Abstract
The established methods for the study of atom-detailed protein structure in the condensed phases, X-ray crystallography and nuclear magnetic resonance spectroscopy, have recently been complemented by new techniques by which nearly or fully desolvated protein structures are probed in gas-phase experiments. Electron capture dissociation (ECD) is unique among these as it provides residue-specific, although indirect, structural information. In this Critical Insight article, we discuss the development of ECD for the structural probing of gaseous protein ions, its potential, and limitations.
Graphical Abstract.
ᅟ
Keywords: Protein structure, Folding, Gas phase, Electron capture dissociation, Mass spectrometry
History
Shortly after their breakthrough discovery of electron capture dissociation (ECD) [1], McLafferty and coworkers reported that electron capture by (M + 34H)34+ ions of carbonic anhydrase (~29 kDa) [2] and (M + 33H)33+ ions of an IgE construct (~49 kDa) [3] gave only signals corresponding to molecular ions with reduced charge [i.e., (M + nH)(n-1)+•, (M + nH)(n-2)+••, etc.] but no products from dissociation [i.e., c or z • fragments from protein backbone cleavage (Scheme 1)]. This experimental finding was attributed to convoluted higher order structures of the protein ions that prevented separation of the c and complementary z • fragments [2]. In subsequent work [4], vibrational ion activation by energetic collisions with N2 gas during exposure to low-energy electrons (“activated ion ECD”) was used to break intramolecular noncovalent bonds in proteins of up to ~42 kDa, which significantly increased the number of c and z • fragments.
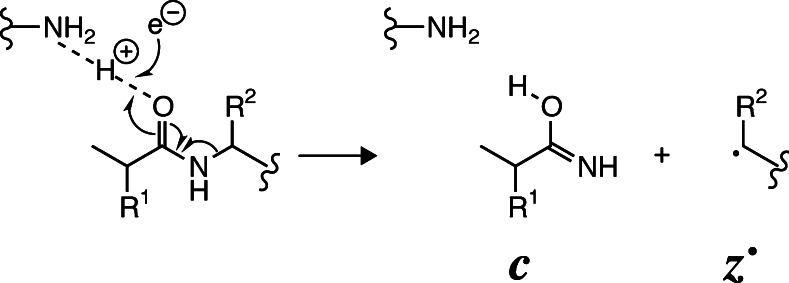
Scheme 1.
Proposed mechanism for the formation of c and z • fragments by ECD [32]; that for the far less common dissociation into a • and y fragments is not discussed here
Importantly, collisional activation or exposure to infrared (IR) photons of (M + 8H)7+• ions of ubiquitin gave no b and y fragments, which are typical products of slow (M + nH)n+ ion heating, but instead, c and z • fragments characteristic for ECD, whose site-specific relative abundances were substantially different from those in the ECD spectrum of (M + 8H)8+ ions [2]. Vibrational activation of (M + nH)(n-1)+• ions can also yield c • and z fragments, presumably from hydrogen transfer between two complementary c and z • fragments that are still held together by noncovalent bonding [5–7]; c • and z ions generally disappear when the (M + nH)n+ ions are unfolded prior to or during reaction with electrons [7, 8]. In yet another study, ~90% dissociation of ubiquitin (M + 7H)6+• ions into c and z • (and c • and z) fragments was obtained by vibrational excitation at far lower energies (~10%) than those required for collisionally activated dissociation (CAD) or infrared multiphoton dissociation (IRMPD) of (M + 7H)7+ ions into b and y fragments, consistent with breaking of noncovalent instead of covalent bonds [9]. This was interpreted as evidence for ECD data reflecting protein structure in the gas phase, in that the observation of separated c and z • fragments indicates the lack of noncovalent bonding between each other [9]. Ever since, the unusual capability of ECD to break covalent N–Cα bonds while maintaining noncovalent bonds that account for the higher order structure of gaseous ions has been used to obtain information about peptide or protein structure in the absence of solvent [7, 9–25]. More recently, this approach has been extended to the study of noncovalently bound protein complexes [18, 26–31].
Formation of ECD Fragments
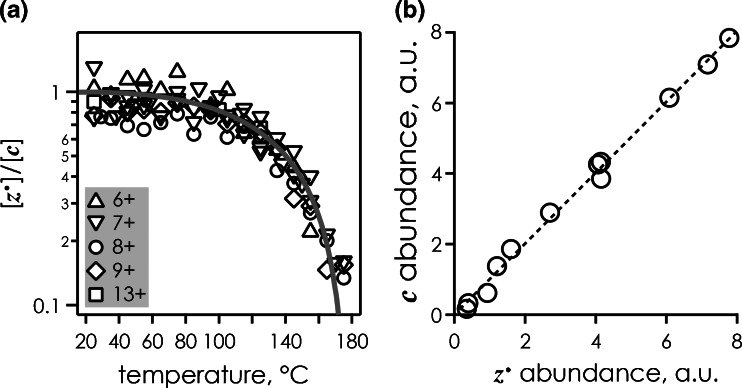
The mechanism of protein backbone bond cleavage into c and complementary z • ions is still debated, but it is generally agreed that ECD involves positively charged sites [1, 2, 33–39]. Moreover, it was shown that heating (from 25 to 125°C) of ubiquitin (M + 13H)13+ ions, which have an extended structure with a collision cross-section (CCS) of ~2115 Å2 at room temperature that is largely unaffected by collisional activation [40], does not appreciably affect the overall or site-specific yields of c ions from ECD [12]. This means that ECD cleavage is equally facile at ion temperatures of 25, 100, and 125°C, although the thermal stability of the c and z • fragment ions is substantially different (Figure 1a). Consistent with their radical nature, z • ions are far less stable than the even-electron c ions, and spontaneously undergo secondary dissociation at higher ion temperatures, which makes them less useful for structural probing in thermal unfolding experiments [9].
Figure 1.
(a) Ratio of z • and c fragments from ECD of ubiquitin (M + nH)n+ ions thermalized by blackbody infrared irradiation (data from references [9, 12]), the solid line is meant to guide the eye; (b) abundances of c versus z • fragments from ECD of KIX (M + nH)n+ ions (n = 7–16) at room temperature (circles, data from reference [19]), dashed line with unit slope indicates equal abundances [for n = 11 and 12, data from ECD of (M + nH)n+ ions from ESI of both “native” and “quasi-native” solutions are included]
Excess Energy in ECD
Is it possible that any excess energy in ECD [i.e., the energy from cation-electron charge recombination (CR) that is not consumed by N–Cα bond cleavage] causes secondary dissociation of the radical z • ions, or even changes in the higher order structure of the protein ions? At room temperature, ECD of linear proteins generally produces very similar yields of c and z • ions (Figure 1b), which shows that any excess energy must be significantly smaller than that required for secondary dissociation of the radical z • ions. For peptides and proteins that comprise cyclic backbone or disulfide bonded structures, secondary radical reactions can, nonetheless, proceed at room temperature [3, 20, 41], most likely driven by the energy released by breaking up the strained cyclic structures.
For energy minimized forms of the ubiquitin crystal structure in the absence of solvent, Williams and coworkers have conservatively estimated that capture of one and two electrons, with assumed CR energies of 6 and 12 eV, is associated with a temperature increase from ~20 to ~85 and ~140°C, respectively [17]. Calculated CR energies for small (~130 to 1115 Da, 18 to 172 atoms) linear peptide ions with m/z values between ~100 and 555 and up to three charges did not exceed 6.6 eV, and generally increased with increasing charge [42]. For example, the adiabatic CR energy for (AHDAL +2H)2+ ions with m/z 264, 5.53 eV, was about twice as high as that for (AHDAL + H)+ ions with m/z 526, 2.93 eV. Vertical CR energies were even smaller by an average factor of 0.8, in good agreement with the CR energy of 4.3 eV for (KYK + 2H)2+ ions at m/z 220 determined in nanocalorimetry experiments [43].
