Abstract
Mutations of GJB2 which encodes connexin 26, contribute to 6–7% of profound deafness in Pakistan. We investigated the involvement of GJB2 mutations in a cohort of 84 pedigrees and 86 sporadic individuals with moderate or severe hearing loss. Individuals in eight consanguineous families and four sporadic cases (9.52% and 4.65%, respectively) were homozygous or compound heterozygous for p.W24X or p. W77X mutations in GJB2. These two variants are also among the most common mutations known to cause profound deafness in South Asia. The association of identical mutations with both profound and less severe phenotype of hearing loss suggests that alleles of other genes modify the phenotype due to these GJB2 nonsense mutations. Our study demonstrates that GJB2 mutations are an important contributor to aetiology of moderate to severe hearing loss in Pakistan.
Keywords: Connexin 26, deafness, hearing loss, GJB2, Pakistan
Introduction
Genetics contributes to hearing loss in more than 50% of individuals affected with prelingual deafness. Mutations in gap junction beta-2, (GJB2) are a major cause of non-syndromic hereditary hearing loss in European, American Caucasians and a few other populations. GJB2 encodes a gap junction protein, connexin 26, which is involved in the movement of small metabolites and ions between the neighboring cells [1]. Some mutations in GJB2 are quite common, but their frequency is variable worldwide [2]. One of the most prevalent mutations in Caucasians as a cause of recessive inherited deafness is c.35delG. This mutation may account for 86% (95 percent confidence interval, 79–91%) of all GJB2 related deafness in the French [3] and 85% (95% confidence interval, 78–93%) in some other European populations [4]. The carrier frequency of the mutation in these populations can be as high as 1.96%, (95%, confidence interval, 1.4–2.5%) [5]. Similarly, c.167delT is the most frequent GJB2 mutation in the Ashkenazi Jews with a carrier frequency of 4.3%, (95% confidence interval, 2.5 to 6.0%), [6].
The contribution of mutations in GJB2 in Pakistan to severe to profound deafness has been reported to be 6.1% (95 percent confidence interval, 3.2–10.4%) in a study involving 196 large pedigrees with multiple affected individuals with hearing loss [7]. Mutations in GJB2 account for 6.9% (95%confidence interval, 3.2–10.4%), of moderate to profound deafness in Pakistanis living in the UK, as inferred from analyses of samples from 123 sib pairs [8]. However, the contribution of GJB2 mutations to deafness in Pakistan was reported to be as high as 53%, (95% confidence interval, 35–71%), in a study involving 30 families with severe to profound deafness [9]. The large discrepancy between the results of this research on 30 families and the estimates of frequency of mutations in GJB2 obtained from previous studies is not clear, although small sample size of these studies may have contributed to the difference. We sought to focus on the identification of GJB2 mutations and assessment of hearing function in a series of families in Pakistan, which were characterized by a high rate of consanguineous marriages. The probands were affected by essentially moderate and severe hearing loss.
Methods
Institutional review board approval was obtained for the study from School of Biological Sciences, University of the Punjab, Lahore. Probands were identified from schools catering to children with different disabilities and with help of audiologists in the Punjab province. Detailed clinical histories were obtained to minimize inclusion of individuals with hearing loss due to syndromes and environmental factors. Audiometry was performed in ambient noise conditions at participants’ homes since sound-proof rooms were not available. Hearing loss was classified as moderate to severe (41 to 70 dB HL), severe (71 to 95 dB HL) and profound (above 95 dB HL) [10]. Blood samples were collected from 84 large families and 86 sporadic participants segregating non-syndromic, recessive, moderate to severe hearing loss. Written informed consent was obtained from the participants or the parents in case of minor children. The single coding exon of GJB2 was sequenced from DNA of all affected individuals and 100 ethnically matched controls.
