Abstract
Mammalian cells have the ability to recognize virus infection and mount a powerful antiviral response. Pattern recognition receptor proteins detect molecular signatures of virus infection and activate antiviral signaling. The RIG-I-like receptor (RLR) proteins are expressed in the cytoplasm of nearly all cells and specifically recognize virus-derived RNA species as a molecular feature discriminating the pathogen from the host. The RLR family is composed of three homologous proteins, RIG-I, MDA5, and LGP2. All RLRs have the ability to detect virus-derived dsRNA and hydrolyze ATP, but display individual differences in enzymatic activity, intrinsic ability to recognize RNA, and mechanisms of activation. Emerging evidence suggests that MDA5 and RIG-I utilize distinct mechanisms to form oligomeric complexes along dsRNA. Aligning of their signaling domains creates a platform capable of propagating and amplifying antiviral signaling responses. LGP2 with intact ATP hydrolysis is critical for the MDA5-mediated antiviral response, but LGP2 lacks the domains essential for activation of antiviral signaling, leaving the role of LGP2 in antiviral signaling unclear. Recent studies revealed a mechanistic basis of synergy between LGP2 and MDA5 leading to enhanced antiviral signaling. This review briefly summarizes the RLR system, and focuses on the relationship between LGP2 and MDA5, describing in detail how these two proteins work together to detect foreign RNA and generate a fully functional antiviral response.
Keywords: antiviral immunity, Rig-I-like receptors, MDA5, LGP2
RIG-I-like Receptors: Domain Structure and Connections to Antiviral Signaling
The antiviral functions of the RLR proteins were first revealed when RIG-I was identified in a function-based cDNA library screen designed to discover proteins that could induce the expression of an IFNβ promoter reporter gene. Sequence alignment studies revealed MDA5 and LGP2 as close homologs to RIG-I, with an overall amino acid sequence identity of approximately 30%. The sequence identity was even higher within key domains, suggesting potential functional similarity (Yoneyama et al., 2005). This potential functionally similarity was confirmed when MDA5 was characterized as an activator of IFN expression in response to RNA virus infection or transfection with the double-stranded RNA analog polyinosinic:polycytidilic acid (poly(I:C)) (Yoneyama et al., 2005). The similarity in structure, function, and sequence homology of all three RLR proteins define them as an evolutionarily conserved gene family (Figure 1) (Cagliani et al., 2014). RLR signaling and domain structure and function have been reviewed in detail previously (for example, (Bruns and Horvath, 2012; 2014; Rawling and Pyle, 2014)).
Figure 1. RIG-I-like Receptor Protein Domain Structure.
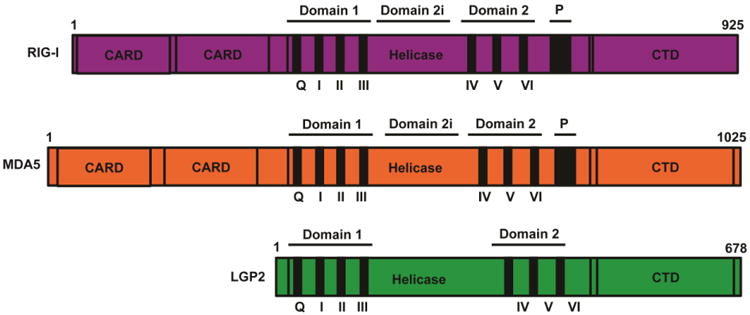
The three RLRs are SF2 type helicase proteins composed of a central DECH-box helicase domain that contains the conserved domains Hel1 (motifs Q, I, II, and III) and Hel2 (motifs IV, V, VI). Between the two helicase domains lies the insertion domain, Hel2i, required for RIG-I autoregulation. All RLRs also contain relatively conserved C-terminal domain (CTD). In RIG-I the CTD uniquely has a positively charged binding pocket for the recognition of 5′-PPP or 5′PP RNA substrates. RIG-I and MDA5 have two N-terminal caspase activation and recruitment domains (CARDs). Alignment of multiple CARDs is essential for interactions with MAVs and initiation of downstream antiviral signaling.
The RLRs share many key structural features, most notably a central DECH-box helicase domain that is homologous to the SF2 superfamily of RNA helicase proteins. Like other SF2 members, the RLRs share a catalytic core composed of two RecA-like domains that encompass up to twelve conserved helicase domain motifs (Cordin et al., 2006; Linder and Jankowsky, 2011). The most highly conserved and prominent helicase domain sequence motifs, Q and I-VI, cluster into either of the two RecA subdomains (referred to as domain 1 and domain 2). The helicase domain generally functions to coordinate RNA binding, ATP hydrolysis, and conformational rearrangements upon RNA recognition. Crystal structures of MDA5 and RIG-I also revealed the presence of an insertion domain (Domain 2i) and pincer (P) within the broader helicase domain structure, both of which are involved in structural rearrangements critical for adoption of the active signaling conformation after dsRNA detection (Civril et al., 2011; Jiang et al., 2011; Jiang and Chen, 2012; Kowalinski et al., 2011; Luo et al., 2011). These elements may be present in LGP2 as well, but structures are needed to confirm if these domains have conservation in LGP2. At their extreme C-terminus, all three RLRs have a regulatory domain (CTD) that has been implicated in regulation of RLR signaling activity (Saito et al., 2007; Yoneyama et al., 2004) and is crucial for the specific recognition of RNA substrate features (Cui et al., 2008, Takahasi et al., 2008). In addition, RIG-I and MDA5 have tandem N-terminal caspase activation and recruitment domain (CARD) regions that are absent in LGP2. The CARDs are protein interaction modules required for mediating antiviral signal transduction downstream of RNA recognition (Saito et al., 2007, Saito et al., 2008, Takahasi et al., 2008). The N-terminal CARDs of RIG-I were isolated in the original cDNA library screen as activators of IFNβ expression, and provided the first connection between the RLRs and antiviral signaling. Our understanding of CARDs as antiviral signaling domains has grown in recent years, as CARD oligomerization has emerged as a key component of antiviral signal transduction (Berke and Modis, 2012; Berke et al., 2013; 2012; Patel et al., 2013; Peisley et al., 2013; Wu et al., 2013).
Both RIG-I and MDA5 utilize CARD alignment to initiate antiviral signaling cascades, but LGP2 lacks the CARDs critical for downstream signaling. Due to its clear structural similarity to RIG-I and MDA5, lack of CARDs, and strong dsRNA-binding affinity, LGP2 was originally characterized as a feedback inhibitor of RLR signaling (Komuro and Horvath, 2006; Rothenfusser et al., 2005; Yoneyama et al., 2005). Other more recent studies have characterized LGP2 as an activator of RLR signaling, revealing opposing roles of LGP2 as both a positive and negative regulator of RLR signaling (Bruns et al., 2014; 2013; Satoh et al., 2010; Venkataraman et al., 2007a). Studies discussed below suggest that the positive and negative regulation of RLR signaling by LGP2 are not mutually exclusive, and propose a resolution to these seemingly contradictory roles of LGP2 (Bruns et al., 2013; 2014).