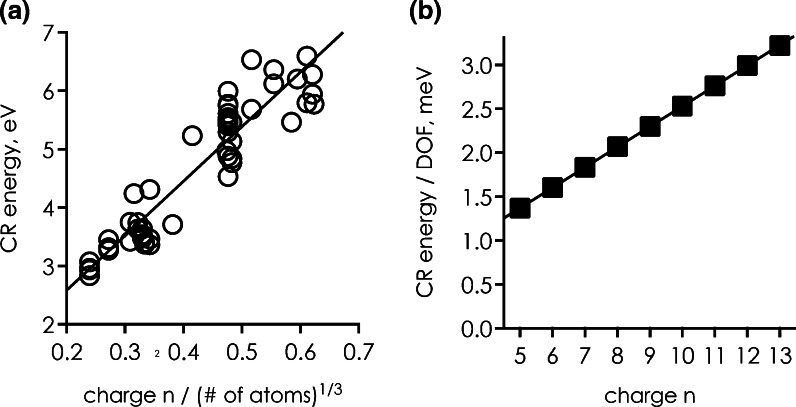
We found that the calculated CR energies increased roughly linearly with peptide ion charge divided by the cube root of the ion’s number of atoms as illustrated in Figure 2a. Extrapolation of the linear fit function to n = 0 gives an intercept of 0.721 ± 0.213 eV, which can be interpreted as the average electron affinity (EA) of gaseous peptides with zero net charge. Other neutral compounds with EA values in this range include inorganic salts such as sodium chloride (0.727 eV [44]) and potassium iodide (0.728 eV [44]), whereas small model compounds that more closely resemble the chemical structure of peptides, such as formamide (0.017 eV [44]), N-methylformamide (0.016 eV [44]), acetic acid (–1.302 to –0.482 eV [45]), and acrylamide (–0.585 eV [46]) generally have substantially lower or even negative electron affinities. Judging from these EA values, it seems possible that gaseous peptides, even with zero net charge, can feature salt bridge structures, but direct experimental evidence for this has so far been demonstrated only for (RR + H)+ ions [47].
Figure 2.
(a) Calculated adiabatic CR energies of small peptide ions from reference [42] versus charge divided by the cube root of the ion’s number of atoms, solid line shows linear fit with r2 = 0.874; (b) estimated CR energies for ubiquitin (M + nH)n+ ions per vibrational degree of freedom (DOF)
Furthermore, can we rationalize the observed linear correlation of CR energy with charge divided by the cube root of the number of atoms (Figure 2a)? Because ion volume should be roughly proportional to the number of atoms, the cube root of the number of atoms can be interpreted as the average value of the ion’s three dimensions. The linear correlation of CR energy with charge divided by the cube root of the number of atoms shown in Figure 2a is thus consistent with simple Coulomb interactions dominating cation-electron charge recombination in roughly spherical ion structures. From the fit in Figure 2a, adiabatic CR energies between 5.1 and 12.0 eV can be estimated for ubiquitin (M + nH)n+ ions with n = 5–13, which corresponds to ~1–3 meV or ~0.1–0.3 kJ/mol per vibrational degree of freedom (DOF, calculated as three times the number of atoms minus 6) as shown in Figure 2b. Nevertheless, this estimate for protein ion CR energies is far higher than that of 4–7 eV by Zubarev [48] who used a thermodynamic cycle that comprised apparent proton affinities of (M + nH)n+ ions [49].
However large or small, the CR energies may not be fully available for potential ion heating as part of it can be consumed by backbone cleavage. N–Cα bond dissociation energies between –0.92 and 1.63 eV were calculated for small radical peptide model systems [42], indicating that the excess energy in ECD can be substantially lower than the energy from charge recombination. Moreover, even the exoergic reactions with negative bond dissociation energies can be slowed by barriers, for which transition state energies between 0.02 and 0.99 eV were calculated [42]. Computing CR and dissociation energies for peptide and protein (M + nH)(n-1)+• ions remains nonetheless challenging, not least because the exact charge locations and structures of most gaseous peptides and essentially all protein ions are unknown.
To illustrate the intricate interplay of charge location and structure in gaseous ions, three examples are given here. First, even for compounds as simple as para-aminobenzoic acid methyl ester, the preferred protonation site changes from the carbonyl oxygen to the amine nitrogen upon relatively small changes in chemical environment (i.e., by increasing the number of attached water molecules from 2 to 3 [50]). Translating this scenario to gaseous peptide or protein ions, even relatively small local structural changes such as sidechain reorientations, especially those involving hydrogen bonds, are likely to significantly affect the proton affinities of individual sites and, therefore, the CR energies. Second, ion mobility spectrometry (IMS) studies showed that simple crown ether binding can, depending on ion net charge, decrease or increase collision cross-sections of gaseous protein ions by more than 30% [51], indicating major structural changes caused by minor local binding events. Third, positioning of negative and positive charge on the N- and C-terminus of polyalanine peptides, respectively, stabilizes α-helical structures through favorable charge–helix dipole interactions, whereas polyalanine peptides with opposite charge locations adopt globular structures [52]. The above findings exemplify the complexity of gaseous protein ions structures [53], which is further increased by the possibility of kinetic trapping of less stable structures from electrospray ionization (ESI) [54].
McLuckey and coworkers have recently investigated the effect of recombination energy by comparing ECD spectra of small peptides to those from electron transfer dissociation (ETD), in which the recombination energy should be smaller than in ECD by the electron affinity of the reagent used, 0.6 eV for azobenzene and 1.7 eV for 1,3-dinitrobenzene [55]. Data interpretation was complicated by the fact that products from sequential electron transfer and H• loss could not be distinguished from products from competitive proton transfer. However, the partitioning among the various c, z • ion channels was remarkably similar in ECD and ETD for most of the model peptides studied, which would argue against any effect of charge recombination energy on peptide ion structure prior to backbone cleavage into c and z • ions.
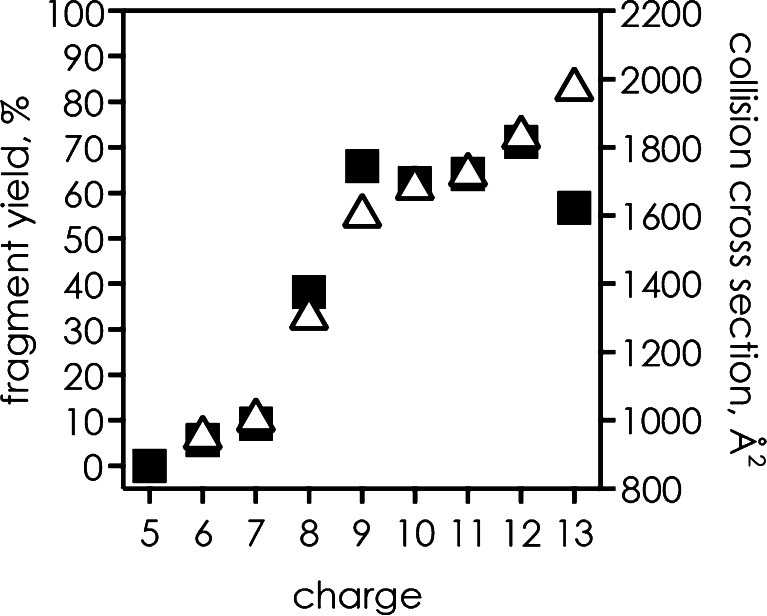
The above discussion considered mostly peptides, but is there any experimental evidence for protein ion unfolding caused by ECD? The strong correlation of c and z • ion yields with collision cross-sections found for ubiquitin [9] (Figure 3) suggests that any excess energy deposited by electron capture does not significantly affect protein ion higher order structure on the timescale of the ECD experiment. This hypothesis is corroborated by data from ECD of different ubiquitin conformers of the same charge, separated by ion mobility, which showed significant differences in both overall and site-specific fragment ion abundances [17]. More specifically, any ubiquitin ion unfolding caused by electron capture must be far slower than the timescale for structural probing by ECD of typically several tenths of milliseconds, or the excess energy must be insufficient to cause significant unfolding, possibly due to fast radiative cooling by emission of infrared or higher-energy photons [57].
