Abstract
Nonsense-mediated mRNA decay (NMD) is a post-transcriptional mechanism that targets aberrant transcripts and regulates the cellular RNA reservoir. Genetic modulation in vertebrates suggests that NMD is critical for cellular and tissue homeostasis, although the underlying mechanism remains elusive. Here, we generate knockout mice lacking Smg6/Est1, a key nuclease in NMD and a telomerase cofactor. While the complete loss of Smg6 causes mouse lethality at the blastocyst stage, inducible deletion of Smg6 is compatible with embryonic stem cell (ESC) proliferation despite the absence of telomere maintenance and functional NMD. Differentiation of Smg6-deficient ESCs is blocked due to sustained expression of pluripotency genes, normally repressed by NMD, and forced down-regulation of one such target, c-Myc, relieves the differentiation block. Smg6-null embryonic fibroblasts are viable as well, but are refractory to cellular reprograming into induced pluripotent stem cells (iPSCs). Finally, depletion of all major NMD factors compromises ESC differentiation, thus identifying NMD as a licensing factor for the switch of cell identity in the process of stem cell differentiation and somatic cell reprograming.
Keywords: cell reprograming, ESC differentiation, NMD, Smg6/Est1, telomere
See also: C-H Lou et al (June 2015)
Introduction
Embryonic stem cells (ESCs) have two distinctive capacities; the first is to proliferate infinitely (self-renewal) and the other is to generate restricted daughter progenies (differentiation) that form all three germ layers: ectoderm, endoderm, and mesoderm. These characteristics dictate the growth and diversification of tissues and cell types during development. Transcription factors, epigenetic changes, and non-coding RNAs are known mechanisms that maintain the status of ESCs, while also promoting their differentiation (Keller, 2005; He et al, 2009; Young, 2011; Zhou et al, 2011). Historically, ESCs were the main source of pluripotent stem cells (which are derived from the inner cell mass (ICM) of mammalian blastocysts). More recently, induced pluripotent stem cells (iPSCs) have been generated by direct reprograming of differentiated cells (Takahashi & Yamanaka, 2006; Buganim et al, 2013). The elucidation of the cellular and molecular mechanisms that govern the self-renewal and differentiation of stem cells is fundamental to the understanding of embryonic development and tissue homeostasis and has potentially major biomedical implications (Keller, 2005).
Nonsense-mediated mRNA decay (NMD) is an ancient and conserved RNA surveillance and regulatory mechanism (Chawla & Azzalin, 2008; Nicholson et al, 2010; Huang & Wilkinson, 2012; Palacios, 2013). Classically, NMD serves to clean RNA species containing a premature termination codon (PTC) located 50–55 nt before the last exon–exon junction. At the PTC site, the NMD complex, which composes of Smg1, Upf1, Upf2, and Upf3, is assembled, in which Smg1 phosphorylates Upf1 and Upf2, which further recruits and activates either the Smg6 endonuclease or Smg5-/Smg7-mediated exonuclease to process mRNAs for degradation. NMD also targets transcripts containing uORFs or other structures (e.g. 3′-UTR) to regulate mRNA levels (Ruiz-Echevarria & Peltz, 2000; Barbosa et al, 2013). Thus, NMD eliminates or modulates the abundance of aberrant mRNAs, which balances and regulates different isoforms within the total mRNA reservoir (Nicholson et al, 2010; Huang & Wilkinson, 2012). The accumulation of aberrant mRNAs, which can be translated into deleterious truncated proteins, may cause pathological symptoms (Frischmeyer & Dietz, 1999; Hwang & Maquat, 2011; Palacios, 2013). Knockdown of the NMD factors Upf1, Upf2, Smg5, or Smg6 in zebrafish (Wittkopp et al, 2009) and knockout of Smg1, Upf1, or Upf2 in mice (Medghalchi et al, 2001; Weischenfeldt et al, 2008; McIlwain et al, 2010) unanimously result in early embryonic lethality. Thus, NMD is thought to be essential for cellular and tissue homeostasis and ultimately organismal survival in vertebrates. However, due to early lethality and the lack of model vertebrate systems, the underlying mechanism remains unknown.
Interestingly, most NMD factors, such as Smg1, Upf1, Upf2, Smg5, Smg6, and Smg7, are also involved in the telomere maintenance (Chawla & Azzalin, 2008; Hwang & Maquat, 2011), which complicates the interpretation of the phenotypic effects resulted from NMD inactivation mutations. Smg5, Smg6, and Smg7 are the mammalian homologs of Ever shorter telomere 1 (Est1) (Snow et al, 2003), which was originally identified as a telomerase-associated factor in yeast (Lundblad & Szostak, 1989). Yeast Est1 mutants showed a progressive loss of telomeres and restricted cell viability (Lundblad & Szostak, 1989). Est1 bridges between telomerase and Cdc13 (homolog of Pot1 in mammals) and directly binds telomerase RNA, which is critical for both telomerase activation and telomere maintenance (Steiner et al, 1996; Qi & Zakian, 2000; Zhou et al, 2000; Evans & Lundblad, 2002). Among Est1 homologues, the human Smg6 is perhaps the most studied and is associated with telomerase and telomeres (Reichenbach et al, 2003; Snow et al, 2003). Similar to yeast Est1, Smg6 is reported to be involved in the regulation of telomere maintenance and viability of human cancer cells. Paradoxically, overexpression or knockdown of SMG6 in human cells causes the shortening or loss of telomeres and cell cycle arrest (Reichenbach et al, 2003; Azzalin et al, 2007; Chawla & Azzalin, 2008). These studies highlight the important function of Smg6 in various cellular processes.
Results
Smg6 is essential for mouse embryonic development
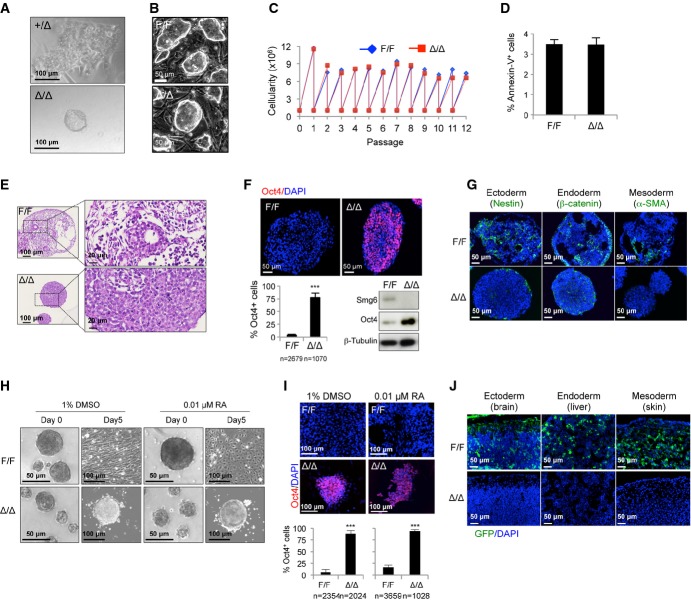
To study the biological function of Smg6, we disrupted the Smg6 locus in the mouse germ line via gene targeting in ESCs (Supplementary Fig S1A). The gene-targeted ESC clones (Smg6+/T) were identified by Southern blotting (Supplementary Fig S1B) and were injected into blastocysts to generate Smg6+/T mice, which were then crossed with FLP transgenic mice to remove the neomycin cassette and generate Smg6+/F mice (Supplementary Fig S1C). Crossing of the Smg6+/F mice with Nestin-Cre transgenic mice generated the deleted allele (Δ) (Supplementary Fig S1C). Intercrossing of Smg6+/Δ mice resulted in no viable Smg6Δ/Δ newborns, but gave rise, albeit rarely, to growth-retarded E7.5 embryos (Supplementary Fig S1D). Although Smg6Δ/Δ blastocysts (E3.5) were morphologically normal, their ICM failed to grow in cultures after 5 days, and thus, no mutant ESCs could be derived (Fig1A).
Figure 1. Smg6-deficient ESCs fail to differentiate in vitro and in vivo.
A In vitro culture of control (+/Δ) and Smg6Δ/Δ (Δ/Δ) blastocysts. Note that there is no ICMs outgrowth of Smg6Δ/Δ blastocysts on day 5 of culture in contrast to the controls.
B Morphology of control (F/F) and Smg6Δ/Δ ESCs.
C Proliferation curve of control (F/F) and Smg6Δ/Δ ESCs.
D Cell death analysis of control (F/F) and Smg6Δ/Δ ESCs by FACS after Annexin-V staining (n = 3 for each genotype).
E H&E staining of control and Smg6Δ/Δ ESCs derived EBs on day 8. Enlarged panels (right) show that control EBs contained differentiated connective tissues and epithelial-like structures and Smg6Δ/Δ EBs contained undifferentiated cells.
F Immunostaining and quantification (top and lower left) and Western blotting (lower right) of Oct4 expression in control and Smg6Δ/Δ ESCs derived EBs on day 8. β-Tubulin was used as a loading control. n, the total number of cells counted from five controls and six Smg6Δ/Δ EBs.
G Immunostaining of ectoderm marker Nestin, endoderm marker β-catenin, and mesoderm marker α-SMA on day 8 of control F/F and Δ/Δ EB samples.
H ESC differentiation upon DMSO and RA treatment. Note that Smg6Δ/Δ ESCs continued to maintain ES-like structure, while control ESCs differentiated into different cell types (fibroblast-like cells by DMSO induction; endothelial cells after RA treatment) on day 5 after plating onto gelatin-coated dishes. The morphology of the EBs before plating was shown (day 0).
I Immunostaining of Oct4 expression in control and Smg6Δ/Δ ESCs derived differentiation cultures treated for DMSO or RA on day 8. The frequency of Oct4+ cells was summarized in low panels. n, the total number of cells used for the quantification. At least three differentiation cultures were used.
J Chimerism assay of the contribution of GFP-labeled control and Smg6Δ/Δ ESC derivatives to different mouse tissues (E12.5 embryos are analyzed). Note the absence of GFP-labeled Smg6Δ/Δ ESC derivatives within chimeras. Representative images of the brain (ectoderm), liver (endoderm), and skin connective tissues (mesoderm) are shown.
Data information: The error bars represent the SEM. Unpaired Student's t-test was used. ***P < 0.001.Source data are available online for this figure.
Smg6Δ/Δ ESCs are viable
To overcome the lethality of the Smg6Δ/Δ embryos, we isolated Smg6F/F blastocysts from the intercrossing of Smg6+/F mice and established conditional Smg6F/F ESC lines (Supplementary Fig S2A). Following the transfection of the GFP-Cre vector into Smg6F/F ESCs, the GFP-positive population was cloned (Supplementary Fig S2A). PCR, Southern blot, and Western blot analyses all confirmed Smg6 deletion in these ESC clones, which were designated as Smg6Δ/Δ ESCs (Supplementary Fig S2B–D). To substantiate this unexpected viability of Smg6Δ/Δ ESCs, we crossed the Smg6+/F mice with CreERT2 mice (Ventura et al, 2007) (Supplementary Fig S1C) and isolated Smg6F/F;CreER+ (designated Smg6-CER) ESCs from the blastocysts derived from the intercrossing of Smg6+/F;CreER+ mice (Supplementary Fig S2A). By the addition of 4-OHT to these cells, we established Smg6Δ/Δ ESCs, which were confirmed by PCR (data not shown) and Western blotting (Supplementary Fig S2E). Smg6Δ/Δ ESCs were morphologically indistinguishable from control ESCs (FigB) and proliferated normally when compared to controls (Fig1C). Furthermore, cell cycle analysis revealed similar frequencies of cells in G1, S, and G2/M phases in control and Smg6Δ/Δ ESC cultures (Supplementary Fig S2F). Finally, Smg6Δ/Δ ESCs did not undergo obvious cell death as measured by FACS analysis, after Annexin-V antibody staining (Fig1D). Thus, we conclude that Smg6 is dispensable for ESC viability and self-renewal.
