SUMMARY
Understanding the mechanism of transcription termination by a eukaryotic RNA polymerase (RNAP) has been limited by lack of a characterizable intermediate that reflects transition from an elongation complex to a true termination event. While other multisubunit RNAPs require multipartite cis-signals and/or ancillary factors to mediate pausing and release of the nascent transcript from the clutches of these enzymes, RNAP III does so with precision and efficiency on a simple oligo(dT) tract, independent of other cis-elements or trans-factors. We report a RNAP III pre-termination complex that reveals termination mechanisms controlled by sequence-specific elements in the non-template strand. Furthermore, the TFIIF-like, RNAP III subunit, C37 is required for this function of the non-template strand signal. The results reveal the RNAP III terminator as an information-rich control element. While the template strand promotes destabilization via a weak oligo(rU:dA) hybrid, the non-template strand provides distinct sequence-specific destabilizing information through interactions with the C37 subunit.
INTRODUCTION
The transcription cycle includes initiation, elongation and termination. Initiation factors bind to promoter DNA elements, recruit RNAP and also stabilize the initial transcribing complex until ~10 nt of nascent transcript is formed as a RNA:DNA hybrid in the active center (Dvir et al., 2001; Keene and Luse, 1999). Once formed, the RNA:DNA hybrid becomes the major component of the stability of the elongation complex (EC) which is capable of long distance processivity (Kireeva et al., 2000; Sidorenkov et al., 1998). ECs remain stable even when deprived of NTPs (Uptain et al., 1997).
At the end of the cycle, the EC must be destabilized to release its transcript and DNA (Gilmour and Fan, 2008). Analogous to initiation, cis-signals and trans-acting factors are required to mediate EC destabilization and release of the transcript from the grip of the RNAP. Yet, detailed mechanisms that mediate such transcript release are lacking for eukaryotic RNAPs I, II & III (Richard and Manley, 2009). A limitation has been lack of a characterizable transition state intermediate of termination.
Eukaryotic RNAP III is required for the synthesis of large amounts of tRNAs and other short essential transcripts. The high productivity of RNAP III is due in part to efficient transition from termination to reinitiation (Arimbasseri et al., 2013a; Dieci and Sentenac, 1996). RNAP III achieves efficient termination despite the apparent simplicity of its cis-acting DNA terminator element, a stretch of five or more T residues on the non-template (NT) strand which directs termination within this site without need for additional cis-elements or trans-acting factors (Arimbasseri et al., 2014; Bogenhagen and Brown, 1981; Braglia et al., 2005; Cozzarelli et al., 1983). This is a striking feat considering that other RNAP ECs require multipartite termination signals and/or ancillary factors to release the RNA from their clutches (Arimbasseri et al., 2013b; Richard and Manley, 2009). Transcript release by bacterial RNAP during intrinsic termination also occurs on a T-rich tract, although an upstream RNA hairpin is also required. Thus, insight into termination by RNAP III is of general interest. Yet, while the cis-element controlling RNAP III termination has been identified, its mechanistic action is unknown (Arimbasseri et al., 2014; Richard and Manley, 2009).
RNAP III is comprised of seventeen subunits (Arimbasseri et al., 2013a). Three of its subunits, Rpc11, Rpc37 and Rpc53 (denoted hereafter as C11, C37 and C53) can be dissociated during purification from a S. cerevisiae C11 mutant, leaving a core enzyme of fourteen subunits that is deficient for termination, known as pol IIIΔ but hereafter referred to as RNAP III-core (Arimbasseri and Maraia, 2013; Landrieux et al., 2006). C11 shares homology with the RNAP II transcript-cleavage, elongation factor, TFIIS, while the hetero-dimeric subunits C53/37 share homology with two subunits of TFIIF (Arimbasseri and Maraia, 2013; Chedin et al., 1998; Landrieux et al., 2006).
Biochemical analyses indicate two distinct mechanistic components of RNAP III termination (Arimbasseri and Maraia, 2013). The mechanism used by the 17-subunit holoenzyme is dependent on C53/C37/C11 and functions at terminators of 5–7 Ts, as found at most tRNA genes. The RNAP III-core mechanism mediates efficient termination independent of C53/C37/C11 but requires 8–9 Ts to do so (Arimbasseri and Maraia, 2013). This sensitivity to terminator length is critical because ~85% of the RNAP III terminators in S. cerevisiae have 7Ts or fewer (Braglia et al., 2005; Iben and Maraia, 2012) and the RNAP III-core enzyme is known to read through 7T terminators (Arimbasseri and Maraia, 2013; Chedin et al., 1998; Landrieux et al., 2006).
C53/C37 promotes termination by slowing the RNAP III elongation rate (Landrieux et al., 2006; see Rijal and Maraia, 2013) and also prevents transcription arrest in the proximal part of the terminator (Arimbasseri and Maraia, 2013). While the dimerization domains of C53/C37 interact with a peripheral lobe of RNAP III, parts of each contact the catalytic center (Kassavetis et al., 2010; Wu et al., 2011), suggesting that they may interact with the termination signal. Although one report indicates transcript cleavage by C11 during termination (Huang et al., 2005), others indicate C11 cleavage-independent roles in RNAP III termination (Arimbasseri and Maraia, 2013; Iben et al., 2011; Landrieux et al., 2006; Rijal and Maraia, 2013). Yet, the mechanisms by which these three subunits direct RNAP III-core to navigate the oligo(dT) signal and function in termination are unknown.
For the present study, we used purified yeast S. cerevisiae RNAP III to isolate a metastable pre-termination complex (PTC). We show that the C53/C37 and C11 subunits are necessary for PTC formation, which is directed by the proximal part of the NT strand of the terminator. PTC formation does not assure transcript release as this is promoted by the fifth T of the NT strand and the C-terminal domain of C37. The data support a model for RNAP III in which the terminator template strand is a primary effector of EC destabilization while the NT strand carries distinct sequence-specific signals for termination that work through the C53/C37/C11 subunits.