Figure 3.
Yield of c, z • fragments from ECD (squares, left axis) and collision cross-sections (triangles, right axis) of ubiquitin (M + nH)n+ ions at room temperature versus charge (data from references [9], [56])
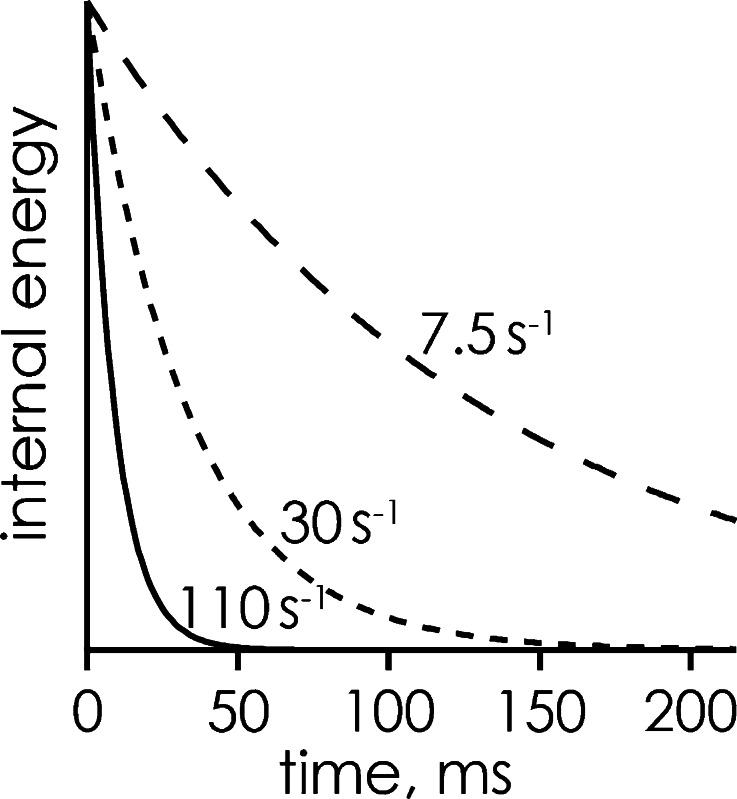
Radiative cooling by emission of infrared photons can be a complex process with rates that generally increase with increasing internal energy of the ions (i.e., hotter ions cool faster). For example, Al4 – clusters showed a decrease in radiation intensity from 40 eV/s at 1127°C to 1 eV/s at 227°C [58]. Nevertheless, Dunbar pointed out that “often it will be a reasonable approximation to think of the cooling as exponential” such that the internal energy decay of a population of superthermal ions can be described in terms of first-order cooling rate constants [59]. For small non-halogenated organic ions, infrared radiative cooling rate constants between 0.5 and 15 s–1 at energies between 0.3 and 4 eV have been measured [59], and those for singly protonated pentapeptide leucine enkephalin ions were deduced from experimental data as 30 s–1 at 165°C [60] and 7.5 ± 0.5 s–1 at ~25°C [61]. The internal energy of (M + H)+ ions of leucine enkephalin at 25°C is ~1 eV [62], which can be increased to ~3 eV (at which the cooling rate constant of 7.5 ± 0.5 s–1 was determined) [63] in tandem MS experiments to overcome the activation energy of 1.09 ± 0.06 eV [64] for dissociation into typical products from slow ion heating [65] within a few milliseconds [66]. From the fit in Figure 2a, CR energies of 2.90 and 5.09 eV can be estimated for electron attachment to (M + H)+ and (M + 2H)2+ ions of leucine enkephalin, respectively, which should be sufficient for dissociation into b and y fragments on a milliseconds timescale. Whereas the uncharged products from ECD of (M + H)+ ions cannot be directly detected, ECD of (M + 2H)2+ ions of leucine enkephalin in fact produced no c or z • but instead a, b, y and w ions [67]. It has been suggested that b ions form after loss of H• from “hot” (M + 2H)+• ions, the probability for which depends on peptide composition, structure, and internal energy [68]. Nevertheless, the formation of b ions in ECD of proteins and most peptides is rare, consistent with nanocalorimetry ECD experiments of partially hydrated (KYK + 2H)2+ ions, which gave both c and b ions, but c ion formation required only ~14% of the energy required for b ion formation [43].
The estimated CR energies of 2.90 and 5.09 eV for leucine enkephalin correspond to 13 and 22 meV deposited per DOF, which is about an order of magnitude higher than the estimated values of ~1–3 meV for ubiquitin ions (Figure 2b). Moreover, larger peptide and protein ions in the rapid energy-exchange (REX) limit should thermally equilibrate to the temperature of the surrounding vacuum system, typically room temperature, at rates that are the same as in the high pressure limit [69, 70]. The corresponding cooling rate constants should by far exceed the ~110 s–1 measured for singly protonated leucine enkephaline at 3.7 × 10–4 mbar and 25°C [60], such that cooling to room temperature of larger peptide and protein ions by emission of infrared photons should be possible within a few milliseconds (Figure 4).
Figure 4.
Internal energy versus time assuming exponential decay, calculated for the cooling rate constants indicated
Conversion of the CR energy, or parts of it, into IR radiation of course requires prior conversion into vibrational energy by intramolecular vibrational redistribution (IVR). It is generally agreed that electron capture involves higher-energy Rydberg states that undergo fast relaxation to lower energy Rydberg and then non-Rydberg (n < 3) electronic states [71], but whether this involves radiative or nonradiative relaxation by IVR, or both, is still unclear. Moreover, nothing is as yet known about radiative cooling of peptide or protein (M + nH)n+• ions by emission of higher-energy photons, but the possibility that radiation from electronically excited states cools the radical ions formed by electron capture cannot generally be excluded [72]. Despite the gain in energy from charge recombination, the protein (M + nH)(n-1)+• ions from electron capture are sufficiently stable for isolation and subsequent dissociation into c and z • (or c • and z) fragments by IR laser radiation [9, 11, 20] at energies that are far lower than those required to dissociate (M + nH)n+ ions into b and y fragments. Possibly the radical (M + nH)(n-1)+• ions formed by electron attachment into a Rydberg orbital with subsequent transfer to π* or σ* orbitals [34] also cool by emission of higher-energy photons from electronically excited states, which raises the “old” question if ECD can be described as a nonergodic process or if it is better described as a statistical process [1, 73, 74]. Anyhow, when cooling rates are significantly faster than the rates of unfolding, any excess energy from charge recombination should have little effect on the protein structures probed by ECD, but this may not be the case for small peptides. For example, Turecek and coworkers have calculated a temperature of ~580°C for a doubly charged pentapeptide after electron capture without radiative relaxation [75].
