Abstract
G-quadruplexes (G4) are polymorphic four-stranded structures formed by certain G-rich nucleic acids, with various biological roles. However, structural features dictating their formation and/or functionin vivo are unknown. InS. cerevisiae, the pathological persistency of G4 within the CEB1 minisatellite induces its rearrangement during leading-strand replication. We now show that several other G4-forming sequences remain stable. Extensive mutagenesis of the CEB25 minisatellite motif reveals that only variants with very short (≤ 4 nt) G4 loops preferentially containing pyrimidine bases trigger genomic instability. Parallel biophysical analyses demonstrate that shortening loop length does not change the monomorphic G4 structure of CEB25 variants but drastically increases its thermal stability, in correlation with thein vivo instability. Finally, bioinformatics analyses reveal that the threat for genomic stability posed by G4 bearing short pyrimidine loops is conserved inC. elegans and humans. This work provides a framework explanation for the heterogeneous instability behavior of G4-forming sequencesin vivo, highlights the importance of structure thermal stability, and questions the prevailing assumption that G4 structures with short or longer loops are as likely to formin vivo.
Keywords: genomic instability, G-quadruplex, minisatellite, Phen-DC3, Pif1
Introduction
G-quadruplexes (G4) are four-stranded structures formed by certain G-rich DNA or RNA sequences consisting in the stacking of multiple ‘G-quartets’ (a planar arrangement of four guanines (Gellertet al, 1962)) coordinated by cations (Williamsonet al, 1989). Intramolecular G4-forming sequences typically contain four tracts of consecutive guanines separated by relatively short-loop regions of the form G3NxG3NxG3NxG3 where N can be any nucleotide, andx is usually 7 or less (Huppert & Balasubramanian, 2005; Guedinet al, 2010). Biophysical and structural studies revealed an impressive diversity of G4 conformations depending on the number of G-quartets, the length of the loops, and their sequences as well as different strand orientation (Burgeet al, 2006) and handedness (Chunget al, 2015). However, how this conformational diversity and the thermodynamic properties of these transient secondary structures modulate their cellular functions remains poorly understood.
Compelling evidence implicates G4 motifs in various biological processes (reviewed in Maizels & Gray, 2013), including regulation of transcription (Siddiqui-Jainet al, 2002; Lawet al, 2011), telomere capping (Paeschkeet al, 2005, 2008), replication initiation at certain origins (Valtonet al, 2014; Foulket al, 2015), programmed genome rearrangements (Cahoon & Seifert, 2009), and accidental genomic instability (Kruisselbrinket al, 2008; Ribeyreet al, 2009; Piazzaet al, 2010, 2012; Lopeset al, 2011) as well as RNA maturation, translation, and transport (Wieland & Hartig, 2007; Decorsiereet al, 2011; Subramanianet al, 2011). During replication, the formation of intramolecular G4 is likely facilitated by the occurrence of single-strand DNA regions, but the determinants that affect the folding and stability of G4in vivo remain to be elucidated.In vitro, several helicases unwind G4 that are strong impediments to the replicative polymerase progression (Woodfordet al, 1994). Consequently, the formation and persistence of G4 in helicase defective cells or upon G4 stabilization with G4 ligands are highly suspected to drive the genomic instability of G4-prone genomic regions (Cheunget al, 2002; Kruisselbrinket al, 2008; Rodriguezet al, 2012; Vannieret al, 2012; Kooleet al, 2014).
In previous studies, we examined the genomic instability of the G4-forming human minisatellite CEB1 in mitotically growingS. cerevisiae cells. In the absence of Pif1, an evolutionary conserved G4 unwinding helicase (Ribeyreet al, 2009; Sanders, 2010; Paeschkeet al, 2013), frequent expansion/contraction of the CEB1 tandem array is observed (Ribeyreet al, 2009). This instability depends on the ability of the CEB1 motif (39 nt) to form G4in vitro and was not observed with the G-mutated array (Ribeyreet al, 2009; Lopeset al, 2011; Piazzaet al, 2012). Consistently, treatment of wild-type (WT) cells with the Phen-DC3 G4-ligand (De Cianet al, 2007; Monchaudet al, 2008; Piazzaet al, 2010) phenocopies thePIF1 deletionin vivo (Piazzaet al, 2010; Lopeset al, 2011). Physical analysis of replication intermediates by 2D-gel revealed that G4 specifically perturbs the leading-strand replication, thus yielding CEB1 internal rearrangements in an orientation-dependent manner (Lopeset al, 2011). Here, we use this sensitive assay to characterize the G4 determinants dictating genomic instability in yeast. We assayed several validated G4-forming sequences and found that some but not all minisatellites exhibit instability. We identified the molecular determinants driving thein vivo instability by extensive mutagenesis of the stable human CEB25 minisatellite motif and parallel biophysical characterization of the resulting G4 structure by UV, CD and NMR spectroscopy. The CEB25 G4 structure has been recently solved by NMR (Amraneet al, 2012); it adopts an all-parallel strand arrangement connected by propeller loops, the first and third loop being a single T residue and the central loop being 9 nt long. Each motif in a CEB25 tandem array adopts the same monomorphic structure, leading to the possibility to form a homogeneous ‘pearl-necklace’ G4 structure in minisatellites (Amraneet al, 2012). Here, we show that only variants with shortened loops (≤ 4 nt) and a maximal total loop length of 5 nt containing pyrimidines exhibit minisatellite instability. Shortening the loop does not alter the monomorphic core structure of the CEB25 G4 but drastically increases its thermal stability, in correlation with itsin vivo behavior. Finally, we performed a bioinformatics analysis of single-nucleotide loop G4 motifs in various model organisms. This enabled us to severely narrow the fraction of potential G4 motifs in theS. cerevisiae, S. pombe,C. elegans, and human genomes that might be ‘at risk’ to trigger genome instability. Strikingly, short pyrimidine loops are clearly under-represented compared to purine loops, but are strongly enriched for DNA damage upon treatment of human cells with the G4-ligand pyridostatin (Rodriguezet al, 2012). This study highlights the conserved threat for genomic stability posed specifically by highly stable G4 structures and alters the prevailing assumptions that G4 structures with short or longer loops are as likely to formin vivo and/or exert phenotypes.
Results
Heterogeneous behavior of chromosomally integrated G4-forming minisatellites
Here, we assayed the rearrangement frequency (also referred to as ‘instability’) of various synthetic minisatellites comprising natural G4 motifs and variant sequences (Supplementary Table S1, Materials and Methods). All arrays were chromosomally inserted near theARS305 replication origin (Materials and Methods), and oriented so that the G-rich strand is template for the leading-strand replication machinery (‘Orientation I’ in Fig1A in Lopeset al (2011)) (Supplementary Table S2; Materials and Methods). This is our most sensitive and best characterized location for the study of G4-induced rearrangements (Lopeset al, 2011).
Figure 1.
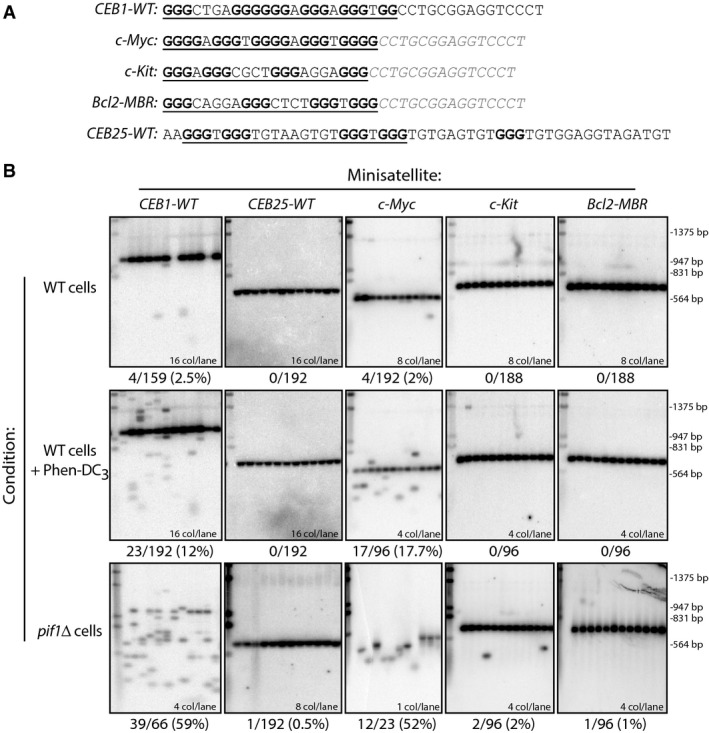
- Motif sequence of different G4-forming tandem repeats. G4 motif is underlined. G-tracts are shown in bold. Thec-Myc,c-Kit, andBcl2-MBR G4-forming sequences have been separated by the neutral CEB1 spacer (in gray) to prevent the formation of irrelevant G4 conformations resulting from the tandem organization. Details about the minisatellite size, number of motifs, and GC content are provided in Supplementary Table S1.
- Southern blot analysis of the G4-forming minisatellitesCEB1-WT (26 motifs; WT: ORT7131;pif1Δ: ORT7137),CEB25-WT (13 motifs; WT: ORT7167;pif1Δ: ORT7175),c-Myc (18 motifs; WT: ORT7338;pif1Δ: ORT7345-8),c-Kit (18 motifs; WT: ORT7339;pif1Δ: ORT7346), andBcl2-MBR (18 motifs; WT: ORT7337;pif1Δ: ORT7344) in WT cells treated for 8 generations with DMSO (control) or the G4-ligand Phen-DC3 (10 μM), and inpif1Δ cells. The number of colonies analyzed per lane and the total rearrangement frequencies are indicated. Each blot may not show all the colonies analyzed to obtain the final rearrangement frequency. DNA was digested with EcoRI that cuts within 20 nt at each side of the minisatellite, and membranes have been hybridized with the appropriate probe. The same molecular ladder (Lambda DNA digested by HindIII/EcoRI) is run in the first lane of each blot. Frequencies and statistical comparison are reported in Supplementary Table S3.
In WT cells, aCEB1-WT array is rather stable (4 rearrangements/159 colonies) but undergoes frequent rearrangements upon addition of 10 μM Phen-DC3 or in the absence of Pif1 (23/192 and 39/66;P-value vs. WT cells = 9.52 × 10−4 and 2 × 10−21, respectively) (Fig1B, Supplementary Table S3). In contrast,CEB25-WT remained stable in both contexts (0/192 and 1/192, respectively), not significantly different from WT cells (0/192) (Fig1B, Table1). Thus, conditions that induced expansion–contraction of CEB1 exert no effect on CEB25. This is not due to an intrinsic inability of CEB25 to rearrange since, like CEB1, it exhibits expansion and contraction in therad27Δ mutant (data not shown).
Table 1.
