Abstract
Bacteria that adhere to the surfaces of implanted medical devices can cause catastrophic infection. Since chemical modifications of materials' surfaces have poor long-term performance in preventing bacterial buildup, approaches using bactericidal physical surface topography have been investigated. The authors used Nanoimprint Lithography was used to fabricate a library of biomimetic nanopillars on the surfaces of poly(methyl methacrylate) (PMMA) films. After incubation of Escherichia coli (E. coli) on the structured PMMA surfaces, pillared surfaces were found to have lower densities of adherent cells compared to flat films (67%–91% of densities on flat films). Moreover, of the E. coli that did adhere a greater fraction of them were dead on pillared surfaces (16%–141% higher dead fraction than on flat films). Smaller more closely spaced nanopillars had better performance. The smallest most closely spaced nanopillars were found to reduce the bacterial load in contaminated aqueous suspensions by 50% over a 24-h period compared to flat controls. Through quantitative analysis of cell orientation data, it was determined that the minimum threshold for optimal nanopillar spacing is between 130 and 380 nm. Measurements of bacterial cell length indicate that nanopillars adversely affect E. coli morphology, eliciting a filamentous response. Taken together, this work shows that imprinted polymer nanostructures with precisely defined geometries can kill bacteria without any chemical modifications. These results effectively translate bactericidal nanopillar topographies to PMMA, an important polymer used for medical devices.
I. INTRODUCTION
Biofilm formation on an implanted medical device can cause persistent infection, eliciting immune response and triggering the release of harmful toxins in the body.1 A biofilm is composed of bacteria, proteins, and cells that adhere and aggregate on the material surface. Biofilm development begins when a single planktonic cell attaches to an available material surface in response to environmental cues, including nutrient availability and physicochemical forces.1,2 Once adhered to the material surface, the bacteria begin to proliferate, secreting extracellular polysaccharide substance and forming multilayer cell clusters on the material surface to create the biofilms.1,3
Most antibiofilm surface coatings use antimicrobial agents to prevent cell proliferation4–6 or employ chemical surface modifications, such as crosslinking with poly(ethylene glycol),7,8 which purportedly prevent bacterial adhesion. However, neither are long-term solutions. Antimicrobial agents can breed resistant bacteria. Additionally, bacterial cells in biofilm are 10- to 1000-fold less susceptible to antimicrobial agents than the planktonic counterparts.9 Meanwhile, surface chemical modifications are readily masked by host and bacteria-produced protein layers.10–12 Finally, introduction of chemical species to the surface increases the burden of biocompatibility testing.
Of great interest, then, are surfaces with micro- and nanoscale surface features that render them intrinsically antibacterial. Size, shape, and pattern of surface structures dictate bacterial response.13 Fabricated high aspect ratio (HAR) nanopost structures (with structure spacings ranging from 0.8 to 2.2 μm) were found to control the spatial patterning of bacteria on substrates.14 HARs could be modified to be antibacterial.15 An engineered microscale surface design inspired by the topography of shark skin (with features 2 μm wide, 3 μm in height) was found to disrupt biofilm formation on patterned poly(dimethylsiloxane) elastomer without the use of bactericidal agents, likely by inhibiting quorum sensing.16–18 Some studies posit superhydrophobicity as a precursor to biofilm reduction, as reduced contact area between bacteria and micro or nanostructured superhydrophobic surfaces ostensibly prevents adhesion. For example, researchers showed decreased bacterial adhesion on superhydrophobic hard plastics with roughened surfaces that have multiscale features transferred from heat-shrunken polyolefin film.19 Along those lines, researchers found that nanorough titanium surfaces produced by electron beam evaporation decreased bacterial adhesion in comparison to unmodified, nanotubular, and nanotextured titanium surfaces.20 In addition, nanophase ceramic surfaces (zinc oxide and titania) decreased bacterial adhesion more than microphase surfaces.21 However, some bacteria do adhere to all the aforementioned surfaces, and any adherent bacteria can proliferate to form biofilm. Therefore, we start from the presumption that the bacteria will adhere, and try to mitigate their detrimental effects.
Recently, a mechanical mechanism for killing adherent bacteria was discovered. It was found that the surface of the cicada wing is covered with 200 nm high pillars spaced approximately 170 nm apart which penetrate and consequently kill Pseudomonas aeruginosa within several minutes of adhesion.22,23 In follow-up studies, the nanopillars on the dragonfly wing were found to kill Gram-positive bacteria as well as yeast.24,25 Similar nanopillars found on specially treated silicon wafers (black silicon)26 had similar effects. According to these researchers, bacterial cells are killed on contact as they stretch over the pillars. Therefore, recruitment of additional cells, biofilm buildup, and eventual fouling are all prevented. Additionally, bacterial proliferation is stemmed so resistance to the nanofeatures cannot evolve. Unfortunately, these discoveries have not been translated to technologically scalable processes.
Here, we propose that biomimetic polymer nanopillars with defined surface patterns can be broadly bactericidal. Notably, we use a scalable process to imprint biomimetic nanostructures onto the surfaces of poly(methyl methacrylate) (PMMA) films. The nanostructured surfaces of cicada wings are replicated via soft lithography, and subsequently, nanoimprint lithography33 is used to imprint nanopillars onto PMMA films from commercially purchased silicon and nickel molds. We show for the first time that nanopillared polymer surfaces are bactericidal, while exploring the roles of geometric parameters of nanopillars on antibacterial properties. These results effectively translate bactericidal nanopillar topographies to PMMA, an important polymer used for medical devices.
II. EXPERIMENT
A. Fabrication of nanostructures on polymer surface via nanoimprint lithography
Nanostructures were fabricated from PMMA, a polymer approved by the United States Food and Drug Administration for use in biomedical implants. First, we fabricated PMMA thin films. Glass cover slips (22 × 22 mm) were pretreated with aminopropyltriethoxysilane to facilitate polymer-glass adhesion. Next, PMMA (M.W. = 120 kDa, Sigma Aldrich, Milwaukee, WI) was dissolved in toluene (5 wt. %) and spin-coated on glass cover slips at 600 rpm for 45 s. Films were annealed on a hot plate at 100 °C prior to imprinting.
