (sub)group, root-mean-square deviations (RMSDs) from the pairwise model comparisons were examined to identify the model with the lowest average RMSD for display. SPH42, whose model differed significantly from other group A:3 members (see below for the naming of clip domains), was moved to group B:2 (1st clip domain) to generate the lowest average RMSD.
2.5. Expression profiling
Manduca Cufflinks Assembly 1.0b (http://darwin.biochem.okstate.edu/blast/blast_links.html) was constructed based on Manduca Genome Assembly 1.0 (http://agripestbase.org/manduca/) and 52 cDNA libraries sequenced by Illumina technology (Cao and Jiang, 2015). These libraries represent mRNA samples isolated from whole larvae, organs, or tissues at various developmental stages. The number of reads mapped onto each transcript in the list of 107 SPs and 18 SPHs using RSEM (Li and Dewey, 2011) was used to calculate FPKM (fragments per kilobase of exon per million fragments mapped) in these libraries. Hierarchical clustering of the log2(FPKM+1) values was performed using MultiExperiment Viewer (v4.9) (http://www.tm4. org/mev.html) with the Pearson correlation-based metric and average linkage clustering method. K-means clustering was performed with Pearson correlation metric and 50 iterations to properly categorize the SP/SPHs based on their expression patterns. Based on the figure of merit (FOM), k was set to six for the final analysis. In each of these six groups, Tukey’s honestly significant difference (HSD) test was applied to further classify the log2(FPKM+1) values with p <0.05.
3. Results and discussion
3.1. Identification of highly reliable M. sexta SP and SPH sequences by a combined approach
The Manduca genome assembly consists of 20,891 scaffolds with N50 at 664 kb (X et al., 2015). Half of the scaffolds are shorter than 1 kb, accounting for 1.70% of the 419 Mb assembly. Some of the 17,000 NNN regions (4.71% of the assembly) contain gene elements. Repetitive elements and other highly similar sequences cause assembling errors. These imperfections pose challenges for quality gene annotation of large gene families, especially.
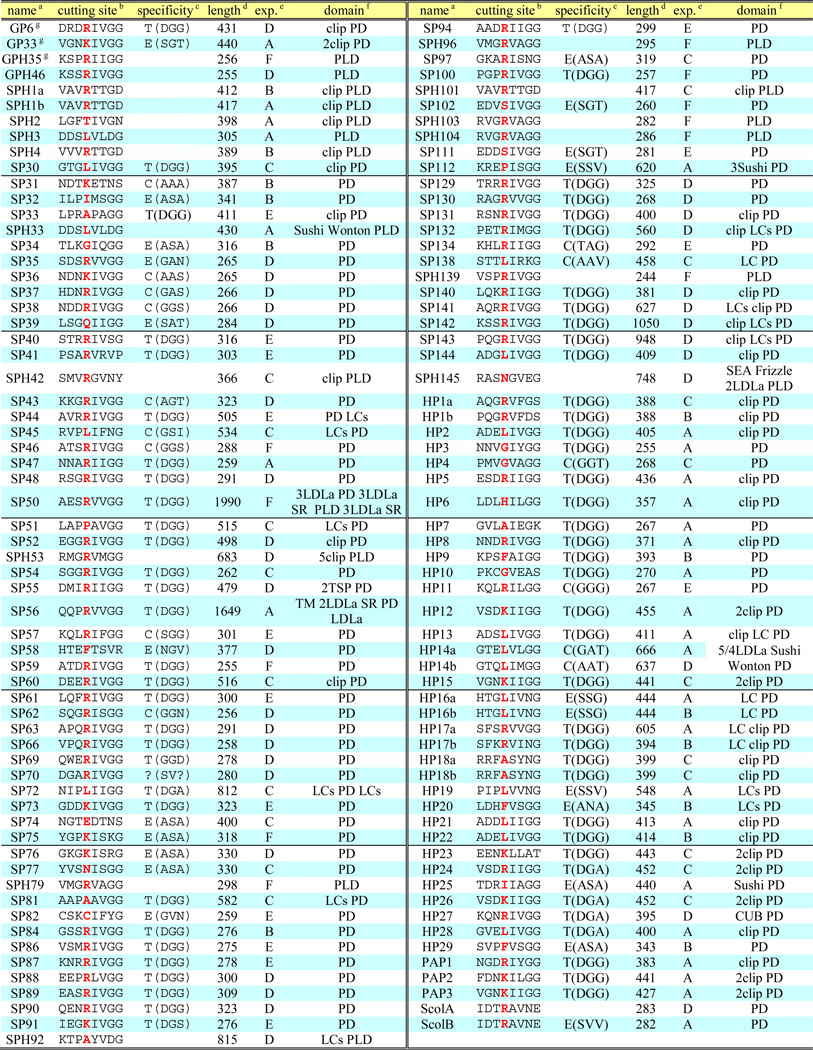
A TBLASTN search of Manduca Cufflinks Assembly 1.0 using the known M. sexta SP and SPH sequences as queries resulted in a long list of 563 homologs. After removal of the redundant hits and genes whose aligned regions are lower than 40% identical or shorter than 30 residues, 417 candidate transcripts were retrieved for ORF identification and domain prediction. During confirmation of the published SP and SPH sequences, we found in several cases parts of a single cloned cDNA located on different scaffolds. For instance, exons 1 through 5 and exons 7 through 10 of the HP2 gene are present on one scaffold and the region between exons 5 and 7 does not contain any unknown sequence, but exon 6 is found on another scaffold. This indicates that some regions of the genome were incorrectly assembled, perhaps due to repetitive sequences or high sequence similarity among certain family members. The Cufflinks sequences, assembled based on the genome sequence, cannot reveal such genes whose exons are not all on the same scaffold. Assuming similar defects exist in other scaffolds/contigs, we decided to use Oases and Trinity to assemble de novo all RNA-Seq data from the 52 cDNA libraries (Cao and Jiang, 2015). The genome-independent assemblies proved to be valuable tools for revealing genome assembly problems and, more commonly, extending Cufflinks sequences that are often disrupted by gaps between genomic fragments. During sequence crosschecks, we found about half of the SP/SPH hits in Manduca Official Gene Set 1.0 need minor-to-major improvements and, by combining the de novo assemblies with highly reliable genome contigs, the accuracy of predicted gene models was improved to 95% or higher. Furthermore, we examined the contigs containing multiple SP/SPH genes in a cluster and uncovered them one by one based on their conserved exon-intron borders. Within a large gene cluster, corresponding exons with high sequence similarity led to numerous “splicing isoforms” in Cufflinks 1.0 and 1.0b assemblies by merging some exons from one gene with other exons in nearby gene(s). Cufflinks 1.0b was constructed based on all the RNA-Seq data, including reads from the 19 cDNA libraries that had not been assembled into Cufflinks 1.0 (Cao and Jiang, 2015). Extensive search of all these sequence databases and painstaking cross-examination yielded a complete collection of SP-related coding sequences whose quality was further improved by removing redundant and fragmented ones. The final list included 193 SPs and SPHs. Based on preliminary analysis of their expression patterns, 68 are classified as gut (serine) proteases (GPs) and their close homologs (GPHs), which are reported in a companion paper (Kuwar et al., 2015). The other 125 SP-related proteins, including hemolymph (serine) proteases (HPs) and hemolymph SPHs, are reported in Table 1. In the rest of this paper, we use “SPs” or “SPHs” to refer to these 125 proteins presumably not involved in food digestion.
Table 1.
Properties of the 125 nondigestive SPs and SPHs in Manduca sexta
|
SP49, SP64, SP71, SP98, SP110, SP118, SP114, SP115, SP116, SPH117, SP119, SPH124, and SP133 are not listed since they are identical to PAP1, GP64, GP66, SP31, GP33, GP33, SP112, SP104, SP97, SPH1a, SP51, SPH101, and GP8, respectively. HP15b, SP68, SPH78, SPH80, SP83, SP85, SP105, SP106, SP107, SP108, SP109, SP113, SP120, SP121, SP122 (=SP123), and SP125 (=SP126) are incomplete genes and not listed either.
Putative activation cleavage site with the P1 site highlighted red;
enzyme specificity predicted based on Perona and Craik (1995). T: trypsin, C: chymotrypsin, E: elastase, blank: not applicable, letters in parentheses: residues determining the primary specificity of S1 pocket;
size of entire protein including signal peptide;
expression patterns;
PD: SP protease domain; PLD: SPH protease-like domain; LDLa: low-density lipoprotein receptor class A repeat; SR: scavenger receptor Cys-rich domain; LC: low complexity;
SP(H)s first identified in the midgut EST project (Pauchet et al., 2010).
