Abstract
It is well established that the plastidic isoform of glutamine synthetase (GS2) is the enzyme in charge of photorespiratory ammonium reassimilation in plants. The metabolic events associated to photorespiratory NH4 + accumulation were analyzed in a Lotus japonicus photorespiratory mutant lacking GS2. The mutant plants accumulated high levels of NH4 + when photorespiration was active, followed by a sudden drop in the levels of this compound. In this paper it was examined the possible existence of enzymatic pathways alternative to GS2 that could account for this decline in the photorespiratory ammonium. Induction of genes encoding for cytosolic glutamine synthetase (GS1), glutamate dehydrogenase (GDH) and asparagine synthetase (ASN) was observed in the mutant in correspondence with the diminishment of NH4 +. Measurements of gene expression, polypeptide levels, enzyme activity and metabolite levels were carried out in leaf samples from WT and mutant plants after different periods of time under active photorespiratory conditions. In the case of asparagine synthetase it was not possible to determine enzyme activity and polypeptide content; however, an increased asparagine content in parallel with the induction of ASN gene expression was detected in the mutant plants. This increase in asparagine levels took place concomitantly with an increase in glutamine due to the induction of cytosolic GS1 in the mutant, thus revealing a major role of cytosolic GS1 in the reassimilation and detoxification of photorespiratory NH4 + when the plastidic GS2 isoform is lacking. Moreover, a diminishment in glutamate levels was observed, that may be explained by the induction of NAD(H)-dependent GDH activity.
Introduction
Nitrogen nutrition is one of the most crucial factors for plant productivity [1]. Nitrate is the main form of nitrogen used by many crops, but most plants are also able to use ammonium, amino acids and urea [2]. Symbiotic N2 fixation in the root nodule of legumes also provides important amount of reduced nitrogen to crops without the need of extensive fertilization [3]. NO3 - uptake by the roots is mediated by different families of transporters that are also involved in the sensing of this molecule [4]. Once inside the cell, NO3 - is first reduced to NO2 - in the cytoplasm by the action of nitrate reductase (NR; EC 1.7.1.1/2) and then further reduced to NH4 + by the action of nitrite reductase in the plastids (NiR; EC 1.7.7.1) [5]. The NH4 + that has been taken up directly from the soil or produced by the reduction of NO3 -, atmospheric N2 in root nodules or urea hydrolysis, is then assimilated by the action of the glutamine synthetase (GS, EC 6.3.1.2)/glutamate synthase (GOGAT, EC 1.4.7.1 and 1.4.1.14) cycle. In addition to primary nitrogen assimilation, other pathways are able to produce NH4 + like amino acid catabolism, phenylpropanoid biosynthesis, asparagine breakdown and photorespiration in C3 plants [6]. Photorespiration is probably the most important of these pathways due to the very high flux and the different interconnections with other metabolic routes [7]. The large amounts of ammonium generated by the photorespiratory cycle are produced in the mitochondria as a result of the conversion of two molecules of glycine into one molecule of serine by the action of glycine decarboxylase (EC 2.1.2.10)/serine hydroxymethyl transferase multienzymatic complex (EC 2.1.2.1). NH4 + is transported to the chloroplast where it is reassimilated by the plastidic GS/GOGAT cycle.
Plants have two types of GS isoenzymes: the cytosolic isoform (or GS1), that is generally encoded by a small gene family composed of 3–5 members, and the plastidic GS isoform (or GS2), that is generally encoded by a single gene, with the exception of Medicago truncatula and poplar where two genes encoding for GS2 have been described [8–10]. While GS1 is the main enzyme involved in primary ammonium assimilation and recycling of the NH4 + generated during senescence and phenylpropanoid biosynthesis [11, 12], GS2 is mainly involved in the reassimilation of the ammonium generated during photorespiration. The key role played by GS2 in the reassimilation of photorespiratory ammonium was first demonstrated by the isolation of photorespiratory barley mutant that specifically lacked of this GS isoform [13, 14]. These mutants were conditionally lethal, since they grew well under a CO2-enriched atmosphere (>0.2% CO2), where photorespiration is suppressed, but showed stress symptoms and accumulated ammonium when transferred to normal CO2 conditions (about 0.04% CO2). Later on, oilseed rape plants with 50–75% lower GS2 activity were obtained using antisense technology [15]. However, these plants had low tissue levels of NH4 + since the residual GS2 activity was enough to efficiently reassimilate the ammonium produced during photorespiration. In our group, the first photorespiratory mutants deficient in GS2 from legume plants were isolated in the model legume Lotus japonicus [16]. These mutants lack completely of plastidic GS2 enzyme activity but have normal levels of cytosolic GS1 and were shown to be specifically affected in the gene encoding for plastidic GS2 [17] and accumulated high levels of ammonium when grown under active photorespiratory conditions.
A recent work from our group made use of a L. japonicus plastidic GS2 mutant called Ljgln2-2 in order to study the transcriptomic and metabolic consequences of the lack of plastidic GS2 both under high and normal CO2 conditions [18]. A coordinate repression of the transcription of most photorespiratory genes was observed when the plants were shifted from high CO2 to normal CO2 conditions, as a way to avoid further accumulation of ammonium under active photorespiratory conditions. However, transcriptomic analysis indicated that important genes involved in N assimilation were modulated in the mutant under normal CO2 conditions, including genes encoding for cytosolic GS1 [18, 19]. This was part of a greater modulation of the transcriptome as well as of the metabolic profile of the mutant. These preliminary results indicated that other enzymes besides plastidic GS2 could be involved in the reassimilation of photorespiratory ammonium in the Ljgln2-2 mutant.
Different enzymatic systems have been described in plants that may use ammonium [20]: 1) the GS/GOGAT cycle previously described. 2) Glutamate dehydrogenase (GDH, EC 1.4.1.2) deaminates glutamate to α-ketoglutarate (α-KG) and ammonium in order to replenish the TCA cycle of intermediates (NAD-dependent GDH activity) [21], but it has been also shown that the reverse reaction, also called aminating NADH-dependent GDH activity, may work in order to detoxify ammonium when the levels of this compounds are very high [22, 23]. In addition, a NADP(H)-GDH (EC 1.4.1.4) of unknown function also exists. 3) Asparagine synthetase (ASN, EC 6.3.5.4), whose main reaction is the incorporation of the amino group of glutamine into aspartate to yield asparagine [24, 25]. 4) Finally, carbamoylphosphate synthetase (CPS, EC 6.3.5.5) produces carbamoylphosphate (CP) from HCO3 -, ATP and either NH4 + or glutamine [20].
The toxic effect that NH4 + produces in most plants is an interesting and still not fully understood phenomenon [26, 27]. For this reason, the study of the reactions involved in the assimilation/detoxification of both the external NH4 + obtained from the soil as well as the endogenous NH4 + produced by routes as the photorespiratory cycle is a topic of particular interest [19]. In this paper we studied the metabolic reactions that may be involved in NH4 + detoxification in the Ljgln2-2 photorespiratory mutant. For this purpose, we carried out measurements of the transcript levels for the different sets of genes encoding for possible NH4 + detoxification enzymes in L. japonicus plants followed by determination of the corresponding polypeptide content and enzyme activities, as well as of the relative levels of the metabolites involved in the corresponding reactions at different time points. The results obtained indicate that, in the absence of plastidic GS2, different enzymes are induced in response to photorespiratory ammonium accumulation and may constitute an alternative way for photorespiratory ammonium reassimilation and detoxification in this plant.
Materials and Methods
Plant material and growth
Lotus japonicus (Regel) Larsen cv. Gifu was initially obtained from Professor Jens Stougaard (Aarhus University, Denmark) and then self-propagated at the University of Seville. WT and Ljgln2-2 mutant plants were grown as described by Pérez-Delgado et al. [18]. For Ljgln2-2, the mutant progeny of two consecutive backcrosses into the WT background was used in this work. WT and mutant seeds were scarified and surface-sterilized, then germinated in 1% (w/v) agar in Petri dishes and transferred to pots using vermiculite as solid support. Five seedlings were planted in each pot and grown during 35 d in a growth chamber under 16 h: 8 h day: night, 20: 18°C, with a photosynthetic photon flux density of 250 μmol m-2 s-1 and a constant humidity of 70%. CO2 was automatically injected to a final concentration of 0.7% (v/v) (high CO2) to allow for normal growth of the Ljgln2-2 mutant in a photorespiration-suppressed atmosphere. Plants were watered with “Hornum” nutrient solution, containing 5 mM NH4NO3 and 3 mM KNO3 [28]. After 35 days of growth under high CO2 atmosphere the plants had an average number of 7 trefoils. At this time, total leaf tissue was harvested for each plant genotype, constituting the time zero point (photorespiration suppressed conditions). All the leaf samples used in this work were harvested 4 hours after the beginning of the light period. Plants were then transferred to normal CO2 conditions (0.04% CO2 v/v) and each plant genotype was sampled at different time points (S1 Fig). Visible symptoms of the air sensitivity phenotype of the Ljgln2-2 mutant plants started at the lower (older) leaves of the plants and progressed up to the younger leaves only after long periods of incubation under normal CO2 conditions that were not considered in this work. In any case, the 2–3 lower trefoils from the plants were always discarded to avoid any effect of senescence of the older leaves in our samples. Therefore, only the healthy leaves from the plants were harvested and pooled, and the corresponding tissue was flash-frozen in liquid N2, grinded with a pestle in a mortar that was pre-cooled with liquid N2 and the powder was stored at -80°C until use. Three independent biological replicates were harvested for each genotype and time point. A biological replicate consisted of tissue pooled from five plants grown in the same pot.