Besides the replicated cicada wing, two other types of pillar arrays were generated. The imprinted samples are referenced by their periodicity, as “P600” and “P300.” P600 surfaces were generated from silicon nanohole molds (Lightsmyth, 8 × 8.3 mm). P300 surfaces were generated from silicone negative molds of a commercially available nickel stamp (HT-AR-02, Holotools GmbH, Freiburg, Germany, 20 × 20 mm). Cicada wing replicates in PMMA, referenced as “P200,” were generated from silicone negative molds of cicada wings. Silicone molds were fabricated from hard polydimethylsiloxane using standard literature procedures.27–29 Before imprinting, silicon molds were cleaned with piranha solution (3:1 sulfuric acid: hydrogen perioxide) and an antistiction coating [perfluorodecyltrichlorosilane (FDTS) from Gelest, Inc.] was applied to the molds by molecular vapor deposition (MVD 100, Applied MST, San Jose, CA). The silicone molds needed no further processing before imprinting. To imprint the silicon molds, the nanoimprinter (Jenoptik, Hex03) was used. The PMMA film was heated to 170 °C, 60 °C above the glass transition temperature of PMMA. The FDTS-treated silicon mold was pressed down against the PMMA film under 4.5 MPa for 5–10 min. The mold was then allowed to cool to 50 °C in 3 min. The glass-supported PMMA film was released from the silicon mold, leaving the imprinted nanostructures on the film surface. A similar protocol was followed to create PMMA structures from the silicone molds using the Tetrahedron Press (Tetrahedron, San Diego CA).
Very large area P200 pillars (cicada wing replicate films) (circular, 15 mm diameter) were impossible to nanoimprint due to the nonuniform surface texture (e.g., veins). For experiments requiring large area surfaces, we dropcasted 5 wt. % PMMA solution in toluene onto silicone negative cicada wing molds and cured the P200 films at 80 °C for 20 min.
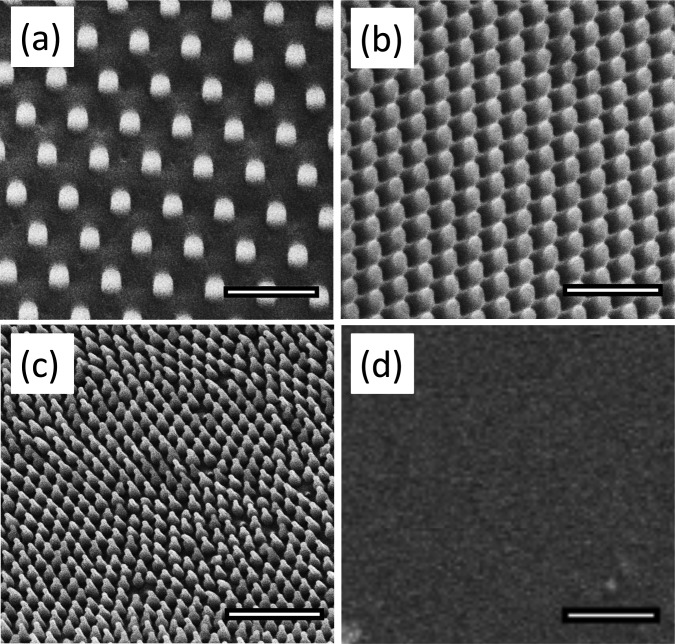
All nanoimprinted surfaces were examined using scanning electron microscopy (SEM) and atomic force microscopy (AFM). SEM images were taken at 5 kV (FEI Quanta 3D, Irvine Materials Research Institute, University of California, Irvine) at varying magnifications. Features on the P200 samples were found to have the same dimensions as those of the native wings. Figure 1 shows representative images of the nanostructures; Table I displays the relevant feature sizes. As evident from the micrographs, the P200 and cicada wing feature size and spacing have some natural variability, so the dimensions below represent the averages.
Fig. 1.
Micrographs of PMMA surfaces taken at 5 kV with the FEI Quanta 3D SEM. We used commercial molds to generate nanopillars in two different sizes, referenced here by periodicity: P600 shown in (a) and P300 shown in (b). We also used a two-step lithography process to replicate the nanostructures on the surface of the cicada wing in PMMA, referenced as P200, shown in (c). Images (a)–(c) were taken at a 30° tilt while the flat control (d) was taken at 0° tilt. All scale bars = 1 μm.
Table I.
Measured dimensions of nanoimprinted features.
| Sample | Feature width (nm) | Array periodicity (nm) | Feature spacing (nm) | Feature height (nm) |
|---|---|---|---|---|
| P600 | 215 | 595 | 380 | 300 |
| P300 | 190 | 320 | 130 | 300 |
| P200 | Cap: 70 | 170 | 100 | 210 |
| Cicada wing | Cap: 60 | 170 | 100 | 210 |
B. Bacteria culture
Escherichia coli (E. coli), a Gram-negative, rod-shaped bacterium (DH5-alpha strain, Life Technologies, Carlsbad, CA) was used to examine antibacterial properties of the PMMA films, both with and without the nanostructures. A glycerol stock solution of E. coli was inoculated in 5 ml Luria Broth (LB) media (Thermo Fisher Scientific) overnight in an air bath shaker at 300 rpm in 37 °C. The bacteria solution was diluted 1000 times in LB. Ten microliters of this diluted solution was added on an LB-agar plate (Thermo Fisher Scientific) and was spread on the agar using a sterile metal loop and a turntable per the spread plate method. The plate was incubated at 37 °C overnight. At the start of each experiment, fresh starter cultures were grown overnight at 37 °C in 5 ml LB media by shaking at 250 rpm.
C. Viability on surfaces
First, viability was examined. Two trials were run. Three biological replicates comparing flat samples against P600 pillar samples were run. Next, two biological replicates comparing flat samples against P300 and P200 pillar samples were run. Bacterial suspensions were adjusted to OD600 = 0.3.22 Each PMMA sample was immersed in an excess of bacteria suspension and incubated for 20 h at 37 °C. Prior to conducting experiments, samples were washed twice with 1X phosphate buffered saline (PBS) at room temperature (25 °C) by streaming the PBS along the side of the sample and gently swirling the well plate ten times. Viability on the unfixed samples was assessed using the BacLight™ Live/Dead staining kit (Molecular Probes®, Life Technologies, Carlsbad, CA). To make the staining solution, components were diluted in PBS at the concentration recommended by the manufacturer. The samples were incubated for 15 min in the staining solution. Samples were rinsed twice with PBS and examined under wide-field fluorescence using a standard green filter set to image live cells and a standard red filter set to image dead cells. Both live and dead populations of cells were counted with imagej software using the Cell Counter plugin from several random regions, each containing no less than 300 cells, on each sample. Cells that exhibited red fluorescence or yellow fluorescence (combined green and red) were included in the dead cell counts. The total numbers of dead and live cells from the regions of each sample were added and divided by the total area imaged to determine each sample's average dead and live densities. Then, the percentage of cells on each sample that were dead was calculated as the ratio of dead divided by total cells.