Smg6Δ/Δ ESCs fail to differentiate in vitro and in vivo
The fact that Smg6Δ/Δ ESCs are viable and Smg6Δ/Δ embryos die soon after implantation raised the possibility that Smg6 deletion blocks ESC differentiation. To investigate this, we utilized several approaches to investigate the differentiation capacity of Smg6Δ/Δ ESCs (Supplementary Fig S3A). Under spontaneous differentiation conditions (the removal of leukemia inhibitory factor (LIF) and feeders), about 50% of Smg6Δ/Δ cultures maintained the ESC colony morphology on day 3, in contrast to 3% in controls (Supplementary Fig S3B and C). On day 6 of differentiation, almost all Smg6Δ/Δ ESC cultures were positive for alkaline phosphatase (AP) (a stem cell marker), whereas only scattered AP-positive colonies were seen in control ESC cultures (Supplementary Fig S3B). Secondly, we applied the in vitro embryoid body (EB) formation assay (Kurosawa, 2007) by culturing the ESCs in Petri dishes without LIF. The Smg6Δ/Δ EB size did increase during differentiation, but they were always smaller than control EBs (Supplementary Fig S3D). This size difference was not caused by impaired proliferation because 5-ethynyl-2′-deoxyuridine (EdU) pulse labeling detected an even higher proliferation rate (seen by EdU-positive cells) in Smg6Δ/Δ EBs (Supplementary Fig S3E). Also, the frequencies of TUNEL-positive and cleaved caspase-3-positive cells were similar in both control and Smg6Δ/Δ EBs, indicating normal apoptosis in Smg6Δ/Δ EBs (Supplementary Fig S3F). Histological analysis of the EBs on day 8 revealed that control ESCs formed an EB structure which contained a characteristic cystic structure and differentiated cells (FigE). In contrast, Smg6Δ/Δ EBs were composed of a high density of cells with a typical undifferentiated cell morphology (Fig1E). These mutant EBs expressed a high level of the stem cell marker Oct4 (Fig1F), but were devoid of the differentiation markers of different germ layers, that is, ectoderm (Nestin) and mesoderm (α-SMA filament), but exhibited an aberrant expression pattern of the endoderm marker β-catenin (Fig1G). In normal wild-type ESC colonies, β-catenin and Oct4 are co-expressed, while in differentiated EBs, β-catenin localizes to the outer layer, and the Oct4 signal is lost (Supplementary Fig S3G). In contrast to control EBs, expression patterns of β-catenin and Oct4 overlapped in mutant EBs in a similar pattern as ESC colonies (Supplementary Fig S3G).
To further confirm that the differentiation block was caused by Smg6 deletion, we forced EB differentiation via DMSO, which triggers fibroblastic or muscle-like cell differentiation, and retinoic acid (RA), which induced both ectoderm and endoderm differentiation in controls. In DMSO-treated samples, all control EBs showed fibroblast-like morphology, whereas all Smg6Δ/Δ EBs maintained ESC colony morphology (Fig1H). After RA treatment, 73% of control EBs differentiated, while only 8% of Smg6Δ/Δ EBs showed minor differentiation (Fig1H). Immunostaining showed that Smg6Δ/Δ EBs contained high levels of Oct4+ cells in comparison with controls, indicative of a failure to undergo differentiation (Fig1I).
To test whether the differentiation defect in Smg6Δ/Δ ESCs is cell autonomous, we performed an in vivo chimera assay. To this end, we injected GFP-labeled control (Smg6-CER ESCs without 4-OHT treatment) or Smg6Δ/Δ ESCs (Smg6-CER ESCs with 5 days of 4-OHT treatment, or constitutive Smg6 deletion) into wild-type blastocysts to generate chimeras (Supplementary Fig S3A). The GFP+ cell contribution within various tissues of different germ layer origins [e.g. brain (ectoderm), liver (endoderm), and skin connective tissues (mesoderm)] can then be scored. In contrast to control ESCs, GFP+ cells were barely detectable in embryos injected with Smg6Δ/Δ ESCs (Fig1J). In total, we injected seven control and six Smg6Δ/Δ ESC lines and detected a GFP signal within a high frequency of chimeric embryos (28/45) from control ESCs and only 1 out of 33 chimeras derived from Smg6Δ/Δ ESCs (Table1). Collectively, these data demonstrate that the deletion of Smg6 blocks ESC differentiation in vitro and in vivo due to an intrinsic mechanism.
Table 1.
Summary of chimerism analysis of control (F/F) and Smg6Δ/Δ (Δ/Δ) ESCs
| Parental ES cell clones | 4-OHT (genotype) | No. of ES clones injected | No. GFP+ embryos/total analyzeda | |
|---|---|---|---|---|
| E12 (Smg6F/F) | − (F/F) | 2 | 8/12 | |
| + (Δ/Δ) | 2 | 0/10 | ||
| E6 (Smg6F/F) | − (F/F) | 3 | 11/18 | |
| + (Δ/Δ) | 2 | 0/10 | ||
| E22 (Smg6-CER) | E22-9 + CAG-GFP | − (F/F) | 1 | 4/9 |
| E22-9 + CAG-GFP | + (Δ/Δ) | 1 | 1/5a | |
| E22-5 + CAG-GFP | − (F/F) | 1 | 5/6 | |
| E22-5 + CAG-GFP | + (Δ/Δ) | 1 | 0/8 | |
| Summary | Control | F/F | 7 | 28/45 (62.2%) |
| Δ/Δ | Δ/Δ | 6 | 1/33 (3%) | |
All embryos were analyzed at E12.5.
One embryo showed a trace amount of GFP signal, possibly due to incomplete deletion by 4-OHT treatment.
Smg6 deletion is compatible with somatic cell survival, but compromises somatic cell reprograming
Because Smg6 deletion blocked stem cell differentiation, somatic cells could not be obtained, thus providing a fitting explanation for the cause of embryonic lethality. To test whether Smg6 is also essential for differentiated cells, we isolated Smg6F/F;CreER+ (Smg6-CER) MEFs from E13.5 embryos. After 4-OHT treatment for 9 days, we established stable MEF clones with Smg6 deletion (designated Smg6Δ/Δ MEFs), as confirmed by both PCR and Western blotting (Supplementary Fig S4A and B). This indicates that Smg6 is dispensable for somatic cell viability.
Given the essential role of Smg6 in the process of ESC differentiation, we asked whether Smg6 also controls the reverse process, namely dedifferentiation of somatic cells. To this end, we used an OSKM (Oct4-Sox2-Klf4-Myc)-expressing vector to induce iPSCs from MEFs (Supplementary Fig S4C). After OSKM transduction, Smg6F/F cultures showed many AP+ ESC colonies, whereas Smg6Δ/Δ MEFs gave rise to only small and scattered ESC appearance in the dish (Fig2A), which tended to be lost after clonal expansion (Fig2A and D and data not shown). These iPSC clones were not fully reprogramed because they had an insufficient reactivation of endogenous Oct4, Sox2, and Nanog (Fig2B and C). The smaller size and less ESC-like appearance of colonies from Smg6Δ/Δ MEFs reflected an inefficient iPSC production, because they contained less proliferating iPSCs and thus formed smaller colonies. Thus, Smg6 deletion severely compromises the reprograming efficiency of MEF cells.
Figure 2. Smg6 deficiency compromises the cellular reprograming of MEFs.
A Representative phase contrast images and AP staining of iPSC cultures from control (Smg6F/F, F/F) and Smg6Δ/Δ (Δ/Δ) MEFs 14 days after OSKM transduction.
B Western blot analysis of individual iPSC clones derived from control (B7, B9, A11) and Smg6Δ/Δ (C2, C12, D4) MEFs. Expression levels of Oct4, Sox2, and Nanog in MEFs after OSKM transduction are shown. Note the reactivation of endogenous Oct4 and Sox2 in control iPSC clones derived from MEFs and to a lesser extent in the mutant clones. MEFs before OSKM transduction and E14.1 ESCs were used as the negative and positive control, respectively. β-Actin was used as a loading control. Endo, endogenous; Exo, exogenous.
C Summary of iPSC generation efficiency characterized by ESC-like morphology and reactivation of endogenous Oct4 expression from either immortalized MEFs.
D Characterization of stable iPSCs after expansion and passaging of individual clones. Note the AP staining of ES-like colonies only in control, but not in Smg6Δ/Δ iPSC cultures (passage 9). H&E staining of teratomas induced by the respective iPSCs was shown on the right. Control (F/F)-derived teratomas contained endoderm-, mesoderm-, and ectoderm-derived tissue structures, while tumors of Smg6Δ/Δ origin contained only poorly differentiated cells. Ecto, ectodermal structure; Meso, mesodermal structure; Endo, endodermal structure.
Source data are available online for this figure.
Next, we investigated whether Smg6 deletion similarly restricts the differentiation of iPSCs. To test this, we performed a teratoma assay using iPSC clones. In stark contrast to the teratomas that formed from control iPSCs, which contained tissues derived from all three germ layers, Smg6Δ/Δ iPSC clones formed tumors that only contained structures characteristic of undifferentiated cells (FigD). From a total collection of five mutant iPSC clones, none of them gave rise to any cellular derivatives of the different germ layers in the teratoma assay (Supplementary Fig S4D). Furthermore, only the control MEFs-derived iPSCs, but not the mutants, contributed to the formation of chimeric mice when injected into blastocysts (Supplementary Fig S4E). These studies further confirmed that Smg6 is an essential factor for differentiation, even in reprogramed pluripotent stem cells.
Smg6Δ/Δ ES and MEF cells are defective in telomere maintenance and NMD
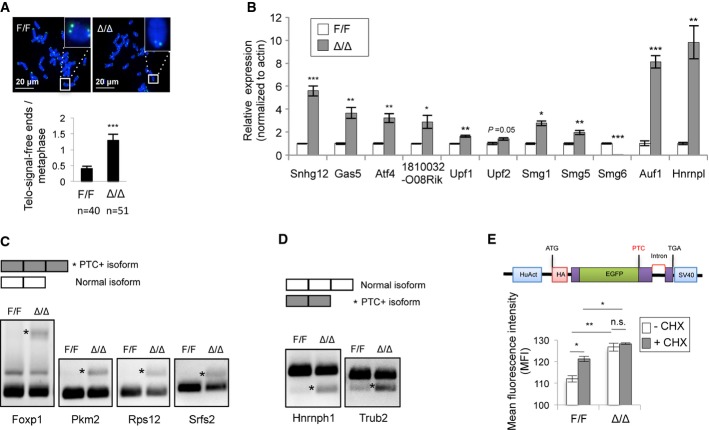
Smg6 functions in both telomere maintenance and NMD (Chawla & Azzalin, 2008; Hwang & Maquat, 2011; Schweingruber et al, 2013). We next studied which possible functions of Smg6 are responsible for the cell identity transition. First, we performed telomere FISH analysis in ESCs. Although Smg6Δ/Δ ESCs have a similar karyotype as compared to control ESCs (Supplementary Fig S2G), they contained a higher number of chromosomes lacking or expressing low telomere signals, that suggests a compromised telomere integrity (Fig3A). This is consistent with the notion that Smg6 is an important regulator in telomere maintenance (Reichenbach et al, 2003; Snow et al, 2003).
Figure 3. Smg6 is required for telomere maintenance and NMD in ESCs.
A Telomere FISH analysis of control (F/F) and Smg6Δ/Δ (Δ/Δ) ESC metaphases. Representative images (up) and quantification of chromosomes lacking telomere signals (telomere-signal-free ends) (lower panel) are shown. n, the number of metaphases analyzed.
B qRT–PCR analysis of NMD target transcripts in control and Smg6Δ/Δ ESCs. The expression levels of the NMD target genes were normalized to β-actin. The data are from three independent biological samples.
C RT–PCR analysis of exon inclusion generated PTC-containing (*PTC+) isoforms in control and Smg6Δ/Δ ESCs.
D RT–PCR analysis of exon exclusion generated PTC-containing (*PTC+) isoforms in control and Smg6Δ/Δ ESCs.