RESULTS
RNAP III pre-termination complex
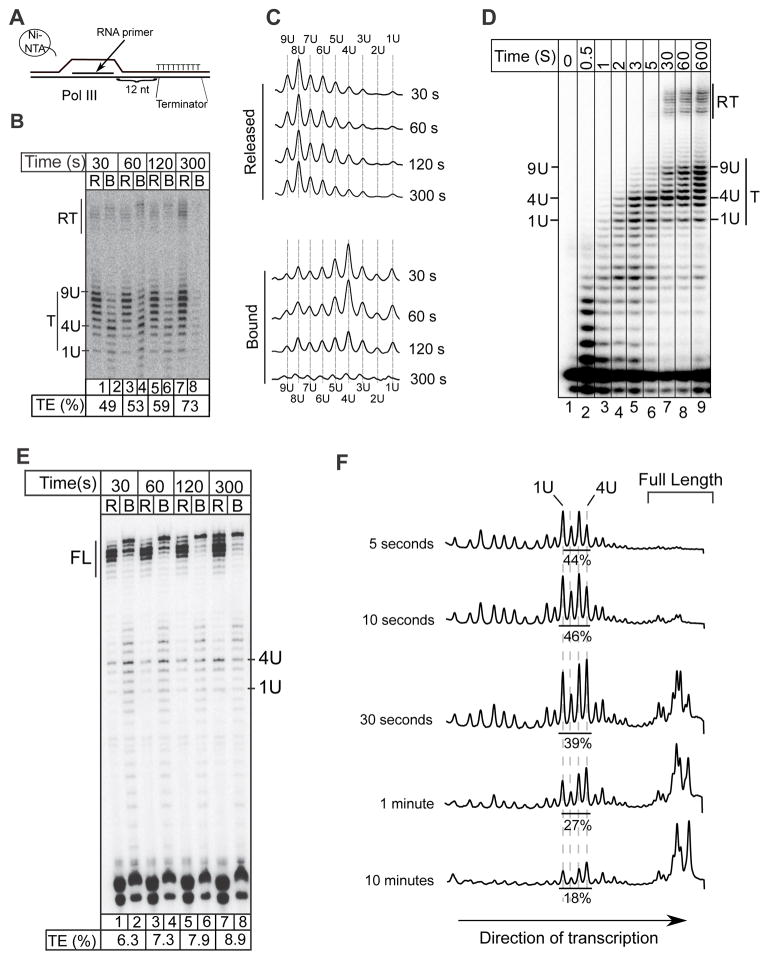
To better understand the holoenzyme mechanism of termination, we analyzed RNAP III ECs assembled on oligo-nucleotide DNA scaffolds with a 9T terminator located downstream of a 10’mer RNA primer (Fig. 1A). For this, the 32P-5′-end labeled RNA is annealed to a single strand of template DNA to which purified Nickel agarose-immobilized RNAP III is added. The NT DNA strand is added and allowed to anneal to form an EC, followed by washing; the reaction is initiated by addition of NTPs (Arimbasseri and Maraia, 2013; Komissarova et al., 2003). An advantage of using a 9T terminator is that it supports termination by RNAP III-holo and RNAP III-core and can be used to study both mechanisms. A time course shows that while the majority of U1–U9 (containing 1 to 9 Us at their 3′ ends) terminated transcripts were released within 30s, a fraction remained bound with preferential retention of U4 transcripts (Fig. 1B, lanes 1 & 2; 1C). The U4 and other RNAP III-bound transcripts decreased with time (Fig. 1B, C) suggesting that they reflect paused complexes that subsequently extended and/or released their transcripts.
Figure 1. Pol III pauses at T1–T4 of a terminator before transcript release.
A) Schematic of the experimental system. 10 nt RNA primer and 81 bp DNA are shown; oligo(dT) on the non-template strand is indicated as a T9 stretch. B) Time course assay for transcript release by RNAP III. R and B above lanes indicate released and bound transcripts respectively. Quantification of termination efficiency (TE) (Methods) reflect percentage of transcripts released at the terminator, below the lanes. RT and T to the sides of gel images represent read-through and terminated transcripts respectively. C) Lane tracings of B. D) Time course of RNAP III transcription in seconds; the 0.5 point is approximate. E) Transcript release assay for a 4T terminator with TE below; FL = full-length. F) Lane trace profiles comparing total transcripts at early time points on a template with a 4T stretch; positions of 1U–4U transcripts are indicated above. Numbers below show the % of total signal at the 4T stretch (see text). See also supplemental figures S1, and S2.
The data suggested among other possibilities that pausing at U1–U4 reflects a pre-termination intermediate. If so, a significant fraction of RNAP III might be found at U1–U4 positions at earlier time points. Thus, we examined total transcripts at earlier times (Fig 1D). Even though pausing was observed at each residue within the T track, the relative paucity of transcripts beyond the U4 position at 2, 3 and 5 seconds indicate that a majority of ECs pause at U4, in the proximal part of the terminator (Fig. 1D, lanes 4–6). To rule out a possible effect of template design on this pattern, we analyzed templates of varied design including ones with 6T and 7T terminators (Fig. S1). The heterogeneity of readthrough (RT) transcripts raised the possibility of transcript slippage within the homopolymeric terminator (Molodtsov et al., 2014; Strathern et al., 2013). However, a template in which the 9T terminator was replaced by a mixed nucleotide sequence showed similar RT heterogeneity (Fig. S1B) likely due to RNAP encountering the template end rather than slippage in the oligo(T) tract (Izban et al., 1995). The data indicate that the terminator proximal pause is an intrinsic property of RNAP III and suggest that it reflects an intermediate: a pre-termination complex (PTC).