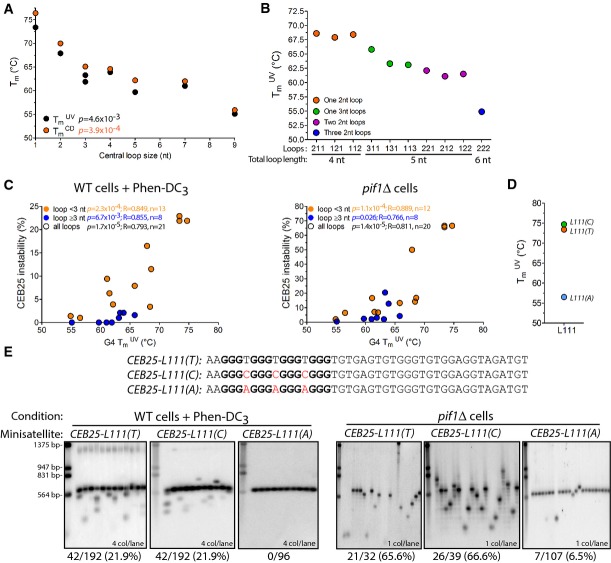
Sequence andin vivo instability ofCEB25 allele variants in different contexts, and thermal stability of their associated G4
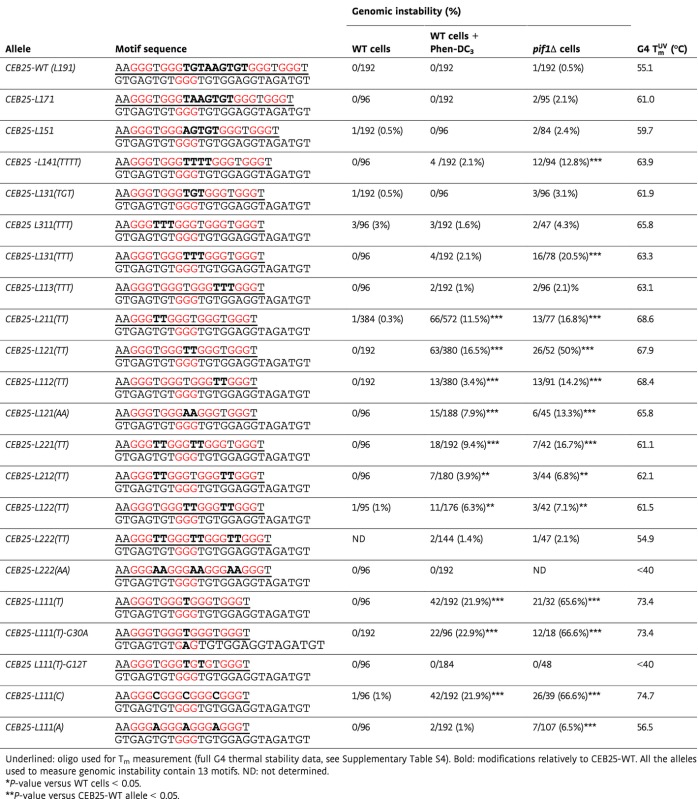 |
To investigate the behavior of other G4-prone sequences, we constructed three other minisatellite arrays each containing 18 identical G4 motifs. The G4-prone sequences were separated from one another by a non-G4 sequence spacer in order to prevent inter-motif G4 formation (Fig1A; spacer italicized in gray; full array information in Supplementary Table S1). We chose the well-characterized G4 motifs present in thec-Myc andc-Kit oncogene promoters, and at the major translocation t(14:18) breakpoint found in follicular lymphoma, in the vicinity of theBcl2 gene (Bcl2-MBR). The c-Myc motif can adopt two different conformations depending on the G-tracts used, both exhibiting three-layered G-quartets and all propeller loops (Phanet al, 2004; Ambruset al, 2005). The c-Kit motif forms a unique G4 structure utilizing an isolated guanine residue and a snapback segment of two guanine residues at the 3′ end of the sequence to complete a pseudo-backbone (Phanet al, 2007; Toddet al, 2007; Weiet al, 2012). The Bcl2-MBR motif forms a three-layered parallel G4 structure (Nambiaret al, 2011). Intriguingly, we found that thec-Myc allele exhibited significant destabilization upon Phen-DC3 treatment andPIF1 deletion (17/96 and 12/23,P-value vs. untreated WT cells = 4.56 × 10−6 and 1.3 × 10−10, respectively), while thec-Kit andBcl2-MBR alleles remained stable in the same conditions (Fig1B, Supplementary Table S3). Thus,c-Myc behaves likeCEB1-WT, whilec-Kit andBcl2-MBR behave likeCEB25-WT. Hence, despite being able to form G4in vitro, only a subset of G4-forming sequences exhibit genomic instability in the same yeast assay.
The 9-nt central loop of the CEB25 G4 is required and sufficient to stabilize the arrayin vivo
The sharp differences in the behavior of the G4-prone sequences prompted us to investigate the underlying molecular basis, using the CEB25 G4 as a model. To achieve this, we assayed the instability of CEB25 allele variants bearing modified G4 motifs (listed in Table1, full allele information in Supplementary Table S1) and performed biophysical analyses of the G4 variants, presented afterward.
A striking structural feature of theCEB25-WT G4 motif is the presence of a long central loop of 9 nt (Fig2A). To address whether this loop account for the stablein vivo behavior ofCEB25-WT (also referred to asL191, with the numbers indicating the sizes of three loops), we first replaced it by a single thymine residue to yield theCEB25-L111(T) variant (Fig2A). Whereas theCEB25-L111(T) array is stable in WT cells (0/96 rearrangements), it became unstable upon addition of Phen-DC3 (42/192) or deletion ofPIF1 (21/32) (Fig2A, Table1). These instabilities are the highest ever measured in our experimental system, especially for such short minisatellites (13 motifs). These results were confirmed with an independent strain bearing a shorterCEB25-L111(T) allele containing 8 motifs (CEB25-L111(T)-8m); it is also highly destabilized in the presence of Phen-DC3 or in the absence of Pif1 (10/94 and 17/38, respectively). Thus, the variantCEB25-L111(T) behaves like CEB1.
Figure 2.
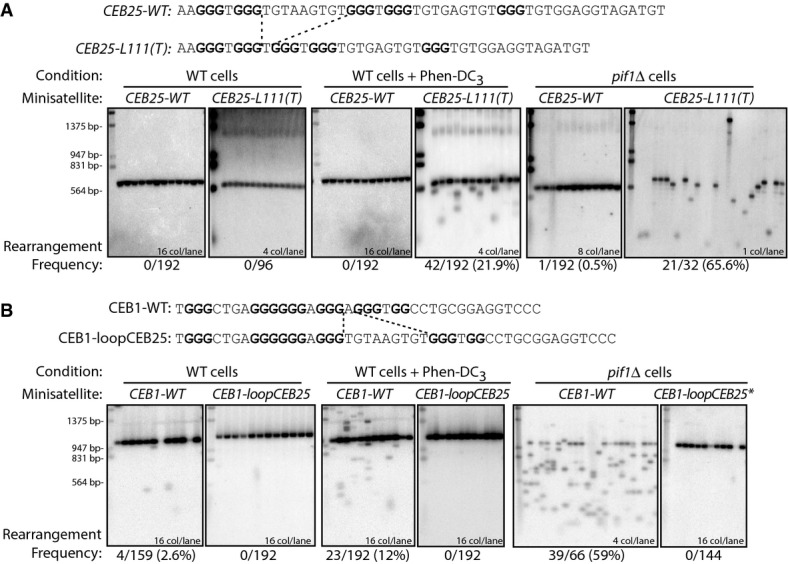
- Replacement of the central 9-nt loop ofCEB25-WT by a single T inCEB25-L111(T) results in the destabilization of the minisatellite in Phen-DC3-treated WT cells (ANT1903), and inpif1Δ cells (ANT1917).
- Replacement of a 1-nt loop ofCEB1-WT by the 9-nt-long central loop ofCEB25-WT inCEB1-loopCEB25 results in the stabilization of the minisatellite in Phen-DC3-treated WT cells (ORT7171), and inpif1Δ cells (ORT7186-5). The parentalCEB1-loopCEB25 allele (*) is 2 motifs shorter in thepif1Δ mutant than in WT cells (24 motifs instead of 26). All other alleles contain 26 motifs. Analysis was done as in Fig1B.
Conversely, we substituted the central single-nucleotide adenine loop within the G4 motif of CEB1 by the 9-nt central loop of CEB25. Strikingly, theCEB1-loopCEB25 allele remained fully stable in both Phen-DC3-treated WT cells and in thepif1Δ mutant (0/192 and 0/144, respectively;P-values vs.CEB1-WT < 2.2 × 10−16) (Fig2B, Supplementary Table S3). The abolishment of the CEB1 instability was confirmed with a second allele bearing a different 9-nt-long loop (Supplementary Fig S1, Supplementary Table S4). Thus, these CEB1 loop size variants behave likeCEB25-WT. Altogether, these results demonstrate that a single long loop within the G4 motif, although not affecting the ability to adopt a G4 structurein vitro, is required and sufficient to stabilize the minisatellitein vivo.
Mutagenesis of the unstableCEB25-L111 variant
The unstable behavior ofCEB25-L111(T) strongly suggests that persistent G4s are formedin vivo. To confirm thatCEB25-L111(T) instability depends on G4 folding, we constructed theCEB25-L111(T)-G12T array (Table1) bearing a single G→T substitution in one of the four G-triplets involved in CEB25 G4 formationin vitro. As expected, this single-point mutation abolished the minisatellite instability in both Phen-DC3-treated WT cells and inpif1Δ cells (Table1, and Supplementary Fig S2). Consistently, the single-point mutation of another G-triplet (G30A) not involved in CEB25 G4 formation (Amraneet al, 2012) had no effect on the rearrangement frequencies: The resultingCEB25-L111(T)-G30A allele exhibited instability levels not significantly different from those ofCEB25-L111(T) in both Phen-DC3-treated (22/96 vs. 42/192, respectively) andpif1Δ cells (12/17 vs. 21/32, respectively) (Table1, and Supplementary Fig S2). These results demonstrate that alike the natural CEB1 minisatellite sequence (Piazzaet al, 2010, 2012), the destabilization of the variantCEB25-L111(T) minisatellite depends on its G4 motif.
Total loop length and position requirements for CEB25 instability in vivo
Next, we investigated the granularity of the loop length effect on CEB25 instability. First, we shortened the 9-nt central loop of CEB25 from the 5′ end to yield theCEB25-L171,CEB25-L151, andCEB25-L131(TGT) variants. Remarkably, these constructs were stable upon Phen-DC3 treatment and in the absence of Pif1 (Fig3A, Table1). Then, we built theCEB25-L121(TT),CEB25-L131(TTT), andCEB25-L141(TTTT) variants homogenized to bear only T in the central loop. Upon treatment of WT cells with Phen-DC3, theCEB25-L141(TTTT) andCEB25-L131(TTT) variants, likeCEB25-L131(TGT), were stable, but strikingly, theCEB25-L121(TT) variant was destabilized (63/380,P-value vs. WT cells = 1.2 × 10−12), suggesting that the CEB25 variants become significantly unstable when the central loop is less than 3 nt in length (Fig3B). Consistently, inpif1Δ cells,CEB25-L121(TT) was also unstable (26/52,P-value vs. WT cells = 2.7 × 10−17) (Table1 and Fig3A). However, in contrast to the stableCEB25-L131(TGT) variant,CEB25-L131(TTT) was clearly destabilized (16/78,P-value vs. WT cells = 1 × 10−6), quantitatively slightly less than theCEB25-L121(TT) and much less than theCEB25-L111(T) variant. As well, theCEB25-L141(TTTT) was slightly unstable (12/94,P-value vs. WT cells = 1.5 × 10−4). These results indicate that the threshold of instability of the CEB25 variants in the presence of Phen-DC3 and in the absence of Pif1 is ≤ 2 and 4 nt in length, respectively, but also depends on the nucleotide composition (see below, Fig3B). This threshold difference between the two conditions might reflect the higher sensitivity of the mutant situation.
Figure 3.
