Abstract
Background
Defective cellular transport processes can lead to aberrant accumulation of trace elements, iron, small molecules and hormones in the cell, which in turn may promote the formation of reactive oxygen species, promoting DNA damage and aberrant expression of key regulatory cancer genes. As DNA damage and uncontrolled proliferation are hallmarks of cancer, including epithelial ovarian cancer (EOC), we hypothesized that inherited variation in the cellular transport genes contributes to EOC risk.
Methods
In total, DNA samples were obtained from 14,525 case subjects with invasive EOC and from 23,447 controls from 43 sites in the Ovarian Cancer Association Consortium (OCAC). Two hundred seventy nine SNPs, representing 131 genes, were genotyped using an Illumina Infinium iSelect BeadChip as part of the Collaborative Oncological Gene-environment Study (COGS). SNP analyses were conducted using unconditional logistic regression under a log-additive model, and the FDR q<0.2 was applied to adjust for multiple comparisons.
Results
The most significant evidence of an association for all invasive cancers combined and for the serous subtype was observed for SNP rs17216603 in the iron transporter gene HEPH (invasive: OR = 0.85, P = 0.00026; serous: OR = 0.81, P = 0.00020); this SNP was also associated with the borderline/low malignant potential (LMP) tumors (P = 0.021). Other genes significantly associated with EOC histological subtypes (p<0.05) included the UGT1A (endometrioid), SLC25A45 (mucinous), SLC39A11 (low malignant potential), and SERPINA7 (clear cell carcinoma). In addition, 1785 SNPs in six genes (HEPH, MGST1, SERPINA, SLC25A45, SLC39A11 and UGT1A) were imputed from the 1000 Genomes Project and examined for association with INV EOC in white-European subjects. The most significant imputed SNP was rs117729793 in SLC39A11 (per allele, OR = 2.55, 95% CI = 1.5-4.35, p = 5.66x10-4).
Conclusion
These results, generated on a large cohort of women, revealed associations between inherited cellular transport gene variants and risk of EOC histologic subtypes.
Introduction
Epithelial ovarian carcinoma (EOC) is the second-most common gynecological malignancy and the leading cause of gynecological cancer-related mortality in the United States and other developed nations [1]. Early stage EOC is accompanied by vague, non-specific symptoms and is difficult to detect. As yet, no EOC screening or early detection strategies have been proven useful in general population [2]. As a consequence, approximately 60% of cases are diagnosed at advanced stages (III-IV), with 5-year overall survival of less than 20% [3]. Given these grim statistics, improved understanding of the etiology of EOC is critical to reducing the associated morbidity and mortality.
An inadequate understanding of genetic and biological etiology of EOC has limited the ability to detect and treat this disease effectively. Disruptions in cellular transport, lead to abnormal levels of trace elements (iron, zinc and copper), hormones and small molecules which impact the expression of key regulatory genes. Aberrant expression of transmembrane transport genes has been associated with increased risk, as well as aggressiveness, of a number of cancers including breast [4–7], prostate [8], liver [9], colorectal and colon [10–12], thyroid [13] and neuroblastoma [14]. Iron is essential for erythropoiesis [15] and the function of the mitochondrial respiratory chain where it plays a key role in electron transport, in the form of iron-sulphur clusters or in heme centers [16]. However, excess iron and copper have been reported to promote the formation of reactive oxygen species (ROS) which damage cellular DNA and support cancer growth [17]. Hydrogen peroxide generated as a byproduct in the mitochondrial respiratory chain, can react with iron or copper and form hydroxyl radicals that are extremely reactive and damaging to the genome [18]. Consistent with these data, transport of the three most abundant transition metals in humans–iron, zinc and copper–has been linked to the etiology of colorectal and liver cancer [9,19] and prostate cancer [20].
Despite the growing evidence that cellular transport processes influence cancer risk, the association of germline genetic variation in cellular transport genes and EOC risk has not been well studied. As histologic subtypes of EOC differ in clinical behavior and biologic and genetic origin [21], we hypothesized that single nucleotide polymorphisms (SNPs) in cellular transport genes are associated with EOC risk and vary by histopathology. This study examined the association of 279 SNPs in 131 cellular transport genes and EOC risk in an international collaboration that included 18,174 case and 26,134 control subjects.
Materials and Methods
Sample and Procedure
The discovery set included DNA samples from 3,761 EOC case subjects and 2,722 control subjects in two ovarian cancer Genome Wide Association Studies (GWAS) in North America and the United Kingdom (UK). Details of these studies have been previously published [22]. In brief, the North American study was comprised of four case-control studies genotyped using the Illumina 610-quad Beadchip Array (1,814 case subjects and 1,867 control subjects) as well as a single case-control study genotyped on the Illumina 317K and 370K arrays (133 case subjects and 142 control subjects). The UK study was comprised of four case-only studies genotyped on the Illumina 610-quad Beadchip Array and two common control sets genotyped on the Illumina 550K array (1,814 case subjects and 713 control subjects).
The replication sample consisted of DNA samples from 14,525 women with invasive EOC and 23,447 control women with European ancestry from 43 sites in the Ovarian Cancer Association Consortium (OCAC). Samples from an additional 1,747 participants with tumors of low malignant potential were also analyzed. Details of the sample QC are provided in Pharoah et al., [22]. This replication set include 89 SER cases and 200 controls of African ancestry, and 249 SER cases and 1574 controls of Asian ancestry.
All research involving human participants has been approved by each study site’s local Institutional Review Board (IRB) according to the principles expressed in the Declaration of Helsinki. Written informed consent was obtained from all of the participants. The permission to use of the data in this study was granted by the University of South Florida (IRB#: Pro00000249).
Gene and SNP Selection
Gene (NCBI) [23], BioCarta [24], GenomeNet [25] and other relevant gene/pathway databases were used to select the candidate genes for inclusion in the transport pathway. Specifically, the search yielded 131 genes involved in transport of trace elements, ions, hormones and small molecules. A total of 5202 SNPs in the selected genes were identified from the Human-610 Quad BeadChip (Illumina). Those SNPs were genotyped in the four ovarian cancer GWAS studies (US GWAS, UK GWAS, COGS and Mayo clinic). The final selection of transport gene SNPs for genotyping in the replication stage was informed according to lowest p-values (cut off p<0.01 was used) by ranking of minimal p-values across four sets of results in the discovery set: 1) North American all histologies, 2) North American serous histology, 3) combined GWAS meta-analysis all histologies, and 4) combined GWAS meta-analysis serous histology. Additional functional SNPs in these genes were also included. In total 299 SNPs were included on the COGS chip of which 279 SNPs (in 131 genes) passed QC (described in detail in Pharoah et al., [22]).
Imputation Analyses
These analyses were based on imputed genotypes from the four ovarian cancer GWAS studies (US GWAS, UK GWAS, COGS and Mayo clinic) with a total of 15,398 invasive EOC case subjects and 30,816 control subjects of white-European ancestry. Imputation of each dataset into the 1000 Genomes Project was performed using IMPUTE2 software [26]. We used the 1000 Genomes Project v3 as the reference with pre-phasing of the data using SHAPEIT [27].