Pausing on a T4 stretch does not assure transcript release
Pausing is prominent at position T4 yet a T4 stretch is not a terminator for S. cerevisiae RNAP III (Braglia et al., 2005; Hamada et al., 2000). We asked if residues beyond T4 play a role in pausing and/or release at T1–T4. We replaced the 9T terminator with 4Ts followed by GTCTC. Fig. 1E shows that while some transcripts paused/arrested in the T4 stretch, their release was limited to ~10% of total transcripts at the end time point, indicating low termination efficiency (TE, under lanes). Quantification of total transcripts at earlier times revealed 46% in the 4T stretch at 10 seconds whereas most extended to full length at later times (Fig. 1F). Thus a four T stretch is sufficient for pausing whereas efficient release requires additional Ts. Importantly, pausing at T4 does not commit the RNAP III EC to termination. Thus, PTC formation in the proximal terminator precedes transcript release but pausing in a T4 stretch does not assure transcript release.
RNAP III PTCs are metastable, active for nucleotide addition and transcript release
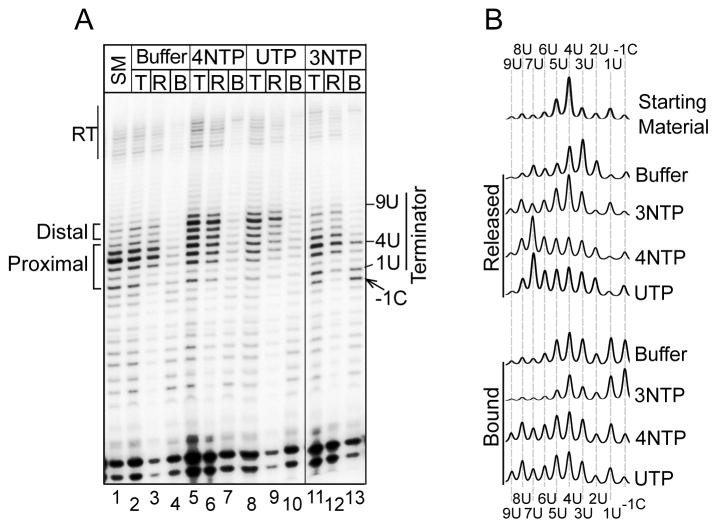
Most pauses do not lead to transcript release. If terminator pauses reflect PTCs, they should be distinct from ECs paused elsewhere on the template. To examine this, we isolated terminator-paused complexes. A transcription reaction was separated into released and bound fractions within 15 seconds. While some release had occurred (not shown), a substantial amount of transcripts was in the bound fraction which was further characterized. Lane 1 of Fig. 2A shows an aliquot of the bound fraction starting material (SM) for analysis in which the terminator transcripts were centered on U4 (lane trace, Fig. 2B). We subjected these complexes to different treatments, each for one minute, after which stop buffer was added to the total (T), released (R) and bound (B) fractions. Buffer alone led to spontaneous release of U1–U4 and longer transcripts while the shorter transcripts paused upstream of the T track were stably bound (Fig. 2A lanes 3 & 4), as expected of stably paused ECs. Addition of all 4 NTPs led to extension in the terminator and release (Fig. 2A compare lanes 5–7 with 2–4). Addition of UTP alone also allowed the transcripts paused in the terminator to be extended; most of these were released (Fig. 2A, compare lanes 3 & 9) while the shorter transcripts remained bound (lane 10).
Figure 2. PTCs are metastable, active for release or nucleotide addition.
A) Isolated PTCs were treated with different reagents as indicated above the lanes (SM = starting material; Buffer alone; 4NTP = 0.5 mM each ATP, CTP, GTP, UTP & 5 mM MgCl2; UTP = 0.5 mM UTP & 5 mM MgCl2; 3NTP = 0.5 mM ATP, GTP, CTP & 5 mM MgCl2. T, R and B above lanes indicate total, released and bound. RT indicates read-through. The -1C with arrow points to a band representing the C nucleotide immediately preceding the terminator. Transcript bands representing the proximal and distal parts of the terminator are indicated by brackets to the left. Vertical line between lanes 10 & 11 indicate that lanes between them in the gel were removed. B) Lane tracings of the starting material (SM), and the released and bound transcripts from panel A.
Upon addition of 3NTPs lacking UTP most terminator-paused transcripts were released without extension, similar to buffer alone, as expected (Fig. 2A lanes 11–13). In this reaction ECs paused upstream of the oligo(dT) extended some of their transcripts to accumulate and stall on -1C immediately preceding the terminator, without release of the -1C transcript (Fig. 2A lanes 12–13, 2B). The observed difference between the stability of the -1C EC and release within the oligo(dT) by the PTC as well as the ability of PTCs to add UTP supports that PTCs are metastable yet catalytically active. The cumulative results confirm that the terminator paused PTC is unlike ECs paused elsewhere on the template and represents a termination intermediate.
Sequence context affects PTC formation
We examined termination on templates that differ in sequence between the primer and terminator. Figs S1A and S2 show that these templates also directed an initial pause in the proximal terminator with the notable difference that in Fig. S2 the pause extended to T5. It is known that sequence context can affect RNAP III termination (Bogenhagen and Brown, 1981; Braglia et al., 2005; Maraia et al., 1992). These results may reflect effects of sequence context on PTC formation.