E Smg6Δ/Δ ESCs are NMD defective. ESCs were transfected with the NMD reporter (upper panel) and analyzed in triplicate by FACS. The GFP signal intensity determines the NMD activity. CHX was used to inhibit NMD. The data represent one of two independent ES clones of each genotype.
Data information: The error bars represent the SEM. Unpaired Student's t-test was used. n.s., not significant, P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001.Source data are available online for this figure.
To examine whether Smg6-null background would affect the NMD function, we first analyzed the expression of endogenous NMD target transcripts, for example, Snhg12,Gas5,Atf4,1810032O08Rik (Weischenfeldt et al, 2008), Smg1,Smg5,Smg6,Upf1, Upf2 (Huang et al, 2011), Auf1, and Hnrnpl (http://www.ensembl.org/Mus_musculus/Info/Index). Quantitative RT–PCR (qRT–PCR) analysis showed a considerable up-regulation of these transcripts within Smg6Δ/Δ ESCs (Fig3B). Alternative splicing events can result in PTC-containing isoforms through either exon inclusion or exon exclusion. We next analyzed the alternative splicing coupled NMD (AS-NMD) in ESCs (Weischenfeldt et al, 2012) by RT–PCR analysis. As shown in Fig3C and D, Smg6 deletion significantly increased the NMD target transcripts with features of both exon inclusion (Foxp1, Pkm2, Rps12, Srfs2) and exon exclusion (Hnrnph1, Trub2). To further test the NMD capacity in these knockout cells, we stably transfected Smg6-CER ESCs with an NMD reporter (Paillusson et al, 2005), in which the NMD activity reversely correlates with the GFP signal intensity. Deletion of Smg6 by 4-OHT treatment significantly enhanced the GFP intensity, similar to control samples that were treated with the NMD inhibitor cycloheximide (CHX) (Fig3F), which indicated a NMD activity deficiency in Smg6Δ/Δ ESCs. Taken together, Smg6 deletion compromises NMD in ESCs.
Similar to that of ESCs, Smg6Δ/Δ MEFs contained a high frequency of chromosomes with little or no telomere signals (Supplementary Fig S4F). qRT–PCR analysis also showed an up-regulation of NMD target genes in Smg6Δ/Δ MEFs (Supplementary Fig S4G), suggesting that Smg6 is involved in the NMD process of differentiated cells. Thus, Smg6 participates in both telomere maintenance and NMD in vivo. The loss of function of Smg6 in telomere maintenance and NMD is still compatible with the viability of both ESCs and MEF cells.
The NMD function of Smg6 licenses ESC differentiation
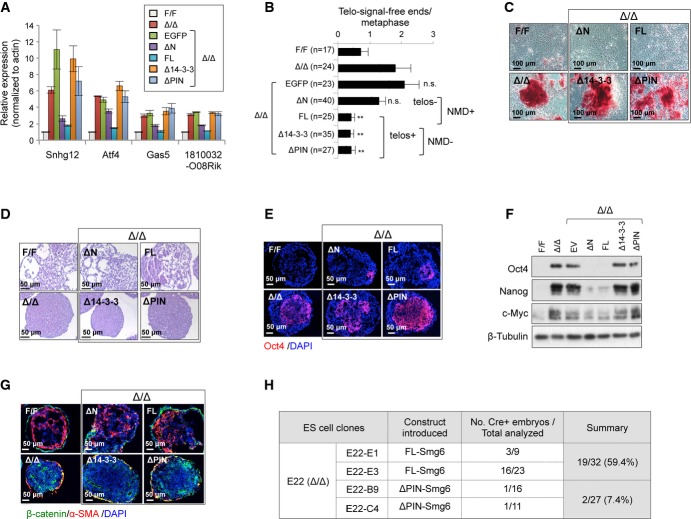
To define precisely which function of Smg6, telomere maintenance or NMD function, controls ESC differentiation, we constructed GFP-tagged either NMD- or telomere-functional domains of Smg6 and ectopically expressed them in Smg6Δ/Δ ESCs (Supplementary Fig S5A–C). qRT–PCR analysis revealed that the NMD-proficient constructs (ΔN-Smg6 and FL-Smg6) efficiently alleviated the transcript increase of NMD targets Snhg12,Gas5,Atf4, and 1810032O08Rik (FigA). However, this repression could not be effected by the reintroduction of either the empty vector (GFP-EV) or NMD-deficient Smg6-truncated vectors (Δ14-3-3-Smg6 and ΔPIN-Smg6) (Fig4A). In addition, ΔN-Smg6 and FL-Smg6 expression corrected the accumulation of the PTC+ isoforms which are generated from either exon inclusion or exon exclusion in Smg6Δ/Δ ESCs (Supplementary Fig S5D and E). Of note, all vectors, which contain the telomere-functional N-terminal domains (FL-Smg6, Δ14-3-3-Smg6, and ΔPIN-Smg6), completely rescued the telomere defects in Smg6Δ/Δ ESCs (Fig4B).
Figure 4. Smg6-mediated NMD function determines ESC differentiation.
A qRT–PCR assay of NMD target gene expression after reconstitution of Smg6Δ/Δ ESCs with NMD-proficient or NMD-deficient Smg6 constructs (see Supplementary Fig S5A). The data are from three independent qRT–PCR experiments.
B Telomere FISH analysis of metaphases of ESCs with indicated genotype and reconstituted with indicated Smg6-expression vectors. The number of telomere-signal-free ends per metaphase is shown. n, the total number of metaphases used for the quantification. NMD+, NMD-proficient vectors; NMD-, NMD-deficient vectors; telos+, telomere-proficient vectors; telos-, telomere-deficient vectors. Statistic analysis was used to compare the Δ/Δ samples and their derived clones with different Smg6 truncation expression vectors.
C Spontaneous differentiation of Smg6Δ/Δ (Δ/Δ) ESCs reconstituted with NMD-proficient (FL and ΔN) or NMD-deficient (Δ14-3-3 and ΔPIN) Smg6 constructs. Representative images of AP staining of cultures at day 6 after the removal of LIF and feeder are shown.
D EB formation assay of Smg6Δ/Δ ESCs reconstituted with NMD-proficient or NMD-deficient Smg6 vectors. The morphology of EB cells and cavity formation at day 8 are indicative of the differentiation status.
E Immunofluorescence analysis of expression of the stem cell marker Oct4 on EBs of reconstituted Smg6Δ/Δ ESCs on day 8.
F Western blotting of Smg6Δ/Δ EB cells reconstituted with various Smg6 truncation vectors on day 8. The expressions of Oct4, Nanog, and c-Myc are shown. β-Tubulin was used as a loading control.
G Immunofluorescence analysis of expression of markers β-catenin and α-SMA on EBs of reconstituted Smg6Δ/Δ ESCs on day 8.
H Chimerism analysis of in vivo rescue of differentiation defects of Smg6Δ/Δ ESCs after reconstitution with NMD-proficient (FL-Smg6) or NMD-deficient (ΔPIN-Smg6) construct. Parental ES clone E22 (Smg6-CER) contained the Cre sequence, which was used to detect ESC contribution in E12.5 embryos by PCR. All ESC clones were pretreated with 4-OHT for 6 days to delete endogenous Smg6 before blastocyst injection.
Data information: The error bars represent the SEM. Unpaired Student's t-test was used. n.s., not significant; **P < 0.01. Source data are available online for this figure.
We then investigated whether Smg6Δ/Δ ESCs reconstituted with NMD-proficient Smg6 constructs can differentiate. After 6 days of culture on feeder-free dishes without LIF, NMD-proficient (ΔN-Smg6 and FL-Smg6), but not NMD-deficient vectors (Δ14-3-3-Smg6 and ΔPIN-Smg6, both of which are proficient in telomere function), could relieve the differentiation block of Smg6Δ/Δ ESCs as judged by AP staining (FigC). When day 8 EBs were analyzed, NMD-proficient Smg6 constructs restored the appearance of cavitation and differentiated cell components in Smg6Δ/Δ EBs (Fig4D, Supplementary Fig S5F), which are associated with a reduced level of Oct4 (FigE). Western blot analysis further confirmed a reduction of the pluripotent markers Oct4, c-Myc, and Nanog in Smg6Δ/Δ EBs after the reintroduction of NMD-proficient Smg6 constructs (Fig4F, see also below). In contrast, reconstitution of Smg6Δ/Δ ESCs with either the empty vector or NMD-deficient constructs (Δ14-3-3-Smg6 and ΔPIN-Smg6) failed to restore the differentiation capacity of these cells, as judged by the lack of differentiated cells and high levels of stemness markers in their EBs (Fig4A–E). Moreover, immunostaining of β-catenin (for endoderm) and α-SMA (for mesoderm) demonstrated that only NMD-proficient constructs generated both endoderm and mesoderm germ layers in Smg6Δ/Δ ESC-derived EBs (Fig4G). qRT–PCR analysis of Smg6Δ/Δ EBs at day 5 revealed that the NMD-proficient Smg6 constructs efficiently repressed abnormally up-regulated RNA transcripts of pluripotency genes Esrrb, Oct4, Nanog, Rex1, as well as the mis-regulated lineage markers Foxa2, Mixl1, and T (Supplementary Fig S5G).
The in vivo chimerism assay further revealed that the NMD-proficient (FL-Smg6) but not the NMD-deficient (ΔPIN-Smg6) construct greatly improved the capacity of Smg6Δ/Δ ESCs to form the tissues of chimeric embryos (FigH). qPCR analysis of the Cre transgene (carried in by injected ESCs) of chimeric embryos revealed that ectopic FL-Smg6 expression gave rise to a much higher efficiency (ΔΔCq = log2 = 6.7, > 100-fold) of Smg6Δ/Δ ESCs that contributed to chimerism (Supplementary Fig S5H and I). Since the ectopic expression of NMD-deficient Smg6 (Δ14-3-3-Smg6 and ΔPIN-Smg6) in Smg6Δ/Δ ESCs could rescue the telomere, but not the differentiation defects, and ΔN-Smg6 (telomere deficient, NMD proficient) failed to correct the telomere defect, but could correct the NMD and differentiation defects of Smg6Δ/Δ ESCs, the telomere maintenance function of Smg6 is unlikely to be a decisive factor for ESC differentiation.
Smg6 regulates the c-Myc expression through its NMD activity
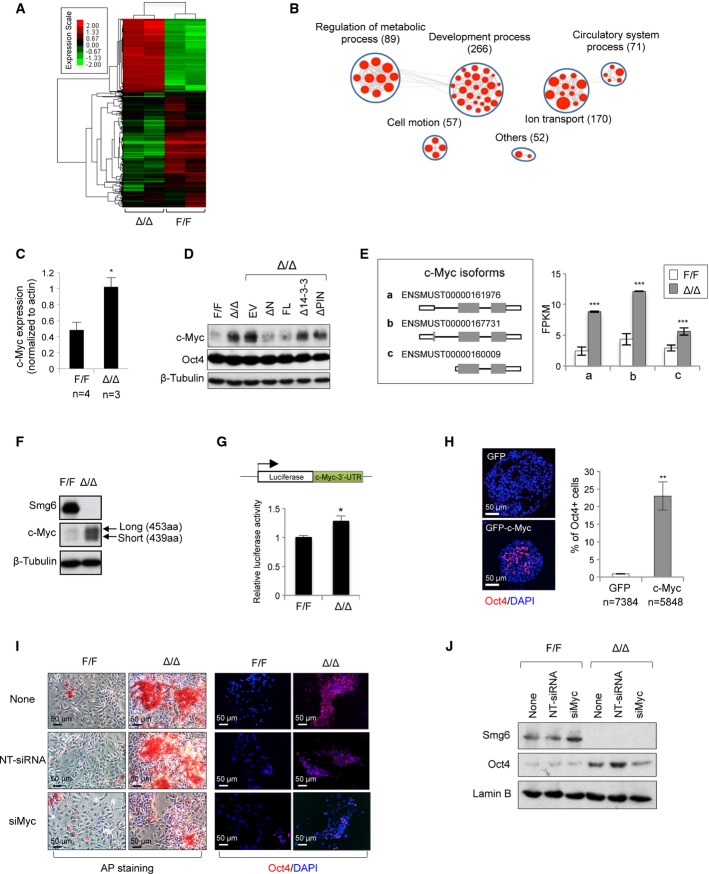
We next investigated the molecular mechanism by which Smg6-mediated NMD may govern ESC differentiation. RNA-seq analysis revealed that gene transcript levels in control and Smg6Δ/Δ ESCs were highly and significantly correlated (Pearson's correlation coefficient r2 = 0.9227; P < 10−16; Supplementary Fig S6A), indicating a general similarity of the transcriptional profiles. However, when compared to control ESCs, 2,449 differentially expressed genes (DEGs) (≥ 2-fold change in expression, P < 10−6, and read number > 50) were enriched in Smg6Δ/Δ ESCs, which represents ∽9.5% of the whole transcriptome (FigA, and Supplementary Tables S1 and S2) and is consistent with the prediction that 3–10% of all gene transcripts depend on NMD (Schweingruber et al, 2013). Based on the Gene Ontology (GO) analysis, 266 of these DEGs were significantly enriched in pathways which were related to embryonic development and differentiation (FigB and Supplementary Table S3).
