Abstract
Purpose
To evaluate whether annual updating of the Prostate Cancer Prevention Trial Risk Calculator (PCPTRC) would improve institutional validation over static use of the PCPTRC alone.
Materials and Methods
Data from five international cohorts, SABOR, Cleveland Clinic, ProtecT, Tyrol, and Durham VA, comprising n = 18,400 biopsies were used to evaluate an institution-specific annual recalibration of the PCPTRC. Using all prior years as a training set and the current year as the test set, the annual recalibrations of the PCPTRC were compared to static use of the PCPTRC in terms of areas underneath the receiver operating characteristic curves (AUC) and Hosmer-Lemeshow goodness-of-fit statistic (H-L).
Results
For predicting high-grade disease, the median AUC (higher is better) of the recalibrated PCPTRC (static PCPTRC) across all test years for the five cohorts were 67.3 (67.5), 65.0 (60.4), 73.4 (73.4), 73.9 (74.1), 69.6 (67.2), respectively, and the median H-L statistics indicated better fit for recalibration compared to the static PCPTRC for Cleveland Clinic, ProtecT and the Durham VA, but not for SABOR and Tyrol. For predicting overall cancer, median AUCs were 63.5 (62.7), 61.0 (57.3), 62.1 (62.5), 66.9 (67.3), and 68.5 (65.5), respectively, and the median H-L statistics indicated better fit for recalibration on all cohorts except for Tyrol.
Conclusions
A simple to implement method has been provided to tailor the PCPTRC to individual hospitals in order to optimize its accuracy for the patient population at hand.
Keywords: Risk calculator, prostate biopsy, prostate-specific antigen, digital rectal examination, family history
INTRODUCTION
The Prostate Cancer Prevention Trial Risk Calculator (PCPTRC) was originally posted online in 2006 and updated in 2014 to predict low- (Gleason grade < 7) versus high-grade disease.1,2 The calculator predicts the likelihood of detecting prostate cancer if a prostate biopsy were to be performed, based on commonly collected clinical risk factors: prostate-specific antigen (PSA), digital rectal exam (DRE), age, race, family history of prostate cancer, and prior biopsy history. The calculator was built using a cohort of patients participating in the Prostate Cancer Prevention Trial (PCPT) during the late 1990s to early 2000s.3 Since this time, a variety of changes in clinical practice, diagnostic techniques, and pathologist grading patterns have occurred. While a 6-core biopsy technique was used in the PCPT, contemporary clinical practice has switched to 10–12 cores which has increased detection of low- and high-grade cancer.4 United States SEER analyses from 2004 to 2010 have revealed an upward shift in the diagnosis of Gleason grade 7 and higher tumors.5 These and more recent modifications in prostate cancer screening and biopsy such as the increasing use of MRI-directed biopsy will require a move away from a static risk prediction tool for outcome on prostate biopsy towards a dynamic one.
Concurrent with these clinical changes has come the recognition that institutions across the United States and Europe vary a great deal in the type of patient referred for biopsy. Analyses of ten international prostate biopsy cohorts from participating institutions of the Prostate Cancer Biopsy Collaborative Group (PBCG) revealed large variations in estimated risk according to prostate-specific antigen, and areas-underneath-the-receiver-operating-characteristic curves (AUC) ranging close to 20 percentage points (from 56.2% to 72.0%) across cohorts.6,7 These statistics further argue against use of a static risk assessment tool for all institutions.
The need for a biopsy risk calculator to evolve dynamically over time in response to a changing clinical landscape as well as to be tailored to disparate patient populations have led us to derive a new method for updating the PCPTRC 2.0 based on standard statistical methods that can be implemented locally. The method is illustrated by application to yearly biopsy data from five international cohorts participating in the PBCG.