Statistical Analysis
For the discovery set, the North American and UK studies were analyzed separately and four sets of results: 1) North American all histologies, 2) North American serous histology, 3) combined GWAS meta-analysis all histologies, and 4) combined GWAS meta-analysis serous histology were conducted. SNP analyses were performed using unconditional logistic regression under a log-additive model. The last two sets were analyzed using the fixed effects meta-analysis. These analyses were carried out in PLINK [28] by combining results across studies by using the Mantel–Haenszel method [29].
For the replication set, demographic and clinical characteristics of case subjects and control subjects were compared using t-test for continuous variables and chi-square test for categorical variables. Unconditional logistic regression was used to evaluate associations between SNPs and ovarian cancer risk. SNPs were modeled using number of minor alleles as ordinal variables (log-additive model). Per-allele log odds ratios and their 95% confidence intervals were estimated. All analyses were done separately by race groups.
In order to adjust for population substructure, intercontinental ancestry was assigned based on genotype frequencies for European, Asian, and African populations using LAMP software [30] Subjects with greater than 90 percent European ancestry were defined as European; those with greater than 80 percent Asian and African ancestry were defined as being Asian and African, respectively. The set of 37,000 unlinked markers was applied to perform principal-components analysis within each major population subgroup [31].The number of principal components was based on the inflection position of the principal components scree plot. The models for white-European subjects were adjusted for study site and for the first five principal components. For the African American (AA) and Asian (AS) study subjects, the first one and five principal components were included in the models, respectively.
We evaluated associations between candidate SNPs and risk of all invasive EOC (INV), each of the four main histological subtypes (serous (SER); endometroid (END); clear cell (CC); and mucinous (MUC)), and tumors of low malignant potential (LMP). Odds ratios for each histologic subtype were estimated by comparing cases of each subtype to all controls as reference. False discovery rate (FDR) q-value was applied for adjusting multiple comparisons [32]. Associations with a p value < .05 and a false discovery rate (FDR) q-value < .20 were considered statistically significant.
For the imputation set analyses, the meta-analysis using an in-house program written in C++ was carried out for combining results across studies. A fixed effect meta-analysis was used, with the log odds ratio being the estimate for each study and the standard error of this estimate determining the weighting. For each SNP, only the studies with valid estimates for that SNP (i.e. r2 > 0.25) were used in the meta-analysis calculation.
Results
Sample characteristics are described in the S1 Table. As expected, significant differences were observed between case and control subjects on EOC risk factors including age, family history of ovarian cancer, age at menarche, body mass index (BMI), history of oral contraceptive use, and number of full term births (p-values<0.05). The proportion of tumors of SER (57.6%) was higher than that of other subtypes (14.2% END, 7.1% CC, 6.5% MUC, and 14.6% other), which is typical for white-European populations.
Among the 279 cellular transport SNPs genotyped in the replication set, 81 SNPs in 48 genes showed nominally significant (p<0.05) associations with at least one histological subtype (S2 Table). All invasive cancers combined (INV), LMP and the four main histological subtypes (SER [n = 8,369]; END [n = 2,067]; CC [n = 1,024]; and MUC [n = 943]) were analyzed.
The strongest evidence of an association for INV EOC was observed for SNP rs17216603 in the iron transporter gene HEPH (OR = 0.85, 95%CI = 0.77–0.93, P = 2.55x10-4; FDR q-value = 0.065), which was also the most significant SNP associated with SER (P = 1.99x10-4; 95%CI = 0.73–0.91, FDR q-value = 0.054), and LMP subtypes (P = 0.0206) (Table 1). The most significant association for END EOC was rs11563251 within the UGT1A gene cluster (OR = 0.82, 95%CI = 0.73–0.92, P = 6.59x10-4; FDR q-value = 0.177). Only these two SNPs were associated with q-values <0.20. The most significant association for MUC subtype was rs681309, near the SLC25A45 gene (OR = 0.89, 95%CI = 0.81–0.97, P = 0.012). This SNP was also associated with the END subtype (P = 0.0035) and INV EOC (P = 0.029). Association with rs9908917, in the intron of SLC39A11, was observed for LMP cases (OR = 0.85, 95%CI = 0.77–0.93, P = 3.9x10-4). This SNP was also associated with the SER subtype (P = 0.0123) and INV EOC (P = 0.0144). The SNP rs1804495 in SERPINA7 was associated with SER, MUC, CC, INV and LMP (P<0.05), but not END (P>0.05).
Table 1. The most significant SNPs in the transport pathway genes and risk of EOC by histology, invasiveness, and race/ethnicity 1 .
| GENE SNP | INV | LMP | SER | CC | END | MUC | Asian (SER) | African-American (SER) |
|---|---|---|---|---|---|---|---|---|
| HEPHrs17216603 | 0.85 (0.77–0.93); 2.55x10 -4 ; q = 0.063 | 0.78 (0.63–0.97); 0.021; | 0.81 (0.73–0.91); 1.99x10 -4 ; q = 0.054 | 0.77 (0.58–1.02); 0.07 | 0.9 (0.74–1.08); 0.26 | 0.92 (0.7–1.21); 0.56 | 1.45 (1.15–1.83) 0.0019 | 0.76 (0.15–3.86); 0.74 |
| SLC39A11rs9908917 | 0.95 (0.92–0.99); 0.014 | 0.85 (0.77–0.93); 3.9x10 -4 | 0.94 (0.9–0.99); 0.010 | 1.01 (0.91–1.13); 0.82 | 0.94 (0.87–1.02); 0.16 | 0.98 (0.87–1.00); 0.7 | 1.25 (1.01–1.56); 0.049 | 1.23 (0.85–1.77); 0.27 |
| SERPINA7Rs1804495 | 1.05 (1.00–1.1); 0.042 | 1.14 (1.03–1.27); 0.016 | 1.06 (1.00–1.12); 0.045 | 1.21 (1.06–1.39); 0.0042 | 1.06 (0.96–1.17); 0.28 | 0.85 (0.73–1.00); 0.045 | 0.81 (0.65–1.00); 0.05 | 1.11 (0.75–1.65); 0.6 |
| SLC25A45Rs681309 | 0.97 (0.94–1.00); 0.029 | 1.04 (0.97–1.12); 0.27 | 1 (0.96–1.04); 0.99 | 0.98 (0.89–1.07); 0.59 | 0.91 (0.85–0.97); 0.0035 | 0.89 (0.81–0.97); 0.012 | 0.78 (0.63–0.98); 0.033 | 0.87 (0.6–1.26); 0.41 |
| UGT1A Rs11563251 | 0.95 (0.91–1.00); 0.05 | 1.08 (0.96–1.2); 0.21 | 0.95 (0.89–1.01); 0.07 | 1.03 (0.89–1.18); 0.73 | 0.82 (0.74–0.92); 6.8x10 -4 ; q = 0.18 | 1.06 (0.91–1.22); 0.49 | 0.73 (0.5–1.06); 0.1 | 1.22 (0.85–1.25); 0.28 |
| MGST1 Rs6488840 | 0.96 (0.93–1.00); 0.048 | 1.0 (0.92–1.09); 0.9 | 0.95 (0.91–1.00); 0.042 | 0.96 (0.86–1.07); 0.41 | 1 (0.92–1.08); 0.95 | 0.96 (0.86–1.08); 0.48 | 0.54 (0.25–1.18); 0.12 | 0.55 (0.37–0.82); 0.0035 |
1 INV: all invasive EOC combined; LMP: low malignant potential / borderline tumors; SER: serous; CC: clear cell; End: endometrioid; Muc: mucinous. Statistically significant associations are indicated in bold (P<0.05). Data format is the following: OR (95% CI); p-value; FDR q-value (white-European women). Only significant FDRs (q<0.2) are shown (HEPH: INV and SER; UGT1A: End).
