Summary
BRD4, a bromodomain and extraterminal domain (BET) family member, is an attractive target in multiple pathological settings, particularly cancer. While BRD4 inhibitors have shown some promise in MYC-driven malignancies such as Burkitt’s Lymphoma (BL), we show that BRD4 inhibitors lead to robust BRD4 protein accumulation, which may account for their limited suppression of MYC expression, modest anti-proliferative activity and lack of apoptotic induction. To address these limitations, we designed ARV-825, a heterobifunctional PROTAC (Proteolysis Targeting Chimera) that recruits BRD4 to the E3 ubiquitin ligase cereblon leading to fast, efficient, and prolonged degradation of BRD4 in all BL cell lines tested. Consequently, ARV-825 more effectively suppresses c-MYC levels and downstream signaling than small molecule BRD4 inhibitors resulting in more effective cell proliferation inhibition and apoptosis induction in BL. Our findings provide strong evidence that cereblon-based PROTACs provide a better and more efficient strategy in targeting BRD4 than traditional small molecule inhibitors.
Introduction
BRD4 belongs to the bromodomain and extraterminal domain (BET) family of proteins, which is characterized by two bromodomains (BD) at the N-terminus and an extraterminal domain (ET domain) at the C-terminus (Belkina and Denis, 2012; Shi and Vakoc, 2014). The two BDs recognize and interact with acetylated lysine residues at the N-terminal tails of histones; the ET domain, which is not yet fully characterized, is largely considered to serve a scaffolding function in recruiting diverse transcriptional regulators (Belkina and Denis, 2012; Shi and Vakoc, 2014). Thus, BRD4 plays a key role in regulating gene expression by recruiting relevant transcription modulators to specific genomic loci. Several recent studies establish that BRD4 is preferentially located at super-enhancer regions, which often reside upstream of important oncogenes, such as c-myc, bcl-xL and bcl-6, and play a key role in regulating their expressions (Chapuy et al., 2013; Loven et al., 2013). Owing to its pivotal role in modulating the expression of essential oncogenes, BRD4 has emerged as a promising therapeutic target in multiple cancer types, including midline carcinoma, AML, MM, BL, and prostate cancer (Asangani et al., 2014; Delmore et al., 2011; French et al., 2008; Loven et al., 2013; Mertz et al., 2011; Wyce et al., 2013; Zuber et al., 2011). Particularly, BRD4 is exciting as an alternative strategy for targeting c-MYC, which contributes to the development and maintenance of a majority of human cancers but has remained undruggable (Baratta et al., 2015; Delmore et al., 2011; Gabay et al., 2014; Mertz et al., 2011). Indeed, the development of small molecule BRD4 inhibitors, such as JQ1, iBET and OTX015, has quickly demonstrated their promising therapeutic potential in preclinical models of various cancers, including BL (Asangani et al., 2014; Baratta et al., 2015; Boi et al., 2015; Chapuy et al., 2013; Delmore et al., 2011; Loven et al., 2013; Mertz et al., 2011; Puissant et al., 2013). Almost all BL cancers contain a c-myc gene translocation that places it under control of a super-enhancer located upstream of IgH, thus driving an abnormally high level of c-MYC expression, crucial to tumor development and maintenance (Klapproth and Wirth, 2010). Excitingly, preclinical studies with BRD4 inhibitors demonstrate their value in suppressing c-MYC and proliferation in BL cell lines, albeit with IC50 values often in the range of 100 nM to 1 µM (Ceribelli et al., 2014; Mertz et al., 2011).
Small molecule inhibitors, either by inhibiting an enzyme activity (such as kinase inhibitors) or by interfering with protein-protein interactions (such as BRD4 inhibitors) have been the cornerstone of oncology drug development (Hoelder et al., 2012). However, given the reversible binding of most small molecule inhibitors, large systemic drug concentrations and continuous exposures are often required to ensure sufficient functional inhibition (Johnson et al., 2010). In a number of cases, it proves challenging to achieve and maintain a drug concentration high enough for efficacy in vivo. As an alternative, approaches to eliminate disease-causing abnormal proteins, either by RNAi or by small molecule-induced protein degradation, are evolving as novel strategies for targeting the “undruggable” or “difficult” targets (Burnett et al., 2011; Howell et al., 2004). We have developed an induced protein degradation strategy that utilizes Proteolysis Targeting Chimerae (PROTACs) that recruit targeted proteins to the E3 ubiquitin ligase Von Hippel Lindau (VHL) for ubiquitination and subsequent proteasome-mediated degradation (Buckley and Crews, 2014; Raina and Crews, 2010; Sakamoto et al., 2001; Sakamoto et al., 2003; Schneekloth and Crews, 2005; Schneekloth et al., 2004). This PROTAC technology provides great potential in harnessing the action of a single E3 ligase toward pathologically important proteins which are currently “undruggable” through conventional strategies for drug development.
In this study, we demonstrate that small molecule BRD4 inhibitors, JQ1 and OTX015, lead to significant BRD4 protein accumulation over time in all BL cell lines tested. Although both inhibitors initially suppress downstream c-MYC level, the suppression is incomplete and requires high drug concentrations. This robust accumulation of BRD4, together with the reversible nature of inhibitor binding to BRD4, may in part account for the modest effect of these inhibitors on downstream c-MYC suppression and cell proliferation inhibition. To circumvent these limitations, we designed a hetero bifunctional molecule, ARV-825, by connecting a small molecule BRD4 binding moiety (OTX015) to an E3 ligase cereblon binding moiety (pomalidomide) using PROTAC technology (Boi et al., 2015; Fischer et al., 2014; Ito et al., 2010). OTX015 is currently in phase I clinical trial and pomalidomide is a potent third-generation immunomodulatory drug (IMiD), which functions through interacting with the E3 ligase cereblon and inducing degradation of essential Ikaros transcription factors in multiple myeloma (Boi et al., 2015; Lu et al., 2014). ARV-825 actively recruits BRD4 to cereblon, resulting in the rapid and efficient degradation of the former via the proteasome. We demonstrate that, compared to the BRD4 inhibitors, ARV-825 treatment results in a strikingly more pronounced effect on the levels of c-MYC, and downstream cell proliferation and apoptosis induction in BL cell lines. These findings strongly support the development of BRD4 PROTACs as a promising novel strategy to efficiently target BRD4. Moreover, this study is the first to describe a potent PROTAC that acts via the recruitment of the E3 ligase cereblon, Our study demonstrates that hijacking the E3 ubiquitin ligase cereblon, or other E3 ligases, through the PROTAC platform, holds great potential for pursuing effective therapeutics.