D. Aqueous suspension colony forming unit counts
Next, we assessed the ability of nanopillared surfaces to decrease bacterial load of aqueous suspensions. P200 were chosen for this study because we determined in the viability study that P200 pillars were most effective. Three biological replicates consisting of three samples each were performed. Bacterial suspensions were adjusted to OD600 = 0.1 and diluted tenfold.26 Large-area (circular, 15 mm diameter) P200 surfaces were seeded with 1.5 ml of the diluted suspension of E. coli for each well of a 12-well plate. Flat PMMA samples made in the same manner were also seeded. At various time points (3, 6, 18, and 24 h), diluted aliquots of cell suspension were spread on agar plates using a sterile loop and turntable per the spread plate method to determine the number of colony forming units (CFUs) in suspension above each sample. The plates were incubated at 37 °C overnight prior to colony counting.
E. Bacterial morphology
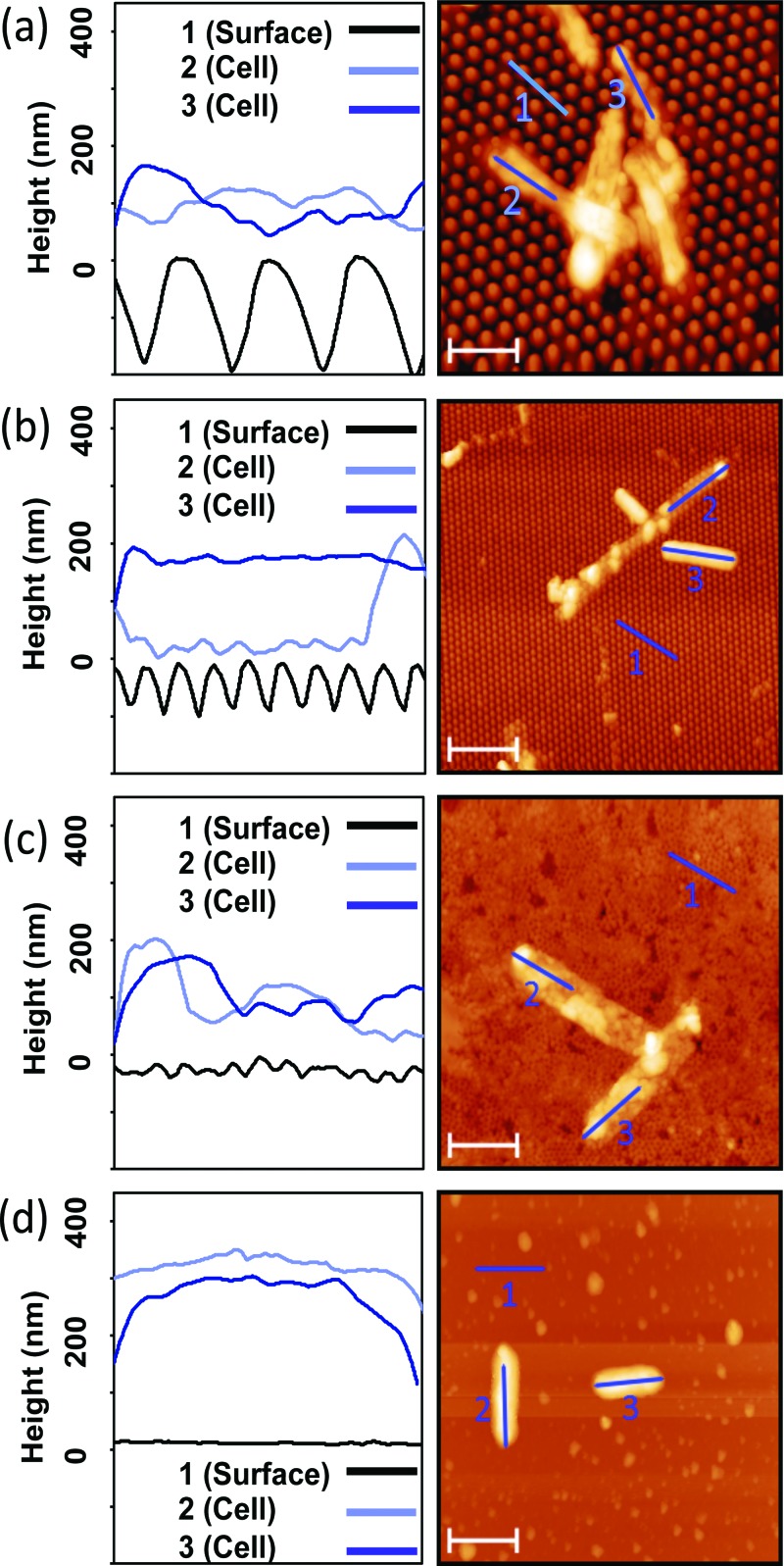
Finally, to examine morphology changes, we imaged fixed bacterial cells under SEM and AFM. Bacterial suspensions were adjusted to OD600 = 0.3.22 Each PMMA sample was immersed in an excess of this bacteria suspension and incubated for 20 h at 37 °C. Samples were rinsed twice with 1X PBS at room temperature (25 °C) by streaming the PBS along the side of the sample and gently swirling the well plate ten times. Samples were fixed with 2.5% glutaraldehyde in PBS (glutaraldehyde from Arcos Organics).30 Fixed bacteria were examined using tapping-mode AFM (NT-MDT, NTEGRA) with a 10 nm radius of curvature tip (ACL-10, Applied NanoStructures, Mountain View, CA), or sputter coated with iridium and imaged under high-vacuum SEM (FEI Quanta 3D). Lengths of individual bacterial cells and their angular orientations with respect to the pillar arrays were measured from the SEM images using imagej software. Height profiles of bacteria on representative AFM scans were measured using gwyddion software. The “height” was defined as the difference between the average height value of a 2 μm segment on the top surface of a bacterial cell and the average tip-height of the pillars in the scan. Between 8 and 21 bacteria were measured on each type of PMMA surface. Average heights and outliers were calculated using R software, with the modified box plot, which is the default.