Probing Protein Ion Structure
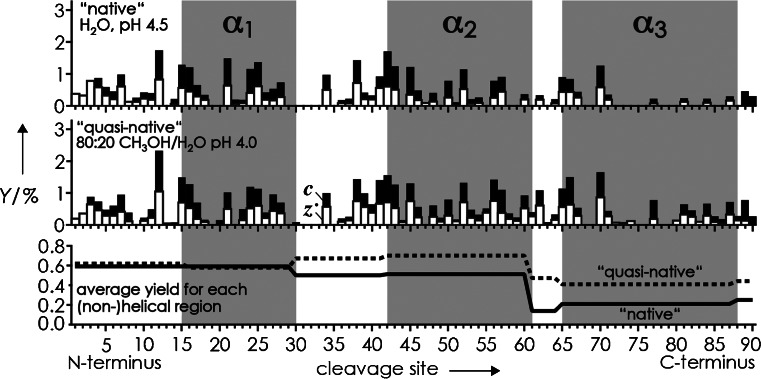
In a previous Critical Insight article, Hall and Robinson asked the important question “Do charge state signatures guarantee protein conformations?” and came to the conclusion that although experimental evidence “points to a consensus that the lowest charge states are the most compact,” none of the available techniques provides sufficient information for elucidation of gaseous protein structures [76]. Nevertheless, ion mobility spectrometry (IMS) studies showed that protein (M + nH)n+ ions with the same net charge but electrosprayed from different solutions can have considerably different collision cross-sections [56]. The first evidence that ECD can probe differences in protein structure that result from differences in solvent composition came from a study of the three-helix bundle protein KIX [19]. ECD of KIX (M + 12H)12+ ions electrosprayed from “native” and slightly denaturing (“quasi-native”) solutions gave total fragment ion yields1 of 37% and 49%, respectively, consistent with a higher extent of noncovalent bonding in ions from “native” solutions resulting in fewer separated fragments. Moreover, site-specific c, z • ion yields revealed that denaturation unfolded the region comprising helices α2 and α3 (residues 30–91), whereas no overall effect was observed in the N-terminal region (Figure 5).
Figure 5.
c (black bars) and z • (open bars) yields versus cleavage site from ECD of KIX (M + 12H)12+ ions from ESI of “native” (top) and “quasi-native” (center) solutions, and average yield for each helical and non-helical region (bottom) for “native” (solid line) and “quasi-native” (dashed line) solutions; data from reference [19]
It is important to note that CAD differs markedly from ECD in this respect. For example, Clemmer and coworkers selected different conformers of ubiquitin (M + nH)n+ ions with n = 8–10 by ion mobility for subsequent dissociation by CAD, and found that the fragment mass spectra for different conformers of the same charge were identical within experimental error, which they attributed to rearrangement into structurally similar transition states prior to dissociation into b and y fragment ions [77]. Because CAD relies solely on vibrational excitation that incrementally increases an ion’s internal energy until dissociation occurs, it is reasonable that noncovalent bonds are broken before covalent bonds, unless the latter are weaker than the former.
Apart from higher order structure, the site-specific c, z • ion yields from ECD are also affected by how positive charge is distributed and locally solvated within the protein ions [78], which gives rise to irregular (“jumpy”) features in plots of site-specific yield versus cleavage site, even when the protein ions are unfolded [9, 12, 19]. This is similar to steric and inductive effects of amino acid sidechains on hydrogen/deuterium exchange of protein backbone amides in solution, for which scalar correction factors have been established [79]. Delineating the effects of charge location and solvation in ECD is less straightforward, but it was recently shown that the “jumpy” features are due to distinct patterns termed charge site spectra [78] that can be retained over a wide range of precursor ion charge states for a given protein sequence. Finally, because ECD requires the presence of positive charge, a lack of c and z • fragments can also be observed in protein regions that lack any basic residues, or in protein regions that are bridged by disulfide bonds [20].
In any case, structural probing by ECD is indirect, meaning that it relies on observing separated c and z • fragments from protein backbone cleavage to reveal the absence of noncovalent bonding between them. Information about the number, types, or strengths of intramolecular interactions that prevent fragment ion separation cannot directly be inferred from a single ECD spectrum. In other words, structural probing by ECD provides an imprint of unfolded structure that shows which regions are unfolded, but not how and to what extent the other regions are folded. This is similar to footprinting techniques in solution in which folded regions can be shielded from, for example, hydroxyl radical attack, but with the major difference that in ECD, a single noncovalent interaction between two non-adjacent residues could prevent separation of all c and z • fragments from backbone cleavage in between these residues, no matter how compact that region is. Nevertheless, ECD provides valuable structural information, especially when data obtained under different experimental conditions are compared with each other (Figure 5), and when it is used to probe protein unfolding or folding [9, 10, 21, 80].
Monitoring Protein Unfolding
Proteins can unfold for a number of different reasons, including solution pH, the presence of organic solvent or “denaturants” such as guanidinium hydrochloride, and elevated temperatures. As evidenced by ECD, the mere removal of solvent during ESI causes spontaneous unfolding of horse and tuna heart cytochromes c and ubiquitin, all of which have highly stable “native” folds in solution [21, 80]. The extent of unfolding in different regions of these proteins, however, was found to depend on the extent of stabilization by electrostatic interactions. Remarkably, the (M + 7H)7+ ions of the three-helix bundle protein KIX were sufficiently stabilized to fully preserve the native fold on a timescale of up to 4 s after transfer into the gas phase, and a gradual loss of higher order structure by sequential unraveling of the helices from their terminal ends was observed with increasing charge of the (M + nH)n+ ions [19]. Thus, ECD can be used to monitor unfolding in solution, or to probe structural changes that occur during the transition from solution into the gas phase, but distinguishing between these two possibilities is not straightforward when the solution structure is not known.
Regardless of whether a compact protein ion structure resembles the solution fold or not, its thermal unfolding in the gas phase can be monitored by ECD. For ubiquitin (M + 6H)6+ to (M + 9H)9+ ions that were thermally equilibrated by blackbody infrared radiation for at least 40 s, the yield of separated c, z • fragments steadily increased with increasing temperature from 25 to 175°C [9]. Importantly, a van’t Hoff analysis of the ECD data revealed a three-state process for ubiquitin (M + nH)n+ ion unfolding in the gas phase, with ΔH values generally decreasing with increasing ion net charge. However, the ΔH values for n = 7 were similar to those for n = 6 at temperatures of up to 100°C, and similar to those for n = 8 at higher temperatures, showing a stepwise decrease in ΔH from ~30 to ~10 kJ/mol, in contrast to the gradual decrease of ΔH with decreasing pH for ubiquitin unfolding in solution [81]. Protein ions can also be unfolded by collisional activation prior to structural probing by ECD [80] or ETD [82], and although in such experiments carried out under multiple low-energy collisions it is usually not possible to determine ion temperatures and derive thermodynamic information, they can be used to establish the order of stability for different regions of a gaseous protein [21].