Imputed Variants
In total, 1785 imputed SNPs in six genes (HEPH, MGST1, SERPINA, SLC25A45, SLC39A11 and UGT1A) were examined for association with INV EOC in white-European subjects only. From these, 274 SNPs were found with p-value<0.05 (S3 Table). Across all six genes, the most significant imputed SNP was rs117729793 in SLC39A11 (per allele, OR = 2.55, 95% CI = 1.5–4.35, p = 5.66x10-4). Interestingly, 190 of 274 (~70%) imputed SNPs with p-values < 0.05 were located in or near SLC39A11.
Results in women of African-American (AA) and Asian (AS) ethnicities
We conducted exploratory analyses for other ethnicities and SER EOC. Fourteen SNPs showed significant associations in AS and AA women. Six of the SNPs in the AS women were also significant in the white-European women, compared to two of the 14 SNPs in the AA women. The top SNP in women of Asian ancestry (rs17216603 in HEPH) was shared with the white-European women. The SLC25A45 rs681309 was also shared. SERPINA7 rs1804495 was borderline significant (P = 0.0503), perhaps due to a small sample size. The most significant association in women of AA ancestry was noted at the SNP rs6488840 near to the microsomal glutathione S-transferase 1 (MGST1) gene. In our study, MGST1 rs6488840 was associated with statistically significantly reduced SER EOC risk in women of AA ancestry (OR = 0.55; P = 0.0035). This SNP was of borderline significance in women of white-European (OR = 0.95; P = 0.042), but not Asian (P>0.05), ancestry. In the groups of AS and AA women, no SNPs had FDR q-value <0.20. Results for the top hits across women of different ancestries are presented in Table 2.
Table 2. Top SNPs associated with SER EOC across racial groups.
| Race | White-European | Asian | African American | ||||||
|---|---|---|---|---|---|---|---|---|---|
| GENE SNP | MAF 1 | p-value 2 | OR (95% CI) 3 | MAF 1 | p-value 2 | OR (95% CI) 3 | MAF 1 | p-value 2 | OR (95% CI) 3 |
| HEPH rs17216603 | A = 0.03 | 2x10 -4 | 0.81 (0.73–0.91) | A<0.01 | 0.002 | 1.45 (1.15–1.83) | A<0.01 | 0.74 | 0.76 (0.15–3.86) |
| SLC39A11 rs9908917 | T = 0.13 | 0.010 | 0.94 (0.9–0.99) | T = 0.2 | 0.049 | 1.25 (1.0–1.56) | T<0.01 | 0.27 | 1.2 (0.85–1.78) |
| SERPINA7 rs1804495 | A = 0.12 | 0.045 | 1.06 (1–1.12) | A = 0.23 | 0.050 | 0.8 (0.65–1.0) | A = 0.1 | 0.60 | 1.1 (0.75–1.65) |
| SLC25A45 rs681309 | G = 0.49 | 0.990 | 1 (0.96–1.04); | G = 0.26 | 0.033 | 0.78 (0.63–0.98) | G = 0.1 | 0.41 | 0.8 (0.57–1.26) |
| UGT1A rs11563251 | T = 0.12 | 0.074 | 0.95 (0.89–1.01) | T = 0.07 | 0.100 | 0.7 (0.5–1.06) | T = 0.42 | 0.281 | 1.2 (0.85–1.75) |
| MGST1 rs6488840 | C = 0.2 | 0.042 | 0.95 (0.91–1) | A = 0.42 | 0.121 | 0.54 (0.25–1.18) | C = 0.35 | 0.004 | 0.55 (0.37–0.82) |
1 MAF, minor allele and its frequency
2 p-value <0.05 are in bold
3 Odds ratio, 95% confidence interval
Discussion
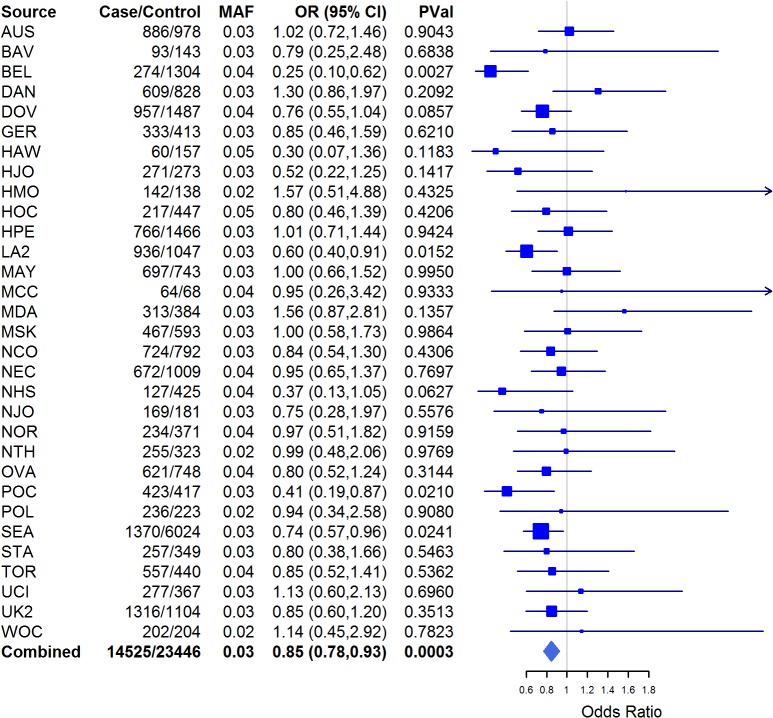
The development and progression of ovarian cancer is accompanied by aberrant cellular metabolism [33]. Central to cellular metabolic processes are the transport of trace elements and hormones through cellular and nuclear membranes. In this study, we aimed to elucidate whether germline SNPs in cellular transport genes were associated with EOC risk and histopathologic subtype. We detected nominal associations (P<0.05) with 81 SNPs and EOC risk in at least one of the histopathologic subtypes (S3 Table). Associations were noted with rs17216603 in HEPH and SER, INV and LMP subgroups as well as in SER and INV cases in women of white-European and Asian ancestries. The Hephaestin (HEPH) gene encodes a transmembrane copper-dependent ferroxidase (HEPH protein) responsible for dietary iron transport from intestinal enterocytes into the blood stream [34–36]. HEPH catalyzes ferrous (F2+) iron reoxidation to its ferric (F3+) state [37–38] that can be utilized by the body. The role of iron homeostasis in cancer progression is yet to be fully understood; however depletion of iron stores in cells induces cell cycle arrest and apoptosis, limits the rate of DNA synthesis, and down-regulates expression of various potentially carcinogenic kinases such as cyclins and cyclin-dependent kinases [39]. Additionally, iron is known to facilitate generation of mutagenic reactive oxygen species (ROS) that may drive cancer development and progression [40] as has been observed in colorectal cancer [41]. In silico analysis of HEPH rs17216603 combining results from Snpnexus, SNPinfo and Annovar [42–44] showed that this variant results in the substitution of Alanine at residue 598 with Threonine and may lead to reduction or loss of HEPH function. Functional analyses of this SNP and gene will be needed to clarify the impact of this finding. There was no evidence of MAF heterogeneity because the MAF range of rs17216603 across the studies is 2–5%. This SNP was significantly associated with Invasive EOC risk in women of white-European descent (P = 0.0003) for the combined results (Fig 1).