III. RESULTS AND DISCUSSION
A. Nanotopographical effects on adhesion and viability
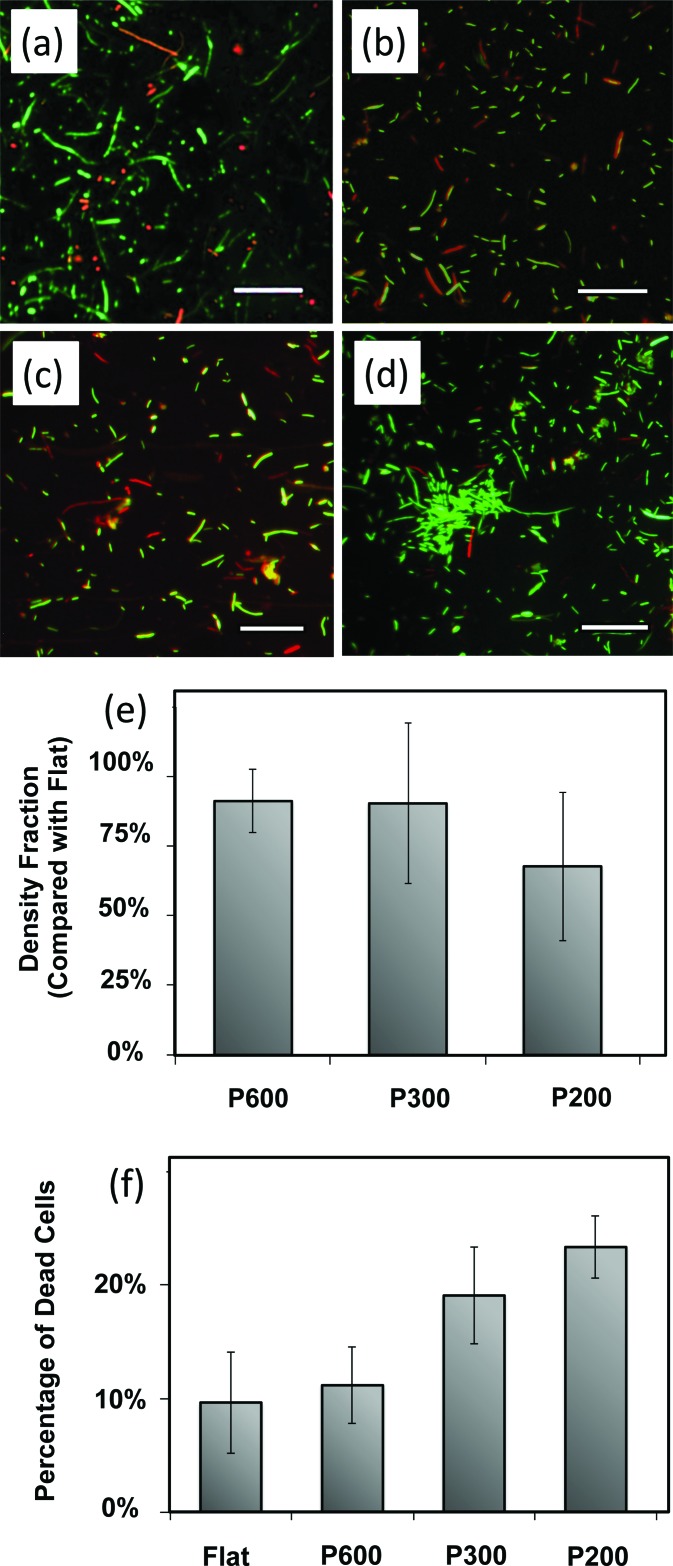
First, we examined the viability of bacterial cells on the surfaces. Representative composite fluorescence microscopy images of the live-dead-stained bacteria are shown in Figs. 2(a)–2(d). On flat films [Fig. 2(d)], many live bacteria (in green) are observed, including some clusters that may be the precursors to biofilm formation. On the pillared films [Figs. 2(a)–2(c)], particularly films with smaller, more closely spaced pillars, more dead bacteria are observed and there do not appear to be as many clusters of live bacteria.
Fig. 2.
Live-dead assay micrographs are representative micrographs on (a) P600, (b) P300, (c) P200, and (d) the flat control. Live cells are tagged with green fluorescent SYTO9 while dead cells are tagged with red fluorescent propidium iodide. Scale bars = 30 μm. (e) There is a decreased cell density on pillared surfaces compared to control flat surfaces, more so on smaller, more closely spaced pillars. Error bars represent the standard deviation of all samples. (f) The percent of cells on the surface that are dead is greater on pillars than on the flat controls. Additionally, the percent of dead cells is higher on smaller, more closely spaced pillared surfaces. Error bars represent the standard deviation of all samples.
The numbers of adherent bacteria were quantified and ranged from 8000 to 22 000 cells/mm2. Within each trial, we consistently measured lower cell densities on pillared surfaces than on the flat surfaces, as shown in Fig. 2(e). We calculated the ratio of cell density on each pillared surface with respect to the flat control in the trial. The results, plotted in Fig. 2(e), show that the cellular density depends on the pillar geometry, as the smaller, more closely spaced pillars of P200 surfaces show greater decreases in cell density compared to flat film.
We also quantified the increases in dead cells seen on the pillared films compared to flat films. We counted the live (green) and dead (red) cells and calculated the fraction of cells on each surface that were dead, as shown in Fig. 2(f). Pillared films had a higher percentage of dead bacteria than the flat films. The results show that smaller and more closely spaced pillars, P300 and P200, are more effective than P600 pillars. The percentage of dead cells increases by 16% on P600 surfaces, while a 97% increase is measured on P300 surfaces and a 114% increase is measured on P200 surfaces (compared with flat controls).
Many dead bacteria were observed on cicada wing surfaces by Ivanova et al. as well. They proposed that as the bacterial cell adsorbs to the pillars, the wetted surface area increases. This stretches the cell wall until it is breached, leading directly to lysis.22,23 These authors produced similar results on gold-coated cicada wings and nanospiked silicon, indicating that surface chemistry was not a key factor. This is unsurprising, since bacterial cells have a variety of protein, lipid and sugar surface moieties adapted for adsorption (through hydrogen bonds) to a variety of material surfaces.1,2 As mentioned, the increase in bacterial dead fractions on P200 compared with P300 surfaces is due to the combination of decreased feature size and decreased spacing. This is consistent with the model developed by Ivanova et al., because the smaller pillars would exert more local stress on the bacterial cell membranes, leading to increased cell death on pillared surfaces. However, in this study, we did not attempt to deconvolute the respective roles of feature size and spacing.