3.2. General structural features of the nondigestive SPs and SPHs
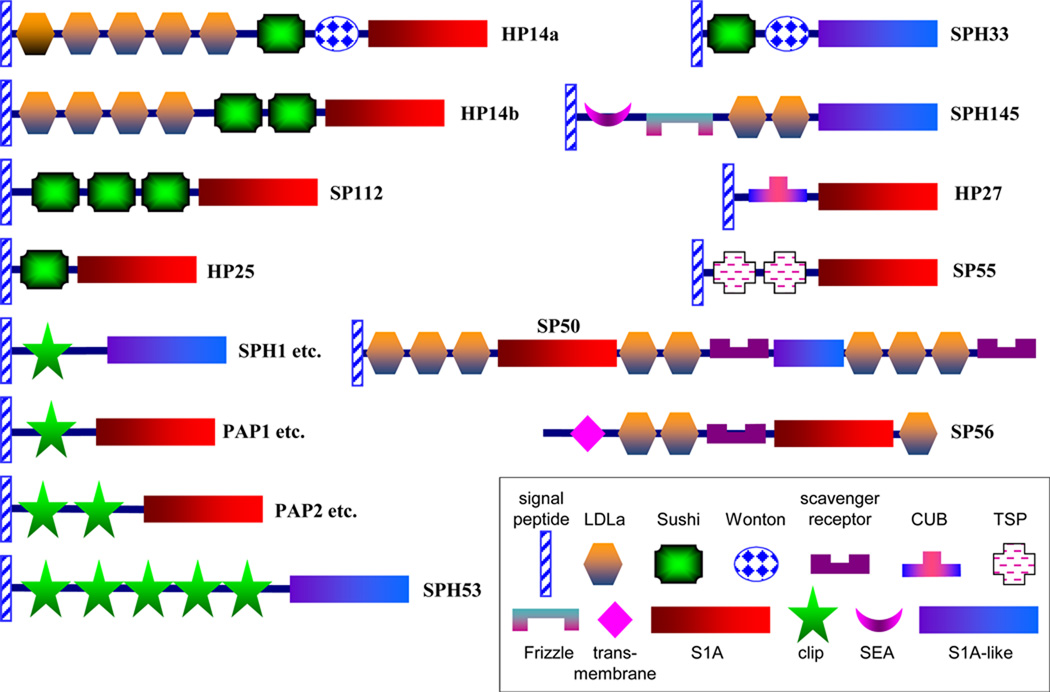
Consistent with their expected extracellular functions, all the sequences (except for SP56) are predicted to have a secretion signal peptide (Table 1). In the S1A family of SPs, the catalytic residues (His, Asp, Ser) are often embedded in conserved motifs of TAAHC, DIAL and GDSGGP, respectively. We identified these motifs in the 107 SP sequences, which are most likely catalytically functional proteases. In contrast, none of the 18 SPHs is anticipated to be catalytically active, because at least one of the catalytic triad is substituted. The overall foldings of the PDs (serine protease domains) and PLDs (SPH protease-like domains) are expected to be conserved, due to their similar primary structures. A typical S1A PD is 235~250 residues long, but these SPs and SPHs range from 255 to 1990 residues, with an average size of 413, indicating that many of them contain domains or other structural additions, which may have regulatory functions. Hence, we examined and identified in 52 of the 125 sequences ten domain types, namely clip (54), LDLa (low-density lipoprotein receptor class A repeat, 22), Sushi (8), Wonton (1), SR (scavenger receptor, 3), TSP (thrombospondin, 2), CUB (C1r/C1s, Uegf, Bmp1, 1), Frizzle (1) and SEA (sperm protein, enterokinase, agrin, 1) (Fig. 1), with the domain numbers indicated in parentheses. These structural modules probably allow their PDs or PLDs to interact with each other and form SP pathways to mediate complex physiological processes, such as embryonic development and immunity. Unlike digestion of dietary proteins, proper domain interactions are necessary in these cases to control the catalytic activities or to localize proteolytic reactions. This notion is supported by the conserved domain organization of certain SPs (e.g. HP14/HP14b/MSP, SP50/Nudel) and SPHs (e.g. SPH53/Masquerade) in a phylogenetically wide range of holometabolous insects including beetles, moths, butterflies, bees, wasps, ants, mosquitos, and flies (Christophides et al., 2002; Ross et al., 2003; Jiang et al., 2010; Park et al., 2010; Waterhouse et al., 2007; Zhao et al., 2010; Zou et al., 2006 and 2007).
Fig. 1. Domain organization of 42 multi-domain SPs and SPHs in M. sexta.
Signal peptide and other structural elements were predicted as described in Section 2.2. There are 27 single clip SPs, 6 single clip SPHs and 8 dual clip SPs represented by PAP1, SPH1 and PAP2, respectively.
Clip domains constitute the largest group of regulatory domains in the SP-related proteins of M. sexta. These disulfide-bridged structures exist in many arthropod SPs and SPHs involved in defense responses and embryonic development (Jiang and Kanost, 2000). In total, 54 clip domains are present in 35 SPs and 7 SPHs of M. sexta (Figs. 1 and 2), accounting for 34% of the nondigestive SPs/SPHs. Most (33) of them contain a single amino-terminal clip domain (27 SPs and 6 SPHs); 8 SPs have two clip domains; SPH53 has five clip domains. SPH53 is orthologous to Masquerade, a Drosophila SPH that affects axonal guidance and taste behavior (Murugasu-Oei et al., 1996). Penta-clip-domain SPHs are identified in all the holometabolous insects with known genome sequences (data not shown). The tandem clip domains may allow the attached SP or SPH domain to interact with multiple partners.
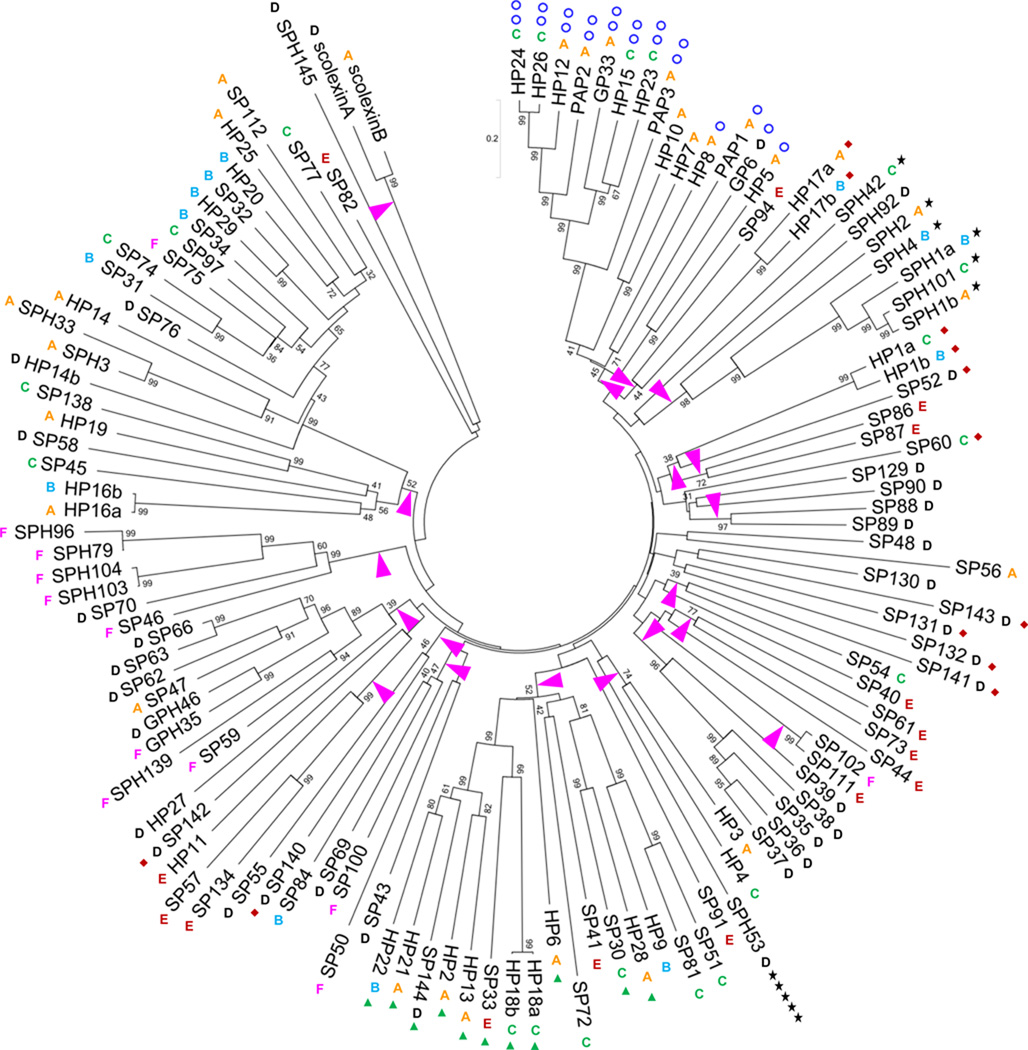
Fig. 2. Phylogenetic relationships of the 125 nondigestive M. sexta SPs and SPHs.
