Abstract
Background & Aims
Rapid induction of β-PDGF receptor (β-PDGFR) is a core feature of hepatic stellate cell activation, but its cellular impact in vivo is not well characterized. We explored the contribution of β-PDGFR-mediated pathway activation to hepatic stellate cell responses in liver injury, fibrogenesis, and carcinogenesis in vivo using genetic models with divergent β-PDGFR activity, and assessed its prognostic implications in human cirrhosis.
Methods
The impact of either loss or constitutive activation of β-PDGFR in stellate cells on fibrosis was assessed following carbon tetrachloride (CCl4) or bile duct ligation. Hepatocarcinogenesis in fibrotic liver was tracked after a single dose of diethylnitrosamine (DEN) followed by repeated injections of CCl4. Genome-wide expression profiling was performed from isolated stellate cells that expressed or lacked β-PDGFR to determine deregulated pathways and evaluate their association with prognostic gene signatures in human cirrhosis.
Results
Depletion of β-PDGFR in hepatic stellate cells decreased injury and fibrosis in vivo, while its auto-activation accelerated fibrosis. However, there was no difference in development of DEN-induced pre-neoplastic foci. Genomic profiling revealed ERK, AKT, and NF-kB pathways and a subset of a previously identified 186-gene prognostic signature in hepatitis C virus (HCV)-related cirrhosis as downstream of β-PDGFR in stellate cells. In the human cohort, the β-PDGFR signature was not associated with HCC development, but was significantly associated with a poorer outcome in HCV cirrhosis.
Conclusions
β-PDGFR is a key mediator of hepatic injury and fibrogenesis in vivo and contributes to the poor prognosis of human cirrhosis, but not by increasing HCC development.
Keywords: HCC, cirrhosis, receptor tyrosine kinase, gene expression signatures, pathway analysis
Introduction
Among mitogenic pathways in stellate cells, signaling by the beta platelet-derived growth factor receptor (β-PDGFR) is the most potent [1, 2]. Expression of PDGF receptors is low in healthy liver, but dramatically increases in stellate cells during injury [2, 3]. In both mice and humans, the PDGF signaling network is comprised of four ligands, PDGF A–D, which transduce their signals through dimeric transmembrane receptors α- and β-PDGFR, which can form hetero- and homodimers [4]. Upon ligand binding, receptor dimerization provokes phosphorylation of the tyrosine residues within the intracellular domain, leading to activation of the Fas-MAPK pathway, signaling through the PI3K-AKT/PKB pathway and activation of PKC family members [5].
Antagonism of β-PDGFR has been an appealing target to treat hepatic fibrosis. Indeed, our previous study and those of others [6–8] have demonstrated that the RTK inhibitor imatinib mesylate (Gleevec®) whose targets include β-PDGFR, inhibits stellate cell activation and reduces fibrosis.
Recent evidence links the behavior of stromal cells, especially driven by the ligand PDGF-B, not only to the pathogenesis of fibrosis, but also to inflammation, regeneration and cancer [9]. Sorafenib (Nexavar®), a multi-receptor tyrosine kinase inhibitor whose targets include β-PDGFR, remains the only drug approved for treatment of advanced, non-resectable HCC [10].
Despite the suggestion that promotion of fibrosis by β-PDGFR can accelerate HCC development, this important question has not been addressed experimentally. Here we have specifically explored the contribution of β-PDGFR signaling by activated hepatic stellate cells to injury, fibrosis and cancer by exploiting complementary genetic models using a Cre-Lox strategy, one in which the receptor was deleted in stellate cells, or another in which it was auto-activated. Importantly, we have examined whether β-PDGFR signaling in hepatic stellate cells is linked in human cirrhosis to overall prognosis.
Methods
For detailed description of methods see Supplementary Methods.
Animals
β-PDGFRfl/fl mice, as previously described [11], (on the 129S4/SvJaeSor background) were crossed with a transgenic FVB line expressing Cre-recombinase under control of the human glial fibrillary acidic protein (GFAP) promoter to generate β-PDGFRfl/fl GFAP-Cre mice with a deletion of β-PDGFR in stellate cells – this GFAP promoter has been successfully validated in prior studies as active in hepatic stellate cells [12, 13]. To create animals with constitutively activated β-PDGFR in stellate cells, β-PDGFRbetaJ/+ mice, as previously described [14], (on the 129S4/B6 background) were also crossed with a transgenic GFAP-Cre line to generate β-PDGFRbetaJ/+ GFAP-Cre mice. These animals harbor hepatic stellate cells with autoactivation of β-PDGFR, owing to an activating mutation knocked into the β-PDGFR locus, plus addition of a lox-stop-lox cassette between the splice acceptor and the initiating codon of the cDNA [14].
Models of Murine Liver injury and Fibrosis
Liver fibrosis was induced either by ligation of the common bile duct (BDL) [15] or by intraperitoneal (i.p.) injections of carbon tetrachloride (CCl4, Sigma, St. Louis, MO) [16]. For acute CCl4 injury studies, mice received a total of 3 i.p. injections (alternating days) of either corn oil or 10% CCl4 (diluted in corn oil) at a dose of 0.5 µl/g body weight. For the chronic injury model, mice received i.p. injections of CCl4 3 times per week for a total of 6 weeks.
Induction of Carcinogenesis
Mice received a single dose of diethylnitrosamine (DEN, Sigma, St. Louis, MO) (25µg/g bw i.p.) at day 15 post-partum. Starting two weeks after DEN, mice received a total of 22 injections of CCl4 (0.5µl/g bw i.p., 1 injection/week) [17]. Mice were sacrificed 48 hours following the last CCl4 injection. Nodule number and size was documented as described by counting and measuring the diameter of each lesion using a caliper.
Primary Hepatic Stellate cell Isolation and Cell Culture
Mouse hepatic stellate cells were isolated from β-PDGFRfl/fl GFAP-Cre negative and β-PDGFRfl/fl GFAP-Cre positive mice by enzymatic pronase and collagenase digestion and density gradient centrifugation as previously described [18]. Cells were cultured with Dulbecco’s modified Eagle medium (DMEM) containing 10% fetal bovine serum. Cells were either treated with or without PDGF-B [10 ng/ml] (Peprotech, Princeton, NJ) diluted in albumin (vehicle) containing serum-free media (DMEM).