MATERIALS AND METHODS
2.1 Participants and biopsy results
Annual biopsies from five cohorts from the PBCG were used, which have been previously described.6 Three screening cohorts targeting healthy men, the San Antonio Biomarker of Risk of cancer study (SABOR), Texas, U.S., ProtecT, UK, and Tyrol, Austria followed primarily a 10-core biopsy scheme. Two clinical cohorts from the U.S., Cleveland Clinic, Ohio and the Durham VA, North Carolina, included patients evaluated in clinical practices of Urology. The biopsy scheme used in these cohorts was mixed, but was primarily 10- to 14-cores. For cohorts where the beginning or ending years had few biopsies, aggregation into the first and last years was performed. Biopsies with PSA values greater than 50 ng/mL were excluded from the analysis in order to match online use of the PCPTRC; in 51 cases Gleason grade was missing, these observations were deleted from the analysis.
2.2 Statistical methods
Two methods—the static PCPTRC and annual recalibration of the PCPTRC—were compared on each of the five institutions separately. For recalibration of the PCPTRC, starting with year 2, all prior years were used as a training set to recalibrate the PCPTRC model (to be described). The two methods were then compared using the current year as test set in terms of the AUC and Hosmer-Lemeshow goodness-of-fit-statistic (H-L) applied to the endpoints of predicting high-grade prostate cancer and any prostate cancer separately. Graphs displaying the AUC and H-L statistics according to year for the two methods were used for comparison; yearly 95% confidence intervals (CI) are additionally displayed in the Supplementary Appendix.
Recalibration of the PCPTRC was conducted by extending the principles by Steyerberg for logistic regression to multinomial logistic regression for the three outcomes on biopsy provided by PCPTRC 2.0: no cancer, low-grade and high-grade cancer.8 The PCPTRC is available online at myprostatecancerrisk.com and accommodates missing values of DRE, family history or prior biopsy history. It was extended herein to accommodate missing race. The recalibration method calculated the PCPTRC relative risks (RRs) of high-grade cancer (versus no cancer as reference) and low-grade cancer (versus no cancer) for each participant in the training set and used the logarithm of these two risks as covariates in a multinomial logistic regression in the training set. The fitted multinomial logistic regression included 6 parameters: an intercept, a slope for the PCPTRC high-risk estimate, and a slope for the PCPTRC low-risk estimate, for the RRs for high- and low-grade prostate cancer relative to no cancer. The PCPTRC model was effectively not re-calibrated when the following conditions held: both intercepts are zero, the high-grade RR slope is 1 for the PCPTRC high-grade RR and 0 for the PCPTRC low-grade RR, and the reverse for the low-grade RR. Otherwise the model was re-calibrated to be tailored to the training set. The static PCPTRC method did not require training, but rather used the online PCPTRC for all test years.
RESULTS
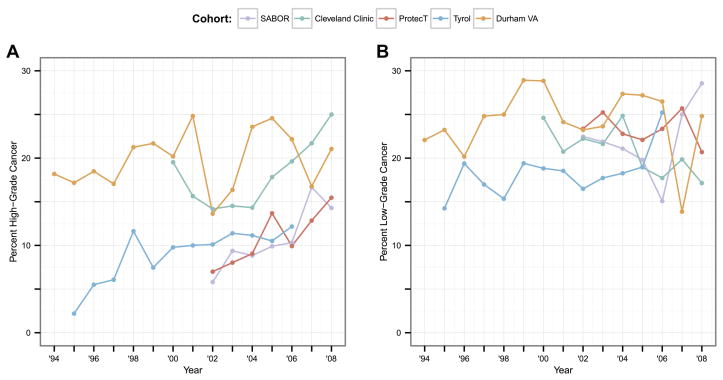
The five PBCG cohorts ranged in size from 900 biopsies in SABOR to 7260 in ProtecT (Table 1). Four of the cohorts included more than one biopsy per patient, which was accounted for with previous biopsy as a risk factor. As expected, two clinically referred cohorts, Cleveland Clinic and Durham VA, had the highest cancer rates, 39% and 44%, respectively, compared to all other primarily screening cohorts, with rates from 28% to 35%. The remaining risk factors, including DRE, family history, African American origin, and prior biopsy history, were either not collected for some of the cohorts or missing. Of note was the very high representation of African Americans (44%) at the Durham VA clinic in comparison with the others. Collection of the cohorts spanned the years 1994 to 2010, with the number of biopsies performed per year ranging from 77 to 1272 (Supplementary Appendix Table 1). Rates of high-grade cancer increased in general over the years of observation for most cohorts; these increases were not monotonic with wide swings in rates over the period in some cohorts (left side of Figure 1). In contrast to high-grade cancers, rates of low-grade cancer were somewhat more constant over the observation periods (right side of Figure 1).