The amino acid sequences of the entire proteins were aligned for constructing the neighbor-joining tree as described in Section 2.2. On the basis of an arbitrary bootstrap value cutoff (>350 out of 1000 trials), arrowheads are placed at nineteen nodes closest to the tree center. These twigs are clockwisely numbered branches 1 through 19. Clip-domain SPs/SPHs in the CLIPA (★), CLIPB (○), CLIPC (▲) and CLIPD (♦) subfamilies are marked with the symbols, whose numbers correspond to the clip domain counts. The expression profiles (A, B, C, D, E and F, see Fig. S1) are indicated next to their names.
Some SPs contain other types of modules with conserved architecture. SP50 and Drosophila Nudel have the same domain structure of 3LDLa-PD-2LDLa-SR-PLD-3LDLa-SR (Fig. 1), suggesting that a partial fusion of two to three protease genes gave rise to an ancestral gene before it radiated to various insect groups to exert an essential function. Drosophila Nudel is an initiator of the SP cascade that determines the dorsal-ventral polarity of embryos. Interestingly, SP50 is also specifically produced by adult ovary in M. sexta (see below) and, like Nudel, may be deposited in eggs as a maternal factor. Other LDLa-domain SPs/SPHs include SP56, SPH145, HP14, and HP14b. M. sexta HP14, T. molitor and D. melanogaster modular serine proteases (MSPs) act as the first enzyme to trigger the prophenoloxidase (proPO) activation system and pro-Spätzle processing (Wang and Jiang, 2006; Kim et al., 2008; Buchon et al., 2009). In addition to the five LDLa repeats and PD domain, HP14, MSP and their homologs in other insects contain a Sushi domain and its variant, Wonton domain. M. sexta HP14b, containing four LDLa, two Sushi and one PD, is substantially expressed only in 0–3 h embryos (see below). Its physiological function in embryonic development is worth exploring in the future.
Sushi-domain SPs play key roles in the human complement system (Whaley and Lemercier, 1993). M. sexta SP112, HP25, and SPH33 contain one or three Sushi domains (Fig. 1). Other disulfide-stabilized domains include SR in SP50 and SP56, TSP in SP55, CUB in HP27, and Frizzle in SPH145. Except for SP56, all these SPs/SPHs contain no transmembrane region, and that does not exclude the possibility some of them associate with cell membrane via other mechanisms.
3.3. Sequence alignment and evolution of the nondigestive SPs and SPHs
Alignment of the 125 complete SP/SPH sequences and construction of the phylogenetic tree (Fig. 2) revealed 19 branches, clades or hypothetical taxonomic units (HTUs), whose root node bootstrap values were greater than 350 in 1000 trials. The low threshold was chosen to reveal significant phylogenetic relationships and preserve deeper thus less reliable ones at the same time (Ross et al., 2003). While HP27, SPH42, SPH53, SPH145, SP60, SP129, SP90, SP48, SP56, SP130, SP143, SP131, SP132, SP40, SP44, SP91, SP43, SP50, SP142 and SP82 are less similar to the other 105 SPs or SPHs, evolutionary relationships of the latter are revealed in these branches. Currently, we do not know the chromosomal locations of these genes. However, based on their positions on scaffolds or contigs (Table S1), we know that some of the 105 genes are in close proximity and probably arose from somewhat recent gene duplication and sequence divergence. These include: HP1a-HP1b (branch 4), SP86–SP87 (branch 5), SP88–SP89 (branch 6), SP35-SP36-SP37-SP38 (branch 10), SP47-SP62-SP63-SP66 and GPH35–GPH46 (branch 16), SPH79–SPH96 (branch 17), HP19-SP138, SP31-SP32-SP97 and HP14b-HP20-HP25-SPH3-SPH33-SP34-SP74-SP75-SP76-SP77 (branch 18), and scolexinA-scolexinB (branch 19). Each gene pair/cluster is located in the same scaffold.
Similar gene duplication events also gave rise to clip-domain SPs and SPHs (i.e. CLIPs) (Fig. 3). HP24, HP26, HP12, PAP2, and PAP3 genes are next to each other on adjacent contigs (Table S1). Whether their close homologs GP33, HP15 and HP23 are nearby on the same chromosome is not yet known. Genes in the following clusters (HP5-HP8-GP6 in branch 1, SPH1a-SPH1b-SPH4-SPH101 in branch 3, HP2-HP13-HP18a-HP18b-HP21-HP22-SP33 in branch 12) all encode a single clip domain and reside in the same or nearby contigs. An analysis of the 125 PDs/PLDs yielded many similar results (data not shown), indicating that gene duplications were responsible for the gene family expansion.
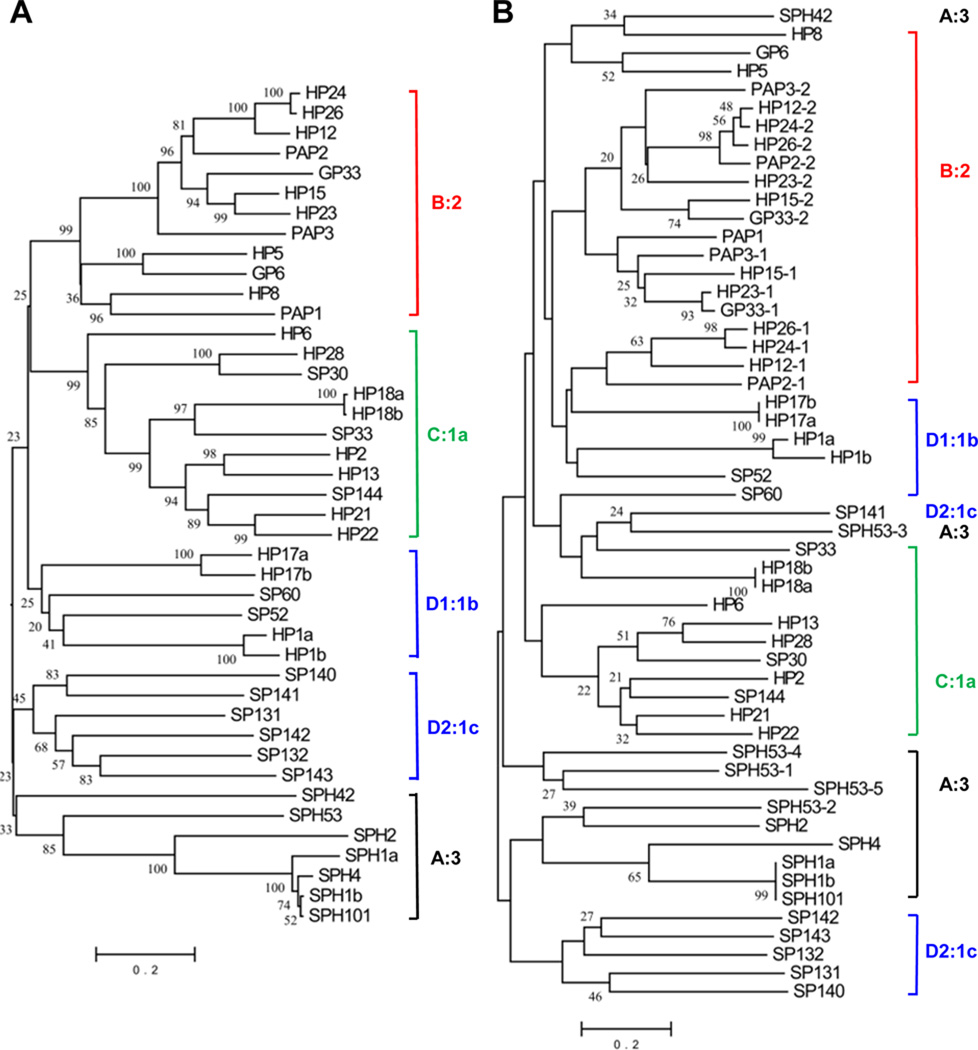
Fig. 3. Phylogenetic relationships of the catalytic/protease-like (A) and clip (B) domains of the 42 M. sexta CLIPs.
Sequence alignments and neighbor-joining trees were constructed as described in Section 2.3. For proteins with multiple clip domains, these domains are numbered 1, 2, 3… Bootstrap values of greater than 200 in 1000 trials are marked at the corresponding nodes. Vertical bars and names (A:3, B:2, C:1a, D1:1b and D2:1c) indicate the CLIP (A to D2) and clip domain (1a to 3) groups.
3.4. Phylogenetic relationships of the clip-domain SPs and SPHs
Intrigued by their functional importance, we further studied the sequences of the 42 CLIPs. A phylogenetic tree based on their PD and PLD sequences (Fig. 3A) indicates that the proteins fall into five clades, which also have interesting similarities in their domain architecture. Clade A consists of six single clip SPHs and one quintuple clip SPH53; clade B includes eight dual clip SPs (HP15, HP23, HP24, HP26, HP12, PAP2, PAP3, GP33) and four single clip SPs (HP5, HP8, GP6, PAP1); clade C has eleven single clip SPs (HP2, HP6, HP13, HP18a, HP18b, HP21, HP22, HP28, SP30, SP33, and SP144); clade D is composed of twelve single clip SPs that form two subgroups D1 (HP1a, HP1b, HP17a, HP17b, SP52 and SP60) and D2 (SP131, SP132, SP140, SP141, SP142 and SP143). These clade names are designated based on their alignment with homologs from D. melanogaster, A. gambiae, and T. castaneum (data not shown). As such, clades A through D correspond to A. gambiae CLIPAs through CLIPDs, respectively. These results suggest that the major groups of CLIP genes were in existence before the radiation of holometabolous insects. M. sexta CLIPA genes are a lot fewer than those in the fly, beetle, and mosquitoes, whereas A. aegypti CLIPB genes are considerably more than the other insects (Table 2).