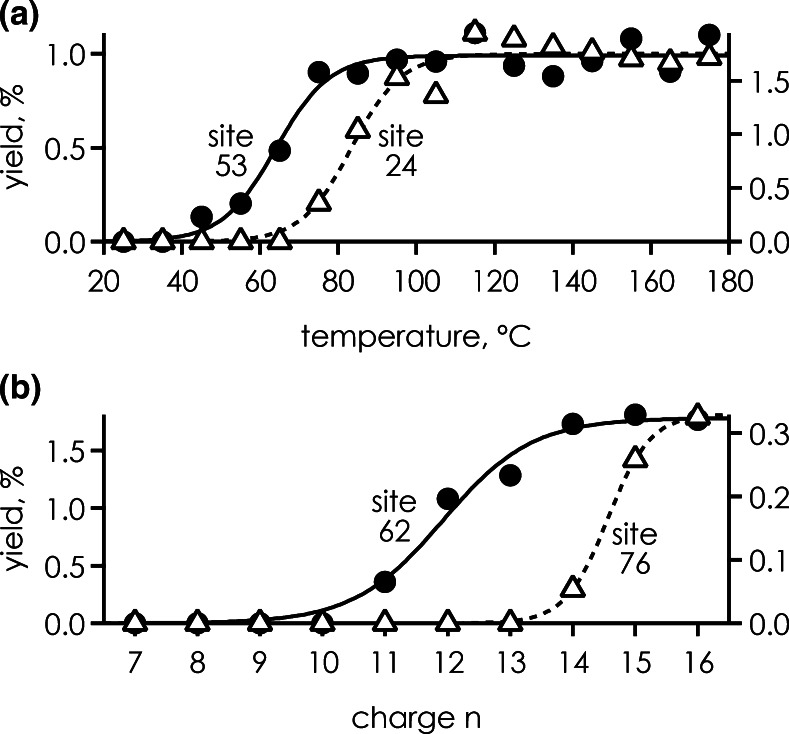
As discussed above, the site-specific fragment ion yields in ECD of protein (M + nH)n+ ions are not only affected by higher order structure but also by how positive charge is distributed and locally solvated within the protein ions studied (i.e., some variation in yield is found among all cleavage sites even in ECD of unfolded ions). Nevertheless, the site-specific c and z • ion yields generally showed sigmoidal behavior for (M + nH)n+ ion unfolding by temperature or charge n (Figure 6), from which melting temperatures [9] or transition charge values [19] can be derived, respectively. Monitoring the c and z • ion yields indicated different melting temperatures for different sites in the thermal unfolding of ubiquitin (M + 7H)7+ ions [e.g., 83 ± 2°C for site 53 and 64 ± 2°C for site 24 (Figure 6a)]. However, because z • ions can undergo secondary dissociation at elevated temperatures (Figure 1a), melting curves derived from only c instead of c and z • ions are more reliable in thermal unfolding experiments [9]. Thermal dissociation of z • ions should not be an issue in experiments in which protein ion unfolding is monitored by measuring site-specific c and z • ion yields with increasing (M + nH)n+ charge at room temperature (Figure 1b). The corresponding data also show sigmoidal transitions, as illustrated in Figure 6b for KIX (M + nH)n+ ions at sites 62 and 76. The scatter in transition charge values for adjacent sites can be substantial [19], which can be attributed to the loss of both local and global interactions. To eliminate the effect of interactions between adjacent residues and characterize the unfolding of protein higher order structure, integrated yields can be used for the determination of transition charge values for specific regions of the protein [19].
Figure 6.
Site-specific yield of c and complementary z • fragments from ECD of (a) ubiquitin (M + 7H)7+ ions versus ion temperature for sites 24 (right axis) and 53 (left axis), data from reference [9], and (b) KIX (M + nH)n+ ions versus charge n for sites 62 (left axis) and 76 (right axis), data from reference [19]; lines represent sigmoidal fit functions
Monitoring Protein Folding
A major drawback of structural probing by ECD is that a single spectrum does not reveal how much structure is retained in regions framed by two residues that are still held together by noncovalent bonding. This means that if gaseous protein ions were to fold by first associating their N- and C-terminal residues, all fragment ions would disappear simultaneously, no matter how compact or extended the structure were of the residues in between. However, for the proteins studied so far, horse and tuna heart cytochromes c and ubiquitin, such behavior has not been observed. Instead, regions in which the site-specific yield of ECD fragments decreased with increasing time allowed for folding (indicating the formation of noncovalent interactions between the complementary c and z • fragments from backbone cleavage in that region) were generally separated by regions in which the yield did not change (no new interactions formed), decreased to a lesser extent (fewer interactions formed), or even increased (loss of interactions) [9, 80]. Moreover, for these proteins, folding in the gas phase is not only slower than folding in solution by several orders of magnitude [10, 12, 80, 83, 84], it is also dominated by nearest neighbor interactions instead of global structural changes, and driven by electrostatic instead of hydrophobic interactions [80].
Experimental Conditions for ECD
An important issue to be considered in ECD experiments for structural probing, as well as for sequencing applications, is secondary dissociation as a result of secondary (or multiple) electron capture. Because the cross-section for electron capture generally increases with the square of the net positive charge of an ion [2], the probability for secondary electron capture is generally higher for larger fragment ions that typically carry more charge than smaller fragments. Thus, under conditions of multiple electron capture, larger c and z • fragments are preferentially depleted, and only smaller fragments from backbone cleavage close to the protein termini are detected, which produces ECD spectra that can mistakenly be interpreted as reflecting noncovalent bonding in the center region of the protein. Such spectra are characterized by missing complementary ions, that is, only small c and z • fragments from cleavage close to the N and C terminus are detected, respectively. An example of such a spectrum can be found in Figure 1 of reference [20], and spectra recorded under conditions of single electron capture that show approximately equal yields of c and z • fragments at each cleavage site can be found in Figure 2 of reference [19].
Open Questions and Future Challenges
ECD of proteins does not provide direct structural information, but the observation of c and z • fragments immediately reveals the absence of noncovalent bonding between them. Moreover, when changes in ECD spectra are followed as the temperature or charge of the protein ions under study is increased, or time is allowed for folding, site-specific data on the structural changes involved can be obtained. A major challenge for comprehensive structural studies is the analysis of complex ECD spectra. Although automated algorithms for spectral interpretation that can correctly assign and quantify the majority (~90%) of signals in an ECD spectrum are available, additional manual interpretation is, in our experience, absolutely essential. Regarding the mechanism of protein backbone bond cleavage into c and complementary z • ions and the implications for structural probing, it remains to be seen if there are lower mass and higher charge limits within which the energy gained by electron capture does not cause significant unfolding, and the role of electronically excited states in radiative cooling needs to be addressed. Finally, because charge locations in peptide and protein ions from ESI are critical to the development of theoretical models of the ECD process, a better understanding of the electrospray process and the structural changes involved is vitally important.
Acknowledgments
This work was supported by the Austrian Science Fund (FWF): Y372 and P27347 to K.B.
Footnotes
ECD fragment ion yields are calculated as percentage values relative to all ECD products, considering that backbone dissociation of a parent ion gives a pair of complementary c and z • or a pair of a • and y ions, 100% = 0.5[c]. + 0.5[a •] + 0.5[z •] + 0.5[y] + [other products], in which other products are reduced molecular ions and products from loss of small neutral species from the latter. In cases where a • and y ions are of low abundance with correspondingly small signal-to-noise ratios, they can be excluded from the analysis.
