Abstract
Members of the apolipoprotein B mRNA editing complex polypeptide 1-like (APOBEC) family of enzymes exhibit inhibitory activity against a variety of exogenous and endogenous retroviruses including retrotransposons, such as long interspersed element 1 (LINE-1). Indeed, human APOBEC3A, APOBEC3B, and APOBEC3F inhibit retrotrans-position of human LINE-1, mouse IAP and MusD retrotransposons. In our study, we examined whether the inhibitory effect of APOBEC3 proteins correlates with APOBEC3 ability to bind the LINE-1 ORF1 protein. We examined the interactions between the LINE-1 ORF1 protein and the most potent LINE-1 retrotransposon inhibitors, human APOBEC3A and APOBEC3B, by immunofluorescence and immunoprecipitation. Although human APOBEC3A shows the highest inhibitory potency against LINE-1 retrotransposon, no direct interactions were identified either by immunofluorescence or by co-immunoprecipitation. APOBEC3B binds to LINE-1 ORF1 protein, yet no co-localization was detected. We concluded that APOBEC3 proteins interfere indirectly with the LINE-1 retrotransposition pathway, probably through interference with RNA targeting.
Keywords: LINE-1, retrotransposon, APOBEC3, transposable elements
Introduction
Retrotransposons constitute almost half of the human genome and are considered to be one of the major driving forces in the evolution of eukaryotic genomes.1,2 They have profoundly shaped the genomes via insertions, deletions, and DNA rearrangements and are important agents in the evolution of genes as well as complex regulatory networks in the organism.3,4 In humans, half a million copies of LINE-1 (L1) retrotransposons constitute 17% of the genome. Only 80 to 100 L1 copies are retrotranspositionally active.5 The 6 kb full-length human L1 element encodes 149 kDa ORF2 protein with endonuclease and reverse transcriptase activities and 40 kDa nucleic acids binding ORF1 protein (reviewed in Ref. 6). LINE-1 retrotransposons entered the eukaryotic genome at the time of origin of deuterostomes and are highly amplified in mammals where they represent the only active retrotransposon.7 When retrotransposons integrate in the genes, they are harmful to their hosts. For this purpose, several mechanisms have evolved to keep them in check. These include transcriptional silencing via DNA methylation,8 posttranscriptional silencing via RNA interference (RNAi),9 and cytidine deamination via the APOBEC3 family of proteins.10 APOBEC3 proteins are recently discovered cytidine deaminases that are capable of DNA and RNA editing. They belong to the apolipoprotein B mRNA editing complex polypeptide 1-like (APOBEC) family of enzymes. The most characterized members of the APOBEC family are APOBEC1,11 an editor of apolipoprotein B mRNA, and activation-induced deaminase (AID), DNA editing enzyme that is a key regulator in somatic hypermutation and class switch recombination, which enables diversification of immunoglobulins in vertebrates.12 APOBEC3 proteins are specific to mammals in which their copy number highly varies: primates have seven APOBEC3 genes, mice have only one.13 The first member of the APOBEC3 family to be discovered as a host restriction factor against HIV-1 Vif deficient virus was human APOBEC3G.14 APOBEC3G incorporates into the virion where it causes extensive deamination of C to U in the newly synthesized viral cDNA during the course of viral replication.15 Human APOBEC3F has also been shown to restrict HIV-1 Vif deficient virus.16 Subsequently, a variety of viruses including retroviruses, the hepadna virus, hepatitis B, and adeno associated virus have been shown to be restricted with different APOBEC3 family members. The importance of APOBEC3 proteins in viral restriction has been proven in vivo. Mice lacking APOBEC3 gene are more susceptible to viral infection than wild-type mice.17,18 In addition to being antiviral host factors, APOBEC3 proteins have also been shown to inhibit retrotransposition of endogenous retroviruses, such as LTR retrotransposons HERVK, MusD, IAP, Ty1,10,19,20 and non-LTR retrotransposons, LINE-1 and Alu.20–24 Human APOBEC3A (A3A) and APOBEC3B (A3B) are the most potent inhibitors of the human LINE-1 retrotransposon and only A3B has anti HIV-1 activity, whereas A3A does not block HIV-1 replication. The mechanisms of antiretroviral and antiretrotransposon potency differ and the latter seems to be independent of the enzymatic activity.23 The exact mechanism of their action is unclear. In our study, we examined whether mammalian APOBEC3 proteins interact with LINE-1 ORF1 protein.