Table 2.
Number of CLIP genes in the five holometabolous insectsa
| CLIP subfamily | M. sexta | D. melanogaster | A. gambiae | A. aegypti | T. castaneum |
|---|---|---|---|---|---|
| A: SPHsb | 6 | 14 | 20 | 10 | 17 |
| B: SPs | 13 | 14 | 20 | 36 | 15 |
| C: SPs | 11 | 7 | 8 | 10 | 6 |
| D: SPs | 12 | 10 | 7 | 7 | 11 |
| total | 42 | 45 | 55 | 63 | 49 |
The counts of D. melanogaster, A. gambiae, and A. aegypti clip-domain SPs and SPHs are based on Waterhouse et al., 2007. The T. castaneum gene counts are from Zou et al., 2007.
Dipteran clip-domain SPHs in subfamilies A and E are combined.
Alignment of the 54 clip domains generally supports the phylogenetic relationships derived from those of the complete and PD/PLD sequences, but the aligned sequences and tree topology reveal remarkable diversity of the regulatory domains (Table 3; Fig. 3B). Nine of the eleven group-3 clip domains in CLIPAs form two clades with root node bootstrap values lower than 20. SPH1a, SPH1b, SPH101, and SPH4 clip domains form a tight subtree with topology similar to that of the PLD sequences to which they are linked (Fig. 3A). Clip domains of SPH53-3-SP141 (D2:1c) and SPH42-HP8 (B:2) form two pairs. Five group-1c clip domains in CLIPD2s form a clade similar in topology to that of the associating PDs. In contrast, sequence hypervariability of the six group-1b clip domains in CLIPD1s greatly affects the reliability of branching orders, when compared with those based on alignments of the corresponding PDs. Seven of the eleven group-1a clip domains in CLIPCs form a close group (Fig. 3B), and so do most of the group-2 clip domains in CLIPBs. The branch orders of the 2nd clip domains in HP12-HP15-HP23-HP24-HP26-PAP2-PAP3-GP33 (Fig. 3B) closely resemble those in Fig. 3A. In summary, despite variability in clip domain sequences, their evolution seems to have followed similar paths as their associated PDs/PLDs. CLIPBs are closer to CLIPCs and CLIPDs than to CLIPAs.
Table 3.
Properties of the 55 clip domains and their structure models
| Clip domaina | C1–C2b | C2–C3 | C3–C4 | C4–C5 | C5–C6 | Groupc | Avg. RMSDd | Grp. RMSDe | Structuref |
| HP24-1 | 9 | 5 | 24 | 9 | 0 | B:2 | 0.68 | PAP2 | |
| HP26-1 | 9 | 5 | 24 | 9 | 0 | B:2 | 0.74 | PAP2 | |
| HP12-1 | 9 | 5 | 24 | 9 | 0 | B:2 | 0.74 | PAP2 | |
| PAP2-1 | 9 | 5 | 24 | 9 | 0 | B:2 | 0.71 | PAP2g | |
| GP33-1 | 9 | 5 | 24 | 9 | 0 | B:2 | 0.69 | PAP2 | |
| HP15-1 | 9 | 5 | 24 | 9 | 0 | B:2 | 0.74 | PAP2 | |
| HP23-1 | 9 | 5 | 24 | 9 | 0 | B:2 | 0.71 | PAP2 | |
| PAP3-1 | 9 | 5 | 23 | 9 | 0 | B:2 | 0.70 | PAP2 | |
| SPH42 | 5 | 5 | 25 | 9 | 0 | A:3 | 1.44 | 0.79 | PAP2 |
| HP24-2 | 9 | 5 | 23 | 8 | 0 | B:2 | 0.59 | PAP2 | |
| HP26-2 | 9 | 5 | 23 | 8 | 0 | B:2 | 0.69 | PAP2 | |
| HP12-2 | 9 | 5 | 23 | 8 | 0 | B:2 | 0.78 | PAP2 | |
| PAP2-2 | 9 | 5 | 23 | 8 | 0 | B:2 | 0.64 | PAP2g | |
| GP33-2 | 9 | 5 | 23 | 8 | 0 | B:2 | 0.65 | PAP2 | |
| HP15-2 | 9 | 5 | 23 | 8 | 0 | B:2 | 0.70 | PAP2 | |
| HP23-2 | 9 | 5 | 23 | 8 | 0 | B:2 | 0.71 | PAP2 | |
| PAP3-2 | 9 | 5 | 23 | 8 | 0 | B:2 | 0.89 | 0.66 | PAP2 |
| HP5 | 8 | 5 | 26 | 9 | 0 | B:2 | 0.72 | PAP2 | |
| GP6 | 8 | 5 | 26 | 9 | 0 | B:2 | 0.83 | PAP2 | |
| HP8 | 5 | 5 | 23 | 10 | 0 | B:2 | 0.80 | PAP2 | |
| PAP1 | 9 | 5 | 24 | 9 | 0 | B:2 | 0.67 | 0.75 | PAP2 |
| 8–9 (8.9) | 5 (5.0) | 23–26 (23.7) | 8–10 (8.7) | 0 (0) | B:2 | 0.73 | |||
| HP6 | 10 | 5 | 17 | 9 | 0 | C:1a | 1.31 | PAP2 | |
| HP28 | 10 | 5 | 16 | 10 | 0 | C:1a | 2.08 | PAP2 | |
| SP30 | 10 | 5 | 16 | 10 | 0 | C:1a | 2.03 | PAP2 | |
| HP18a | 8 | 5 | 15 | 9 | 0 | C:1a | 1.61 | PAP2 | |
| HP18b | 8 | 5 | 15 | 9 | 0 | C:1a | 1.32 | PAP2 | |
| HP2 | 8 | 5 | 15 | 9 | 0 | C:1a | 1.47 | PAP2 | |
| HP13 | 8 | 5 | 15 | 9 | 0 | C:1a | 1.26 | PAP2 | |
| SP144 | 8 | 5 | 16 | 9 | 0 | C:1a | 1.33 | PAP2 | |
| HP21 | 9 | 5 | 15 | 9 | 0 | C:1a | 1.32 | PAP2 | |
| HP22 | 8 | 5 | 16 | 9 | 0 | C:1a | 1.57 | PAP2 | |
| SP33 | 8 | 5 | 26 | 18 | 0 | C:1a | 3.04 | PAP2 | |
| 8–10 (8.6) | 5 (5.0) | 15–17 (15.6) | 9–10 (9.2) | 0 (0) | C:1a | 1.67 | |||
| HP1a | 9 | 5 | 16 | 9 | 0 | D1:1b | 1.59 | PAP2 | |
| HP1b | 9 | 5 | 16 | 9 | 0 | D1:1b | 1.58 | PAP2 | |
| HP17a | 9 | 5 | 24 | 9 | 0 | D1:1b | 1.37 | PAP2 | |
| HP17b | 9 | 5 | 24 | 9 | 0 | D1:1b | 1.57 | PAP2 | |
| SP52 | 9 | 5 | 14 | 9 | 0 | D1:1b | 1.80 | PAP2 | |
| SP60 | 9 | 5 | 22 | 13 | 0 | D1:1b | 2.15 | 1.68 | PAP2 |
| 9 (9.0) | 5 (5.0) | 14–24 (19.3) | 9 (9.0) | 0 (0) | D1:1b | 1.68 | |||
| SP131 | 8 | 5 | 10 | 7 | 0 | D2:1c | 4.38 | HNP1 | |
| SP132 | 8 | 5 | 10 | 8 | 0 | D2:1c | 3.89 | MBD8 | |
| SP140 | 8 | 5 | 10 | 7 | 0 | D2:1c | 4.55 | No | |
| SP141 | 5 | 5 | 10 | 6 | 0 | D2:1c | 4.50 | MBD8 | |
| SP142 | 14 | 5 | 10 | 8 | 0 | D2:1c | 4.21 | MBD8 | |
| SP143 | 10 | 5 | 10 | 8 | 0 | D2:1c | 4.48 | 4.39 | MBD8 |
| 8–10 (8.5) | 5 (5.0) | 10 (10.0) | 6–8 (7.3) | 0 (0) | D2:1c | 4.39 | |||
| SPH1a | 9 | 5 | 23 | 6 | 0 | A:3 | 2.21 | PAP2 | |
| SPH1b | 9 | 5 | 23 | 6 | 0 | A:3 | 2.38 | PPAFII | |
| SPH2 | 1 | 5 | 30 | 6 | 0 | A:3 | 3.85 | anthopleurin A | |
| SPH4 | 9 | 5 | 22 | 6 | 0 | A:3 | 2.89 | PPAFII | |
| SPH101 | 9 | 5 | 23 | 6 | 0 | A:3 | 2.37 | 2.74 | PPAFII |
| SPH53-1 | 3 | 8 | 8 | 5 | 0 | A:3 | 4.95 | No h | |
| SPH53-2 | 3 | 8 | 9 | 5 | 0 | A:3 | 5.23 | No | |
| SPH53-3 | 3 | 8 | 8 | 5 | 0 | A:3 | 4.57 | No h | |
| SPH53-4 | 3 | 9 | 9 | 6 | 0 | A:3 | 4.91 | No | |
| SPH53-5 | 3 | 8 | 11 | 6 | 0 | A:3 | 5.67 | 5.07 | No |
| 9 (9.0) | 5 (5.0) | 22–30 (24.3) | 5–6 (5.7) | 0 (0) | A:3 | 3.91 | |||
Clip domain with the lowest average root-mean-square deviation (RMSD) in each (sub)group is shaded light green.