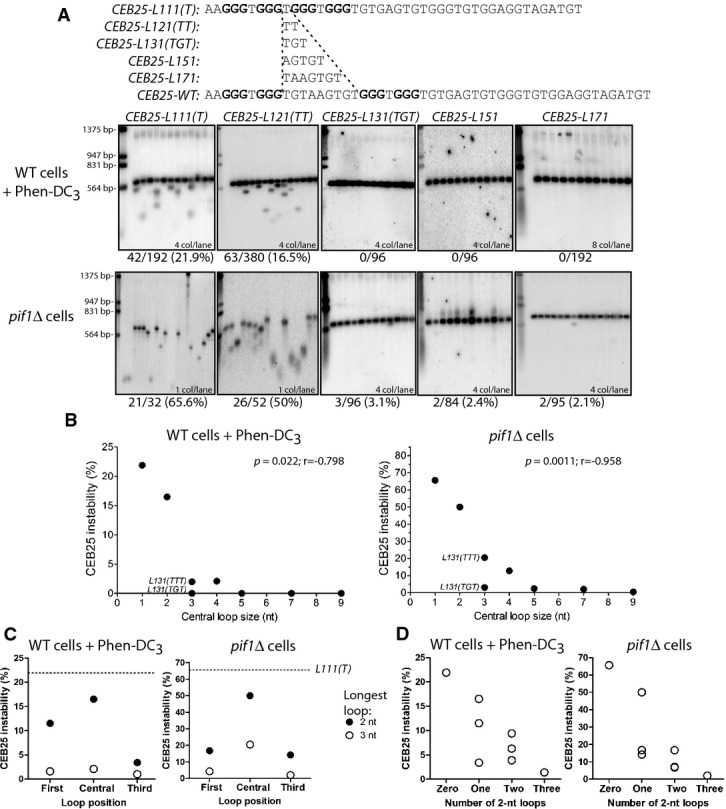
- Southern blot analysis of CEB25 allele variants with shortened central loop length in WT cells treated with Phen-DC3 (top panel) andpif1Δ cells (bottom panel). From left to right: WT strains are ANT1903, ANT1904, ORT7333, ORT7334, and ANT1901;pif1Δ strains are ANT1917, ANT1918, ORT7340, ORT7341, and ANT1902. All the alleles contain 13 motifs. * indicates incompletely digested DNA. Analysis was done as in Fig1B.
- Graphic representation of the instability measurement of central loop length CEB25 variants in WT cells treated with Phen-DC3 (left panel) and inpif1Δ cells (right panel). Instability is inversely correlated to the central loop length in both contexts (two-tailed Spearman correlation test). Alleles bearing sequence modifications other than the central loop (side loops, or intervening sequence) have not been plotted.
- Position effect of a single loop of 2 (filled circles) or 3 nt (open circles) in Phen-DC3-treated WT cells (left panel), and inpif1Δ cells (right panel). Other loops are single residues, and all the nucleotides in loops are thymine. The dotted line denotes the instability of theCEB25-L111(T) allele.
- Effect of the number of 2-nt-long loops (zero, one, two or three) on the CEB25 instability in Phen-DC3-treated WT cells (left panel), and inpif1Δ cells (right panel). Loops that are not 2-nt-long are single residues (consequently the ‘zero’ value corresponds to theCEB25-L111(T) allele). All loops are thymine.
Next, we asked whether the position of the longer loop within the G4 motif would affect the instability of CEB25. For this purpose, we built theCEB25-L311(TTT) andCEB25-L113(TTT) variants in which the 3-nt loop has been moved in the first and third position, respectively. In Phen-DC3-treated cells andpif1Δ cells, both constructs were stable (Fig3C, Table1). Similarly, we moved the 2-nt loop in first or third position inCEB25-L211(TT) andCEB25-L112(TT), respectively. Both alleles exhibited a significant increase of instability upon Phen-DC3 treatment (66/572 and 13/380,P-values vs. WT cells = 1.6 × 10−14 and 6.1 × 10−3, respectively) or in the absence of Pif1 (13/77 and 13/91,P-values vs. WT cells = 3.7 × 10−10 and 2.1 × 10−7) (Fig3C, Table1). Thus, a single 2-nt loop located at any position within the G4 motif limits but does not preclude CEB25 instability. Quantitatively, bearing the 2- and 3-nt loops in lateral positions is more innocuous for the stability of the array than in the central position (Fig3C).
Moreover, we examined the impact of the combinatorial presence of several loops of variable length. The addition of a second 2-nt loop (TT) in theCEB25-L221(TT),CEB25-L212(TT), andCEB25-L122(TT) variants did not abolish CEB25 instability, but decreased it on average ≈two- to threefold compared to the variants bearing only one 2-nt loop in both Phen-DC3 andpif1Δ context, respectively (Fig3D, Table1). However, theCEB25-L222(TT) variant bearing three 2 nt loops became stable in these conditions (Fig3D, Table1). Hence, each 2-nt loop contributes to a decrease in the destabilizing potential of the G4 motif (Fig3D).
Altogether, the above experiments uncovered a drastic decrease of the CEB25 G4-dependent instability with an incremental increase of a single loop from 1 to 3 nt and outlined the subtle combinatorial burden of each loop, above which CEB25 remains stable.
All variant sequences form intra-molecular parallel G4 resembling native CEB25
To rationalize the observations above, we investigated the conformational and thermodynamic properties of CEB25 variant oligonucleotides (sequences underlined in Table1), including: (i) several mutants to probe the effect of central loop shortening by replacing loop sequence with poly-thymine, that is, CEB25-L111(T), CEB25-L121(TT), CEB25-L131(TTT), and CEB25-L141(TTTT) or by truncating natural loop residues from the 5′ side, that is, CEB25-L131(TGT), CEB25-L151, and CEB25-L171 (folds later shown in Fig4E); (ii) two mutants to assess positional consequence of 3-nt propeller loop within the structure, that is,CEB25-L311(TTT) and CEB25-L113(TTT); (iii) five mutants to address the position and number of 2-nt loops, that is,CEB25-L211(TT), CEB25-L112(TT), CEB25-L221(TT), CEB25-L212(T), CEB25-L122(TT), and CEB25-L222(TT) (Fig4F-I); (iv) two mutants to measure the stability of all 1-nt loops with all C or A residues, that is,CEB25-L111(C), andL111(A); and (v) one mutant bearing a mutated G-tract, that is,L111(T)-G12T.
Figure 4.
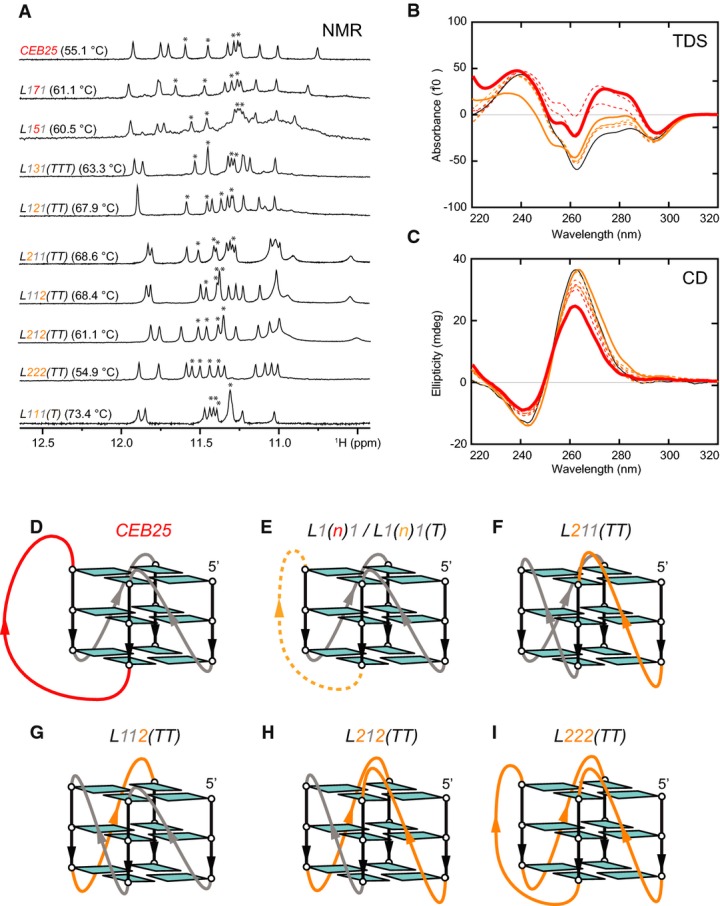
G4 formed by CEB25 native and representative variant sequences
A Imino proton spectra of CEB25 and mutants in potassium solution. Except for the CEB25 spectra, recorded in ≥ 20 mM K+ solution, all other spectra were obtained in 1 mM KPi buffer. UV-derived melting temperatures are shown in brackets. Solvent-exchange protected imino proton peaks are marked by asterisks.
B, C Thermal difference spectra (TDS)(B) and CD spectra (C) of CEB25 and mutants in potassium solution. Samples were dissolved in 1 mM KPi buffer at ˜ 4 μM DNA strand concentrations. TDS and CD spectra are in colors associated with those in (A). Spectra plotted in broken lines are originated from native [-L1(n)1] or poly-T [L1(n)1T] loop sequences.
D–I G4 folding topologies: (D) CEB25 comprising an extended central loop of 9 nt; (E) mutants involving central loops of variable length (n from 1 to 7 nt) and sequence (native loop sequence or poly-thymine); (F, G)L211(TT) andL112(TT) containing thymine loops of 2 and 1 nt at indicated positions; (H)L212(TT) consisting of two thymine loops of 2 nt and one central thymine loop of 1-nt; (I)L222(TT) consisting of three thymine loops of 2 nt. Tetrad-bound guanines and backbones are colored cyan and black, respectively. 1-nt thymine central loop is in gray; 9-nt natural central loop, red; 5–7-nt central loop of native or thymine sequence, red (broken-line); 1-, 2-, and 3-nt poly-thymine central loop, orange (broken-line); 2-nt thymine side loop, orange.
CEB25 (CEB25) forms a parallel-stranded three-layered G4 with three propeller loops of 1, 9, and 1 nt, respectively (Fig4D) (Amraneet al, 2012). Thein vitro formation of a single three-layered G4 structure for all variant sequences was confirmed by NMR spectra showing twelve major imino proton peaks (four for each G-tetrad layer) at ∼10–12 ppm (Fig4A; Supplementary Fig S3) (Adrianet al, 2012). Thermal difference UV absorption spectra (TDS) and CD spectroscopy were used to support G4 formation (Mergnyet al, 2005) and to identify their strand orientations (Grayet al, 2008), respectively (Fig4B and C; Supplementary Fig S4). When dissolved in 1 mM KPi buffer, TDS of each mutant generally showed typical pattern of a G4 structure with a negative minimum at 295 nm and two positive maxima at 240 and 275 nm (Fig4B; Supplementary Fig S4) (Mergnyet al, 2005). Concurrently, CD spectrum of each mutant displayed a positive maximum at 260 nm and a negative minimum at 240 nm, characteristic of a parallel-stranded G-quadruplex (Fig4C; Supplementary Fig S4) (Grayet al, 2008). The stoichiometry of G4 was deduced based on solvent-exchange protection pattern of its imino proton peaks. For each of the mutants, there were four peaks left after one hour exposure in D2O solvent (which are associated with one well-protected middle G-tetrad layer within a three-layered G4) (Fig4A; Supplementary Fig S3), thus implying monomeric nature of folded G4. Supported by NMR, UV, and CD data, all variant sequences form intra-molecular parallel-stranded G4 structures, similar to that of nativeCEB25. Thus, the differential behavior of the CEB25 variantsin vivo cannot be explained by conformational change of the G4.