Materials and Methods
Plasmids
Plasmid encoding full length L1 element with neo cassette was previously described.34 For expression of A3A and A3B proteins pKA3A.HA and pKA3B.HA plasmids were used.22 Construct-expressing mouse A3 was prepared from previously reported plasmid pcDNA3.1mA3.V517 by cloning mA3 fragment into pcDNA6 plasmid (Invitrogen). pFLAG.mA3 vector was constructed by cloning mA3 fragment into pFLAG plasmid.35 ORF1 protein was amplified from JM10125 plasmid and cloned into pcDNA3.1 Topo Cloning vector (Invitrogen) to yield pcDNA3.1ORF1.V5 plasmid.
Retrotransposition Assay
HeLa cells were plated at 2 × 105 cells per well in 6-well dishes and within 24 h were co-transfected with 1 μg of plasmid encoding retrotransposon JM10125 and 1 μg of plasmid encoding A3 protein or an empty vector (pcDNA6) using 6 μl of Fugene 6 Transfection Reagent (Roche) according to manufacturers instructions. Three days posttransfection, cells were selected with G418 (500 μg/μl) for 10 days, fixed, stained with Trypan Blue and counted. Data is the result of at least two experiments.
Immunoprecipitation
293T cells were transfected with 5 μg of pcDNA3.1ORF1.V5 and 5 μg of plasmid encoding A3 protein. Forty-eight hours after transfection, cells were lysed in IP lysis buffer (1%NP-40, 10 mmol Tris-HCl, 150 mmol NaCl, 2 mmol EDTA), Protease inhibitor Coctail (Sigma, St. Louis, MO) and Rnase inhibitor RNAsin (Promega, Madison, WI) for 40 min at 4°C. After removing nuclei and unbroken cells, anti-HA antibodies (Santa Cruz Biotech, Santa Cruz, CA) or anti-FLAG (Sigma) was added to the supernatant and mixed 2 h at 4°C. Thereafter, protein A-beads (GE Healthcare, Piscataway, NJ) were added and incubated overnight at 4°C. Next, beads were washed and bound complexes eluted with 2x gel loading buffer. Proteins were detected by Western blotting using anti-V5 monoclonal mouse antibody (Invitrogen, Carlsbad, CA). Half of the cell samples were lysed in IP lysis buffer with RNase A (Roche Applied Science, Indianapolis, IN) and Protease Inhibitor Cocktail (Sigma) and processed as samples without RNase A treatment.
Immunofluorescence
HeLa cells were seeded on cover slips into 6-well plates a day before transfection. Following day, cells were transfected with 1 μg of plasmid DNA coding L1 ORF1 protein or A3 proteins. For co-localization studies 1 μg of each plasmid was used for transfection. Forty-eight hours after transfection, cells were washed, fixed in 4% paraformaldehyde (Electron Microscopy Science, Hatfield, PA) in PBS, and incubated 30 minutes at room temperature. Than cells were washed in phosphate-buffered saline (PBS) and permeabilized with 0.2% Triton X-100 in PBS for 30 minutes at room temperature. Cells were washed in PBA (PBS + BSA 1 mg/ml), followed by incubation with 1:500 diluted primary mouse monoclonal anti-V5 (Invitrogen) and 1:500 primary rabbit anti-HA (Sigma) antibody over night at 4°C. The next day after extensive washing, cells were incubated with 1:1000 diluted secondary goat antimouse antibody conjugated with Alexa-Fluor 488 (Molecular Probes, Eugene, OR) and 1:1000 diluted secondary goat antirabbit antibody conjugated with Alexa-Fluor 568 (Molecular Probes). Finally, cells were washed, transferred into a ProLong Gold antifade reagent with DAPI (Molecular Probes) and fluorescence microscopy was performed on Nikon Eclipse E800 microscope.