Results
Small molecule BET domain inhibitors lead to significant BRD4 protein accumulation and inefficient c-MYC suppression
As recently discovered, BL cells are responsive to BRD4 inhibitors, mostly due to their dependence on the c-myc oncogene that is translocated and brought under the control of upstream IgH super-enhancers regulated by BRD4 (Klapproth and Wirth, 2010; Mertz et al., 2011). Interestingly, we found that both JQ1, the most frequently used inhibitor in published studies, and OTX015, the most advanced inhibitor in clinical development, led to significant BRD4 protein accumulation in a dose-dependent manner in all BL cell lines tested (Figure 1A and Supp. Figure 1). This is consistent with the previously reported observation that JQ1 treatment results in BRD4 up-regulation in some lung cancer cell lines (Shimamura et al., 2013). Moreover, the accumulation of BRD4 is rapid, resulting in a multi-fold increase of BRD4 within 24 hours of treatment (Figure 1B). We reasoned that the BRD4 inhibitor-induced increase in BRD4 protein level may incur a higher barrier for BRD4 to be effectively suppressed. Indeed, even at concentrations up to ten-fold over their IC50 values, JQ1 and OTX015 were unable to suppress c-MYC levels fully (Figure 1C). Even though higher BRD4 level does not necessarily warrant higher BRD4 nuclear localization under these conditions, this finding implies cancer cells may quickly restore their transcriptional activation upon the withdrawal of bromodomain inhibitors. Indeed, c-MYC levels were rapidly restored to normal upon the withdrawal of JQ1 and OTX015 from the culture media (Figure 1D). This is consistent with previous findings in AML that c-MYC is repressed by JQ1 treatment, but rebounds quickly upon JQ1 withdrawal (Mertz et al., 2011).
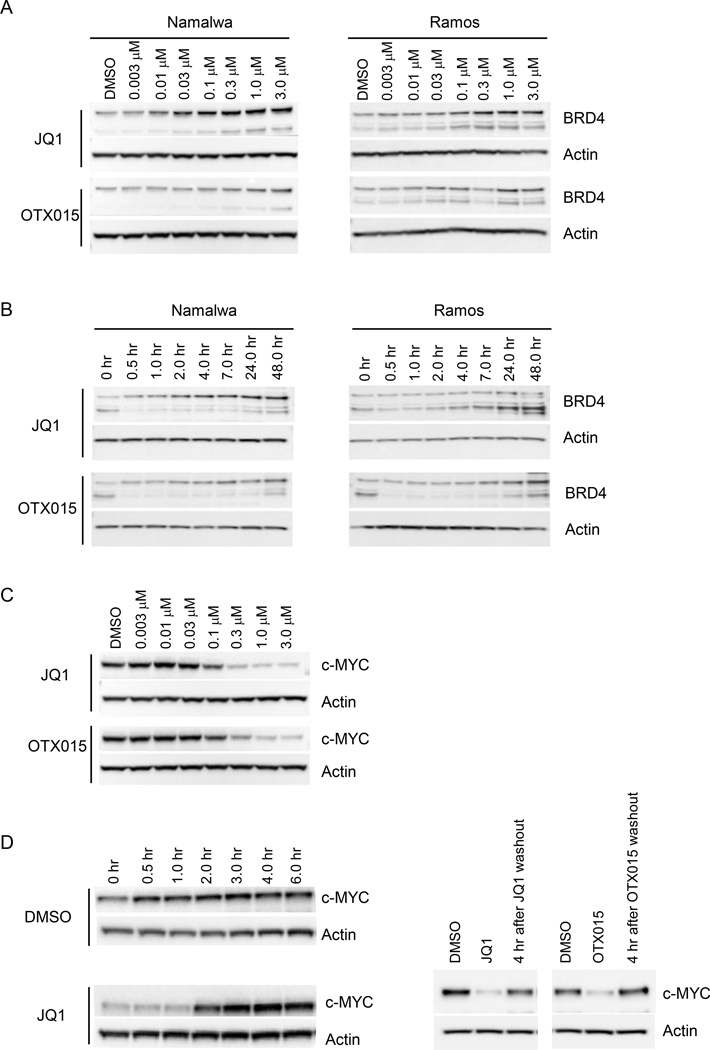
Figure 1. Small molecule BRD4 inhibitors lead to significant BRD4 accumulation and inefficient c-MYC suppression.
(A) Small molecule BRD4 inhibitors lead to significant BRD4 accumulation. Namalwa and Ramos cells were treated overnight with increasing doses of JQ1 and OTX015. Lysates were collected and subjected to immunoblot analysis with antibodies for BRD4 and actin.
(B) Small molecule BRD4 inhibitors lead to rapid BRD4 accumulation. Namalwa and Ramos cells were treated with 0.3 µM of JQ1 or OTX015 for various times as indicated, lysates were collected and analyzed by immunoblot for BRD4 and actin.
(C) Small molecule BRD4 inhibitors lead to downstream c-MYC suppression, but not efficiently. Namalwa cells were treated overnight with increasing doses of JQ1 and OTX015, lysates were collected and analyzed by immunoblot with antibodies for c-MYC and actin.
(D) Loss of c-MYC suppression shortly after BRD4 inhibitors withdrawal. (left panel) Namalwa cells were treated with JQ1 (1.0 µM) for 24 hours, followed by three washes to remove compound. Cells were re-seeded for lysates collection at various time points, c-MYC level was determined by immunoblot; (right panel) Ramos cells were treated with JQ1 (1.0 µM) or OTX015 (1.0 µM) for 24 hours, followed by compounds removal and re-seeding in fresh medium for 4 hours; lysates were subjected for immunoblot with c-MYC and actin antibodies.
Hijacking the E3 Ubiquitin Ligase Cereblon to create PROTAC to efficiently degrade BRD4
We noted the rapid and robust accumulation of BRD4 associated with inhibitor treatments, and the resulting moderate effects on downstream c-MYC suppression and proliferation. To circumvent these limitations, we utilized PROTAC technology to design the chimeric small molecule ARV-825 (Figure 2A). ARV-825 consists of a classic BRD4 binding moiety of the triazolo-diazepine acetamide class seen in OTX015, and pomalidomide, a known cereblon binding moiety of the IMiD class (Fischer et al., 2014), connected by a flexible polyethyleneglycol linker. This linker was chosen because of its high conformational flexibility that would maximize the chance of the PROTAC associating with both BRD4 and cereblon. We subsequently demonstrated that ARV-825 demonstrates only slightly reduced binding affinities to bromodomain 1 and 2 compared to those of OTX015 (Table 1).