Thermal stability of variant sequences is dependent on loop sizes
The thermal stability of CEB25 and variant G4s was measured from the melting temperatures (Tm) in heating/cooling experiments performed by UV and CD spectroscopy (Table1, Supplementary Table S4). Parallel G4 containing all 1-nt propeller loops of a pyrimidine residue is known to be extremely stable in physiological salt condition at ∼100 mM K+ (Rachwalet al, 2007a; Guedinet al, 2010). Indeed, the melting temperature ofL111(T) was above 80°C and could not be accurately determined even at relatively low concentration of potassium cations in 5–20 mM KPi buffer. For this reason, all sequences were dissolved in 1 mM KPi buffer to yield melting temperatures within the sensitive temperature region of CD or UV heating/cooling experiments.
Compared with the nativeCEB25-L191 sequence that was characterized with a Tm of 55.1°C, a drastic increase of TmUV to 73.4°C was recorded for CEB25-L111(T) (Fig5A, Table1) (Rachwalet al, 2007a). TheCEB25-L121(TT),CEB25-L131(TTT) andL141(TTTT) sequences were found to have TmUV of 67.9°C, 63.3°C, and 63.9°C, respectively (Fig5A, Table1). Adding one thymine to a 1- and 2-nt poly-thymine central loop monotonically decreases melting temperatures by ΔTmUV (1 -> 2 nt) = −5.5°C and ΔTmUV (2 -> 3 nt) = −4.6°C, respectively. Interestingly, the 4-nt poly-thymine central loop in theL141(TTTT) marginally stabilizes the structure relative to theL131(TTT), that is, ΔTmUV (3 -> 4 nt) = 0.6°C. It may result from inter-residue interaction within the longer loop inL141(TTTT). These results were confirmed independently by CD spectroscopy (Fig5A, Supplementary Table S4).
Figure 5.
CEB25 variant instability correlates to thermal stability of its associated G4
A, B Thermal stability dependence on loop length and position as measured by UV and CD spectroscopy. All melting temperatures (Tm) were obtained in 1 mM KPi buffer at ˜ 4 μM DNA strand concentrations. (A) Thermal stability of CEB25 G4 variants is inversely correlated to the central loop length (P-values obtained using the Spearman correlation test). Other loops are single thymine. The two TmUV values for a central loop of 3 nt correspond toL131(TGT) andL131(TTT), respectively. (B) Effect of the position of a single 2- or 3-nt-long loop and permutation of two or three 2-nt-long loops on the thermal stability of CEB25 G4 variants. All loop residues are thymine.
C In vivo instability of CEB25 allele variants plotted as a function of the melting temperature of the corresponding G4 measured by UV spectroscopy, in WT cells treated with Phen-DC3 (left panel) and inpif1Δ cells (right panel).P-values and correlation coefficients were obtained using a two-tailed Spearman correlation test.
D Sequence effect of single loop residue substitutions on the thermal stability of theCEB25-L111 G4. Melting temperatures (Tm) were obtained in 1 mM KPi buffer at ˜ 4 μM DNA strand concentrations.
E Sequence effect of three 1-nt-long loops on the CEB25 instability in WT cells treated with Phen-DC3 (left panel, strains ANT1903, ANT1953, and ANT1936) and inpif1Δ cells (right panel, strains ANT1917, ANT1974, and ANT1980). Analysis was done as in Fig1B.
Other variants CEB25-L131(TTT),L151, andL171 conserved the 3′ end sequence of natural loop, whose two residues (GT) were found to render base pairing interaction with flanking residues at 5′ end of the strand (Amraneet al, 2012). Containing 3-nt central loop of TGT sequence,L131(TGT) has slightly lower TmUV of 61.9°C compared to those ofL131(TTT) (ΔTmUV = −1.4°C). Addition of two purine residues to construct a 5-nt central loop of AGTGT sequence such as inL151 lowered TmUV to 59.7°C. Elongation of the central loop to 7 nt with TAAGTGT sequence inL171 produced TmUV of 61.0 °C, comparable to those ofL151 (Table 2, Fig5A). Notably, at least one Watson–Crick base pair presumably between A2 and T16 similar to that observed inCEB25-L191 was formed inL171 (Supplementary Fig S5). Indeed, additional hydrogen bond interactions from base pair formation have been shown to raise the thermal stability ofCEB25 (Amraneet al, 2012).
The all-thymine loop position (of 2 or 3 nt length) within the G4 barely affects the thermal stability of the structure (Fig5B, Table1). The inclusion of double 2-nt thymine loops at different positions such as inL221(TT),L212(TT), andL122 (TT) similarly lowered TmUV to 61.1°C, 62.1°C, and 61.5°C, respectively (Fig5B, Table1 and Supplementary Table S4). Dramatic reduction of thermal stability was observed in all 2-nt thymine loopL222(TT) with TmUV of 54.9°C. As loop position only moderately affects thermal stability, the difference in melting temperatures ofL222(TT) andL121(TT) (ΔTmUV = −13.0°C) can be attributed to additive effect of 2-nt thymine loops at three loop positions (Fig5B, Table1).
Phen-DC3 similarly binds and stabilizes CEB25 G4 variants bearing different loop length
Phen-DC3 exhibits a high affinity and an exceptional selectivity for G4 over dsDNA (De Cianet al, 2007) but poorly discriminates between different G4 conformations (Largyet al, 2011). The recently published NMR structure of the ligand in a 1:1 complex with the c-Myc Pu24T G4 provides the basis for this universal G4 recognition (Chunget al, 2014). Using the FRET melting method on oligonucleotides [L191,L131(TTT),L121(TT), andL111(T)] labeled with fluorescein and tetramethylrhodamine at the 3′ and 5′ ends, respectively, we verified that Phen-DC3 binds and stabilized similarly CEB25 G4 variants bearing different central loop length: While the thermal stabilities of the G4 formed by the labeled oligonucleotides are very close to the values measured by UV and CD spectroscopy, addition of 1 molar equivalent of Phen-DC3 resulted in a stabilization (ΔTm) of 9.6°C [forL191 andL111(T)] to 13°C [L121(TT)] and 14°C [L131(TTT)] (Supplementary Table S4). This similar increase in stability indicates that Phen-DC3 binds and stabilizes G4 bearing different central loop length to similar extents. Since Phen-DC3 inhibits G4 unwinding by Pif1in vitro (Piazzaet al, 2010), this similar recognition of G4 variants by Phen-DC3 is consistent with the treatment of WT cells that quantitatively phenocopies the absence of Pif1 (Supplementary Fig S6).
CEB25 variant instability is correlated with thermal stability of their G4
The above results reveal a striking correlation between the G4 thermal stability and thein vivo genomic instability of the cognate minisatellite (Fig5C). However, the total loop length (and hence the overall volume of the structure or the amount of ssDNA in the loops) could be a confounding factor since it is also negatively correlated to the thermal stability (P < 5 × 10−3). To address whether the G4 thermal stability dictates minisatellite instabilityin vivo independently of the loop length, we substituted all the single thymine residues in theCEB25-L111(T) allele by either cytosine or adenine to yield theCEB25-L111(C) andCEB25-L111(A) sequences, respectively (Fig5E). While T-to-C substitutions inCEB25-L111(C) had no effect on the thermal stability of the structure (TmUV = 74.7°C), thymine-to-adenine substitutions in all 1-nt loop structure inCEB25-L111(A) plummeted its melting temperature to 56.5°C [ΔTmUV = −16.9°C relative toCEB25-L111(T) values] (Fig5D, Table1). It highlights the tremendous destabilization effect of purine residue inclusion into G4 short loops (Rachwalet al, 2007b; Guedinet al, 2008). Strikingly, while theCEB25-L111(C) allele exhibited genomic instability levels very similar to those observed forCEB25-L111(T) in Phen-DC3-treated WT cells (42/192 in both cases) and inpif1Δ cells (26/39 vs. 21/32), T-to-A substitutions inCEB25-L111(A) abolished the instability in WT-treated cells (2/192) and drastically decreased it in apif1Δ mutant (7/107) (P-value vs.CEB25-L111(T) = 1.1 × 10−11 and 1.9 × 10−11, respectively) (Fig5E, Table1). To further test the effect of the loop base composition, we also generated theCEB25-L121(AA) variant containing AA in the central loop. This 2-nt substitution decreased the TmUV of the structure by 2.1°C compared toL121(TT) (Table1). Consistently, this variant was unstable in both the WT Phen-DC3-treated cells and in the absence of Pif1, but two- to fourfold less thanCEB25-L121(TT) (15/188 vs. 63/380 (P = 4.4 × 10−3) and 6/45 vs. 26/52 (P = 1.8 × 10−4), respectively) (Table1). Consistently withCEB25-L222(TT) being stable in any conditions, theCEB25-L222(AA) allele exhibited no instability (Table1). We conclude that the base composition of the loop is another determinant that affects G4-dependent CEB25 instability. The lower thermal stability of the G4 folds containing A instead of C or T residues strongly suggests that G4 thermal stability, but not the overall volume or amount of ssDNA in loops, is a direct determinant of the sequence instabilityin vivo.
Single pyrimidine loop G4 motifs are particularly ‘at risk’ for genomic stability in other eukaryotic genomes
Our study inS. cerevisiae points to G4 motifs bearing short pyrimidine (C or T) loops as being at higher risk for genomic stability than those bearing short purine loops. It prompted us to examine the diversity of the potential G4 motifs in other organisms. We determined single-nucleotide loop G4 motifs (hereafter referred to as G4L1 motifs, listed in Supplementary Table S5) and studied their base composition (Supplementary Table S6) in theS. cerevisiae,S. pombe,C. elegans, and human genomes.
The classical consensus (G3-5N1-7G3-5N1-7G3-5N1-7G3-5) used to mine genomes for G4-prone sequences (Huppert & Balasubramanian, 2005; Toddet al, 2005) identifies 27 and 30 motifs in theS. cerevisiae and theS. pombe genomes, respectively (Supplementary Fig S7A and B, Supplementary Table S5). Among those, only 3 and 2, respectively, bear single-nucleotide loops only, that all contain the most innocuous purine loops (Supplementary Fig S7B). Consequently, both yeast genomes are devoid of the most detrimental G4L1 motifs. TheC. elegans genome contains 2,226 G4-prone sequences, among which 1,172 match the G4L1 motif (Fig6A). Strikingly, the peculiarly high prevalence of mono-G-runs in theC. elegans genome accounts for 98% (1,153/1,172) of these motifs (956 perfect and 197 imperfect (e.g., bearing a single interrupting nucleotide)). Poly-G sequence G15 has been shown to form a propeller-type parallel G-quadruplex containing three single-residue guanine loops (Sengaret al, 2014). Overall, theC. elegans genome contains only 10 G4L1 motifs bearing ≥ 2 pyrimidines, two of which are in essential genes (Fig6A, Supplementary Table S5). This is 117-fold less than purine-rich monoG G4L1 motifs. In the human genome, among the 376,000 G4 motifs identified (Huppert & Balasubramanian, 2005; Toddet al, 2005), 18,153 are G4L1 motifs. With the same base probabilities (mean human genome GC content of 41%), G4L1 motifs containing only A loops are 11.1-fold more prevalent than those bearing only T loops, and G-containing motifs are 4-fold more prevalent than those bearing only C loops (Fig6B). The trend is the same for G4L1 motifs bearing non-identical loops 3.7-fold more G4 motifs containing purine loops only over those bearing pyrimidine loops only (Fig6B). This depletion is more pronounced in the repeated regions (Supplementary Fig S7C). In conclusion, the more detrimental pyrimidine-containing G4L1 motifs are either absent (S. cerevisiae andS. pombe) or strongly under-represented compared to the purine-containing ones (C. elegans and human).