Current Overview of RLR-mediated Antiviral Signaling
Much of the current understanding of RLR signaling is based on extensive studies of RIG-I, the first recognized RLR protein (Yoneyama et al., 2004), and more recent discoveries about MDA5 contribute to the current model of RLR signaling (simplified model illustrated in Figure 2). Upon virus infection, interaction with virus-derived RNA ligands leads to a conformational change exposing the active CARDs, allowing association with an essential mitochondrial antiviral signaling protein, MAVS (also known as IPS-1) (Kawai et al., 2005; Kumar et al., 2006; Seth et al., 2005; Sun et al., 2006). When stimulated by MDA5 or RIG-I CARD alignment, MAVS acts as a polymeric signaling scaffold by assembling into filamentous structures along the outer mitochondrial membrane (Figure 2 C,H,J) (Hou et al., 2011; Jacobs and Coyne, 2013; Xu et al., 2014). The MAVS scaffold then enables the assembly and activation of signaling proteins including TRAF2, TRAF5, and TRAF6, and associated serine kinases (TBK1 and the IKK family) (Figure 2D) (Liu et al., 2013). These kinases activate master transcription factors IRF3 and NFκB, that drive the expression of diverse antiviral genes including IFNβ, the major antiviral cytokine (Figure 2E) (Freaney et al., 2013; Honda et al., 2006). Secreted IFNβ acts in an autocrine and paracrine fashion via the JAK-STAT signaling pathway, initiating antiviral programs in the infected cell and neighboring cells that provide an effective barrier against virus replication (Au-Yeung et al., 2013; Fink and Grandvaux, 2013).
Figure 2. Simplified Model: RIG-I-like Receptor Activation and Signaling.
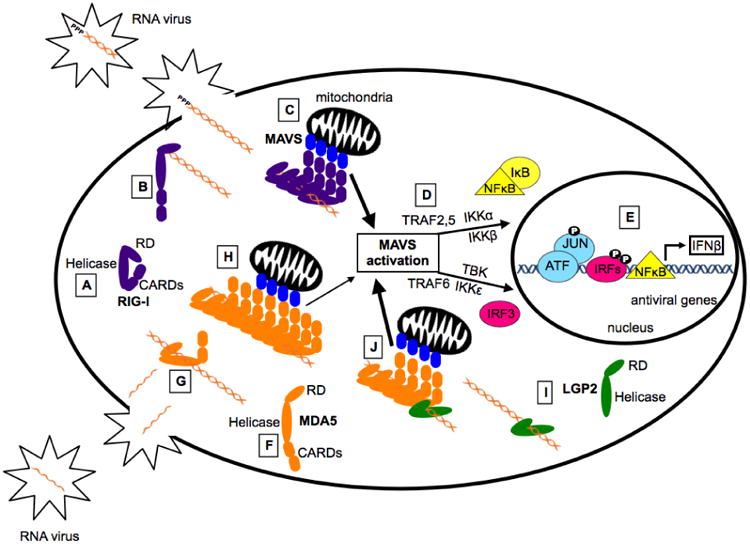
Much of our understanding of RLR signaling comes from studies characterizing RIG-I, the prototype RLR. In the cytoplasm RIG-I is present in an autoinhibited conformation (A), in which the CARDs interact with Hel2i, leaving them unavailable for downstream signaling interactions. During virus infection, non-self RNA species accumulate in the cytoplasm. RIG-I is activated when the CTD engages the appropriate non-self 5′-PPP or 5′-PP dsRNA ligand (B). This structural rearrangement transforms the helicase domain into an active ATPase and allows the formation of filamentous structures along the RNA (C). This alignment of multiple CARDs stimulates MAVS activation (D), leading to the assembly of a signaling scaffold for TRAF2, TRAF5, and TRAF6, and their associated kinases (TBK, IKKα, IKKβ, IKKε) that result in phosphorylation-mediated activation of latent IRF3 and NFκB transcription factors. These transcription factors, along with ATF/JUN, assemble on the IFNβ gene proximal enhancer and recruit RNA polymerase to induce transcription of IFNβ and other antiviral genes (E). MDA5 is present in the cytoplasm in a flexible conformation in which the CARDs are not inhibited (F). This is consistent with studies demonstrating that overexpression of MDA5 alone can stimulate the IFNβ promoter. Unlike RIG-I, MDA5 does not recognize specific RNA end modifications but binds slowly, with low affinity, internally along the dsRNA stem (G). Following nucleation, additional MDA5 monomers bind the RNA forming filaments stabilized by protein-protein interactions on adjacent MDA5 monomers (H). The alignment of MDA5 CARDs stimulates MAVS, ultimately leading to the transcription of IFNβ and other antiviral mediators. LGP2 binds with high affinity to dsRNA regardless of RNA length or end modifications, but lacks the CARDs critical for downstream signaling (I). Recent studies have demonstrated the LGP2 increases the initial rate of MDA5-RNA interaction and stabilizes the formation of shorter MDA5 filaments (J). Because LGP2 increases the rate of MDA5-RNA interaction, these shorter stable filaments are formed more rapidly and greater MAVS activation and antiviral signaling is achieved (J).
Generally, the RLR proteins share the ability to detect molecular signatures of virus infection, but differ in both their RNA recognition specificity and signaling properties (Ramos and Gale, 2011; Yoneyama et al., 2005). This review contains a brief summary of RIG-I as the prototype RLR, and describes in greater detail the RNA recognition properties of MDA5 and LGP2, and the role of enzymatic ATP hydrolysis activity as it relates to RNA detection and the receptor's ability to form active antiviral signaling complexes.
RIG-I
RIG-I was the first RLR to be characterized in antiviral innate immunity and became the prototype for our understanding of RLR signaling. Many recent studies have demonstrated that dsRNA detection by RIG-I can be conceptualized as having two phases, the initial interaction of the first protein monomer with dsRNA, often referred to as nucleation, followed by propagation of a protein filament along the dsRNA (Patel et al., 2013; Peisley et al., 2013; 2014). Filament formation results in the alignment of multiple CARDs, interacting with the CARD region of MAVS and seeding formation of MAVS filaments (Hou et al., 2011; Patel and García-Sastre, 2014; Xu et al., 2014). Current information suggests that the initiation of MAVS prion-like assembly is a central outcome of RNA recognition by the RLR proteins, and is critical for the efficient activation of downstream antiviral signaling cascades.
Initial RNA Recognition
Various studies have investigated initial RIG-I-RNA interactions. While under certain contexts, RIG-I has been shown to interact with RNAs of various lengths, sequences, and end modifications (Schlee and Hartmann, 2010), the consensus is that RIG-I is initially activated by the CTD binding to RNAs bearing a triphosphate (PPP) moiety and blunt-ended base pairing at the 5′ end (Goubau et al., 2013; Hornung et al., 2006; Schlee, 2013; Schlee et al., 2009; Schmidt et al., 2009). Recent studies have demonstrated that RIG-I can also be activated by the CTD binding to dsRNAs bearing a 5′-diphosphate (PP) (Goubau et al., 2014). Like the 5′-PPP generated by viral polymerases, recognition of 5′-PP dsRNA serves to discriminate self from non-self as 5′-PP RNAs are found in some viruses, such as the reovirus genome, but are not found in uninfected cells (Goubau et al., 2014). This initial CTD-mediated recognition of 5′-end modifications found on virus-derived RNAs induces conformational changes leading to RIG-I activation.
Activation and Structural Rearrangement
In the current model of RIG-I signaling, RIG-I is present in the cytoplasm in an autoinhibited ligand-free state (Figure 2A), and is only activating by the CTD binding to the appropriate 5′-PPP or 5′-PP dsRNA ligand (Figure 2B, 3A). In this ligand free conformation, the two CARDs are interacting with the Hel2i insertion domain, leaving the CARDS unavailable for downstream signaling while the CTD retains a high degree of flexibility to sample the cellular environment for foreign RNAs. When the CTD recognizes an appropriate 5′-PPP or 5′-PP dsRNA end modification, a conformational change is initiated in which the dsRNA helix displaces the CARDs, allowing the helicase domains to form a ring-like structure around the RNA helix, and freeing the CARDs for downstream signaling interactions with MAVS (Figure 2C) (Civril et al., 2011; Jiang et al., 2011; Kowalinski et al., 2011; Luo et al., 2011). This structural rearrangement of the helicase motifs transforms RIG-I into an active ATPase, suggesting that ATP binding may also promote CARD exposure (Jiang and Chen, 2012). The CARDs are critical for downstream antiviral signaling and their RNA binding-induced exposure provides one mechanism of RIG-I regulation. The CARDs are also modified and regulated in vivo by CARD ubiquitination, phosphorylation, and via interaction with other regulatory proteins (Arimoto et al., 2007; Friedman et al., 2008; Gack et al., 2007; Inn et al., 2011; Oshiumi et al., 2010; Zeng et al., 2010; Zhang et al., 2008).