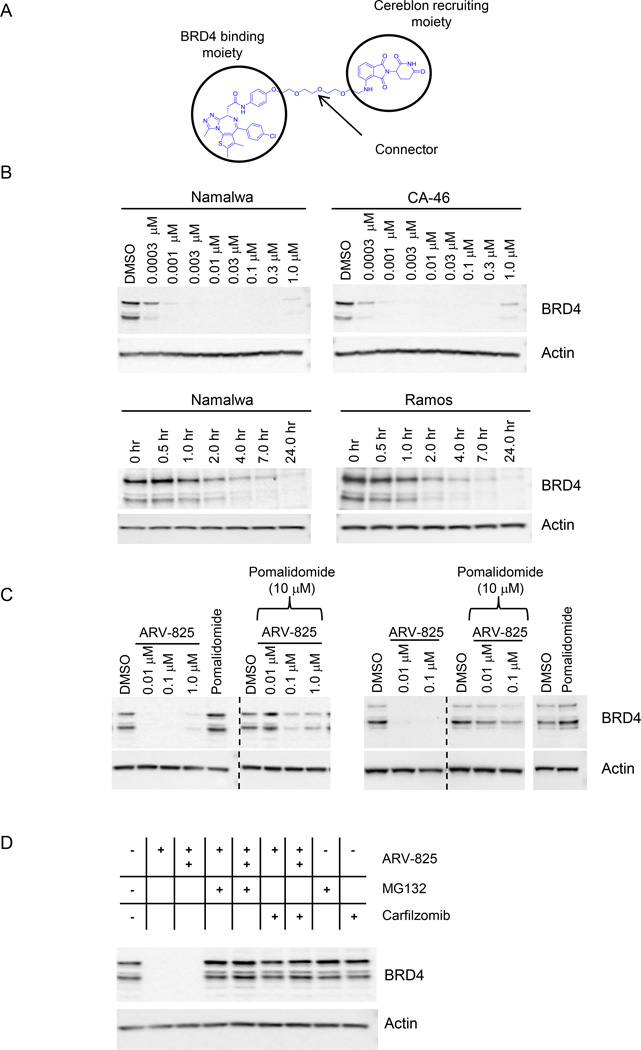
Figure 2. Hijacking the E3 Ubiquitin Ligase Cereblon to create PROTAC to efficiently degrade BRD4.
(A) A schematic representation of bifunctional PROTAC ARV-825.
(B) ARV-825 leads to fast and efficient degradation of BRD4. (top panel) Namalwa and CA-46 cells were treated overnight with increasing doses of ARV-825, lysates were analyzed for BRD4 levels by immunoblot with actin serving as loading control; (bottom panel) Namalwa and Ramos cells were treated with ARV-825 (0.1 µM) for indicated time points, lysates were collected and subjected to immunoblot analysis with antibodies for BRD4 and actin.
(C) Confirmation of cereblon-based mechanism in driving BRD4 degradation upon ARV-825 treatment. Namalwa (left panel) and Ramos (right panel) cells were treated overnight with various concentrations of ARV-825 or pomalidomide (10 µM), or combination of ARV-825 and pomalidomide; lysates were analyzed by immunoblot for BRD4 and actin.
(D) Confirmation of proteasome-based mechanism in driving BRD4 degradation upon ARV-825 treatment. Namalwa cells were treated overnight with ARV-825 (+/0.01 µM and ++/0.1 µM) alone, MG132 (5 µM) or carfilzomib (5 µM) alone, or combination of ARV-825 with MG132 or with carfizomib; lysates were collected and analyzed by immunoblot for BRD4 and actin.
Table 1. Affinity to BD1 and BD2 of BRD4 by ARV-825, JQ1 and OTX015.
Affinity of compounds with Bromodomain 1 and 2 of BRD4 was determined with BROMOscan™ by DiscoverX.
| BD1 | BD2 | |
|---|---|---|
| ARV-825 | 90 nM | 28 nM |
| JQ1 | 12 nM | 10 nM |
| OTX015 | 14 nM | 3.5 nM |
Next, we showed that treatment of BL cell lines with ARV-825 results in almost complete BRD4 protein degradation, with DC50 (50% of maximum degradation) below 1 nM (Figure 2B). Given that BRD4 and cereblon binding moieties in ARV-825 have Kds of 28–90 nM and ~3 µM to their respective targets (Table 1) (Lopez-Girona et al., 2012), this suggests that ARV-825 acts in a substoichiometric way in mediating BRD4 degradation. Interestingly, we observed a bell-shaped dose-dependence of BRD4 degradation by ARV-825 treatment, with some BRD4 protein remaining at the high concentration of 1 µM (Figure 2B). This phenomenon argues for a “BRD4/ARV-825/cereblon” trimer complex as the active species in driving efficient BRD4 degradation. High concentrations of ARV-825 would be predicted to result in the formation of non-functional “BRD4/ARV-825” and “ARV-825/cereblon” dimers that compete with formation of the active trimer, resulting in lower BRD4 degradation (Supp figure 2). The BRD4 degradation induced by ARV-825 occurs rapidly, resulting in more than 50% of protein being lost within 2 hours of compound treatment (Figure 2B).
To confirm that BRD4 degradation induced by ARV-825 is mediated by cereblon, we treated BL cells with ARV-825 in presence of an excess of the cereblon ligand pomalidomide. As expected, the excess pomalidomide was able to reduce ARV-825 induced BRD4 protein degradation, confirming a cereblon-mediated mechanism. Pomalidomide treatment by itself in the same experiment had no impact on BRD4 protein levels (Figure 2C). Furthermore, co-treatment with a proteasome inhibitor (MG132 or carfilzomib) completely blocked the BRD4 degradation induced by ARV-825 (Figure 2D). Taken together, these data demonstrate that ARV-825 leads to fast and efficient BRD4 degradation in a cereblon-mediated and proteasome-dependent mechanism.
ARV-825 leads to more significant and longer lasting c-MYC suppression than small molecule inhibitors
Next, we performed a head-to-head comparison between ARV-825 and the small molecule inhibitors JQ1 and OTX015, to determine their effects on BRD4 and c-MYC levels. JQ1 or OTX015 treatment consistently leads to robust BRD4 accumulation and inefficient c-MYC suppression (Figure 3A). In contrast, ARV-825 results in significant BRD4 degradation and a more pronounced downregulation of c-MYC when compared to that seen with JQ1 and OTX015 treatment at higher concentrations (Figure 3A). Moreover, the suppression of BRD4 and c-MYC protein levels by ARV-825 is long-lasting (Figure 3B). Namalwa cells were treated with ARV-825, JQ1 or OTX015 for 24 hrs, and then washed with fresh medium three times to remove compounds. c-MYC recovers to the control level 4 hours after the removal of JQ1 and OTX015 (Figure 3B). In contrast, ARV-825 maintains both BRD4 degradation and c-MYC suppression up to 24 hours (Figure 3B). Consistent with c-MYC suppression, its downstream target gene, SLC19A1, is suppressed more substantially with ARV-825 than with JQ1 and OTX015 treatments. Moreover, SLC19A1 recovers to control treatment level at 6 hours post removal of JQ1 and OTX015, while ARV-825 sustains its effect on SLC19A1 repression significantly longer (Figure 3C).