Table 1.
Biopsy characteristics from the 5 PBCG cohorts.
| SABOR | Cleveland Clinic | ProtecT | Tyrol | Durham VA | |
|---|---|---|---|---|---|
| Number of patients | 722 | 2603 | 7260 | 3628 | 1674 |
| Number of biopsies | 900 | 3257 | 7260 | 4788 | 2195 |
| Years of observation | 2001–2010 | 2000–2008 | 2001–2008 | 1995–2007 | 1994–2009 |
| Age median (range) | 63.9 (36, 89) | 64.0 (50, 75) | 62.6 (50, 72) | 62.0 (50, 75) | 63.9 (50, 75) |
| PSA median (range) | 3.3 (0.1, 49.8) | 5.7 (0.2, 49.9) | 4.3 (3.0, 49.7) | 4.0 (0.2, 49.6) | 5.0 (0.1, 49.5) |
| < 3 ng/ml | 385 (43%) | 337 (10%) | 0 (0%) | 1519 (32%) | 288 (13%) |
| ≥ 3 ng/ml | 515 (57%) | 2920 (90%) | 7260 (100%) | 3269 (68%) | 1907 (87%) |
| DRE result | |||||
| Normal | 603 (67%) | 3057 (94%) | 0 (0%) | 4425 (92%) | 876 (40%) |
| Abnormal | 234 (26%) | 200 (6%) | 0 (0%) | 363 (8%) | 251 (11%) |
| Unknown | 63 (7%) | 0 (0%) | 7260 (100%) | 0 (0%) | 1068 (49%) |
| Family history | |||||
| No | 246 (27%) | 1679 (52%) | 5692 (78%) | 0 (0%) | 0 (0%) |
| Yes | 295 (33%) | 371 (11%) | 453 (6%) | 0 (0%) | 0 (0%) |
| Unknown | 359 (40%) | 1207 (37%) | 1115 (15%) | 4788 (100%) | 2195 (100%) |
| African origin | |||||
| No | 796 (88%) | 2799 (86%) | 6878 (95%) | 0 (0%) | 1114 (51%) |
| Yes | 104 (12%) | 412 (13%) | 31 (0%) | 0 (0%) | 969 (44%) |
| Unknown | 0 (0%) | 46 (1%) | 351 (5%) | 4788 (100%) | 112 (5%) |
| Prior biopsy | |||||
| Yes | 306 (34%) | 1089 (33%) | 0 (0%) | 1434 (30%) | 550 (25%) |
| No | 594 (66%) | 2168 (67%) | 7260 (100%) | 3354 (70%) | 1645 (75%) |
| Cancer | 287 (32%) | 1265 (39%) | 2507 (35%) | 1320 (28%) | 973 (44%) |
| Biopsy Gleason grade | |||||
| < 7 | 193 (67%) | 668 (53%) | 1695 (68%) | 860 (65%) | 529 (54%) |
| ≥ 7 | 92 (32%) | 597 (47%) | 812 (32%) | 421 (32%) | 434 (45%) |
| Unknown | 2 (1%) | 0 (0%) | 0 (0%) | 39 (3%) | 10 (1%) |
Figure 1.
For the 5 PBCG cohorts, yearly rates of a.) high-grade and b.) low-grade cancer.