References
- 1.Zubarev RA, Kelleher NL, McLafferty FW. Electron capture dissociation of multiply charged protein cations. A nonergodic process. J. Am. Chem. Soc. 1998;120:3265–3266. doi: 10.1021/ja973478k. [DOI] [Google Scholar]
- 2.Zubarev RA, Horn DM, Fridriksson EK, Kelleher NL, Kruger NA, Lewis MA, Carpenter BK, McLafferty FW. Electron capture dissociation for structural characterization of multiply charged protein cations. Anal. Chem. 2000;72:563–573. doi: 10.1021/ac990811p. [DOI] [PubMed] [Google Scholar]
- 3.Zubarev RA, Kruger NA, Fridriksson EK, Lewis MA, Horn DM, Carpenter BK, McLafferty FW. Electron capture dissociation of gaseous multiply-charged proteins is favored at disulfide bonds and other sites of high hydrogen atom affinity. J. Am. Chem. Soc. 1999;121:2857–2862. doi: 10.1021/ja981948k. [DOI] [Google Scholar]
- 4.Horn DM, Ge Y, McLafferty FW. Activated ion electron capture dissociation for mass spectral sequencing of larger (42 kDa) proteins. Anal. Chem. 2000;72:4778–4784. doi: 10.1021/ac000494i. [DOI] [PubMed] [Google Scholar]
- 5.O’Connor PB, Lin C, Cournoyer JJ, Pittman JL, Belyayev M, Budnik BA. Long-lived electron capture dissociation product ions experience radical migration via hydrogen abstraction. J. Am. Soc. Mass Spectrom. 2006;17:576–585. doi: 10.1016/j.jasms.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 6.Savitski MM, Kjeldsen F, Nielsen ML, Zubarev RA. Hydrogen rearrangement to and from radical z fragments in electron capture dissociation of peptides. J. Am. Soc. Mass Spectrom. 2007;18:113–120. doi: 10.1016/j.jasms.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 7.Lin C, Cournoyer JJ, O’Connor PB. Probing the gas-phase folding kinetics of peptide ions by IR activated DR-ECD. J. Am. Soc. Mass Spectrom. 2008;19:780–789. doi: 10.1016/j.jasms.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ledvina AR, McAlister GC, Gardner MW, Smith SI, Madsen JA, Schwartz JC, Stafford GC, Syka JEP, Brodbelt JS, Coon JJ. Infrared photoactivation reduces peptide folding and hydrogen-atom migration following ETD tandem mass spectrometry. Angew. Chem. 2009;48:8526–8528. doi: 10.1002/anie.200903557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breuker K, Oh HB, Horn DM, Cerda BA, McLafferty FW. Detailed unfolding and folding of gaseous ubiquitin ions characterized by electron capture dissociation. J. Am. Chem. Soc. 2002;124:6407–6420. doi: 10.1021/ja012267j. [DOI] [PubMed] [Google Scholar]
- 10.Horn DM, Breuker K, Frank AJ, McLafferty FW. Kinetic intermediates in the folding of gaseous protein ions characterized by electron capture dissociation mass spectrometry. J. Am. Chem. Soc. 2001;123:9792–9799. doi: 10.1021/ja003143u. [DOI] [PubMed] [Google Scholar]
- 11.Oh H, Breuker K, Sze SK, Ge Y, Carpenter BK, McLafferty FW. Secondary and tertiary structures of gaseous protein ions characterized by electron capture dissociation mass spectrometry and photofragment spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 2002;99:15863–15868. doi: 10.1073/pnas.212643599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Breuker K, Oh HB, Lin C, Carpenter BK, McLafferty FW. Nonergodic and conformational control of the electron capture dissociation of protein cations. Proc. Natl. Acad. Sci. U.S.A. 2004;101:14011–14016. doi: 10.1073/pnas.0406095101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mihalca R, Kleinnijenhuis AJ, McDonnell LA, Heck AJR, Heeren RMA. Electron capture dissociation at low temperatures reveals selective dissociations. J. Am. Soc. Mass Spectrom. 2004;15:1869–1873. doi: 10.1016/j.jasms.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 14.Adams CM, Kjeldsen F, Zubarev RA, Budnik BA, Haselmann KF. Electron capture dissociation distinguishes a single D-amino acid in a protein and probes the tertiary structure. J. Am. Soc. Mass Spectrom. 2004;15:1087–1098. doi: 10.1016/j.jasms.2004.04.026. [DOI] [PubMed] [Google Scholar]
- 15.Polfer NC, Haselmann KF, Langridge-Smith PRR, Barran PE. Structural investigation of naturally occurring peptides by electron capture dissociation and AMBER force field modeling. Mol. Phys. 2005;103:1481–1489. doi: 10.1080/00268970500095998. [DOI] [Google Scholar]
- 16.Geels RBJ, van der Vies SM, Heck AJR, Heeren RMA. Electron capture dissociation as structural probe for noncovalent gas-phase protein assemblies. Anal. Chem. 2006;78:7191–7196. doi: 10.1021/ac060960p. [DOI] [PubMed] [Google Scholar]
- 17.Robinson EW, Leib RD, Williams ER. The role of conformation on electron capture dissociation of ubiquitin. J. Am. Soc. Mass Spectrom. 2006;17:1469–1479. doi: 10.1016/j.jasms.2006.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang H, Cui WD, Wen JZ, Blankenship RE, Gross ML. Native electrospray and electron-capture dissociation in FTICR mass spectrometry provide top-down sequencing of a protein component in an intact protein assembly. J. Am. Soc. Mass Spectrom. 2010;21:1966–1968. doi: 10.1016/j.jasms.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Breuker K, Brüschweiler S, Tollinger M. Electrostatic stabilization of a native protein structure in the gas phase. Angew. Chem. 2011;50:873–877. doi: 10.1002/anie.201005112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ganisl B, Breuker K. Does electron capture dissociation cleave protein disulfide bonds? ChemistryOpen. 2012;1:260–268. doi: 10.1002/open.201200038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skinner OS, McLafferty FW, Breuker K. How ubiquitin unfolds after transfer into the gas phase. J. Am. Soc. Mass Spectrom. 2012;23:1011–1014. doi: 10.1007/s13361-012-0370-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan JX, Heath BL, Jockusch RA, Konermann L. Structural interrogation of electrosprayed peptide ions by gas-phase H/D exchange and electron capture dissociation mass spectrometry. Anal. Chem. 2012;84:373–378. doi: 10.1021/ac202730d. [DOI] [PubMed] [Google Scholar]
- 23.Perot-Taillandier M, Zirah S, Rebuffat S, Linne U, Marahiel MA, Cole RB, Tabet JC, Afonso C. Determination of peptide topology through time-resolved double-resonance under electron capture dissociation conditions. Anal. Chem. 2012;84:4957–4964. doi: 10.1021/ac300607y. [DOI] [PubMed] [Google Scholar]
- 24.Le TN, Poully JC, Lecomte F, Nieuwjaer N, Manil B, Desfrancois C, Chirot F, Lemoine J, Dugourd P, van der Rest G, Gregoire G. Gas-phase structure of amyloid-beta (12–28) peptide investigated by infrared spectroscopy, electron capture dissociation, and ion mobility mass spectrometry. J. Am. Soc. Mass Spectrom. 2013;24:1937–1949. doi: 10.1007/s13361-013-0722-x. [DOI] [PubMed] [Google Scholar]
- 25.Kalapothakis JMD, Berezovskaya Y, Zampronio CG, Faull PA, Barran PE, Cooper HJ. Unusual ECD fragmentation attributed to gas-phase helix formation in a conformationally dynamic peptide. Chem. Commun. 2014;50:198–200. doi: 10.1039/C3CC46356G. [DOI] [PubMed] [Google Scholar]