RNA extraction, cDNA synthesis and qRT-PCR
RNA extraction was carried out using the hot borate method as described by [29]. cDNA synthesis was carried out with the Superscript III reverse transcriptase (Invitrogen) exactly as described by [18]. qRT-PCR was carried out in a LightCycler 480 thermal cycler (Roche) using the SensiFAST SYBR No-ROX Kit (Bioline) also using the same conditions described by [18]. Expression levels were quantified by determining the CT cycle of each reaction with the LightCycler 480 software version 1.5.0. Expression data were standardized to the geometric mean of four housekeeping genes. The housekeeping genes were LjGPI-anchored protein (probeset: chr3.CM0047.42), LjPp2A (probeset: chr2.CM0310.22), LjUbc10 (probeset: chr1.TM0487.4) and LjUbq4 (probeset: chr5.CM0956.27); these genes were selected amongst the most stably expressed genes in plants [30]. A list of all the primers used for qRT-PCR is provided in the online version of this article (S1 Table).
Preparation of crude extracts
Frozen leaf tissue was extracted with a pellet homogeniser in 5 ml/g fresh weight of extraction buffer. The homogenate was centrifuged for 15 min at 15,000 g and 4°C and the supernatant constituted the crude extract. The extraction buffer for GS activity assay and immunoblot was prepared according to [16]. For GDH activity and immunoblot the extraction buffer was 100 mM Tris·HCl pH 8 supplemented with 1% of polyvinylpolypyrrolidone. The extraction buffer for CPS activity assay was prepared according to [31]. Crude extracts for the assay of ASN activity were prepared in TrisˑHCl 50 mM pH 6.0.
Western Blot
Western blots from denaturing PAGE were performed using the ECL Western blotting system (GE Healthcare) according to the manufacturer instructions. SDS-PAGE was performed according to the method of [32] using a Mini-Protean Tetra Cell system (Bio-Rad). An acrylamide concentration of 12%, 10% and 12.5% (w/v) was used in the SDS-PAGE running gels for immunodetection of GS, GDH and ASN respectively. Stacking gels were always prepared at 4% (w/v) acrylamide. 10 μg of total protein from leaf crude extracts were loaded in each well of the gels. Protein concentration was quantified using the Bradford protein assay (Bio-Rad). The polypeptide molecular weights were estimated using the broad range molecular weight standards (Bio-Rad).
Anti-GS primary antibodies were raised in rabbit immunised with the recombinant homopolymeric α GS from Phaseolus vulgaris purified by metal-affinity chromatography [33] and used at 1:1,000 dilution. Anti-GDH primary antibodies (a gift from Prof. K.A. Roubelakis-Angelakis, University of Crete) were raised in rabbit using purified NADH-GDH from grapevine [34]. Anti-ASN antibodies (a gift from Prof. FM Cánovas and RA Cañas, University of Málaga) were raised in rabbit using purified ASN from Scots pine (Pynus sylvestris L.) as described by [35]. Anti-rabbit secondary antibodies labelled with peroxidase were obtained from GE Healthcare.
Detection of peroxidase signal in the membranes was carried out with a quimioluminescence detection system (Fujifilm LAS 3000 mini, Fujifilm). Relative quantification of the polypeptide levels was carried out for three different biological replicates for each time point and genotype. Densitometry measurements were performed with the ImageJ software (http://rsb.info.nih.gov/ij/).
Enzyme activity assays
Biosynthetic GS enzyme activity was determined by measuring ATP hydrolysis as described by [36]. GDH aminating and deaminating activities were measured as described by [37], except that 0.1 mM of NADH or NADPH was used in the aminating reaction. CPS enzyme activity was determined according to [38]. Assays for ASN enzyme activity were carried out according to [39].
Metabolite profiling analysis
Metabolite profiling analysis was carried out using the dataset generated by Pérez-Delgado et al. [18]. Statistical differences between metabolite levels were assessed with two-way ANOVA at P < 0.001 using the Multiexperiment Viewer version 3.1 [40] and the factors “genotype” and “time under normal CO2”. Only metabolites that showed significant differences according to this analysis were considered. The relative metabolite levels presented in this paper are the direct normalized responses of metabolite pool measures, that is, mass detector signals in arbitrary units normalized to internal standard and sample fresh weigh.
Ammonia determination was carried out according to the method of [41] with some modifications as described by [16]. CP content in crude extracts was determined with the same colorimetric method used for CPS activity but using undialyzed crude extracts.
Statistical analysis
All differences described in the text among WT and mutant plants and/or different time periods were confirmed to be significantly distinct using Student’s t test (p < 0.05)
Results
Ammonium accumulation and induction of cytosolic GS1 in the Ljgln2-2 mutant
The levels of ammonium and of several other metabolites involved in NH4 + assimilation and metabolism like Glu, Gln, α-KG, Asp, Asn, Gly, Ser, Lys and CP were measured in leaves of WT and Ljgln2-2 mutant plants at different time points of the transfer from high CO2 (suppressed photorespiration) to normal CO2 (active photorespiration) (Table 1). The levels of glycine and serine were indicative of the existence of a manifest photorespiratory carbon flow both in WT and mutant plants. As it would be expected, an important increase in ammonium levels could be observed in the mutant plants compared to the WT, which was due to the lack of plastidic GS2 in the mutants. The presence of normal levels of plastidic GS2 in the WT plants made them able to efficiently reassimilate the photorespiratory ammonium in contrast to the Ljgln2-2 mutant plants. However, interestingly, a gradual decline of these ammonium levels was detected in the mutants after the maximum reached at day 3. qRT-PCR analysis showed a coordinate repression of the genes involved in photorespiration in the Ljgln2-2 mutant after the transfer to active photorespiratory conditions [18], suggesting that the mutant may down regulate the photorespiratory pathway in order to avoid further accumulation of ammonium. In addition, the sudden drop in ammonium levels observed in Ljgln2-2 mutant plants after three days of the transfer to normal air was suggestive of the induction of alternative routes for the reassimilation of photorespiratory ammonium that was accumulated in this genotype. The analysis of these routes was further explored.
Table 1. Metabolite levels in WT and mutant plants.
| Days in normal CO2 | ||||||||
|---|---|---|---|---|---|---|---|---|
| 0 | 2 | 3 | 4 | 6 | 8 | 10 | ||
| NH4+ | ||||||||
| WT | 0.0 ± 0.0 | 0.4 ± 0.1 | 0.4 ± 0.1 | 2.6 ± 2.8 | 0.7 ± 0.1 | 0.3 ± 0.0 | 0.5 ± 0.2 | |
| Ljgln2-2 | 3.0 ± 2.0 | 45.6 ± 0.5 | 64.4 ± 1.1 | 52.7 ± 1.0 | 30.9 ± 8.2 | 21.7 ± 2.3 | 26.0 ± 3.4 | |
| Glu | ||||||||
| WT | 12.9 ±3.1 | 11.1 ± 3.0 | 9.5 ± 0.5 | 12.0 ± 2.0 | 10.5 ± 0.2 | 13.6 ± 2.8 | 11.7 ± 1.5 | |
| Ljgln2-2 | 9.7 ± 1.1 | 4.8 ± 0.5 | 4.4 ± 0.7 | 5.9 ± 0.8 | 7.4 ± 0.5 | 7.3 ± 3.1 | 5.5 ± 0.9 | |
| Gln | ||||||||
| WT | 6.8 ± 2.7 | 3.1 ± 2.4 | 4.2 ± 0.6 | 1.9 ± 0.6 | 2.4 ± 0.0 | 3.8 ± 1.3 | 2.3 ± 0.6 | |
| Ljgln2-2 | 3.7 ± 1.4 | 6.8 ± 2.5 | 8.0 ± 1.3 | 13.0 ± 2.8 | 20.1 ± 8.7 | 23.4 ± 5.7 | 16.5 ± 1.6 | |
| α-KG | ||||||||
| WT | 0.9 ± 0.4 | 0.7± 0.4 | 0.8 ± 0.7 | 0.4 ± 0.1 | 0.2 ± 0.0 | 0.5 ± 0.2 | 0.4 ± 0.1 | |
| Ljgln2-2 | 0.5 ± 0.1 | 13.9 ± 2.7 | 21.4 ± 5.7 | 9.1 ± 2.7 | 3.3 ± 0.7 | 4.4 ± 1.5 | 2.5 ± 0.7 | |
| Asp | ||||||||
| WT | 14.6 ± 4.9 | 13.2 ± 3.1 | 10.1 ± 0.6 | 12.8 ± 0.7 | 16.3 ± 1.7 | 17.5 ± 3.8 | 12.9 ± 2.2 | |
| Ljgln2-2 | 7.9 ± 0.7 | 4.7 ± 0.4 | 3.2 ± 0.1 | 4.6 ± 0.8 | 10.4 ± 4.7 | 7.8 ± 3.5 | 5.3 ± 0.4 | |
| Asn | ||||||||
| WT | 14.2 ± 5.6 | 8.5 ± 3.2 | 10.8 ± 2.9 | 6.3 ± 0.3 | 6.5 ± 0.1 | 9.4 ± 1.9 | 7.9 ± 1.4 | |
| Ljgln2-2 | 8.5 ± 1.2 | 5.7 ± 0.8 | 8.4 ± 0.2 | 10.9 ± 0.7 | 12.1 ± 2.0 | 13.6 ± 5.3 | 12.1 ± 1.0 | |
| Gly | ||||||||
| WT | 0.6 ± 0.1 | 0.7 ± 0.3 | 0.6 ± 0.1 | 0.5 ± 0.1 | 0.4 ±0.1 | 0.7 ± 0.2 | 0.4 ± 0.2 | |
| Ljgln2-2 | 0.7 ± 0.1 | 0.9 ± 0.1 | 1.7 ± 0.3 | 1.3 ± 0.2 | 1.3 ± 0.2 | 1.3 ± 0.2 | 0.8 ± 0.1 | |
| Ser | ||||||||
| WT | 7.9 ± 1.2 | 11.9 ± 2.2 | 10.2 ± 1.2 | 10.0 ± 0.7 | 8.3 ± 0.1 | 13.8 ± 3.6 | 9.5 ± 1.3 | |
| Ljgln2-2 | 11.5 ± 2.2 | 16.7 ± 1.0 | 22.9 ± 3.8 | 15.0 ± 1.8 | 19.7 ± 6.0 | 18.2 ± 4.8 | 12.0 ± 0.9 | |
| Lys | ||||||||
| WT | 2.3 ± 0.3 | 1.8 ± 0.4 | 1.9 ± 0.6 | 1.5 ± 0.5 | 2.0 ± 0.7 | 3.5 ± 2.6 | 2.1 ± 0.7 | |
| Ljgln2-2 | 1.7 ± 0.5 | 9.1 ± 2.0 | 5.9 ± 3.5 | 5.4 ± 0.5 | 5.3 ± 1.1 | 5.1 ± 0.9 | 3.9 ± 0.2 | |
| CP * | ||||||||
| WT | 0.15± 0.08 | 0.35 ± 0.11 | 0.28 ± 0.10 | 0.21 ± 0.13 | 0.19 ± 0.15 | 0.17 ± 0.14 | 0.18 ± 0.12 | |
| Ljgln2-2 | 0.05± 0.02 | 0.07 ± 0.04 | 0.01 ± 0.01 | 0.01 ± 0.03 | 0.05 ± 0.01 | 0.04 ± 0.02 | 0.01 ± 0.01 | |
The levels of different metabolites were determined from leaves of WT and Ljgln2-2 mutant plants grown in high CO2 conditions (0 days) and after different periods of time under normal CO2 conditions. Metabolite levels are expressed as relative units with the exception of NH4 + and CP that are expressed in μmol·g FW-1. Numbers in bold represent significant differences between WT and mutant genotypes at this time point according to Student’s t test (p<0.05).