Fig 1. Forest plot for HEPH rs17216603 across studies.
Squares represent the estimated per-allele odds ratio (OR) for each study. Lines indicate the 95% confidence intervals. Diamond represents the OR estimate and confidence limits. Invasive EOC risk in women of white-European descent only; MAF in controls.
The SNP rs9908917 lies within an intron of the SLC39A11 gene, and was associated with SER, all INV and LMP EOC. In addition, of the 1785 SNPs in six genes (HEPH, MGST1, SERPINA, SLC25A45, SLC39A11 and UGT1A) imputed from the 1000 Genomes Project and examined for association with INV EOC in white-European subjects, the most significant imputed SNP was rs117729793 in SLC39A11 (per allele, OR = 2.55, 95% CI = 1.5–4.35, p = 5.66x10-4). In The Cancer Genome Atlas (TCGA) data [45], expression of SLC39A11 was significantly higher in ovarian tumors compared to normal tissues (P = 9.99x10-8). Taken together, these data highlight a potential role for this gene in EOC pathogenesis. The solute carrier family 39, member 11 (SLC39A11) is a poorly studied gene belonging to a family of metal ion transporters. SLC39A11 may act as a zinc-influx transporter, although the exact functions of the A11 gene have yet to be experimentally established [46]. Other members of the SLC39 family transport metal ions, such as iron, copper, cadmium and manganese [47].
SNP rs681309, in the intergenic region near SLC25A45, showed significant associations with all INV, MUC, and END and was the most significant SNP among the MUC subtype. The solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 45 (SLC25A45) belongs to the family of membrane proteins that catalyze the transport of solutes across the inner mitochondrial membrane [48]. While substrates for the SLC25 family carriers include ADP/ATP, amino acids, malate, ornithine, and citruline [49], the predominant substrate(s) for SLC23A45 have not yet been characterized [50], although sequence similarity to SLC25A29 suggests that this protein may be involved in the transport of long-chain fatty acids such as palmitoylcarnitine and acylcarnitine [51]. SLC25A45 is expressed in skeletal muscle, intestine, brain, and testis and is downregulated during ovarian cancer progression [50]. Taken together, these data suggest that additional studies are warranted on the role of SLC25A4 in particular, as well as mitochondria in general, in EOC etiology.
The SNP, rs1804495 in SERPINA7 was associated with all INV, SER, MUC, CC and LMP subtypes, and was the most statistically significant association among the CC subtype. The serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 (SERPINA7), also known as thyroxine-binding globulin (TBG), is a protein that binds thyroid hormones thyroxin (T4) and 3,5,3’-triiodothyronine (T3) in circulation [52]. Numerous mutations in SERPINA7 have been identified [53–54] leading to partially or completely absent TBG function. The hallmark of TBG deficiency is abnormally low T3 and T4 combined with normal thyroid stimulating hormone (TSH) values [55]. The specific role of SERPINA7 in cancer etiology has not been established; however, thyroid hormones may support cancer growth [56]. Thus, it is conceivable that altered TBG production over many years may modulate growth of early-stage ovarian cancer cells. The index SNP, rs1804495, is coding, but the resulting missense change is predicted to have neutral impact on the protein function, suggesting a linked variant may be the causal allele at this locus. We note that in our analyses, rs1804495 confers statistically significantly increased risk for LMP, INV, SER and CC, but decreased risk for mucinous EOC. This observation highlights observations from previously published studies that reveal differences between MUC and other EOCs [57–59].
The most significant association among the END subtype was with UGT1A1 rs11563251. UDP glucuronosyltransferase 1 family, polypeptide A cluster (UGT1A) represents a complex locus which encodes nine human UDP-glucuronosyltransferases. UDP-glucuronosyltransferase (UGT) enzymes are localized to endoplasmic reticulum (ER) and catalyze glucuronidation, which is involved in the elimination of bilirubin, steroids, bile acids, toxic dietary components, and several drugs, including morphine, and irinotecan [60–61]. Genetic variation in UGT1A1 is involved in inherited disorders of bilirubin metabolism such as Crigler-Najjar syndrome, which is manifested in complete absence (type 1) or diminished (types 2–3) bilirubin glucuronidation and resulting impaired bilirubin excretion. Previously, the UGT1A7*3 allele exhibited modestly significant association with colorectal cancer (OR = 2.39; P = 0.02) [57], lung cancer [62], endometrial cancer [63] and pancreatic cancer (OR = 1.98; P = 0.003) [64] with a particularly strong association in smokers with pancreatic carcinoma who were younger than 55 years (OR = 4.7; P = 0.0009), suggesting the magnitude of the observed associations may be modified by environmental interactions. Down-regulation of UGT1A appears to be an early event in carcinogenesis [65]; it is postulated that constitutive expression of UGT1A family genes in normal mucosa protects organs from carcinogens released in the bladder or absorbed from the diet in the colon. The rs11563251 variant lies within the 3’UTR of the UGT1A1,-A6 and–A10 genes, and so could feasibly impact the RNA stability of these transcripts. Alternatively, since this SNP also lies within intronic sequences of other UGT1A genes, this SNP could possibly be involved in cis-regulation of expression of one or more genes in this cluster.
In this study we conducted exploratory analyses in AS and AA subjects. However, the power to detect associations in women of non-European ancestries was limited due to small sample size and only the SER subtype of EOC was investigated for risk associations. The top SNPs in the AS ancestry group (rs17216603 in HEPH and rs1552846 in SLC39A11) were also significant in white-European women. In women of AA ancestry, the most significant SNP rs6488840 (P = 0.0035) was close to the microsomal glutathione S-transferase 1 (MGST1) gene, which encodes a protein that catalyzes the conjugation of glutathione to electrophiles and the reduction of lipid hydroperoxides. This protein is localized to the endoplasmic reticulum and outer mitochondrial membrane where it is thought to protect these membranes from oxidative stress. The product of this gene is involved in cellular defenses against toxic, carcinogenic, and pharmacologically active electrophilic compounds [66]. MGST1 overexpression has been demonstrated in various cancers (e.g., prostate cancer and lung cancer [67–68]) and has been associated with high metastatic potential and chemoresistance [69]. MGST1 is abundantly expressed in EOC primary tumors, metastases and effusions [66]. Other significant SNPs in women of AA ancestry include various members of the SLC39 family: SLC39A11 (rs9905659 and rs16977431) and SLC39A8 (rs233807).