C53/C37 and C11 are required for PTC formation
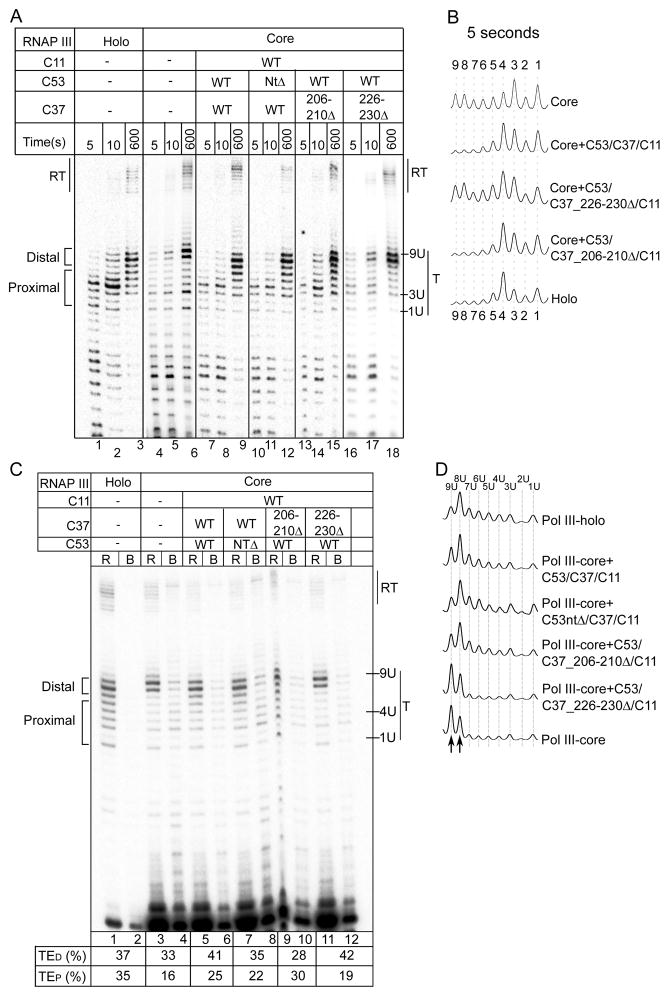
We asked if C53/C37 and C11 play a role(s) in PTC formation. We compared RNAP III-core lacking C53/C37/C11 with the RNAP III-holoenzyme (Fig. 3A lanes 1–3 and 4–6). RNAP III-core produced a relative paucity of 4U–7U bands as well as a prominent distal band at 9U and a proximal band at 3U, as compared to 8U and 4U respectively for RNAP III-holo (Fig. 3A lanes 1–3 and 4–6, 3B, S3A–B). The RNAP III-core U3 band reflects transcription arrest rather than pause because most U3 transcripts were neither released nor chased while the 8–9U transcripts were released early and efficiently (Fig. S3C) as observed previously (Arimbasseri and Maraia, 2013).
Figure 3. C53/C37 and C11 are required for PTC formation.
A) Time course transcription by RNAP III-holo and RNAP II-core, the latter with and without recombinant C53/C37 and C11 as indicated above the lanes. Labeling conventions are same as in Fig 1. Vertical lines between lanes indicate non-contiguous sections; lanes 4–6 and 16–18 were from a different gel than the others. B) Lane tracings of the 5 second time-points in A. C) Release and bound assay for the components indicted above the lanes. Transcripts from the proximal and distal parts of the terminator are indicated to the left; quantification of termination efficiency in the proximal and distal parts of the terminator are shown under the lanes as TEP and TED respectively (Experimental procedures). D) Lane tracings of ‘released’ lanes in C; vertical arrows below help asses ratios of 8U to 9U transcripts (text). See also supplemental figure S3.
The data comparing RNAP III-holo and RNAP III-core suggest that C53/C37/C11 promote transcript release from the proximal terminator. The holoenzyme pattern was reconstituted from RNAP III-core plus recombinant C53/C37/C11 (Fig 3A lanes 1–9, Fig. 3B). This reestablished a RNAP III-holo major pause at T4, an increase in the 4U–7U bands, and a shift in the major distal transcript from 9U to 8U (Fig 3A lanes 1–9, 3B).
A specific C-terminal region of C37 is required for proper PTC function
As noted above regions of C53 and C37 had been localized to the RNAP III catalytic center. We tested select mutants of C53 and C37 for PTC formation. An N-terminal truncated C53 that retains only its dimerization domain reconstituted the PTC and restored the transcript release pattern similar to full length C53 although a fraction was arrested in the terminator (Fig. 3A lanes 10–12, Fig S3F, I) in agreement with prior results (Arimbasseri and Maraia, 2013). Significantly, deletion of 5 amino acids (226–230) in the C-terminal region of C37 that crosslink near the catalytic center (Wu et al., 2011) and are hot spots in termination-specific mutants in S. pombe (Rijal and Maraia, 2013), decreased the U3 band and reestablished the U4 pause at early time points (Fig 3A lanes 16–17; 3B). Importantly however, these proximal paused transcripts were chased at later times, yielding a release pattern with relative paucity of U4–U7 transcripts, somewhat similar to RNAP III-core (Fig 3A lanes 18 and 6; Fig 3C lanes 11–12). Examination of the released and bound transcripts produced in the C53/37 mutants indicate that the C37 226–230 mutation reproducibly uncouples the pause at U1–U4 from transcript release at U4–U7 (Fig. 3C, D and data not shown). The deficiency of C37 226–230 is most readily apparent in the released fraction lane tracings in Fig. 3D. By contrast, a 5 amino acid deletion in a nearby region, C37(206–210) was reproducibly much more like C37-WT (Fig. 3A lanes 13–15, Fig. 3B; Fig. 3C–D, Fig S3, and data not shown). The data indicate C37 region 226–230 functions to promote transcript release after the proximal pause.