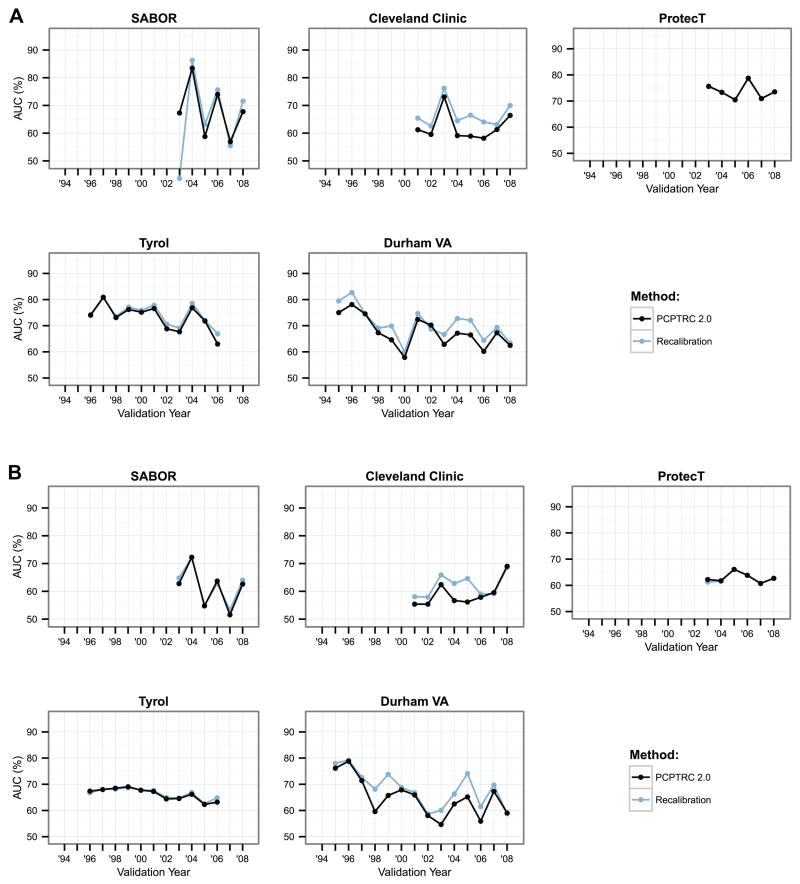
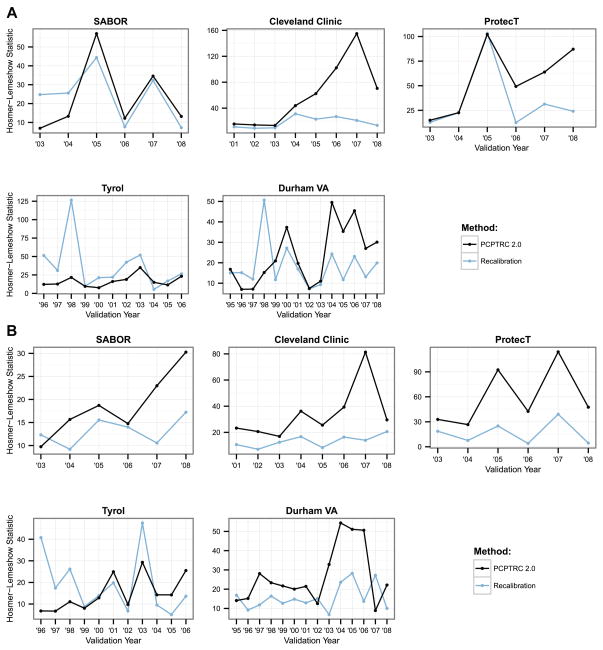
Annual AUCs for both high-grade and overall cancer detection were only slightly higher (better) for recalibration of the PCPTRC for most years across the cohorts, and the differences were likely statistically insignificant (Figure 2, Table 2). The AUC measures the ability of a risk prediction tool to discriminate a random case from a control and the lack of difference seen may be more indicative of the robustness of the measure than lack of effect of recalibration. The H-L statistics shown in Figure 3 measure the closeness of predicted risks to observed risks with smaller values indicating closer agreement; see Supplementary Appendix Figure 1 for confidence intervals. These indicate better accuracy for recalibrating the PCPTRC tool in most cases (Table 2; Supplementary Appendix Figure 1). The recalibration method is sensitive to a sudden spike in the outcome as can be seen by the sudden uptick in high-grade cases observed in Tyrol in 1998 (Figure 1). This results in poor calibration of the recalibrated model that year since it was trained with the lower prevalence of high-grade disease observed in Tyrol up through 1997 (Figure 3a).