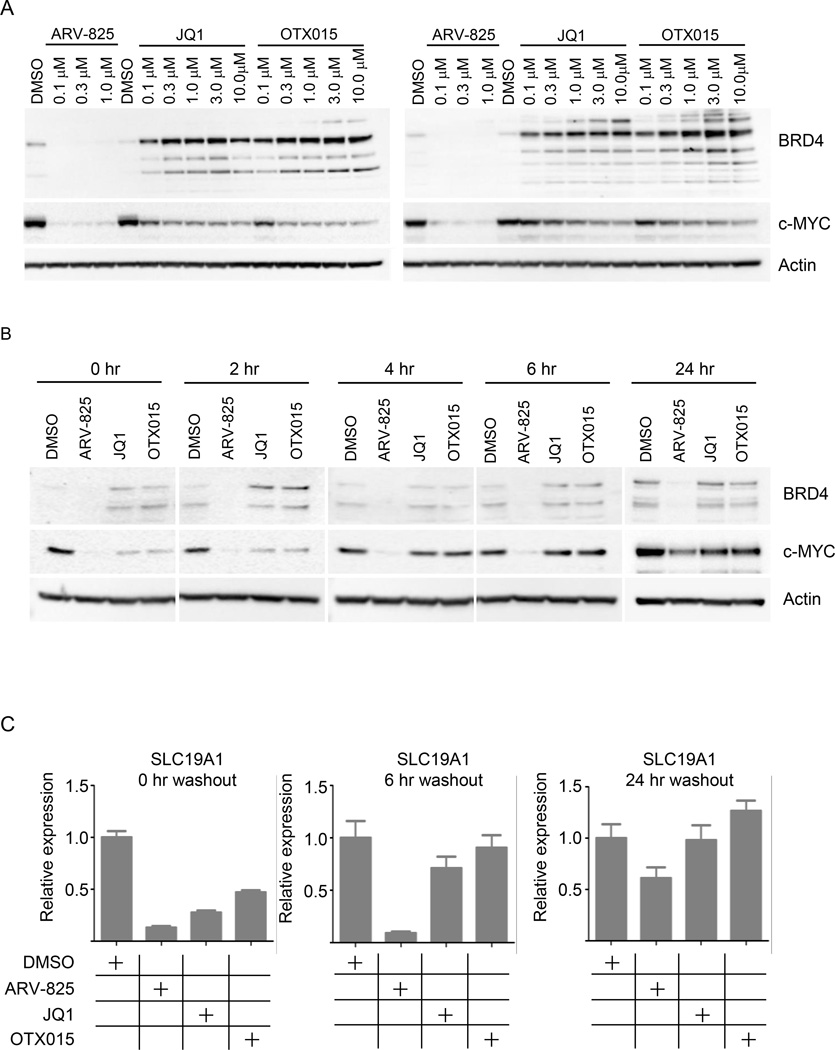
Figure 3. ARV-825 leads to more significant and longer lasting c-MYC suppression than small molecule inhibitors.
(A) Namalwa (left) and Ramos (right) cells were treated overnight with increasing doses of ARV-825 (up to 1.0 µM), or JQ1 (up to 10.0 µM), or OTX015 (up to 10.0 µM); lysates were analyzed by immunoblot for BRD4, c-MYC and actin.
(B) Namalwa cells were treated for 24 hours with ARV-825 (0.1 µM), JQ1 (1.0 µM) and OTX015 (1.0 µM M), followed by three washes to remove compounds, and re-seeded in fresh medium for various time points. Lysates were collected and analyzed by immunoblot for BRD4, c-MYC and actin.
(C) Namalwa cells were treated as in Figure 3B, RNA was extracted at 0, 6 and 24 hours post compounds removal, reverse-transcribed into cDNA and quantified by qPCR with SLC19A1 specific primers; GAPDH serves as internal control.
ARV-825 leads to a superior effect on BL cells proliferation suppression than BRD4 inhibitors
BL cells are known to be sensitive to BRD4 inhibitors, which suppress c-MYC signaling and inhibit cell proliferation (Mertz et al., 2011). Because ARV-825 treatment results in prolonged BRD4 down-regulation and downstream signaling suppression compared to BRD4 inhibitors, we hypothesized that it would provide superior functional effects compared to the inhibitors. Indeed, ARV-825 leads to more pronounced suppression of proliferation than both JQ1 and OTX015 in all BL cell lines tested (Figure 4A). Moreover, the proliferation suppression effect of ARV-825 is sustained longer than that of JQ1 and OTX015 after the removal of compounds following a 24-hr treatment (Figure 4B). This is consistent with our earlier findings that ARV-825 provides long-lasting effect on BRD4 degradation and downstream signaling repression (Figure 3B and 3C). As demonstrated in Figure 2C, presence of excess cereblon ligand, pomalidomide, is able to prevent efficient BRD4 degradation induced by ARV-825 due to competition of cereblon binding. Consistently, the presence of excessive pomalidomide rescued the proliferation suppression effect of ARV-825 in BL cells in a dose-dependent manner (Figure 4C). Importantly, pomalidomide alone didn’t show any significant effects on the proliferation of these cell lines (Figure 4D).
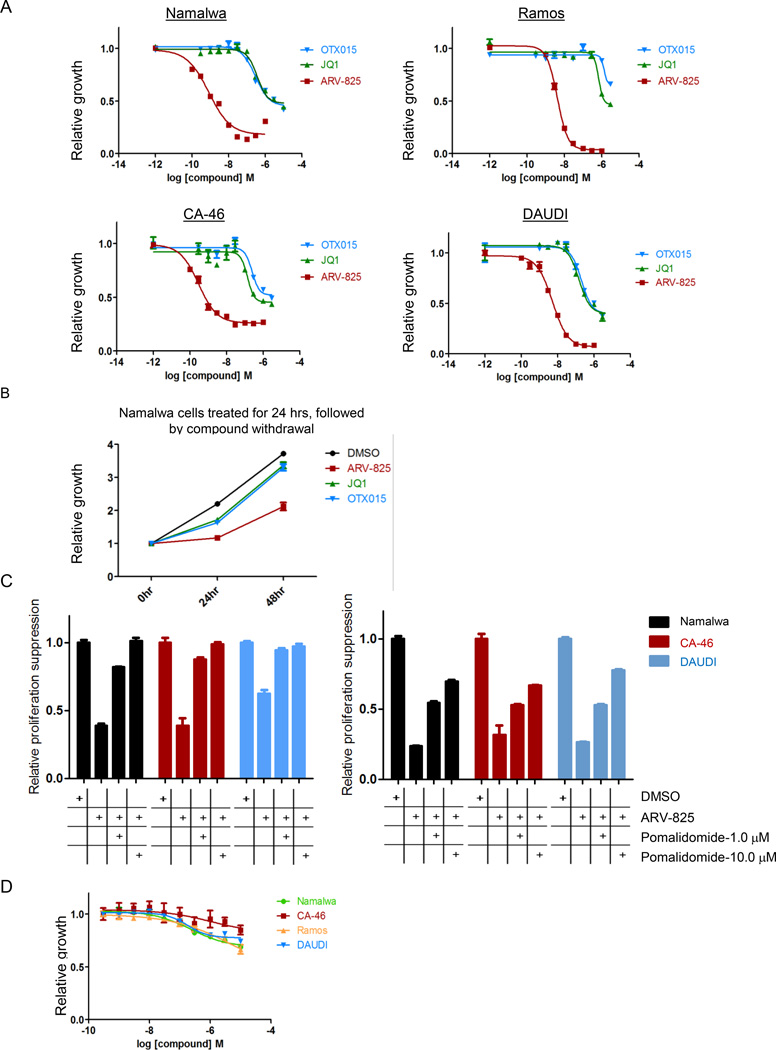
Figure 4. ARV-825 leads to a superior effect on BL cells proliferation suppression than BRD4 inhibitors.