*CP levels have two decimal digits since they were measured using a different technique compared to the other metabolites.
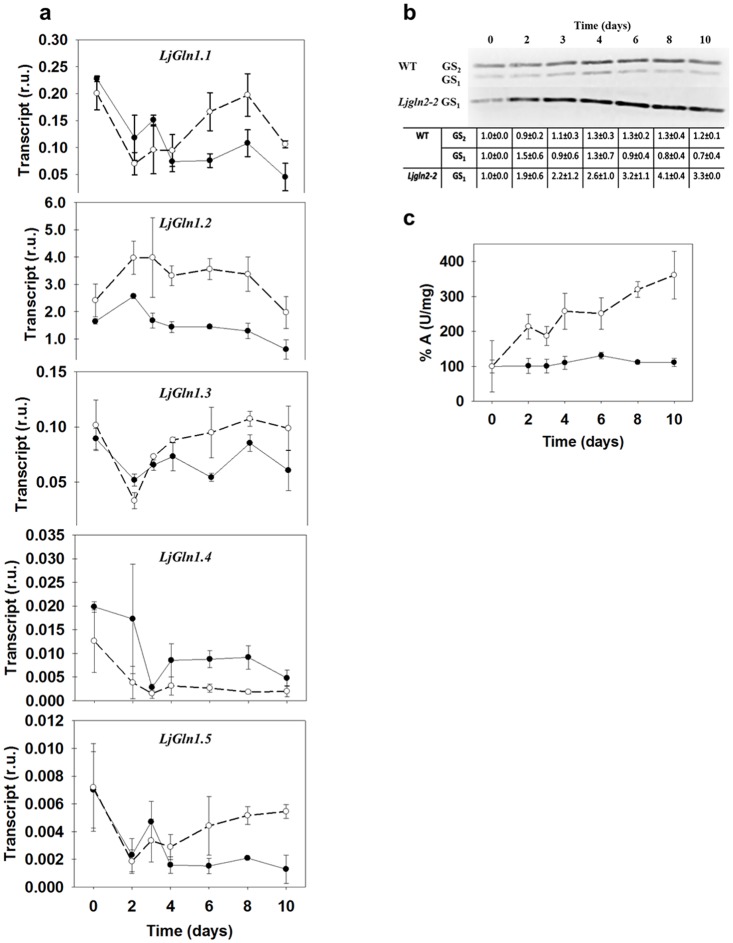
The levels of expression of the different GS1 genes were determined both for WT and mutant plants at different time points under active photorespiratory conditions. In order to do that, all the sequences corresponding to GS1 were retrieved from the Kazusa database (www.kazusa.or.jp/e/). A total of five unique gene sequences encoding for cytosolic GS1 were found (see S1 Table for the accession numbers). Gene specific oligonucleotides were used in order to measure the mRNA levels in WT and mutant plants by qRT-PCR at the different time points considered. LjGln1.2 was the most highly expressed GS1 gene in leaves and was induced almost two-fold in the mutant, with a maximum in transcript levels at day 3 followed by a slow decrease (Fig 1a). LjGln1.1, LjGln1.3 and LjGln1.5 genes showed similar expression profiles, with a sudden decrease in transcript levels in both genotypes, followed by a recovery in transcript levels at days 6–8 exclusively in the mutant. Finally, LjGln1.4 was expressed at very low levels and was repressed in both genotypes after the shift to normal CO2 conditions. All these results indicated that the different genes encoding for cytosolic GS1 were modulated by the transfer to active photorespiratory conditions in both genotypes.
Fig 1. Changes in GS gene expression and activity in WT and Ljgln2-2 mutant plants.
(a) Expression of the genes for cytosolic GS1 in WT and mutant plants. Transcript levels were quantified by qRT-PCR in WT (black dots, solid line) and mutant (white dots, dashed line) leaves at the time points indicated. Relative transcript levels are reported for each genotype compared to the housekeeping genes. (b) Immunodetection of cytosolic GS1 and plastidic GS2 in leaf crude extracts from WT and mutant plants. 10 μg of total protein were loaded on each lane. The position of the molecular weight marker (kDa) is indicated at the right side of the blot. The table reports the densitometric quantification of the GS1 and GS2 bands relative to their values at time zero which were taken as 1. Data are the mean ± SD of three independent biological replicates. The blot shown in the Figure is only one of the different replicates analyzed. (c) The GS specific activity was determined in crude extracts from leaves of WT (black dots, solid line) and Ljgln2-2 (white dots, dashed line). GS activity is shown as specific activity (U/mg of total protein). Data are the mean ± S.D. of three independent biological replicates.
In order to determine if these changes in gene expression were paralleled by changes in polypeptide levels, a Western blot analysis was carried out using anti-GS antibodies that recognize both cytosolic and plastidic GS. Cytosolic GS1 polypeptide levels were increased in the mutant during the time-course of the experiment. The maximum levels of GS1 polypeptide were observed after eight days under normal CO2 conditions, where the polypeptide levels were about four times higher than in the WT (Fig 1b).
Total GS enzyme activity was also increased exclusively in Ljgln2-2 in good agreement with GS1 polypeptide levels, indicating that the increased amount of GS1 was represented by active enzyme (Fig 1c). Moreover, glutamine levels were also increased in the mutant in parallel with GS1 polypeptide levels and total GS activity (Table 1). These data suggest a role for cytosolic GS1 as an alternative enzyme for photorespiratory NH4 + reassimilation and detoxification in GS2-deficient plants. In contrast, a drop in glutamine levels was observed in WT plants after the transfer to active photorespiratory conditions (Table 1).
Induction of different GDH activities in the Ljgln2-2 mutant
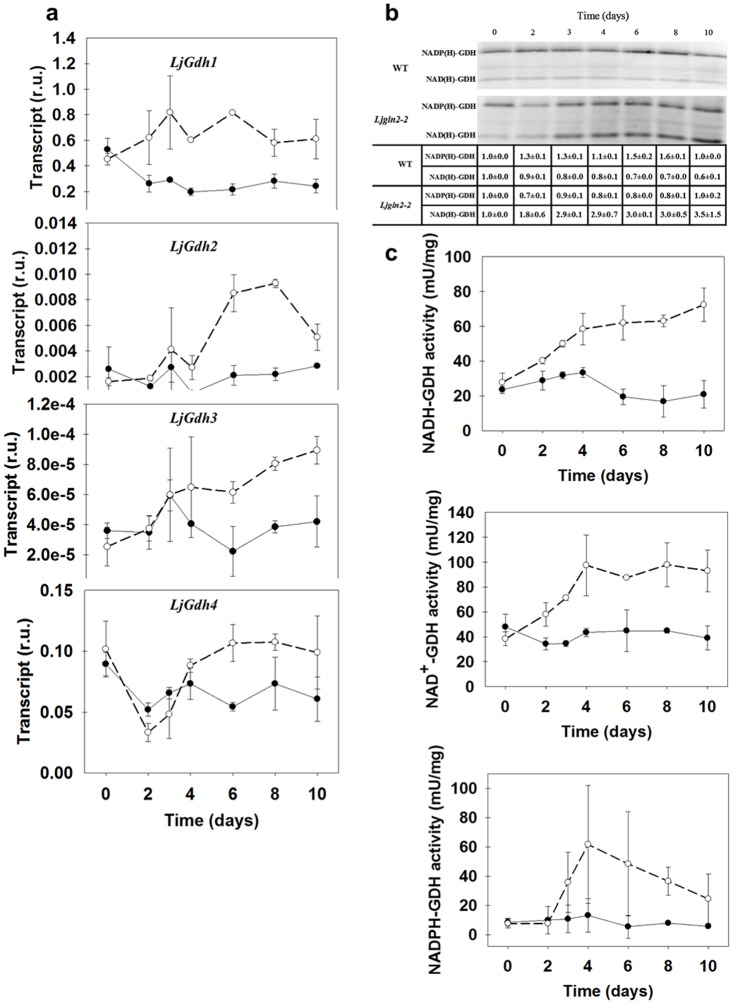
Three different gene sequences encoding for NAD(H)-GDH (LjGdh1, LjGdh2 and LjGdh3) as well as a fourth gene encoding for a putative NADP(H)-dependent GDH enzyme (LjGdh4) were found in the L. japonicus genome. The experimental molecular weights of the corresponding polypeptides depicted from the gel were of about 45 kDa for the NAD(H)-GDH isoforms and of about 71 kDa for the NADP(H)-GDH one, that are in good agreement with the theoretical molecular weights of about 44.7 kDa and 70.7 kDa calculated for the NAD(H)-dependent and NADP(H)-dependent isoforms respectively (S2 Fig). Two of the three genes encoding for NAD(H)-GDH were induced in the mutant and were not significantly modulated in the WT after four days under normal CO2 conditions (LjGdh2 and LjGdh3, Fig 2a). On the other hand, the most highly expressed GDH gene in leaves (LjGdh1) was significantly induced in the mutant after the transfer to normal CO2 conditions and it was repressed about three-fold in the WT genotype. LjGdh4 transcript levels showed in both genotypes a sudden drop after 2 days under active photorespiration followed by a clear increase taking place in the mutant plants, with a trend that was almost opposite to NH4 + levels.