The main strength of our study is a large sample size of white-European women that afforded sufficient statistical power to detect modest risk differences. Weaknesses, however, include the lack of functional or metabolic studies to establish biological significance of the observed associations. Another weakness is a small sample size in AA and AS women. The contribution of genetic and/or biological differences to EOC among different ethnic groups is unclear. However, because ovarian cancer health disparities are observed along the whole continuum of the disease globally and in the U.S. [70], this topic is without a doubt important and deserves its own dedicated studies.
In summary, we have found that genetic variation in transmembrane transport genes appear to be associated with EOC risk across various histologic subtypes (Table 1). These data suggest that disruptions in cellular transport (trace elements, hormones and small molecules) may play roles in EOC pathogenesis. Functional and metabolic studies are needed to support these findings.
Supporting Information
(DOCX)
Significant associations are bolded.
(DOCX)
SNPs are sorted by p-values.
(DOCX)
Acknowledgments
Individual acknowledgments by study: We thank all the individuals who took part in this study and all the researchers, clinicians and technical and administrative staff who have made possible the many studies contributing to this work. In particular, we thank: D. Bowtell, A. deFazio, D. Gertig, A. Green, P. Parsons, N. Hayward, P. Webb and D. Whiteman (AUS); G. Peuteman, T. Van Brussel and D. Smeets (BEL); the staff of the genotyping unit, S LaBoissiere and F Robidoux (Genome Quebec); U. Eilber and T. Koehler (GER); L. Gacucova (HMO); P. Schurmann, F. Kramer, W. Zheng, T. W. Park, Simon, K. Beer- Grondke and D. Schmidt (HJO); S. Windebank, C. Hilker and J. Vollenweider (MAY); the state cancer registries of AL, AZ, AR, CA, CO, CT, DE, FL, GA, HI, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, and WYL (NHS); L. Paddock, M. King, L. Rodriguez-Rodriguez, A. Samoila, and Y. Bensman (NJO); M. Sherman, A. Hutchinson,N. Szeszenia—‐ Dabrowska, B. Peplonska, W. Zatonski, A. Soni, P. Chao and M. Stagner (POL); C. Luccarini,P. Harrington the SEARCH team and ECRIC (SEA); R. Royer, S. Zhang (TOR); I. Jacobs, M. Widschwendter, E. Wozniak, N. Balogun, A. Ryan and J. Ford (UKO); Carole Pye (UKR); A. Amin Al Olama, K. Michilaidou, K. Kuchenbaker (COGS). Special thanks to the members of the AOCS management group: Penny Webb (QIMR Berghofer Medical Research Institute, Brisbane, Australia); David Bowtell (Peter MacCallum Cancer Centre, Melbourne, VIC); and Anna deFarzio (Westmead Hospital, Westmead, NSW).
Main funding: The scientific development and funding for this project were funded by the following: NIH R01 CA-1491429 (Phelan PI); the US National Cancer Institute (R01-CA076016); the COGS project is funded through a European Commission's Seventh Framework Programme grant (agreement number 223175 HEALTH F2 2009–223175); the Genetic Associations and Mechanisms in Oncology (GAME‐ON): a NCI Cancer Post-GWAS Initiative (U19-CA148112); the Ovarian Cancer Association Consortium is supported by a grant from the Ovarian Cancer Research Fund thanks to donations by the family and friends of Kathryn Sladek Smith (PPD/RPCI.07).
Investigator-specific funding: G.C.-T. is supported by the National Health and Medical Research Council; B.K. holds an ACS Early Detection Professorship (SIOP—‐06—‐258—‐01—‐COUN); L.E.K. is supported by a Canadian Institute of Health Research New Investigator Award (MSH—-87734). AWL is supported by NIEHS T32 training grant (T32ES013678).
Funding of included studies: Funding of the constituent studies was provided by the California Cancer Research Program (00—‐01389V—‐20170, N01—‐CN25403, 2II0200); the Canadian Institutes of Health Research (MOP—-86727); Cancer Australia; Cancer Council Victoria; Cancer Council Queensland; Cancer Council New South Wales; Cancer Council South Australia; Cancer Council Tasmania; Cancer Foundation of Western Australia; the Cancer Institute of New Jersey; Cancer Research UK (C490/A6187, C490/A10119, C490/A10124); the Danish Cancer Society (94—‐222—‐52); the ELAN Program of the University of Erlangen—‐Nuremberg; the Eve Appeal; the Helsinki University Central Hospital Research Fund; Helse Vest; the Norwegian Cancer Society; the Norwegian Research Council; the Ovarian Cancer Research Fund; Nationaal Kankerplan of Belgium; Grant—‐in—‐Aid for the Third Term Comprehensive 10—‐Year Strategy For Cancer Control from the Ministry of Health Labour and Welfare of Japan; the L & S Milken Foundation; the Polish Ministry of Science and Higher Education (4 PO5C 028 14, 2 PO5A 068 27); the Roswell Park Cancer Institute Alliance Foundation; the US National Cancer Institute (K07—‐CA095666, K07—‐CA143047,K22—‐CA138563, N01—‐CN55424, N01—-PC67001, N01—‐PC067010, N01—‐PC035137, P01—‐CA017054, P01—‐CA087696, P30—-CA072720, P50—‐CA105009, P50-CA136393, R01—‐CA014089, R01—‐CA016056, R01—‐CA017054, R01—‐CA049449, R01—‐CA050385, R01—‐CA054419, R01—‐ CA058598, R01—‐CA058860, R01—‐CA061107, R01—‐CA061132, R01—‐CA067262, R01—‐CA071766, R01—‐CA074850, R01—‐CA080742, R01—‐CA080978, R01—‐CA083918, R01—‐CA087538, R01—‐CA092044, R01—‐095023, R01—‐CA122443, R01—‐CA112523, R01—‐CA114343, R01—‐CA126841, R01—‐CA136924, R03—‐CA113148, R03—‐CA115195, U01—‐CA069417, U01—‐CA071966 and Intramural research funds); the US Army