- 26.Yin S, Loo JA. Elucidating the site of protein-ATP binding by top-down mass spectrometry. J. Am. Soc. Mass Spectrom. 2010;21:899–907. doi: 10.1016/j.jasms.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 27.Clarke DJ, Murray E, Hupp T, Mackay CL, Langridge-Smith PRR. Mapping a noncovalent protein–peptide interface by top-down FTICR mass spectrometry using electron capture dissociation. J. Am. Soc. Mass Spectrom. 2011;22:1432–1440. doi: 10.1007/s13361-011-0155-3. [DOI] [PubMed] [Google Scholar]
- 28.Enyenihi AA, Yang HQ, Ytterberg AJ, Lyutvinskiy Y, Zubarev RA. Heme binding in gas-phase holo-myoglobin cations: distal becomes proximal? J. Am. Soc. Mass Spectrom. 2011;22:1763–1770. doi: 10.1007/s13361-011-0182-0. [DOI] [PubMed] [Google Scholar]
- 29.Heath BL, Jockusch RA. Ligand migration in the gaseous insulin-Cb7 complex-a cautionary tale about the use of ECD-MS for ligand binding site determination. J. Am. Soc. Mass Spectrom. 2012;23:1911–1920. doi: 10.1007/s13361-012-0470-3. [DOI] [PubMed] [Google Scholar]
- 30.Zhang H, Cui WD, Gross ML, Blankenship RE. Native mass spectrometry of photosynthetic pigment–protein complexes. FEBS Lett. 2013;587:1012–1020. doi: 10.1016/j.febslet.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li HL, Wolff JJ, Van Orden SL, Loo JA. Native top-down electrospray ionization-mass spectrometry of 158 kDa protein complex by high-resolution Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 2014;86:317–320. doi: 10.1021/ac4033214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Breuker K, McLafferty FW. Native electron capture dissociation for the structural characterization of noncovalent interactions in native cytochrome c. Angew. Chem. 2003;42:4900–4904. doi: 10.1002/anie.200351705. [DOI] [PubMed] [Google Scholar]
- 33.Simons J. Mechanisms for S–S and N–C–alpha bond cleavage in peptide ECD and ETD mass spectrometry. Chem. Phys. Lett. 2010;484:81–95. doi: 10.1016/j.cplett.2009.10.062. [DOI] [Google Scholar]
- 34.Simons J, Ledvina AR. Spatial extent of fragment-ion abundances in electron transfer dissociation and electron capture dissociation mass spectrometry of peptides. Int. J. Mass Spectrom. 2012;330:85–94. doi: 10.1016/j.ijms.2012.07.014. [DOI] [Google Scholar]
- 35.Turecek F, Moss CL, Chung TW. Correlating ETD fragment ion intensities with peptide ion conformational and electronic structure. Int. J. Mass Spectrom. 2012;330:207–219. doi: 10.1016/j.ijms.2012.08.001. [DOI] [Google Scholar]
- 36.Ben Hamidane H, He H, Tsybin OY, Emmett MR, Hendrickson CL, Marshall AG, Tsybin YO. Periodic sequence distribution of product ion abundances in electron capture dissociation of amphipathic peptides and proteins. J. Am. Soc. Mass Spectrom. 2009;20:1182–1192. doi: 10.1016/j.jasms.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 37.Jones AW, Cooper HJ. Probing the mechanisms of electron capture dissociation mass spectrometry with nitrated peptides. Phys. Chem. Chem. Phys. 2010;12:13394–13399. doi: 10.1039/c0cp00623h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sohn CH, Chung CK, Yin S, Ramachandran P, Loo JA, Beauchamp JL. Probing the mechanism of electron capture and electron transfer dissociation using tags with variable electron affinity. J. Am. Chem. Soc. 2009;131:5444–5459. doi: 10.1021/ja806534r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoo HJ, Wang N, Zhuang SY, Song HT, Håkansson K. Negative-ion electron capture dissociation: radical-driven fragmentation of charge-increased gaseous peptide anions. J. Am. Chem. Soc. 2011;133:16790–16793. doi: 10.1021/ja207736y. [DOI] [PubMed] [Google Scholar]
- 40.Koeniger SL, Clemmer DE. Resolution and structural transitions of elongated states of ubiquitin. J. Am. Soc. Mass Spectrom. 2007;18:322–331. doi: 10.1016/j.jasms.2006.09.025. [DOI] [PubMed] [Google Scholar]
- 41.Leymarie N, Costello CE, O'Connor PB. Electron capture dissociation initiates a free radical reaction cascade. J. Am. Chem. Soc. 2003;125:8949–8958. doi: 10.1021/ja028831n. [DOI] [PubMed] [Google Scholar]
- 42.Turecek F, Julian RR. Peptide radicals and cation radicals in the gas phase. Chem. Rev. 2013;113:6691–6733. doi: 10.1021/cr400043s. [DOI] [PubMed] [Google Scholar]
- 43.Prell JS, O'Brien JT, Holm AIS, Leib RD, Donald WA, Williams ER. Electron capture by a hydrated gaseous peptide: effects of water on fragmentation and molecular survival. J. Am. Chem. Soc. 2008;130:12680–12689. doi: 10.1021/ja8022434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Available at: http://webbook.nist.gov. Accessed 14 Jan 2015
- 45.Valadbeigi Y, Farrokhpour H. DFT, CBS-Q, W1BD, and G4MP2 calculation of the proton and electron affinities, gas phase basicities, and ionization energies of saturated and unsaturated carboxylic acids (C1–C4) Int. J. Quantum Chem. 2013;113:1717–1721. doi: 10.1002/qua.24391. [DOI] [Google Scholar]
- 46.Zhan CG, Nichols JA, Dixon DA. Ionization potential, electron affinity, electronegativity, hardness, and electron excitation energy: molecular properties from density functional theory orbital energies. J. Phys. Chem. A. 2003;107:4184–4195. doi: 10.1021/jp0225774. [DOI] [Google Scholar]
- 47.Prell JS, O’Brien JT, Steill JD, Oomens J, Williams ER. Structures of protonated dipeptides: the role of arginine in stabilizing salt bridges. J. Am. Chem. Soc. 2009;131:11442–11449. doi: 10.1021/ja901870d. [DOI] [PubMed] [Google Scholar]
- 48.Zubarev RA, Haselmann KF, Budnik B, Kjeldsen F, Jensen F. Towards an understanding of the mechanism of electron-capture dissociation: a historical perspective and modern ideas. Eur. J. Mass Spectrom. 2002;8:337–349. doi: 10.1255/ejms.517. [DOI] [Google Scholar]
- 49.Zhang X, Cassady CJ. Appararent gas-phase acidities of multiply protonated peptide ions: ubiquitin, insulin B, and renin substrate. J. Am. Soc. Mass Spectrom. 1996;7:1211–1218. doi: 10.1016/S1044-0305(96)00110-9. [DOI] [PubMed] [Google Scholar]
- 50.Chang TM, Prell JS, Warrick ER, Williams ER. Where’s the charge? Protonation sites in gaseous ions change with hydration. J. Am. Chem. Soc. 2012;134:15805–15813. doi: 10.1021/ja304929h. [DOI] [PubMed] [Google Scholar]
- 51.Warnke S, von Helden G, Pagel K. Protein structure in the gas phase: the influence of side-chain microsolvation. J. Am. Chem. Soc. 2013;135:1177–1180. doi: 10.1021/ja308528d. [DOI] [PubMed] [Google Scholar]
- 52.Johnson AR, Dilger JM, Glover MS, Clemmer DE, Carlson EE. Negatively-charged helices in the gas phase. Chem. Commun. 2014;50:8849–8851. doi: 10.1039/C4CC03257H. [DOI] [PubMed] [Google Scholar]
- 53.Breuker K, McLafferty FW. Stepwise evolution of protein native structure with electrospray into the gas phase, 10–12 to 102 s. Proc. Natl. Acad. Sci. U.S.A. 2008;105:18145–18152. doi: 10.1073/pnas.0807005105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Silveira JA, Fort KL, Kim D, Servage KA, Pierson NA, Clemmer DE, Russell DH. From solution to the gas phase: stepwise dehydration and kinetic trapping of substance P reveals the origin of peptide conformations. J. Am. Chem. Soc. 2013;135:19147–19153. doi: 10.1021/ja4114193. [DOI] [PubMed] [Google Scholar]