Results
The affected participants of the study were born in consanguineous unions to unaffected parents. Hearing loss in affected individuals in 80% of the participating families could be classified as moderate to severe. Intrafamilial phenotypic variability was observed in 20% of the families in which 1–3 individuals were affected with severe to profound deafness, while the majority of the individuals had moderate to severe hearing loss. All affected individuals were born in consanguineous unions to unaffected parents. Sequence analysis revealed two disease causing mutations in GJB2 for some of the participants (Table 1). Eight out of 84 families had nonsense mutations in GJB2 (c.71G>A, p.W24X or C.231G>A, p.W77X). Among the non-familial cases of hearing loss, homozygous mutations in GJB2 were identified in 4 out of 86 individuals. The p.W24X mutation was most frequently observed among the participants (Table 1). Samples from all other participating familial and non-familial participants were negative for homozygous, or compound heterozygous, mutations in GJB2.
Table 1. Description of families, phenotype and GJB2 mutations.
Results are tabulated for 8 of 84 families and 4 of 86 sporadic individuals positive for mutations in GJB2.
| ID | Hearing Loss, PTA500-4000*, dB HL | Mutation | Comment |
|---|---|---|---|
| Familial Cases | |||
| HLRB1 | Moderate to severe 54,65, 71,103 |
p.W77X/p.W77X | 4 affected individuals in 3 loops (1 affected individual has a profound HL) |
| HLRB8 | Moderate to severe 70, 78, 78 |
p.W24X/p.W77X | 6 affected individuals in 2 loops |
| HLAI-02 | Severe and profound 81, 86, 88, 98, 103, 104 |
p.W24X/p.W24X | 8 affected individuals in 5 loops (3 individuals have profound deafness) |
| HLAI-12 | Moderate to severe 59, 71, 90 |
p.W24X/p.W24X | 3 affected individuals |
| HLAM09 | Moderate to severe 61 |
p.W24X/p.W77X | 3 affected individuals |
| HLGM25 | Moderate to severe 70, 70, 94 |
p.W24X/p.W24X | 5 affected individuals in 2 loops (1 affected individual has profound HL) |
| HLMS16** | Severe 74,79 |
p.W24X/p.W24X | 2 affected individuals |
| HLMS34** | Severe 75, 78 |
p.W24X/p.W24X | 2 affected individuals in 2 loops |
| Sporadic cases | |||
| HLMS11 | Severe 84 |
p.W77X/p.W77X | NA |
| HLMS25 | Severe 84 |
p.W24X/p.W24X | NA |
| SA15 | Severe 89 |
p.W24X/p.W24X | NA |
| HLRB15# | Severe 82 |
p.W24X/p.W24X | NA |
Pure-tone air conduction averages, db HL, for hearing thresholds at 500, 1000, 2000 and 4000 Hz (PTA500-4000) of the better hearing ear is provided for all affected participants, except for those who refused the evaluation.
The only two families in the entire collection of 84 large pedigrees with inherited hearing loss which had 2 affected individuals in one or two consanguineous marriages. The other 82 families had multiple (3 or more) affected individuals in a single, or several marriage loops.
Although family HLRB15 had four affected individuals, sequencing revealed that only one participant had a homozygous mutation in GJB2. Therefore, the sample from this family is considered as a sporadic case. HL, Hearing Loss, NA, Not applicable.
We observed compound heterozygous mutations (p.W24X, p.W77X) in samples of affected individuals in two large families, HLRB8 and HLAM09, although the subjects were born in consanguineous marriages. This may be due to the high carrier frequencies of these founder mutations in the population. However, we did not detect these two mutations in 200 chromosomes from ethnically matched controls.