Medical Research and Material Command (DAMD17—‐01—‐1—‐0729, DAMD17—‐02—‐1—‐0666, DAMD17—‐02—‐1—‐0669, W81XWH—-07—-0449, W81XWH—‐10—‐1—‐02802); the US Public Health Service (PSA—-042205); The National Health and Medical Research Council of Australia (199600 and 400281); the German Federal Ministry of Education and Research of Germany Programme of Clinical Biomedical Research (01GB 9401); the State of Baden—‐Wurttemberg through Medical Faculty of the University of Ulm (P.685); the Minnesota Ovarian Cancer Alliance; the Mayo Foundation; the Fred C. and Katherine B. Andersen Foundation; the Lon V. Smith Foundation (LVS—‐39420); the Oak Foundation; the OHSU Foundation; the Mermaid I project; the Rudolf—‐Bartling Foundation; the UK National Institute for Health Research Biomedical Research Centres at the University of Cambridge, Imperial College London, University College Hospital “Womens Health Theme” and the Royal Marsden Hospital; WorkSafeBC 14.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The scientific development and funding for this project were funded by the following: NIH R01 CA-1491429 (Phelan PI); the US National Cancer Institute (R01-CA076016); the COGS project is funded through a European Commission's Seventh Framework Programme grant (agreement number 223175 HEALTH F2 2009-223175); the Genetic Associations and Mechanisms in Oncology (GAME‐ON): a NCI Cancer Post-GWAS Initiative (U19-CA148112); the Ovarian Cancer Association Consortium is supported by a grant from the Ovarian Cancer Research Fund thanks to donations by the family and friends of Kathryn Sladek Smith (PPD/RPCI.07). Investigator-specific funding: GCT is supported by the National Health and Medical Research Council; BK holds an ACS Early Detection Professorship (SIOP--‐06--‐258--‐01--‐COUN); LEK is supported by a Canadian Institute of Health Research New Investigator Award (MSH---87734). AWL is supported by NIEHS T32 training grant (T32ES013678). Funding of included studies: Funding of the constituent studies was provided by the California Cancer Research Program (00--‐01389V--‐20170, N01--‐CN25403, 2II0200); the Canadian Institutes of Health Research (MOP---86727); Cancer Australia; Cancer Council Victoria; Cancer Council Queensland; Cancer Council New South Wales; Cancer Council South Australia; Cancer Council Tasmania; Cancer Foundation of Western Australia; the Cancer Institute of New Jersey; Cancer Research UK (C490/A6187, C490/A10119, C490/A10124); the Danish Cancer Society (94--‐222--‐52); the ELAN Program of the University of Erlangen--‐Nuremberg; the Eve Appeal; the Helsinki University Central Hospital Research Fund; Helse Vest; the Norwegian Cancer Society; the Norwegian Research Council; the Ovarian Cancer Research Fund; Nationaal Kankerplan of Belgium; Grant--‐in--‐Aid for the Third Term Comprehensive 10--‐Year Strategy For Cancer Control from the Ministry of Health Labour and Welfare of Japan; the L & S Milken Foundation; the Polish Ministry of Science and Higher Education (4 PO5C 028 14, 2 PO5A 068 27); the Roswell Park Cancer Institute Alliance Foundation; the US National Cancer Institute (K07--‐CA095666, K07--‐CA143047,K22--‐CA138563, N01--‐CN55424, N01---PC67001, N01--‐PC067010, N01--‐PC035137, P01--‐CA017054, P01--‐CA087696, P30---CA072720, P50--‐CA105009, P50-CA136393, R01--‐CA014089, R01--‐CA016056, R01--‐CA017054, R01--‐CA049449, R01--‐CA050385, R01--‐CA054419, R01--‐ CA058598, R01--‐CA058860, R01--‐CA061107, R01--‐CA061132, R01--‐CA067262, R01--‐CA071766, R01--‐CA074850, R01--‐CA080742, R01--‐CA080978, R01--‐CA083918, R01--‐CA087538, R01--‐CA092044, R01--‐095023, R01--‐CA122443, R01--‐CA112523, R01--‐CA114343, R01--‐CA126841, R01--‐CA136924, R03--‐CA113148, R03--‐CA115195, U01--‐CA069417, U01--‐CA071966 and Intramural research funds); the US Army Medical Research and Material Command (DAMD17--‐01--‐1--‐0729, DAMD17--‐02--‐1--‐0666, DAMD17--‐02--‐1--‐0669, W81XWH---07---0449, W81XWH--‐10--‐1--‐02802); the US Public Health Service (PSA---042205); The National Health and Medical Research Council of Australia (199600 and 400281); the German Federal Ministry of Education and Research of Germany Programme of Clinical Biomedical Research (01GB 9401); the State of Baden--‐Wurttemberg through Medical Faculty of the University of Ulm (P.685); the Minnesota Ovarian Cancer Alliance; the Mayo Foundation; the Fred C. and Katherine B. Andersen Foundation; the Lon V. Smith Foundation (LVS--‐39420); the Oak Foundation; the OHSU Foundation; the Mermaid I project; the Rudolf--‐Bartling Foundation; the UK National Institute for Health Research Biomedical Research Centres at the University of Cambridge, Imperial College London, University College Hospital “Womens Health Theme” and the Royal Marsden Hospital; WorkSafeBC 14.
References
- 1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011). Global cancer statistics. CA Cancer J Clin 61: 69–90. 10.3322/caac.20107 [DOI] [PubMed] [Google Scholar]
- 2. Buys SS, Partridge E, Black A, Johnson CC, Lamerato L, Isaacs C, et al. (2011) Effect of screening on ovarian cancer mortality: the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Randomized Controlled Trial. JAMA 305: 2295–2303. 10.1001/jama.2011.766 [DOI] [PubMed] [Google Scholar]
- 3.Ries LAG, Young Jr JL, Keel GE, Eisner MP, Lin YD, Horner MJD. (2007) Cancer survival among adults: US SEER program, 1988–2001. Patient and tumor characteristics SEER Survival Monograph Publication: 07–6215.