Non-template strand residues T3 and T4 are required for efficient PTC formation
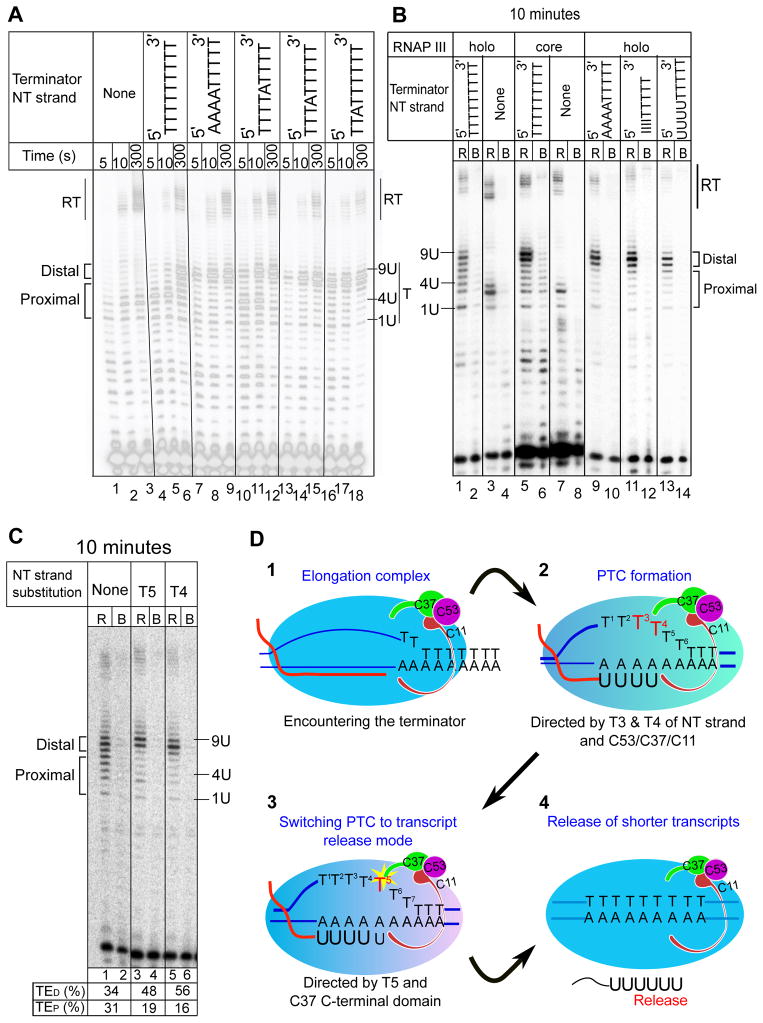
Crosslink analyses had shown that C37 lies in close proximity to the NT strand within the transcription bubble and other components of the EC (Bartholomew et al., 1993; Braun et al., 1992; Kassavetis et al., 2010). We tested the DNA components for effects on PTC formation and transcript release. RNAP III-holo lacking the entire NT strand produced a U3 transcript that was efficiently released, but without pause or release in the distal part of the dA tract (Fig. 4A lanes 1–3; 4B, lanes 3–4). Fig. 4B lanes 1–4 show that these U1–U4 transcripts are very efficiently released in the absence of the NT strand. This suggests that the RNAP III EC is quite sensitive to a short rU:dA hybrid and that the NT strand is an important component of a proper functioning RNAP III termination signal.
Figure 4. Non-template (NT) strand bases T3, T4 and T5 are required for PTC formation and transcript release.
A) Time course of transcription by RNAP III-holo with full-length NT strands containing the terminator NT sequences above the lanes. No NT strand was added to reaction lanes 1–3. B) Transcript release assay for RNAP III-holo and RNAP III-core with terminator NT strand sequences as indicated above the lanes; I = deoxyinosine (dI) and U = deoxyuridine (dU). No NT strand was added to reaction lanes 3–4 and 7–8. The vertical lines between some lanes indicate non-contiguous sections. C) Transcript release assay for single base substitutions in the terminator NT strand. T5 and T4 indicate T->A substitution at NT strand positions 5 and 4 respectively. Labeling conventions are as in Fig 1. Proximal and distal parts of the terminator are indicated by brackets. Transcripts from the proximal and distal parts of the terminator are indicated to the left; termination efficiency in the proximal and distal parts of the terminator are shown under the lanes as TEP and TED respectively. D) A model for the RNAP III holoenzyme mechanism of transcription termination. 1) The C53/C37/C11 subcomplex transforms the RNAP III EC to a 2) PTC upon addition of 4 Us. T3 and T4 of the NT strand constitute a signal for PTC formation. 3) C37 amino acids 226–230 and T5 of NT strand contribute to switching the PTC to 4) transcript release mode upon nucleotide addition. See also supplemental figure S4.
Importantly, the termination patterns of RNAP III-holo and RNAP III-core are almost indistinguishable in the absence of the NT strand (Fig 4B lanes 3–4, 7–8). This indicates that C53/C37/C11 do not influence RNAP III termination in oligo(dA) in the absence of an NT strand. This further suggests that sensitivity to a short rU:dA hybrid is a property of the RNAP III-core mechanism that can be modulated by the NT strand. Experiments below focus on specific effects of the NT strand and involvement of C53/37/11.
Nucleotide substitutions in the NT strand were analyzed. T-to-A substitutions of the proximal 4 nucleotides of the NT strand diminished the U4 PTC (Fig. 4A lanes 7–9) with release mostly limited to 7U–9U transcripts (Fig. 4B lanes 9–10). The most prominent band observed for the AAAATTTTT NT strand at 5 seconds was U8 (Fig. 4A lanes 7–9) suggesting fast elongation through the terminator, strikingly similar to RNAP III-core (Fig. 3A lanes 4–6). Because the substituted residues should be single-stranded in the transcription bubble, loss of potential base-pairing with the oligo(dA) template was not expected to be a determinant of the effects observed. Consistent with this expectation, restoring the potential for base pairing by substituting deoxyinosine or deoxyuridine, rather than dA in the NT strand did not restore termination in the proximal terminator (Fig. 4B lanes 11–12 & 13–14). These data argue that NT base identity contributes to an important function of the proximal terminator, PTC formation. Similar effects were observed with substitutions at T4 or T3 (Fig. 4A lanes 13–18, Fig. 4C) indicating that Ts at these positions are functional components of the RNAP III termination signal.