Figure 5. Smg6-NMD targeting c-Myc expression dictates ESC differentiation.
A Heat map showing gene expression profiles from control and Smg6Δ/Δ ESCs (two of each genotype). A total of 2,449 DEGs were subjected to this analysis. Red and green colors depict high and low gene expression levels, respectively, based on the log2 P-values of their RPKMs.
B All these DEGs were subjected to GO analysis using DAVID. An enrichment map was constructed using Cytoscape installed with the Enrichment Map plug-in. Each enriched GO pathway is represented by a red node (P < 0.05; FDR < 0.05; overlap cutoff > 0.5). Note that the size of nodes is proportional to the total number of genes in each pathway. Edge thickness represents the number of overlapping genes between nodes. GO pathways of similar function are sorted into one cluster, marked with a circle and label. Gene number in each cluster is in parentheses.
C Relative c-Myc mRNA level in control (F/F) and Smg6Δ/Δ (Δ/Δ) ESCs by qRT–PCR analysis. n: number of ESC lines analyzed. Unpaired Student's t-test was used for the statistical analysis. *P < 0.05.
D Western blotting of c-Myc protein expression in Smg6Δ/Δ ESCs reconstituted with different Smg6 truncations. Note, only NMD-proficient constructs (ΔN-Smg6 and FL-Smg6) could repress the c-Myc protein expression.
E Expression of c-Myc isoforms (schemes is shown on the left panel) in control (F/F) and Smg6Δ/Δ (Δ/Δ) ESCs. The expression level is shown with FPKM calculated with RNA-seq data (n = 2 for each genotype) (right panel). ***P < 0.001.
F Expression of c-Myc protein isoforms in control (F/F) and Smg6Δ/Δ (Δ/Δ) ESCs.
G c-Myc 3′-UTR is a NMD target. A luciferase reporter fused with the c-Myc 3′-UTR was used to determine the mRNA stability in control (F/F) and Smg6Δ/Δ (Δ/Δ) ESCs. The data are summarized from three independent experiments. *P < 0.05.
H Overexpression of c-Myc in wild-type (E14.1) ESCs blocks differentiation. c-Myc overexpressed EBs (day 5) retained a high number of cells expressing Oct4. The percentage of Oct4+ cells in EBs is shown on the right panel. n, the number of cells scored. **P < 0.01. Unpaired Student's t-test was used for the statistical analysis.
I AP staining (left) and Oct4 immunostaining (right) of the day 5 culture of control and Smg6Δ/Δ ESCs in the spontaneous differentiation assay.
J Western blot analysis of Oct4 in c-Myc siRNA-treated Smg6Δ/Δ ESCs at day 7 in spontaneous differentiation assay. Non-targeted siRNA (NT-siRNA) is a control treatment.
Data information: The error bars represent the SEM.Source data are available online for this figure.
After mining the RNA-seq data, we found c-Myc to be one of the top hits among pluripotency genes that were expressed at a very high level in Smg6Δ/Δ ESCs (Supplementary Tables S1 and S2). qRT–PCR and Western blot analyses further confirmed the up-regulation of both the c-Myc transcript and protein in Smg6Δ/Δ ESCs as compared to controls (FigC and D). After the analysis of c-Myc targets (Chen et al, 2008) by mining the RNA-seq dataset, we found that 95 out of the 2,449 Smg6 DEGs were regulated by c-Myc (Supplementary Tables S4 and S5). Although many cell cycle and apoptosis genes are c-Myc targets in somatic cells (Dang, 1999), qRT–PCR analysis revealed that these representative gene transcripts were not significantly changed in Smg6-null ESCs (Supplementary Fig S6B), consistent with the normal cell cycle profile and apoptosis of mutant ESCs (see FigC and D and Supplementary Fig S2F).
It has been shown that high levels of c-Myc promote the ESC status as well as iPSC production (Cartwright et al, 2005; Knoepfler, 2008). c-Myc positively regulates the stemness genes (Kim et al, 2010; Smith & Dalton, 2010; Varlakhanova et al, 2010; Nie et al, 2012). We were then prompted to investigate whether the NMD activity of Smg6 is responsible for dysregulated c-Myc expression and whether the modulation of c-Myc expression would counteract Smg6-NMD deficiency-induced ESC differentiation defects. Interestingly, the introduction of NMD-proficient ΔN-Smg6 and FL-Smg6 vectors, but not the deficient Δ14-3-3-Smg6 nor ΔPIN-Smg6, evidently repressed the c-Myc expression in Smg6Δ/Δ ESCs (Fig5D) and their derived EBs (Fig4F), which concurrently rescued Smg6Δ/Δ ESCs differentiation (Fig4C–E). These data indicate that c-Myc is a target of Smg6-NMD in ESCs.
Failure to down-regulate c-Myc by NMD deficiency prevents differentiation of Smg6-deficient ESCs
To further characterize how Smg6 regulates c-Myc mRNA stability and its expression, the relative abundance of c-Myc isoforms (represented by FPKM; sequences of c-Myc isoforms are from http://www.ensembl.org) in ESCs, three c-Myc isoforms from the RNA-seq database were identified. mRNA isoforms (ENSMUST00000161976, ENSMUST00000160009, and ENSMUST00000167731), encoding two c-Myc protein products, with sizes of 439aa and 453aa, respectively, were overrepresented in Smg6Δ/Δ ESCs (Fig5E and F). Interestingly, these three c-Myc isoforms all have a 3′-UTR structure, which may destabilize c-Myc mRNA (Jones & Cole, 1987; Yeilding et al, 1996; Singh et al, 2008). In order to test whether c-Myc is stabilized in the NMD-deficient background, we transfected a c-Myc 3′-UTR luciferase reporter (pRL-c-Myc-3′-UTR) (Kumar et al, 2007), which should efficiently trigger NMD, into control and Smg6Δ/Δ ESCs. A significant increase in luciferase activity was observed in Smg6Δ/Δ ESCs, indicating a stabilization of luciferase mRNA in Smg6-deficient ESCs (Fig5G). These data suggest that Smg6 likely de-stabilize c-Myc mRNAs through their 3′-UTR.
Furthermore, we investigated whether c-Myc up-regulation in Smg6-deficient ESCs is responsible for the differentiation block. To this end, we stably overexpressed c-Myc (GFP-c-Myc, without 3′-UTR and thus NMD insensitive) in wild-type ESCs (Supplementary Fig S6C). Of note, c-Myc overexpression in ESCs did not impair NMD in ESCs because we found a similar level of NMD target transcripts (Supplementary Fig S6D). Nonetheless, these ESCs showed impaired differentiation as indicated by smaller EB size and significantly higher proportion of Oct4+ cells in the EBs on differentiation day 5, as compared to control GFP-transfected ESCs (FigH, and data not shown). Because the TGF-beta signaling pathway, via phospho-Smad2/3, regulates the pluripotency genes Oct4, Nanog, and c-Myc transcriptionally (Dang, 2012), we next treated ESCs with the TGF-beta inhibitor SB431542 and found a reduction of c-Myc within Smg6Δ/Δ ESCs (Supplementary Fig S6E). Concurrently, the TGF-beta inhibitor treatment partially relieved the differentiation block of Smg6-deficient ESCs, as judged by the lack of AP staining in the differentiation culture (Supplementary Fig S6F) and the cavitation of Smg6-deficient EBs, as well as Oct4 down-regulation (Supplementary Fig S6G and data not shown). To ultimately study whether elevated c-Myc mRNA is causal for Smg6-NMD deficiency-associated differentiation block, we knocked down c-Myc by siRNA in Smg6-mutant ESCs (Supplementary Fig S6H). Strikingly, siMyc treatment greatly reduced both AP-positive cells (Fig5I) and Oct4 expression (Fig5I and J) in Smg6Δ/Δ ESC differentiation cultures of 7 days, indicating that a selective down-regulation of c-Myc by Smg6-mediated NMD is necessary for ESC differentiation. These data suggest that c-Myc regulation by the Smg6-mediated NMD is responsible for the differentiation blockage seen in Smg6-deficient ESCs.
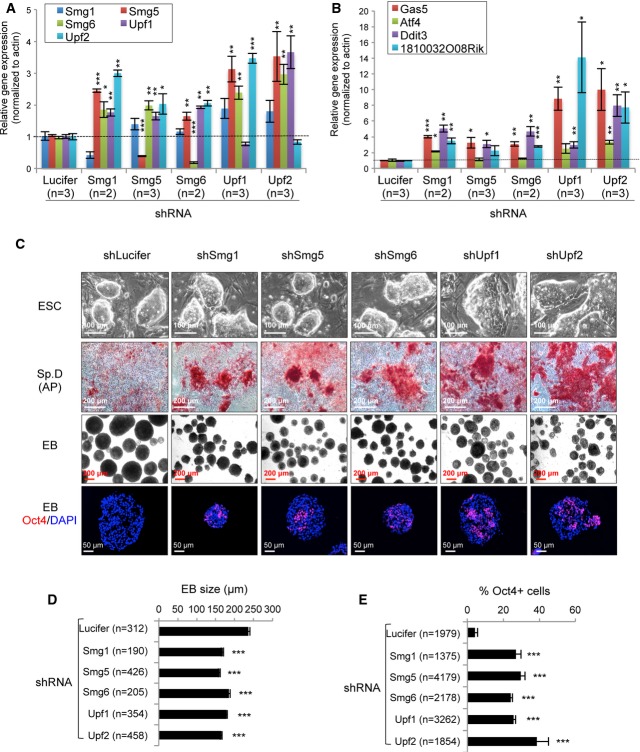
Knockdown of NMD factors results in an ESC differentiation defect
To test the generality of the NMD function in ESC maintenance and differentiation, we used shRNA vectors to knock down other NMD factors, Smg1, Smg5, Upf1, and Upf2 in mouse ESCs (E14.1). qRT–PCR analysis and Western blotting (using available antibodies) confirmed the reduction in the mRNA or protein levels of the respective NMD factors in these ESC clones (FigA and Supplementary Fig S7A). As it was previously reported that NMD factors are NMD targets (Huang et al, 2011), we noticed an up-regulation of mRNA transcripts of other NMD factors if one NMD factor was knocked down (FigA). In addition, the NMD target gene transcripts of Atf4, Gas5, Ddit3, and 1810032O08Rik were all up-regulated in knockdown ESC clones (Fig6B), indicative of a NMD defect. To our surprise, and not as previously reported (Chawla & Azzalin, 2008; McIlwain et al, 2010), knockdown of NMD factors did not compromise the survival and self-renewal of mouse ESCs. However, under spontaneous differentiation conditions (the removal of LIF and feeders), these NMD-knockdown ESC clones were refractory to differentiation as they showed a strong AP activity after 5 days, which was in contrast to control shLuciferase-transfected ESCs (Fig6C). Furthermore, the EB formation assay revealed that all NMD-knockdown ESC-derived EBs were smaller when compared to controls (Fig6C and D). While control ESCs formed EBs containing no detectable Oct4 expression, EBs derived from ESCs with knockdown of NMD factors had a significantly higher frequency of Oct4+ cells (Fig6C and E). These data indicate that NMD plays a general role in the ESC differentiation process. Of note, only the stable knockdown of Upf1 and Upf2 increased c-Myc (Supplementary Fig S7B), suggesting that c-Myc is unlikely a main target for the Smg1- and Smg5-NMD pathways.