Fig 2. Changes in GDH gene expression and activity in WT and Ljgln2-2 mutant plants.
(a) Expression of the genes for NAD(H)-GDH (LjGdh1, LjGdh2 and LjGdh3) and NADP(H)-GDH (LjGdh4) in WT and mutant plants. Transcript levels were quantified by qRT-PCR in WT (black dots, solid line) and mutant (white dots, dashed line) leaves at the time points indicated. Relative transcript levels are reported for each genotype compared to the housekeeping genes. (b) Immunodetection of NAD(H)-GDH and NADP(H)-GDH in leaf crude extracts from WT and mutant plants. 10 μg of total protein were loaded on each lane. The position of the molecular weight markers (kDa) is indicated at the right side of the blot. The table reports the densitometric quantification of the NAD(H)-GDH and NADP(H)-GDH bands relative to their values at time zero which were taken as 1. Data are the mean ± SD of three independent biological replicates. The blot shown in the Figure is only one of the different replicates analyzed. (c) GDH specific activity was determined in crude extracts from leaves of WT (black dots, solid line) and Ljgln2-2 (white dots, dashed line). GDH activity was measured in both directions using either NAD+, NADH, NADP+ or NADPH as co-factors. NADP+-dependent GDH activity was undetectable in both genotypes and is not shown here. GDH activity is shown as specific activity (mU/mg of total protein). Data are the mean ± S.D. of three independent biological replicates.
Two main immunoreactive bands were detected by Western blot analysis using anti NAD(H)-GDH antibodies from grapevine: one compatible with the size expected for the NAD(H)-GDH polypeptides (about 45 kDa for all the three possible isoforms) and another one with a higher molecular weight that was compatible with the size of the putative NADP(H)-GDH polypeptide (about 71 kDa). Cross-reactivity between NAD(H)-GDH antibodies and NADPH-GDH may be expected. Sequence alignment of the L. japonicus NADP(H)-GDH protein (LjGDH4) with the other three NAD(H)-GDH protein sequences showed the presence of two highly conserved domains between the two types of GDH: a dehydrogenase dimerization domain (pfam02812), between the amino acid residues 240 and 360 of LjGDH4, and the GDH NAD(P)-binding domain, from the amino acid residue 379 until the end of the LjGDH4 sequence (S2 Fig). These highly conserved regions between NAD(H)-GDH and NAD(P)H-GDH may account for the cross-reactivity observed for the anti NAD(H)-GDH antibody. Quantification of the immunoblots indicated an increase of up to three times in NAD(H)-GDH polypeptide levels in the mutant, while in the WT they were almost unchanged (Fig 2b). On the other hand, NADP(H)-GDH polypeptide levels were higher in the WT while in the mutant plants they showed an initial decrease followed by a substantial recovery along the time in parallel with the levels found for the LjGdh4 transcript.
The levels of NAD(H)-GDH enzyme activity were determined both in the aminating and deaminating directions. Both assays indicated an increase in activities of about 3–4 times exclusively in the mutant, in very good agreement with the polypeptide levels. On the other hand, the NADP(H)-GDH enzyme activity was also induced in the mutant, although with more variation between biological replicates compared to the NADH-GDH and NAD+-GDH activities (Fig 2c). In this case, only the aminating NADPH-dependent GDH enzyme activity was detectable, showing an increase in parallel with the increase in transcript levels observed for the corresponding LjGdh4 gene. This increase in NADPH-dependent GDH activity took place concomitantly with the decrease in ammonium levels observed in Ljgln2-2 mutants after three days of the transfer to normal CO2 (Table 1). The NADP+-dependent deaminating activity was undetectable in WT and mutant extracts (not shown). These results are very interesting considering that NADP(H)-GDH is still a poorly characterized enzyme in plants.
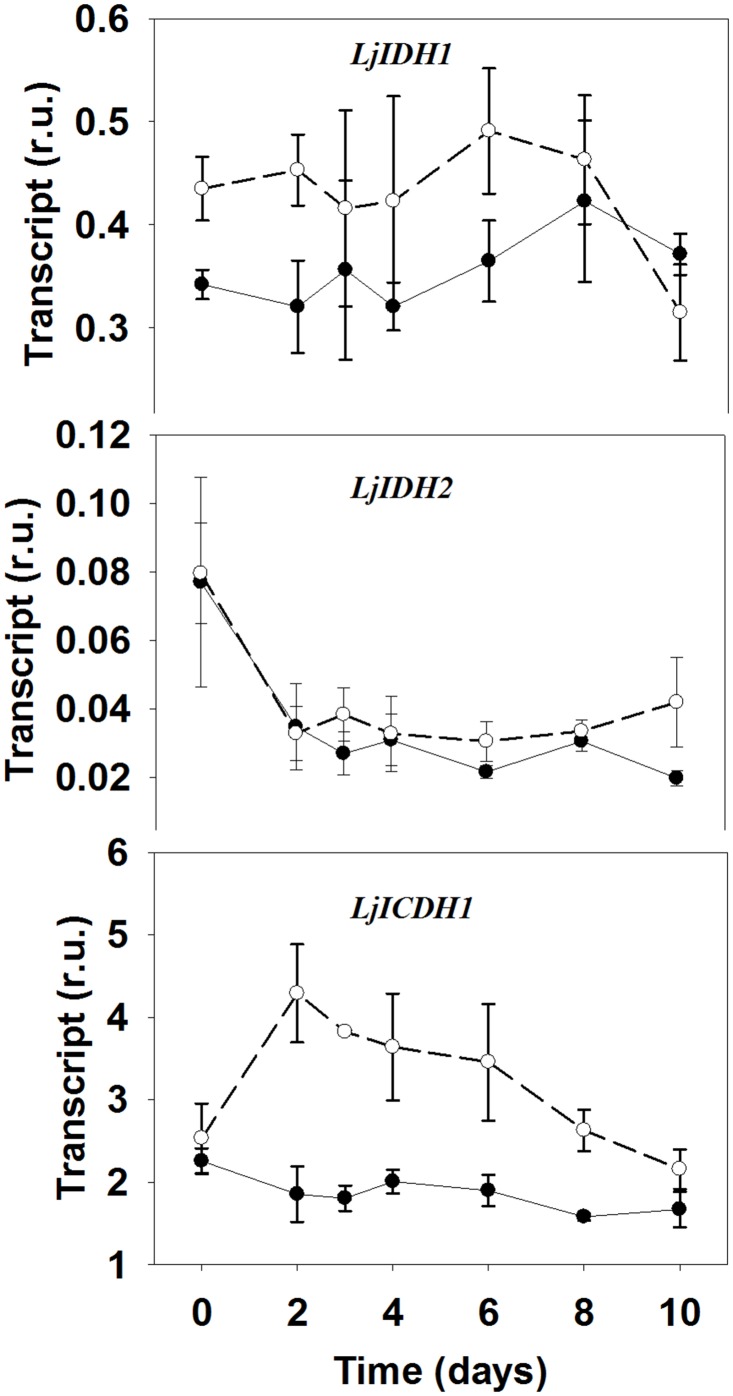
Analysis of the metabolite levels corresponding to the substrates and products of GDH reaction showed that the levels of α-KG were increased in the mutant, with a trend over time very similar to the ammonium one (Table 1). On the other hand, glutamate leaf content decreased about two-fold in the mutant genotype after the shift to active photorespiratory conditions. These data suggested that GDH activity may contribute to provide the carbon skeletons necessary for the reassimilation of photorespiratory NH4 + by means of Glu deamination. However, α-KG can be also produced by other metabolic reactions, most importantly by the oxidative decarboxylation of isocitrate by the action of isocitrate dehydrogenase (called IDH or ICDH depending on whether the cofactor is NADH or NADPH, [42]). Moreover, Lys catabolism has been also proved able to evolve α-KG as a terminal product [43]. The latter option seemed improbable since Lys accumulated about 3–5 times in the mutant under active photorespiratory conditions (Table 1). However, qRT-PCR analysis of transcript levels for different genes encoding for isocitrate dehydrogenase showed that LjICDH1, which encodes for a NADP-dependent ICDH predicted to be localized in the cytosol, was highly induced in the mutant (Fig 3). Interestingly, the transcript levels for LjICDH1 were parallel to the α-KG metabolite ones, suggesting that this gene may also contribute to the increase in α-KG detected in the mutant. On the other hand, the most transcribed gene encoding for the NAD-dependent isocitrate dehydrogenase isoform, LjIDH1, did not show any clear regulation after the transfer to active photorespiratory conditions, while LjIDH2, the second gene encoding for IDH in L. japonicus, was repressed in both genotypes under the same conditions (Fig 3).
Fig 3. Expression levels of genes encoding for different IDH and ICDH isoforms in WT and Ljgln2-2 mutant plants.
Transcript levels were quantified by qRT-PCR in WT (black dots, solid line) and mutant (white dots, dashed line) leaves at the time points indicated. Relative transcript levels are reported for each genotype compared to the housekeeping genes. Data are the mean ± S.D. of three independent biological replicates.