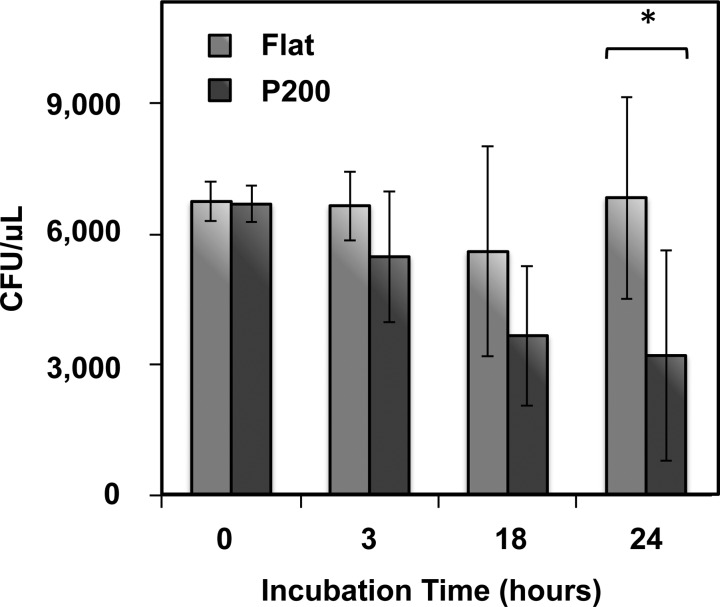
As shown in Fig. 2(e), the densities of adherent bacteria are diminished on pillared films compared to flat films. This implies one of two scenarios. If bacteria adhere to all surfaces at the same rate, then the pillared films must be killing and releasing some of these adherent cells over time. Otherwise, there must be decreased rates of adhesion on the pillared films. In order to distinguish between these scenarios, we seeded E. coli on the most effective pillared surfaces—the P200 pillars—and on flat films and tracked the CFU counts of the supernatant over time. A decreased CFU count above pillared films compared with flat films would imply that the pillared surfaces are killing bacteria. Meanwhile, if the pillared surfaces are merely repelling bacteria, the CFU counts in the supernatant are expected to be comparable. The results (Fig. 3) show that by 24 h, there is a statistically significant decrease of 50% in CFU counts above the P200 pillars compared to the flat film. This confirms that bacterial cells are dying upon the surface rather than being repelled, and indicates the potential for this surface to decrease bacterial loads in aqueous environments. Therefore, we can say that nanopillared polymer surfaces are bactericidal, killing adherent bacteria on contact.
Fig. 3.
Concentrations of colony forming units in suspension above flat and P200 samples. CFU counts remain roughly constant over 24 h above the flat films, while CFU counts decrease by 50% above the P200 pillars. Error bars represent the standard deviation. A “*” indicates statistical significance: Student's t-test <0.05.
B. Nanotopographical effects on cell morphology
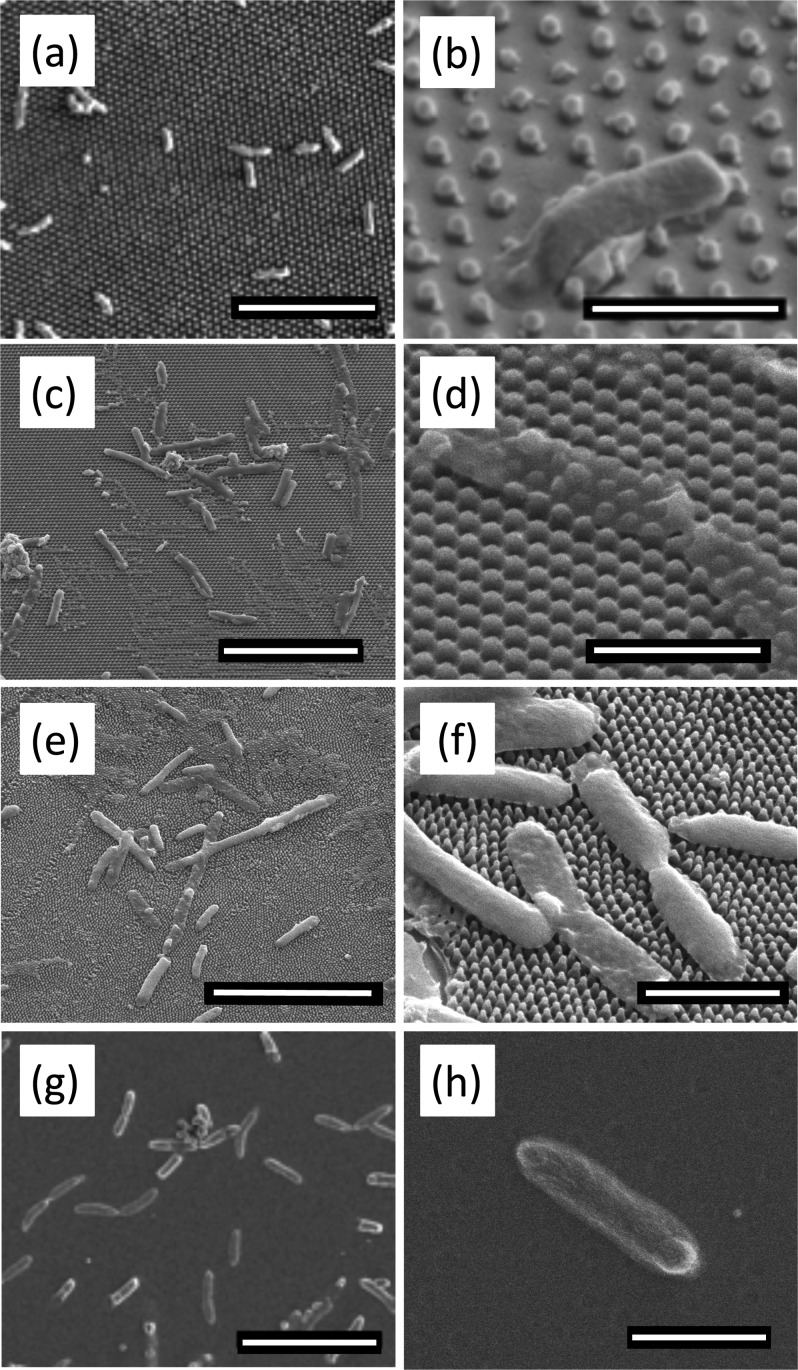
We examined the morphology and distribution of bacteria on the flat and nanopillared PMMA samples using SEM, as shown in Fig. 4. We fixed the bacterial cells with glutaraldehyde (as mentioned in Sec. II) prior to SEM imaging, which crosslinks the proteins, making the cell more resistant to changes under vacuum. On the flat PMMA control surface, bacteria were rod-shaped, the normal morphology of E. coli. On nanopillared structures, bacteria appeared deflated as they stretched over several pillars. The irregular (lumpy) sacculi of bacteria on pillars indicate that the cells have been ruptured and the turgor pressure has been lost.
