Abstract
The mechanisms by which multi-potent stem cells switch their program to become functional differentiated cells have intrigued biologists for decades. Most focus has been on transcriptional pathways, but whether they have sufficient dynamic range to cause discrete shifts in cell state is not clear. Because the steady-state level of RNAs is also dictated by their decay rate, an attractive possibility is that specific RNA decay mechanisms also have a role in promoting differentiation mechanisms. In this issue of The EMBO Journal, Li et al (2015) obtained evidence that a highly conserved RNA degradation pathway called nonsense-mediated RNA decay (NMD) is critical for the differentiation of embryonic stem (ES) cells.
See also: T Li et al (June 2015)
NMD was originally identified as a quality control pathway that rapidly degrades aberrant transcripts harboring premature stop (nonsense) codons (Schweingruber et al, 2013). Recent studies have shown that NMD also degrades normal mRNAs with in-frame stop codon if in an appropriate context; for example, upstream of an exon–exon junction or a long 3′ untranslated region (UTR). The discovery that NMD is not only an RNA surveillance pathway but a regulator of normal gene expression has raised the possibility that NMD regulates normal biological processes, including development, a scientific problem that Li et al addresses. Their studies focus on SMG6, an endonuclease that degrades a subset of NMD target mRNAs near the site of the in-frame stop codon (Schweingruber et al, 2013). To study its role, Li et al generated knockout mice lacking SMG6. Consistent with earlier studies on mice harboring debilitating mutations in other NMD factor genes (Hwang & Maquat, 2011), Li et al found that Smg6-null mice suffer from early embryonic lethality. To examine the underlying mechanism, the authors turned to mouse ES cells. An earlier study observed that stable depletion of the central NMD factor, UPF1, had no measurable deleterious effects on undifferentiated mouse ES cells (Hurt et al, 2013). In agreement with this, Li et al found that undifferentiated Smg6-null ES cells were normal in every way that they examined, including proliferation rate and expression of pluripotency markers. However, they observed that the loss of SMG6 caused a profound defect in the ability of these cells to differentiate (Fig1A). In vitro experiments showed that Smg6-null ES cells cultured under conditions that normally promote embryoid body formation (removal of LIF) or lineage-specific differentiation events (addition of retinoic acid or DMSO) failed to acquire differentiation markers and instead maintained high levels of pluripotency markers, such as OCT4. In vivo chimera analysis showed that Smg6-null ES cells lost the ability to differentiate into all three germ layers (Fig1A). Together, these experiments indicated that SMG6 acts in a cell-autonomous manner in early development and it suggested that the early embryonic lethality in Smg6-null mice might result from a failure of early embryonic cells to properly undergo differentiation.
Figure 1. The NMD pathway influences differentiation decisions.
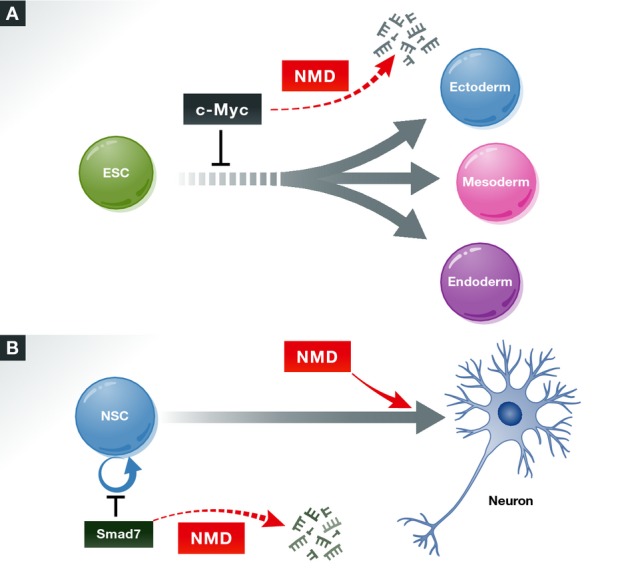
(A) NMD promotes the differentiation of embryonic stem cells (ESCs) into the three primary germ layers, in part, by promoting the decay of c-mycmRNA, which encodes an anti-differentiation/pro-proliferation factor (Li et al, 2015). (B) NMD factors have complex effects on the differentiation of neural lineage cells. The central NMD factor, UPF1, inhibits the differentiation of neural stem cells through its ability to promote the decay of the mRNA encoding the pro-neural differentiation factor SMAD7 (Lou et al, 2014). A NMD factor required for normal human cognition—UPF3B—appears to promote neural precursor differentiation and is required for neural maturation (Jolly et al, 2013).
While these data clearly showed that SMG6 is critical for ES cell differentiation in vitro, can one conclude that NMD is responsible? This answer is ‘no’, because, like most NMD factors, SMG6 has biochemical activities in addition to NMD. Most notably, SMG6 is known to interact with telomeric regions of chromosomes and promote telomerase activity (Sealey et al, 2011). To distinguish between SMG6 acting through its telomerase- or NMD-promoting activity to drive differentiation, Li et al performed ‘rescue experiments’ with mutant forms of SMG6. They found that mutants lacking telomerase-promoting activity, but not those lacking NMD-promoting activity, rescued ES cell differentiation. This provided evidence that the NMD-promoting activity of SMG6 is responsible for driving ES differentiation. As further evidence, the authors found that depletion of several NMD factors in addition to SMG6 (UPF1, UPF2, SMG1, and SMG5) caused a defect in ES cell differentiation. To our knowledge, this is the first case in which decay and non-RNA decay activities of a NMD factor have been functionally dissected. This is a major breakthrough for the field; it may lead to similar efforts by other investigators to dissect the various biological and biochemical roles of NMD factors.