Figure 3. Distinct Mechanisms of RIG-I and MDA5 Filament Formation.
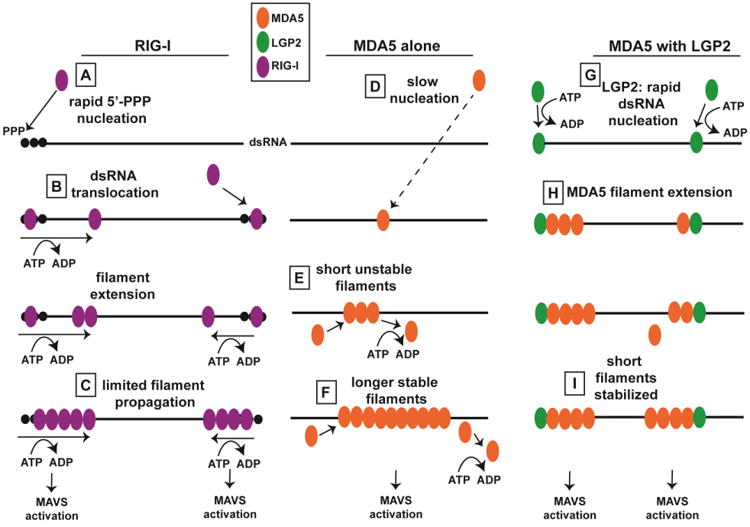
Figure illustrates distinct mechanisms of RIG-I and MDA5 filament formation, and depicts how LGP2 may regulate MDA5 filament assembly. (A) RIG-I CTD recognizes 5′-triphosphorylated dsRNA. (B) ATP-dependent dsRNA translocation by RIG-I aids in filament assembly, leading to limited filament propagation (C). MDA5 nucleation occurs very slowly (D), and short filaments are unstable (E), while filament extension leads to stabilization via protein-protein interactions on adjacent MDA5 monomers (F). LGP2 may nucleate MDA5 filaments by binding dsRNA termini (G). MDA5 filaments extend (H), and LGP2 stabilizes the formation of these shorter filaments (I).
ATP Hydrolysis and Filament Formation
After this initial structural rearrangement into the enzymatically active conformation, RIG-I can then utilize the energy from ATP hydrolysis to translocate along the dsRNA (Figure 3B) (Jiang and Chen, 2012; Myong et al., 2009). The rate of RIG-I translocation along dsRNA is dependent on the concentration of ATP, and the presence of CARD domains inhibits translocation. However, when the dsRNA substrate contained a 5′-PPP modification, the presence of the CARD domains no longer had an inhibitory effect on the rate of RIG-I translocation. Based on these studies, activation of the RIG-I ATPase and subsequent dsRNA translocation require both a 5′-PPP and double stranded regions of RNA (Myong et al., 2009). This ATP-dependent translocation was found to increase the frequency or efficiency of RIG-I assembly into high molecular weight aggregates on 5′-PPP RNA templates (Patel et al., 2013). In the absence of ATP, RIG-I binds to dsRNA ends as a monomer, but with the addition of ATP it assembles into a filament starting at the dsRNA end and elongating toward the interior (Figure 3C) (Peisley et al., 2013). These filaments were able to form in vitro and stimulate MAVS filament formation by both ubiquitin independent and ubiquitin dependent mechanisms (Peisley et al., 2013; 2014). Interestingly, these filaments did not entirely coat long dsRNAs, but were rather observed as multiple shorter filaments present at the ends of the RNA, and occasionally observed internally along the length of the RNA. In these in vitro experiments, a dsRNA as short as 62bp, a length that can accommodate only 4-5 RIG-I molecules, was capable of activating MAVS oligomerization in vitro (Peisley et al., 2013). This demonstrates that very long filaments containing numerous aligned CARDs are likely not essential for MAVS activation. The importance of this will be highlighted below regarding a potential mechanism of LGP2-mediated regulation of MDA5 filament formation.
MDA5
RNA Binding Properties
While the ligand for RIG-I can be summarized as a 5′-PPP or 5′-PP dsRNA, specific chemical features of RNA required for recognition by MDA5 have not been identified. The lack of detailed information regarding MDA5 substrates is in part due to the apparently weak RNA binding activity of MDA5 for the RNAs tested. However, a few studies have elucidated potential RNA features recognized by MDA5. Using an RNA substrate that had been enzymatically digested or sheared to create populations of discrete lengths, MDA5 was most effectively activated by RNA molecules longer than 2kbp, whereas RIG-I was efficiently activated by shorter molecules (Kato et al., 2008). In addition to RNA substrate length, secondary structure may play a role in RNA recognition by MDA5. High molecular weight RNAs extracted from virus-infected cells were shown to preferentially activate MDA5-mediated signaling (Pichlmair et al., 2009), suggesting that structural features, such as RNA branches found in RNAs with both single-stranded and double-stranded regions might be required for recognition by MDA5. This is consistent with the notion that poly(I:C), the synthetic dsRNA analogue, which forms branches due to annealing of poly I and poly C strands of non-defined length, is primarily detected by MDA5 in vitro and in vivo (Gitlin et al., 2006; Kato et al., 2006). In addition to a preference for secondary structure, MDA5 may also discriminate 2′-O-methylation as a feature distinguishing self from virus-derived RNAs (Daffis et al., 2010; Zust et al., 2011). Because MDA5 has no preference for dsRNA end modifications, it recognizes viruses that are evading RIG-I detection by masking the 5′-PPP moiety with a capping protein, such as encephalomyocarditis virus (EMCV). The lack of RNA feature specificity and requirement for longer dsRNAs is related to the ability of MDA5 to form filamentous assemblies along dsRNA (discussed below).
ATP Hydrolysis Activity: Mutational Analysis and Increased RNA Dissociation
Like RIG-I, MDA5 also derives its ATP hydrolysis activity from the central SF2-like helicase domain. MDA5 ATP hydrolysis is strictly dependent on dsRNA stimulation and displays greater ATPase activation by longer dsRNA substrates. In the presence of optimal dsRNA substrates, MDA5 exhibits a lower rate of ATP hydrolysis than RIG-I (Pollpeter, 2011). A mutational analysis of all core helicase domains (I-VI) was conducted on MDA5, and all mutations were found to abrogate ATP hydrolysis activity (Bamming and Horvath, 2009). Four of the MDA5 mutants (motif II, IV, V, and VI) resulted in proteins that were no longer able to signal to the IFNβ promoter, but no trans inhibition was observed for any of the mutated proteins. Two other proteins with mutations to helicase motif I (ATP binding site) or motif III, produced MDA5 proteins with constitutive RNA-independent hyperactive signal transduction activity, despite their inability to hydrolyze ATP. These phenotypes clearly show that ATP hydrolysis is not strictly required for MDA5 signaling activity, but rather that ATPase may play role in regulating MDA5-mediated antiviral signaling. Because the phenotypes of the homologous mutations were different between RIG-I and MDA5, these studies suggest the regulatory role for ATP hydrolysis in RLR signal transduction that may be idiosyncratic to the individual receptor and its signaling context (Bamming and Horvath, 2009). The conclusions of this mutational analysis are supported by recent studies illustrating the clinical importance of ATP hydrolysis-mediated regulation of MDA5 signaling. In these studies, point mutations in the MDA5 protein (in humans and murine models) disrupted ATP hydrolysis leading to constitutive activation of MDA5 and autoimmune disease (Funabiki et al., 2014; Rice et al., 2014). These mutations and clinical outcomes will be discussed in detail below (Antiviral Signaling and Beyond).