- 4. Macheda ML, Rogers S, Best JD (2005) Molecular and cellular regulation of glucose transporter (GLUT) proteins in cancer. J Cell Physiol 202: 654–662. [DOI] [PubMed] [Google Scholar]
- 5. Guilbert A, Gautier M, Dhennin-Duthille I, Haren N, Sevestre H, Ouadid-Ahidouch H. (2009) Evidence that TRPM7 is required for breast cancer cell proliferation. Am J Physiol Cell Physiol 297: C493–502. 10.1152/ajpcell.00624.2008 [DOI] [PubMed] [Google Scholar]
- 6. Pinheiro C, Sousa B, Albergaria A, Paredes J, Dufloth R, Vieira D, et al. (2011) GLUT1 and CAIX expression profiles in breast cancer correlate with adverse prognostic factors and MCT1 overexpression. Histol Histopathol 26: 1279–1286. [DOI] [PubMed] [Google Scholar]
- 7. Ota I, Sakurai A, Toyoda Y, Morita S, Sasaki T, Chishima T, et al. (2010) Association between Breast Cancer Risk and the Wild-type Allele of Human ABC Transporter ABCC11. Anticancer Research 30(12): 5189–5194 [PubMed] [Google Scholar]
- 8. Pertega-Gomes N, Vizcaino JR, Miranda-Goncalves V, Pinheiro C, Silva J, Pereira H, et al. (2011) Monocarboxylate transporter 4 (MCT4) and CD147 overexpression is associated with poor prognosis in prostate cancer. BMC Cancer 11: 312 10.1186/1471-2407-11-312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen J, Chloupkova M (2009) Abnormal iron uptake and liver cancer. Cancer Biol Ther 8: 1699–1708. [DOI] [PubMed] [Google Scholar]
- 10. Kurzawski M, Droździk M, Suchy J, Kurzawski G, Białecka M, Górnik W, et al. (2005) Polymorphism in the P-glycoprotein drug transporter MDR1 gene in colon cancer patients. European Journal of Clinical Pharmacology 61(5–6): 389–394 [DOI] [PubMed] [Google Scholar]
- 11. Zhang H, Liao L- H, Liu S- M, Lau KW, Lai AK, Zhang JH, et al. (2007) Microsomal glutathione S-transferase gene polymorphisms and colorectal cancer risk in a Han Chinese population. International Journal of Colorectal Disease 22(10): 1185–1194 [DOI] [PubMed] [Google Scholar]
- 12. Cross AJ, Sinha R, Wood RJ, Xue X, Huang WY, et al. (2011) Iron homeostasis and distal colorectal adenoma risk in the prostate, lung, colorectal, and ovarian cancer screening trial. Cancer Prev Res (Phila) 4: 1465–1475. 10.1158/1940-6207.CAPR-11-0103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Filetti S, Bidart JM, Arturi F, Caillou B, Russo D, Schlumberger M. (1999) Sodium/iodide symporter: a key transport system in thyroid cancer cell metabolism. Eur J Endocrinol 141: 443–457. [DOI] [PubMed] [Google Scholar]
- 14. Ishihara T, Inoue J, Kozaki K, Imoto I, Inazawa J (2011) HECT-type ubiquitin ligase ITCH targets lysosomal-associated protein multispanning transmembrane 5 (LAPTM5) and prevents LAPTM5-mediated cell death. J Biol Chem 286: 44086–44094. 10.1074/jbc.M111.251694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nemeth E (2008) Iron regulation and erythropoiesis. Curr Opin Hematol 15: 169–175. 10.1097/MOH.0b013e3282f73335 [DOI] [PubMed] [Google Scholar]
- 16. Lenaz G, Baracca A, Barbero G, Bergamini C, Dalmonte ME, Del Sole M, et al. (2010) Mitochondrial respiratory chain super-complex I-III in physiology and pathology. Biochim Biophys Acta 1797: 633–640. 10.1016/j.bbabio.2010.01.025 [DOI] [PubMed] [Google Scholar]
- 17. Foy SP, Labhasetwar V (2011) Oh the irony: Iron as a cancer cause or cure? Biomaterials 32: 9155–9158. 10.1016/j.biomaterials.2011.09.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iron Atlas. Available: http://wwwironatlascom.
- 19. Huang X (2003) Iron overload and its association with cancer risk in humans: evidence for iron as a carcinogenic metal. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis 533(1-2): 153–171 [DOI] [PubMed] [Google Scholar]
- 20. Lin SF, Wei H, Maeder D, Franklin RB, Feng P (2009) Profiling of zinc-altered gene expression in human prostate normal vs. cancer cells: a time course study. J Nutr Biochem 20: 1000–1012. 10.1016/j.jnutbio.2008.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kurian AW, Balise RR, McGuire V, Whittemore AS (2005) Histologic types of epithelial ovarian cancer: have they different risk factors? Gyn Oncol 96: 520–530. [DOI] [PubMed] [Google Scholar]
- 22. Pharoah PD, Tsai YY, Ramus SJ, Phelan CM, Goode EL, Lawrenson K, et al. (2013) GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat Genet 45: 362–370, 370e361-362. 10.1038/ng.2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.“Gene” database by NCBI: http://www.ncbi.nlm.nih.gov/gene
- 24.“BioCarta” database: http://www.biocarta.com/
- 25.“GenomeNet” database: http://www.genome.jp/
- 26. Howie BN, Donnelly P, Marchini J (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genetics 5(6): e1000529 10.1371/journal.pgen.1000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR (2012) Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nature Genetics 44(8): 955–959. 10.1038/ng.2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. September 2007;81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Petitti D. Meta-analysis decision analysis and cost-effectiveness analysis New York: Oxford; 1994. [Google Scholar]
- 30. Sankararaman S, Sridhar S, Kimmel G, Halperin E. Estimating local ancestry in admixed populations. Am. J. Hum. Genet. February 2008;82(2):290–303. 10.1016/j.ajhg.2007.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. August 2006;38(8):904–909. [DOI] [PubMed] [Google Scholar]
- 32. Storey JD. A direct approach to false discovery rates. Journal of the Royal Statistical Society: Series B 2002;64:479–98. [Google Scholar]
- 33. Anderson AS, Roberts PC, Frisard MI, McMillan RP, Brown TJ, Lawless MH (2013) Metabolic changes during ovarian cancer progression as targets for sphingosine treatment. Exp Cell Res. 10;319(10):1431–42. 10.1016/j.yexcr.2013.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fleming RE, Sly WS (2003) The iron gatekeeper: keys to the front and back doors. Blood 102: 1567–1567. [Google Scholar]
- 35. Vulpe CD, Kuo YM, Murphy TL, Cowley L, Askwith C, Libina N, et al. (1999) Hephaestin, a ceruloplasmin homologue implicated in intestinal iron transport, is defective in the sla mouse. Nat Genet 21: 195–199. [DOI] [PubMed] [Google Scholar]
- 36. Li L, Vulpe CD, Kaplan J (2003) Functional studies of hephaestin in yeast: evidence for multicopper oxidase activity in the endocytic pathway. Biochem J 375: 793–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sebastiani G, Pantopoulos K (2010) Iron Metabolism and Disease In: Zalups RK, Koropatnick DJ, editors. Cellular and molecular biology of metals: CRC Press. [Google Scholar]
- 38. Yeh KY, Yeh M, Glass J (2011) Interactions between ferroportin and hephaestin in rat enterocytes are reduced after iron ingestion. Gastroenterology 141: 292–299. e291. 10.1053/j.gastro.2011.03.059 [DOI] [PubMed] [Google Scholar]
- 39. Merlot AM, Kalinowski DS, Richardson DR (2013) Novel chelators for cancer treatment: where are we now? Antioxid Redox Signal 18: 973–1006. 10.1089/ars.2012.4540 [DOI] [PubMed] [Google Scholar]