The NT strand residue T5 acts to signal transcript release
In contrast to substitutions at T1–T4, T3 and T4, a single NT substitution at T5 supported the major pause at T1–T4 at 5 seconds (Fig 4A lane 10 vs. 4). However, while the T5A NT proximal pauses were chased at later times, these RNAP III-holo complexes read-through the 5–7 positions with efficient release only at T8–T9 (Fig. 4A lane 12, Fig. 4C lanes 3–4) very similar to C37 226–230 (Fig 3). A T5A substitution in a 7T terminator decreased termination efficiency supporting the importance of this residue in the holoenzyme mechanism (Fig. S4A). PTCs formed with the T5A NT and subsequently isolated, spontaneously released their terminator transcripts while the shorter transcripts remained bound (Fig. S4B) as expected.
The data led to the proposal that T5 is important to signal transcript release by the U4 PTC after nucleotide addition. That the NT T5A substitution and the C37 226–230 mutation led to similar effects (compare Fig. 3A lane 18 with Fig. 4A lane 12, and Fig. 3C lane 11 with Fig. 4C lane 3) suggests that the C-terminal region of C37 works through the T5 nucleotide to enable transcript release.
DISCUSSION
The oligo(dT) RNAP III termination signal functions to halt the polymerase, define the 3′ ends of its structural RNA products, and disassemble the complex, and it does so with high efficiency and readiness for efficient reinitiation (Arimbasseri et al., 2013a; Dieci and Sentenac, 1996). The results here provide mechanistic insight into the problem of how an apparently simple sequence element can mediate such multifarious effects. The RNAP III termination signal is an information-rich control element, of which both its template and non-template strands constitute functionally distinct components. The data extend prior results that the RNAP III holoenzyme mechanism operates in the proximal terminator and is dependent on C53/37/11, and the core mechanism is operative in the distal terminator independent of C53/37/11 (Arimbasseri and Maraia, 2013). We believe that the two component mechanistic system, RNAP III-core and RNAP III-holo, helps us understand this in the context of other multisubunit RNAPs that require bipartite termination signals (e.g., an upstream hairpin preceding a T-rich tract) or more complex signals. We envision that lack of a hairpin component of the RNAP III termination signal is compensated by the holo component of RNAP III, i.e., the NT strand and C37/53/11.
In the absence of a NT strand, the oligo(dA) in the template strand constitutes a termination signal. Experiments using substituted NT strands and C37 mutants indicate that the NT provides distinct sequence-specific signals that work via C37. A model that emerges (Fig. 4D) is that the proximal Ts of the terminator NT strand are recognized as part of a mechanism that leads to a metastable PTC in a C53/C37/C11-dependent manner. PTC fate is determined by NT T5 which along with C37 signals transcript release.
This model is supported by very similar defects in termination caused by altering either the NT strand sequence or the 226–230 region of C37, which was known to interact with the RNAP III active center and to be important for termination (Bartholomew et al., 1994; Bartholomew et al., 1993; Rijal and Maraia, 2013; Wu et al., 2011). Our findings also fit with previous results on S. pombe C37, in which case this region was a hot spot for mutations that caused in vivo read-through of 5T, 6T and 7T terminators (Rijal and Maraia, 2013).
We believe that transcription termination is likely activated by instability of the innate weakness of the rU:dA hybrid (Martin and Tinoco, 1980). The inherent sensitivities of RNAP III-core and RNAP III-holo to the rU:dA hybrid is reflected by efficient release of U1–U4 RNAs in the absence of a NT strand (Fig. 4B). The results support the idea that RNAP III is intrinsically and unusually sensitive to a rU:dA hybrid since other RNAPs do not terminate at this motif in the absence of other cis-elements or trans-acting factors.
Bioinformatic analysis of RNAP III terminators support the importance of the first 4 Ts of the terminator. Although the oligo(dT) length requirement for termination differs among species (Hamada et al., 2000), genome-wide analyses of tRNA gene terminators using the sequence Logo algorithm, whose output reflects the information content of a sequence motif (Schneider and Stephens, 1990), indicates that the first four nucleotides of the oligo(dT) signal carry the most information (Fig S4C).
An interaction between C37 and the NT strand has precedent in the bacterial NusG family of proteins, factors that recognize the non base paired NT strand of the transcription bubble with varying degrees of sequence-specificity to regulate pausing and termination (Tomar and Artsimovitch, 2013). Elongation factor Spt5, which is a NusG homolog in eukaryotes, also binds in close proximity to the NT strand in the RNAP II transcription bubble (Martinez-Rucobo et al., 2011). Of further note is that termination mutagenesis screens of the second largest subunits of yeast RNAPs III and II (Kubicek et al., 2013; Shaaban et al., 1995) uncovered regions conserved in both, suggesting that their mechanisms of transcript release may be similar.
EXPERIMENTAL PROCEDURES
All oligos were purchased either from IDT or Operon and were PAGE purified. Oligos are listed in supplemental Table S1.
RNA polymerase III (-holo and -core) from Saccharomyces cerevisiae and recombinant subunits were purified as described (Arimbasseri and Maraia, 2013).
EC assembly and transcriptions
Elongation complex assembly on scaffolds and the transcription reactions are described in detail elsewhere (Arimbasseri and Maraia, 2015).