Figure 6.
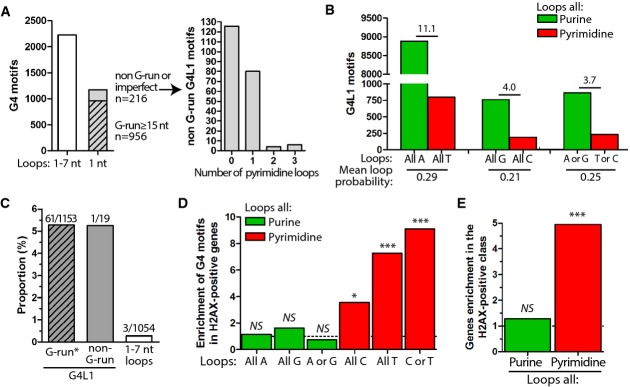
- Analysis of the long- and short-loop G4 motifs in theC. elegans genome. Left panel: number of G4 motifs bearing individual loops up to 7 nt, or single-nucleotide loops only (referred as to ‘G4L1 motifs’). Perfect mono-G-runs (≥ 15 nt, dashed) account for 81.6% (956/1,172) of the G4L1 motifs, imperfect mono-G-runs (with a single loop being different from a G) account for another 16.8% (197/1,172). Only 1.6% (19/1,172) of G4L1 motifs do not belong to the mono-G microsatellite class. Right panel: Pyrimidine loops content among the imperfect and non-G-runs G4L1 motifs (n = 216).
- Composition of the loops of the G4L1 motifs in the human genome. Pairwise comparison between G4L1 motifs bearing exclusively purines (green) and pyrimidines (red) has been performed only for bases with the same probability (e.g., A vs. T), given a mean GC content of 41% for the human genome. We separately analyzed G4L1 motifs bearing identical loops (‘all A’, ‘all T’, etc. as in ourL111 series) from those bearing non-identical loops (e.g., combination of C and T for pyrimidines and A and G for purines), because G4L1 motifs with identical loops are much more prevalent than any of the non-identical G4L1 motifs.
- The 66 non-redundant 100–200-bp deletions mapped in theC. elegans genome upon deletion of thedog-1 helicase (data obtained from Kruisselbrinket al, 2008) are localized at G4L1 motifs. The G4L1 motifs belonging to the G-run* (perfect or imperfect) and the non-G-run classes were equally affected by deletions (5.3% of the sequences in each class), 18-fold more than G4 motifs identified with the least stringent loop length constraint (1–7 nt long). Detailed sequence analysis of these G4 motifs revealed that they still bear short loops (two loops of 1 nt and one loop of 2–4 nt, see Supplementary Fig S7D).
- Fold enrichment of G4L1 motifs by loop composition in γH2AX-positive vs. γH2AX-negative genes following pyridostatin treatment in SV40-infected MRC-5 human fibroblasts (data obtained from Rodriguezet al, 2012). As in (B), we separately analyzed G4L1 motifs bearing identical loops from those bearing non-identical loops. *P < 0.05, ***P < 0.001, NS: non-significant.
- Fold enrichment of genes in the γH2AX-positive vs. γH2AX-negative class depends on the presence of G4L1 motifs bearing pyrimidine loops, but not purine loops. ***P < 0.001, NS: non-significant.
Then, we tested our prediction that pyrimidine-containing G4L1 motifs would be more prevalent at sites of damage or rearrangement than purine-containing ones or than G4 motifs bearing longer loops. First, we mapped the location of the 100–200-bp deletions that arise inC. elegans animals deficient for thedog-1 (DeletionOfG-rich DNA-1) helicase, ortholog of the G4-unwinding FANC-J helicase (Kruisselbrinket al, 2008). The authors identified a total of 69 deletions (among which 65 were non-recurrent), all present at G4 motifs. The majority (62/65) fell at G4L1 motifs: 61 at perfect or almost perfect mono-G-runs (61/1,153, 5.3%) and one at a non mono-G motif (1/19, 5.3%) (Fig7A). The 3 remaining deletions occurred at G4 motifs that had two single-nt loops and one loop ≥ 1 nt (3/1,054, 0.3%) (Supplementary Fig S7D). Thus, the G4L1 motifs are 18.6-fold more often affected by deletions than G4 motifs bearing a single loop ≥ 1 nt (P-value = 1.8 × 10−14, two-tailed Fisher's exact test), consistent with our findings in yeast.
Figure 7.
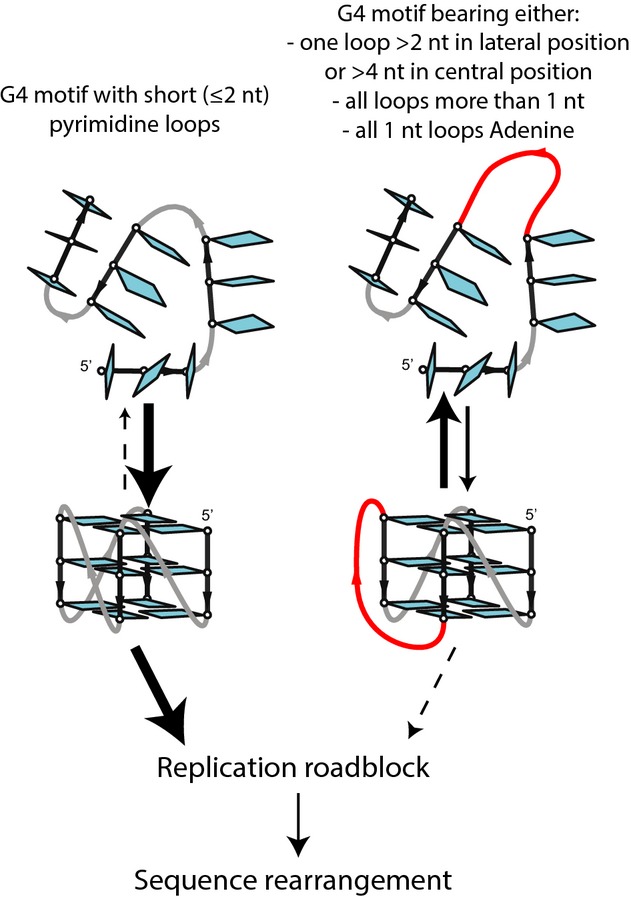
Summary of the G4 loop parameters dictating sequence instability
(i) The length of a single loop that connects the G-strands: Most variants bearing a single loop length of ≥ 3 nt remain stable, while those with a 2- and 1-nt loop exhibit a gradual increase of instability, respectively. Importantly, in the WT Phen-DC3-treated cells and in the absence of Pif1, the trend is highly correlated (Fig3B), although with a slightly different threshold (CEB25-L131(TTT) andCEB25-L141(TTTT) exhibit instability inpif1Δ cells only). It may reflect the higher sensitivity of thepif1Δ assay and/or the biochemical loop length sensitivity of the Pif1 helicase that unwinds the Phen-DC3-bound G4 in WT cells. In the absence of Pif1, the G4 might be processed by another helicase, although the similar effect of the Phen-DC3 ligand in WT cells makes it less likely. (ii) The position of the longest loop: Having the longest loop in the central position yields a higher frequency of rearrangements (for example, compareCEB25-L131(TTT) vs.CEB25-L113(TTT), Fig3C). (iii) The total number of nucleotide in the loops: Each 2-nt loop contributes to a decrease in the destabilizing potential of the G4 motif (Fig3D). (iv) The base composition of the loop is a drastic determinant of sequence instability. Most remarkably, theCEB25-L111 variants with three single pyrimidine loops (T or C) are extremely unstable in WT Phen-DC3-treated andpif1Δ cells but become fully stable upon substitution with adenine (Fig5A). Hence, the large spectrum of rearrangement frequencies observed with the CEB25 variants demonstrates the important role of the G4 loops in modulating the instability.
The second G4-related study that we re-analyzed concerns the location of the DNA damage signaling marker phospho-γH2AX in human cells treated with the G4-ligand pyridostatin (Rodriguezet al, 2012). Precisely, we mined the G4L1 motifs loop composition in the 1,214 genes (proto-oncogenes and tumor suppressor genes) analyzed for the presence of γH2AX ChIP-Seq peaks (290 γH2AX-positive and 924 γH2AX-negative genes, see Materials and Methods, Supplementary Table S7). In agreement with our prediction, pyrimidine-containing G4L1 are strongly enriched in the γH2AX-positive versus γH2AX-negative genes, while purine-containing G4L1 motifs are not (Fig6D). This is true for G4L1 with both identical loops (7.3- and 3.6-fold for T- and C-containing loops (P = 1.33 × 10−9 and 0.033, respectively) vs. 1.1- and 1.6-fold for A- and G-containing loops (P = 0.37 and 0.24, respectively)) and non-identical loops (9.1- vs. 0.8-fold for pyrimidine- versus purine-containing loops,P = 1.58 × 10−4 and 0.81, respectively) (Fig6D). Conversely, γH2AX-positive genes were strongly enriched over γH2AX-negative genes for pyrimidine-containing (4.9-fold increase,P = 1.13 × 10−8) but not purine-containing (1.3-fold,P = 0.18) G4L1 motifs (Fig6E).
Thus, our analysis of the prevalence and loop composition of single-nt loop G4 motifs in these eukaryotic genomes and their association with DNA damage and genome rearrangement phenotypes inC. elegans and human cells upon inhibition of G4 unwinding show that the rules dictating the instability of a G4 motifs determined in our model yeast system can be generalized to other evolutionary distant organisms.
Discussion
The G4 loops modulate minisatellite instability
In this study, we sought to decipher the heterogeneous instability phenotype of several G4-forming arrays in yeast. LikeCEB1, we found that thec-Myc tandem array was frequently rearranged but not theCEB25-WT,c-Kit, andBcl2-MBR sequences that also form G4in vitro (Phanet al, 2004, 2007; Ambruset al, 2005; Toddet al, 2007; Kumar & Maiti, 2008; Nambiaret al, 2011; Amraneet al, 2012; Weiet al, 2012). The molecular determinants of this behavioral discrepancy reside in the G4 loops. Extensive mutagenesis of CEB25 G4 motif uncovered four determinants (detailed in Fig7) that dictate sequence instabilityin vivo: (i) the length of a single loop that connects the G-stretches, (ii) the position of the longest loop, (iii) the total number of nucleotide in the loops, and (iv) the base composition of the loops.
The CEB25 rules are consistent with the unstable behavior of theCEB1 andc-Myc G4 sequences that exclusively contain loops of 1 or 2 nt (Phanet al, 2004; Ambruset al, 2005; Adrianet al, 2014) and the stability of thec-Kit andBcl2-MBR sequences that have two loops ≥ 4 nt (Fig1A)( Phanet al, 2007; Toddet al, 2007; Nambiaret al, 2011; Weiet al, 2012). Hence, our extensive mutagenesis study narrows the fraction of destabilizing G4-forming sequence to those matching the following consensus: G3NxG3NyG3NzG3, where N are preferentially pyrimidines,x,z ≤ 2,y ≤ 4, andx + y + z ≤ 7 nt.