- 55.Mentinova M, Crizer DM, Baba T, McGee WM, Glish GL, McLuckey SA. Cation recombination energy/Coulomb repulsion effects in ETD/ECD as revealed by variation of charge per residue at fixed total charge. J. Am. Soc. Mass Spectrom. 2013;24:1676–1689. doi: 10.1007/s13361-013-0606-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li JW, Taraszka JA, Counterman AE, Clemmer DE. Influence of solvent composition and capillary temperature on the conformations of electrosprayed ions: unfolding of compact ubiquitin conformers from pseudonative and denatured solutions. Int. J. Mass Spectrom. 1999;187:37–47. doi: 10.1016/S1387-3806(98)14135-0. [DOI] [Google Scholar]
- 57.Lifshitz C. Kinetic shifts. Eur. J. Mass Spectrom. 2002;8:85–98. doi: 10.1255/ejms.476. [DOI] [Google Scholar]
- 58.Toker, Y., Aviv, O., Eritt, M., Rappaport, M.L., Heber, O., Schwalm, D., Zajfman, D.: Radiative cooling of Al4– clusters. Phys. Rev. A 76, (2007)
- 59.Dunbar RC. Infrared radiative cooling of gas-phase ions. Mass Spectrom. Rev. 1992;11:309–339. doi: 10.1002/mas.1280110403. [DOI] [Google Scholar]
- 60.Black DM, Payne AH, Glish GL. Determination of cooling rates in a quadrupole ion trap. J. Am. Soc. Mass Spectrom. 2006;17:932–938. doi: 10.1016/j.jasms.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 61.Peltz C, Drahos L, Vekey K. SORI excitation: collisional and radiative processes. J. Am. Soc. Mass Spectrom. 2007;18:2119–2126. doi: 10.1016/j.jasms.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 62.Meot-Ner M, Somogyi A. A thermal extrapolation method for the effective temperatures and internal energies of activated ions. Int. J. Mass Spectrom. 2007;267:346–356. doi: 10.1016/j.ijms.2007.04.007. [DOI] [Google Scholar]
- 63.Herrmann KA, Somogyi A, Wysocki VH, Drahos L, Vekey K. Combination of sustained off-resonance irradiation and on-resonance excitation in FT-ICR. Anal. Chem. 2005;77:7626–7638. doi: 10.1021/ac050828+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schnier PD, Price WD, Strittmatter EF, Williams ER. Dissociation energetics and mechanisms of leucine enkephalin (M + H)+ and (2 M + X)+ ions (X = H, Li, Na, K, and Rb) measured by blackbody infrared radiative dissociation. J. Am. Soc. Mass Spectrom. 1997;8:771–780. doi: 10.1016/S1044-0305(97)84129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McLuckey SA, Goeringer DE. Slow heating methods in tandem mass spectrometry. J. Mass Spectrom. 1997;32:461–474. doi: 10.1002/(SICI)1096-9888(199705)32:5<461::AID-JMS515>3.0.CO;2-H. [DOI] [Google Scholar]
- 66.Schnier PD, Jurchen JC, Williams ER. The effective temperature of peptide ions dissociated by sustained off-resonance irradiation collisional activation in Fourier transform mass spectrometry. J. Phys. Chem. B. 1999;103:737–745. doi: 10.1021/jp9833193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Antoine R, Broyer M, Chamot-Rooke J, Dedonder C, Desfrancois C, Dugourd P, Gregoire G, Jouvet C, Onidas D, Poulain P, Tabarin T, van der Rest G. Comparison of the fragmentation pattern induced by collisions, laser excitation and electron capture. Influence of the initial excitation. Rapid Commun. Mass Spectrom. 2006;20:1648–1652. doi: 10.1002/rcm.2489. [DOI] [PubMed] [Google Scholar]
- 68.Cooper HJ. Investigation of the presence of b ions in electron capture dissociation mass spectra. J. Am. Soc. Mass Spectrom. 2005;16:1932–1940. doi: 10.1016/j.jasms.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 69.Price WD, Schnier PD, Jockusch RA, Strittmatter EF, Williams ER. Unimolecular reaction kinetics in the high-pressure limit without collisions. J. Am. Chem. Soc. 1996;118:10640–10644. doi: 10.1021/ja961812r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Price WD, Williams ER. Activation of peptide ions by blackbody radiation: factors that lead to dissociation kinetics in the rapid energy exchange limit. J. Phys. Chem. A. 1997;101:8844–8852. doi: 10.1021/jp9722418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Simons J. Analytical model for rates of electron attachment and intramolecular electron transfer in electron transfer dissociation mass spectrometry. J. Am. Chem. Soc. 2010;132:7074–7085. doi: 10.1021/ja100240f. [DOI] [PubMed] [Google Scholar]
- 72.Antoine R, Dugourd P. Visible and ultraviolet spectroscopy of gas phase protein ions. Phys. Chem., Chem. Phys. 2011;13:16494–16509. doi: 10.1039/c1cp21531k. [DOI] [PubMed] [Google Scholar]
- 73.Lifshitz C. Intramolecular Vibrational Energy Distribution and Ergodicity of Biomolecular Dissociation. In: Principles of Mass Spectrometry Applied to Biomolecules. Hoboken: Wiley; 2006. [Google Scholar]
- 74.Zubarev RA. Reactions of polypeptide ions with electrons in the gas phase. Mass Spectrom. Rev. 2003;22:57–77. doi: 10.1002/mas.10042. [DOI] [PubMed] [Google Scholar]
- 75.Turecek F, Chung TW, Moss CL, Wyer JA, Ehlerding A, Holm AIS, Zettergren H, Nielsen SB, Hvelplund P, Chamot-Rooke J, Bythell B, Paizs B. The histidine effect. Electron transfer and capture cause different dissociations and rearrangements of histidine peptide cation-radicals. J. Am. Chem. Soc. 2010;132:10728–10740. doi: 10.1021/ja907808h. [DOI] [PubMed] [Google Scholar]
- 76.Hall Z, Robinson CV. Do charge state signatures guarantee protein conformations? J. Am. Soc. Mass Spectrom. 2012;23:1161–1168. doi: 10.1007/s13361-012-0393-z. [DOI] [PubMed] [Google Scholar]
- 77.Badman ER, Hoaglund-Hyzer CS, Clemmer DE. Dissociation of different conformations of ubiquitin ions. J. Am. Soc. Mass Spectrom. 2002;13:719–723. doi: 10.1016/S1044-0305(02)00374-4. [DOI] [PubMed] [Google Scholar]
- 78.Skinner OS, Breuker K, McLafferty FW. Charge site mass spectra: conformation-sensitive components of the electron capture dissociation spectrum of a protein. J. Am. Soc. Mass Spectrom. 2013;24:807–810. doi: 10.1007/s13361-013-0603-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins: Struct., Funct., Genet. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schennach M, Breuker K. Proteins with highly similar native folds can show vastly dissimilar folding behavior when desolvated. Angew. Chem. 2014;53:164–168. doi: 10.1002/anie.201306838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Breuker K. Protein Structure and Folding in the Gas Phase: Ubiquitin and Cytochrome c. In: Principles of Mass Spectrometry Applied to Biomolecules. Hoboken: Wiley; 2006. [Google Scholar]
- 82.Zhang Z, Browne SJ, Vachet RW. Exploring salt bridge structures of gas-phase protein ions using multiple stages of electron transfer and collision induced dissociation. J. Am. Soc. Mass Spectrom. 2014;25:604–613. doi: 10.1007/s13361-013-0821-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Myung S, Badman ER, Lee YJ, Clemmer DE. Structural transitions of electrosprayed ubiquitin ions stored in an ion trap over ~10 ms to 30 s. J. Phys. Chem. A. 2002;106:9976–9982. doi: 10.1021/jp0206368. [DOI] [Google Scholar]
- 84.Badman ER, Myung S, Clemmer DE. Evidence for unfolding and refolding of gas-phase cytochrome c ions in a Paul trap. J. Am. Soc. Mass Spectrom. 2005;16:1493–1497. doi: 10.1016/j.jasms.2005.04.013. [DOI] [PubMed] [Google Scholar]