Electron microscopy, ATP hydrolysis assays, and electrophoretic mobility shift assays have demonstrated that MDA5 ATP hydrolysis is correlated with MDA5 dissociation from RNA (Peisley et al., 2011). Surprisingly, physiological levels of ATP (1-5mM) actually inhibit cooperative MDA5-RNA interactions (Berke and Modis, 2012). These findings were confirmed by single molecule RNA-binding experiments, quantitatively demonstrating that the addition of ATP increases the MDA5 off rate from both 25bp and 44bp dsRNA substrates (Bruns et al., 2014). The concept that ATP hydrolysis activity results in MDA5-RNA disassembly is also consistent with a model in which ATPase plays a regulatory role in MDA5-medaited antiviral signaling (Figure 3E). Synthesis of the currently available studies of both RIG-I and MDA5 suggests the importance of ATP hydrolysis for proper RLR-mediated antiviral signaling is related to the distinct mechanisms by which RIG-I and MDA5 form filamentous assemblies along dsRNA, and subsequently activate downstream signaling (Berke and Modis, 2012; Berke et al., 2012; 2013; Jiang et al., 2011; Luo et al., 2011; Motz et al., 2013; Wu et al., 2013).
Initial RNA Recognition, Conformational Changes, and Filament Assembly
RNA detection by MDA5 leading to activation of MAVS and antiviral signaling involves multiple conformational changes. Prior to RNA recognition, ligand-free MDA5 is present in the cytoplasm (Figure 2F). When foreign cytoplasmic RNA is detected, MDA5 binds initially as a monomer to dsRNA (Figure 2G), nucleating filament propagation down the length of the dsRNA (Figure 2H). This multiple CARD alignment via filament formation is necessary to seed formation of MAVS oligomers (Hou et al., 2011; Patel and García-Sastre, 2014; Xu et al., 2014). MAVS filaments have been observed in cells along the mitochondria in the context of virus infection, and appear to be critical for a fully functional antiviral response (Xu et al., 2014).
In the ligand-free state, MDA5 does not adopt a strictly autoinhibited conformation like RIG-I (Figure 2F). The region of Hel2i that binds to and autoinhibits the CARDs in RIG-I is shorter in MDA5, and missing a key phenylalanine residue, suggesting Hel2i does not autoinhibit the MDA5 CARDs in the absence of RNA (Berke and Modis, 2012). These findings demonstrate that MDA5, unlike RIG-I, retains a flexible uninhibited conformation in the absence of RNA. This is consistent with the observation that overexpression of MDA5 (even in the absence of RNA) significantly stimulates IFNβ promoter transcriptional activity (Bamming and Horvath, 2009; Rodriguez and Horvath, 2013). Small angle x-ray scattering and protein modeling show that full-length ligand-free MDA5 adopts multiple extended structures, none of which reveal CARD inhibition (Berke et al., 2013), suggesting alternative mechanisms of CARD regulation. Many studies have investigated MDA5 CARD regulation in vivo, demonstrating various levels of regulation by ubiquitination, phosphorylation, and direct or indirect association with other antiviral mediators (Belgnaoui et al., 2011; Hiscott and Ware, 2011; Jiang et al., 2012; Paz et al., 2011; Wies et al., 2013)
While the salient feature of initial RNA recognition by RIG-I can be easily identified as the 5′-PPP moiety, the difficulty in identifying key RNA features involved in the initial detection of dsRNA by MDA5 is related to important structural differences between RIG-I and MDA5. For example, while the CTD of RIG-I contains a positively charged 5′-PPP (or 5′-PP) binding pocket, the analogous surface on the CTD of MDA5 is flatter and has a lower affinity for dsRNA (Li et al., 2009). The CTD of MDA5 is instead involved in cooperative binding of MDA5 to dsRNA (Berke and Modis, 2012; Peisley et al., 2011). This cooperative binding allows MDA5 to form filamentous assemblies along dsRNA.
Despite the difficulty in detecting and analyzing initial monomeric binding to MDA5 to dsRNA, recent electron microscopy studies revealed that MDA5 forms filaments along dsRNA, with ringlike asymmetric units that form helical twists (Berke and Modis, 2012; Berke et al., 2012; Motz et al., 2013; Peisley et al., 2011; 2012; Wu et al., 2013). The first monomer of MDA5 binds slowly to RNA, with low affinity, but unlike RIG-I this initial RNA-interaction does not occur at the ends of dsRNAs. Rather, MDA5 nucleates filament formation at any location along the dsRNA helix (Figure 3D), followed by assembly of long head-to-tail filaments. These filaments are stabilized by protein-protein interactions on adjacent MDA5 monomers (Figure 3F) (Wu et al., 2013). While RIG-I filaments require ATP hydrolysis and RNA translocation to form, surprisingly, MDA5 filaments are actually destabilized by ATP hydrolysis (Figure 3E) (Berke and Modis, 2012; Feng et al., 2012; Wu et al., 2013). Because ATP hydrolysis is involved in MDA5 dissociation from dsRNA, these long helical filament structures are most readily observed in the presence of a stabilizing ATP transition state analogue, ADP-AlF4. MDA5 filament propagation does not appear to be directional, and it has been speculated that ATP hydrolysis allows MDA5 to dissociate in order to repair breaks when two elongating filaments collide, resulting in the formation of one continuous longer filament structure (Peisley et al., 2012).
Recent studies have revealed that these long MDA5 filaments are not requisite for antiviral signaling. MDA5 mutant proteins that display defects in filament formation retain ∼75% of wild-type antiviral signaling activity (Bruns et al., 2014). These findings suggest that long MDA5 filaments observed in the presence of ADP-AlF4 may represent a captured transition state formed during MDA5 signaling, rather than a stable intracellular structure. The idea that long MDA5-RNA filaments are not required for signaling is supported by additional experiments investigating the role of LGP2 in MDA5-RNA interactions, discussed below.
LGP2
RNA Binding and LGP2 as a Regulator of RLR-mediated Antiviral Signaling
LGP2 has the highest RNA binding affinity of the RLRs, and has the ability to recognize diverse dsRNAs, irrespective of 5′-PPP or RNA length (Figure 2I) (Bamming and Horvath, 2009; Takahasi et al., 2009). Despite strong dsRNA binding, LGP2 lacks the CARDs necessary for downstream signaling, historically leaving the exact role of LGP2 in RNA recognition and antiviral signaling unresolved. This lack of clarity is in part due to apparently opposing actions of LGP2 as both a positive and negative regulator of RLR-mediated signaling depending on the experimental approaches used.
Exogenous overexpression of LGP2 protein in cells effectively inhibits RLR-mediated IFN induction (Komuro and Horvath, 2006; Rothenfusser et al., 2005; Yoneyama et al., 2005). LGP2 is thought to be in low abundance at steady state, but is strongly inducible by antiviral stimulation, including poly(I:C) transfection, IFNα treatment, or virus infection. Based on these findings, and the absence of CARD signaling motifs, LGP2 was originally described as a feedback inhibitor for RLR signaling (Komuro et al., 2008; Rothenfusser et al., 2005; Saito et al., 2007). Several mechanisms for LGP2 feedback inhibition were proposed. Due to its strong RNA binding ability, LGP2 was originally suggested to sequester viral RNA from detection by RIG-I (Yoneyama et al., 2005). However, LGP2 proteins that are defective for RNA binding can still inhibit RLR signaling, indicating that RNA competition does not explain the effect of LGP2 expression (Bamming and Horvath, 2009). Based on titration experiments, it was suggested that CARD-independent LGP2 interaction with MAVS allowed LGP2 to compete with an essential signaling kinase, preventing its recruitment to the MAVS signaling scaffold. This model may account for the ability of LGP2 to inhibit antiviral signaling triggered in the absence of RNA through tandem CARD or MAVS expression (Komuro and Horvath, 2006).