Results and Discussion
Retrotransposition Assay
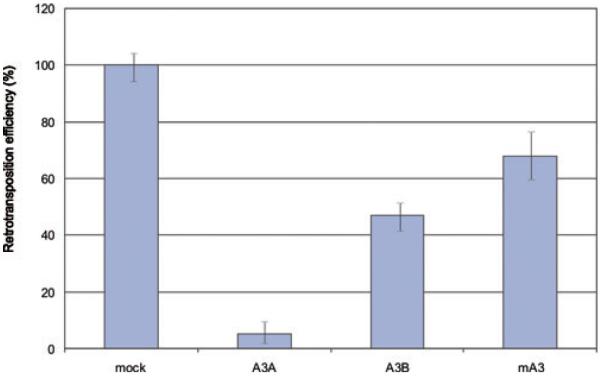
To evaluate the antiretrotransposon activity of human A3A, A3B, and mouse APOBEC3 (mA3) we first performed a cell culture based retrotransposition assay.25 In this assay, a full length LINE-1 element was marked with a neo gene interrupted by an intron in the opposite direction. Only, after transcription of the full length LINE-1, retrotransposition, splicing, and reverse transcription of LINE-1 RNA, the neomycin marker is expressed.25 HeLa cells were cotransfected with LINE-1 retrotransposon (JM101) and human A3 proteins or a control empty plasmid. Retrotransposition was inhibited by A3A and A3B, whereas mA3 had no impact on L1 retrotransposition as previously reported (Fig. 1).20–24
Figure 1.
Neo-based retrotransposition assay. HeLa cells were cotransfected with L1 construct (JM101) and respective A3 protein. Five days after transfection, cells were subjected to G418 selection for 12 days. For each experiment colonies were counted from two dishes and retrotransposition efficiency was determined relative to the control, which was set to 100%.
Localization of L1 ORF1 Protein and A3 Proteins
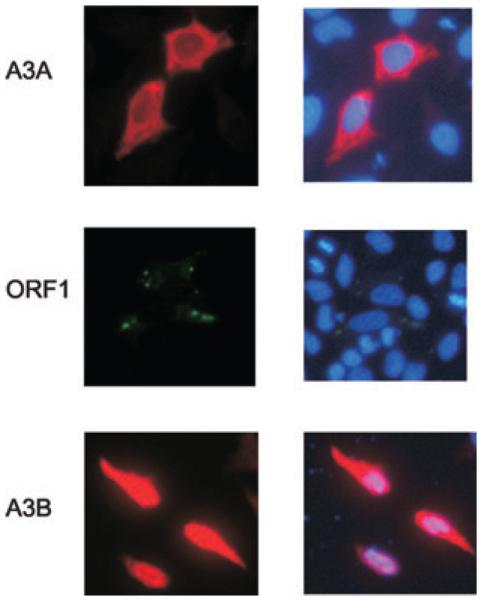
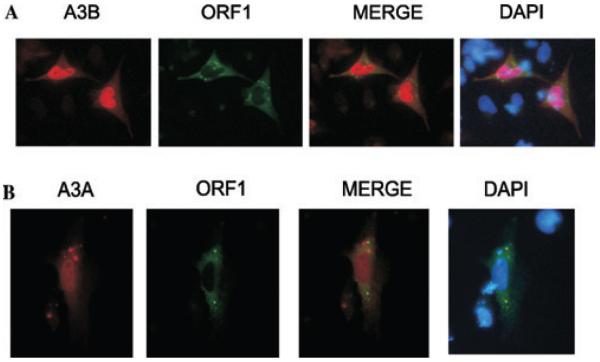
A3 proteins localize differently in the cell, A3G and A3F are found only in the cytoplasm, A3A and A3C are distributed throughout the cell and A3B is concentrated only in the nucleus.21,26 A3G and A3F have been shown to localize to punctuate cytoplasmic bodies and co-localize with proteins constituting processing bodies (P bodies).27 Under stress conditions A3G and A3F proteins are redistributed to stress granules.28 L1 retrotransposon encode ORF1 protein that is required for retrotrans-position.25 ORF1 protein assembles into RNP particles and has been also identified to localize to punctuate cytoplasmic bodies and to co-localize with stress granule proteins.29 This has prompted further analysis of whether or not A3A or A3B proteins co-localize with ORF1 protein in the cell. To verify that ORF1 and A3 proteins do not influence each others localization in the cell, immunostaining was performed in HeLa cells transfected only with tagged LINE-1 ORF1 protein, A3A or A3B protein (Fig. 2). Forty-eight hours after transfection cells were fixed, stained with anti-HA or anti-V5 antibody, we confirmed that A3A was distributed in the nucleus and the cytoplasm, whereas A3B was found only in the nucleus. LINE-1 ORF1 protein localized to cytoplasmic foci as previously reported.29 When A3 and ORF1 proteins were coexpressed the localizations of the proteins remained unchanged suggesting that A3 proteins do not redistribute ORF1 proteins or vice versa (Fig. 3). Although, A3A is evenly distributed in the cell, it does not co-localize with ORF1 protein. As expected, A3B, being a nuclear protein, did not co-localize with mainly cytoplasmic ORF1 protein. A3 proteins and ORF1 proteins do not co-localize to the same ribonucleoprotein (RNP) complexes, at least under normal conditions. Because the composition of ribonucleoprotein particles varies with the cell conditions, it would be interesting to test whether exposing cells to stress impacts the ORF1/A3 localization. It is plausible to believe that stress would redistribute all ORF1, A3A, and A3B proteins and they could co-localize with the same cytoplasmic bodies. In addition, stress also impacts the RNA pool of the cell, which may be crucial for L1 RNP formation and therefore A3 and ORF1 protein localization. Further studies of L1 RNP and A3 protein localization under stress conditions may reveal whether or not L1 proteins play a role in A3 inhibitory process.