Transcriptional modulation of ASN genes
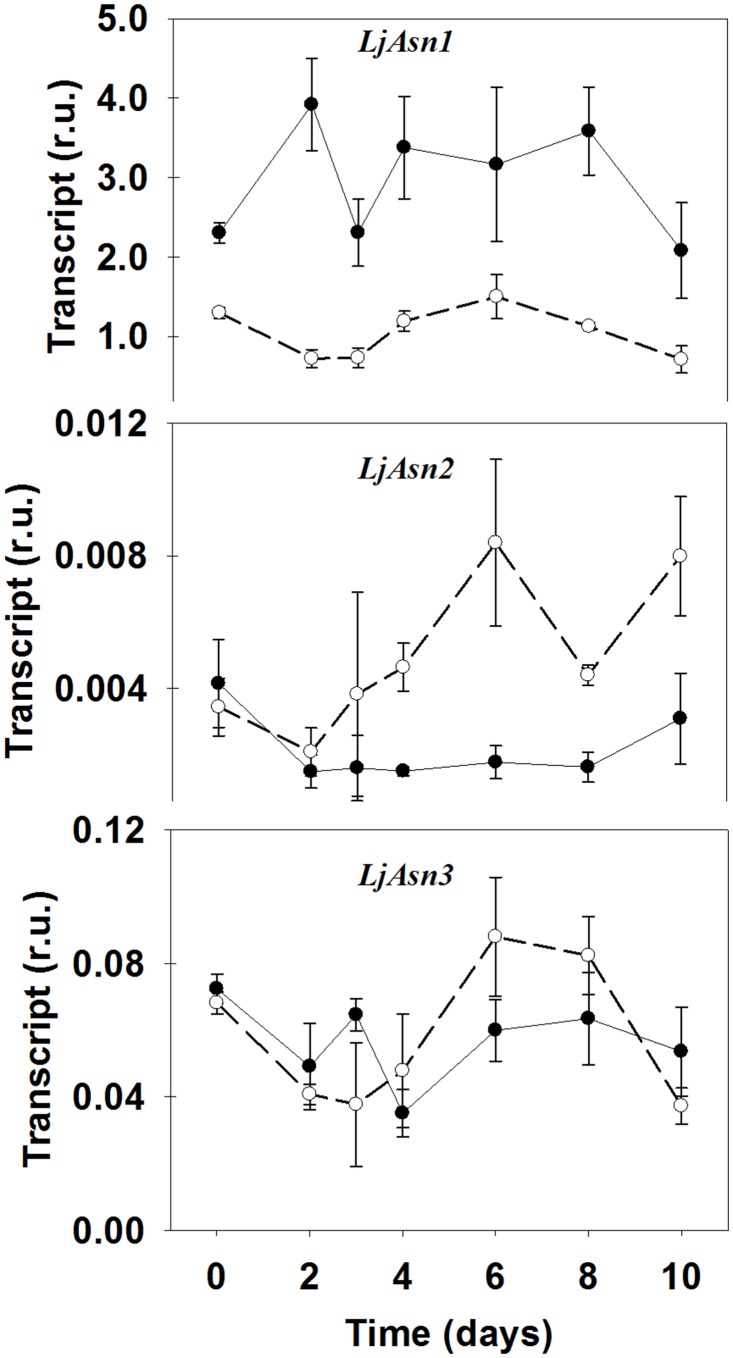
Two genes were initially described as encoding for ASN in L. japonicus [44]: LjAS1 and LjAS2. Here we will call these two genes LjAsn1 and LjAsn2 respectively according to the current nomenclature for asparagine synthetase genes. However, mining of the available database information led to the identification of a third Asn gene, that we will call LjAsn3.
Information about the role of ASN in plants came mainly from measurements of gene expression and metabolite levels. This is because of the difficulty in determining ASN polypeptide levels and enzyme activity due to the rapid turnover of the enzyme, to the presence of asparaginase activity and of ASN inhibitors in the extracts [45]. This was also the case of L. japonicus leaves extracts since it was not possible to determine ASN enzyme activity and no immunoreactive bands were found using anti-ASN primary antibodies (results nor shown). However, qRT-PCR analysis showed a clear induction of the LjAsn2 gene, the less expressed of the three ASN genes, exclusively in the mutant (Fig 4). LjAsn1, the most expressed one, did not show a clear trend over the time-course of the experiment but it was always more expressed in the WT than in the mutant, even at time zero (high CO2 conditions) where most genes showed similar expression levels in the two genotypes. No significant differences between WT and Ljgln2-2 mutants were detected for the expression of the LjAsn3 gene except for a minor increase observed in the mutants at days 6 and 8.
Fig 4. Expression levels of genes encoding for different ASN isoforms in WT and Ljgln2-2 mutant plants.
Transcript levels were quantified by qRT-PCR in WT (black dots, solid line) and mutant (white dots, dashed line) leaves at the time points indicated. Relative transcript levels are reported for each genotype compared to the housekeeping genes. Data are the mean ± S.D. of three independent biological replicates.
Interestingly, a significant accumulation of asparagine was observed in the Ljgln2-2 mutant at four, six and ten days of permanence under active photorespiratory conditions (Table 1), suggesting that asparagine biosynthesis may be increased in the mutant plants in accordance with the data obtained on LjAsn2 gene expression (Fig 4). On the other hand, aspartate levels were always lower in the mutant plants compared to the WT thus suggesting that aspartate may be used for the synthesis of asparagine and/or other uses in the mutant plants.
CPS gene expression and CP content in WT and Ljgln2-2
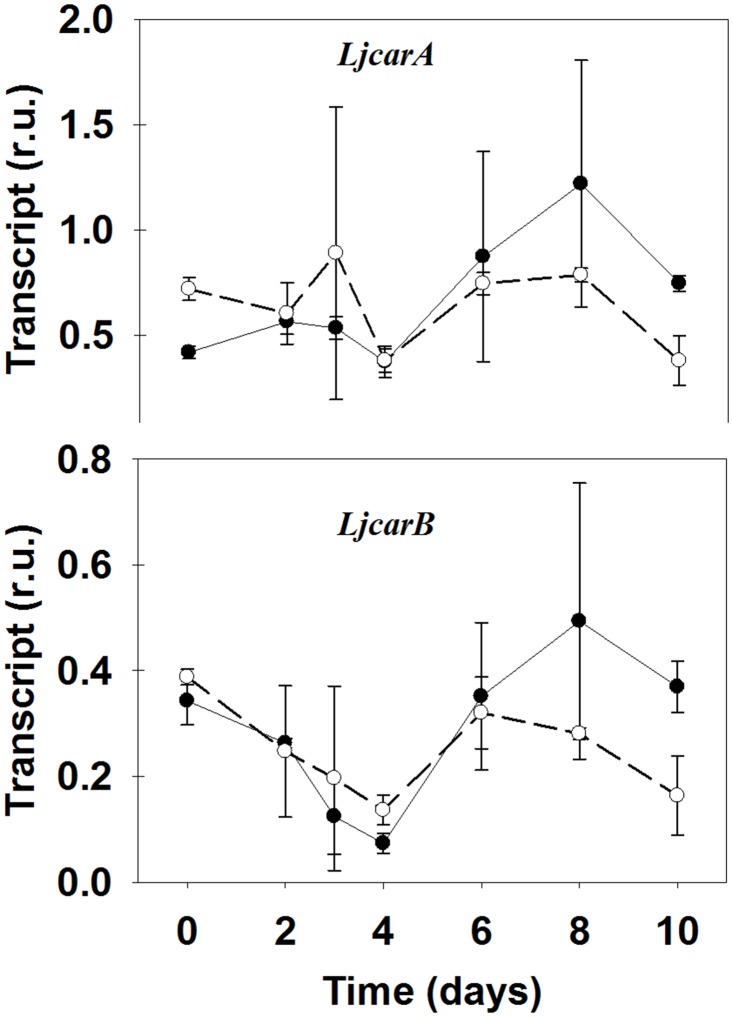
Plant CPS is similar to the bacterial enzyme and is formed by a large and a small subunit [46]. Most plants have two genes encoding for the small and large CPS subunits respectively, that in Arabidopsis are called carA and carB [20]. In L. japonicus, two homologous genes for CPS were found: LjCarA and LjCarB. The two genes showed similar levels of expression in L. japonicus leaves (Fig 5). The transfer to active photorespiratory conditions produced a repression of about three times in the expression of LjCarB gene after four days of active photorespiration followed by a recovery of transcript levels in both genotypes while no clear differences were obtained in the expression levels of LjCarA gene. No detectable levels of CPS activity were found in L. japonicus leaf extracts, either in WT or mutant plants. However, it was found that the levels of carbamoylphosphate were always much higher in the WT than in the Ljgln2-2 mutant plants and, in the case of WT plants, CP increased more than two-fold with a peak at day 2 followed by a sudden decrease (Table 1).
Fig 5. Expression levels of genes encoding for CPS in WT and Ljgln2-2 mutant plants.
Transcript levels for the genes encoding for the CPS small subunit (LjCarA) and for the large subunit (LjCarB) were quantified by qRT-PCR in WT (black dots, solid line) and mutant (white dots, dashed line) leaves at the time points indicated. Relative transcript levels are reported for each genotype compared to the housekeeping genes. Data are the mean ± S.D. of three independent biological replicates.
Discussion
Ljgln2-2 mutant plants accumulated high levels of NH4 + under active photorespiratory conditions followed by a sudden decrease in the content of this compound. This was suggestive of a regulation of photorespiratory metabolism and/or of enzymes involved in secondary NH4 + assimilation in the Ljgln2-2 mutants. Since the transcriptional regulation of photorespiratory and photosynthetic genes has been already described [18], the present paper was focused on the analysis of the different sets of enzymes that may be involved in ammonium reassimilation and detoxification of photorespiratory NH4 + and could account for the drop in this compound observed in the Ljgln2-2 mutant.