Figure 6. Stable knockdown of the NMD factors in ESCs.
A, B qRT–PCR analysis of expression of NMD factors (A) and NMD target transcripts (B) in stable knockdown ESCs. The expression levels of the NMD factors (A) (Smg1, Smg5, Smg6, Upf1, and Upf2) and NMD target genes (B) (Gas5, Atf4, Ddit3, and 1810032O08Rik) were normalized to β-Actin. The mRNA levels of these genes in control (shLucifer) were defined as “1”. n, the number of independent clones analyzed.
C NMD deficiency compromises ESC differentiation. ESC colonies of control (shLucifer) and NMD-knockdown ESCs growing on feeder and LIF+ medium are shown in the upper panel. Spontaneous differentiation (Sp. D) and embryoid body formation on day 5 (EB) from ESCs after removal of feeders and LIF are shown (middle panels). AP staining was used to detect stem cell identity. Oct4 antibody staining (lower panel) was used to determined the stem cell identify in EBs.
D Quantification of the EB size (C, mid panel). n, the number of EBs for quantification.
E Quantification of the Oct4 expression cells in EBs (C, lower panel). n, the number of cells scored.
Data information: The error bars represent the SEM. *P < 0.05; **P < 0.01; ***P < 0.001. Unpaired Student's t-test was used for the statistical analysis.
Discussion
In the present study, we show that Smg6 is essential for embryonic development, not because of its telomere function, but rather due to its vital role as an NMD factor in controlling ESC differentiation. In this regard, the knockout of the telomerase complex (Tert) or its RNA component (Terc) is compatible with the survival of cells and mice (Blasco et al, 1997; Blasco, 2007; Tumpel & Rudolph, 2012). The loss of telomere limits cell proliferation capacity in primary somatic cells because of DNA damage response activation, involving ATM and p53 pathways, which induces either senescence or cell death (Karlseder et al, 1999; Herbig et al, 2004; Sperka et al, 2012). However, Tert−/− ESCs are viable and can be maintained, despite progressive loss of telomere length and chromosome instability (Wang et al, 2005). This suggests that ESCs may harbor a different mechanism to handle telomere integrity-mediated cellular response since ESCs are characterized by a short G1 and lack a strong cell cycle checkpoint (Orford & Scadden, 2008; Abdelalim, 2013).
We further show that Smg6-NMD is a fundamental process to remove the PTC-containing mRNA and to regulate the expression of gene products in eukaryotes and thus dictates ESC differentiation (Chawla & Azzalin, 2008; Nicholson et al, 2010; Lou et al, 2014). Previously, it has been shown that genetic modulation of NMD factors causes severe cellular and organismal defects, as seen by the loss of cellular viability and embryonic lethality (Wittkopp et al, 2009; Hwang & Maquat, 2011). However, we were able to generate a panel of NMD factor-knockdown ESCs and to completely delete the NMD factor Smg6 in ESCs, EBs, and MEFs. Smg6-knockout ESCs and MEFs are thus viable, but NMD deficient. These findings allow us to conclude that NMD per se is not required for the self-renewal of the ESCs, as well as for MEFs. The reason for the discrepancy between our observations and others is unknown, but the compensation scenario, that is, the up-regulation of other NMD factors, seems unlikely to be because of general increase of NMD target transcripts (FigB and Supplementary Fig S4G; see also Supplementary Table S4) and the reduced NMD activity (Fig3E) in Smg6-null cells. However, we cannot rule out the possibility that the lethal phenotype of Smg1 and Upf1 knockout cells seen in previous studies could attribute to other functions of these factors within genotoxic stress and DNA replication (Brumbaugh et al, 2004; Chawla & Azzalin, 2008; Luke & Lingner, 2009). Nevertheless, the essential function of Smg6-NMD in ESC differentiation may provide a plausible mechanistic explanation for the lethality of NMD factor-knockout animal models, for example, zebrafish and mice (Medghalchi et al, 2001; Weischenfeldt et al, 2008; Wittkopp et al, 2009; McIlwain et al, 2010).
Intriguingly, without Smg6, ESCs cannot differentiate as was verified by the rigorous differentiation assays in vitro and in vivo. Smg6-mutant ESCs are completely blocked for differentiation, even at early passages, which is in contrast to the mild differentiation defect seen in telomerase-knockout ESCs, which appears only at very late passages (more than 67 passages) (Pucci et al, 2013). Telomere defects are unlikely to be responsible for the differentiation block of Smg6-deficient ESCs because ectopic expression of NMD-deficient Smg6 (Δ14-3-3-Smg6 and ΔPIN-Smg6) in Smg6Δ/Δ ESCs could rescue the telomere, but not the differentiation, defects (Fig4). Also, the loss of the differentiation capacity of ESCs by Smg6 deletion is not due to impaired proliferation or greater apoptosis (Fig1C and D). In fact, the EdU pulse labeling of EBs revealed more proliferating cells in Smg6-mutant EBs as compared to control EBs (Supplementary Fig S3E), which can likely be attributed to the proliferation status of undifferentiated Smg6Δ/Δ cells within these EBs. We can rule out the possibility that the differentiation block of Smg6-null ESCs is a consequence of cell transformation because: (i) these mutant ESCs maintain a normal karyotype, and (ii) after reintroduction of Smg6-NMD-proficient vectors, they can be rescued for their differentiation failure and form various cell types in vivo.
Rescue of the Smg6-null ESC differentiation block by NMD-proficient constructs (ΔN-, and full-length Smg6), but not by telomere function-proficient vectors, strongly argues that the differentiation block of Smg6-deficient ESCs is solely due to a cell-autonomous defect in NMD. The essential function of NMD is not restricted only to Smg6, because knockdown of the upstream of the NMD complexes, Smg1, Upf1, Upf2, as well as both the Smg5/7 and Smg6 branches, all blocked ESC differentiation (Fig6C). It is interesting to note that knockout ESCs of Dicer 1 and DGCR8, which are involved in the miRNA–mRNA decay pathway, are defective in differentiation as characterized by the inefficient repression of stem cell markers and compromised expression of differentiation markers in an EB assay (Kanellopoulou et al, 2005; Wang et al, 2007). However, Smg6 seems to work differently in the regulation of ESC differentiation, because Smg6-deficient EBs contain an up-regulation of differentiation genes, while maintaining a high level of stem cell markers (Supplementary Fig S5G). Nonetheless, NMD activity restoration by both ΔN- and FL-Smg6 vectors can rescue the expression pattern and rectify the differentiation defects. Our data suggest that the repression of stem cell genes is necessary to remove the differentiation block of ESCs, consistent with the general notion that a failure to repress the stemness gene expression disturbs the differentiation program (Keller, 2005; Ivanova et al, 2006; Gabut et al, 2011). In addition, Smg6-NMD is required for efficient cellular reprograming, that is, from MEFs to iPSCs. Taken together, NMD per se is not required for the steady state and self-renewal of stem or somatic cells, but is a licensing factor for cell identity switching during both differentiation and reprograming processes.
Differentiation and cell reprograming require an orchestrated regulation of expression of developmental genes or transcription factors, as well as epigenetic modulations (Garneau et al, 2007; Orkin & Hochedlinger, 2011; Kervestin & Jacobson, 2012; Buganim et al, 2013; Shu et al, 2013). How is NMD involved in these processes? Our pathway analysis indicates that Smg6-NMD is required for the differentiation program, in response to differentiation cues and for reactivation of the endogenous pluripotency network in response to OSKM. Given the essential function of NMD in the productive output of transcripts and in general post-transcriptional control (Garneau et al, 2007; Huang & Wilkinson, 2012; Kervestin & Jacobson, 2012), NMD is likely to be at the top of several cascades, to regulate pluripotency genes in self-renewal and the differentiation of ESCs. The pluripotency factor c-Myc is abnormally accumulated in Smg6-null ESCs (Fig5C–E). In normal mouse ESCs, there are two isoforms of c-Myc, both of which are repressed in differentiation. However, both isoforms persist at a high level in Smg6-null ESCs and EBs. Interestingly, the ectopic expression of NMD-proficient Smg6 constructs can efficiently repress the elevated c-Myc, which simultaneously corrected the Smg6-null ESC differentiation defect (Figs4F and 5D and F). Mechanistically, the 3′-UTR of c-Myc mRNA is responsible for its stability against NMD (Fig5E and G). This is consistent with previous observations that c-Myc mRNA has a short half-life and could be stabilized in Xenopus oocytes (a natural mRNA decay deficiency condition) or after NMD inhibitor CHX treatment (Wisdom & Lee, 1991; Wright et al, 1991). Our findings, together with previous studies, strongly suggest that c-Myc mRNA is a target of, and regulated, by Smg6-NMD in ESC differentiation.
c-Myc is a master regulator of stem cell pluripotency (Kim et al, 2010; Smith & Dalton, 2010; Varlakhanova et al, 2010; Nie et al, 2012), and overexpression of c-Myc promotes ESC self-renewal and iPSC production efficiency (Knoepfler, 2008; Hanna et al, 2009). A high c-Myc level inhibits NMD in B lymphocytes (Wang et al, 2011), and overexpression of c-Myc in ESCs blocks differentiation (Cartwright et al, 2005; Lin et al, 2009). These studies suggest a c-Myc-NMD feedback loop. However, when we overexpressed c-Myc in wild-type ESCs, NMD did not seem to be affected (Supplementary Fig S6D). The reason behind the operational differences of the c-Myc-NMD feedback loop in these cells is currently unknown, but it may be cell type specific (highly proliferative ESCs versus lower proliferative B cells) or explained by the threshold of c-Myc expression that dictates the NMD inhibition efficiency (Wang et al, 2011). Nevertheless, wild-type ESCs, which are NMD proficient and overexpress c-Myc, are refractory to differentiation (Fig5H). Despite a high level of c-Myc and a low NMD, Smg6-null ESCs exhibit a normal proliferation and apoptotic response and are devoid of transformation. These are interesting observations given the fact that c-Myc plays an important role in somatic cell proliferation and cell death (Dang, 1999) and that the loss-of-function mutation of Upf1, an upstream factor of Smg6-NMD, is associated with human pancreatic adenosquamous carcinomas (Liu et al, 2014). ESC proliferation seems to be insensitive to the change of c-Myc or NMD activities. However, repression of the elevated c-Myc by siRNA and ectopic expression of NMD-proficient Smg6 constructs can largely reverse the ESC differentiation defects imposed by Smg6 deletion (see Figs4 and 5I). These results demonstrate that a dedicated regulation of key pluripotency regulators, such as c-Myc, by NMD is a novel mechanism to orchestrate the network of self-renewal and differentiation of ESCs and also in iPSC production.
In summary, our study uncovers the NMD as a novel mechanism, adding to other well-known transcription factors, epigenetics and non-coding RNAs, in ESC differentiation and in developmental programing.
Materials and Methods
Knockdown of NMD factors in ESCs
shRNA against NMD factors, Smg1, Smg5, Smg6, Upf1, and Upf2, were cloned into the shRNA vector as previously described (Zhou et al, 2013). For control shRNA, the luciferase sequence (5′-GGCTTGCCAGCAACTTACA-3′) was used to generate the shLucifer vector. shRNA vectors were linearized with ApaL1 and electroporated into E14.1 (129/Sv/Ola background) mouse ESCs. After selection against G418 (275 μg/ml), the stable ESC clones were picked, expanded, and further characterized by qRT–PCR and Western blotting. The targeting sequences and qRT–PCR primers for NMD factors are available upon request.