Number of residues between adjacent cysteines, with the ones differ significantly from others in each (sub)group shown in red font. The range and average (in parenthesis) number for each group (excluding the red ones) are enlisted.
Group names of cSPs/cSPHs (B, C, D1, D2, and A) and corresponding clip domains (2, 1a, 1b, 1c, and 3) are indicated.
average of the RMSD values (Å) of a clip domain structure model pairwisely compared with the other models in the same (sub)group.
Group and subgroup averages of average RMSDs, excluding SP33 and SPH42 (highlighted yellow), demonstrate variations of structure models in the same (sub)group.
Most similar three-dimensional structure: PAP2, the two clip domains of M. sexta proPO activating protease-2 (Huang et al., 2007), PPAFII, a clip-domain SPH named proPO activating factor II from Holotrichia diomphalia (Piao et al., 2005), HNP-1, human neutrophil peptide-1 (i.e. human α-defensin 1) (Zhang et al., 2010), MBD8, mouse β-defensin 8 (Bauer et al., 2001), and anthopleurin A, a cardiac stimulant from the sea anemone Anthopleura xanthogrammica (Pallaghy et al., 1995).
Average RMSDs between the models and experimentally determined structures (Huang et al., 2007) are 0.821 Å and 0.636 Å for M. sexta PAP2 clip domain-1 and -2, respectively.
The structures of metallothionein and Amaranthus caudatus AMP2 are most similar to the models of SPH53-1 and -3 respectively, but the locations of Cys residues and linkages of disulfide bonds are remarkably different from those in the two clip domains. In addition to that, the disulfide linkages of HNP-1 (C1–C6, C2–C4, C3–C5) (Zhang et al., 2010) are different from those in the known clip domain structures (C1–C5, C2–C4, C3–C6) (Huang et al., 2007; Piao et al., 2005).
Except for GP33, all the 35 clip-domain SPs are predicted to have trypsin-like specificity by cleaving next to a positively charged residue (Arg, Lys or His), since their primary substrate binding pockets are composed of Asp-Gly-Ala in HP24, HP26 and HP28 or Asp-Gly-Gly in the others (Table 1) (Perona and Craik, 1995). With Ser-Gly-Thr located in the equivalent positions, GP33 may be an elastase-like protease, cleaving after small residues (e.g. Ala). Although the same trypsin-like specificity cannot be correlated with the different CLIP groups, their proteolytic activation sites exhibit interesting patterns: all the six CLIPD2s are probably activated by cleavage between Arg and Ile; the six CLIPD1s are cut between Arg and Val/Ile. In contrast, seven of the eleven CLIPCs are predicted to be cleaved between Leu and Ile; SP33, HP18a and HP18b after Ala; HP6 after His (Table 1). All four single (GP6, PAP1, HP5 and HP8) and one dual clip (HP24) CLIPBs are activated by cutting between Arg and Ile, and the other seven (dual clip) CLIPBs between Lys and Ile/Leu. These findings are consistent with the general notion that initiator SPs (e.g. HP14/MSP) with chymotrypsin-like specificity cuts CLIPCs next to Leu and the active CLIPCs then activates CLIPBs by cleavage next to Lys/Arg (Jiang and Kanost, 2000; An et al., 2009; Kellenberger et al., 2011).
3.5. Structural features of the clip-domains deduced from molecular modelling
We proposed that clip domain may interact with a regulatory protein or anchor the enzyme to the surface of an invading organism (Jiang and Kanost, 2000). While 3D-structures of three clip domains have been solved (Piao et al., 2005; Huang et al., 2007), little is known about their functions in relation to the structures. With all the clip domains identified from the genome project, we attempted to predict their tertiary structures for use as a basis to understand their functions and evolution. Using I-TASSER server, we built models of the 54 clip domain structures based on multiple-threading alignments and iterative template assembly simulations (Roy et al., 2010). Results of the phylogenetic analysis (Fig. 3) allowed us to align and compare the predicted structures in the eight groups, despite of the limitations of molecular modeling.
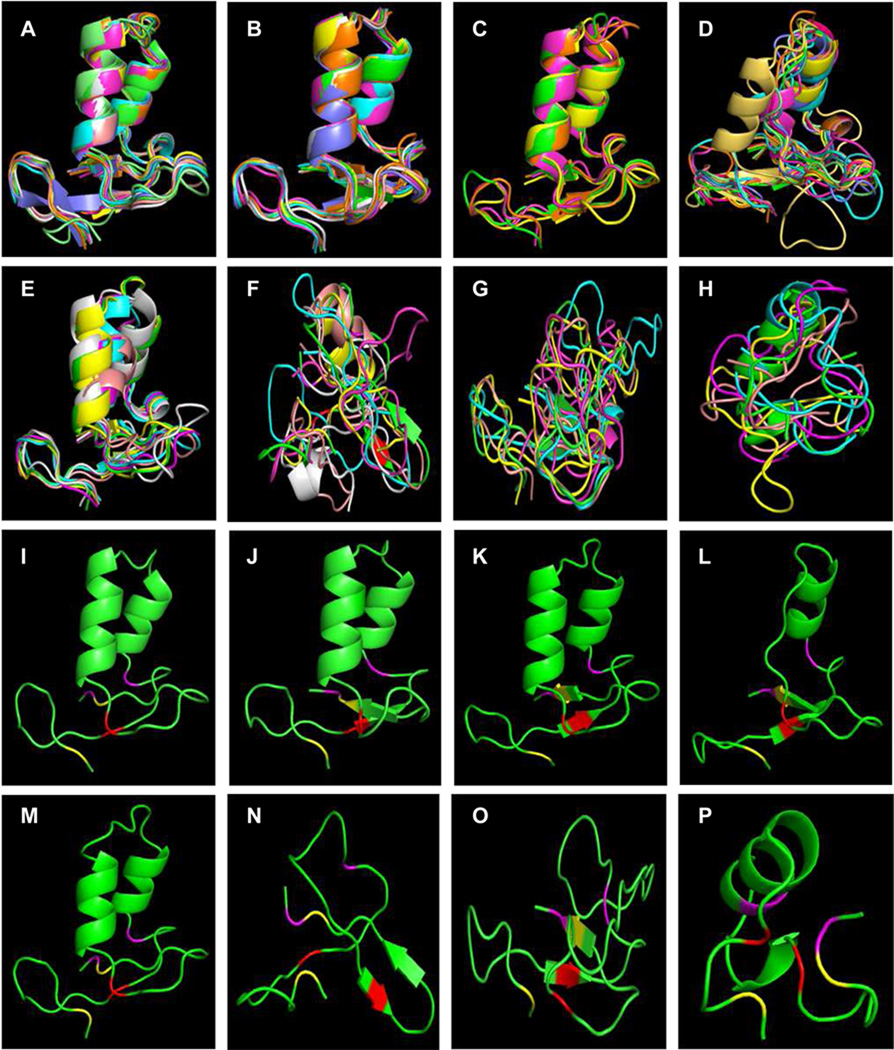
The first clip domains in the dual clip SPs (Fig. 4A) are remarkably similar to each other. HP24-1 (Fig. 4I) best represents this group, since the average root-mean-square deviations (RMSD) for the other eight models compared with HP24-1 is the lowest (0.68 Å) (Table 3). The average RMSDs against the other models range from 0.68 to 1.44 Å with a group average at 0.79 Å. The RMSD between PAP2-1 model and actual structure (Huang et al., 2007) is 0.64 Å. Likewise, the second clip domains in the same proteins are highly similar to each other, which are best represented by HP24-2 (Fig. 4, B and J). The lowest, range, and average RMSDs for this group are 0.59, 0.59 to 0.89, and 0.66 Å, respectively. The RMSD between PAP2-2 model and actual structure is 0.64 Å. Since the PAP2-1 and PAP2-2 models are constructed not based on their known structures, the structural resemblances (0.64 and 0.64 Å) being so close to the group RMSDs (0.79 and 0.66 Å) strongly suggest that all these models are reliable. So are those for the clip domains in the four single clip SPs (Fig. 4, C and K). PAP1 best represents this group and the lowest, range and average RMSDs are 0.67, 0.67 to 0.83, and 0.75 Å (Table 3), respectively. These group-2 clips in CLIPBs may adopt the same structures: a three-stranded antiparallel β-sheet flanked by two α-helices. Interestingly, the clip domain in SPH42 is predicted to have a structure similar to the group-2 clips, especially the clip domain-1s (Fig. 4A and Table 3). This is consistent with the clip domain tree where the “group-3” clip is together with the group-2 clips of HP8, GP6 and HP5 (Fig. 3C).