While the high accumulation of NH4 + observed in Ljgln2-2 clearly indicates that the enzyme involved in the re-assimilation of photorespiratory NH4 + is plastidic GS2, the induction of cytosolic GS1 activity, together with the increase in Gln levels, suggested that GS1 may be induced in order to contribute to the reassimilation and detoxification of this compound. This would be in agreement with previous reports that have shown that overexpression of cytosolic GS1 in leaf mesophyll cells of tobacco resulted in a reduction of free ammonium levels in leaves under active photorespiratory conditions [47]. In addition, particular isoforms of cytosolic GS1 such as GLN1.2 in Arabidopsis were suggested to be essential for ammonium detoxification [48]. Other recent data also indicate that GS1 could have an important role under ammonium nutrition [49, 50]. It has to be pointed out that cytosolic GS1, as well as ASN and GDH are induced in old senescing leaves in order to reassimilate the NH4 + generated by protein degradation [5]. However, we exclude the possibility that the changes observed in the present paper may be due to the onset of the senescence because young plants and healthy leaves were always analyzed at the different time periods examined and no proper symptoms of senescence were detected (no protein or nucleic acid degradation or other senescence parameters were observed at 10 days of transfer). In fact, most of the modulations in gene expression and enzyme activities observed in this paper started to take place at a very short time after the transfer from high CO2 to normal air conditions making it very unlikely that they could be attributed to senescence. The sudden modulation of gene expression and enzyme activities upon transfer from high CO2 to normal air conditions, rather support the idea that the events described in this paper represent an early response to the increased levels of photorespiratory NH4 +. Moreover, the peak in NH4 + level and the subsequent drop were clearly observed before any symptoms of the air-sensitivity phenotype can be detected.
It has to be taken into consideration that the different products of the cytosolic GS1 gene family are generally located in different cells of the vascular tissue, while plastidic GS2 is normally located in mesophyll cells where photorespiratory metabolism occurs [11, 51]. If this is the case in L. japonicus, then some transport or diffusion mechanism between photosynthetic and vascular tissues should function in order to shift the accumulated NH4 + in the mutant to the tissues were cytosolic GS1 is present. However, it should be also taken into consideration that under certain stress conditions the presence of cytosolic GS1 in mesophyll cells has been detected [52]. Further studies carried out with a higher cellular resolution will be needed in order to get more insight into the intercellular traffic of N metabolites, and especially of photorespiratory NH4 +, in this model legume.
Glutamate dehydrogenase can catalyze in vitro both the NADH-dependent incorporation of ammonium into α-KG to give glutamate (aminating reaction) and the NAD-dependent deamination of glutamate to give α-KG and ammonium (deaminating reaction). However, recent evidences indicate that the enzyme works in vivo in the direction of glutamate deamination in order to provide α-KG to the Krebs cycle and to control glutamate homeostasis [53, 54]. It may be possible that an increased deamination of glutamate by NAD-GDH activity is needed in order to provide carbon skeletons for the reassimilation of photorespiratory NH4 + in the mutant. The metabolite profiles presented in this work for the Ljgln2-2 mutant support this hypothesis, since glutamate levels drop after the shift to active photorespiratory conditions at the same time that α-KG levels are increased. Moreover, the induction of the LjICDH1 gene also suggested that a cytosolic ICDH isoform may also contribute to the production of α-KG for the reassimilation of NH4 + in this genotype. Despite of the central role of α-KG in both carbon and nitrogen metabolism, it is still unclear where is the major site of production of this metabolite [55]. Several reports suggested that cytosolic NADP-dependent isocitrate dehydrogenase may synthesize the α-KG necessary for the GS/GOGAT cycles ([42] and references therein). However, no conclusive evidence supporting this idea has been presented up to the date. The induction of LjICDH1 described here, together with the similar trends of LjICDH1 transcript levels and α-KG metabolite levels support the idea that cytosolic ICDH may at least in part provide carbon skeletons for NH4 + re-assimilation in the mutant.
A gene sequence encoding for a putative NADP(H)-GDH was detected in the L. japonicus genome. When Ljgln2-2 mutant plants were transferred to active photorespiratory conditions, an induction of NADPH-dependent GDH aminating activity was observed. In addition, a band of the expected size (about 71 kDa) corresponding to NADP(H)-GDH was also detected in the immunoblots. This is, to our knowledge, the first ever identification of the NADP(H)-GDH protein by Western Blot in higher plants. In contrast, when the NADP-dependent deaminating activity was assayed, no detectable enzyme activity was found. These results on the induction of NADP(H)-GDH gene expression, enzyme activity and polypeptide levels in L. japonicus are very interesting since there is no clear evidence for the presence of an active NADP(H)-dependent GDH in other plant species since very low levels of this enzyme have been found [56]. This may explain also the lack of other reports of any possible cross-reactivity among NAD(H)-GDH antibodies and NADP(H)-GDH. In fact, previous work carried out with Arabidopsis could only detect very low levels of NADP-dependent enzyme activity by in-gel GDH activity staining of root, leaves and stem extracts [56]. However, the activity bands observed coincided with these of NAD-GDH ones, suggesting that NADP(H)-dependent GDH is not expressed as an active protein in this plant [56]. The data shown in the present work strongly suggest that NADPH-GDH enzyme is active in L. japonicus and that this activity is induced in parallel with the lowering of photorespiratory NH4 + accumulation in the Ljgln2-2 mutant. Considering that only the aminating and not the deaminating enzyme activity of chloroplastic NADP(H)-GDH was detected, a role of this particular isoenzyme in ammonium reassimilation and/or detoxification in L. japonicus Ljgln2-2 mutants can not be excluded. It could be possible that NADPH-GDH might have an auxiliary role in ammonium detoxification and/or glutamate homeostasis in L. japonicus chloroplasts, at least when plastidic GS2 is absent in this organelle. Further work will be still required to determine the precise role of NADP(H)-GDH in this plant.
Of the three genes encoding for ASN found in the L. japonicus genome, LjAsn2 was highly induced in the mutant after the transfer to normal CO2 conditions. Considering that the trend in asparagine over time was very similar to the trend in expression level of LjAsn2 in both genotypes (Fig 4, Table 1), these data suggest that LjAsn2 may be responsible for the increase in asparagine levels observed in L. japonicus after the transfer to active photorespiratory conditions. This is consistent with the induction of LjAsn2 exclusively in the mutant and suggests a role for the corresponding enzyme in supporting NH4 + detoxification by using part of the Gln formed by the action of GS1 in this genotype.
The results shown in this paper indicate that GS1/GDH/ASN may have a physiological role in the mutant genotype with the aim of reducing the excessive levels of ammonium. However, in WT plants the normal levels of plastidic GS2 seem to be sufficient to efficiently reassimilate the photorespiratory ammonium. Further work would be still required on the diurnal profiling of GS1/GDH/ASN that would probably help to get insights into the question whether these genes do show a certain kind of short-term or intermitted response in the WT and therefore would strengthen the physiological role/sense of these alternative enzymes.
Finally, carbamoylphosphate synthase provides carbamoylphosphate for both pyrimidine synthesis pathway [46] and for arginine biosynthesis [57]. Recent studies have shown that CPS can also play a role in NH4 + detoxification by supporting Arg production [20]. In fact, the CP produced by CPS enzyme activity can be transformed to citrulline that is a precursor of Arg [20]. Of the two genes encoding for CPS in L. japonicus, only LjCarB, which encodes for the large CPS subunit, was significantly modulated by the transfer to active photorespiratory conditions, with a sudden drop in transcript levels followed by a subsequent recovery in both genotypes. CP levels were increased about two times exclusively in the WT genotype by the transfer to active photorespiratory conditions, and were always higher in this genotype compared to the mutant (Table 1). However, no significant differences in gene expression levels were detected among WT and Ljgln2-2. Since it was not possible to measure CPS enzyme activity in L. japonicus leaf extracts, further evidence is needed to support the possible role of CPS in ammonium assimilation and/or detoxification in L. japonicus plants.
In this work we have analyzed the different enzyme activities and metabolic pathways that may be involved in the reassimilation and detoxification of the photorespiratory NH4 + that is accumulated in the Ljgln2-2 mutant. The increase observed for various enzymes and amino acids exclusively in Ljgln2-2 mutants suggests that nitrogen metabolism is not a limiting factor for the fitness of the mutant and, simultaneously, may constitute a way for ammonium detoxification. Our interpretation of the different observations described in this paper could be summarized as follows: in the absence of plastidic GS2 activity, Gln can not be synthesized in the mutant chloroplasts. This may contribute to the increase in α-KG that takes place during the first three days of the transfer from high CO2 to normal CO2 conditions (Table 1), because of the lack of a functional GS2/GOGAT cycle. This may led to an increased cytosolic Gln synthesis by means of cytosolic GS1 induction once this Gln is started to be synthesized in the cytosol. Gln should be then redirected to the chloroplast for the purpose of Glu biosynthesis by means of a combined GS1/GOGAT cycle. This may explain the gradual drop in α-KG levels observed after day 3 (Table 1). This should contribute to Glu homeostasis that is fundamental for the plant since Glu is the precursor of several amino acids [58]. The increased cytosolic Gln produced from photorespiratory NH4 + may also support Asn biosynthesis by the action of ASN. This is important since Asn plays a fundamental role in nitrogen transport and remobilization in L. japonicus [59], and may also contribute to the drop in Glu levels observed in the mutant, since Asp, one of the substrates of ASN, is synthesized from oxaloacetate and Glu. The induction of genes encoding for different NAD(H)-dependent GDH isoforms and the increase of the corresponding polypeptide levels and enzyme activity is very probably aimed to provide carbon skeletons for photorespiratory NH4 + reassimilation and, on the other hand, to fuel the Krebs cycle under conditions in which many photosynthetic genes are repressed [18]. This catabolic role of GDH would be in accordance with the most recent results proposed for this enzyme in plants [53, 54] and would account also for the drop in Glu observed in the Ljgln2-2 mutant plants. A canalization of the C contained in the amino acids to the Krebs cycle and finally a higher production of transport and reserve amino acids such as Gln and Asn would be expected. Our data also suggested that a cytosolic ICDH isoform may contribute to the production of carbon skeletons for the re-assimilation of NH4 +. On the other hand, another interesting feature of the present paper is the finding of an active chloroplastic NADP(H)-GDH which may have an auxiliary role in ammonium detoxification and/or glutamate biosynthesis/homeostasis in L. japonicus chloroplasts. The results presented in this work make a significant improvement in the study of nitrogen reassimilation in L. japonicus and of the regulation of the key enzymes of this process.