Our biophysical studies demonstrated that all the CEB25 variant sequences having loops of 1 to 9 nt retained a single major intramolecular parallel G4 conformation (Fig4). Thus, their distinct and continuousin vivo behavior cannot be explained by a drastic conformational change in the structure of the G4. Rather, we uncovered that their thermodynamic stability greatly differed (varying over 25°C, for the CEB25 variants in 1 mM K+) in a trend inversely correlated with the loop length (Guedinet al, 2010) (Fig5A). Overall, our CEB25in vitro data regarding the loop length and sequence, as well as the effect of Phen-DC3 binding on G4 stability, are consistent with previous observations on other G4 (Rachwalet al, 2007a; Guedinet al, 2008, 2010; Agrawalet al, 2013; Tippanaet al, 2014). Thus, we conclude that the G4 thermodynamic stability is a key determinant for their formation and persistencein vivo and thereof of their capacity to trigger the genomic instability of the arrays during replication by acting as a stable roadblock for the replicative polymerase (Lopeset al, 2011).
Notably, most of the CEB25 variants bearing a single loop of 3 nt remain stablein vivo, even though their associated G4 Tm are slightly higher than those of the unstable CEB25 variants bearing two loops of 2 nt (Fig5B, and compare orange and blue instabilities in Fig5C). This observation may suggest the existence of additionalin vivo factors ensuring the genomic stability of the underlying sequence when a loop ≥ 3 nt is present. We envision that a G4-induced phenotype (in our case genomic instability) can be regulated by subtle changes in the G4 loops, either smoothly when acting on the structure stability below a certain loop length (≤ 2 nt), or more sharply when it exceeds this threshold. This can make G4 both versatile switches and fine-tunable regulators of discrete processes at an evolutionary time scale.
Narrowing the fraction of G4 motifs ‘at risk’ for genomic stability
The present study strongly suggests that the threat posed by short-loop G4 to genomic stability is a recurrent feature and is not limited to tandem repeats. In yeasts, the rarity of G4L1 could be explained by an evolutionary counter-selection. In contrast, the remaining presence of robust G4 motifs containing short loops in theC. elegans and human genomes (Huppert & Balasubramanian, 2005) suggests their beneficial role in other essential processes such as the regulation of gene expression. Perhaps to be evolutionary maintained, they preferentially require specialized binding or unwinding proteins to temper their potential to generate damage and rearrangements during replication. Differently, the presence of G4L1 motifs in tandem arrays aggravates the risk of instability (Lopeset al, 2011; Piazzaet al, 2012). Likewise G4-forming microsatellites of the form (GGGN)>8 (related to our CEB25-L111 series) are particularly under-represented in the human genome, and the decreasing number of (GGGA)>8 > (GGGT)>8 > (GGGC)>8 (539, 4 and zero occurrences, respectively) (Bacollaet al, 2008) correlates with the decreasing level of G4-induced instability in our yeast system (Fig5E). This under-representation of (GGGN)>8 sequences suggests that, at the evolutionary time scale, tandem arrays of such structures are prone to rearrange even in cells proficient for their unwinding, and drift toward shorter arrays with greater stability.
Notably, the telomeric sequence of almost all eukaryotes is tandem repeats, up to several kb in length, bearing the conserved ability to form G4in vitro (Tranet al, 2011) but composed of a G-triplet accompanied by 2–4 other nucleotides, never single nucleotides. In light of our study, this conserved ability to form telomeric G4 of moderate stability (Tranet al, 2011) provides a useful compromise between the requirement for the structure in the biology of telomeres (as documented for ciliates, reviewed in Lipps & Rhodes, 2009) and the threat it may pose for the stability of the array. It might explain why, despite a considerable enrichment for G4 motifs at telomeres, G4 ligands such as pyridostatin did not induce a high level of damage at telomeres compared to interstitial clusters of G4 motifs (Rodriguezet al, 2012). On the contrary, sequences forming highly stable G4 are mostly present in a non-repeated fashion (Huppert & Balasubramanian, 2005; Bacollaet al, 2008), likely limiting their propensity to induce genome rearrangements.
Remarkably, G4-induced instability could in some instances be positively selected, as it may be exploited as a rudimentary inducer of genetic diversity: For example, the only short-loop G4 (identical to the one inCEB25-L121(TT)) in the genome of the bacteriaNeisseria gonorrhoeae is located in the promoter of the pilin expression locuspilE and stimulates its recombination on polymorphicpilS pseudogenes, thus promoting antigenic variation (Cahoon & Seifert, 2009).
Having delineated the fraction of G4 motifs that are the most ‘at risk’ to trigger genome instability raises the question of how robust is our overall capacity to predict the existence of G4 structures from genomic sequences. Mostly based on biophysical studies on G4 structures formed by oligonucleotidein vitro, the G4 consensus motif of the form G≥3NxG≥3NyG≥3NzG≥3, wherex,y, andz define the loop length, has largely been used in G4 prediction algorithms (Hazelet al, 2004; Huppert & Balasubramanian, 2005; Toddet al, 2005; Rachwalet al, 2007a; Kumar & Maiti, 2008; Guedinet al, 2010). A reasonable compromise between sensitivity and robustness consisted in restricting each loop to 7 nt (Huppert & Balasubramanian, 2005; Toddet al, 2005; Guedinet al, 2010), which identifies only 27 potentially G4-forming sequences in theS. cerevisiae genome. Differently, Capraet al, by relaxing the loop length constraint to 25 nt each, identified 552 and 446 potential G4 sequences in theS. cerevisiae (Capraet al, 2010; Paeschkeet al, 2011) andS. pombe (Sabouriet al, 2014) genomes, respectively. On the opposite side, the present data, allowing a maximum loop length of 3 nt, would call for only four G4 motifs in each yeast, all being isolated sequences bearing the most innocuous purine loops (Supplementary Fig S7). Thus, how manyS. cerevisiae andS. pombe sequences really form a G4 able to create a replication impediment remains uncertain, but likely very few. If so, the enrichment of Pif1/Pfh1 binding at numerous potential G4 sequences defined with loops of 25 nt (i.e., 138 and 90) in theS. cerevisiae andS. pombe genomes, respectively (Paeschkeet al, 2011; Sabouriet al, 2014), would suggest that other prominent factors than G4-forming capacity are at play. Along the same line, the genome-wide mapping of fragile sites in yeast cells exhibiting reduced levels of Polα revealed no association with potential G4 motifs (Songet al, 2014) with loops ≤ 7 nt or ≤ 12 nt each. However, a significant association was found using up to 25 nt as loop length, even when the sequences from the more stringent datasets were removed (Songet al, 2014), suggesting that a non-G4 confounding factor causes fragility.
In conclusion, we described the heterogeneous behavior of G4-forming sequences in yeast and identified their underlying structural and biophysical specificities. G4 loops, in correlation with the thermodynamic stability of the structure, appear as the main determinants. We also highlighted the risk of assuming the reliance of a phenotype on G4 structures solely based on the ability of a sequence to adopt such structurein vitro or be called by a relaxed bioinformatics prediction. Our efforts strongly advocate for more analytical G4 prediction algorithms and a thorough validation of the G4-dependent phenotype by combining, for example, mutagenesis of the G4 motif and enhancement of the phenotype with specific G4-stabilizing molecules.
Materials and Methods
Media
Liquid synthetic complete (SC) and solid yeast–peptone–dextrose (YPD) media have been prepared according to standard protocols (Treco & Lundblad, 2001). SC media containing Phen-DC3 at 10 μM have been prepared as described previously (Piazzaet al, 2010).
Strains
Relevant genotypes of theSaccharomyces cerevisiae strains used in this study are listed in Supplementary Table S2. Strains with minisatellites inserted nearARS305 were derived from SY2209 (W303RAD5+ background)(Fachinettiet al, 2010) by regular lithium acetate transformation, as described in Lopeset al (2011). Briefly, minisatellites have been inserted nearARS305, in the intergenic region betweenYCL048w andYCL049c (precisely at chrIII:41801-41840, yielding a small deletion of 39 bp), by replacement of aURA3-hphMX cassette in the strain ORT6143-13 (WT) or ORT7178-5 (pif1Δ). The minisatellite is oriented on the chromosome in order to have its G-rich strand on the Crick molecule (e.g., template for the leading machinery of forks emanating fromARS305, see orientation I in Fig1A in Lopeset al (2011)). Correct integration and minisatellite size are verified by Southern blot. Alternatively, thePIF1 gene was deleted by transformation of apif1::HIS3 cassette after integration of the minisatellite. CorrectPIF1 deletion is verified by Southern blot using a probe external to the transforming fragment. The presence of the parental minisatellite size is also verified by Southern blot in the transformant.
Minisatellite synthesis
TheCEB1-WT (CEB1-WT-1.0 in Piazzaet al (2012)),CEB1-loopCEB25,CEB1-loopCEB25-m, andCEB25-WT (CEB25-WT-0.7 in Piazzaet al (2012)) minisatellites have been synthesized using homemade PCR-based method described in Ribeyreet al (2009). Other minisatellites have been synthesized by GenScript. Minisatellites size, sequence, and GC content are listed in Supplementary Table S1.
Measurement of minisatellite instability
Minisatellite instability during vegetative growth has been measured as previously described in WT cells andpif1Δ cells (Ribeyreet al, 2009), and Phen-DC3-treated WT cells (Lopeset al, 2011). Briefly, untreated WT cells andpif1Δ cells from a fresh patch of cells made from a single colony bearing the parental allele size (checked by Southern blot) are diluted in 5 mL of YPD (2 × 105 cells/ml), grown for 8 generations at 30°C with shaking, and spread as single colonies on YPD plates. The instability measurement in these cells thus corresponds to the rearrangement frequency after 45–50 generations. To measure minisatellite instability upon Phen-DC3 treatment, WT cells from a fresh patch on YPD were grown for 8 generations at 30°C in liquid SC containing Phen-DC3 at 10 μM (Lopeset al, 2011). Isolated colonies or pools of colonies are analyzed by Southern blot using the EcoRI digestion that cut at each side of the minisatellite. The membranes are hybridized with a probe corresponding to the minisatellite of interest. The signals are detected with a Typhoon PhosphorImager (Molecular Dynamics). The elimination of potential early clonal events (that occurred early during the colony growth before liquid culture) has been performed as described in Lopeset al (2011). In mutant strains with very high minisatellite instability (for example,CEB25-L111(T) in thepif1Δ mutant), the probability of obtaining two independent rearrangements of the same size is high. Therefore, the removal of rearrangements of the same size (suspected early clonal events) leads to an underestimation of the real rearrangement frequency. To more accurately determine the minisatellite instability in these highly unstable trains, the rearrangement frequency has been determined with fewer colonies (12–24) but on a higher number of independent clones.
DNA oligonucleotide preparation
DNA oligonucleotides (sequences see Table1 or Supplementary Table S4) were chemically synthesized on an ABI 394 DNA/RNA synthesizer. Oligonucleotides were purified and dialyzed successively against potassium chloride solution and water. Oligonucleotides were dissolved both in 1 mM potassium phosphate buffer (pH 7) and in 20 mM potassium phosphate buffer containing 70 mM potassium chloride (pH 7). DNA concentration was expressed in strand molarity using a nearest-neighbor approximation for the absorption coefficients of the unfolded species (Cantoret al, 1970).
Thermal difference spectra
Thermal difference spectra (TDS) were obtained by taking the difference between the absorbance spectra from unfolded and folded oligonucleotides that were, respectively, recorded much above (90°C) and below (20°C) its melting temperature (Tm). TDS provide specific signatures of different structural conformations (Mergnyet al, 2005). The DNA oligonucleotides at approximately 4 μM strand concentrations were prepared in 1 mM potassium phosphate buffer (pH 7). Spectra were recorded between 220 and 320 nm on a JASCO V-650 UV/Vis spectrophotometer using 1-cm pathlength quartz cuvettes. For each experiment, an average of three scans was taken, and the data were zero-corrected at 320 nm.