Histologic and Immunohistochemical Studies
Liver samples were formalin-fixed, paraffin-embedded, sectioned at 4 µm, and processed routinely for H&E staining. Sirius Red, combined with morphometry, was used to quantify collagen using Bioquant image analysis software (Bioquant Image Analysis Corporation, Nashville, TN). Immunohistochemical staining of αSMA and desmin was performed on formalin-fixed, paraffin-embedded liver sections with a rabbit polyclonal antibody (Abcam, Cambridge, England). A pathologist blindly scored 5 random areas per slide for necrosis, inflammation and dysplasia.
Genome-wide expression profiling
Genome-wide gene expression profiling of mouse primary hepatic stellate cells was performed, in triplicate, by using MouseWG-6 v2.0 Expression BeadChip (Illumina) according to the manufacturer’s protocol. Raw scanned data were normalized by using cubic spine algorithm implemented in the GenePattern genomic analysis toolkit (www.broadinstitute.org/genepattern) [19]. Probe-level expression data were collapsed into gene-level by calculating the median of multiple probes, and converted to human genes based on an orthologous mapping table provided by the Jackson laboratory (www.informatics.jax.org). The dataset (GSE#52253) is available at NCBI Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo).
Bioinformatics and Statistical Analysis
Enrichment of molecular pathways was evaluated by Gene Set Enrichment Analysis (GSEA) [20] on a comprehensive gene set collection in Molecular Signatures Database (see Supplementary Methods).
Results
β-PDGFR Expression is Induced Upon Liver Injury In Vivo and In Vitro
We generated a mouse line in which the expression of β-PDGFR was deleted in hepatic stellate cells by crossing β-PDGFRfl/fl mice with animals expressing Cre-recombinase under the human glial fibrillary acidic protein promoter (GFAP-Cre) (Suppl. Fig. 1A) [13, 21].
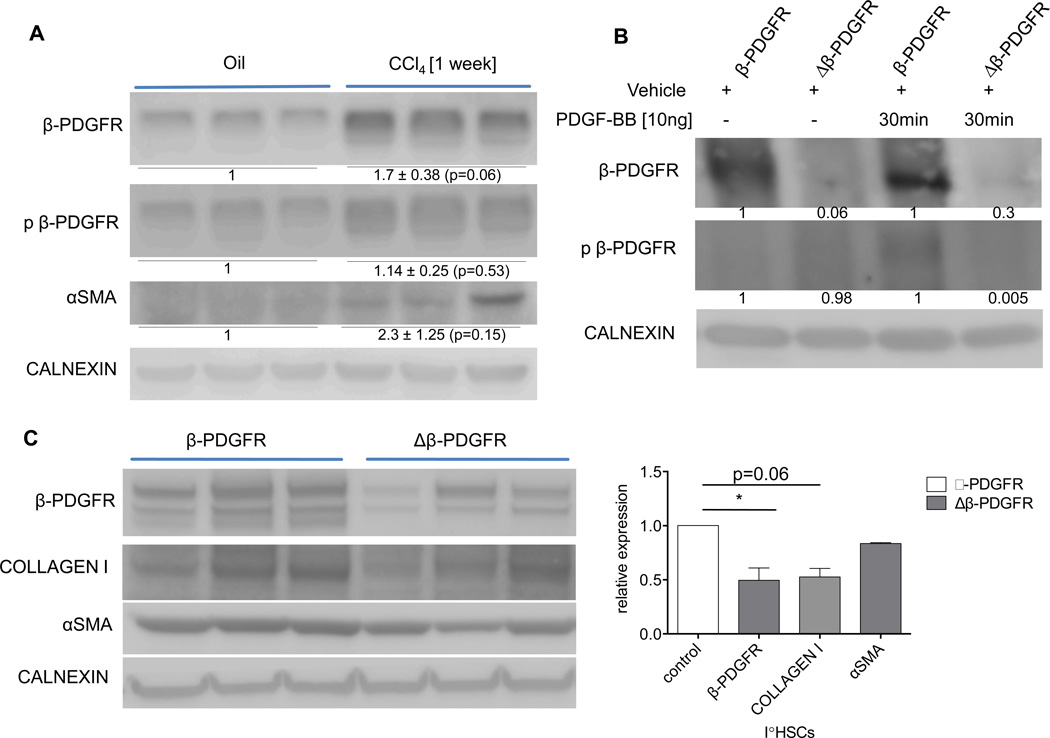
To first confirm the induction of β-PDGFR following acute injury, β-PDGFRfl/fl GFAP-Cre negative animals were treated with CCl4 in three doses in one week. Whole liver lysates contained increased β-PDGFR expression and phosphorylation, as well as up-regulation of αSMA (Fig. 1A).
Fig. 1. β-PDGFR expression correlates with activation of mouse hepatic stellate cells in vivo and in vitro.
(A) Mice were injected three times with either oil or CCl4 to induce acute liver injury and were sacrificed two days after the last injection. Immunoblot of whole liver lysates demonstrating increased expression of β-PDGFR, phospho-β-PDGFR and αSMA upon injury.
(B, C) Δβ-PDGFR mice and control littermates were injected once with CCl4 followed by isolation of HSCs 48 hours thereafter. Primary HSCs were kept in culture for 6 days.
(B) Immunoblot demonstrates phosphorylation of the receptor in control animals upon ligand exposure and lack of receptor activation in HSCs of Δβ-PDGFR animals.
(C) Immunoblot showing decreased HSC activation of Δβ-PDGFR mice in culture compared to wild type β-PDGFR mice, with reduced expression of Collagen I and αSMA. Graph indicating densitometric analysis of each band, verifying significant knock down of β-PDGFR in primary HSCs of Δβ-PDGFR mice and decreased expression of HSC activation markers.
Data represent the mean value of at least 3 separate experiments (*p<0.05, error bars indicate SEM). 3 animals per condition were used in each experiment. Protein ratios (normalized to calnexin) were used to quantify the fold change relative to control, and are shown below each blot.
We next analyzed isolated hepatic stellate cells from β-PDGFRfl/fl Cre negative mice (β-PDGFR) as well as their β-PDGFRfl/fl Cre expressing littermates (Δβ-PDGFR) (Fig. 1B, C). Isolated stellate cells were maintained in primary culture for six days following isolation. To first validate the knock down and diminished activation of β-PDGFR in the Cre expressing population, cell lysates were analyzed via immunoblot after incubation with either vehicle or PDGF-BB (Fig. 1B). Stellate cells from Δβ-PDGFR mice displayed attenuated receptor expression and lack of activation at baseline and after 30 minutes of ligand exposure. Next, the expression of collagen I and αSMA were compared in primary stellate cells of the two groups. Stellate cells isolated and cultured for 6 days displayed significant knock down of the receptor in the Δβ-PDGFR group, with substantially reduced expression levels of collagen I and αSMA compared to stellate cells from β-PDGFR animals (Fig. 1C). β-PDGFR expression was upregulated during hepatic injury and correlates with stellate cell activation in vitro.