Fig. 4.
Representative SEM micrographs of bacteria on flat and patterned PMMA surfaces. Scale bars on left column = 10 μm. Scale bars on right column = 2 μm. The morphology and spread of bacterial cells were observed on [(a) and (b)] P600; [(c) and (d)] P300; [(e) and (f)] P200; and [(g) and (h)] flat control. While the bacteria remain rod-shaped on the flat PMMA, the bacteria on the pillars deflate as they drape across several pillars. There is evidence of leakage of cytoplasm in (b). Images (a), (b), (g), and (h) were taken at 2 kV. All other images were taken at 5 kV.
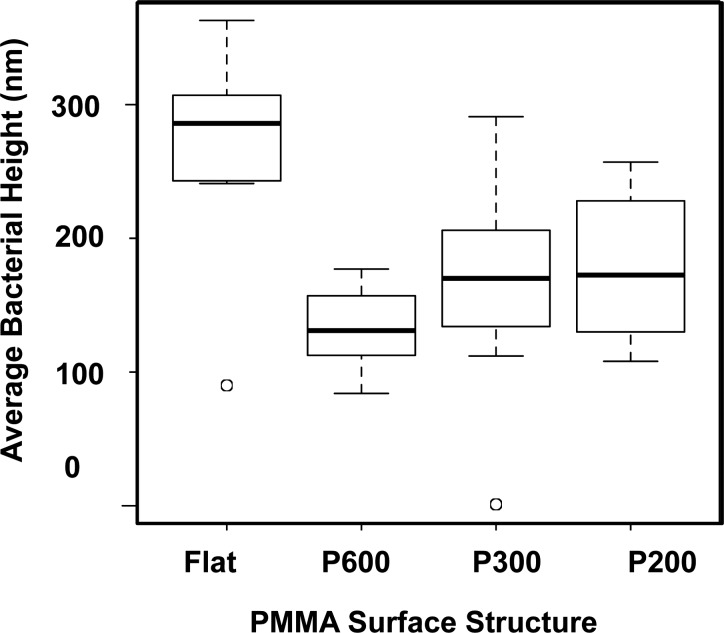
Due to the concern of the potential for high vacuum to change the cellular morphology, we also performed AFM of samples prepared in the same manner and saw very similar morphologies: deflated bacterial cells with lower heights that appeared stretched over the tops of the pillars. AFM images and height profiles of bacterial cells on nanopillar arrays and flat films are shown in Fig. 5. Bacteria imaged on the nanostructured PMMA surfaces were longer in length and less round in morphology, corroborating the SEM data. We also determined that the cells' thicknesses decreased as the cells deflated, indicating that some of the cytoplasm leaked out of the cell. In many cases, these deflated bacteria conformed to the shape of the pillared arrays. This is especially evident in Figs. 5(a-3), 5(b-2), and 5(c-3). As visualized in Figs. 5(b) and 5(c), nonuniform deflation was observed on the P300 and P200 pillars. Not all bacteria on the pillared samples exhibited deflation, which may be due to their orientation on the pillar arrays. As mentioned in Sec. II, we measured average heights of cells on all the surfaces, and the results are presented in Fig. 6. The average heights of cells on all pillar geometries were at least 30% lower than the heights of the cells on flat films, further confirming the observations made in the discussion of Fig. 5.
Fig. 5.
Representative AFM micrographs and height profiles of bacteria on (a) P600, (b) P300, (c) P200, and (d) flat film. Left column: x axes widths =2 μm. Right column: Scale bars = 2 μm. Deflated bacteria can be seen on the P600, P300, and P200 surfaces. The tallest observed bacteria, measured on the flat samples, were 300 nm. Nonuniform deflation is observed in cell 2 on the P600 surface. Variance in the P600 bacteria height is caused by cell clumping.
Fig. 6.
Box plot of the average heights of bacteria on pillared and flat PMMA substrates. The average heights of bacterial cells stretched over pillared surfaces were found to be lower. Flat: n = 7, P600: n = 8, P300: n = 21, and P200: n = 8.
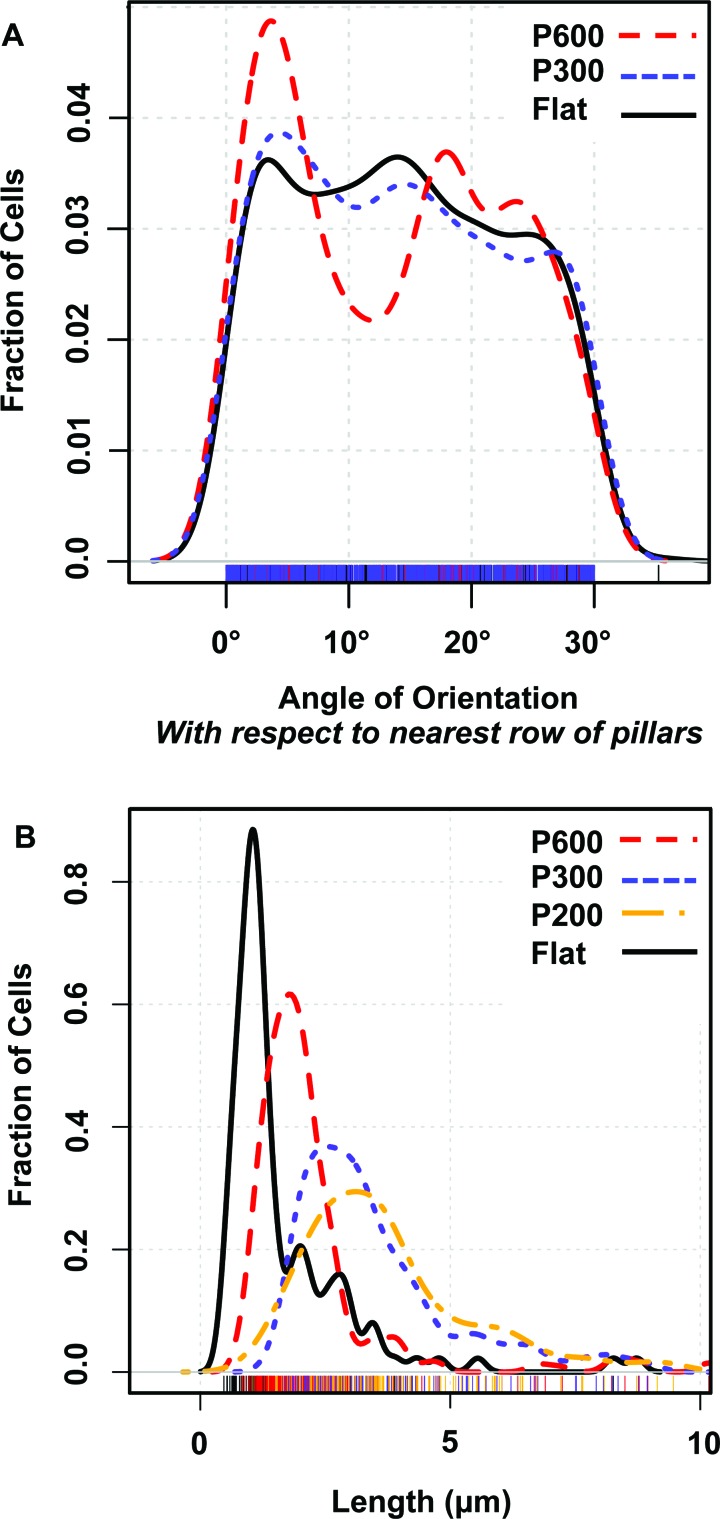
We have shown that the E. coli generally deflate and die atop nanopillared surfaces. Their behavior may also be altered while they are viable. On a flat surface, the E. coli cell displays twitching movement induced by pili.31 Possibly, the pillars entrap the twitching bacteria, as suggested by Meel et al.31 Figures 4(a), 4(b), and 5(a) show bacterial cells that appear to have settled between the rows of pillars. This phenomenon would be expected to lead to some degree of alignment of bacterial cells with the nanoscale features, as was found by Epstein et al.15 If bacteria are able to adopt orientations that minimize contact with pillars, they may be less likely to be penetrated and killed. Orientations of bacterial cells with respect to the nearest row of nearest neighbor pillars [Fig. 7(a)] were measured. These show that most bacteria on P600 films adopt one of several preferential orientations, while bacteria on flat and smaller, more closely spaced pillar arrays (P300 films) show no preferred orientation. Rows of P600 pillars are spaced only 380 nm apart, while a typical E. coli cell is 500 nm wide, so in any orientation it will contact pillars, but the number of contacts with pillars on adherent cells varies with the cell's orientation. Since the different contacts give rise to a nonrandom distribution of orientations, it is reasonable that the pillars' bactericidal ability will be variable as well. As shown in Fig. 7(a), a random distribution of orientations is found on the P300 pillar arrays (spacing = 130 nm), indicating that these pillars likely affect cells adhesion and viability more uniformly. Thus, the dimensions of these two pillar arrays define the range for the maximum optimal interpillar spacing, i.e., between 130 and 380 nm.