(A) Various BL cell lines were seeded at 50000 cells/100ul in 96-well plates and then treated with increasing doses of ARV-825, JQ1 and OTX015; relative proliferation was determined by CellTiter-Glow (CTG) assay 72 hours later.
(B) ARV-825 leads to longer lasting proliferation suppression than small molecule inhibitors Namalwa cells were treated for 24 hours with ARV-825 (0.1 µM), JQ1 (1.0 µM) and OTX015 (1.0 µM), followed by three washes to remove compounds, and re-seeded in fresh medium in 96-well plate, relative proliferation was determined by CTG assay at 24 hours and 48 hours after re-seeding.
(C) Pomalidomide partially rescued the effect on proliferation suppression by ARV-825 treatment. Different BL cell lines were treated with ARV-825 (0.01 µM on left panel or 0.1 µM on right panel) alone, or together with pomalidomide (1.0 µM or 10.0 µM) for 72 hours, relative cell proliferation was determined by CTG assay.
(D) Pomalidomide does not have significant effect on BL cell proliferation. Different BL cell lines were treated with increasing doses of pomalidomide (up to 10.0 µM) for 72 hours, relative proliferation was determined by CTG assay.
ARV-825 leads to a superior effect on BL cells apoptosis induction than small molecule inhibitors
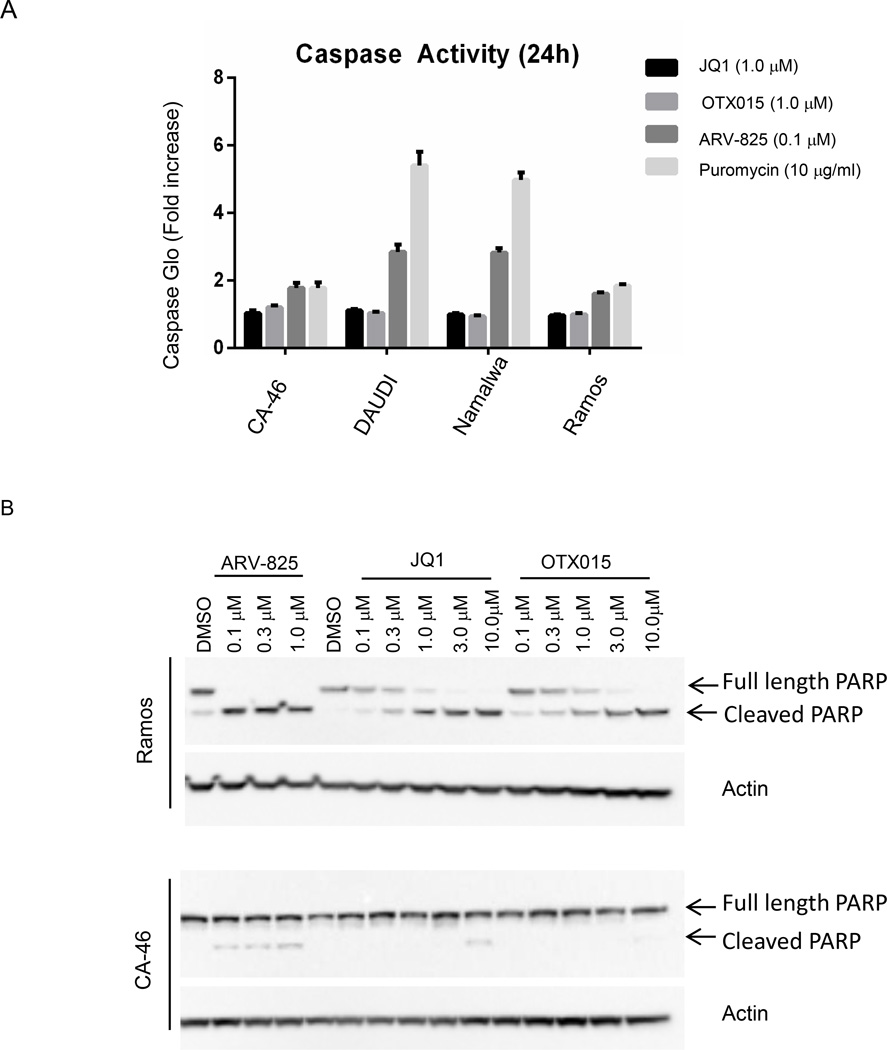
c-MYC is a pleiotropic oncoprotein involved in many hallmarks of cancer, including cell cycle, senescence, proliferation and apoptosis, depending on the tumor (Gabay et al., 2014). We have observed inhibition of proliferation in all BL lines tested following treatment with both BRD4 inhibitors, and ARV-825 (Figure 4). However, the effects on apoptosis are markedly different, depending on the treatment. We observed increased caspases 3/7 activity after a 24 hour treatment of all four BL cell lines with ARV-825, but not with high dose of JQ1 and OTX015 (Figure 5A). By 48 hours, Ramos cells demonstrated significant apoptosis with 0.1 µM ARV-825, as evidenced by prominent PARP cleavage (Figure 5B). In contrast, significantly higher dose of inhibitors, JQ1 and OTX015, are needed to elicit similar level of apoptosis in corresponding cell lines, likely due to their inefficiency of BRD4 inhibition and downstream c-MYC repression. Taken together, these findings strongly corroborate that PROTAC-mediated BRD4 degradation is an effective strategy in targeting BRD4 in BLs.
Figure 5. ARV-825 leads to a superior effect on BL cells apoptosis induction than small molecule inhibitors.
(A) Various BL cell lines were treated with ARV-825 (0.1 µM), or JQ1 (1.0 µM), or OTX015 (1.0 µM), or puromycin (10 mg/ml) as positive control of apoptosis induction, for 24 hours, caspase 3/7 activity was measured by Caspase 3/7- Glow assay.
(B) Ramos and CA-46 cells were treated with increasing doses of ARV-825 (up to 1.0 µM), or JQ1 and OTX015 (up to 10.0 µM) for 48 hours. Lysates were collected and analyzed by immunoblot for PARP cleavage with actin as loading control.