Deletion of β-PDGFR in Stellate Cells Attenuates Liver Fibrosis In Vivo
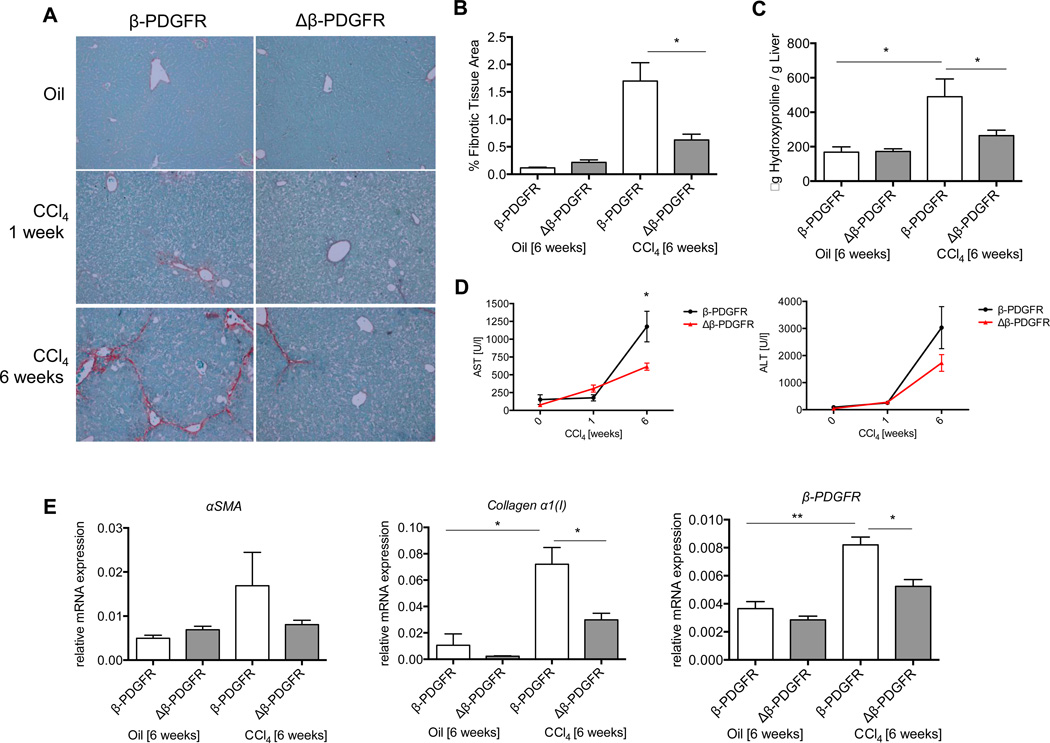
Since mice responded to acute liver injury with up-regulation of β-PDGFR on stellate cells associated with fibrogenic markers, we analyzed control and Δβ-PDGFR animals (Suppl. Fig. 1A) following acute (1 week) and chronic (6 weeks) liver injury. To do so, we injected both groups with either oil or CCl4 (3 i.p. injections weekly).
Macroscopically, there were no differences in the livers between control and Δβ-PDGFR littermates after treatment (Suppl. Fig. 1B, C), but less necrosis was present at the early time point (Suppl. Fig. 2A, B). Although there was a trend in reduced inflammation in the livers of Δβ-PDGFR mice after one week of injections (Suppl. Fig. 2B), comprehensive flow cytometry did not confirm any differences in the numbers or subsets of lymphocytes at baseline or after injury between wild type and Δβ-PDGFR littermates (data not shown). β-PDGFR-deficiency led to significantly decreased collagen deposition after chronic hepatic injury as assessed by Sirius Red morphometry and hydroxyproline assay (Fig. 2 A, B, C). Serum transaminase levels (AST and ALT) were increased, especially after 6 weeks (Fig. 2D). As expected, livers with β-PDGFR deletion in stellate cells expressed less αSMA and significantly less collagen α1(I) mRNAs (Fig. 2E). In order to validate the effect of diminished β-PDGFR expression on the expansion of activated HSCs following CCl4 treatment, we quantified the αSMA-positive tissue area in both groups (Suppl. Fig. 3A). Both after a short-term, and more strikingly, after long-term injury, mice of the Δβ-PDGFR group had reduced expansion of activated HSCs versus controls. This finding was further validated using whole liver lysates of livers after 6 weeks of CCl4 treatment (Suppl. Fig. 3B). Here, the decreased expression of β-PDGFR resulted in a diminished expression of αSMA. As an additional marker for HSCs and to measure the expansion of HSCs during injury within the liver, desmin staining was performed on tissue sections following 1 week of CCl4 treatment (Suppl. Fig. 4). In these mice, there was a significant decrease in desmin-positive tissue quantification in Δβ-PDGFR mice versus controls.
Fig. 2. Loss of β-PDGFR on HSCs leads to decreased collagen deposition in vivo.
(A–E) Δβ-PDGFR and control mice were injected with CCl4 over either one or six weeks to induce acute or chronic liver injury.
(A) Sirius Red staining of paraffin embedded liver sections following acute or chronic liver injury depicts significantly lower collagen deposition after chronic injury (magnification 200×).
(B) Graph displays the percentage of liver area positive for Sirius Red staining measured by morphometric analysis. The area of fibrotic tissue is significantly reduced within Δβ-PDGFR animals compared to controls.
(C) Measurement of hydroxyproline content per gram of whole liver after 6 weeks of CCl4 reflects reduced hydroxyproline content in livers of Δβ-PDGFR versus controls.
(D) Levels of serum AST and ALT during acute and chronic injury.
(E) Whole liver mRNA expression of Collagen α1(I), αSMA, β-PDGFR after 6 weeks of CCl4 treatment confirm increased expression of stellate cell activation genes within control animals upon injury, as well as lack of increase within the Δβ-PDGFR group.
All figures represent the mean of at least n=3 animals per experimental group. mRNA is expressed normalized to Gapdh (*p < 0.05, **p< 0.001; error bars indicate SEM).
Similar but less striking results were apparent in the bile duct ligation model (see Suppl. Fig. 5).