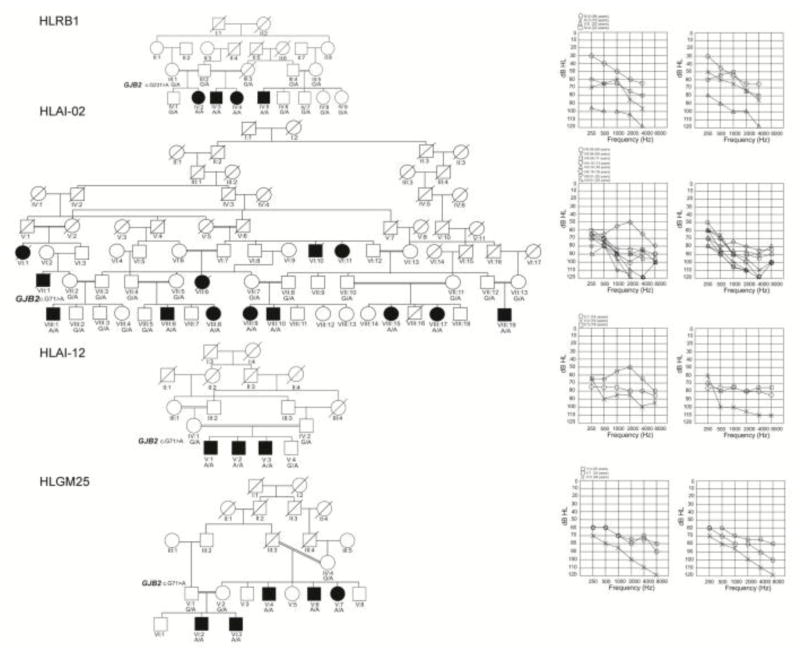
In 4 families (HLRB1, HLAI-02, HLAI-12 and HLGM25), there was marked intrafamilial variability of the phenotype (Table 1 and Fig. 1). The majority of the affected subjects in these pedigrees had moderate to severe hearing loss, while 1 to 3 individuals in each family had profound deafness. There was no correlation with age or environment which could account for the worse hearing loss of these individuals as compared to the other affected individuals with relatively better hearing in their families.
Figure 1. Pedigrees of four families in which the affected individuals exhibit intrafamilial differences in hearing thresholds.
Black circle’s and squares represent affected female and male participants, respectively. The double horizontal lines denote consanguineous marriages. The genotype for the mutation in GJB2 is provided below each symbol representing the participant. The audiograms on the right side of each pedigree represent the hearing thresholds of the left and the right ears of the affected individuals, respectively.
Discussion
The present study revealed that GJB2 mutations contribute to 9.52% (95% confidence interval, 3.2 to 15.7%) of moderate to severe hearing loss among familial participants and 4.65% (95% confidence interval, 0.2 to 9.1%) among sporadically affected individuals. The lower contribution of 4.65% in the sporadic cases as compared to that with a clearly demonstrated recessive mode of inherited deafness, may reflect the inclusion of some individuals in this cohort who are either affected due to non-genetic causes, or have hearing loss as a result of de novo mutations in other deafness causing genes.
Previously, truncating mutations in GJB2, present together with non-truncating mutations (i.e. compound heterozygote) have been reported to result in a less severe hearing loss [11,12]. To date, c.35delG is the only known deletion mutation in GJB2 which has been widely reported to lead to less severe hearing loss in individuals who are homozygous for the allele. However, we found that the two nonsense mutations in the Pakistani population, p.W24X and p.W77X, present either in the homozygous state or in compound heterozygosity, may similarly result in moderate to severe hearing loss in a large number of affected individuals. Interestingly, p.W24X and p.W77X are also the most common mutations involved in profound deafness in Pakistan [7]. The variability in hearing thresholds of individuals with p.W24X and p.W77X mutations is similar to the phenotypic difference exhibited by the individuals homozygous for the c.35delG allele.
The association of p.W24X and p.W77X mutations with both profound and less severe phenotype of hearing loss suggests the effect of modifier genes that lead to variation in hearing thresholds [13] and is similar to that suspected for c.35delG related deafness. So far, for lack of a sufficiently large family, it has not been possible to map a genetic modifier, which reduces severity of hearing loss caused by GJB2 mutations [14]. Whole exome or genome sequencing and a sufficiently large number of individuals in nuclear consanguineous families with GJB2 pathogenic alleles showing carefully documented variation in hearing thresholds, as well as matched controls, might allow for the identification of enhancers and suppressors of GJB2 hearing loss. This will broaden our understanding of audition and may help in devising eventual treatments or cures.