Figure 2.
Localization of L1 ORF1, A3A, and A3B proteins. HeLa cells were transfected with plasmids encoding L1 ORF1 (pcDNA3.1.ORF1.V5), A3A (pKA3A.HA), or A3B (pKA3B.HA) proteins. At 48 h after transfection, cells were fixed and stained with rabbit anti-HA or mouse monoclonal anti-V5 antibody and a mouse-specific secondary antibody conjugated to Alexa-Fluor 488 or rabbit specific secondary antibody conjugated to Alexa-Fluor 568. Samples were viewed by fluorescence microscopy. (Left) Immunofluorescence with primary anti-HA (red), or primary anti-V5 antibodies (green). (Right) DNA visualized by DAPI staining, merge with immunofluorescence is shown.
Figure 3.
Co-localization studies ORF1, A3A, and A3B proteins. HeLa cells were cotransfected with plasmids encoding L1 ORF1 (pcDNA3.1ORF1.V5) and A3A (pKA3A.HA) (A) and L1 ORF1 and A3B (pKA3B.HA) proteins (B). Samples were processed as in Figure 2. Merged views are displayed on the right.
Co-immunoprecipitation of L1 ORF1 Protein
Several RNA binding proteins involved in RNA metabolism (such as P bodies and stress granules) have been shown to associate with A3G and A3F proteins suggesting that A3 proteins have a role in regulation of cellular RNA function.27,28 On the other hand, L1 ORF1 protein is a nucleic acid binding protein.25 To examine the interactions between A3A, A3B, and L1 ORF1 proteins, 293T cells were transiently transfected with plasmid encoding L1 ORF1 protein (pcDNA3.1 ORF1.V5) and HA tagged plasmid for A3A (pKA3A.HA) and A3B (pKA3B.HA). As a control, mouse A3 (pFLAG.mA3) without an inhibitory effect against L1 retrotransposons was also included in the experiment. ORF1 proteins co-precipitated with A3B (Fig. 4, lanes 7 and 8) and mouse A3 (Fig. 4, lanes 11 and 12) but not with A3A (Fig. 4, lanes 9 and 10) as detected by immunoblot using anti-V5 antibodies. The same interactions were also detected in the reciprocal experiments when proteins were precipitated with anti-V5 antibodies (data not shown). The interactions between A3B and L1 ORF1 proteins were affected by RNase treatment (Fig. 4, lane 8) suggesting that A3B binds to ORF1 protein in an RNA dependent manner. Similarly, the interactions between mA3 and ORF1 protein were also resilient to RNA removal (Fig. 4, lane 12). Interestingly, in our co-localization study, no co-localization between ORF1p and A3B was detected (Fig. 3), probably due to different compartmentization of both proteins. There are several explanations why A3B co-precipitates with ORF1 protein. First, ORF1 protein was also faintly detected in the nucleus29 and the immunoprecipitation results could be a consequence of these rare events that could not be detected by the immunofluorescence experiment. More likely, ORF1 protein and A3B protein bind during the lysis, after cells and part of the nuclei are disrupted. To verify these hypotheses further studies using only nuclear or cytoplasmic fractions for immunoprecipitation are needed. Surprisingly, ORF1 did not co-precipitate with A3A protein. A3A and ORF1 are both nucleic acid binding proteins, therefore, one would expect them to bind via RNA interactions even though the interactions were not specific. One possible explanation is that A3A and ORF1 proteins bind different RNA targets, at