Loss of β-PDGFR in Stellate Cells Diminishes Their Proliferative Response In Vivo
To determine whether increased fibrosis upon hepatic injury was due to β-PDGFR mediated stellate cell proliferation, we performed in vivo labeling of dividing stellate cells. Mice of both groups received a total of three i.p. injections of CCl4 every 48 hours; 44 hours after the last injection, mice received a single i.p. injection of BrdU (1.5mg/150µl PBS), and cells were isolated four hours thereafter. Isolated cells were stained for CD45 and BrdU and then analyzed by flow cytometry. UV autofluorescence was used to distinguish stellate cells from non-fluorescent cells, while the expression of CD45 was used to discriminate CD45− stellate cells from other non-parenchymal cells (Suppl. Fig. 6A). Based on this analysis, 44.3% of stellate cells from β-PDGFR mice had incorporated nuclear BrdU compared to 16.3% in Δβ-PDGFR cells (Suppl. Fig. 6B), indicating that β-PDGFR expression of stellate cells correlates with significantly increased proliferation during hepatic injury.
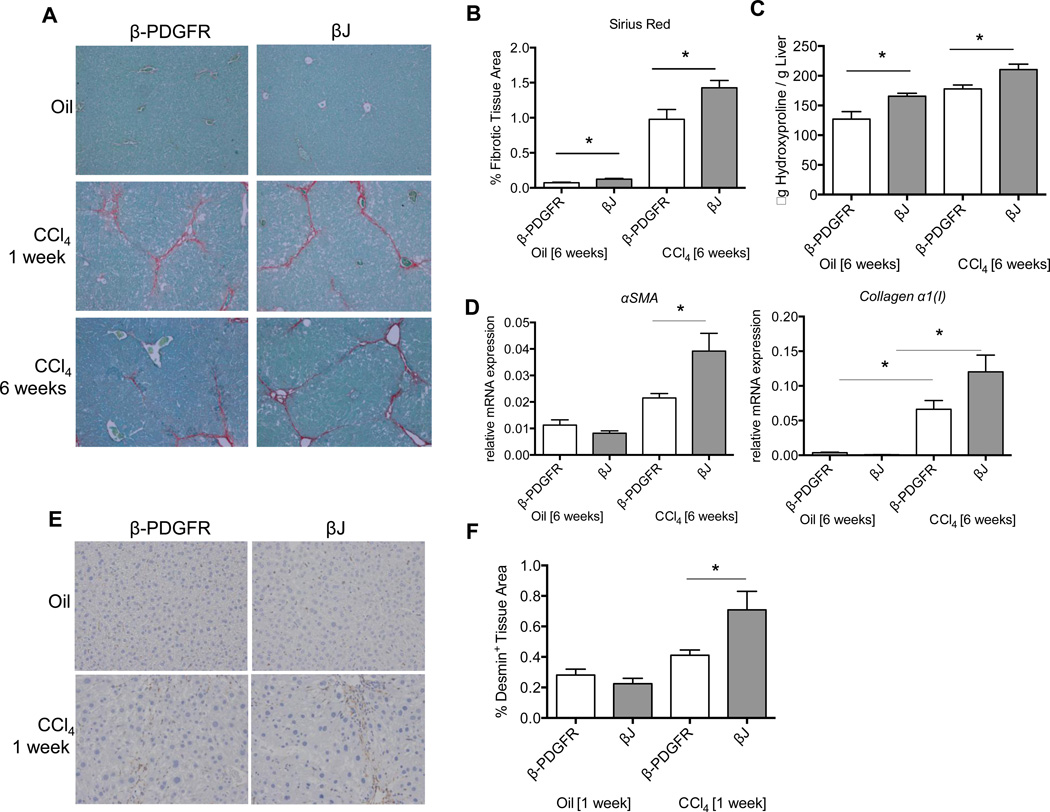
Constitutively Activated β-PDGFR in Stellate Cells Amplifies Liver Fibrosis In Vivo
To confirm the vital role of β-PDGFR during hepatic fibrosis, we analyzed a mouse line in which the expression of a β-PDGFR auto-activating mutant was induced in hepatic stellate cells by crossing β–PDGFRbetaJ/+ mice with GFAP-Cre transgenic animals (Suppl. Fig. 7A). We compared PDGFRbetaJ/+ Cre negative (β-PDGFR) and their PDGFRbetaJ/+ Cre positive (βJ) littermates following three weekly i.p. injections of either corn oil or CCl4 (0.5 µl/g body weight) over one (acute) or six (chronic) weeks (Fig. 3). The auto-activating βJ group displayed differences in fibrotic tissue area versus control animals upon oil injections (Fig. 3A, B, C). Differences were also significant following 6 weeks of CCl4, with βJ mice displaying significantly increased collagen deposition (Fig. 3A, B, C). Consistent with these findings, mutant βJ expression on stellate cells led to significantly more collagen α1(I) and αSMA mRNA expression upon CCl4 treatment (Fig. 3D).
Fig. 3. The ‘βJ’ constitutively activating mutant of β-PDGFR on stellate cells leads to increased collagen deposition upon injury in vivo.
(A–D) βJ and control mice were injected with CCl4 over either one or six weeks to induce acute or chronic liver injury.
(A) Sirius Red staining of paraffin embedded liver sections following acute or chronic liver injury depicts significantly higher collagen deposition after acute and chronic injury (magnification 200×).
(B) Graph shows the percentage of liver area positive for Sirius Red staining measured by morphometry. The area of fibrotic tissue is significantly increased in livers of βJ animals compared to controls.
(C) Increased hydroxyproline content per gram of whole liver after 6 weeks of CCl4 in livers of βJ mice compared to controls.
(D) Whole liver mRNA expression of αSMA and Collagen α1(I) after 6 weeks of CCl4 treatment confirms increased expression of stellate cell activation genes in βJ animals upon liver injury.
(E) Desmin staining of paraffin embedded liver sections following acute liver injury depicts increased stellate cell expansion within livers of βJ mice versus controls.
(F) Graph shows percentage of tissue area positive for desmin measured by morphometry.
All figures represent the mean of at least n=5 animals per experimental group. mRNA is expressed normalized to Gapdh (*p < 0.05, **p< 0.001; error bars indicate SEM).
There were no macroscopic differences between control and mutant βJ littermates before or after treatment (Suppl. Fig. 7B) but there was an increase in liver to body weight ratio in both treatment groups after 1 and 6 weeks (Suppl. Fig. 7C). Serum transaminase levels increased proportionate to treatment duration, without differences between β-PDGFR and βJ littermates (Suppl. Fig. 7D). Both groups displayed increased inflammation and necrosis (Suppl. Fig. 8A, B).