Figure 2.
AUCs using all past data as training and the next year as the validation year (x-axis) for discriminating a.) high-grade versus low-grade or no cancer, and b.) overall (high- and low-grade cancer) versus no cancer. Higher values indicate better discrimination of the risk tool.
Table 2.
Median, minimum and maximum AUCs and Hosmer-Lemeshow (H-L) test-statistics across all test years for each cohort for predicting high-grade cancer (top) and overall cancer (bottom).
| Method | AUC | Hosmer-Lemeshow | |
|---|---|---|---|
| SABOR | PCPTRC 2.0 | 67.5 (56.9, 83.4) | 13.2 (6.9, 57.1) |
| Recalibration | 67.3 (43.7, 86.3) | 25.2 (7.2, 44.4) | |
|
| |||
| Cleveland Clinic | PCPTRC 2.0 | 60.4 (58.2, 73.1) | 53.2 (13.4, 155.0) |
| Recalibration | 65.0 (62.6, 76.2) | 17.3 (9.0, 31.3) | |
|
| |||
| ProtecT | PCPTRC 2.0 | 73.4 (70.5, 78.8) | 56.6 (15.0, 102.2) |
| Recalibration | 73.4 (70.5, 78.6) | 23.4 (12.6, 103.3) | |
|
| |||
| Tyrol | PCPTRC 2.0 | 74.1 (63.0, 80.9) | 15.0 (7.7, 35.0) |
| Recalibration | 73.9 (66.9, 80.7) | 26.5 (5.5, 126.6) | |
|
| |||
| Durham VA | PCPTRC 2.0 | 67.2 (57.9, 78.1) | 20.3 (7.0, 49.5) |
| Recalibration | 69.6 (60.0, 82.7) | 15.1 (7.2, 50.7) | |
| Method | AUC | Hosmer-Lemeshow | |
|---|---|---|---|
| SABOR | PCPTRC 2.0 | 62.7 (51.5, 72.3) | 17.2 (9.8, 30.3) |
| Recalibration | 63.5 (53.3, 72.2) | 13.2 (9.2, 17.2) | |
|
| |||
| Cleveland Clinic | PCPTRC 2.0 | 57.3 (55.4, 69.0) | 27.6 (16.9, 81.3) |
| Recalibration | 61.0 (57.9, 68.6) | 13.2 (7.1, 20.6) | |
|
| |||
| ProtecT | PCPTRC 2.0 | 62.5 (60.7, 66.1) | 45.1 (26.6, 114.1) |
| Recalibration | 62.1 (60.9, 66.1) | 13.1 (4.0, 39.1) | |
|
| |||
| Tyrol | PCPTRC 2.0 | 67.3 (62.3, 69.0) | 12.9 (6.7, 29.3) |
| Recalibration | 66.9 (62.6, 68.7) | 13.9 (5.1, 47.5) | |
|
| |||
| Durham VA | PCPTRC 2.0 | 65.5 (54.7, 78.8) | 21.9 (8.9, 54.4) |
| Recalibration | 68.5 (58.7, 79.2) | 14.2 (6.8, 28.3) | |
Figure 3.
Hosmer-Lemeshow test statistics using all past data as training and the next year as the validation year (x-axis) for predicting a.) high-grade versus low-grade or no cancer, and b.) overall (high- and low-grade cancer) versus no cancer. Lower values indicate better fit, closer agreement between observed and predicted risks.