Fig. 7.
(a) Density plot of distribution of cells' orientation with respect to a row of nearest-neighbor pillars. On P600 films (interpillar spacing 1/4 380 nm), the bacteria adopt preferential orientation, while on P300 films (interpillar spacing 1/4 130 nm) and on flat films, the bacteria adopt no preferential orientation. (b) Density plot of distribution of cells' lengths. Cells on pillared films tend to be longer and have a wider distribution of lengths. The smallest pillars, P200, have the largest effect.
Some bacterial cells, especially those on pillared surfaces appear under SEM and AFM to be more elongated than a typical E. coli cell, which are 2 μm long. When quantified, the length data [Fig. 7(b)] shows that cells tend to be longer on P600 than on flat films, and cells are even longer on the smaller, more closely spaced pillar arrays (P300 and P200 films) than on P600 films. Additionally, the distribution of lengths is broader on P300 and P200 films than on P600 or flat films, indicating that more variability in cell lengths is induced by smaller pillar arrays. Filamentous growth of E. coli indicates that cells cannot divide normally. This phenomenon occurs under stress conditions such as nutrient shortage, oxidative damage, or DNA damage.32 Therefore, the elongation of E. coli on the pillared films indicates that the nanopillars are inducing stress, corroborating our finding that the surface is bactericidal, and perhaps indicating a secondary mechanism.
The combined results of this work chart a course for optimization. We have shown that on small, closely spaced nanopillared surfaces, bacteria cannot preferentially orient, and therefore, they are forced to contact multiple pillars. Smaller tip radii generate higher stress fields on adsorbing bacterial sacculi, which should increase the likelihood of rupture. Therefore, we hypothesize that fabricating polymer surfaces with smaller, more closely spaced pillars will improve bactericidal performance. Such smaller, sharper nanopillars found on dragonfly wings and on black silicon have also been proven effective in killing Gram-positive bacteria and yeast cells.24,26
IV. SUMMARY AND CONCLUSIONS
This study shows that a commercially available polymer, PMMA, imprinted with nanopillars is effective in killing Gram-negative E. coli bacteria, with 16%–141% increases in the fraction of adherent bacteria that are dead (compared to flat controls). We were able to distinguish the nanopillars' bactericidal mechanism from a hypothetical anti-adhesion mechanism by showing that the bacterial load in aqueous suspension above pillared surfaces decreases by 50% over 24 h as bacteria are killed. Smaller, more closely spaced pillars were more effective, possibly because bacteria on these surfaces both contact more nanopillars and feel higher stresses at these contact points. These results were borne out by length data, which indicated that nanopillars cause stress on the cells, eliciting the filamentous response. AFM and SEM characterization confirms substantial changes morphology of bacteria on pillared surfaces. Utilizing nanoimprint lithography to pattern bactericidal nanopillars on consumer-grade materials such as PMMA diminishes the burden of proving long-term biocompatibility. Thus, results from this study pave the way for a safe and effective antibacterial surface coating for biomedical implants.