Generation of conditional and conventional Smg6-knockout allele
To disrupt the Smg6 gene in the mouse germ line, the targeting vector was constructed as shown in Supplementary Fig S1A. Briefly, the gene-targeting vector, in which Smg6 exons 2–4 were flanked by two LoxP sites and the neomycin (neo) cassette, flanked by Frt sites, was introduced into intron 2 and was electroporated into E14.1 ESCs that were cultured in ES medium [DMEM, 15% FCS, 1× sodium pyruvate, 1× Pen/Strep, 1× glutamine, 1× non-essential amino acids, 1 μM 2-mercaptoethanol, 1,000 units/ml LIF (ESGRO®; Merck-Millipore, Schwalbach, Germany)]. Following selection with G418, the gene-targeting events (Smg6+/T ESCs) were confirmed by Southern blotting using an external probe upstream of exon 1 after NcoI digestion of the genomic DNA. After injection of Smg6+/T ESCs into blastocysts, germ line offspring were obtained and were then bred with FLP transgenic mice to remove the neo cassette and to generate Smg6+/F mice. Smg6+/F mice were further bred with Nestin-Cre transgenic mice to delete exons 2–4 to derive Smg6+/Δ mice or bred with CreERT2 transgenic mice (Ventura et al, 2007) to generate Smg6F/F;CreER+ (Smg6-CER) mice. For the genotyping of mutant mice, the following primers were used: Smg6-1R: gaatctctgtcatttactatctcatcc; Smg6-2F: ctgaataggagccgggtgt; and Smg6-4R: tcagatccaatgcctccttc. The sizes of the wild-type (WT), flox (F), and deleted (Δ) alleles were 621, 667 and 514 bp, respectively. All mice in this study were from 129/Sv/C57BL/6 mixed background generated in the FLI animal facility. All mice were kept in pathogen-free conditions. All animal experiments were conducted according to German animal welfare legislation.
Generation of Smg6-deficient ESCs and MEFs
Two approaches were used to generate Smg6-deficient ESCs. Firstly, Smg6F/F ESCs were transiently transfected with pCAG-GFP-Cre (#23776; Addgene, Cambridge, MA, USA). The GFP+ populations were FACS sorted and single clones were isolated to establish Smg6Δ/Δ ESCs. Secondly, Smg6F/F;CreER+ ESCs were treated with 1 μM 4-OHT (Sigma-Aldrich, Munich, Germany) for 5 days in culture to induce Smg6 deletion. MEFs were isolated from E13.5 Smg6F/F;CreER+ embryos and immortalized using p19ARF shRNA as previously described (Herkert et al, 2010). To generate Smg6Δ/Δ MEFs, Smg6F/F;CreER+ MEFs were treated with 1 μM 4-OHT for 9 days in culture to induce Smg6 deletion. Deletion of Smg6 was characterized by Southern blotting, PCR, or Western blotting.
Characterization of Smg6-null ES cells
For the proliferation assay, 1 million control or Smg6-null ESCs were plated on a feeder layer in a six-well plate. Every 3 days, ESC clones were dissociated and cell numbers were counted for 12 passages. The cell death was measured by staining ESCs with an anti-Annexin-V-APC antibody (eBioscience, Frankfurt am Main, Germany) and with propidium iodide (100 μg/ml), followed by FACS analysis. For cell cycle analysis, cells were transiently labeled with BrdU for 1 h and fixed in 70% ethanol and followed with anti-BrdU-APC antibody staining (eBioscience) as per the company protocol.
Construction of Smg6 truncation vectors
In order to stably express the different domains of Smg6 in ESCs, pEGFP-C1 was engineered by replacing the CMV promoter with the hEF1α promoter (human elongation factor-1-alpha promoter) and renamed pEGFP-C1-EF1α. For the generation of the full-length Smg6 cDNA, total RNAs were isolated from E14.1 ESCs and cDNA was synthesized using an AffinityScript Multiple Temperature cDNA Synthesis kit (Agilent Technologies, Santa Clara, CA, USA). The full length of Smg6 was further amplified with Phusion® high-fidelity DNA polymerase (New England Biolabs, Frankfurt, Germany) by using gene-specific primers (Smg6-F: atggcggaggggttggagcg; Smg6-R: gcccacctgggcccacgtaa) and subcloned into the pEGFP-C1-EF1α vector. The truncated Smg6-expression vectors (listed below) were further constructed based on the full-length Smg6 cDNA. These vectors were electroporated into ESCs, and stable clones were obtained after selection with G418.
| Construct | Description |
|---|---|
| EGFP | GFP gene for the ESC labeling |
| FL-Smg6 | Full length of Smg6 |
| ΔN-Smg6 | Smg6 with deletion of the N-terminus (1–575 aa) |
| Δ14-3-3-Smg6 | Smg6 with deletion of the 14-3-3 domain (576–815 aa) |
| ΔPIN-Smg6 | Smg6 with deletion of the PIN domain (1239–1418 aa) |
qRT–PCR assay
For the quantification of NMD target gene expression, total RNA was isolated and cDNA was synthesized using the Affinity Script Multiple Temperature cDNA Synthesis kit or with SuperScript® III Reverse Transcriptase (Invitrogen, Darmstadt, Germany). qRT–PCR was performed in triplicate for each sample using Platinum SYBR Green qPCR SuperMix-UDG and a CFC96 Touch™ Real-Time PCR Detection System (Bio-Rad, Munich, Germany). The primers used for the PCR amplification of Snhg12, Atf4, Gas5, 1810032O08Rik, Ddit3, and β-Actin were synthesized according to a previous publication (Weischenfeldt et al, 2008). For the RT–PCR analysis to identify the normal and the PTC+ isoforms of Pkm2, Srfs2, Hnrnph1, Rps12, Trub2, the primer sequences were adopted from the previous study (Weischenfeldt et al, 2012). qRT–PCR primers for the stemness and differentiation genes are reported in a previous study (Lyashenko et al, 2011). qPCR primers for cyclin A2, D1, E1, E2, and cdk1 were published in Pandit et al (2012). qPCR primers for PAI-1, PAI-2, p16, p21, and p19 were described in Mudhasani et al (2008). β-actin was used as the reference control for all of the study.
In addition, the following primers designed in this study are:
Hnrnp1: Fwd, tcgcagtgtatgtttgatggg; Rev, ctggcgtttgttggggttac
Foxp1: Fwd, tggttcacacgaatgtttgc; Rev, ggaagcctgtaaagctgcat
Eif2α: Fwd, caccacacttcacagaaagca; Rev, ggcaaacaatgtcccatcct
Eif4e: Fwd, ctggctagagacactgctgt; Rev,tgtgtgtgactgcatctctg
p53: Fwd, ggaagactccagtgggaa; Rev, tcttctgtacggcggtctct
c-Myc: Fwd, ctgctgtcctccgagtcct; Rev, gcctcttctccacagacacc
Quantification of the qRT–PCR data was performed by the ΔΔCq method using β-Actin as an internal control.
Spontaneous differentiation assay
Control and Smg6-deficient ESCs were plated onto gelatin-coated culture dishes and cultured in ES medium without LIF. For AP staining, ESC differentiation cultures (on day 6) were fixed with 4% PFA for 5 min at room temperature and then stained using an AP staining kit (Sigma-Aldrich) at 37°C for at least 30 min.
EB formation assay
For in vitro differentiation, 2 × 106 ESCs were plated onto 10-cm bacterial grade Petri dishes in ES medium without LIF. On the second day, EBs were pelleted and 1/10 of the EBs was used for each 10-cm Petri dish for continuous culture. EBs were analyzed at the indicated days for sectioning and immunostaining and also collected for immunoblotting.
Induced differentiation of ESCs
For induced differentiation by either DMSO or RA (Sigma-Aldrich), 3 × 105 cells were plated per 10-cm bacterial grade Petri dish and cultured in differentiation medium (DMEM supplemented with 10% FCS, 1× Pen/Strep, 1 μM 2-mercaptoethanol). 1% DMSO or 0.01 μM RA was supplied as the inducer. After inducer treatment, EBs were split into 3 × 6-cm Petri dishes preloaded with gelatin-coated coverslips. The cultures were fixed at the indicated times and stained with antibodies against stem cell or differentiation markers.
Chimerism assay of embryos
ESCs transfected by GFP-tagged vectors were injected into C57BL/6 blastocysts and transplanted into pseudo-pregnant recipients. The embryos were collected at E12.5 for chimerism analysis by either GFP+ cells or qPCR genotyping of Cre transgenes. The primers for the Cre transgene detection were Cre1, 5′-gcattaccggtcgatgcaacgagtgatgag-3′; or Cre2, 5′-gagtgaacgaacctggtcgaaatcagtgcg-3′. For qPCR analysis of the contribution of the ESC derivatives within chimeras, the following primers were used: Cre21, 5′-ctgatttcgaccaggttcgt-3′; Cre22, 5′-attctcccaccgtcagtacg-3′. β-Actin was used as the internal control.
Immunoblot analysis
For the detection of protein expression in ESCs and EBs, samples were lysed in RIPA buffer supplied with 1 mM PMSF (Li & Wang, 2011). 40–80 μg of cell lysates was processed with SDS–PAGE. The following primary antibodies were used: rabbit anti-Upf1 (1:1,000, Bethyl Laboratories, Montgomery, TX, USA), rabbit anti-Upf2 (1:1,000, New England Biolabs), rabbit anti-Oct4 (1:1,000; New England Biolabs), mouse anti-Oct4 (1:6,000; Santa Cruz, Heidelberg, Germany), rabbit anti-Nanog (1:5,000; Merck-Millipore), rabbit anti-Smg6/Est1A (1:1,500; Abcam, Cambridge, UK), rabbit anti-p-Smad2/3 (1:1,000; New England Biolabs), rabbit anti-β-Catenin (1:800; Sigma-Aldrich), mouse anti-β-Tubulin (1:2,000; Sigma-Aldrich), and mouse anti-Actin (1:10,000; Sigma-Aldrich). The secondary antibodies used in these studies were HRP-conjugated goat anti-rabbit IgG or goat anti-mouse IgG (1:2,000; DAKO, Hamburg, Germany). ECL Western blotting substrates (Thermo Scientific, Rockford, IL, USA) were used for HRP detection.
Histological analysis and immunofluorescent staining
EBs and E12.5 embryos were fixed overnight with 4% paraformaldehyde (PFA) (pH 7.2) and processed into paraffin sections. 5-μm-thick sections were stained with hematoxylin and eosin (H&E). For the detection of stem cell and differentiation markers, EBs and embryos were fixed overnight with 4% PFA before transfer to 30% sucrose. EBs and embryos were embedded with Neg-50 frozen medium (Richard-Allan Scientific, Kalamazoo, MI, USA). 8- to 12-μm-thick frozen sections were used for immunostaining as previously described (Gruber et al, 2011). The primary antibodies used were: mouse anti-Oct4 (1:400; Santa Cruz, Heidelberg, Germany), mouse anti-α-SMA (1:200; Santa Cruz), rabbit anti-GFP-Alexa 488 conjugated (1:200; Invitrogen), rabbit anti-β-Catenin (1:800; Sigma-Aldrich), and mouse anti-Nestin (1:200; Millipore). The cell death assays were conducted by the TUNEL assay and by staining the EB sections with rabbit anti-Cleaved Caspase-3 (1:200, Cell Signaling). The secondary antibodies used were: sheep anti-mouse IgG (Cy3 or FITC conjugated), goat anti-rabbit IgG (Cy3 or FITC conjugated) (1:400; Sigma-Aldrich), streptavidin-Cy3 (1:500, Sigma-Aldrich). In all cases, sections were mounted with Prolong® Gold Antifade medium with DAPI (Invitrogen). EdU staining was conducted on cryosections using a Click-iT EdU Alexa Fluor 647 Imaging kit (Life Technologies, Carlsbad, CA, USA) following the manufacturer's instructions.