Conclusions
An increase in the corresponding enzyme activity and/or transcription was found for several genes encoding for enzymes that may constitute an alternative way for photorespiratory ammonium reassimilation and detoxification in L. japonicus plants in the absence of plastidic GS2. The induction of cytosolic GS1 suggests that, this key enzyme of N metabolism may also be involved in the reassimilation of photorespiratory ammonium when plastidic GS2 is lacking and may be responsible for the increase in the levels of glutamine observed in the mutant plants. GDH enzyme activity and ASN gene expression were also higher in the mutant, indicating that these enzymes may act in conjunction with GS1 to cope with the process of photorespiratory ammonium accumulation in the plants. In addition, the increase in the polypeptide levels and aminating enzyme activity of NADP(H)-GDH discovered in the Ljgln2-2 mutant plants under photorespiratory active conditions, constitutes a very novel and important finding of the present work.
Supporting Information
WT and mutant plants were grown under high CO2 conditions (0.7% v/v) for 35 days and then transferred to normal CO2 conditions (0.04% v/v). Leaf samples from three different biological replicates were taken at time 0 (high CO2) and at the time points in days that are indicated under normal CO2.
(PPT)
The alignment was carried out using ClustalW with the Gonnet protein weight matrix and a gap open penalty of 10. The theoretical molecular weights of the polypeptides are of 44.74, 44.72, 44.66 and 70.74 kDa for LjGDH1, LjGDH2, LjGDH3 and LjGDH4 respectively. A dehydrogenase dimerization domain (pfam02812) between the amino acids 240 and 360 of LjGDH4 and the NAD(P)-binding domain of GDH from the amino acid 379 to the end of LjGDH4 were detected using the BLASTP program at NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
(PPT)
(DOC)
Acknowledgments
We thank Dr. K.A. Roubelakis-Angelakis (University of Crete) for the gift of anti-GDH antibodies, Dr. F.M. Cánovas and Dr. R. Cañas (University of Málaga) for the gift of anti-AS antibodies and for inspiring discussions on nitrogen assimilation. We would also like to thank D.H. Sánchez (University of Cambridge) for assistance with metabolomic data analysis. CMP acknowledges the support of PIF and V Plan Propio fellowships from the University of Seville.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by project P1O-CVI-6368 and BIO-163 support from Consejería de Economía, Innovación y Ciencia, Junta de Andalucía (http://www.juntadeandalucia.es/organismos/economiainnovacioncienciayempleo.htmThis work was supported by project P1O-CVI-6368 and BIO-163 support from Consejería de Economía, Innovación y Ciencia, Junta de Andalucía (http://www.juntadeandalucia.es/organismos/economiainnovacioncienciayempleo.html) and by the project AGL2014-54413-R from Spanish Ministerio de Economía y Competitividad (http://www.mineco.gob.es/). CMP acknowledges the support of PIF and V Plan Propio fellowships from the University of Seville (www.us.es). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Gutiérrez RA. System biology for enhanced plant nitrogen nutrition. Science. 2012;336: 1673–1675. 10.1126/science.1217620 [DOI] [PubMed] [Google Scholar]
- 2. Andrews M, Raven JA, Lea PJ. Do plants need nitrate? The mechanism by which nitrogen form affects plants. Ann Appl Biol. 2013;163: 174–199. [Google Scholar]
- 3. Udvardi MK, Poole PS. Transport and metabolism in legume-rhizobia symbiosis. Annu Rev Plant Biol. 2013;64: 781–805. 10.1146/annurev-arplant-050312-120235 [DOI] [PubMed] [Google Scholar]
- 4. Krapp A, David LC, Chardin C, Grin T, Marmagne A, Leprince AS, et al. Nitrate transport and signalling in Arabidopsis. J Exp Bot. 2014;65: 789–798. 10.1093/jxb/eru001 [DOI] [PubMed] [Google Scholar]
- 5. Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, Chardon F, Gaufichon L, Suzuki A. Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann Bot. 2010;105: 1141–1157. 10.1093/aob/mcq028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Betti M, García-Calderón M, Pérez-Delgado CM, Credali A, Estivill G, Galván F, et al. Glutamine synthetase in legumes: recent advances in enzyme structure and functional genomics. Int J Mol Sci. 2012;13: 7994–8024. 10.3390/ijms13077994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fernie AR, Bauwe H, Eisenhut M, Florian A, Hanson DT, Hagemann M, et al. Perspectives on plant photorespiratory metabolism. Plant Biol. 2013;15: 748–753. 10.1111/j.1438-8677.2012.00693.x [DOI] [PubMed] [Google Scholar]
- 8. Seabra AR, Vieira CP, Cullimore JV, Carvalho HG. Medicago truncatula contains a second gene encoding a plastid located glutamine synthetase exclusively expressed in developing seeds. BMC Plant Biol. 2010;10: 183 10.1186/1471-2229-10-183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Castro-Rodríguez V, García-Gutiérrez A, Cañas RA, Pascual M, Avila C, Cánovas FM. Redundancy and metabolic function of the glutamine synthetase gene family in poplar. BMC Plant Biol. 2015;15: 20 10.1186/s12870-014-0365-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Castro-Rodríguez V, García-Gutiérrez A, Canales J, Avila C, Kirby EG, Cánovas FM. The glutamine synthetase gene family in Populus. BMC Plant Biol. 2011;11: 119 10.1186/1471-2229-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bernard SM, Habash DZ. The importance of cytosolic glutamine synthetase in nitrogen assimilation and recycling. New Phytol. 2009;182: 608–620. 10.1111/j.1469-8137.2009.02823.x [DOI] [PubMed] [Google Scholar]
- 12. Thomsen HC, Eriksson D, Møller IS, Schjoerring JK. Cytosolic glutamine synthetase: a target for improvement of crop nitrogen use efficiency? Trends Plant Sci. 2014;19: 656–663. 10.1016/j.tplants.2014.06.002 [DOI] [PubMed] [Google Scholar]
- 13. Blackwell RD, Murray AJS, Lea PJ. Inhibition of photosynthesis in barley with decreased levels of chloroplast glutamine synthetase activity. J Exp Bot. 1987;38: 1799–1809. [Google Scholar]
- 14. Wallsgrove RM, Turner JC, Hall NP, Kendall AC, Bright SWJ. Barley mutants lacking chloroplast glutamine synthetase—biochemical and genetic analysis. Plant Physiol. 1987;83: 155–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Husted S, Mattsson M, Mollers C, Wallbraun M, Schjoerring JK. Photorespiratory NH4+ production in leaves of wild-type and glutamine synthetase 2 antisense oilseed rape. Plant Physiol. 2002;130: 989–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Orea A, Pajuelo P, Pajuelo E, Quidiello C, Romero J, Márquez AJ. Isolation of photorespiratory mutants from Lotus japonicus deficient in glutamine synthetase. Physiol Plant. 2002;113: 193–202. [DOI] [PubMed] [Google Scholar]
- 17. Betti M, Arcondéguy T, Márquez AJ. Molecular analysis of two mutants from Lotus japonicus deficient in plastidic glutamine synthetase: functional properties of purified GLN2 enzymes. Planta. 2006;224: 1068–1079. [DOI] [PubMed] [Google Scholar]
- 18. Pérez-Delgado CM, García-Calderón M, Sánchez DH, Udvardi MK, Kopka J, Márquez AJ, et al. Transcriptomic and metabolic changes associated with photorespiratory ammonium accumulation in the model legume Lotus japonicus . Plant Physiol. 2013;162: 1834–1848. 10.1104/pp.113.217216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Betti M, García-Calderón M, Pérez-Delgado CM, Credali A, Pal’Ove-Balang P, Estivill G, et al. Reassimilation of ammonium in Lotus japonicus . J Exp Bot. 2014;65: 5557–5566. 10.1093/jxb/eru260 [DOI] [PubMed] [Google Scholar]
- 20. Potel F, Valadier M-H, Ferrario-Méry S, Grandjean O, Morin H, Gaufichon L, et al. Assimilation of excess ammonium into amino acids and nitrogen translocation in Arabidopsis thaliana–roles of glutamate synthases and carbamoylphosphate synthase. FEBS J. 2009;276: 4061–4076. 10.1111/j.1742-4658.2009.07114.x [DOI] [PubMed] [Google Scholar]
- 21. Tercé-Laforgue T, Bedu M, Dargel-Grafin C, Dubois F, Gibon Y, Restivo FM, et al. Resolving the role of plant glutamate dehydrogenase: II. Physiological characterization of plants overexpressing the two enzyme subunits individually or simultaneously. Plant Cell Physiol. 2013;54: 1635–1647. 10.1093/pcp/pct108 [DOI] [PubMed] [Google Scholar]
- 22. Skopelitis DS, Paranychianakis NV, Paschalidis KA, Pliakonis ED, Delis ID, Yakoumakis DI, et al. Abiotic stress generates ROS that signal expression of anionic glutamate dehydrogenase to form glutamate for proline synthesis in tobacco and grapevine. Plant Cell. 2006;18: 2767–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Setién I, Fuertes-Mendizabal T, González A, Aparicio-Tejo PM, González-Murua C, González-Moro MB, et al. High irradiance improves ammonium tolerante in wheat plants by increasing N assimilation. J Plant Physiol. 2013;170: 758–771. 10.1016/j.jplph.2012.12.015 [DOI] [PubMed] [Google Scholar]