least under our experimental conditions. Interestingly, human A3A has been found to bind LINE-1 RNA and this was supposed to shift A3A from small to the high molecular mass complex in the cell.30 On the other hand, ORF1 protein was shown to cofractionate with L1 RNA in a large ribonucleoprotein complex, yet so far no direct interaction between ORF1 protein and L1 RNA have been detected.31,32 The composition of both large ribonucleoprotein complexes may differ or vary depending on the cell cycle. To exclude the possibility that the absence of a shared RNA molecule could have caused the lack of A3A/ORF1 protein interactions, a detail analysis of ORF1 ribonuclear particles in the presence of L1 RNA and A3 proteins is in progress. Our results indicate that there are no direct interactions between ORF1 protein and A3 proteins, yet A3B and mA3 bind to ORF1 protein via an RNA bridge. Our data imply that no correlation between A3 and ORF1 binding and its antiretrotransposition activity exists. Taken together with previously reported data, A3A protein indirectly interferes with L1 metabolism, probably by binding LINE-1 RNA.
Figure 4.
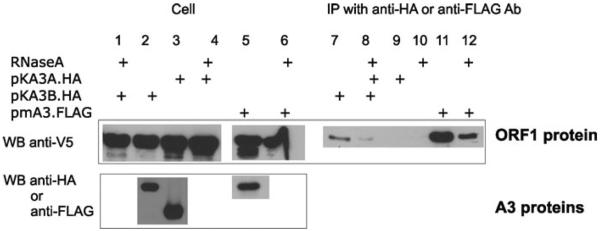
Co-immunoprecipitation of L1 ORF1 and A3 proteins. 293T cells were cotransfected with plasmids encoding L1 ORF1 (pcDNA3.1ORF1.V5) and A3A (pKA3A.HA), A3B (pKA3B.HA), mA3 (pFLAG.mA3) proteins. At 48 h after transfection cells were lysed and immunoprecipitation with anti-HA or anti-FLAG antibodies. Immunobloting was performed with anti-V5 antibodies. Addition of RNase A during the immunoprecipitation is indicated.
Conclusions
The potency of A3 proteins to inhibit retrotransposition of L1 retrotransposon could not simply be explained by the interactions between A3 and the L1 ORF1 proteins or their co-localization. A3A protein that inhibits retrotransposition does not bind ORF1 protein, and mA3 that does not inhibit retrotransposition binds to ORF1 protein. Results of our study, together with previous reports, indicate that A3 proteins interfere with L1 life cycle indirectly. A3A and A3B proteins probably overcompete for ORF1 protein binding factors that are crucial for the L1 life cycle. Numerous potential targets that interact with both L1 and A3 proteins and play a crucial role in the L1 life cycle exist. For instance, RNA granule proteins, P bodies, stress granules, and polysomes have been recognized to interact with ORF1 protein as well as with human A3G and A3F proteins. Alternatively, A3 proteins could also bind the L1 ORF2 protein that has reverse transcriptase activity. Unfortunately, L1 ORF2 has been shown to be very hard to detect in the cell and that hypothesis is currently difficult to examine.33 This directs future efforts towards the identification of mutual steps of A3 proteins and retrotransposons in the cell. Nonetheless, besides blocking the exogenous retroviruses, editing enzymes have also evolved to inhibit the replication of the major editors of our genome, transposable elements.
Acknowledgments
We thank John Moran and Bryan Cullen for reagents. This research was funded by the National Institute of Health and in part by the Slovenian Research Agency.
Footnotes
Conflicts of Interest The authors declare no conflicts of interest.