How does NMD drive ES cell differentiation? Because NMD elicits the decay of specific subsets of mRNAs, an intriguing possibility is that NMD promotes the decay of mRNAs encoding pluripotency factors. In general agreement with this, Li et al found—through RNA-seq analysis—that transcripts encoding factors important for embryonic development and differentiation were overrepresented among those differentially expressed in response to the loss of SMG6. Among the pluripotency genes differentially expressed was c-myc, which they regarded as a particularly attractive candidate to be an effector of NMD for several reasons. First, C-MYC is a major pluripotency factor with well-defined actions that functions in a wide variety of cellular contexts (Chappell & Dalton, 2013). Second, the authors found that many SMG6-regulated mRNAs had previously been shown to also be regulated by C-MYC in other contexts. Third, they observed that c-myc mRNA regulation was specifically elicited by SMG6 mutants with the NMD-promoting domain intact. To assess whether C-MYC has a functional role downstream of SMG6, the authors performed both mimic and rescue experiments. They found that overexpressing C-MYC in normal ES cells inhibited their ability to differentiate, thereby mimicking the differentiation defect in Smg6-null cells, which express elevated levels of C-MYC. Reversing the abnormally high expression of C-MYC in Smg6-null ES cells (using RNA interference) caused these cells to acquire the ability to differentiate. Together with evidence that c-myc mRNA is a direct NMD target, the authors' data supported the existence of a NMD-based molecular circuit involving c-myc that is critical for ES cell differentiation (Fig1A). This follows the recent discovery of other NMD-based circuits—one critical for maintaining the neural stem cell state and the other shaping the unfolded protein response (UPR)—through decay of the mRNAs encoding the TGF-β signaling inhibitor SMAD7 and the conserved UPR sensor IRE1α, respectively (Lou et al, 2014; Karam et al, 2015).
Are the findings of Li et al in differentiating mouse ES cells generalizable to other developmental systems? While time will tell, early indications are both ‘yes’ and ‘no’. On the one hand, evidence suggests that NMD drives the maturation of neural cells (Fig1B) and possibly muscle lineage cells (Gong et al, 2009; Jolly et al, 2013). However, another study demonstrated that NMD inhibits neural differentiation and instead is critical for maintaining the stem-like state of multi-potent and neural cells (Lou et al, 2014) (Fig1B). While seemingly contradictory, these studies likely reflect that the influence of NMD on developmental decisions depends on the developmental stage of the cells and the specific transcriptome it acts on. Indeed, evidence suggests that NMD has branches that act in a tissue- and cell type-specific manner (Huang et al, 2011). It will be fascinating in the future to elucidate the yin and yang roles of NMD in specific differentiation events and how this is subverted in disease.
References
- Chappell J, Dalton S. Roles for MYC in the establishment and maintenance of pluripotency. Cold Spring Harb Perspect Med. 2013;3:a014381. doi: 10.1101/cshperspect.a014381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Kim YK, Woeller CF, Tang Y, Maquat LE. SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs. Genes Dev. 2009;23:54–66. doi: 10.1101/gad.1717309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Lou CH, Chan W, Shum EY, Shao A, Stone E, Karam R, Song HW, Wilkinson MF. RNA homeostasis governed by cell type-specific and branched feedback loops acting on NMD. Mol Cell. 2011;43:950–961. doi: 10.1016/j.molcel.2011.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt JA, Robertson AD, Burge CB. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res. 2013;23:1636–1650. doi: 10.1101/gr.157354.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J, Maquat LE. Nonsense-mediated mRNA decay (NMD) in animal embryogenesis: to die or not to die, that is the question. Curr Opin Genet Dev. 2011;21:422–430. doi: 10.1016/j.gde.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly LA, Homan CC, Jacob R, Barry S, Gecz J. The UPF3B gene, implicated in intellectual disability, autism, ADHD and childhood onset schizophrenia regulates neural progenitor cell behaviour and neuronal outgrowth. Hum Mol Genet. 2013;22:4673–4687. doi: 10.1093/hmg/ddt315. [DOI] [PubMed] [Google Scholar]
- Karam R, Lou CH, Kroeger H, Huang L, Lin JH, Wilkinson MF. The unfolded protein response is shaped by the NMD pathway. EMBO Rep. 2015;16:599–609. doi: 10.15252/embr.201439696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Shi Y, Wang P, Guachalla LM, Sun B, Joerss T, Chen YS, Groth M, Krueger A, Platzer M, Yang YG, Rudolph KL, Wang ZQ. Smg6/Est1 licenses embryonic stem cell differentiation via nonsense-mediated mRNA decay. EMBO J. 2015 doi: 10.15252/embj.201489947. doi: 10.15252/embj.201489947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou CH, Shao A, Shum EY, Espinoza JL, Huang L, Karam R, Wilkinson MF. Posttranscriptional control of the stem cell and neurogenic programs by the nonsense-mediated RNA decay pathway. Cell Rep. 2014;6:748–764. doi: 10.1016/j.celrep.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweingruber C, Rufener SC, Zünd D, Yamashita A, Mühlemann O. Nonsense-mediated mRNA decay - mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim Biophys Acta. 2013;1829:612–623. doi: 10.1016/j.bbagrm.2013.02.005. [DOI] [PubMed] [Google Scholar]
- Sealey DCF, Kostic AD, LeBel C, Pryde F, Harrington L. The TPR-containing domain within Est1 homologs exhibits species-specific roles in telomerase interaction and telomere length homeostasis. BMC Mol Biol. 2011;12:45. doi: 10.1186/1471-2199-12-45. [DOI] [PMC free article] [PubMed] [Google Scholar]