Circular dichroism
Circular dichroism (CD) spectra were recorded on a JASCO-810 spectropolarimeter using 1-cm pathlength quartz cuvettes. The DNA oligonucleotides at approximately 4 μM strand concentration were prepared in 1 mM potassium phosphate buffer (pH 7). For each experiment, an average of three scans was taken, the spectrum of the buffer was subtracted, and the data were zero-corrected at 320 nm.
UV/CD melting experiments
The thermal stability of G4 structures formed by oligonucleotides was characterized in heating/cooling experiments by recording the UV absorbance at 295 nm and the CD ellipticity at 260 nm as a function of temperature (Mergnyet al, 1998) using a JASCO V-650 UV/Vis spectrophotometer and a JASCO-810 spectropolarimeter, respectively. UV/CD melting experiments were conducted as previously described in Mergny and Lacroix (2003) at constant DNA strand concentrations of approximately 4 μM in 1 mM potassium phosphate buffer (pH 7). The heating and cooling rates were 0.2°C/min. Experiments were performed with 1-cm pathlength quartz cuvettes.
NMR spectroscopy
NMR experiments were performed on 600 MHz Bruker spectrometers at 25°C. The strand concentration of the NMR samples was typically 0.2–0.6 mM both in 1 mM potassium phosphate buffer (pH 7) and 20 mM potassium phosphate buffer containing 70 mM potassium chloride (pH 7). NMR spectra were zero-referenced to resonance of DSS compound.
FRET melting
Stabilization of compounds with quadruplex structure via FRET melting assay was performed in a 1.4-ml quartz cell in a fluorescence Cary Eclipse spectrophotometer with a 4-position Peltier effect thermostated cell holder. FRET melting assay was carried out with oligonucleotides equipped with FRET partners at each extremity: fluorescein/FAM molecule at 5′ end and tetramethylrhodamine (TAMRA) at 3′ end. G4-DNA oligonucleotides were prepared by heating the corresponding sequence at 90°C for 5 min in a 10 mM lithium cacodylate buffer (pH 7.4) with 1 mM KCl/99 mM LiCl, and cooling in ice for 30 min to favor the intramolecular folding by kinetic trapping. After addition of Phen-DC3 (0.2 μM), the final volume is 800 μl. Measurements were made with excitation at 492 nm and detection at 516 nm while heating at 25°C for 5 min and then from 25°C to 95°C at a 1°C/min rate.
Bioinformatics analyses of G4L1 motifs
The G4L1 motifs in theC. elegans (assembly 235, accessed from Ensembl on 01/30/2015) and human (GRCh38, accessed from the USCS Web site on 01/20/2015) genomes were determined using custom scripts (available upon request) under R 2.13.1 (R Development Core Team, 2011). To avoid bias induced by the high prevalence in the human genome of tandem repeats of the form (GGGN)≥8 (that match two or more G4L1 motifs), especially (GGGA)≥8 (539 occurrences) (Bacollaet al, 2008), we distinguished G4L1 motifs belonging to unique regions versus repeated regions of the genome (3,542 and 13,438 in human, respectively, and 1,173 overlapping the junction of the two regions) (Supplementary Fig S7C). Repeated sequences were determined by UCSC with RepeatMasker and Tandem Repeats Finder with periodicities ≥ 12 bp and soft-masked in the GRCh38 genome assembly. We only counted non-overlapping identical G4L1 motifs, and we did not merge identical overlapping motifs (for example, the (GGGA)7GGG sequence will be scored as two consecutive G4L1 motifs, not a single merged one nor five partially overlapping ones). However, overlapping motifs with different loop sequences are both scored (for example, GGGAGGGAGGGTGGGAGGG will count for two G4L1 motifs, one with loops A-A-T and one with loops A-T-A). Overlaps are indicated for each G4L1 motif (Supplementary Table S5). The G4L1 motif loop composition of theC. elegans and human genomes is provided in Supplementary Table S6. Overall, in bothC. elegans and human, a minor fraction of G4L1 motifs (16%) is considered overlapping (190/1,172 in theC. elegans genome and 2,859/18,153 in the human genome). Most of these overlaps occur in tandem repeats: In theC. elegans and human genomes, respectively, all (190/190) and 87% (2,485/2,859) of the overlapping sequences felt in the repeated portion of the genome, or at junctions between unique and repeated regions. InC. elegans, 188/190 are monoG-runs.
We also provide in Supplementary Table S5 the lists of G4 motifs with individual loops of 1–7 nt, which have been downloaded from QuadDB (now offline) (Wonget al, 2010) on 04/03/2012 (S. cerevisiae assembly 62) and 03/08/2012 (C. elegans assembly 180 andH. sapiens GRCh36). We determined theS. pombe G4 motifs using QGRS mapper (Kikinet al, 2006) using the assembly 294 on 01/31/2015.
Re-analysis of deletion breakpoint location indog-1-deficientC. elegans
We used the list of G4 motifs and monoG-runs found at deletion breakpoints provided in Table S1 in the original study by Kruisselbrinket al (2008). We manually determined the smaller possible G4 motif in each sequence. Non-monoG-run G4 motif sequences are presented in Supplementary Fig S7D. A two-tailed Fisher's exact test was used to compare the proportion of affected monoG-runs and consensus G4 motif.
Re-analysis of pyridostatin-induced γH2AX signal
Phospho-γH2AX ChIP-Seq data following pyridostatin treatment of SV40-infected MRC-5 fibroblast cells have been obtained from Rodriguezet al (2012). The study focused on a subset of 1,224 genes (482 proto-oncogenes and 742 tumor suppressors) for which a qualitative H2AX score was attributed (‘yes(**)’, ‘yes(*)’, ‘yes’, ‘yes/no’, and ‘no’; Supplementary Dataset 2 in Rodriguezet al (2012)). Using gene names of Supplementary Dataset 2 and custom scripts, we could retrieve the GRCh37 coordinates and G4 motif content from Supplementary Dataset 3 in Rodriguezet al (2012). Next, these coordinates were lifted-over to the GRCh38 release using the online Ensembl lift-over tool, and duplicated entries were manually curated to obtain a final list of 1,214 genes (479 proto-oncogenes and 735 tumor suppressors) and their associated coordinates, density of G4 motifs (or PQS for potential quadruplex sequence, loops 1–7 nt), and H2AX score (Supplementary Table S7). We then measured the intersection between G4L1 motifs of different loop composition and H2AX-positive (‘yes’ to ‘yes(**)’, score 1–3) and H2AX-negative (‘no’ and ‘yes/no’, score 0) genes, in order to determine (i) the enrichment for certain G4L1 motifs in γH2AX-positive vs. γH2AX-negative genes (Fig6D) and (ii) the enrichment for H2AX signal in genes containing G4L1 motifs bearing certain loops (Fig6E). For simplicity, we considered only G4L1 motifs bearing either 3 purine loops or 3 pyrimidine loops. In each case, enrichment was normalized to the total size of the genes. Proportion of G4L1 motifs or of G4L1 motif-containing genes in the γH2AX-positive and γH2AX-negative classes were compared using a two-tailed Fisher's exact test.
Statistical analysis
Rearrangement frequencies have been compared using a two-tailed Fisher's exact test. Correlations between Tm andin vivo instability, loop size and Tm, and between instabilities were determined using a two-tailed Spearman non-parametric correlation test. Statistical cutoff has been set to 0.05. All statistical tests have been performed using R2.13.1 (R Development Core Team, 2011) or GraphPad Prism 4.03.
Acknowledgments
We thank past and present members of our laboratories for helpful discussions. AP received fellowships from the Ministère de l'Education Nationale, de la Recherche, et de la Technologie (MENRT), and from the Association pour la Recherche sur le Cancer (ARC). AS was supported by a postdoctoral fellowship from the Fondation pour la Recherche Médicale (FRM). MA was supported by the Yousef Jameel scholarship. This research was supported by Grant ANR-12-BSV6-0002 (to AN and MPTF), Singapore Ministry of Education Academic Research Fund Tier 3 (MOE2012-T3-1-001) and Nanyang Technological University grants (to ATP) and the Singapore-France Merlion grant (to ATP and AN).
Author contributions
AP, MA, FS, AS, JL, FH, ATP, and AN designed the experiments. AP, MA, FS, BH, FH, AS, and JL performed the experiments. AP, MA, FS, BH, FH, AS, JL, MPTF, AH, ATP, and AN analyzed the data. AP performed the bioinformatics analyses. AP, MA, and AN wrote the manuscript with contributions by BH, FH, ATP, and MPTF.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information for this article is available online: http://emboj.embopress.org
References
- Adrian M, Ang DJ, Lech CJ, Heddi B, Nicolas A, Phan AT. Structure and conformational dynamics of a stacked dimeric G-quadruplex formed by the human CEB1 minisatellite. J Am Chem Soc. 2014;136:6297–6305. doi: 10.1021/ja4125274. [DOI] [PubMed] [Google Scholar]
- Adrian M, Heddi B, Phan AT. NMR spectroscopy of G-quadruplexes. Methods. 2012;57:11–24. doi: 10.1016/j.ymeth.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Agrawal P, Hatzakis E, Guo K, Carver M, Yang D. Solution structure of the major G-quadruplex formed in the human VEGF promoter in K+: insights into loop interactions of the parallel G-quadruplexes. Nucleic Acids Res. 2013;41:10584–10592. doi: 10.1093/nar/gkt784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrus A, Chen D, Dai J, Jones RA, Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- Amrane S, Adrian M, Heddi B, Serero A, Nicolas A, Mergny JL, Phan AT. Formation of pearl-necklace monomorphic G-quadruplexes in the human CEB25 minisatellite. J Am Chem Soc. 2012;134:5807–5816. doi: 10.1021/ja208993r. [DOI] [PubMed] [Google Scholar]
- Bacolla A, Larson JE, Collins JR, Li J, Milosavljevic A, Stenson PD, Cooper DN, Wells RD. Abundance and length of simple repeats in vertebrate genomes are determined by their structural properties. Genome Res. 2008;18:1545–1553. doi: 10.1101/gr.078303.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon LA, Seifert HS. An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science. 2009;325:764–767. doi: 10.1126/science.1175653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor CR, Warshaw MM, Shapiro H. Oligonucleotide interactions. 3. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers. 1970;9:1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- Capra JA, Paeschke K, Singh M, Zakian VA. G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae. PLoS Comput Biol. 2010;6:e1000861. doi: 10.1371/journal.pcbi.1000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung I, Schertzer M, Rose A, Lansdorp PM. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat Genet. 2002;31:405–409. doi: 10.1038/ng928. [DOI] [PubMed] [Google Scholar]
- Chung WJ, Heddi B, Hamon F, Teulade-Fichou MP, Phan AT. Solution structure of a G-quadruplex bound to the bisquinolinium compound Phen-DC(3) Angew Chem Int Ed Engl. 2014;53:999–1002. doi: 10.1002/anie.201308063. [DOI] [PubMed] [Google Scholar]
- Chung WJ, Heddi B, Schmitt E, Lim KW, Mechulam Y, Phan AT. Structure of a left-handed DNA G-quadruplex. Proc Natl Acad Sci USA. 2015;112:2729–2733. doi: 10.1073/pnas.1418718112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cian A, Delemos E, Mergny JL, Teulade-Fichou MP, Monchaud D. Highly efficient G-quadruplex recognition by bisquinolinium compounds. J Am Chem Soc. 2007;129:1856–1857. doi: 10.1021/ja067352b. [DOI] [PubMed] [Google Scholar]