Acknowledgments
We thank all the individuals who participated in the research. We are grateful to the various schools, audiologists and doctors for their help. We express our gratitude to Usman, Khalid and Arif for assistance in collection of samples from the participants. We are indebted to Dr. Thomas Friedman for reviewing the manuscript. This research was supported by grant R01TW007608 from the Fogarty International Center and National Institute of Deafness and other Communication Disorders, National Institutes of Health, USA to S.N.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- 1.Thonnissen E, Rabionet R, Arbones ML, Estivill X, Willecke K, Ott T. Human connexin26 (GJB2) deafness mutations affect the function of gap junction channels at different levels of protein expression. Hum Genet. 2002;111 (2):190–197. doi: 10.1007/s00439-002-0750-2. [DOI] [PubMed] [Google Scholar]
- 2.Kenneson A, Van Naarden Braun K, Boyle C. GJB2 (connexin 26) variants and nonsyndromic sensorineural hearing loss: a HuGE review. Genet Med. 2002;4 (4):258–274. doi: 10.1097/00125817-200207000-00004. [DOI] [PubMed] [Google Scholar]
- 3.Denoyelle F, Marlin S, Weil D, Moatti L, Chauvin P, Garabedian EN, Petit C. Clinical features of the prevalent form of childhood deafness, DFNB1, due to a connexin-26 gene defect: implications for genetic counselling. Lancet. 1999;353 (9161):1298–1303. doi: 10.1016/S0140-6736(98)11071-1. [DOI] [PubMed] [Google Scholar]
- 4.Estivill X, Fortina P, Surrey S, Rabionet R, Melchionda S, D’Agruma L, Mansfield E, Rappaport E, Govea N, Mila M, Zelante L, Gasparini P. Connexin-26 mutations in sporadic and inherited sensorineural deafness. Lancet. 1998;351 (9100):394–398. doi: 10.1016/S0140-6736(97)11124-2. [DOI] [PubMed] [Google Scholar]
- 5.Gasparini P, Rabionet R, Barbujani G, Melchionda S, Petersen M, Brondum-Nielsen K, Metspalu A, Oitmaa E, Pisano M, Fortina P, Zelante L, Estivill X. High carrier frequency of the 35delG deafness mutation in European populations. Genetic Analysis Consortium of GJB2 35delG. Eur J Hum Genet. 2000;8 (1):19–23. doi: 10.1038/sj.ejhg.5200406. [DOI] [PubMed] [Google Scholar]
- 6.Morell RJ, Kim HJ, Hood LJ, Goforth L, Friderici K, Fisher R, Van Camp G, Berlin CI, Oddoux C, Ostrer H, Keats B, Friedman TB. Mutations in the connexin 26 gene (GJB2) among Ashkenazi Jews with nonsyndromic recessive deafness. N Engl J Med. 1998;339 (21):1500–1505. doi: 10.1056/NEJM199811193392103. [DOI] [PubMed] [Google Scholar]
- 7.Santos RL, Wajid M, Pham TL, Hussan J, Ali G, Ahmad W, Leal SM. Low prevalence of Connexin 26 (GJB2) variants in Pakistani families with autosomal recessive non-syndromic hearing impairment. Clin Genet. 2005;67 (1):61–68. doi: 10.1111/j.1399-0004.2005.00379.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoong SY, Mavrogiannis LA, Wright J, Fairley L, Bennett CP, Charlton RS, Spencer N. Low prevalence of DFNB1 (connexin 26) mutations in British Pakistani children with non-syndromic sensorineural hearing loss. Arch Dis Child. 2011;96 (9):798–803. doi: 10.1136/adc.2010.209262. [DOI] [PubMed] [Google Scholar]