Material and Methods
Reagents
Namalwa, Ramos, CA-46 and DAUDI cells were purchased from ATCC and maintained as instructed. Antibodies against BRD4 (#E2A7X), BRD2 (#5848), c-MYC (#D84C12), PARP (#46D11) were purchased from Cell Signaling Technology. Actin (#A5441) antibody was purchased from SigmaAldrich. BRD3 (#2088c3a) antibody was purchased from Santa Cruz Biotechnology. Secondary antibodies (#7074, #7076) were purchased from Cell Signaling Technology. MG132 (#M7449) was purchased from SigmaAldrich. Carfilzomib (#S2853) was purchased from Selleck. JQ1, OTX015, pomalidomide were synthesized according to methods published.
Western Blot Analysis
Cultured cells were collected in lysis buffer containing 40 mM HEPES (pH 7.4), 140 mM NaCl, 2.5 mM EDTA, 1% NP-40, 0.1% SDS and protease inhibitor cocktail. After 10 minutes of centrifugation (14000 rpm), supernatant was collected for protein concentration determination by BCA method and subjected for immunoblotting by standard protocol. Western blot results were visualized using Bio-Rad Clarity ECL Western Blotting Substrate on Bio-Rad ChemiDoc™ MP imaging system.
RT-QPCR
RNA extraction was performed with Aurum™ Total RNA Mini Kit (#732-6820) from Bio-Rad. First-strand cDNA from total RNA was synthesized with High-Capacity cDNA Reverse Transcription Kit (#4368813) from Life Technologies according to manufacturer’s instruction. Quantitative PCR was performed using Bio-Rad SsoAdvanced™ Universal SYBR® Green Supermix (#172-5271). Primers used are GAPDH-F: GAAGGTGAAGGTCGGAGTC, GAPDH-R: GAAGATGGTGATGGGATTTC, SLC19A1-F: ATGGCCCCCAAGAAGTAGAT, SLC19A1-R: GTCAACACGTTCTTTGCCAC
Proliferation and caspase 3/7 assays
To assess the effect of compounds on proliferation, cells (50000/100ul) were seeded in 96-well tissue culture plates followed by addition of compound at indicated concentration. After 72 hours, 100 µL per well of reconstituted CellTiter-Glo reagent (#G7572 from Promega) was added and read on Cytation 3 imaging reader from BioTek. Relative cell growth was determined by comparing assay readings of treated cells with control DMSO treated cells. For apoptosis induction, Caspase-Glo® 3/7 Assay (#G8091 from Promega) was used following manufacturer’s instruction.
Compound synthesis
The synthetic scheme and detailed experimental procedure for the preparation of ARV-825 is described in supplementary section. The structure of ARV-825 was confirmed by 1H-NMR and LC/MS. Purity of ARV-825 was more than 97% based on HPLC analysis.
Kd determination
Affinity of compounds with Bromodomain 1 and 2 of BRD4 was determined with BROMOscan™ by DiscoverX.
Discussion
BRD4 has captured considerable attention from academia and the pharmaceutical industry alike due to its great potential as a novel target in multiple disease settings, particularly in cancer. One key advantage of BRD4 as an oncology target is that it is identified as preferentially clustering at super-enhancer regions in control of pivotal oncogenes such as c-myc, bcl-xl and pax5 (Chapuy et al., 2013; Loven et al., 2013), and thus offers an alternative strategy in targeting those oncoproteins which are difficult to inhibit by traditional strategies. Moreover, BRD4’s distinct high occupancy of genomic loci proximal to specific oncogenes provides the potential for a therapeutic window that could allow specific targeting of tumor cells while sparing normal tissues. Indeed, BRD4 inhibitors have shown anti-tumor activities with good tolerability in different mouse tumor models (Asangani et al., 2014; Baratta et al., 2015; Boi et al., 2015; Ceribelli et al., 2014; Chapuy et al., 2013; Loven et al., 2013; Mertz et al., 2011; Shimamura et al., 2013; Wyce et al., 2013). And, not surprisingly, high sensitivity to BRD4 inhibitors, such as JQ1, has been associated with high level of either c-MYC or n-MYC in different tumor types, including c-MYC driven BL (Baratta et al., 2015; Loosveld et al., 2014; Mertz et al., 2011; Puissant et al., 2013). Currently, four BET Bromodomain inhibitors are in Phase I clinical trials with focus largely on midline carcinoma and hematological malignancies (CPI-0610, NCT01949883; GSK525762, NCT01587703; OTX015, NCT01713582; TEN-010, NCT01987362). In this report, we found that the BRD4 inhibitors JQ1 and OTX015 lead to fast and robust accumulation of BRD4 protein in all BL cell lines tested. Similar observations have been found in a panel of lung and prostate cancer cell lines (Shimamura et al., 2013). One possible explanation is that the binding of inhibitors to BRD4 results in a conformational change which leads to increased thermodynamic stability of the protein. Similarly, inhibitor binding could hinder BRD4 accessibility to the endogenous cellular degradation machinery, thus rendering it kinetically stable. Alternatively, the BRD4 inhibitors may be interrupting a BRD4-mediated negative feedback loop that regulates BRD4 protein levels. Nevertheless, this prominent increase of BRD4 levels, together with the reversible nature of inhibitor binding, could prevent efficient BRD4 inhibition. Indeed, both preclinical and clinical studies have shown that the effects of BRD4 inhibitors are largely cytostatic, with apoptosis limited to a few cell lines and tumors from phase I patients (Chapuy et al., 2013; Delmore et al., 2011; Shao et al., 2014). This could significantly limit the potential benefit of patients at clinically achievable concentrations of BRD4 inhibitors. One strategy to achieve more effective BRD4 inhibition is to design irreversible/covalent inhibitors, which have revived significant interest in recent years, as they may achieve the desired pharmacological effect at lower drug concentrations (Johnson et al., 2010). However, covalent inhibitors have their own limitations, most notably the potential immunogenicity of protein adduct and high hurdle of selectivity (Johnson et al., 2010).
Here, we designed a novel chimera molecule, ARV-825, using the PROTAC platform to efficiently degrade BRD4, as an alternative strategy of targeting BRD4. In the process, we also demonstrated for the first time the incorporation of the E3 ligase cereblon into the PROTAC technology paradigm (Fischer et al., 2014). We successfully achieved rapid and prominent BRD4 degradation by ARV-825, which leads to robust and long-lasting downstream c-MYC suppression. Most importantly, ARV-825 results in more significant proliferation suppression, and robust apoptosis induction, than even high concentrations of both JQ1 and OTX015. The improved functional effects of BRD4 degrader over inhibitors could be partially attributed to the more complete and sustained suppression on c-MYC, a driver oncoprotein in BLs. It is also possible that BRD4 possess scaffolding functions, as it is a large protein with many binding partners, many of which remain to be identified and elucidated. Understandably, eliminating BRD4 would elicit a more profound effect than mere inhibition of its binding to acetyl-lysine containing partners. A comparison of phenotypes of BRD4 knockout or knockdown (such as by CRISPR or shRNAs) with that of BRD4 inhibition by inhibitors would help inform this possibility. Such an analysis, however, is beyond the scope of this report.