- 40. Galaris D, Pantopoulos K (2008) Oxidative stress and iron homeostasis: mechanistic and health aspects. Crit Rev Clin Lab Sci 45: 1–23. 10.1080/10408360701713104 [DOI] [PubMed] [Google Scholar]
- 41.E Abu Z Dayem Ullah, Nicholas R Lemoine and Claude Chelala, A practical guide for the functional annotation of genetic variations using SNPnexus <http://bib.oxfordjournals.org/content/early/2013/02/08/bib.bbt004.abstract>, Briefings in Bioinformatics, 2013, 10.1093/bib/bbt004 [DOI] [PubMed]
- 42.Zongli Xu, Jack A. Taylor (2009). SNPinfo: Integrating GWAS and Candidate Gene Information into Functional SNP Selection for Genetic Association Studies. Nucleic Acids Research [DOI] [PMC free article] [PubMed]
- 43.Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from next-generation sequencing data <http://nar.oxfordjournals.org/content/38/16/e164> Nucleic Acids Research, 38:e164, 2010 [DOI] [PMC free article] [PubMed]
- 44. Brookes MJ, Hughes S, Turner FE, Reynolds G, Sharma N, Ismail T, et al. (2006) Modulation of iron transport proteins in human colorectal carcinogenesis. Gut 55: 1449–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.The Cancer Genome Atlas: http://cancergenome.nih.gov/
- 46. Eide DJ (2004) The SLC39 family of metal ion transporters. Pflugers Arch 447: 796–800. [DOI] [PubMed] [Google Scholar]
- 47. Taylor KM, Morgan HE, Smart K, Zahari NM, Pumford S, Ellis IO, et al. (2007) The emerging role of the LIV-1 subfamily of zinc transporters in breast cancer. Mol Med 13: 396–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Palmieri F (2004) The mitochondrial transporter family (SLC25): physiological and pathological implications. Pflugers Arch 447: 689–709. [DOI] [PubMed] [Google Scholar]
- 49. Haitina T, Lindblom J, Renstrom T, Fredriksson R (2006) Fourteen novel human members of mitochondrial solute carrier family 25 (SLC25) widely expressed in the central nervous system. Genomics 88: 779–790. [DOI] [PubMed] [Google Scholar]
- 50. Palmieri F (2013) The mitochondrial transporter family SLC25: identification, properties and physiopathology. Mol Aspects Med 34: 465–484. 10.1016/j.mam.2012.05.005 [DOI] [PubMed] [Google Scholar]
- 51. Iacobazzi V, Invernizzi F, Baratta S, Pons R, Chung W, Garavaqlia B, et al. (2004) Molecular and functional analysis of SLC25A20 mutations causing carnitine-acylcarnitine translocase deficiency. Hum Mutat 24: 312–320. [DOI] [PubMed] [Google Scholar]
- 52. Law RH, Zhang Q, McGowan S, Buckle AM, Silverman GA, Wong W, et al. (2006) An overview of the serpin superfamily. Genome Biol 7: 216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Domingues R, Font P, Sobrinho L, Bugalho MJ (2009) A novel variant in Serpina7 gene in a family with thyroxine-binding globulin deficiency. Endocrine 36: 83–86. 10.1007/s12020-009-9202-2 [DOI] [PubMed] [Google Scholar]
- 54.41. Lacka K, Nizankowska T, Ogrodowicz A, Lacki JK (2007) A Novel Mutation (del 1711 G) in the TBG Gene as a Cause of Complete TBG Deficiency. Thyroid 17(11): 1143–1146. [DOI] [PubMed] [Google Scholar]
- 55. Kobayashi H, Sakurai A, Katai M, Hashizume K (1999) Autosomally Transmitted Low Concentration of Thyroxine-Binding Globulin Thyroid 9(2): 159–163. [DOI] [PubMed] [Google Scholar]
- 56. Cristofanilli M, Yamamura Y, Kau SW, Bevers T, Strom S, Patangan M, et al. (2005) Thyroid hormone and breast carcinoma. Primary hypothyroidism is associated with a reduced incidence of primary breast carcinoma. Cancer 103: 1122–1128. [DOI] [PubMed] [Google Scholar]
- 57. Frumovitz M, Schmeler KM, Malpica A, Sood AK, Gershenson DM (2010) Unmasking the complexities of mucinous ovarian carcinoma. Gynecol Oncol 117: 491–496. 10.1016/j.ygyno.2010.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shimada M, Kigawa J, Ohishi Y, Yasuda M, Suzuki M, Hiura M, et al. (2009) Clinicopathological characteristics of mucinous adenocarcinoma of the ovary. Gynecol Oncol 113: 331–334. 10.1016/j.ygyno.2009.02.010 [DOI] [PubMed] [Google Scholar]
- 59. Risch HA, Marrett LD, Jain M, Howe GR. Differences in Risk Factors for Epithelial Ovarian Cancer by Histologic Type. Am J Epidemiol 1996; 144(4):363–72. [DOI] [PubMed] [Google Scholar]
- 60. Levesque E, Girard H, Journault K, Lepine J, Guillemette C (2007) Regulation of the UGT1A1 bilirubin-conjugating pathway: role of a new splicing event at the UGT1A locus. Hepatology 45: 128–138. [DOI] [PubMed] [Google Scholar]
- 61. Tukey RH, Strassburg CP (2000) Human UDP-glucuronosyltransferases: metabolism, expression, and disease. Annu Rev Pharmacol Toxicol 40: 581–616. [DOI] [PubMed] [Google Scholar]
- 62. Araki J, Kobayashi Y, Iwasa M, Urawa N, Gabazza EC, Taquchi O, et al. (2005) Polymorphism of UDP-glucuronosyltransferase 1A7 gene: a possible new risk factor for lung cancer. Eur J Cancer 41: 2360–2365. [DOI] [PubMed] [Google Scholar]
- 63. Duguay Y, McGrath M, Lepine J, Gagne JF, Hankinson SE, Colditz GA, et al. (2004) The functional UGT1A1 promoter polymorphism decreases endometrial cancer risk. Cancer Res 64: 1202–1207. [DOI] [PubMed] [Google Scholar]
- 64. Ockenga J, Vogel A, Teich N, Keim V, Manns MP, Strassburg CP. (2003) UDP glucuronosyltransferase (UGT1A7) gene polymorphisms increase the risk of chronic pancreatitis and pancreatic cancer. Gastroenterology 124: 1802–1808. [DOI] [PubMed] [Google Scholar]
- 65. Giuliani L, Ciotti M, Stoppacciaro A, Pasquini A, Silvestri I, De Matteis A, et al. (2005) UDP-glucuronosyltransferases 1A expression in human urinary bladder and colon cancer by immunohistochemistry. Oncol Rep 13: 185–191. [PubMed] [Google Scholar]
- 66. Morgenstern R, Zhang J, Johansson K (2011) Microsomal glutathione transferase 1: mechanism and functional roles. Drug Metab Rev 43: 300–306. 10.3109/03602532.2011.558511 [DOI] [PubMed] [Google Scholar]
- 67. Nicolle M. Linnerth, Kelly Sirbovan, Roger A. Moorehead. Use of a transgenic mouse model to identify markers of human lung tumors. International Journal of Cancer Volume 114, Issue 6, pages 977–982, 10 May 2005. [DOI] [PubMed] [Google Scholar]
- 68. H Chaib EK Cockrell, MA Rubin, JA Macoska. Profiling and verification of gene expression patterns in normal and malignant human prostate tissues by cDNA microarray analysis. Neoplasia 3(1), 2001, p 42–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Thea Eline Hetland Dag Andre Nymoen, Emilsen Elisabeth, Kaern J, et al. (2012) MGST1 expression in serous ovarian carcinoma differs at various anatomic sites, but is unrelated to chemoresistance or survival. Gyn oncol 126 (3):460–5 10.1016/j.ygyno.2012.05.029 [DOI] [PubMed] [Google Scholar]
- 70. Chornokur G, Amankwah EK, Schildkraut JM, Phelan CM (2013) Global ovarian cancer health disparities. Gynecol Oncol 129: 258–264. 10.1016/j.ygyno.2012.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Significant associations are bolded.
(DOCX)
SNPs are sorted by p-values.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.