Fig. 4. Molecular modeling of the clip domains in the 35 SPs and 7 SPHs of M. sexta.
The 54 clip domains were modeled and compared as described in Section 2.4. (Sub)groups of the models and their representatives are shown in A through H and I through P, respectively. Panel A: the 1st clip domain (group-2) in the CLIPB proteases GP33, HP12, HP15, HP23, HP24, HP26, PAP2, PAP3, and SPH42; Panel B: the 2nd clip domain (group-2) in the same eight CLIPB proteases; Panel C: the clip domains (group-2) in the CLIPB proteases GP6, HP5, HP8 and PAP1; Panel D: the group-1a clip domains in the CLIPC proteases HP2, HP6, HP13, HP18a, HP18b, HP21, HP22, HP28, SP30, SP33 and SP144; Panel E: the group-1b clip domains in CLIPD1 proteases HP1a, HP1b, HP17a, HP17b, SP52 and SP60; Panel F: the group-1c clip domains in the CLIPD2 proteases SP131, SP132, SP140, SP141, SP142 and SP143; Panel G: the group-3 clip domains of SPH1a, SPH1b, SPH2, SPH4 and SPH101 (CLIPAs); Panel H: the five group-3 clip domains in SPH53 (CLIPA); Panels I through P: clip domains with the lowest RMSD in each (sub)group are HP24 clip domain-1 (I), HP24 clip domain-2 (J), PAP1 (K), HP13 (L), HP17a (M), SP132 (N), SPH1a (O) and SPH53-3 (P). The six Cys residues predicted to form three disulfide bonds (Cys1-Cys5, Cys2-Cy4, and Cys3-Cys6) are colored yellow, red, and magenta, respectively.
The group-1a clip domains in CLIPCs seem to be conserved in the three-stranded β-sheet and one α-helix (Fig. 4, D and L). The C-terminal α-helix either is missing or has only one turn, which may be caused by shorter sequences (15 to 17 residues) between Cys-3 and Cys-4 than those (23 to 26 residues) in group-2 clip domains (Table 3). HP13 best represents this group. The lowest, range and average RMSDs are 1.26, 1.26 to 3.04, and 1.67 Å, respectively. Similarly, the group-1b clip domains in CLIPD1s may also contain a small, antiparallel β-sheet flanked by two short α-helices (Fig. 4, E and M). The regions between Cys-3 and Cys-4 are intermediate: 14 to 24 residues long. HP17a, the best representative of this group, has an average RMSD of 1.37 Å when compared with the other five models (Table 3). The range and average RMSDs are 1.37 to 2.15 and 1.68 Å. The clip domains in CLIPBs, CLIPCs and CLIPD1s appear to be similar in 3D-structure to the PAP2-1 and PAP2-2. In contrast, those in CLIPD2s and CLIPAs somewhat resemble mouse β-defensin-8 and PPAFII, respectively (Fig. 4, F to H, N to P). Their high group RMSD values (4.39 and 2.74 Å) and ranges (3.89 to 4.55 Å and 2.21 to 3.85 Å) suggest major structural divergences. An extreme example is the five clip domains in SPH53, which either have no resemblance to any known protein or have low similarity to structures with different disulfide linkage patterns (Table 3). These structural modeling results are mostly consistent with the evolutionary relationships based on the sequence comparison (Fig. 3).
3.6. Expression profiling
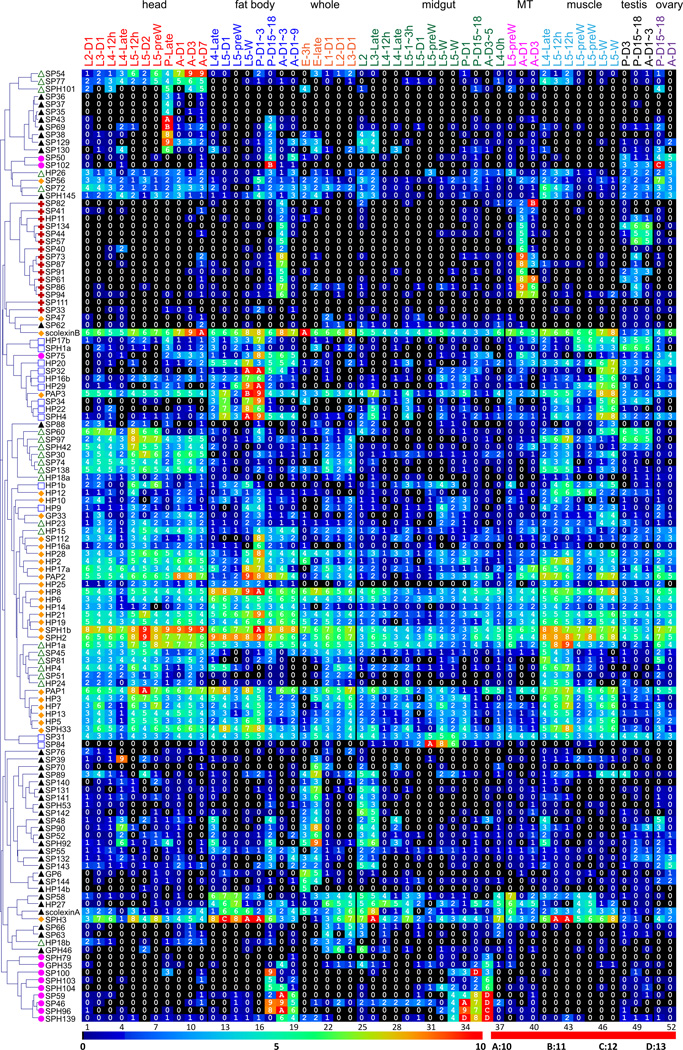
As a step toward understanding roles of the 125 nondigestive SPs and SPHs, we examined their mRNA levels in the 52 tissue samples from M. sexta at various developmental stages. Cluster analysis of the expression profiles resulted in six groups (Fig. S1). Group A included 15 CLIPs and 13 other SPs/SPHs, whose average log2(FPKM+1) value (3.1) of the 28 genes in the 52 libraries was higher than those of the other groups (B: 1.6, C: 1.5, D: 0.7, E: 0.4, and F: 0.8). The mRNA levels for genes in group A were significantly higher in fat body of the wandering larvae (5.0) and early pupae (6.2) and muscles of day 0.5, 5th instar larvae (4.8, 4.8) than in the other libraries. Group B had thirteen members including five CLIPs. Their mRNA levels were also highest in the wandering larval (5.7) and early pupal (5.5) fat body samples, and the levels in the muscles of wandering larvae (4.5, 5.1) were also significantly higher than in the other 48 libraries. Group C consisted of 11 CLIPs and 10 other SPs, whose transcripts were most abundant in heads of larvae, pupae and adults as well as muscles of the late 4th and early 5th instar larvae. Group D includes 10 CLIPs and 26 other SPs/SPHs. Characteristically high mRNA levels were found in heads of the late pupae (2.0), in the late embryos (2.8), and in midgut of the 2nd (2.7) and 3rd (2.3) instar larvae. Group E was composed of 1 CLIP and 14 SPs, with mRNA levels significantly higher in adult fat body (4.1) and Malpighian tubules (4.8, 2.8), pupal and adult testis (2.5, 1.5) than the other libraries. Group F included 6 SPs and 6 SPHs. Their highest transcript levels were found in late pupal and early adult fat body (4.9, 4.3) and midgut (4.4, 6.4). Some of these expression patterns may reflect transcriptional co-regulation.
We also identified unique expression patterns in certain SP genes (Table S2). For instance, the mRNA levels or log2(FPKM+1) values of SP56 (A: 6.6) and SP102 (F: 12.1) were highest in ovaries of late pupae, SP84 (B: 9.7 and 8.3) was at high levels in midgut of pre-wandering and wandering larvae, SP39 (D: 8.8) in heads of late 4th instar larvae, and SP75 (F: 8.1) in fat body of early pupae, compared other tissues (Fig. 5). Such information on tissue/stage-specific expression is useful for their cDNA isolation and functional analysis.
Fig. 5. Transcript profiles of the nondigestive SPs/SPHs in the 52 M. sexta tissue samples.