The more complete and longer-lasting disruption of BRD4 function by PROTACs or other means, on the one hand, will benefit future patients because of the more effective targeting and less frequent dosing regimen. On the other hand, it could also associate with more severe side effects as BRD4 regulates diverse biological process in a cellular context dependent manner (Bolden et al., 2014; Fernandez et al., 2014). Recent findings using an inducible and reversible transgenic BRD4 targeting RNAi mouse model have elegantly shed some light on the potential limitations of potent and sustained BRD4 targeting, including epidermal hyperplasia, alopecia, and reduced cellular diversity and stem cell loss in the small intestine (Bolden et al., 2014). This represents a dilemma commonly seen in drug development, namely, determining whether and how an optimal therapeutic index can be achieved in the clinic. Importantly, these on-target side effects of Brd4 suppression appear to be reversible, giving hope that there will be a therapeutic window through better understanding and managing BRD4 targeted therapeutics (Bolden et al., 2014).
Due to the high homology of BET family BD1 and BD2 domains, current BET inhibitors in clinical development, including OTX015, are not selective in binding and inhibiting BRD2/3/4/T (Filippakopoulos et al., 2010; Noel et al., 2014). We also examined the effect of ARV-825 on BRD2 and BRD3 (BRDT is only expressed in testis). Not surprisingly, both BRD2 and BRD3, like BRD4, can be degraded by ARV-825 (Supp Figure 3) owing to the similar affinity of OTX015 to all BET family members, and a large number of overlapping surface lysines on all BET members. However, PROTACs can provide more opportunities of achieving better selectivity through the careful design of the “linker” region that could increase/decrease affinities to various binding targets, or could position its E3 ligase moiety for optimal surface lysine presentation of specific targets. These hypotheses need extensive investigation both through modeling and experimental testing. If true, PROTACs, as a new class of drug molecule, could greatly benefit future patients across multiple disease areas.
Here we report the first success PROTAC that acts via hijacking the E3 ligase cereblon by utilizing a clinically approved IMiD pomalidomide. To further confirm the degradation mechanism of ARV-825 is through cereblon, we prepared a direct analog of ARV-825 by methylating the NH group in the piperidione fragment. Since this NH group is crucial in the binding to cereblon His380 (PDB 4CI3), N-methylation is predicted to cause significant loss of cereblon binding affinity. As expected, N-methylated ARV-825 does not degrade BRD4 (Supp Figure 4), Cereblon is a ubiquitously expressed protein, thus this approach can potentially apply to multiple disease settings in various organs/tissues when different target protein recruiting ligands are attached. Downregulation of cereblon has been proposed as a mechanism of resistance to IMiDs in multiple myeloma (Lopez-Girona et al., 2012). In situations like this, alternative E3 ligases with known binding molecule could be utilized in designing PROTAC therapies.
Binding affinity of OTX015 and pomalidomide to their respective target, BRD4 and cereblon, are ~10 nM and ~3 µM, respectively. Excitingly, ARV-825, which is based on these two ligands, achieves a DC50 for BRD4 below 1 nM. This strongly suggests that a BRD4 PROTAC is able to engage in multiple cycles of protein degradation and opens up opportunities to develop functional degraders consisting of target ligands with insufficient affinity to provide a functional effect. Therefore, many targets, which lack enzymatic active sites, but can be bound by small molecules in shallow pockets, may become “druggable” with PROTAC-mediated degradation.
In summary, we report a novel strategy to efficiently target BRD4 by creating a potent BRD4 degrader through PROTAC technology utilizing the E3 ligase cereblon. This breaks ground for a new class of drug molecules which actively recruits an E3 ligase to target specific pathological proteins for degradation, thus rendering many drug targets refractory to traditional small molecule approaches now “druggable”.
Supplementary Material
Significance.
In this study, we designed a novel bifunctional PROTAC molecule, which contains a BRD4 recruiting moiety and an E3 Ligase cereblon recruiting moiety. Our BRD4 PROTAC actively degrades BRD4, leading to significant and persistent downstream c-MYC suppression and, most importantly, resulting in robust proliferation inhibition and apoptosis induction in BLs. BRD4 PROTAC represents a new strategy to efficiently block BRD4, which is a promising target in multiple cancers. Moreover, this study serves an example that PROTAC-mediated protein degradation provides a promising strategy to target “undruggable” pathological proteins.
Highlights.
-
-
First cereblon-based BRD4 PROTAC was successfully developed;
-
-
BRD4 PROTAC achieved rapid and potent degradation of BRD4;
-
-
It provides a novel and effective strategy to target BRD4;
Acknowledgments
This research was partially supported by the NIH AI084140. We also thank Drs. Lianghe Mei and Luyan Zhang of Suzhou Medinoah Co., Ltd. for their assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: J.L and K.C designed the experiments; J.L, M.A, K.R, J.H performed the experiments; YM.Q, HQ.D, J.W and A.P.C designed and modeled compounds; J.L, K.C, J.D.W, A.P.C and C.M.C. analyzed the data; J.L wrote the manuscript; K.R, YM.Q, K.C, J.D.W, A.P.C and C.M.C. edited the manuscript.