We quantified the desmin-positive area, as a reflection of stellate cell expansion (Fig. 3E). Using this approach, mice with auto-activating β-PDGFR displayed increased desmin-positive tissue area upon acute injury (Fig. 3F). The auto-activating mutation of β-PDGFR led to increased stellate cell proliferation and hepatic fibrosis upon acute and chronic injury.
β-PDGFR Deficiency in Stellate Cells has no Impact on the Carcinogenic Response
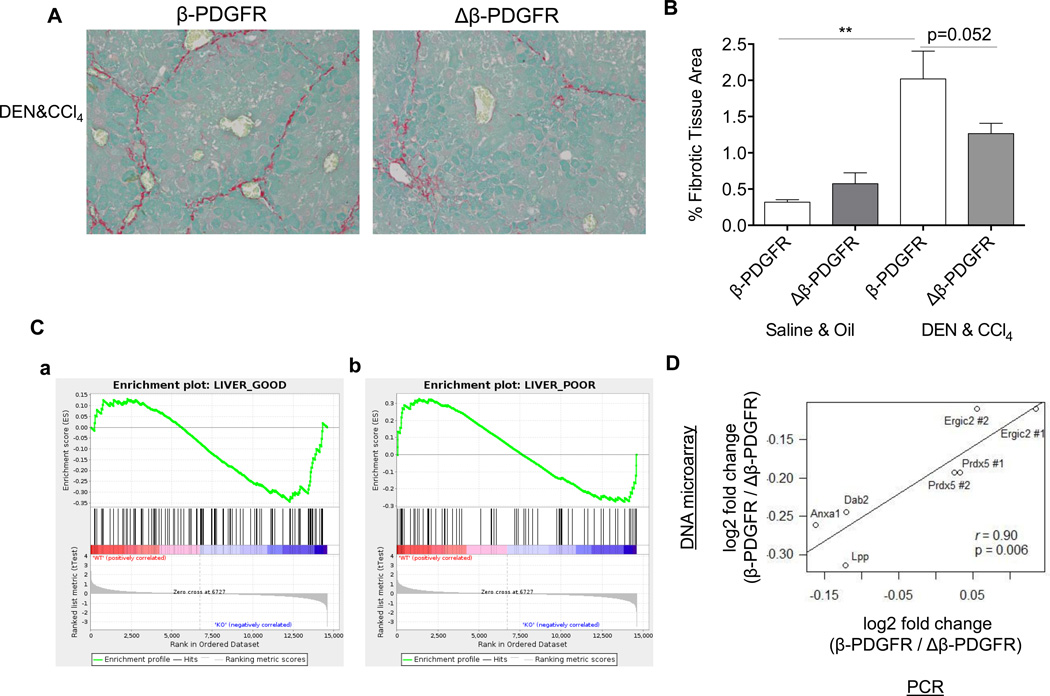
Since the majority of HCCs develop within cirrhotic livers, we assessed the contribution of β-PDGFR signaling in stellate cells to the development of dysplastic nodules following DEN plus CCl4 (Suppl. Fig. 9A) [17]. This regimen reportedly mimics the permissive environment from which regenerative nodules and ultimately HCCs arise [12, 17].
After long-term treatment with both a carcinogen as well as CCl4, Δβ-PDGFR accumulated less collagen as assessed by Sirius Red morphometry (Fig. 4A, B), associated with reduced collagen α1(I) mRNA (Suppl. Fig. 9G). However, loss of Δβ-PDGFR had no impact on the total number of lesions, the maximal nodule size, or the liver weight to body weight ratio (Suppl. Fig. 9B, C, D, E). Of note, careful expert analysis revealed the lesions to be dysplastic nodules, not true HCCs. Histologically, liver sections showed a similar appearance of dysplastic nodules between both β-PDGFR and Δβ-PDGFR mice as assessed by a blinded pathologist (Suppl. Fig. 9F).
Fig. 4. Deletion of β-PDGFR reduces fibrosis but not tumor burden in mice treated with DEN and chronic CCl4, and is linked to better outcome in patients with cirrhosis.
Mice were treated with a single dose of DEN at day 15, followed by weekly injections with CCl4 and sacrificed 48h after the last of 22 injections with CCl4.
(A) Paraffin embedded liver sections were stained with Sirius Red to assess fibrotic tissue content (magnification 200×).
(B) Decreased collagen area measured by morphometry in livers of Δβ-PDGFR mice. The bar graph represents the mean of n=5 animals per experimental group. (**p< 0.001; error bars indicate SEM).
(C) GSEA plots demonstrate β-PDGFR-dependent association with gene signatures for either a good or poor prognosis in an HCV cirrhotic patient cohort.
Gene array samples of primary hepatic stellate cells isolated from either β-PDGFRfl/fl GFAP-Cre negative (indicated as WT) or β-PDGFRfl/fl GFAP-Cre positive (indicated as KO) mice were correlated with gene signatures for good or poor overall prognosis of a human HCV cirrhosis cohort.
(a) Association of the β-PGDFR-knockout gene signature with a good outcome in liver cirrhosis. Enrichment of the β-PGDFR-knockout gene signature was evaluated in association with the risk of overall mortality in 216 HCV-related cirrhosis patients with early-stage cirrhosis (n=216). NES=−1.14, nominal p=0.21, FDR=0.22.
(b) Association of the β-PGDFR gene signature with a poor outcome in HCV cirrhosis. Enrichment of the β-PGDFR gene signature was evaluated in association with the risk of overall mortality in 216 HCV-related cirrhosis patients with early-stage cirrhosis (n=216). NES=1.11, nominal p=0.25, FDR=0.23.
Genes were evaluated using GSEA.
(D) Comparison of β-PDGFR knockout-mediated differential gene expression between DNA microarray and RT-qPCR was performed choosing five of the top differentially expressed genes (Anxa1, Dab2, Ergic2, Lpp, and Prdx5) between wild type and β-PDGFR-knockout mice as selected from DNA microarray data. To verify the differential expression in RT-qPCR, 7 pairs of primers (2 pairs for Ergic and Prdx5) were designed and the same RNA aliquots were assayed in triplicate.
These data indicate that the lack of β-PDGFR expression by stellate cells attenuates fibrosis progression in chronic injury from CCl4, but does not protect against the development of dysplastic nodules when combined with a carcinogen.