Of note for both the AUC and H-L statistics for both the recalibrated and static PCPTRC is that within each of the cohorts, they did not improve over time, i.e., the accumulation of more data within the cohort did not improve either discrimination or calibration. This reflects the limit of the predictive power of the established risk factors for prostate cancer and high-grade disease, and the continued need for new markers to significantly improve early detection.
DISCUSSION
Until quite recently, the concept of static risk calculators built on accumulated clinical data has prevailed. In 2013, when new American guidelines based statin reimbursement on predicted risk of atherosclerotic events from a risk calculator built on a cohort that was several decades old, it was shown by validation on a contemporary US population that the outdated prediction model, still widely used in the US, would greatly over-estimate the risk of a cardiac event with the result of over-medicating the public.9 The past five years have witnessed the updating of commonly used risk tools or tailoring to a new country in a variety of medical settings.10–18 The same data collection is required for either updating an existing risk tool or building a new one de novo. When the changes required are extreme and the sample size is large enough, building a new model de novo will outperform a process of changing the prior risk tool.
Many of the current online prostate biopsy risk calculators are based on data from cohorts assembled during the late 1990s and early 2000s. PSA screening patterns, biopsy techniques and pathologic grading patterns have changed dramatically during this time. For the purpose of providing a patient with his individual risk of prostate cancer on biopsy, the denominator of interest is not all patients, but rather those seen in that particular clinic. The differences in outcomes of biopsy can be substantially different from institution to institution or region to region. The intensity of PSA screening in the local population and racial differences in screening intensity (e.g., is it more intense in a Caucasian or in an African American population) will substantially affect the risk of patients at risk of biopsy. Similarly, the referral behavior of primary physicians who evaluate patients with a PSA result (e.g., refer for PSA > 2.5 ng/mL, for PSA > 4.0 ng/mL, for increase in PSA at even lower levels, or a hesitancy to refer for any level of PSA), will dramatically affect the risk of patients entering a Urology clinical setting. In the Urology clinic, the sensitivity for DRE is highly-variable, further affecting decision-making.19 Patient factors related to the local institutional environment (socioeconomic status, insurance, native language) may further influence the patient’s desire to undergo biopsy. Finally, the Urology setting (e.g., academic, managed care, etc.) will affect whether the patient receives a biopsy recommendation. Temporal changes such as national guidelines on screening or changes in reimbursement may cause abrupt changes in the types of patients undergoing biopsy. All of these factors argue for an institution-specific tool that can detect these changes in real time to provide the most updated assessment of the patient’s risk of cancer or high-grade cancer on prostate biopsy.
This study has provided evidence of gains in accuracy, though not necessarily discrimination, that could be experienced from updating risk tools commonly used in Urology, not just the PCPTRC, but also the European Randomized Study of Screening for Prostate Cancer (ERSPC) study or the many other nomograms available online. A limitation of the study is that in its comprehensive serial investigation, entailing 44 statistical evaluations accounting for all the individual testing years in five cohorts, it only investigated the most fundamental components of validation: discrimination and accuracy. Calibration curves are a graphical display of the building blocks of the H-L statistic and show at what regions of risk the accuracy most fails (the H-L statistic is an average of the calibration curves). We have inspected calibration curves and found no systematic explanation as to how the updating method improves calibration, e.g., either by improving for high or low risks. Net benefit curves are an alternative metric that measure the advantages of using a risk model to no model at all for deciding when to refer a patient to biopsy. They also require inspection of an entire graph, with results depending on the cutoff used to refer to biopsy, and have not been extensively studied for two outcomes (low and high grade prostate cancer risk) so have not been shown here. Finally, this study has not addressed the optimal time to update, whether it be every year, every two years or more so, but rather just shown the effect of cumulative updating, so that the gains to be made by waiting multiple years can be seen by just moving that far along the graphs. One needs sufficient sample sizes, in particular prostate cancer events, in order to update effectively.