In contrast to these examples of negative regulation by LGP2, targeted gene disruption studies have indicated a positive role for LGP2 in IFNβ expression and antiviral signaling (Moresco and Beutler, 2010; Pollpeter et al., 2011; Satoh et al., 2010). Mice lacking LGP2 exhibited decreased IFN-β production in response to various RNA viruses, but overexpression of the CARD domains of RIG-I and MDA5 in LGP2-/- cells rescued IFN-β promoter activity, suggesting LGP2 may act upstream of MDA5 and RIG-I (Satoh et al., 2010). The absence of LGP2 also reveals a role in the recognition of Listeria monocytogenes DNA viruses, and cytosolic DNA (Pollpeter et al., 2011), suggesting a diverse positive effect of LGP2 in anti-microbial signaling.
Evidence Connecting LGP2 and MDA5
A connection between LGP2 and MDA5 has long been speculated, but the exact mechanism by which LGP2 and MDA5 work together in virus detection and antiviral signaling was unknown. Mice with a targeted disruption in the LGP2 locus are more susceptible to infection by the picornaviruses, encephalomyocarditis virus (EMCV) and poliovirus, two viruses thought to be recognized by MDA5 (Kato et al., 2006) rather than RIG-I (Satoh et al., 2010; Venkataraman et al., 2007b). Mouse embryonic fibroblast cells lacking MDA5 and LGP2 failed to produce IFNβ in response to EMCV infection. The exogenous expression of either MDA5 or LGP2 alone in these cells did not restore IFNβ signaling, but the addition of both MDA5 and LGP2 restored signaling to wild type levels, suggesting LGP2 and MDA5 may work together in virus detection and signal transduction (Satoh et al., 2010). Supplementing LGP2 -/- mice with an enzymatically inactive LGP2 mutant via knock-in did not reconstitute defective positive signaling responses or IFNβ production in vivo (Satoh et al., 2010). LGP2 without ATP hydrolysis has the same phenotype as lacking LGP2 altogether, demonstrating the importance of ATP hydrolysis in LGP2-mediated positive regulation of antiviral signaling.
Evidence for LGP2 co-activation of MDA5 is accumulating from investigation of virus-RLR interactions. For example, an LGP2-associated EMCV RNA was found to act as a physiological agonist of MDA5-dependent signaling (Deddouche et al., 2014). Additionally, interferon antagonist proteins encoded by paramyxoviruses target both MDA5 and LGP2, disrupting their ATP hydrolysis activity (Parisien et al., 2009). Single molecule RNA binding experiments and biochemical analysis revealed that ATP hydrolysis activity is required to enable LGP2 to efficiently engage diverse dsRNA species, and is requisite for enhancement of MDA5 signaling (Bruns et al., 2013). This evidence supports a strong connection between MDA5 and enzymatically active LGP2 in RNA virus detection and antiviral signal transduction, suggesting a mechanism for LGP2 enhancement of MDA5-mediated antiviral signaling.
MDA5-LGP2 Synergy
A growing body of evidence supports the idea that LGP2 positively regulates MDA5-mediated signaling, but the lack of knowledge regarding MDA5-RNA interactions and antiviral signal activation precluded an investigation of how LGP2 might be assisting MDA5-RNA recognition or enhancing MDA5-mediated antiviral signaling. The recent surge in mechanistic understanding of MDA5-RNA interactions and filament assembly enabled a detailed investigation of how LGP2 may be influencing these processes, revealing a potential mechanism for the positive regulation of MDA5 by LGP2 (Bruns et al., 2014).
In vitro biochemistry, single molecule RNA-binding, and cellular signaling assays were combined to investigate the effect of LGP2 on MDA5-RNA interactions and antiviral signaling (Bruns et al., 2014). Mutational analysis demonstrated that the co-activation of MDA5 by LGP2 requires LGP2 with intact ATP hydrolysis and RNA-binding activities, as well as MDA5 with a functional oligomeric interface. The discovery that LGP2 could not enhance signaling activity from MDA5 proteins that were defective for filament formation suggested that LGP2 might play a role in MDA5-RNA interaction or filament formation. In vitro RNA-binding experiments revealed that MDA5 accumulates slowly on dsRNA (Figure 3D), as expected, but surprisingly the addition of LGP2 catalytically increases the initial rate and stability of MDA5-dsRNA interactions (Figure 3G,H). While LGP2 was not observed to form filaments along dsRNA like MDA5, when MDA5-dsRNA complexes were formed in the presence of LGP2, a greater number of MDA5 filaments of attenuated length were observed. LGP2 both facilitates initial MDA5-RNA interactions, leading to the nucleation of MDA5 filament assembly (Figure 3H), and regulates the extent of filament progression, resulting in the rapid formation of a greater number of shorter MDA5-RNA complexes (Figure 3I). These shorter MDA5-RNA filaments formed in the presence of LGP2 were transfected into cells in the context of an IFNβ luciferase assay and shown to generate equivalent or greater signaling activity in vivo than the longer filaments containing only MDA5. These findings again support the theory that long MDA5 filaments are not required for antiviral signaling, and provide a potential mechanism for LGP2 coactivation of MDA5 in antiviral responses (Bruns et al., 2014).
Of particular importance, titration of LGP2 by plasmid transfection clearly showed that LGP2 only enhances MDA5-mediated signaling when LGP2 is present at relatively low cellular concentrations (Bruns et al., 2013; 2014; Pippig et al., 2009; Satoh et al., 2010). When the transfected concentration of LGP2 is very high, the previously reported inhibition of MDA5 signaling is observed. We propose LGP2 acts as a concentration dependent biphasic switch. At the beginning of an infection, when LGP2 is present at low cellular concentration, LGP2 enhances MDA5-mediated antiviral signaling. But as the infection progresses, IFN production activates LGP2 expression causing LGP2 to accumulate within the cell, and at these higher concentrations LGP2 now inhibits MDA5 signaling, providing negative feedback. Mutant LGP2 proteins defective for ATP hydrolysis, RNA binding, or both failed to enhance MDA5 signaling, but at high concentrations retained their ability to inhibit MDA5-mediated signaling. This demonstrates that the ATP hydrolysis and RNA binding activities of LGP2 are required for positive effects on MDA5 signaling, but are dispensable for MDA5 inhibition.
LGP2 and MDA5: Antiviral Signaling and Beyond
In addition to their major role in detecting RNA virus infection and initiating antiviral responses, MDA5 and LGP2 have been implicated in other biological functions. LGP2 not only mediates cellular responses related to viral RNA recognition and antiviral signaling, but also participates in antiviral T cell regulation (Suthar et al., 2012), responses triggered by cytosolic dsDNA (Pollpeter et al., 2011), and cancer cell resistance to ionizing radiation (Widau et al., 2014). In cancer, elevated levels of LGP2 are associated with more adverse clinical outcomes. This study demonstrated that ionizing radiation causes cytotoxic stress that induces IFNβ and enhances the expression of LGP2. This increased expression of LGP2 suppresses IFN stimulated genes normally associated with cytotoxic stress by turning off the expression of IFNβ. These findings are consistent with the model of LGP2 acting as a concentration dependent biphasic switch (Bruns et al., 2013; 2014; Rodriguez et al., 2014). In the context of cancer and ionizing radiation therapy, LGP2 only begins to negatively affect IFN stimulated gene expression when present at high cellular concentrations, paralleling its negative regulation of antiviral signaling at high concentrations.