Constitutively Activation of β-PDGFR in Stellate Cells does not lead to an Increased Carcinogenic Response
To further reflect these findings, we used βJ mice and their control littermates to assess their response towards the combined treatment with DEN and CCl4 (Suppl. Fig. 10). Macroscopically, there was no difference in overall quantification of dysplastic nodule formation (Suppl. Fig. 10), regarding tumor number, the largest tumor diameter and overall liver weight to body weight ratio (Suppl. Fig. 10B, C, D, E). Histologically, the grade of dysplasia was not different among βJ and control mice (Suppl. Fig. 10F). Overall, the activation level of β-PDGFR did not correlate with formation of dysplastic nodules in this murine model.
Prognostic Relevance of β-PDGFR-mediated Signaling in Human Cirrhosis
We previously identified and validated a 186-gene signature in liver that predicts prognosis of patients with liver cirrhosis [22, 23]. The signature is assumed to reflect signals of molecular deregulation from multiple cell types in the cirrhotic tissue microenvironment that drive disease progression. We hypothesized that activation of β-PDGFR signaling in stellate cells contributes at least a part of the prognostic gene signature. Using this dataset, we examined whether the stellate cell-derived β-PDGFR or Δβ-PDGFR gene signature was present in the human cirrhosis cohort and determined its prognostic association. To do so, we performed Gene set enrichment analysis (GSEA) to evaluate induction of the 186-gene signature in isolated stellate cells from our β-PDGFR-wild-type and knockout mice. Of note, we observed a statistically significant enrichment of poor-prognosis- and good-prognosis-correlated signature genes in the wild-type and knockout stellate cells, respectively (false discovery rate <0.25) (Fig. 4C a, b). Genes up-regulated in the β-PDGFR knockout cells (i.e., genes suppressed by β-PDGFR pathway activation) (Supplementary Table 2), were associated with a good prognosis (Fig. 4C a). Likewise, genes up-regulated by β-PDGFR activation showed an association with a high overall risk of mortality in HCV cirrhotic patients (Fig. 4C b), suggesting that the pathway activation itself is partially linked to the risk of liver disease and fibrosis progression. Genes involved in inflammation and cell survival such as NFKB2, IER3, IFI30, and BCL2 contributed to the enrichment within the poor prognosis signature (Table 1). However, genes previously implicated in hepatocarcinogenesis such as EGF [24] were not involved in the enrichment, suggesting that β-PDGFR signaling in stellate cells is not directly involved in the process of hepatocarcinogenesis. We verified the expression levels of the top five differentially regulated genes of β-PDGFR-wild-type and knockout mice as selected by DNA microarray using quantitative real-time PCR (Fig. 4D). These data provide evidence for a strong correlation between microarray data as well as PCR from hepatic stellate cells isolated from mice of both groups. For Ingenuity Pathway Analysis of the 186-gene signature within hepatic stellate cells of β-PDGFR control compared to knockout mice, see Suppl. Table 2. Collectively, these results suggest that activation of β-PDGFR signaling in stellate cells has downstream consequences that drive disease progression and poor prognosis of human cirrhosis, but not through increased risk of HCC development.
Table 1.
Induction of 186-gene human prognostic gene signature in primary stellate cells from wild-type and β-Pdgfr knockout mice.
| a) Poor-prognosis-associated genes induced in wild-type mouse stellate cells | ||
|---|---|---|
| Human gene symbol | Mouse gene symbol | t-statistic |
| DAB2 | Dab2 | 1,35 |
| LPP | Lpp | 1,28 |
| ANXA1 | Anxa1 | 1,28 |
| NFKB2 | Nfkb2 | 0,96 |
| EPM2AIP1 | Epm2aip1 | 0,90 |
| SERPINB2 | Serpinb2 | 0,90 |
| ITGA9 | Itga9 | 0,83 |
| IQGAP1 | Iqgap1 | 0,81 |
| IER3 | Ier3 | 0,74 |
| CCDC6 | Ccdc6 | 0,54 |
| COL4A1 | Col4a1 | 0,53 |
| IFI30 | Ifi30 | 0,46 |
| ANXA3 | Anxa3 | 0,44 |
| BCL2 | Bcl2 | 0,40 |
| SERPINB8 | Serpinb8 | 0,37 |
| b) Good-prognosis-associated genes induced in Pdgfrb-knockout mouse stellate cells | ||
|---|---|---|
| Human gene symbol | Mouse gene symbol | t-statistic |
| DAD1 | Dad1 | −0,30 |
| VPS41 | Vps41 | −0,30 |
| ERCC5 | Ercc5 | −0,32 |
| RRM1 | Rrm1 | −0,32 |
| ACOT2 | Acot2 | −0,35 |
| GCGR | Gcgr | −0,38 |
| RFC2 | Rfc2 | −0,40 |
| TIMM8A | Timm8a1 | −0,40 |
| TXN2 | Txn2 | −0,51 |
| GGCX | Ggcx | −0,53 |
| ZNF185 | Zfp185 | −0,54 |
| HSPE1 | Hspe1 | −0,55 |
| FAM129A | Fam129a | −0,56 |
| F9 | F9 | −0,58 |
| PSMB3 | Psmb3 | −0,60 |
| MSH6 | Msh6 | −0,61 |
| XPA | Xpa | −0,67 |
| ZER1 | Zer1 | −0,69 |
| GHR | Ghr | −0,80 |
| ATP5D | Atp5d | −0,88 |
| PMM1 | Pmm1 | −0,90 |
| NENF | Nenf | −0,94 |
t-statistic indicates degree of differential expression between wild-type and Pdgfrb-knockout mouse stellate cells. Positive values indicate up-regulation in wild-type and negative values indicate up-regulation in knockout.
Discussion
In the current study, we established two divergent genetic mouse models, by using a knockout or auto-activation of β-PDGFR, to assess the impact of titrating β-PDGFR expression on stellate cell and its contribution to liver injury and fibrosis. Deleting β-PDGFR on hepatic stellate cells impairs their fibrogenic potential in vivo, leading to decreased expression of αSMA and collagen α1(I), and reducing their proliferation upon injury. Although previous studies have not directly linked β-PDGFR to collagen expression, here we demonstrate that the lack of β-PDGFR on primary hepatic stellate cells leads to decreased expression of collagen I after treatment with PDGF-B. Our data establish that β-PDGFR activation increases fibrosis accumulation at least in part through increased stellate cell numbers based on BrdU incorporation in stellate cells in vivo and subsequent FACS analysis. We also emphasize that while specificity of human GFAP-Cre expression has been questioned [25], both this study and separate reports [12, 13] underscore its utility to drive stellate cell cre expression. Moreover, it has been shown that β-PDGFR is only expressed in activated hepatic stellate cells and some vascular endothelial cells in liver thereby minimizing potential hepatic off-target effects of the GFAP-Cre expression since only stellate cells will be affected by deletion of β-PDGFR. One major issue is that differences in the recombination of different floxed alleles or Cre reporters cannot be ruled out completely.