- Decorsiere A, Cayrel A, Vagner S, Millevoi S. Essential role for the interaction between hnRNP H/F and a G quadruplex in maintaining p53 pre-mRNA 3′-end processing and function during DNA damage. Genes Dev. 2011;25:220–225. doi: 10.1101/gad.607011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachinetti D, Bermejo R, Cocito A, Minardi S, Katou Y, Kanoh Y, Shirahige K, Azvolinsky A, Zakian VA, Foiani M. Replication termination at eukaryotic chromosomes is mediated by Top2 and occurs at genomic loci containing pausing elements. Mol Cell. 2010;39:595–605. doi: 10.1016/j.molcel.2010.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulk MS, Urban JM, Casella C, Gerbi SA. Characterizing and controlling intrinsic biases of Lambda exonuclease in nascent strand sequencing reveals phasing between nucleosomes and G-quadruplex motifs around a subset of human replication origins. Genome Res. 2015;25:725–735. doi: 10.1101/gr.183848.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc Natl Acad Sci USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray DM, Wen JD, Gray CW, Repges R, Repges C, Raabe G, Fleischhauer J. Measured and calculated CD spectra of G-quartets stacked with the same or opposite polarities. Chirality. 2008;20:431–440. doi: 10.1002/chir.20455. [DOI] [PubMed] [Google Scholar]
- Guedin A, De Cian A, Gros J, Lacroix L, Mergny JL. Sequence effects in single-base loops for quadruplexes. Biochimie. 2008;90:686–696. doi: 10.1016/j.biochi.2008.01.009. [DOI] [PubMed] [Google Scholar]
- Guedin A, Gros J, Alberti P, Mergny JL. How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 2010;38:7858–7868. doi: 10.1093/nar/gkq639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazel P, Huppert J, Balasubramanian S, Neidle S. Loop-length-dependent folding of G-quadruplexes. J Am Chem Soc. 2004;126:16405–16415. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikin O, D'Antonio L, Bagga PS. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34:W676–W682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koole W, van Schendel R, Karambelas AE, van Heteren JT, Okihara KL, Tijsterman M. A Polymerase Theta-dependent repair pathway suppresses extensive genomic instability at endogenous G4 DNA sites. Nat Commun. 2014;5:3216. doi: 10.1038/ncomms4216. [DOI] [PubMed] [Google Scholar]
- Kruisselbrink E, Guryev V, Brouwer K, Pontier DB, Cuppen E, Tijsterman M. Mutagenic capacity of endogenous G4 DNA underlies genome instability in FANCJ-defectiveC. elegans. Curr Biol. 2008;18:900–905. doi: 10.1016/j.cub.2008.05.013. [DOI] [PubMed] [Google Scholar]
- Kumar N, Maiti S. A thermodynamic overview of naturally occurring intramolecular DNA quadruplexes. Nucleic Acids Res. 2008;36:5610–5622. doi: 10.1093/nar/gkn543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Largy E, Hamon F, Teulade-Fichou MP. Development of a high-throughput G4-FID assay for screening and evaluation of small molecules binding quadruplex nucleic acid structures. Anal Bioanal Chem. 2011;400:3419–3427. doi: 10.1007/s00216-011-5018-z. [DOI] [PubMed] [Google Scholar]
- Law MJ, Lower KM, Voon HP, Hughes JR, Garrick D, Viprakasit V, Mitson M, De Gobbi M, Marra M, Morris A, Abbott A, Wilder SP, Taylor S, Santos GM, Cross J, Ayyub H, Jones S, Ragoussis J, Rhodes D, Dunham I, et al. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell. 2011;143:367–378. doi: 10.1016/j.cell.2010.09.023. [DOI] [PubMed] [Google Scholar]
- Lipps HJ, Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Lopes J, Piazza A, Bermejo R, Kriegsman B, Colosio A, Teulade-Fichou MP, Foiani M, Nicolas A. G-quadruplex-induced instability during leading-strand replication. EMBO J. 2011;30:4033–4046. doi: 10.1038/emboj.2011.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizels N, Gray LT. The G4 genome. PLoS Genet. 2013;9:e1003468. doi: 10.1371/journal.pgen.1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergny JL, Lacroix L. Analysis of thermal melting curves. Oligonucleotides. 2003;13:515–537. doi: 10.1089/154545703322860825. [DOI] [PubMed] [Google Scholar]
- Mergny JL, Li J, Lacroix L, Amrane S, Chaires JB. Thermal difference spectra: a specific signature for nucleic acid structures. Nucleic Acids Res. 2005;33:e138. doi: 10.1093/nar/gni134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergny JL, Phan AT, Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- Monchaud D, Allain C, Bertrand H, Smargiasso N, Rosu F, Gabelica V, De Cian A, Mergny JL, Teulade-Fichou MP. Ligands playing musical chairs with G-quadruplex DNA: a rapid and simple displacement assay for identifying selective G-quadruplex binders. Biochimie. 2008;90:1207–1223. doi: 10.1016/j.biochi.2008.02.019. [DOI] [PubMed] [Google Scholar]
- Nambiar M, Goldsmith G, Moorthy BT, Lieber MR, Joshi MV, Choudhary B, Hosur RV, Raghavan SC. Formation of a G-quadruplex at the BCL2 major breakpoint region of the t(14;18) translocation in follicular lymphoma. Nucleic Acids Res. 2011;39:936–948. doi: 10.1093/nar/gkq824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paeschke K, Bochman ML, Garcia PD, Cejka P, Friedman KL, Kowalczykowski SC, Zakian VA. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013;497:458–462. doi: 10.1038/nature12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paeschke K, Capra JK, Zakian VA. DNA replication through G-quadruplex motifs is promoted by theSaccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145:678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paeschke K, Juranek S, Simonsson T, Hempel A, Rhodes D, Lipps HJ. Telomerase recruitment by the telomere end binding protein-beta facilitates G-quadruplex DNA unfolding in ciliates. Nat Struct Mol Biol. 2008;15:598–604. doi: 10.1038/nsmb.1422. [DOI] [PubMed] [Google Scholar]
- Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structuresin vivo. Nat Struct Mol Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- Phan AT, Kuryavyi V, Burge S, Neidle S, Patel DJ. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J Am Chem Soc. 2007;129:4386–4392. doi: 10.1021/ja068739h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan AT, Modi YS, Patel DJ. Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J Am Chem Soc. 2004;126:8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza A, Boule JB, Lopes J, Mingo K, Largy E, Teulade-Fichou MP, Nicolas A. Genetic instability triggered by G-quadruplex interacting Phen-DC compounds inSaccharomyces cerevisiae. Nucleic Acids Res. 2010;38:4337–4348. doi: 10.1093/nar/gkq136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza A, Serero A, Boule JB, Legoix-Ne P, Lopes J, Nicolas A. Stimulation of gross chromosomal rearrangements by the human CEB1 and CEB25 minisatellites in saccharomyces cerevisiae depends on G-quadruplexes or Cdc13. PLoS Genet. 2012;8:e1003033. doi: 10.1371/journal.pgen.1003033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2011. ISBN 3-900051-07-0, Available at: http://www.R-project.org/ [Google Scholar]
- Rachwal PA, Brown T, Fox KR. Effect of G-tract length on the topology and stability of intramolecular DNA quadruplexes. Biochemistry. 2007a;46:3036–3044. doi: 10.1021/bi062118j. [DOI] [PubMed] [Google Scholar]
- Rachwal PA, Brown T, Fox KR. Sequence effects of single base loops in intramolecular quadruplex DNA. FEBS Lett. 2007b;581:1657–1660. doi: 10.1016/j.febslet.2007.03.040. [DOI] [PubMed] [Google Scholar]
- Ribeyre C, Lopes J, Boule JB, Piazza A, Guedin A, Zakian VA, Mergny JL, Nicolas A. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009;5:e1000475. doi: 10.1371/journal.pgen.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez R, Miller KM, Forment JV, Bradshaw CR, Nikan M, Britton S, Oelschlaegel T, Xhemalce B, Balasubramanian S, Jackson SP. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat Chem Biol. 2012;8:301–310. doi: 10.1038/nchembio.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabouri N, Capra JA, Zakian VA. The essential Schizosaccharomyces pombe Pfh1 DNA helicase promotes fork movement past G-quadruplex motifs to prevent DNA damage. BMC Biol. 2014;12:101. doi: 10.1186/s12915-014-0101-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders CM. Human Pif1 helicase is a G-quadruplex DNA-binding protein with G-quadruplex DNA-unwinding activity. Biochem J. 2010;430:119–128. doi: 10.1042/BJ20100612. [DOI] [PubMed] [Google Scholar]
- Sengar A, Heddi B, Phan AT. Formation of G-quadruplexes in poly-G sequences: structure of a propeller-type parallel-stranded G-quadruplex formed by a G(15) stretch. Biochemistry. 2014;53:7718–7723. doi: 10.1021/bi500990v. [DOI] [PubMed] [Google Scholar]
- Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Dominska M, Greenwell PW, Petes TD. Genome-wide high-resolution mapping of chromosome fragile sites inSaccharomyces cerevisiae. Proc Natl Acad Sci USA. 2014;111:E2210–E2218. doi: 10.1073/pnas.1406847111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian M, Rage F, Tabet R, Flatter E, Mandel JL, Moine H. G-quadruplex RNA structure as a signal for neurite mRNA targeting. EMBO Rep. 2011;12:697–704. doi: 10.1038/embor.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tippana R, Xiao W, Myong S. G-quadruplex conformation and dynamics are determined by loop length and sequence. Nucleic Acids Res. 2014;42:8106–8114. doi: 10.1093/nar/gku464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd AK, Haider SM, Parkinson GN, Neidle S. Sequence occurrence and structural uniqueness of a G-quadruplex in the human c-kit promoter. Nucleic Acids Res. 2007;35:5799–5808. doi: 10.1093/nar/gkm609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PL, Mergny JL, Alberti P. Stability of telomeric G-quadruplexes. Nucleic Acids Res. 2011;39:3282–3294. doi: 10.1093/nar/gkq1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treco DA, Lundblad V. Preparation of yeast media. Curr Protocols Mol Biol. 2001 doi: 10.1002/0471142727.mb1301s23. Chapter 13: Unit 13.1. [DOI] [PubMed] [Google Scholar]
- Valton AL, Hassan-Zadeh V, Lema I, Boggetto N, Alberti P, Saintome C, Riou JF, Prioleau MN. G4 motifs affect origin positioning and efficiency in two vertebrate replicators. EMBO J. 2014;33:732–746. doi: 10.1002/embj.201387506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MI, Ding H, Boulton SJ. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012;149:795–806. doi: 10.1016/j.cell.2012.03.030. [DOI] [PubMed] [Google Scholar]
- Wei D, Parkinson GN, Reszka AP, Neidle S. Crystal structure of a c-kit promoter quadruplex reveals the structural role of metal ions and water molecules in maintaining loop conformation. Nucleic Acids Res. 2012;40:4691–4700. doi: 10.1093/nar/gks023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland M, Hartig JS. RNA quadruplex-based modulation of gene expression. Chem Biol. 2007;14:757–763. doi: 10.1016/j.chembiol.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Williamson JR, Raghuraman MK, Cech TR. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- Wong HM, Stegle O, Rodgers S, Huppert JL. A toolbox for predicting G-quadruplex formation and stability. J Nucleic Acids. 2010 doi: 10.4061/2010/564946. doi: 10.4061/2010/564946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodford KJ, Howell RM, Usdin K. A novel K(+)-dependent DNA synthesis arrest site in a commonly occurring sequence motif in eukaryotes. J Biol Chem. 1994;269:27029–27035. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.