ACKNOWLEDGMENTS
The authors sincerely thank Markelle Gibbs, Ph.D., Susan C. Wu, M.S., and Nicole Leilani Ing for fabrication expertise and important discussions, Kamran Ali for his advice on the bacteria experiments and providing the bacteria strain for this study, Jian-Guo Zheng, Ph.D. at the Irvine Materials Research Institute at UC Irvine for advice on electron microscopy, and Roger Steinert, M.D., M. Cristina Kenney, M.D., and Marjan Farid, M.D. for providing clinical insight. The project was partially supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant No. UL1 TR000153. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The authors also acknowledge the Allergan Foundation for partial funding. The FEI Quanta 3D used for SEM work is funded in part by the National Science Foundation Center for Chemistry at the Space-Time Limit (CHE-082913). The Jenoptik nanoimprinter is funded in part by an NSF MRI grant.
References
- 1. Katsikogianni M. and Missirlis Y. F., Eur. Cells Mater. 8, 37 (2004). [DOI] [PubMed] [Google Scholar]
- 2. Toole G. O., Kaplan H. B., and Kolter R., Annu. Rev. Microbiol. 54, 49 (2000). 10.1146/annurev.micro.54.1.49 [DOI] [PubMed] [Google Scholar]
- 3. An Y. H. and Friedman R. J., J. Biomed. Mater. Res. 43, 338 (1998). [DOI] [PubMed] [Google Scholar]
- 4. Sondi I. and Salopek-Sondi B., J. Colloid Interface Sci. 275, 177 (2004). 10.1016/j.jcis.2004.02.012 [DOI] [PubMed] [Google Scholar]
- 5. Kim J. S. et al. , Nanomedicine 3, 95 (2007). 10.1016/j.nano.2006.12.001 [DOI] [PubMed] [Google Scholar]
- 6. Banerjee I., Pangule R. C., and Kane R. S., Adv. Mater. 23, 690 (2011). 10.1002/adma.201001215 [DOI] [PubMed] [Google Scholar]
- 7. Saldarriaga Fernandez I. C., van der Mei H. C., Lochhead M. J., Grainger D. W., and Busscher H. J., Biomaterials 28, 4105 (2007). 10.1016/j.biomaterials.2007.05.023 [DOI] [PubMed] [Google Scholar]
- 8. Park K. D. et al. , Biomaterials 19, 851 (1998). 10.1016/S0142-9612(97)00245-7 [DOI] [PubMed] [Google Scholar]
- 9. Davies D., Nat. Rev. Drug Discovery 2, 114 (2003). 10.1038/nrd1008 [DOI] [PubMed] [Google Scholar]
- 10. Gristina A. G., Science 237, 1588 (1987). 10.1126/science.3629258 [DOI] [PubMed] [Google Scholar]
- 11. Neu T. R., Microbiol. Rev. 60, 151 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bos R., van der Mei H. C., and Busscher H. J., FEMS Microbiol. Rev. 23, 179 (1999). 10.1016/S0168-6445(99)00004-2 [DOI] [PubMed] [Google Scholar]
- 13. Anselme K., Davidson P., Popa A. M., Giazzon M., Liley M., and Ploux L., Acta Biomater. 6, 3824 (2010). 10.1016/j.actbio.2010.04.001 [DOI] [PubMed] [Google Scholar]
- 14. Hochbaum A. I. and Aizenberg J., Nemo Lett. 10, 3717 (2010). 10.1021/nl102290k [DOI] [PubMed] [Google Scholar]
- 15. Epstein A. K., Hochbaum A. I., Kim P., and Aizenberg J., Nanotechnology 22, 494007 (2011). 10.1088/0957-4484/22/49/494007 [DOI] [PubMed] [Google Scholar]
- 16. Carman M. L., Estes T. G., Feinberg A. W., Schumacher J. F., Wilkerson W., Wilson L. H., Callow M. E., Callow J. A., and Brennan A. B., Biofouling 22, 11 (2006). 10.1080/08927010500484854 [DOI] [PubMed] [Google Scholar]
- 17. Chung K. K., Schumacher J. F., Sampson E. M., Burne R. A., Antonelli P. J., and Brennan A. B., Biointerphases 2, 89 (2007). 10.1116/1.2751405 [DOI] [PubMed] [Google Scholar]
- 18. Reddy S. T., Chung K. K., McDaniel C. J., Darouiche R. O., Landman J., and Brennan A. B., J. Endourol. 25, 1547 (2011). 10.1089/end.2010.0611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Freschauf L. R., McLane J., Sharma H., and Khine M., PLoS One 7, e40987 (2012). 10.1371/journal.pone.0040987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Puckett S. D., Taylor E., Raimondo T., and Webster T. J., Biomaterials 31, 706 (2010). 10.1016/j.biomaterials.2009.09.081 [DOI] [PubMed] [Google Scholar]
- 21. Colon G., Ward B. C., and Webster T. J., J. Biomed. Mater. Res. Part A 78A, 595 (2006). 10.1002/jbm.a.30789 [DOI] [PubMed] [Google Scholar]
- 22. Ivanova E. P. et al. , Small 8, 2489 (2012). 10.1002/smll.201200528 [DOI] [PubMed] [Google Scholar]
- 23. Pogodin S. et al. , Biophys. J. 104, 835 (2013). 10.1016/j.bpj.2012.12.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ivanova E. P. et al. , PLoS One 8, e67893 (2013). 10.1371/journal.pone.0067893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nowlin K., Boseman A., Covell A., Lajeunesse D., and Lajeunesse D., J. R. Soc. Interface 12, 20140999 (2014). 10.1098/rsif.2014.0999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ivanova E. P. et al. , Nat. Commun. 4, 2838 (2013). 10.1038/ncomms3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Odom T. W., Love J. C., Wolfe D. B., Paul K. E., and Whitesides G. M., Langmuir 18, 5314 (2002). 10.1021/la020169l [DOI] [Google Scholar]
- 28. Kang H., Lee J., Park J., and Lee H. H., Nanotechnology 17, 197 (2006). 10.1088/0957-4484/17/1/032 [DOI] [Google Scholar]
- 29. Zhang G., Zhang J., Xie G., Liu Z., and Shao H., Small 2, 1440 (2006). 10.1002/smll.200600255 [DOI] [PubMed] [Google Scholar]
- 30. Chao Y. and Zhang T., Appl. Microbiol. Biotechnol. 92, 381 (2011). 10.1007/s00253-011-3551-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Young K. D., Microbiol. Mol. Biol. Rev. 70, 660 (2006). 10.1128/MMBR.00001-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Meel C., Kouzel N., Oldewurtel E. R., and Maier B., Small 8, 530 (2012). 10.1002/smll.201101362 [DOI] [PubMed] [Google Scholar]
- 33. Guo L. J., Adv. Mater. 19, 495 (2007). 10.1002/adma.200600882 [DOI] [Google Scholar]