- 9.Shafique S, Siddiqi S, Schraders M, Oostrik J, Ayub H, Bilal A, Ajmal M, Seco CZ, Strom TM, Mansoor A, Mazhar K, Shah ST, Hussain A, Azam M, Kremer H, Qamar R. Genetic spectrum of autosomal recessive non-syndromic hearing loss in Pakistani families. PLoS One. 2014;9 (6):e100146. doi: 10.1371/journal.pone.0100146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martini A, Mazzoli M, Stephens D, Read A. Audiological terms. . Definitions, protocols & guidelines in genetic hearing impairment. Whurr publishers, John Wiley & Sons, Ltd; 2001. [Google Scholar]
- 11.Liu XZ, Pandya A, Angeli S, Telischi FF, Arnos KS, Nance WE, Balkany T. Audiological features of GJB2 (connexin 26) deafness. Ear Hear. 2005;26 (3):361–369. doi: 10.1097/00003446-200506000-00011. [DOI] [PubMed] [Google Scholar]
- 12.Snoeckx RL, Huygen PL, Feldmann D, Marlin S, Denoyelle F, Waligora J, Mueller-Malesinska M, Pollak A, Ploski R, Murgia A, Orzan E, Castorina P, Ambrosetti U, Nowakowska-Szyrwinska E, Bal J, Wiszniewski W, Janecke AR, Nekahm-Heis D, Seeman P, Bendova O, Kenna MA, Frangulov A, Rehm HL, Tekin M, Incesulu A, Dahl HH, du Sart D, Jenkins L, Lucas D, Bitner-Glindzicz M, Avraham KB, Brownstein Z, del Castillo I, Moreno F, Blin N, Pfister M, Sziklai I, Toth T, Kelley PM, Cohn ES, Van Maldergem L, Hilbert P, Roux AF, Mondain M, Hoefsloot LH, Cremers CW, Lopponen T, Lopponen H, Parving A, Gronskov K, Schrijver I, Roberson J, Gualandi F, Martini A, Lina-Granade G, Pallares-Ruiz N, Correia C, Fialho G, Cryns K, Hilgert N, Van de Heyning P, Nishimura CJ, Smith RJ, Van Camp G. GJB2 mutations and degree of hearing loss: a multicenter study. Am J Hum Genet. 2005;77 (6):945–957. doi: 10.1086/497996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McHugh RK, Friedman RA. Genetics of hearing loss: Allelism and modifier genes produce a phenotypic continuum. Anat Rec A Discov Mol Cell Evol Biol. 2006;288 (4):370–381. doi: 10.1002/ar.a.20297. [DOI] [PubMed] [Google Scholar]
- 14.Hilgert N, Huentelman MJ, Thorburn AQ, Fransen E, Dieltjens N, Mueller-Malesinska M, Pollak A, Skorka A, Waligora J, Ploski R, Castorina P, Primignani P, Ambrosetti U, Murgia A, Orzan E, Pandya A, Arnos K, Norris V, Seeman P, Janousek P, Feldmann D, Marlin S, Denoyelle F, Nishimura CJ, Janecke A, Nekahm-Heis D, Martini A, Mennucci E, Toth T, Sziklai I, Del Castillo I, Moreno F, Petersen MB, Iliadou V, Tekin M, Incesulu A, Nowakowska E, Bal J, Van de Heyning P, Roux AF, Blanchet C, Goizet C, Lancelot G, Fialho G, Caria H, Liu XZ, Xiaomei O, Govaerts P, Gronskov K, Hostmark K, Frei K, Dhooge I, Vlaeminck S, Kunstmann E, Van Laer L, Smith RJ, Van Camp G. Phenotypic variability of patients homozygous for the GJB2 mutation 35delG cannot be explained by the influence of one major modifier gene. Eur J Hum Genet. 2009;17 (4):517–524. doi: 10.1038/ejhg.2008.201. [DOI] [PMC free article] [PubMed] [Google Scholar]