Log2(FPKM+1) values for the SP-related mRNAs are shown in the gradient heat map from blue (0) to red (≥10). The values of 0~0.49, 0.50~1.49, 1.50~2.49 … 8.50~9.49, 9.50~10.49 10.50~11.49, 11.50~12.49 and 12.50~13.49 are labeled as 0, 1, 2 … 9, A, B, C and D, respectively. Cluster analysis has revealed six distinct patterns. A (♦), B (□), C (△), D (▲), E (+), and F (●). The 52 cDNA libraries (1 through 52) represent the following tissues and stages: heads [1. 2nd (instar) L (larvae), d1 (day 1); 2. 3rd L, d1; 3. 4th L, d0.5; 4. 4th L, late; 5. 5th L, d0.5; 6. 5th L, d2; 7. 5th L, pre-W (pre-wandering); 8. P (pupae), late; 9. A (adults), d1; 10. A, d3; 11. A, d7], fat body (12. 4th L, late; 13. 5th L, d1; 14. 5th L, pre-W; 15. 5th L, W; 16. P, d1–3; 17. P, d15–18; 18. A, d1–3; 19. A, d7–9), whole animals [20. E (embryos), 3h; 21. E, late; 22. 1st L; 23. 2nd L; 24. 3rd L), midgut (25. 2nd L; 26. 3rd L; 27. 4th L, 12h; 28. 4th L, late; 29. 5th L, 1–3h; 30. 5th L, 24h; 31. 5th L, pre-W; 32–33. 5th L, W; 34. P, d1; 35. P, d15–18; 36. A, d3–5; 37. 4th L, 0h), MT (38. 5th L, pre-W; 39. A, d1; 40. A, d3), muscle (41. 4th L, late; 42–43. 5th L, 12h; 44–45. 5th L, pre-W; 46–47. 5th L, W), testes (48. P, d3; 49. P, d15–18; 50. A, d1–3), and ovaries (51. P, d15–18; 52. A, d1).
3.7. Concluding remarks
Serine proteases and their homologs constitute one of the largest protein families in insects. Here, we have analyzed 125 of the 193 SPs and SPHs in M. sexta and established a framework of knowledge which is expected to facilitate research on their functions in processes not related to food digestion. Over half of the 105 SPs/SPHs have arisen by lineage-specific gene duplications, making it difficult to identify orthologs in closely related insects. Nevertheless, the extensive RNA-Seq data not only ensure the quality of gene annotation but also reveal their temporospatial expression patterns. Based on the correct sequences, we have identified ten types of regulatory domains. Interestingly, the classification of CLIPs and evolutionary relationships deduced from sequence alignments, in the most part agree well with structural diversity of the clip domain models and conservation of the putative proteolytic activation sites. This information, along with the predicted specificity of PDs, will guide our biochemical elucidation of the SP-SPH network that mediates or modulates immune mechanisms.
Supplementary Material
FPKM values were calculated for 193 SP-related genes as described in Section 2.5. Cluster analysis indicated that 68 of them were midgut-specific SP/SPH genes and, therefore, further analyzed in another study (Kuwar et al., 2015). The remaining 125 genes exhibited six distinct expression patterns, as shown in panels A through F with the 28, 13, 21, 36, 15, and 12 names indicated, respectively. The relative expression levels or log2(FPKM+1) values for individual genes are connected by grey lines, and their means by a pink line in each panel. Samples in the same tissue groups are underlined alternately. The libraries are numbered as described in Fig. 5.
Highlights.
Identify 125 nondigestive SPs/SPHs, 52 with 1–10 cystine-stabilized domains;
Reveal phylogenetic relationships among members in each of the 19 branches;
Build reliable 3D-models for the group-2, -1a, and -1b clip domains;
Profile mRNA levels of the 125 SPs/SPHs in tissues at different stages.
Acknowledgements
This work was supported by NIH grants GM58634 (to H. Jiang) and GM41247 (to M. Kanost), NSFC grant 31272367 (to Z. Zou), Chinese IPM1304 (to Z. Zou), and a DARPA grant (to G. Blissard). This work was approved for publication by the Director of Oklahoma Agricultural Experimental Station, and supported in part under project OKLO2450 (to H. Jiang). Computation for this project was performed at OSU High Performance Computing Center at Oklahoma State University supported in part through the National Science Foundation grant OCI–1126330.
Abbreviations
- SP
serine protease
- SPH
serine protease homolog
- HP
hemolymph (serine) protease
- GP
gut (serine) protease
- GPH
gut (serine) protease homolog
- PD
SP catalytic domain
- PLD
SPH protease-like domain
- LDLa
low-density lipoprotein receptor class A repeat
- SR
scavenger receptor
- TSP
thrombospondin
- CUB
C1r/C1s, Uegf, Bmp1
- SEA
sperm protein, enterokinase, agrin
- LC
low complexity
- MSP
modular serine protease
- PSP
plasmatocyte spreading peptide
- HTU
hypothetical taxonomic units
- CLIP
clip-domain SP or SPH
- PO and proPO
phenoloxidase and its precursor
- PAP
proPO activating protease
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- An C, Ishibashi J, Ragan EJ, Jiang H, Kanost MR. Functions of Manduca sexta hemolymph proteinases HP6 and HP8 in two innate immune pathways. J Biol Chem. 2009;284:19716–19726. doi: 10.1074/jbc.M109.007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barillas-Mury C. CLIP proteases and Plasmodium melanization in Anopheles gambiae. Trends Parasitol. 2007;23:297–299. doi: 10.1016/j.pt.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Bauer F, Schweimer K, Klüver E, Conejo-Garcia JR, Forssmann WG, Rösch P, Adermann K, Sticht H. Structure determination of human and murine β-defensins reveals structural conservation in the absence of significant sequence similarity. Protein Sci. 2001;10:2470–2479. doi: 10.1110/ps.24401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N, Poidevin M, Kwon HM, Guillou A, Sottas V, Lee BL, Lemaitre B. A single modular serine protease integrates signals from pattern-recognition receptors upstream of the Drosophila Toll pathway. Proc Natl Acad Sci USA. 2009;106:12442–12447. doi: 10.1073/pnas.0901924106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Jiang H. Integrated modeling of protein-coding genes in the Manduca sexta genome using RNA-Seq data from the biochemical model insect. Insect Biochem Mol Biol. 2015 doi: 10.1016/j.ibmb.2015.01.007. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophides GK, Zdobnov E, Barillas-Mury C, Birney E, Blandin S, Blass C, Brey PT, Collins FH, Danielli A, Dimopoulos G, Hetru C, Hoa NT, Hoffmann JA, Kanzok SM, Letunic I, Levashina EA, Loukeris TG, Lycett G, Meister S, Michel K, Moita LF, Müller HM, Osta MA, Paskewitz SM, Reichhart JM, Rzhetsky A, Troxler L, Vernick KD, Vlachou D, Volz J, von Mering C, Xu J, Zheng L, Bork P, Kafatos FC. Immunity-related genes and gene families in Anopheles gambiae. Science. 2002;298:159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- Finnerty CM, Karplus PA, Granados RR. The insect immune protein scolexin is a novel serine proteinase homolog. Protein Sci. 1999;8:242–248. doi: 10.1110/ps.8.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunaratna R, Jiang H. A comprehensive analysis of the Manduca sexta immunotranscriptome. Dev Com Immunol. 2013;39:388–398. doi: 10.1016/j.dci.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Lu Z, Dai H, Vander Velde D, Prakash O, Jiang H. The solution structure of the clip domains from Manduca sexta prophenoloxidase activating proteinase-2. Biochemistry. 2007;46:11431–11439. doi: 10.1021/bi7010724. [DOI] [PubMed] [Google Scholar]
- Kellenberger C, Leone P, Coquet L, Jouenne T, Reichhart JM, Roussel A. Structure-function analysis of grass clip serine protease involved in Drosophila Toll pathway activation. J Biol Chem. 2011;286:12300–12307. doi: 10.1074/jbc.M110.182741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CH, Kim SJ, Kan H, Kwon HM, Roh KB, Jiang R, Yang Y, Park JW, Lee HH, Ha NC, Kang HJ, Nonaka M, Söderhäll K, Lee BL. A three-step proteolytic cascade mediates the activation of the peptidoglycan-induced toll pathway in an insect. J Biol Chem. 2008;283:7599–75607. doi: 10.1074/jbc.M710216200. [DOI] [PubMed] [Google Scholar]
- Krem MM, Di Cera E. Evolution of enzyme cascades from embryonic development to blood coagulation. Trends Biochem Sci. 2002;27:67–74. doi: 10.1016/s0968-0004(01)02007-2. [DOI] [PubMed] [Google Scholar]
- Kuwar, et al. Evolution and regulation of digestive enzyme genes in Manduca sexta. Insect Biochem Mol Biol. 2015 manuscript in preparation. [Google Scholar]