References
- 1.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Brosius J. The contribution of RNAs and retroposition to evolutionary novelties. Genetica. 2003;118:99–116. [PubMed] [Google Scholar]
- 3.Feschotte C. Transposable elements and the evolution of regulatory networks. Nat. Rev. Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: For better or worse, in sickness and in health. Genome Res. 2008;18:343–358. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- 5.Sassaman DM, et al. Many human L1 elements are capable of retrotransposition. Nat. Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- 6.Goodier JL, Kazazian HH., Jr. Retrotransposons revisited: The restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 7.Kordis D, Lovsin N, Gubensek F. Phylogenomic analysis of the L1 retrotransposons in Deuterostomia. Syst. Biol. 2006;55:886–901. doi: 10.1080/10635150601052637. [DOI] [PubMed] [Google Scholar]
- 8.Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- 9.Kanellopoulou C, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esnault C, et al. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature. 2005;433:430–433. doi: 10.1038/nature03238. [DOI] [PubMed] [Google Scholar]
- 11.Navaratnam N, et al. The p27 catalytic subunit of the apolipoprotein B mRNA editing enzyme is a cytidine deaminase. J. Biol. Chem. 1993;268:20709–20712. [PubMed] [Google Scholar]
- 12.Honjo T. Does AID need another aid? Nat. Immunol. 2002;3:800–801. doi: 10.1038/ni0902-800. [DOI] [PubMed] [Google Scholar]
- 13.Jarmuz A, et al. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- 14.Sheehy AM, et al. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 15.Harris RS, et al. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 16.Zheng YH, et al. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 2004;78:6073–6076. doi: 10.1128/JVI.78.11.6073-6076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okeoma CM, et al. APOBEC3 inhibits mouse mammary tumour virus replication in vivo. Nature. 2007;445:927–930. doi: 10.1038/nature05540. [DOI] [PubMed] [Google Scholar]
- 18.Santiago ML, et al. Apobec3 encodes Rfv3, a gene influencing neutralizing antibody control of retrovirus infection. Science. 2008;321:1343–1346. doi: 10.1126/science.1161121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bogerd HP, et al. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 2006;34:89–95. doi: 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen H, et al. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr. Biol. 2006;16:480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 21.Muckenfuss H, et al. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J. Biol. Chem. 2006;281:22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- 22.Bogerd HP, et al. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl. Acad. Sci. USA. 2006;103:8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stenglein MD, Harris RS. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J. Biol. Chem. 2006;281:16837–16841. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- 24.Kinomoto M, et al. All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res. 2007;35(9):2955–2964. doi: 10.1093/nar/gkm181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moran JV, et al. High frequency retrotrans-position in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 26.Alce TM, Popik W. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J. Biol. Chem. 2004;279:34083–34086. doi: 10.1074/jbc.C400235200. [DOI] [PubMed] [Google Scholar]
- 27.Gallois-Montbrun S, et al. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J. Virol. 2007;81:2165–2178. doi: 10.1128/JVI.02287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wichroski MJ, Robb GB, Rana TM. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS. Pathog. 2006;2:e41. doi: 10.1371/journal.ppat.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodier JL, et al. LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol. Cell. Biol. 2007;27:6469–6483. doi: 10.1128/MCB.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niewiadomska AM, et al. Differential inhibition of long interspersed element 1 by APOBEC3 does not correlate with high-molecular-mass-complex formation or P-body association. J. Virol. 2007;81:9577–9583. doi: 10.1128/JVI.02800-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hohjoh H, Singer MF. Ribonuclease and high salt sensitivity of the ribonucleoprotein complex formed by the human LINE-1 retrotransposon. J. Mol. Biol. 1997;271:7–12. doi: 10.1006/jmbi.1997.1159. [DOI] [PubMed] [Google Scholar]
- 32.Kulpa DA, Moran JV. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat. Struct .Mol. Biol. 2006;13:655–660. doi: 10.1038/nsmb1107. [DOI] [PubMed] [Google Scholar]
- 33.Goodier JL, et al. A potential role for the nucleolus in L1 retrotransposition. Hum. Mol. Genet. 2004;13:1041–1048. doi: 10.1093/hmg/ddh118. [DOI] [PubMed] [Google Scholar]
- 34.Feng Q, et al. Human L1 retrotransposon encodes a conserved endonuclease required for retro-transposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 35.Kohoutek J, Blazek D, Peterlin BM. Hexim1 sequesters positive transcription elongation factor b from the class II transactivator on MHC class II promoters. Proc. Natl. Acad. Sci. USA. 2006;103:17349–17354. doi: 10.1073/pnas.0603079103. [DOI] [PMC free article] [PubMed] [Google Scholar]