A major issue and limiting factor in developing such an individualized risk assessment tool is cost. To locally implement a recalibration of the PCPTRC would require collection of PCPTRC risk factors and the subsequent biopsy result. These data may come from different departments of the hospital and need careful quality assurance editing, for example, making sure the PSA and DRE results are related temporally to the biopsy, before conducting the multinomial regression of biopsy outcomes on output of the PCPTRC (which could be computed online). With the rapid adoption of multi-institutional compatible Electronic Medical Record (EMR) systems, where data are automatically downloaded to a central database as they are generated, such an updating tool will become more practical. Remaining procedures required for the tailoring the PCPTRC could be automated, including data quality checks that would censor erroneous data.20 Such a system would require start-up costs and minor human monitoring.
Our analysis revealed that dynamically recalibrating the PCPTRC significantly improved its accuracy compared to the static PCPTRC, but not the discrimination. The gain in calibration was realized already at the first year of updating, but then fluctuated without necessarily gaining further ground during the remaining years. This reflects the limited predictive ability of the existing risk factors for predicting low- and high-grade prostate cancer and the need for new disease markers to further enhance discrimination and accuracy.
CONCLUSIONS
A simple-to-implement method has been developed to customize the PCPTRC 2.0 to an individual institution as well as to update the calculator periodically as trends in prostate cancer diagnosis and technology change with time.
Supplementary Material
Supplementary Figure 1. Hosmer-Lemeshow test statistics using all past data as training and the next year as the validation year (x-axis) for predicting a.) high-grade versus low-grade or no cancer, and b.) overall (high- and low-grade cancer) versus no cancer. Lower values indicate better fit, closer agreement between observed and predicted risks. Error bars represent the 95% confidence intervals of the test statistic.
Supplementary Appendix Table 1: Number of biopsies performed by year.
Acknowledgments
Support
This work was sponsored by grants U01CA86402, 5P30 CA0541474-19, UM1 CA182883, P01 CA108964-06, the Prostate Cancer Foundation, the Sidney Kimmel Center for Prostate and Urologic Cancers, P50-CA92629, P30-CA008748 and 1R01CA179115-01A1.
Footnotes
Financial disclosure declaration:
No authors have a financial conflict of interest with this manuscript.
References
- 1.Thompson IM, Ankerst DP, Chi C, et al. Assessing prostate cancer risk: results from the Prostate Cancer Prevention Trial. J Natl Cancer Inst. 2006;98(8):529. doi: 10.1093/jnci/djj131. [DOI] [PubMed] [Google Scholar]
- 2.Ankerst DP, Hoefler J, Bock S, et al. Prostate Cancer Prevention Trial risk calculator 2.0 for the prediction of low- vs. high-grade prostate cancer. Urology. 2014;83(6):1362. doi: 10.1016/j.urology.2014.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thompson IM, Goodman PJ, Tangen CM, et al. The influence of finasteride on the development of prostate cancer. N Engl J Med. 2003;349(3):215. doi: 10.1056/NEJMoa030660. [DOI] [PubMed] [Google Scholar]
- 4.Ankerst DP, Till C, Boeck A, et al. The impact of prostate volume, number of biopsy cores and American Urological Association symptom score on the sensitivity of cancer detection using the Prostate Cancer Prevention Trial risk calculator. J Urol. 2013;190(1):70. doi: 10.1016/j.juro.2012.12.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weiner AB, Patel SG, Etzioni R, et al. National trends in the management of low- and intermediate-risk prostate cancer in the United States. J Urol. 2014 doi: 10.1016/j.juro.2014.07.111. to appear. [DOI] [PubMed] [Google Scholar]