A recent surge of information has also contributed to a new appreciation of mouse and human genetic abnormalities in MDA5 that contribute to autoimmune syndromes. Genome-wide association revealed the MDA5 locus as a risk factor for type 1 diabetes, providing a link between MDA5 and autoimmune diseases (Smyth et al., 2006). The connection between MDA5 and autoimmune disease was strengthened by the discovery of MDA5's involvement in lupus susceptibility (Robinson et al., 2011), and other diseases characterized by chronic IFN stimulation (Ivashkiv and Donlin, 2014).
Molecular insights into the role of MDA5 in autoimmune signaling were acquired recently from a study of mutagenized mice that acquired a lupus-like phenotype, including widespread lymphocyte infiltration, chronic multi-organ inflammation, and unregulated cytokine expression. Mice with mutated MDA5 protein displayed widespread IFN and IFN-stimulated gene expression, and this IFN hyperactivation was shown to require MDA5-mediated MAVS signaling (Funabiki et al., 2014). A single point mutation in MDA5 (G821S) was found to produce an MDA5 protein constitutively active for signal transduction, but defective in ATP hydrolysis activity. Mouse cells homozygous for this MDA5 mutation display constitutive activation of IFNβ, but do not respond appropriately when challenged with EMCV infection or dsRNA transfection. This suggests the MDA5 mutation may cause a conformational change in the protein that both exposes the CARDs (resulting in IFNβ hyperactivation), and simultaneously prohibits formation of a functional ATPase or RNA binding domain (causing the poor response to dsRNA or EMCV infection). These findings are in agreement with the previous mutational analysis (discussed above) in which mutation to the core helicase motifs generated MDA5 proteins with constitutive and hyperactive IFN signaling in the absence of RNA binding and ATP hydrolysis (Bamming and Horvath, 2009). This MDA5 mutation leading to autoimmunity (Funabiki et al., 2014) is also consistent with the idea that ATP hydrolysis by MDA5 plays a regulatory role in MDA5-mediated antiviral signaling, by aiding in filament disassembly and reducing the amount of signaling-competent MDA5 CARD oligomers.
These murine mutations to MDA5 clearly demonstrated a connection between MDA5 and autoimmune diseases. The recent discovery of MDA5 mutations in human patients suffering from autoimmune diseases served to highlight the clinical importance of MDA5 in autoimmunity. Aicardi-Goutieres Syndrome (AGS) is a lupus-like hereditary autoimmune disease that features early onset of neurological degeneration and deterioration of myelinated nerve fibers (Miner and Diamond, 2014). AGS is also characterized by constitutive expression of IFN response genes, supporting the connection between MDA5 and autoimmunity. Whole-exome sequencing revealed gain-of-function mutations in MDA5 as the molecular basis for previously genetically uncharacterized cases of AGS in humans. Analysis of these mutant MDA5 proteins determined they are competent for ATP hydrolysis, but have significantly increased RNA binding activity. This suggests yet another mechanism of MDA5 dysregulation leading to disease. The increased RNA binding activity of these MDA5 mutant proteins causes inappropriate interactions with RNA (either host or pathogen derived) resulting in over-activation of immune signaling and disease.
Looking Forward
These discoveries regarding the roles of LGP2 in T cell development and cancer therapy response, as well as various MDA5 mutations leading to autoimmune diseases via different mechanisms all serve to underscore the importance of understanding the biochemical, structural, and enzymatic properties of these proteins. Recent studies revealed novel functions of enzymatic LGP2, utilizing ATP hydrolysis to engage diverse dsRNAs, increasing the initial rate MDA5-RNA interactions, and regulating MDA5 filament assembly, ultimately leading to enhanced MDA5-mediated antiviral signaling (Bruns et al., 2013; 2014). All of these properties of LGP2 were dependent on ATP hydrolysis, providing the first mechanistic basis for the previously observed synergy between LGP2 and MDA5. Despite these recent insights, some aspects of MDA5 and LGP2 in antiviral innate immunity remain mysterious, and additional questions arise regarding the details and impact of the phenomena described here.
Important missing pieces include a structure of ligand-free MDA5, full length MDA5 bound to dsRNA, and an RNA-bound and unbound crystal structure of LGP2. These structures will give valuable insights into conformational changes induced by MDA5 RNA binding and will also help with interpretation of disease-related mutations in MDA5. The structures of LGP2 will uncover the mechanistic details remaining to explain how LGP2 interacts with RNA and how it modulates MDA-RNA interaction. Together these structures will reveal possible interfaces for protein-protein interaction, and if the structure of RNA-bound LGP2 is conducive to stabilization of MDA5-RNA interactions. Analysis of post-translational modifications of LGP2 and MDA5 will undoubtedly reveal additional layers of in vivo regulation in response to virus infection.
Our understanding of RLR-mediated antiviral responses is rapidly evolving, and it will be exciting moving forward to see if the broad principles uncovered in the last five years, including the importance of dsRNA translocation, CARD-containing oligomers serving as a signaling platform, and the differential roles of ATP hydrolysis in regulating these processes will apply to other members of the SF2 family of helicase proteins. Our understanding of the RIG-I-like receptors will continue to grow as we uncover additional layers of RLR regulation and cooperation, cell type and virus-specific nuances, and novel roles in other cellular processes.
Acknowledgments
Research on RLRs in the Horvath lab was supported by NIH grants AI073919 and AI50707 to C.M.H. A.M.B. was supported by a pre-doctoral fellowship from the NIH Cellular and Molecular Basis of Disease Training Grant T32GM008061 and by the Sidney & Joan Pestka Graduate Award for Excellence in Interferon Research. We are grateful to Dr. John Marko for assistance with single-molecule experiments and programming analysis software. Our collaborators Dr. George Leser and Dr. Robert Lamb are a Research Specialist and an Investigator, respectively, of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arimoto KI, Takahashi H, Hishiki T, Konishi H, Fujita T, Shimotohno K. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc Natl Acad Sci USA. 2007;104:7500–7505. doi: 10.1073/pnas.0611551104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au-Yeung N, Mandhana R, Horvath CM. Transcriptional regulation by STAT1 and STAT2 in the interferon JAK-STAT pathway. Jakstat. 2013;2:e23931. doi: 10.4161/jkst.23931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamming D, Horvath CM. Regulation of signal transduction by enzymatically inactive antiviral RNA helicase proteins MDA5, RIG-I, and LGP2. J Biol Chem. 2009;284:9700–9712. doi: 10.1074/jbc.M807365200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgnaoui SM, Paz S, Hiscott J. Orchestrating the interferon antiviral response through the mitochondrial antiviral signaling (MAVS) adapter. Curr Opin Immunol. 2011;23:564–572. doi: 10.1016/j.coi.2011.08.001. [DOI] [PubMed] [Google Scholar]
- Berke IC, Modis Y. MDA5 cooperatively forms dimers and ATP-sensitive filaments upon binding double-stranded RNA. Embo J. 2012;31:1714–1726. doi: 10.1038/emboj.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke IC, Li Y, Modis Y. Structural basis of innate immune recognition of viral RNA. Cell Microbiol. 2013;15:386–394. doi: 10.1111/cmi.12061. [DOI] [PubMed] [Google Scholar]