Our data begin to define nuanced and very specific links between some pathways of stellate cell activation, and prognosis of cirrhosis or incidence of HCC, but not others. For example, recent studies strongly implicate EGFR signaling and clinical outcomes in human HCC [24, 26, 27], yet in contrast to a previous report [28] our findings do not establish a similar link for β-PDGFR signaling, despite its clear contribution to fibrosis. EGF was not among the 14 genes that lead to an enrichment of the phenotype for poor prognosis in GSEA downstream of β-PDGFR signaling (Table 1, Suppl. Table 2). Although sorafenib, which is the only approved molecular therapy for HCC, targets multiple receptor tyrosine kinases, including β-PDGFR, our data suggest that the antineoplastic benefit of sorafenib may not be due to its impact on β-PDGFR signaling alone, but rather on other RTK pathways, especially Ras/Raf kinase, HGF and VEGF signaling [29]. We also emphasize that expert histologic assessment failed to confirm the presence of true HCCs but rather only preneoplastic, dysplastic nodules, undermining the potential value of this model in generating true HCCs, in contrast to what has been previously proposed [28].
β-PDGFR activity in stellate cells clearly contributes to progressive disease during chronic liver injury, consistent with the finding that the β-PDGFR gene signature in primary hepatic stellate cells correlates with poor overall outcome in our human cohort. However, the β-PDGFR-related difference in human outcomes is not due to enhanced tumor development, consistent with the findings in our murine model. Thus, β-PDGFR signaling may contribute to poor outcomes in HCV cirrhosis through its impact on fibrosis but not on carcinogenesis. These findings indicate that while β-PDGFR is an attractive anti-fibrotic target, it may not be a suitable direct target to inhibit the development of neoplasia in cirrhotic liver, in contrast to other RTK pathways, especially EGFR signaling [30], where prophylactic receptor antagonism is a viable strategy for reducing the risk of cancer more directly.
Supplementary Material
Acknowledgements
The authors thank Dr. Marie Berres (Oncological Sciences, Icahn School of Medicine at Mount Sinai) and Hsin Chou for technical support. RNA seq and DNA microarray experiments were performed by the Genomics Core, preprocessing was performed by Dr. Ke Hao and Dr. Antonio Fabio Di Narzo (Genetics and Genomic Science, Icahn School of Medicine at Mount Sinai).
Financial Support
This work has been supported by grants from the Deutsche Forschungsgemeinschaft (DFG) (KO 4086/1-1 to P.K, LE 2794/1-1 to Y.A.L.), The Levine Family Foundation (to A. D.), as well as the NIH DK56621 and AA020709 (to S.L.F.), and the NIH DK099558 and European Commission 7th Framework Programme FP7-Health 2010 (Heptromic) (to Y.H.).
Abbreviations
- β-PDGFR
beta platelet-derived growth factor receptor
- HCV
hepatitis C virus
- CCl4
carbon tetrachloride
- DEN
diethyl nitrosamine
- GFAP
glial fibrillary acidic protein
- BDL
bile duct ligation
- bw
body weight
- AST
aspartate aminotransferase
- ALT
alanine aminotransferase
- mRNA
messenger ribonucleic acid
- PCR
polymerase chain reaction
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- αSMA
alpha smooth muscle actin
- H&E
hematoxylin and eosin
- BrdU
5-bromo-2'-deoxyuridine
- CD45
cluster of differentiation 45
- UV
ultraviolet
- PBS
phosphate buffered saline
- HCC
hepatocellular carcinoma
- IL1R
interleukin 1 receptor
- GSEA
gene set enrichment analysis
- FACS
fluorescence-activated cell sorting
- VEGF
vascular endothelial growth factor
- HGF
hepatocyte growth factor
- RTK
receptor tyrosine kinase
- EGFR
epidermal growth factor receptor
- NES
normalized enrichment score
- FDR
false discovery rate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest:
The authors disclose no conflicts.
Author contributions
P.K. co-designed research plan, performed experiments, analyzed data, generated figures and drafted manuscript, A.L. and Y.A.L. performed experiments, analyzed data and performed validation of gene array results, A.D. performed flow-cytometric analysis, X.S. performed statistical analysis on data generated through gene array, M.I.F.: performed histological grading, S.T. performed histological grading, C.A. designed and supervised flow-cytometric analysis, P.S. generated original genetic mouse models, designed experiments, interpreted data, Y.H. provided statistical analysis and advice on gene array data, performed bioinformatic analysis and interpretation of data, S.L.F. co-designed research plan, reviewed and edited manuscript
Contributor Information
Peri Kocabayoglu, Email: peri.kocabayoglu@uk-essen.de.
Abigale Lade, Email: Abigale.lade@mssm.edu.
Youngmin A. Lee, Email: youngmin.lee@mssm.edu.
Ana-Cristina Dragomir, Email: anacristinadragomir@yahoo.com.
Xiaochen Sun, Email: xiaochen.sun@mssm.edu.
M. Isabel Fiel, Email: mariaisabel.fiel@mountsinai.org.
Swan Thung, Email: swan.thung@mountsinai.org.
Costica Aloman, Email: aloman@uic.edu.
Philippe Soriano, Email: philippe.soriano@mssm.edu.
Yujin Hoshida, Email: yujin.hoshida@mssm.edu.
Scott L. Friedman, Email: scott.friedman@mssm.edu.