- 6.Vickers AJ, Cronin AM, Roobol MJ, et al. The relationship between prostate-specific antigen and prostate cancer risk: The Prostate Biopsy Collaborative Group. Clin Cancer Res. 2010;16(17):4374. doi: 10.1158/1078-0432.CCR-10-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ankerst DP, Boeck A, Freedland SJ, et al. Evaluating the PCPT risk calculator in ten international biopsy cohorts: results from the Prostate Biopsy Collaborative Group. World J Urol. 2012;30(2):181. doi: 10.1007/s00345-011-0818-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steyerberg EW. Clinical Prediction Models: A Practical Approach to Development, Validation, and Updating. New York: Springer; 2009. [Google Scholar]
- 9.Ridker PM, Cook NR. Statins: new American guidelines for prevention of cardiovascular disease. Lancet. 2013;382(9907):1762. doi: 10.1016/S0140-6736(13)62388-0. [DOI] [PubMed] [Google Scholar]
- 10.Janssen KJ, Moons KG, Kalkman CJ, et al. Updating methods improved the performance of a clinical prediction model in new patients. J Clin Epidemiol. 2008;61(1):76. doi: 10.1016/j.jclinepi.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 11.Schuetz P, Koller M, Christ-Crain M, et al. Predicting mortality with pneumonia severity scores: importance of model recalibration to local settings. Epidemiol Infect. 2008;136(12):1628. doi: 10.1017/S0950268808000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen L, Tonkin AM, Moon L, et al. Recalibration and validation of the SCORE risk chart in the Australian population: the AusSCORE chart. Eur J Cardiovasc Prev Rehabil. 2009;16(5):562. doi: 10.1097/HJR.0b013e32832cd9cb. [DOI] [PubMed] [Google Scholar]
- 13.Genders TS, Steyerberg EW, Alkadhi H, et al. A clinical prediction rule for the diagnosis of coronary artery disease: validation, updating, and extension. Eur Heart J. 2011;32(11):1316. doi: 10.1093/eurheartj/ehr014. [DOI] [PubMed] [Google Scholar]
- 14.Van Hoorde K, Vergouwe Y, Timmerman D, et al. Simple dichotomous updating methods improved the validity of polytomous prediction models. J Clin Epidemiol. 2013;66(10):1158. doi: 10.1016/j.jclinepi.2013.04.014. [DOI] [PubMed] [Google Scholar]
- 15.Kong CH, Guest GD, Stupart DA, et al. Recalibration and validation of a preoperative risk prediction model for mortality in major colorectal surgery. Dis Colon Rectum. 2013;56(7):844. doi: 10.1097/DCR.0b013e31828343f2. [DOI] [PubMed] [Google Scholar]
- 16.Visser IH, Hazelzet JA, Albers MJ, et al. Mortality prediction models for pediatric intensive care: comparison of overall and subgroup specific performance. Intensive Care Med. 2013;39(5):942. doi: 10.1007/s00134-013-2857-4. [DOI] [PubMed] [Google Scholar]
- 17.Minne L, Eslami S, de Keizer N, et al. Effect of changes over time in the performance of a customized SAPS-II model on the quality of care assessment. Intensive Care Med. 2012;38(1):40. doi: 10.1007/s00134-011-2390-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minne L, Eslami S, de Keizer N, et al. Statistical process control for validating a classification tree model for predicting mortality — A novel approach towards temporal validation. J Biomed Inform. 2012;45(1):37. doi: 10.1016/j.jbi.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 19.Smith DS, Catalona WJ. Interobserver variability of digital rectal examination in detecting prostate cancer. Urology. 1995;45:70. doi: 10.1016/s0090-4295(95)96812-1. [DOI] [PubMed] [Google Scholar]
- 20.Taylor JM, Park Y, Ankerst DP, et al. Real-time individual predictions of prostate cancer recurrence using joint models. Biometrics. 2013;69(1):206. doi: 10.1111/j.1541-0420.2012.01823.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Hosmer-Lemeshow test statistics using all past data as training and the next year as the validation year (x-axis) for predicting a.) high-grade versus low-grade or no cancer, and b.) overall (high- and low-grade cancer) versus no cancer. Lower values indicate better fit, closer agreement between observed and predicted risks. Error bars represent the 95% confidence intervals of the test statistic.
Supplementary Appendix Table 1: Number of biopsies performed by year.