References
- Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, Escara-Wilke J, Wilder-Romans K, Dhanireddy S, Engelke C, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510:278–282. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baratta MG, Schinzel AC, Zwang Y, Bandopadhayay P, Bowman-Colin C, Kutt J, Curtis J, Piao H, Wong LC, Kung AL, et al. An in-tumor genetic screen reveals that the BET bromodomain protein, BRD4, is a potential therapeutic target in ovarian carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:232–237. doi: 10.1073/pnas.1422165112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkina AC, Denis GV. BET domain co-regulators in obesity, inflammation and cancer. Nature reviews Cancer. 2012;12:465–477. doi: 10.1038/nrc3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boi M, Gaudio E, Bonetti P, Kwee I, Bernasconi E, Tarantelli C, Rinaldi A, Testoni M, Cascione L, Ponzoni M, et al. The BET Bromodomain inhibitor OTX015 affects pathogenetic pathways in pre-clinical B-cell tumor models and synergizes with targeted drugs. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015 doi: 10.1158/1078-0432.CCR-14-1561. [DOI] [PubMed] [Google Scholar]
- Bolden JE, Tasdemir N, Dow LE, van Es JH, Wilkinson JE, Zhao Z, Clevers H, Lowe SW. Inducible in vivo silencing of Brd4 identifies potential toxicities of sustained BET protein inhibition. Cell Rep. 2014;8:1919–1929. doi: 10.1016/j.celrep.2014.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley DL, Crews CM. Small-molecule control of intracellular protein levels through modulation of the ubiquitin proteasome system. Angewandte Chemie. 2014;53:2312–2330. doi: 10.1002/anie.201307761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett JC, Rossi JJ, Tiemann K. Current progress of siRNA/shRNA therapeutics in clinical trials. Biotechnology journal. 2011;6:1130–1146. doi: 10.1002/biot.201100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceribelli M, Kelly PN, Shaffer AL, Wright GW, Xiao W, Yang Y, Mathews Griner LA, Guha R, Shinn P, Keller JM, et al. Blockade of oncogenic IkappaB kinase activity in diffuse large B-cell lymphoma by bromodomain and extraterminal domain protein inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:11365–11370. doi: 10.1073/pnas.1411701111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MG, Qi J, Rahl PB, Sun HH, Yeda KT, Doench JG, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer cell. 2013;24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez P, Scaffidi P, Markert E, Lee JH, Rane S, Misteli T. Transformation resistance in a premature aging disorder identifies a tumor-protective function of BRD4. Cell Rep. 2014;9:248–260. doi: 10.1016/j.celrep.2014.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer ES, Bohm K, Lydeard JR, Yang H, Stadler MB, Cavadini S, Nagel J, Serluca F, Acker V, Lingaraju GM, et al. Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. Nature. 2014;512:49–53. doi: 10.1038/nature13527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French CA, Ramirez CL, Kolmakova J, Hickman TT, Cameron MJ, Thyne ME, Kutok JL, Toretsky JA, Tadavarthy AK, Kees UR, et al. BRD-NUT oncoproteins: a family of closely related nuclear proteins that block epithelial differentiation and maintain the growth of carcinoma cells. Oncogene. 2008;27:2237–2242. doi: 10.1038/sj.onc.1210852. [DOI] [PubMed] [Google Scholar]
- Gabay M, Li Y, Felsher DW. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harbor perspectives in medicine. 2014;4 doi: 10.1101/cshperspect.a014241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelder S, Clarke PA, Workman P. Discovery of small molecule cancer drugs: successes, challenges and opportunities. Molecular oncology. 2012;6:155–176. doi: 10.1016/j.molonc.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell SJ, Johnston SR, Howell A. The use of selective estrogen receptor modulators and selective estrogen receptor down-regulators in breast cancer. Best practice & research Clinical endocrinology & metabolism. 2004;18:47–66. doi: 10.1016/j.beem.2003.08.002. [DOI] [PubMed] [Google Scholar]
- Ito T, Ando H, Suzuki T, Ogura T, Hotta K, Imamura Y, Yamaguchi Y, Handa H. Identification of a primary target of thalidomide teratogenicity. Science. 2010;327:1345–1350. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- Johnson DS, Weerapana E, Cravatt BF. Strategies for discovering and derisking covalent, irreversible enzyme inhibitors. Future medicinal chemistry. 2010;2:949–964. doi: 10.4155/fmc.10.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapproth K, Wirth T. Advances in the understanding of MYC-induced lymphomagenesis. British journal of haematology. 2010;149:484–497. doi: 10.1111/j.1365-2141.2010.08159.x. [DOI] [PubMed] [Google Scholar]
- Loosveld M, Castellano R, Gon S, Goubard A, Crouzet T, Pouyet L, Prebet T, Vey N, Nadel B, Collette Y, et al. Therapeutic targeting of c-Myc in T-cell acute lymphoblastic leukemia, T-ALL. Oncotarget. 2014;5:3168–3172. doi: 10.18632/oncotarget.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Girona A, Mendy D, Ito T, Miller K, Gandhi AK, Kang J, Karasawa S, Carmel G, Jackson P, Abbasian M, et al. Cereblon is a direct protein target for immunomodulatory and antiproliferative activities of lenalidomide and pomalidomide. Leukemia. 2012;26:2326–2335. doi: 10.1038/leu.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G, Middleton RE, Sun H, Naniong M, Ott CJ, Mitsiades CS, Wong KK, Bradner JE, Kaelin WG., Jr The myeloma drug lenalidomide promotes the cereblondependent destruction of Ikaros proteins. Science. 2014;343:305–309. doi: 10.1126/science.1244917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, Bergeron L, Sims RJ., 3rd Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel J, Iwata K, Ooike S, Sugahara K, Nakamura H, Daibata M. Development of the BET bromodomain inhibitor OTX015. Molec Can Therapeutics. 2014;12:C244-C [Google Scholar]
- Puissant A, Frumm SM, Alexe G, Bassil CF, Qi J, Chanthery YH, Nekritz EA, Zeid R, Gustafson WC, Greninger P, et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer discovery. 2013;3:308–323. doi: 10.1158/2159-8290.CD-12-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina K, Crews CM. Chemical inducers of targeted protein degradation. The Journal of biological chemistry. 2010;285:11057–11060. doi: 10.1074/jbc.R109.078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto KM, Kim KB, Kumagai A, Mercurio F, Crews CM, Deshaies RJ. Protacs: chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:8554–8559. doi: 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto KM, Kim KB, Verma R, Ransick A, Stein B, Crews CM, Deshaies RJ. Development of Protacs to target cancer-promoting proteins for ubiquitination and degradation. Mol Cell Proteomics. 2003;2:1350–1358. doi: 10.1074/mcp.T300009-MCP200. [DOI] [PubMed] [Google Scholar]
- Schneekloth JS, Jr, Crews CM. Chemical approaches to controlling intracellular protein degradation. Chembiochem : a European journal of chemical biology. 2005;6:40–46. doi: 10.1002/cbic.200400274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneekloth JS, Jr, Fonseca FN, Koldobskiy M, Mandal A, Deshaies R, Sakamoto K, Crews CM. Chemical genetic control of protein levels: selective in vivo targeted degradation. Journal of the American Chemical Society. 2004;126:3748–3754. doi: 10.1021/ja039025z. [DOI] [PubMed] [Google Scholar]
- Shao Q, Kannan A, Lin Z, Stack BC, Jr, Suen JY, Gao L. BET protein inhibitor JQ1 attenuates Myc-amplified MCC tumor growth in vivo. Cancer research. 2014;74:7090–7102. doi: 10.1158/0008-5472.CAN-14-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Molecular cell. 2014;54:728–736. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamura T, Chen Z, Soucheray M, Carretero J, Kikuchi E, Tchaicha JH, Gao Y, Cheng KA, Cohoon TJ, Qi J, et al. Efficacy of BET bromodomain inhibition in Kras-mutant non-small cell lung cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:6183–6192. doi: 10.1158/1078-0432.CCR-12-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyce A, Degenhardt Y, Bai Y, Le B, Korenchuk S, Crouthame MC, McHugh CF, Vessella R, Creasy CL, Tummino PJ, et al. Inhibition of BET bromodomain proteins as a therapeutic approach in prostate cancer. Oncotarget. 2013;4:2419–2429. doi: 10.18632/oncotarget.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.