- Berke IC, Yu X, Modis Y, Egelman EH. MDA5 assembles into a polar helical filament on dsRNA. Proc Natl Acad Sci USA. 2012;109:18437–18441. doi: 10.1073/pnas.1212186109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns AM, Horvath CM. Activation of RIG-I-like receptor signal transduction. Crit Rev Biochem Mol Biol. 2012;47:194–206. doi: 10.3109/10409238.2011.630974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns AM, Horvath CM. Antiviral RNA recognition and assembly by RLR family innate immune sensors. Cytokine Growth Factor Rev. 2014;25:507–512. doi: 10.1016/j.cytogfr.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns AM, Leser GP, Lamb RA, Horvath CM. The innate immune sensor LGP2 activates antiviral signaling by regulating MDA5-RNA interaction and filament assembly. Mol Cell. 2014;55:771–781. doi: 10.1016/j.molcel.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns AM, Pollpeter D, Hadizadeh N, Myong S, Marko JF, Horvath CM. ATP hydrolysis enhances RNA recognition and antiviral signal transduction by the innate immune sensor, laboratory of genetics and physiology 2 (LGP2) J Biol Chem. 2013;288:938–946. doi: 10.1074/jbc.M112.424416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagliani R, Forni D, Tresoldi C, Pozzoli U, Filippi G, Rainone V, De Gioia L, Clerici M, Sironi M. RIG-I-like receptors evolved adaptively in mammals, with parallel evolution at LGP2 and RIG-I. J Mol Biol. 2014;426:1351–1365. doi: 10.1016/j.jmb.2013.10.040. [DOI] [PubMed] [Google Scholar]
- Civril F, Bennett M, Moldt M, Deimling T, Witte G, Schiesser S, Carell T, Hopfner KP. The RIG-I ATPase domain structure reveals insights into ATP-dependent antiviral signalling. EMBO Rep. 2011;12:1127–1134. doi: 10.1038/embor.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin TY, Schneller S, Zust R, Dong H, et al. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deddouche S, Goubau D, Rehwinkel J, Chakravarty P, Begum S, Maillard PV, Borg A, Matthews N, Feng Q, van Kuppeveld FJM, et al. Identification of an LGP2-associated MDA5 agonist in picornavirus-infected cells. Elife. 2014;3:e01535. doi: 10.7554/eLife.01535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Hato SV, Langereis MA, Zoll J, Virgen-Slane R, Peisley A, Hur S, Semler BL, van Rij RP, van Kuppeveld FJM. MDA5 detects the double-stranded RNA replicative form in picornavirus-infected cells. Cell Rep. 2012;2:1187–1196. doi: 10.1016/j.celrep.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink K, Grandvaux N. STAT2 and IRF9: Beyond ISGF3. Jakstat. 2013;2:e27521. doi: 10.4161/jkst.27521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freaney JE, Kim R, Mandhana R, Horvath CM. Extensive cooperation of immune master regulators IRF3 and NFκB in RNA Pol II recruitment and pause release in human innate antiviral transcription. Cell Rep. 2013;4:959–973. doi: 10.1016/j.celrep.2013.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman CS, O'Donnell MA, Legarda-Addison D, Ng A, Cardenas WB, Yount JS, Moran TM, Basler CF, Komuro A, Horvath CM, et al. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Rep. 2008;9:930–936. doi: 10.1038/embor.2008.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki M, Kato H, Miyachi Y, Toki H, Motegi H, Inoue M, Minowa O, Yoshida A, Deguchi K, Sato H, et al. Autoimmune disorders associated with gain of function of the intracellular sensor MDA5. Immunity. 2014;40:199–212. doi: 10.1016/j.immuni.2013.12.014. [DOI] [PubMed] [Google Scholar]
- Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell R, Diamond MS, Colonna M. Essential role of mda-5 in type I IFN responses to polyriboinosinic-polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci USA. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubau D, Deddouche S, Reis e Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubau D, Schlee M, Deddouche S, Pruijssers AJ, Zillinger T, Goldeck M, Schuberth C, Van der Veen AG, Fujimura T, Rehwinkel J, et al. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature. 2014;514:372–375. doi: 10.1038/nature13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscott J, Ware C. Cytokines. Curr Opin Immunol. 2011;23:561–563. doi: 10.1016/j.coi.2011.09.001. [DOI] [PubMed] [Google Scholar]
- Honda K, Takaoka A, Taniguchi T. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity. 2006;25:349–360. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inn KS, Gack MU, Tokunaga F, Shi M, Wong LY, Iwai K, Jung JU. Linear ubiquitin assembly complex negatively regulates RIG-I- and TRIM25-mediated type I interferon induction. Mol Cell. 2011;41:354–365. doi: 10.1016/j.molcel.2010.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs JL, Coyne CB. Mechanisms of MAVS regulation at the mitochondrial membrane. J Mol Biol. 2013;425:5009–5019. doi: 10.1016/j.jmb.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Ramanathan A, Miller MT, Tang GQ, Gale M, Patel SS, Marcotrigiano J. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479:423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang QX, Chen ZJ. Structural insights into the activation of RIG-I, a nanosensor for viral RNAs. EMBO Rep. 2012;13:7–8. doi: 10.1038/embor.2011.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Kinch LN, Brautigam CA, Chen X, Du F, Grishin NV, Chen ZJ. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity. 2012;36:959–973. doi: 10.1016/j.immuni.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Komuro A, Horvath CM. RNA- and virus-independent inhibition of antiviral signaling by RNA helicase LGP2. J Virol. 2006;80:12332–12342. doi: 10.1128/JVI.01325-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro A, Bamming D, Horvath CM. Negative regulation of cytoplasmic RNA-mediated antiviral signaling. Cytokine. 2008;43:350–358. doi: 10.1016/j.cyto.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- Kumar H, Kawai T, Kato H, Sato S, Takahashi K, Coban C, Yamamoto M, Uematsu S, Ishii KJ, Takeuchi O, et al. Essential role of IPS-1 in innate immune responses against RNA viruses. J Exp Med. 2006;203:1795–1803. doi: 10.1084/jem.20060792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Lu C, Stewart M, Xu H, Strong RK, Igumenova T, Li P. Structural basis of double-stranded RNA recognition by the RIG-I like receptor MDA5. Arch Biochem Biophys. 2009;488:23–33. doi: 10.1016/j.abb.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12:505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- Liu S, Chen J, Cai X, Wu J, Chen X, Wu YT, Sun L, Chen ZJ. MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. Elife. 2013;2:e00785–e00785. doi: 10.7554/eLife.00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Ding SC, Vela A, Kohlway A, Lindenbach BD, Pyle AM. Structural insights into RNA recognition by RIG-I. Cell. 2011;147:409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miner JJ, Diamond MS. MDA5 and autoimmune disease. Nat Genet. 2014;46:418–419. doi: 10.1038/ng.2959. [DOI] [PubMed] [Google Scholar]
- Moresco EMY, Beutler B. LGP2: Positive about viral sensing. Proc Natl Acad Sci USA. 2010;107:1261–1262. doi: 10.1073/pnas.0914011107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motz C, Schuhmann KM, Kirchhofer A, Moldt M, Witte G, Conzelmann KK, Hopfner KP. Paramyxovirus V proteins disrupt the fold of the RNA sensor MDA5 to inhibit antiviral signaling. Science. 2013;339:690–693. doi: 10.1126/science.1230949. [DOI] [PubMed] [Google Scholar]
- Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner KP, Ha T. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiumi H, Miyashita M, Inoue N, Okabe M, Matsumoto M, Seya T. The Ubiquitin Ligase Riplet Is Essential for RIG-I-Dependent Innate Immune Responses to RNA Virus Infection. Cell Host & Microbe. 2010;8:496–509. doi: 10.1016/j.chom.2010.11.008. [DOI] [PubMed] [Google Scholar]