References
- 1.Pinzani M. PDGF and signal transduction in hepatic stellate cells. Front Biosci. 2002;7:d1720–d1726. doi: 10.2741/A875. [DOI] [PubMed] [Google Scholar]
- 2.Pinzani M, Milani S, Herbst H, DeFranco R, Grappone C, Gentilini A, et al. Expression of platelet-derived growth factor and its receptors in normal human liver and during active hepatic fibrogenesis. Am J Pathol. 1996;148(3):785–800. [PMC free article] [PubMed] [Google Scholar]
- 3.Wong L, Yamasaki G, Johnson RJ, Friedman SL. Induction of beta-platelet-derived growth factor receptor in rat hepatic lipocytes during cellular activation in vivo and in culture. J Clin Invest. 1994;94(4):1563–1569. doi: 10.1172/JCI117497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seifert RA, Hart CE, Phillips PE, Forstrom JW, Ross R, Murray MJ, et al. Two different subunits associate to create isoform-specific platelet-derived growth factor receptors. J Biol Chem. 1989;264(15):8771–8778. [PubMed] [Google Scholar]
- 5.Donovan J, Abraham D, Norman J. Platelet-derived growth factor signaling in mesenchymal cells. Front Biosci (Landmark Ed) 2013;18:106–119. doi: 10.2741/4090. [DOI] [PubMed] [Google Scholar]
- 6.Borkham-Kamphorst E, Kovalenko E, van Roeyen CR, Gassler N, Bomble M, Ostendorf T, et al. Platelet-derived growth factor isoform expression in carbon tetrachloride-induced chronic liver injury. Lab Invest. 2008;88(10):1090–1100. doi: 10.1038/labinvest.2008.71. [DOI] [PubMed] [Google Scholar]
- 7.Reichenbach V, Fernandez-Varo G, Casals G, Oro D, Ros J, Melgar-Lesmes P, et al. Adenoviral dominant-negative soluble PDGFRbeta improves hepatic collagen, systemic hemodynamics, and portal pressure in fibrotic rats. J Hepatol. 2012;57(5):967–973. doi: 10.1016/j.jhep.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 8.Kim Y, Fiel MI, Albanis E, Chou HI, Zhang W, Khitrov G, et al. Anti-fibrotic activity and enhanced interleukin-6 production by hepatic stellate cells in response to imatinib mesylate. Liver Int. 2012;32(6):1008–1017. doi: 10.1111/j.1478-3231.2012.02806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kocabayoglu P, Friedman SL. Cellular basis of hepatic fibrosis and its role in inflammation and cancer. Front Biosci (Schol Ed) 2013;5:217–230. doi: 10.2741/s368. [DOI] [PubMed] [Google Scholar]
- 10.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359(4):378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 11.Schmahl J, Rizzolo K, Soriano P. The PDGF signaling pathway controls multiple steroid-producing lineages. Genes Dev. 2008;22(23):3255–3267. doi: 10.1101/gad.1723908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lujambio A, Akkari L, Simon J, Grace D, Tschaharganeh DF, Bolden JE, et al. Non-cell-autonomous tumor suppression by p53. Cell. 2013;153(2):449–460. doi: 10.1016/j.cell.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hernandez-Gea V, Ghiassi-Nejad Z, Rozenfeld R, Gordon R, Fiel MI, Yue Z, et al. Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology. 2012;142(4):938–946. doi: 10.1053/j.gastro.2011.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olson LE, Soriano P. PDGFRbeta signaling regulates mural cell plasticity and inhibits fat development. Dev Cell. 2011;20(6):815–826. doi: 10.1016/j.devcel.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kountouras J, Billing BH, Scheuer PJ. Prolonged bile duct obstruction: a new experimental model for cirrhosis in the rat. Br J Exp Pathol. 1984;65(3):305–311. [PMC free article] [PubMed] [Google Scholar]
- 16.Proctor E, Chatamra K. High yield micronodular cirrhosis in the rat. Gastroenterology. 1982;83(6):1183–1190. [PubMed] [Google Scholar]
- 17.Dapito DH, Mencin A, Gwak GY, Pradere JP, Jang MK, Mederacke I, et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell. 2012;21(4):504–516. doi: 10.1016/j.ccr.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blomhoff R, Berg T. Isolation and cultivation of rat liver stellate cells. Methods Enzymol. 1990;190:58–71. doi: 10.1016/0076-6879(90)90009-p. [DOI] [PubMed] [Google Scholar]
- 19.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38(5):500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 20.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genomewide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang L, Jung Y, Omenetti A, Witek RP, Choi S, Vandongen HM, et al. Fate-mapping evidence that hepatic stellate cells are epithelial progenitors in adult mouse livers. Stem Cells. 2008;26(8):2104–2113. doi: 10.1634/stemcells.2008-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoshida Y, Villanueva A, Sangiovanni A, Sole M, Hur C, Andersson KL, et al. Prognostic gene expression signature for patients with hepatitis C-related early-stage cirrhosis. Gastroenterology. 2013;144(5):1024–1030. doi: 10.1053/j.gastro.2013.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoshida Y, Villanueva A, Kobayashi M, Peix J, Chiang DY, Camargo A, et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med. 2008;359(19):1995–2004. doi: 10.1056/NEJMoa0804525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abu Dayyeh BK, Yang M, Fuchs BC, Karl DL, Yamada S, Sninsky JJ, et al. A functional polymorphism in the epidermal growth factor gene is associated with risk for hepatocellular carcinoma. Gastroenterology. 2011;141(1):141–149. doi: 10.1053/j.gastro.2011.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mederacke I, Hsu CC, Troeger JS, Huebener P, Mu X, Dapito DH, et al. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat Commun. 2013;4:2823. doi: 10.1038/ncomms3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong JH, You XM, Gong WF, Ma L, Zhang Y, Mo QG, et al. Epidermal growth factor gene polymorphism and risk of hepatocellular carcinoma: a meta-analysis. PLoS One. 2012;7(3):e32159. doi: 10.1371/journal.pone.0032159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blivet-Van Eggelpoel MJ, Chettouh H, Fartoux L, Aoudjehane L, Barbu V, Rey C, et al. Epidermal growth factor receptor and HER-3 restrict cell response to sorafenib in hepatocellular carcinoma cells. J Hepatol. 2012;57(1):108–115. doi: 10.1016/j.jhep.2012.02.019. [DOI] [PubMed] [Google Scholar]
- 28.Maass T, Thieringer FR, Mann A, Longerich T, Schirmacher P, Strand D, et al. Liver specific overexpression of platelet-derived growth factor-B accelerates liver cancer development in chemically induced liver carcinogenesis. Int J Cancer. 2011;128(6):1259–1268. doi: 10.1002/ijc.25469. [DOI] [PubMed] [Google Scholar]
- 29.Wilhelm SM, Adnane L, Newell P, Villanueva A, Llovet JM, Lynch M. Preclinical overview of sorafenib, a multikinase inhibitor that targets both Raf and VEGF and PDGF receptor tyrosine kinase signaling. Mol Cancer Ther. 2008;7(10):3129–3140. doi: 10.1158/1535-7163.MCT-08-0013. [DOI] [PubMed] [Google Scholar]
- 30.Fuchs BC, Hoshida Y, Fujii T, Wei L, Yamada S, Lauwers GY, et al. Epidermal growth factor receptor inhibition attenuates liver fibrosis and development of hepatocellular carcinoma. Hepatology. 2014;59(4):1577–1590. doi: 10.1002/hep.26898. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.