For reconstitution using RNAP III-core and C53/C37/C11 subunits, the recombinant proteins were incubated with RNAP III-core before immobilization (incubation after TEC assembly gives identical results). When C53/C37 or C11 were used alone, these were added after EC assembly to prevent dissociation during washes. Additional details are in the Supplemental materials.
Analysis of transcript release
ECs (50μl) were subjected to transcription for the times indicated followed by separation of released from bound transcripts by centrifugation. Beads were washed once with 100μl buffer and the wash was pooled with released fraction. Transcription stop buffer was added to both fractions followed by phenol chloroform purification.
Termination efficiency was calculated from quantified data as follows: TE (%) = (Transcripts released at terminator/total transcripts) X 100. TEP (%) = Transcripts released at T1–T6/total transcripts. TED (%) = Transcripts released at T7–T9/total transcripts. Total transcripts = (transcripts released at terminator + released read through transcripts + bound transcripts at terminator + bound read through transcripts).
Isolation of PTCs and chases
ECs (50μl) were transferred to filter cups (Costar Spin-X tubes) followed by addition of 10μl 6X NTP mix. After 9 seconds the tube was centrifuged for 6 seconds in a centrifuge at room temperature immediately after which 100μl of reagent indicated in Fig 2 and Fig S4B was added and incubated for 1 minute.
Supplementary Material
Acknowledgments
We thank J. Iben (NICHD) for terminator LOGOs, M. Kashlev, M. Kireeva (NCI) and D. Luse (Cleveland Clinic, Cleveland) for discussion, and I. Artsimovitch (Ohio State University, Columbus) for comments. This work was supported by the Intramural Research Program (HD000412-24 PGD) of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health.
Footnotes
AUTHOR CONTRIBUTIONS
AGA and RM conceived experiments, analyzed data and wrote the paper. AGA performed the experiments. RM supervised the project.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arimbasseri AG, Kassavetis GA, Maraia RJ. Comment on “Mechanism of eukaryotic RNA polymerase III transcription termination”. Science. 2014;345:524. doi: 10.1126/science.1253783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimbasseri AG, Maraia RJ. Distinguishing core and holoenzyme mechanisms of transcription termination by RNA polymerase III. Mol Cell Biol. 2013;33:1571–1581. doi: 10.1128/MCB.01733-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimbasseri AG, Maraia RJ. Biochemical Analysis of Transcription Termination by RNA Polymerase III from Yeast Saccharomyces cerevisiae. Methods Mol Biol. 2015;1276:185–198. doi: 10.1007/978-1-4939-2392-2_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimbasseri AG, Rijal K, Maraia RJ. Comparative overview of RNA polymerase II and III transcription cycles, with focus on RNA polymerase III termination and reinitiation. Transcription. 2013a:4. doi: 10.4161/trns.27369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimbasseri AG, Rijal K, Maraia RJ. Transcription termination by the eukaryotic RNA polymerase III. Biochim Biophys Acta. 2013b;1829:318–330. doi: 10.1016/j.bbagrm.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomew B, Braun BR, Kassavetis GA, Geiduschek EP. Probing close DNA contacts of RNA polymerase III transcription complexes with the photoactive nucleoside 4-thiodeoxythymidine. J Biol Chem. 1994;269:18090–18095. [PubMed] [Google Scholar]
- Bartholomew B, Durkovich D, Kassavetis GA, Geiduschek EP. Orientation and topography of RNA polymerase III in transcription complexes. Mol Cell Biol. 1993;13:942–952. doi: 10.1128/mcb.13.2.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen DF, Brown DD. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell. 1981;24:261–270. doi: 10.1016/0092-8674(81)90522-5. [DOI] [PubMed] [Google Scholar]
- Braglia P, Percudani R, Dieci G. Sequence context effects on oligo(dT) termination signal recognition by Saccharomyces cerevisiae RNA polymerase III. J Biol Chem. 2005;280:19551–19562. doi: 10.1074/jbc.M412238200. [DOI] [PubMed] [Google Scholar]
- Braun BR, Kassavetis GA, Geiduschek EP. Bending of the Saccharo-myces cerevisiae 5S rRNA gene in transcription factor complexes. J Biol Chem. 1992;267:22562–22569. [PubMed] [Google Scholar]
- Chedin S, Riva M, Schultz P, Sentenac A, Carles C. The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination. Genes Dev. 1998;12:3857–3871. doi: 10.1101/gad.12.24.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzarelli NR, Gerrard SP, Schlissel M, Brown DD, Bogenhagen DF. Purified RNA polymerase III accurately and efficiently terminates transcription of 5S RNA genes. Cell. 1983;34:829–835. doi: 10.1016/0092-8674(83)90540-8. [DOI] [PubMed] [Google Scholar]
- Dieci G, Sentenac A. Facilitated recycling pathway for RNA polymerase III. Cell. 1996;84:245–252. doi: 10.1016/s0092-8674(00)80979-4. [DOI] [PubMed] [Google Scholar]
- Dvir A, Conaway JW, Conaway RC. Mechanism of transcription initiation and promoter escape by RNA polymerase II. Curr Opin Genet Dev. 2001;11:209–214. doi: 10.1016/s0959-437x(00)00181-7. [DOI] [PubMed] [Google Scholar]
- Gilmour DS, Fan R. Derailing the locomotive: transcription termination. J Biol Chem. 2008;283:661–664. doi: 10.1074/jbc.R700032200. [DOI] [PubMed] [Google Scholar]
- Hamada M, Sakulich AL, Koduru SB, Maraia RJ. Transcription termination by RNA polymerase III in fission yeast. A genetic and biochemically tractable model system. J Biol Chem. 2000;275:29076–29081. doi: 10.1074/jbc.M003980200. [DOI] [PubMed] [Google Scholar]