- Parisien JP, Bamming D, Komuro A, Ramachandran A, Rodriguez JJ, Barber G, Wojahn RD, Horvath CM. A shared interface mediates paramyxovirus interference with antiviral RNA helicases MDA5 and LGP2. J Virol. 2009;83:7252–7260. doi: 10.1128/JVI.00153-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel JR, García-Sastre A. Three-stranded antiviral attack. Elife. 2014;3:e02369–e02369. doi: 10.7554/eLife.02369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel JR, Jain A, Chou YY, Baum A, Ha T, García-Sastre A. ATPase-driven oligomerization of RIG-I on RNA allows optimal activation of type-I interferon. EMBO Rep. 2013;14:780–787. doi: 10.1038/embor.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz S, Vilasco M, Werden SJ, Arguello M, Joseph-Pillai D, Zhao T, Nguyen TLA, Sun Q, Meurs EF, Lin R, et al. A functional C-terminal TRAF3-binding site in MAVS participates in positive and negative regulation of the IFN antiviral response. Cell Res. 2011;21:895–910. doi: 10.1038/cr.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley A, Jo MH, Lin C, Wu B, Orme-Johnson M, Walz T, Hohng S, Hur S. Kinetic mechanism for viral dsRNA length discrimination by MDA5 filaments. Proc Natl Acad Sci USA. 2012;109:E3340–E3349. doi: 10.1073/pnas.1208618109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley A, Lin C, Wu B, Orme-Johnson M, Liu M, Walz T, Hur S. Cooperative assembly and dynamic disassembly of MDA5 filaments for viral dsRNA recognition. Proc Natl Acad Sci USA. 2011;108:21010–21015. doi: 10.1073/pnas.1113651108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley A, Wu B, Xu H, Chen ZJ, Hur S. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature. 2014;509 doi: 10.1038/nature13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley A, Wu B, Yao H, Walz T, Hur S. RIG-I forms signaling-competent filaments in an ATP-dependent, ubiquitin-independent manner. Mol Cell. 2013;51:573–583. doi: 10.1016/j.molcel.2013.07.024. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Rehwinkel J, Kato H, Takeuchi O, Akira S, Way M, Schiavo G, Reis E Sousa C. Activation of MDA5 Requires Higher-Order RNA Structures Generated during Virus Infection. J Virol. 2009;83:10761–10769. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pippig DA, Hellmuth JC, Cui S, Kirchhofer A, Lammens K, Lammens A, Schmidt A, Rothenfusser S, Hopfner KP. The regulatory domain of the RIG-I family ATPase LGP2 senses double-stranded RNA. Nucleic Acids Res. 2009;37:2014–2025. doi: 10.1093/nar/gkp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollpeter D, Komuro A, Barber GN, Horvath CM. Impaired cellular responses to cytosolic DNA or infection with Listeria monocytogenes and vaccinia virus in the absence of the murine LGP2 protein. PLoS ONE. 2011;6:e18842. doi: 10.1371/journal.pone.0018842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos HJ, Gale M. RIG-I like receptors and their signaling crosstalk in the regulation of antiviral immunity. Curr Opin Virol. 2011;1:167–176. doi: 10.1016/j.coviro.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawling DC, Pyle AM. Parts, assembly and operation of the RIG-I family of motors. Curr Opin Struct Biol. 2014;25C:25–33. doi: 10.1016/j.sbi.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, del Toro Duany Y, Jenkinson EM, Forte GMA, Anderson BH, Ariaudo G, Bader-Meunier B, Baildam EM, Battini R, Beresford MW, et al. Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nat Genet. 2014;46:503–509. doi: 10.1038/ng.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson T, Kariuki SN, Franek BS, Kumabe M, Kumar AA, Badaracco M, Mikolaitis RA, Guerrero G, Utset TO, Drevlow BE, et al. Autoimmune disease risk variant of IFIH1 is associated with increased sensitivity to IFN-α and serologic autoimmunity in lupus patients. J Immunol. 2011;187:1298–1303. doi: 10.4049/jimmunol.1100857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez KR, Horvath CM. Amino acid requirements for MDA5 and LGP2 recognition by paramyxovirus V proteins: a single arginine distinguishes MDA5 from RIG-I. J Virol. 2013;87:2974–2978. doi: 10.1128/JVI.02843-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez KR, Bruns AM, Horvath CM. MDA5 and LGP2: Accomplices and Antagonists of Antiviral Signal Transduction. J Virol. 2014 doi: 10.1128/JVI.00640-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenfusser S, Goutagny N, DiPerna G, Gong M, Monks BG, Schoenemeyer A, Yamamoto M, Akira S, Fitzgerald KA. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J Immunol. 2005;175:5260–5268. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale M. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci USA. 2007;104:582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, Tsujimura T, Fujita T, Akira S, Takeuchi O. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci USA. 2010;107:1512–1517. doi: 10.1073/pnas.0912986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M. Master sensors of pathogenic RNA - RIG-I like receptors. Immunobiology. 2013;218:1322–1335. doi: 10.1016/j.imbio.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M, Hartmann G. The Chase for the RIG-I Ligand-Recent Advances. Mol Ther. 2010:1–9. doi: 10.1038/mt.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G. Recognition of 5′ Triphosphate by RIG-I Helicase Requires Short Blunt Double-Stranded RNA as Contained in Panhandle of Negative-Strand Virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Schwerd T, Hamm W, Hellmuth JC, Cui S, Wenzel M, Hoffmann FS, Michallet MC, Besch R, Hopfner KP, et al. 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc Natl Acad Sci USA. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Smyth DJ, Cooper JD, Bailey R, Field S, Burren O, Smink LJ, Guja C, Ionescu-Tirgoviste C, Widmer B, Dunger DB, et al. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat Genet. 2006;38:617–619. doi: 10.1038/ng1800. [DOI] [PubMed] [Google Scholar]
- Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, Chen ZJ. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Suthar MS, Ramos HJ, Brassil MM, Netland J, Chappell CP, Blahnik G, McMillan A, Diamond MS, Clark EA, Bevan MJ, et al. The RIG-I-like Receptor LGP2 Controls CD8(+) T Cell Survival and Fitness. Immunity. 2012;37:235–248. doi: 10.1016/j.immuni.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahasi K, Kumeta H, Tsuduki N, Narita R, Shigemoto T, Hirai R, Yoneyama M, Horiuchi M, Ogura K, Fujita T, et al. Solution structures of cytosolic RNA sensor MDA5 and LGP2 C-terminal domains: identification of the RNA recognition loop in RIG-I-like receptors. J Biol Chem. 2009;284:17465–17474. doi: 10.1074/jbc.M109.007179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman T, Valdes M, Elsby R, Kakuta S, Caceres G, Saijo S, Iwakura Y, Barber GN. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J Immunol. 2007a;178:6444–6455. doi: 10.4049/jimmunol.178.10.6444. [DOI] [PubMed] [Google Scholar]
- Venkataraman T, Valdes M, Elsby R, Kakuta S, Caceres G, Saijo S, Iwakura Y, Barber GN. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J Immunol. 2007b;178:6444–6455. doi: 10.4049/jimmunol.178.10.6444. [DOI] [PubMed] [Google Scholar]
- Widau RC, Parekh AD, Ranck MC, Golden DW, Kumar KA, Sood RF, Pitroda SP, Liao Z, Huang X, Darga TE, et al. RIG-I-like receptor LGP2 protects tumor cells from ionizing radiation. Proc Natl Acad Sci USA. 2014;111:E484–E491. doi: 10.1073/pnas.1323253111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wies E, Wang MK, Maharaj NP, Chen K, Zhou S, Finberg RW, Gack MU. Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity. 2013;38:437–449. doi: 10.1016/j.immuni.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Peisley A, Richards C, Yao H, Zeng X, Lin C, Chu F, Walz T, Hur S. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152:276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- Xu H, He X, Zheng H, Huang LJ, Hou F, Yu Z, de la Cruz MJ, Borkowski B, Zhang X, Chen ZJ, et al. Structural basis for the prion-like MAVS filaments in antiviral innate immunity. Elife. 2014;3:e01489. doi: 10.7554/eLife.01489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M, Akira S, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen ZJ. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Wu X, Lee AJ, Jin W, Chang M, Wright A, Imaizumi T, Sun SC. Regulation of IkappaB kinase-related kinases and antiviral responses by tumor suppressor CYLD. J Biol Chem. 2008;283:18621–18626. doi: 10.1074/jbc.M801451200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zust R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]