Cytogenetic and telomere FISH analyses
Metaphase preparation from ESCs and MEFs was as previously described (Li & Wang, 2011), but with minor modification. Briefly, ESC or MEF cultures were treated with 1 μg/ml colcemid (Sigma-Aldrich) for 3 h before harvesting. The procedure of telomere FISH with FAM-labeled TelG probe (Eurogentec, Cologne, Germany) followed the protocol outlined by DAKO. Chromosome metaphase and telomere FISH images were captured with a Zeiss M1 microscope (Zeiss, Jena, Germany) and analyzed with Volocity Imaging software (PerkinElmer, Waltham, MA, USA).
NMD reporter assay
The NMD reporter vector (pβ510-HA-TCRβ-GFP PTC+) (Paillusson et al, 2005) was kindly provided by Prof. Oliver Mühlemann (University of Bern, Switzerland). The NMD reporter was electroporated into Smg6-CER ESCs, and the stable integration of the vector was selected for with G418. For the positive control, ESCs with the NMD reporter were pretreated with 100 μg/ml CHX (Sigma-Aldrich) for 3 h. NMD efficiency was determined by GFP fluorescence signal intensity by FACS analysis with a FACScanto flow cytometer equipped with FACSDiva software (Becton Dickson, Mountain View, CA, USA).
RNA-seq and bioinformatic analysis
RNA was isolated from two clones of each genotype of ESCs (biological replicates) using TRI® reagent (Sigma-Aldrich) following the manufacturer's manual. RNA integrity was checked using an Agilent Bioanalyzer 2100 (Agilent Technologies, Waldbronn, Germany). All samples showed a RIN (RNA integrity number) of higher than 9. Approximately 2.5 μg of total RNA was used for library preparation using a TruSeq™ RNA Sample Prep Kit v2 (Illumina, San Diego, CA, USA) according to the manufacturer's protocol. The libraries were sequenced using HiSeq2000 (Illumina) in single-read mode, which created reads with a length of 50 bp. Sequencing chemistry v2 (Illumina) was used and samples were multiplexed in two samples per lane. The sequencing approach resulted in 233,776,656, 222,691,017, 214,742,494, and 218,424,305 reads for samples E12_CS2 (Smg6Δ/Δ), E12_CS3 (Smg6Δ/Δ), E12_GS3 (Smg6 control), and E12_GS4 (Smg6 control), respectively. RNA-seq reads of 50 bp were aligned against the mouse genome (mm9) references with TopHat2 (Langmead et al, 2009; Trapnell et al, 2009, 2012). Using uniquely mapped reads (mapping quality larger than or equal to 20), read counts per exon and for all exons within a gene for all genes in the UCSC mm9 refFlat table were obtained. 191,196,227, 178,576,062, 170,911,761, and 172,431,332 uniquely mapped reads with mapping quality larger than or equal to 20 were obtained for E12_CS2, E12_CS3, E12_GS3, and E12_GS4 samples, respectively. RPKM (reads per kilobase per million) for each sample was computed as the number of reads which map per kilobase of exon model per million mapped reads for each gene. DEGs between control and Smg6Δ/Δ ESC samples were determined using R-package DEGseq with the method MARS (MA-plot-based method with random sampling model), fold change cutoff = 2, P-value cutoff = 10−6, and absolute change bigger than 50 reads (Wang et al, 2010). GO analysis of DEGs was performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov/) (Huang et al, 2009a,b). GO terms with P < 0.05 were determined to be statistically significant.
TGF-beta inhibitor treatment of ESCs and differentiation assay
Control (Smg6F/F) and Smg6Δ/Δ ESCs were grown on gelatin-coated culture dishes in LIF-free ESC medium supplemented with TGF-beta inhibitor SB431542 (10 μM; Sigma-Aldrich). On day 5 of inhibitor treatment, the cultures were stained with the AP staining kit to determine the differentiation status of ESCs. To test the inhibitor effect on differentiation in the EB assay, EBs were cultured with TGF-beta inhibitor until day 8, when EBs were sectioned for histological examination and quantified for the presence of cystic EBs.
Cell reprograming by OSKM factors
The StemCCA lentiviral vector under a constitutive EF1α promoter is used to reprogram MEFs to iPSCs (Sommer et al, 2009). Briefly, lentiviruses were produced by transfecting 293T cells with three plasmids—pMD2.G, psPAX2, and StemCCA lentiviral vector. Viruses were collected 36 h post-transfection and applied onto 1 × 105 MEFs in 6-well plates. To determine the reprograming efficiency, the iPSC cultures were stained with the AP staining kit on the indicated days. Single clones were selected for expansion and then further characterized for the expression of stem cell markers by Western blotting. The following antibodies were used: rabbit anti-Oct4 (1:1,000; Cell Signaling), rabbit anti-Sox2 (1:1,000; Abcam), rabbit anti-Nanog (1:1,000; Merck-Millipore), and anti-β-Actin (1:10,000; Sigma-Aldrich). For teratoma induction, each iPSC line was inoculated subcutaneously four points (1 × 106 cells/point) in 4- to 5-week-old male immune-deficient CD1 nude mice. Eighteen days later, tumors were fixed overnight in 4% PFA, sectioned, and stained with H&E to determine derivatives of different germ layers based on the morphological characteristics of cells in tumors/teratomas. iPSC clones were injected into NMR1 blastocysts to generate chimeric mice.
3′-UTR-c-Myc luciferase assay
To determine the 3′-UTR in c-Myc mRNA stability, a luciferase reporter plasmid (pRL-c-Myc 3′-UTR, Addgene number: 14806, a kind gift from Tyler Jacks) was co-transfected with a firefly luciferase reporter plasmid (pGL3) into ESCs (Kumar et al, 2007). The luciferase activity was measured 48 h after transfection with Dual-Luciferase® Reporter Assay system (Promega).
Genetic modulation of c-Myc in ESCs
To overexpress c-Myc in ESCs, mouse c-Myc cDNA was amplified from the StemCCA vector and subsequently subcloned into pEGFP-C1-EF1α to generate the pEGF-C1-EF1α-c-Myc, which was electroporated into E14.1 ESCs. ESC clones with stable c-Myc expression were chosen by G418 selection and expanded for the EB formation assay. To knockdown c-Myc in ESCs, control (non-target, NT-siRNA) and mouse c-Myc siRNA pools (Invitrogen) were transfected by Lipofectamine® RNAiMAX reagent (Invitrogen) into Smg6-proficient and Smg6-deficient ESCs with a reverse transfection method on gelatin-coated 24-well dishes. The ESCs were maintained in the LIF-free differentiation condition. After 48 h, cells were re-plated onto gelatin-coated coverslips filled 6-well dishes for the analysis indicated in the manuscript. RNAi efficiency was investigated by Western blotting 60 h after the siRNA transfection.
Statistical analysis
The unpaired Student's t-test was used in this study. The statistical analysis was performed with GraphPad Prism version 5.00 for Windows (GraphPad Software, San Diego, CA, USA, http://www.graphpad.com). The chi-square test was used for Supplementary Fig S5F following the instructions on the GraphPad Software website (http://graphpad.com/quickcalcs/chisquared1.cfm).
Accession number of RNA-seq data
The NCBI Gene Expression Omnibus accession number for the RNA-seq data reported in this paper is GSE49844.
Acknowledgments
We thank Dr Oliver Mühlemann, University of Bern, Switzerland for the NMD reporter. We also thank Dominique Galendo, Cathrin Mueller, and Christof Birch-Hirschfeld for their excellent assistance in the maintenance of the animal colonies. Furthermore, we thank Ivonne Goerlich and Ivonne Heinze for diligent technical assistance in Illumina sequencing. We are grateful to Erwin Wagner and Denise Barlow for their critical reading and comments of the manuscript and to all other members of the Wang Laboratory for helpful discussions. Z.-Q. W. is partly supported by the Deutsche Forschungsgemeinschaft (DFG), Germany.
Author contributions
TL performed the majority of the experiments, analyzed data, and prepared the figures and the manuscript; YS, BS and Y-SC performed the bioinformatics analysis; LMG conducted the gene targeting in ESCs; PW carried out the transfection and immunoblotting of MEFs/iPSCs; TJ performed the blastocyst injections; AK isolated the ESCs; MG conducted the RNA sequencing; MP, Y-GY and KLR designed experiments and discussed data; Z-QW designed experiments, analyzed data, and composed the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information for this article is available online: http://emboj.embopress.org
References
- Abdelalim EM. Molecular mechanisms controlling the cell cycle in embryonic stem cells. Stem Cell Rev. 2013;9:764–773. doi: 10.1007/s12015-013-9469-9. [DOI] [PubMed] [Google Scholar]
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- Barbosa C, Peixeiro I, Romao L. Gene expression regulation by upstream open reading frames and human disease. PLoS Genet. 2013;9:e1003529. doi: 10.1371/journal.pgen.1003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- Blasco MA. Telomere length, stem cells and aging. Nat Chem Biol. 2007;3:640–649. doi: 10.1038/nchembio.2007.38. [DOI] [PubMed] [Google Scholar]
- Brumbaugh KM, Otterness DM, Geisen C, Oliveira V, Brognard J, Li X, Lejeune F, Tibbetts RS, Maquat LE, Abraham RT. The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol Cell. 2004;14:585–598. doi: 10.1016/j.molcel.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Jaenisch R. Mechanisms and models of somatic cell reprogramming. Nat Rev Genet. 2013;14:427–439. doi: 10.1038/nrg3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright P, McLean C, Sheppard A, Rivett D, Jones K, Dalton S. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development. 2005;132:885–896. doi: 10.1242/dev.01670. [DOI] [PubMed] [Google Scholar]
- Chawla R, Azzalin CM. The telomeric transcriptome and SMG proteins at the crossroads. Cytogenet Genome Res. 2008;122:194–201. doi: 10.1159/000167804. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung WK, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SK, Lundblad V. The Est1 subunit of Saccharomyces cerevisiae telomerase makes multiple contributions to telomere length maintenance. Genetics. 2002;162:1101–1115. doi: 10.1093/genetics/162.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischmeyer PA, Dietz HC. Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet. 1999;8:1893–1900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- Gabut M, Samavarchi-Tehrani P, Wang X, Slobodeniuc V, O'Hanlon D, Sung HK, Alvarez M, Talukder S, Pan Q, Mazzoni EO, Nedelec S, Wichterle H, Woltjen K, Hughes TR, Zandstra PW, Nagy A, Wrana JL, Blencowe BJ. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell. 2011;147:132–146. doi: 10.1016/j.cell.2011.08.023. [DOI] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- Gruber R, Zhou Z, Sukchev M, Joerss T, Frappart PO, Wang ZQ. MCPH1 regulates the neuroprogenitor division mode by coupling the centrosomal cycle with mitotic entry through the Chk1-Cdc25 pathway. Nat Cell Biol. 2011;13:1325–1334. doi: 10.1038/ncb2342. [DOI] [PubMed] [Google Scholar]
- Hanna J, Markoulaki S, Mitalipova M, Cheng AW, Cassady JP, Staerk J, Carey BW, Lengner CJ, Foreman R, Love J, Gao Q, Kim J, Jaenisch R. Metastable pluripotent states in NOD-mouse-derived ESCs. Cell Stem Cell. 2009;4:513–524. doi: 10.1016/j.stem.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Nakada D, Morrison SJ. Mechanisms of stem cell self-renewal. Annu Rev Cell Dev Biol. 2009;25:377–406. doi: 10.1146/annurev.cellbio.042308.113248. [DOI] [PubMed] [Google Scholar]
- Herbig U, Jobling WA, Chen BP, Chen DJ, Sedivy JM. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a) Mol Cell. 2004;14:501–513. doi: 10.1016/s1097-2765(04)00256-4. [DOI] [PubMed] [Google Scholar]