- Huang Y, Intine RV, Mozlin A, Hasson S, Maraia RJ. Mutations in the RNA polymerase III subunit Rpc11p that decrease RNA 3′ cleavage activity increase 3′-terminal oligo(U) length and La-dependent tRNA processing. Mol Cell Biol. 2005;25:621–636. doi: 10.1128/MCB.25.2.621-636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iben JR, Maraia RJ. Yeast tRNAomics: tRNA gene copy number variation and codon use provide bioinformatics evidence of a new wobble pair in a eukaryote. RNA. 2012;18:1358–1372. doi: 10.1261/rna.032151.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iben JR, Mazeika JK, Hasson S, Rijal K, Arimbasseri AG, Russo AN, Ma-raia RJ. Point mutations in the Rpb9-homologous domain of Rpc11 that impair transcription termination by RNA polymerase III. Nucleic Acids Res. 2011;39:6100–6113. doi: 10.1093/nar/gkr182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izban MG, Samkurashvili I, Luse DS. RNA polymerase II ternary complexes may become arrested after transcribing to within 10 bases of the end of linear templates. J Biol Chem. 1995;270:2290–2297. doi: 10.1074/jbc.270.5.2290. [DOI] [PubMed] [Google Scholar]
- Kassavetis GA, Prakash P, Shim E. The C53/C37 subcomplex of RNA polymerase III lies near the active site and participates in promoter opening. J Biol Chem. 2010;285:2695–2706. doi: 10.1074/jbc.M109.074013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene RG, Luse DS. Initially transcribed sequences strongly affect the extent of abortive initiation by RNA polymerase II. J Biol Chem. 1999;274:11526–11534. doi: 10.1074/jbc.274.17.11526. [DOI] [PubMed] [Google Scholar]
- Kireeva ML, Komissarova N, Waugh DS, Kashlev M. The 8-nucleotide-long RNA:DNA hybrid is a primary stability determinant of the RNA poly-merase II elongation complex. J Biol Chem. 2000;275:6530–6536. doi: 10.1074/jbc.275.9.6530. [DOI] [PubMed] [Google Scholar]
- Komissarova N, Kireeva ML, Becker J, Sidorenkov I, Kashlev M. Engineering of elongation complexes of bacterial and yeast RNA polymerases. Methods in Enzymology. 2003;370:233–251. doi: 10.1016/S0076-6879(03)71017-9. [DOI] [PubMed] [Google Scholar]
- Kubicek CE, Chisholm RD, Takayama S, Hawley DK. RNA polymer-ase II mutations conferring defects in poly(A) site cleavage and termination in Saccharo-myces cerevisiae. G3 (Bethesda, Md) 2013;3:167–180. doi: 10.1534/g3.112.004531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrieux E, Alic N, Ducrot C, Acker J, Riva M, Carles C. A sub-complex of RNA polymerase III subunits involved in transcription termination and reinitiation. EMBO J. 2006;25:118–128. doi: 10.1038/sj.emboj.7600915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraia RJ, Chang DY, Wolffe AP, Vorce RL, Hsu K. The RNA pol-ymerase III terminator used by a B1-Alu element can modulate 3′ processing of the intermediate RNA product. Mol Cell Biol. 1992;12:1500–1506. doi: 10.1128/mcb.12.4.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin FH, Tinoco I. DNA-RNA hybrid duplexes containing oligo(dA:rU) sequences are exceptionally unstable and may facilitate termination of transcription. Nucleic Acids Res. 1980;8:2295–2299. doi: 10.1093/nar/8.10.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Rucobo FW, Sainsbury S, Cheung ACM, Cramer P. Architecture of the RNA polymerase–Spt4/5 complex and basis of universal transcription processivity. The EMBO journal. 2011;30:1302–1310. doi: 10.1038/emboj.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molodtsov V, Anikin M, McAllister WT. The presence of an RNA:DNA hybrid that is prone to slippage promotes termination by T7 RNA polymerase. J Mol Biol. 2014;426:3095–3107. doi: 10.1016/j.jmb.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P, Manley JL. Transcription termination by nuclear RNA poly-merases. Genes Dev. 2009;23:1247–1269. doi: 10.1101/gad.1792809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijal K, Maraia RJ. RNA polymerase III mutants in TFIIFα-like C37 cause terminator readthrough with no decrease in transcription output. Nucleic Acids Research. 2013;41:139–155. doi: 10.1093/nar/gks985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaaban SA, Krupp BM, Hall BD. Termination-altering mutations in the second-largest subunit of yeast RNA polymerase III. Mol Cell Biol. 1995;15:1467–1478. doi: 10.1128/mcb.15.3.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorenkov I, Komissarova N, Kashlev M. Crucial role of the RNA:DNA hybrid in the processivity of transcription. Mol Cell. 1998;2:55–64. doi: 10.1016/s1097-2765(00)80113-6. [DOI] [PubMed] [Google Scholar]
- Strathern J, Malagon F, Irvin J, Gotte D, Shafer B, Kireeva M, Lubkowska L, Jin DJ, Kashlev M. The fidelity of transcription: RPB1 (RPO21) mutations that increase transcriptional slippage in S. cerevisiae. J Biol Chem. 2013;288:2689–2699. doi: 10.1074/jbc.M112.429506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomar SK, Artsimovitch I. NusG-Spt5 proteins-Universal tools for transcription modification and communication. Chemical reviews. 2013;113:8604–8619. doi: 10.1021/cr400064k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uptain SM, Kane CM, Chamberlin MJ. Basic mechanisms of transcript elongation and its regulation. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- Wu CC, Lin YC, Chen HT. The TFIIF-like Rpc37/53 dimer lies at the center of a protein network to connect TFIIIC, Bdp1, and the RNA polymerase III active center. Mol Cell Biol. 2011;31:2715–2728. doi: 10.1128/MCB.05151-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.