- 24. Masclaux-Daubresse C, Reisdorf-Cren M, Pageau K, Lelandais M, Grandjean O, Kronenberger J, et al. Glutamine synthetase-glutamate synthase pathway and glutamate dehydrogenase play distinct roles in the sink-source nitrogen cycle in tobacco. Plant Physiol. 2006;140: 444–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lea PJ, Sodek L, Parry MAJ, Shewry PR, Halford NG. Asparagine in plants. Ann Appl Biol. 2007;150: 1–26. [Google Scholar]
- 26. Britto DT, Kronzuker HJ. NH4+ toxicity in higher plants: a critical review. J Plant Physiol. 2002;159: 567–584. [Google Scholar]
- 27. Li B, Li G, Kronzucker HJ, Baluska F, Shi W. Ammonium stress in Arabidopsis: signaling, genetic loci, and physiological targets. Trends Plant Sci. 2014;19: 107–114. 10.1016/j.tplants.2013.09.004 [DOI] [PubMed] [Google Scholar]
- 28. Handberg K, Stougaard J. Lotus japonicus, an autogamous, diploid legume species for classical and molecular genetics. Plant J. 1992;2: 487–496. [Google Scholar]
- 29. Sánchez DH, Lippold F, Redestig H, Hannah MA, Erban A, Krämer U, et al. Integrative functional genomics of salt acclimatation in the model legume Lotus japonicus . Plant J. 2008;53: 973–987. [DOI] [PubMed] [Google Scholar]
- 30. Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK. Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J. 2004;38: 366–379. [DOI] [PubMed] [Google Scholar]
- 31. Pierson DL. A rapid colorimetric assay for carbamyl phosphate synthetase I. J Biochem Biophys Meth.1980;3: 31–37. [DOI] [PubMed] [Google Scholar]
- 32. Laemmli UK. Cleavage of structural protein during the assembly of head of bacteriophage T4. Nature. 1970;251: 614–616. [DOI] [PubMed] [Google Scholar]
- 33. Betti M, Márquez AJ, Yanes C, Maestre A. ATP binding to purified homopolymeric plant glutamine synthetase studied by isothermal titration calorimetry. Thermochim Acta. 2002;394: 63–71. [Google Scholar]
- 34. Loulakakis CA, Roubelakis-Angelakis KA. Immunocharacterization of NADH-glutamate dehydrogenase from Vitis vinifera L. Plant Physiol. 1990;94: 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cañas RA, de la Torre F, Cánovas FM, Cantón FR. High levels of asparagine synthetase in hypocotyls of pine seedlings suggest a role of the enzyme in re-allocation of seed-stored nitrogen. Planta. 2006;224: 83–95. [DOI] [PubMed] [Google Scholar]
- 36. Márquez AJ, Betti M, García-Calderón M, Estivill G, Credali A, Pajuelo P, et al. Nitrate and ammonium assimilatory enzymes In Márquez AJ, editor. Lotus japonicus handbook. Dordrecht:Springer;2005. p. 315–328. [Google Scholar]
- 37. Loulakakis KA, Roubelakis-Angelakis KA. Plant NAD(H)-glutamate dehydrogenase consists of two subunit polypeptides and their participation in the seven isoenzymes occurs in an ordered ratio. Plant Physiol. 1991;97: 104–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Doremus HD. Organization of the pathway of de Novo pyrimidine nucleotide biosynthesis in Pea (Pisum sativum L. cv Progress No. 9) leaves. Arch Biochem Biophys. 1986;250: 112–119. [DOI] [PubMed] [Google Scholar]
- 39. Sheng S, Kraft JJ, Schuster SM. A specific quantitative colorimetric assay for L-asparagine. Anal Biochem. 1993;211: 242–249. [DOI] [PubMed] [Google Scholar]
- 40. Saeed AI, Hagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, et al. TM4 microarray software suite. Methods Enzymol. 2006;411: 134–193. [DOI] [PubMed] [Google Scholar]
- 41. Solorzano L. Determination of ammonia in natural waters by the phenolhypoclorite methos. Ocean Limno Oceanogr. 1969;14: 799–801. [Google Scholar]
- 42. Hodges M. Enzyme redundancy and the importance of 2-oxoglutarate in plant ammonium assimilation. J. Exp Bot. 2002;53: 905–916. [DOI] [PubMed] [Google Scholar]
- 43. Araújo WL, Ishizaki K, Nunes-Nesi A, Larson TR, Tohge T, Krahnert I, et al. Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell 2010;22: 1549–1563. 10.1105/tpc.110.075630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Waterhouse RM, Smyth AJ, Massoneau A, Prosser IM, Clarkson DT. Molecular cloning and characterisation of asparagine synthetase from Lotus japonicus: Dynamics of asparagine synthesis in N-sufficient conditions. Plant Mol Biol. 1996;30: 883–897. [DOI] [PubMed] [Google Scholar]
- 45. Azevedo RA, Lancien M, Lea PJ. The aspartic acid metabolic pathway, an exciting and essential pathway in plants. Amino Acids. 2006;30:143–162. [DOI] [PubMed] [Google Scholar]
- 46. Slocum RD. Genes, enzymes and regulation of arginine biosynthesis in plants. Plant Physiol Biochem. 2005;43: 729–745. [DOI] [PubMed] [Google Scholar]
- 47. Oliveira IC, Brears T, Knight TJ, Clark A, Coruzzi GM. Overexpression of cytosolic glutamine synthetase. Relation to nitrogen, light and photorespiration. Plant Physiol. 2002;129: 1170–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lothier J, Gaufichon L, Sormani R, Lemaitre T, Azopardi M, Morin H, et al. The cytosolic glutamine synthetase GLN1;2 plays a role in the control of plant growth and ammonium homeostasis in Arabidopsis rosettes when nitrate supply is not limiting. J Exp Bot. 2011;62: 1375–1390. 10.1093/jxb/erq299 [DOI] [PubMed] [Google Scholar]
- 49. Sarasketa A, González-Moro MB, González-Murúa C, Marino D. Exploring ammonium tolerance on a large panel of Arabidopsis thaliana natural accessions. J Exp Bot. 2014;65: 6023–6033. 10.1093/jxb/eru342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yamaya T, Kusano M. Evidence supporting distinct functions of the cytosolic glutamine synthetases and two NADH-glutamate synthases in rice. J Exp Bot. 2014;65: 5519–5525. 10.1093/jxb/eru103 [DOI] [PubMed] [Google Scholar]
- 51. Seabra AR, Silva LS, Carvalho HS. Novel aspects of glutamine synthetase (GS) regulation revealed by a detailed expression analysis of the entire GS gene family of Medicago truncatula under different physiological conditions. BMC Plant Biol. 2013;13: 137 10.1186/1471-2229-13-137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pérez-García A, Pereira S, Pissarra J, García-Gutiérrez A, Cazorla FM, Salema R, et al. Cytosolic localization in tomato mesophyll cells of a novel glutamine synthetase induced in response to bacterial infection or phosphinothricin treatment. Planta 1998;206: 426–434. [Google Scholar]
- 53. Fontaine J-X, Tercé-Laforgue T, Armengaud P, Clément G, Renou J-P, Pelletier S, et al. Characterization of a NADH-dependent glutamate dehydrogenase mutant of Arabidopsis demonstrates the key role of this enzyme in root carbon and nitrogen metabolism. Plant Cell. 2012;24: 4044–4065. 10.1105/tpc.112.103689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Labboun S, Tercé-Laforgue T, Roscher A, Bedu M, Restivo FM, Velanis CN, et al. Resolving the role of plant glutamate dehydrogenase I. in vivo real time nuclear magnetic resonance spectroscopy experiments. Plant Cell Physiol. 2009;50: 1761–1773. 10.1093/pcp/pcp118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Araújo WL, Martins AO, Fernie AR, Tohge T. 2-Oxoglutarate: linking TCA cycle function with amino acid, glucosinolate, flavonoid, alkaloid, and gibberellin biosynthesis. Front. Plant Sci. 2014;15: 552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fontaine J-X, Tercé-Laforgue T, Bouton S, Pageau K, Lea PJ, Dubois F, et al. Further insights into the isoenzyme composition and activity of glutamate dehydrogenase in Arabidopsis thaliana . Plant Signal Behav. 2013; 8:e23329 10.4161/psb.23329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mollá-Morales A, Sarmiento-Mañús R, Robles P, Quesada V, Pérez-Pérez JM, González-Bayón R, et al. Analysis of ven3 and ven6 reticulate mutants reveals the importance of arginine biosynthesis in Arabidopsis leaf development. Plant J. 2010;65: 335–345. 10.1111/j.1365-313X.2010.04425.x [DOI] [PubMed] [Google Scholar]
- 58. Forde BG, Lea PJ. Glutamate in plants: metabolism, regulation and signalling. J Exp Bot. 2007;58: 2339–2358. [DOI] [PubMed] [Google Scholar]
- 59. Credali A, García-Calderón M, Dam S, Perry J, Diaz-Quintana A, Parniske M, et al. The K+-dependent asparaginase NSE 1 is crucial for plant growth and seed production in Lotus japonicus . Plant Cell Physiol. 2013;54: 107–118. 10.1093/pcp/pcs156 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
WT and mutant plants were grown under high CO2 conditions (0.7% v/v) for 35 days and then transferred to normal CO2 conditions (0.04% v/v). Leaf samples from three different biological replicates were taken at time 0 (high CO2) and at the time points in days that are indicated under normal CO2.
(PPT)
The alignment was carried out using ClustalW with the Gonnet protein weight matrix and a gap open penalty of 10. The theoretical molecular weights of the polypeptides are of 44.74, 44.72, 44.66 and 70.74 kDa for LjGDH1, LjGDH2, LjGDH3 and LjGDH4 respectively. A dehydrogenase dimerization domain (pfam02812) between the amino acids 240 and 360 of LjGDH4 and the NAD(P)-binding domain of GDH from the amino acid 379 to the end of LjGDH4 were detected using the BLASTP program at NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
(PPT)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.