- Herkert B, Dwertmann A, Herold S, Abed M, Naud JF, Finkernagel F, Harms GS, Orian A, Wanzel M, Eilers M. The Arf tumor suppressor protein inhibits Miz1 to suppress cell adhesion and induce apoptosis. J Cell Biol. 2010;188:905–918. doi: 10.1083/jcb.200908103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang L, Lou CH, Chan W, Shum EY, Shao A, Stone E, Karam R, Song HW, Wilkinson MF. RNA homeostasis governed by cell type-specific and branched feedback loops acting on NMD. Mol Cell. 2011;43:950–961. doi: 10.1016/j.molcel.2011.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Wilkinson MF. Regulation of nonsense-mediated mRNA decay. Wiley Interdiscip Rev RNA. 2012;3:807–828. doi: 10.1002/wrna.1137. [DOI] [PubMed] [Google Scholar]
- Hwang J, Maquat LE. Nonsense-mediated mRNA decay (NMD) in animal embryogenesis: to die or not to die, that is the question. Curr Opin Genet Dev. 2011;21:422–430. doi: 10.1016/j.gde.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Jones TR, Cole MD. Rapid cytoplasmic turnover of c-myc mRNA: requirement of the 3′ untranslated sequences. Mol Cell Biol. 1987;7:4513–4521. doi: 10.1128/mcb.7.12.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlseder J, Broccoli D, Dai Y, Hardy S, de Lange T. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science. 1999;283:1321–1325. doi: 10.1126/science.283.5406.1321. [DOI] [PubMed] [Google Scholar]
- Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 2005;19:1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- Kervestin S, Jacobson A. NMD: a multifaceted response to premature translational termination. Nat Rev Mol Cell Biol. 2012;13:700–712. doi: 10.1038/nrm3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS. Why myc? An unexpected ingredient in the stem cell cocktail. Cell Stem Cell. 2008;2:18–21. doi: 10.1016/j.stem.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- Kurosawa H. Methods for inducing embryoid body formation: in vitro differentiation system of embryonic stem cells. J Biosci Bioeng. 2007;103:389–398. doi: 10.1263/jbb.103.389. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Wang ZQ. Point mutation at the Nbs1 Threonine 278 site does not affect mouse development, but compromises the Chk2 and Smc1 phosphorylation after DNA damage. Mech Ageing Dev. 2011;132:382–388. doi: 10.1016/j.mad.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Lin CH, Jackson AL, Guo J, Linsley PS, Eisenman RN. Myc-regulated microRNAs attenuate embryonic stem cell differentiation. EMBO J. 2009;28:3157–3170. doi: 10.1038/emboj.2009.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Karam R, Zhou Y, Su F, Ji Y, Li G, Xu G, Lu L, Wang C, Song M, Zhu J, Wang Y, Zhao Y, Foo WC, Zuo M, Valasek MA, Javle M, Wilkinson MF, Lu Y. The UPF1 RNA surveillance gene is commonly mutated in pancreatic adenosquamous carcinoma. Nat Med. 2014;20:596–598. doi: 10.1038/nm.3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou CH, Shao A, Shum EY, Espinoza JL, Huang L, Karam R, Wilkinson MF. Posttranscriptional control of the stem cell and neurogenic programs by the nonsense-mediated RNA decay pathway. Cell Rep. 2014;6:748–764. doi: 10.1016/j.celrep.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B, Lingner J. TERRA: telomeric repeat-containing RNA. EMBO J. 2009;28:2503–2510. doi: 10.1038/emboj.2009.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V, Szostak JW. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- Lyashenko N, Winter M, Migliorini D, Biechele T, Moon RT, Hartmann C. Differential requirement for the dual functions of beta-catenin in embryonic stem cell self-renewal and germ layer formation. Nat Cell Biol. 2011;13:753–761. doi: 10.1038/ncb2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlwain DR, Pan Q, Reilly PT, Elia AJ, McCracken S, Wakeham AC, Itie-Youten A, Blencowe BJ, Mak TW. Smg1 is required for embryogenesis and regulates diverse genes via alternative splicing coupled to nonsense-mediated mRNA decay. Proc Natl Acad Sci USA. 2010;107:12186–12191. doi: 10.1073/pnas.1007336107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, Dietz HC. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum Mol Genet. 2001;10:99–105. doi: 10.1093/hmg/10.2.99. [DOI] [PubMed] [Google Scholar]
- Mudhasani R, Zhu Z, Hutvagner G, Eischen CM, Lyle S, Hall LL, Lawrence JB, Imbalzano AN, Jones SN. Loss of miRNA biogenesis induces p19Arf-p53 signaling and senescence in primary cells. J Cell Biol. 2008;181:1055–1063. doi: 10.1083/jcb.200802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Muhlemann O. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell Mol Life Sci. 2010;67:677–700. doi: 10.1007/s00018-009-0177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, Zhao K, Levens D. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orford KW, Scadden DT. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat Rev Genet. 2008;9:115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- Orkin SH, Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillusson A, Hirschi N, Vallan C, Azzalin CM, Muhlemann O. A GFP-based reporter system to monitor nonsense-mediated mRNA decay. Nucleic Acids Res. 2005;33:e54. doi: 10.1093/nar/gni052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios IM. Nonsense-mediated mRNA decay: from mechanistic insights to impacts on human health. Brief Funct Genomics. 2013;12:25–36. doi: 10.1093/bfgp/els051. [DOI] [PubMed] [Google Scholar]
- Pandit SK, Westendorp B, Nantasanti S, van Liere E, Tooten PC, Cornelissen PW, Toussaint MJ, Lamers WH, de Bruin A. E2F8 is essential for polyploidization in mammalian cells. Nat Cell Biol. 2012;14:1181–1191. doi: 10.1038/ncb2585. [DOI] [PubMed] [Google Scholar]
- Pucci F, Gardano L, Harrington L. Short telomeres in ESCs lead to unstable differentiation. Cell Stem Cell. 2013;12:479–486. doi: 10.1016/j.stem.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Zakian VA. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev. 2000;14:1777–1788. [PMC free article] [PubMed] [Google Scholar]
- Reichenbach P, Hoss M, Azzalin CM, Nabholz M, Bucher P, Lingner J. A human homolog of yeast Est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr Biol. 2003;13:568–574. doi: 10.1016/s0960-9822(03)00173-8. [DOI] [PubMed] [Google Scholar]
- Ruiz-Echevarria MJ, Peltz SW. The RNA binding protein Pub1 modulates the stability of transcripts containing upstream open reading frames. Cell. 2000;101:741–751. doi: 10.1016/s0092-8674(00)80886-7. [DOI] [PubMed] [Google Scholar]
- Schweingruber C, Rufener SC, Zund D, Yamashita A, Muhlemann O. Nonsense-mediated mRNA decay - mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim Biophys Acta. 2013;1829:612–623. doi: 10.1016/j.bbagrm.2013.02.005. [DOI] [PubMed] [Google Scholar]
- Shu J, Wu C, Wu Y, Li Z, Shao S, Zhao W, Tang X, Yang H, Shen L, Zuo X, Yang W, Shi Y, Chi X, Zhang H, Gao G, Shu Y, Yuan K, He W, Tang C, Zhao Y, et al. Induction of pluripotency in mouse somatic cells with lineage specifiers. Cell. 2013;153:963–975. doi: 10.1016/j.cell.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Rebbapragada I, Lykke-Andersen J. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol. 2008;6:e111. doi: 10.1371/journal.pbio.0060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K, Dalton S. Myc transcription factors: key regulators behind establishment and maintenance of pluripotency. Regen Med. 2010;5:947–959. doi: 10.2217/rme.10.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow BE, Erdmann N, Cruickshank J, Goldman H, Gill RM, Robinson MO, Harrington L. Functional conservation of the telomerase protein Est1p in humans. Curr Biol. 2003;13:698–704. doi: 10.1016/s0960-9822(03)00210-0. [DOI] [PubMed] [Google Scholar]
- Sommer CA, Stadtfeld M, Murphy GJ, Hochedlinger K, Kotton DN, Mostoslavsky G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperka T, Wang J, Rudolph KL. DNA damage checkpoints in stem cells, ageing and cancer. Nat Rev Mol Cell Biol. 2012;13:579–590. doi: 10.1038/nrm3420. [DOI] [PubMed] [Google Scholar]
- Steiner BR, Hidaka K, Futcher B. Association of the Est1 protein with telomerase activity in yeast. Proc Natl Acad Sci USA. 1996;93:2817–2821. doi: 10.1073/pnas.93.7.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumpel S, Rudolph KL. The role of telomere shortening in somatic stem cells and tissue aging: lessons from telomerase model systems. Ann N Y Acad Sci. 2012;1266:28–39. doi: 10.1111/j.1749-6632.2012.06547.x. [DOI] [PubMed] [Google Scholar]
- Varlakhanova NV, Cotterman RF, deVries WN, Morgan J, Donahue LR, Murray S, Knowles BB, Knoepfler PS. myc maintains embryonic stem cell pluripotency and self-renewal. Differentiation. 2010;80:9–19. doi: 10.1016/j.diff.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- Wang Y, Erdmann N, Giannone RJ, Wu J, Gomez M, Liu Y. An increase in telomere sister chromatid exchange in murine embryonic stem cells possessing critically shortened telomeres. Proc Natl Acad Sci USA. 2005;102:10256–10260. doi: 10.1073/pnas.0504635102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- Wang D, Wengrod J, Gardner LB. Overexpression of the c-myc oncogene inhibits nonsense-mediated RNA decay in B lymphocytes. J Biol Chem. 2011;286:40038–40043. doi: 10.1074/jbc.M111.266361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J, Damgaard I, Bryder D, Theilgaard-Monch K, Thoren LA, Nielsen FC, Jacobsen SE, Nerlov C, Porse BT. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev. 2008;22:1381–1396. doi: 10.1101/gad.468808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J, Waage J, Tian G, Zhao J, Damgaard I, Jakobsen JS, Kristiansen K, Krogh A, Wang J, Porse BT. Mammalian tissues defective in nonsense-mediated mRNA decay display highly aberrant splicing patterns. Genome Biol. 2012;13:R35. doi: 10.1186/gb-2012-13-5-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisdom R, Lee W. The protein-coding region of c-myc mRNA contains a sequence that specifies rapid mRNA turnover and induction by protein synthesis inhibitors. Genes Dev. 1991;5:232–243. doi: 10.1101/gad.5.2.232. [DOI] [PubMed] [Google Scholar]
- Wittkopp N, Huntzinger E, Weiler C, Sauliere J, Schmidt S, Sonawane M, Izaurralde E. Nonsense-mediated mRNA decay effectors are essential for zebrafish embryonic development and survival. Mol Cell Biol. 2009;29:3517–3528. doi: 10.1128/MCB.00177-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S, Mirels LF, Calayag MC, Bishop JM. Premature termination of transcription from the P1 promoter of the mouse c-myc gene. Proc Natl Acad Sci USA. 1991;88:11383–11387. doi: 10.1073/pnas.88.24.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeilding NM, Rehman MT, Lee WM. Identification of sequences in c-myc mRNA that regulate its steady-state levels. Mol Cell Biol. 1996;16:3511–3522. doi: 10.1128/mcb.16.7.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Hidaka K, Futcher B. The Est1 subunit of yeast telomerase binds the Tlc1 telomerase RNA. Mol Cell Biol. 2000;20:1947–1955. doi: 10.1128/mcb.20.6.1947-1955.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Kim J, Yuan X, Braun T. Epigenetic modifications of stem cells: a paradigm for the control of cardiac progenitor cells. Circ Res. 2011;109:1067–1081. doi: 10.1161/CIRCRESAHA.111.243709. [DOI] [PubMed] [Google Scholar]
- Zhou ZW, Liu C, Li TL, Bruhn C, Krueger A, Min W, Wang ZQ, Carr AM. An essential function for the ATR-activation-domain (AAD) of TopBP1 in mouse development and cellular senescence. PLoS Genet. 2013;9:e1003702. doi: 10.1371/journal.pgen.1003702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.