- Jang IH, Nam HJ, Lee WJ. CLIP-domain serine proteases in Drosophila innate immunity. BMB Rep. 2008;41:102–107. doi: 10.5483/bmbrep.2008.41.2.102. [DOI] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Gu Y, Guo X, Zou Z, Scholz F, Trenczek TE, Kanost MR. Molecular identification of a bevy of serine proteinases in Manduca sexta hemolymph. Insect Biochem Mol Biol. 2005;35:931–943. doi: 10.1016/j.ibmb.2005.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Kanost MR. Pro-phenol oxidase activating proteinase from an insect, Manduca sexta : a bacteria-inducible protein similar to Drosophila easter. Proc Natl Acad Sci USA. 1998;95:12220–12225. doi: 10.1073/pnas.95.21.12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Kanost MR. Four serine proteinases expressed in Manduca sexta haemocytes. Insect Mol Biol. 1999;8:39–53. doi: 10.1046/j.1365-2583.1999.810039.x. [DOI] [PubMed] [Google Scholar]
- Jiang H, Kanost MR. The clip-domain family of serine proteinases in arthropods. Insect Biochem Mol Biol. 2000;30:95–105. doi: 10.1016/s0965-1748(99)00113-7. [DOI] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Yu X-Q, Kanost MR. Prophenoloxidase-activating proteinase-2 (PAP-2) from hemolymph of Manduca sexta: a bacteria-inducible serine proteinase containing two clip domains. J Biol Chem. 2003a;278:3552–3561. doi: 10.1074/jbc.M205743200. [DOI] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Yu X-Q, Zhu Y, Kanost MR. Prophenoloxidase-activating proteinase-3 (PAP-3) from Manduca sexta hemolymph: a clip-domain serine proteinase regulated by serpin-1J and serine proteinase homologs. Insect Biochem Mol Biol. 2003b;33:1049–1060. doi: 10.1016/s0965-1748(03)00123-1. [DOI] [PubMed] [Google Scholar]
- Jiang H, Vilcinskas A, Kanost MR. Söderhäll K, editor. Immunity in lepidopteran insects. Invertebrate Immunity. 2010;708:181–204. doi: 10.1007/978-1-4419-8059-5_10. Adv Exp Med Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugasu-Oei B, Balakrishnan R, Yang X, Chia W, Rodrigues V. Mutations in masquerade, a novel serine-protease-like molecule, affect axonal guidance and taste behavior in Drosophila. Mech Dev. 1996;57:91–101. doi: 10.1016/0925-4773(96)00537-0. [DOI] [PubMed] [Google Scholar]
- Pallaghy PK, Scanlon MJ, Monks SA, Norton RS. Three-dimensional structure in solution of the polypeptide cardiac stimulant anthopleurin-A. Biochemistry. 1995;34:3782–3794. doi: 10.1021/bi00011a036. [DOI] [PubMed] [Google Scholar]
- Park JW, Kim CH, Rui J, Park KH, Ryu KH, Chai JH, Hwang HO, Kurokawa K, Ha NC, Söderhäll I, Söderhäll K, Lee BL. Söderhäll K, editor. Beetle immunity. Invertebrate Immunity. 2010;708:163–180. doi: 10.1007/978-1-4419-8059-5_9. Adv Exp Med Biol. [DOI] [PubMed] [Google Scholar]
- Pauchet Y, Wilkinson P, Vogel H, Nelson DR, Reynolds SE, Heckel DG, ffrench-Constant RH. Pyrosequencing the Manduca sexta larval midgut transcriptome: messages for digestion, detoxification and defense. Insect Mol Biol. 2010;19:61–75. doi: 10.1111/j.1365-2583.2009.00936.x. [DOI] [PubMed] [Google Scholar]
- Perona JJ, Craik CS. Structural basis of substrate specificity in the serine proteases. Protein Sci. 1995;4:337–360. doi: 10.1002/pro.5560040301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Piao S, Song YL, Kim JH, Pak SY, Park JW, Lee BL, Oh BH, Ha NC. Crystal structure of a clip-domain serine protease and functional roles of the clip domains. EMBO J. 2005;24:4404–4414. doi: 10.1038/sj.emboj.7600891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien D, McVey J. Blood coagulation, inflammation, and defense. In: Sim E, editor. The Natural Immune System, Humoral Factors. New York: IRL Press; 1993. pp. 257–280. [Google Scholar]
- Rawlings RD, Barrett AJ. Evolutionary families of peptidases. Biochem J. 1993;290:205–218. doi: 10.1042/bj2900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J, Jiang H, Kanost MR, Wang Y. Serine proteases and their homologs in the Drosophila melanogaster genome: an initial analysis of sequence conservation and phylogenetic relationship. Gene. 2003;304:117–131. doi: 10.1016/s0378-1119(02)01187-3. [DOI] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- Shen HB, Chou KC. Signal-3L: A 3-layer approach for predicting signal peptides. Biochem Biophys Res Commun. 2007;363:297–303. doi: 10.1016/j.bbrc.2007.08.140. [DOI] [PubMed] [Google Scholar]
- Waterhouse RM, Kriventseva EV, Meister S, Xi Z, Alvarez KS, Bartholomay LC, Barillas-Mury C, Bian G, Blandin S, Christensen BM, Dong Y, Jiang H, Kanost MR, Koutsos AC, Levashina EA, Li J, Ligoxygakis P, Maccallum RM, Mayhew GF, Mendes A, Michel K, Osta MA, Paskewitz S, Shin SW, Vlachou D, Wang L, Wei W, Zheng L, Zou Z, Severson DW, Raikhel AS, Kafatos FC, Dimopoulos G, Zdobnov EM, Christophides GK. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science. 2007;316:1738–1743. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jiang H. Interaction of β-1,3-glucan with its recognition protein activates hemolymph proteinase 14, an initiation enzyme of the prophenoloxidase activation system in Manduca sexta. J Biol Chem. 2006;281:9271–9278. doi: 10.1074/jbc.M513797200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zou Z, Jiang H. An expansion of the dual clip-domain serine proteinase family in Manduca sexta: gene organization, expression, and evolution of prophenoloxidase-activating proteinase-2, hemolymph proteinase 12, and other related proteinases. Genomics. 2006;87:399–409. doi: 10.1016/j.ygeno.2005.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whaley K, Lemercier C. The complement system. In: Sim E, editor. The Natural Immune System, Humoral Factors. New York: IRL Press; 1993. pp. 121–150. [Google Scholar]
- X, et al. The genome of Manduca sexta. 2015 manuscript in preparation. [Google Scholar]
- Yu X-Q, Jiang H, Wang Y, Kanost MR. Nonproteolytic serine proteinase homologs are involved in prophenoloxidase activation in the tobacco hornworm, Manduca sexta. Insect Biochem Mol Biol. 2003;33:197–208. doi: 10.1016/s0965-1748(02)00191-1. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Doherty T, Li J, Lu W, Barinka C, Lubkowski J, Hong M. Resonance assignment and three-dimensional structure determination of a human alpha-defensin, HNP-1, by solid-state NMR. J Mol Biol. 2010;397:408–422. doi: 10.1016/j.jmb.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Zhang X, Gunaratna R, Najar F, Wang Y, Roe B, Jiang H. Pyrosequencing-based expression profiling and identification of differentially regulated genes from Manduca sexta a lepidopteran model insect that lacks genome sequence. Insect Biochem Mol Biol. 2011;41:733–746. doi: 10.1016/j.ibmb.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P, Wang GH, Dong ZM, Duan J, Xu PZ, Cheng TC, Xiang ZH, Xia QY. Genome-wide identification and expression analysis of serine proteases and homologs in the silkworm Bombyx mori. BMC Genomics. 2010;11:405. doi: 10.1186/1471-2164-11-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z, Najar F, Wang Y, Roe B, Jiang H. Pyrosequence analysis of expressed sequence tags for Manduca sexta hemolymph proteins involved in immune responses. Insect Biochem Mol Biol. 2008;38:677–682. doi: 10.1016/j.ibmb.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z, Lopez D, Kanost MR, Evans JD, Jiang H. Comparative analysis of serine protease-related genes in the honeybee genome: possible involvement in embryonic development and innate immunity. Insect Mol Biol. 2006;15:603–614. doi: 10.1111/j.1365-2583.2006.00684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z, Evans J, Lu Z, Zhao P, Williams M, Sumathipala N, Hetru C, Hultmark D, Jiang H. Comparative genome analysis of the Tribolium immune system. Genome Biol. 2007;8:R177. doi: 10.1186/gb-2007-8-8-r177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z, Shin SW, Alvarez KS, Kokoza V, Raikhel AS. Distinct melanization pathways in the mosquito Aedes aegypti. Immunity. 2010;32:41–53. doi: 10.1016/j.immuni.2009.11.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FPKM values were calculated for 193 SP-related genes as described in Section 2.5. Cluster analysis indicated that 68 of them were midgut-specific SP/SPH genes and, therefore, further analyzed in another study (Kuwar et al., 2015). The remaining 125 genes exhibited six distinct expression patterns, as shown in panels A through F with the 28, 13, 21, 36, 15, and 12 names indicated, respectively. The relative expression levels or log2(FPKM+1) values for individual genes are connected by grey lines, and their means by a pink line in each panel. Samples in the same tissue groups are underlined alternately. The libraries are numbered as described in Fig. 5.