Abstract
Facioscapulohumeral muscular dystrophy (FSHD) is most often associated with variegated expression in somatic cells of the normally repressed DUX4 gene within the D4Z4 repeat array. The most common form, FSHD1, is caused by a D4Z4 repeat array contraction to a size of 1-10 units (normal range 10–100 units). The less common form, FSHD2, is characterized by D4Z4 CpG hypomethylation and is most often caused by loss of function mutations in the structural maintenance of chromosomes hinge domain 1 (SMCHD1) gene on chromosome 18p. The chromatin modifier SMCHD1 is necessary to maintain a repressed D4Z4 chromatin state. Here we describe two FSHD2 families with a 1.2 Mb deletion encompassing the SMCHD1 gene. Numerical aberrations of chromosome 18 are relatively common and the majority of 18p deletion syndrome (18p-) cases have, like these FSHD2 families, only one copy of SMCHD1. Our finding therefore raises the possibility that 18p- cases are at risk of developing FSHD. To address this possibility, we combined genome wide array analysis data with D4Z4 CpG methylation and repeat array sizes in individuals with 18p- and conclude that approximately 1:8 18p- cases might be at risk of developing FSHD.
Keywords: FSHD, SMCHD1, epigenetic modifier, D4Z4, epiallele
Individuals with facioscapulohumeral muscular dystrophy (FSHD; MIM# 158900 [FSHD1] and MIM# 158901 [FSHD2]) have progressive and often asymmetric muscle weakness affecting the face, shoulder, and upper arms, followed by involvement of the distal and proximal lower extremities (Padberg, 1982; Tawil and van der Maarel, 2006). These symptoms are associated with a higher probability of skeletal muscle expression of the DUX4 retrogene (MIM# 606009) which is normally repressed in somatic tissue (Snider et al., 2010). There are at least two genetically distinct causes of FSHD that are clinically indistinguishable. The most common form, FSHD1, is caused by a variant of the polymorphic D4Z4 macrosatellite repeat array on chromosome 4 that is between 1 and 10 units (Wijmenga et al., 1992). In unaffected individuals this polymorphic repeat array ranges between 11-100 D4Z4 units and is heavily CpG methylated in somatic tissue thereby repressing expression of the region (van Overveld et al., 2003). The shorter array in individuals with FSHD1 is insufficient to maintain the repressed chromatin state necessary for sustaining repression of the DUX4 retrogene expression. The less common form, FSHD2, is also characterized by a partial loss of CpG methylation at D4Z4 but with repeat array sizes > 10 units and is usually caused by mutations in the structural maintenance of chromosomes hinge domain 1 gene (SMCHD1; MIM# 614982) on chromosome 18p (Lemmers et al., 2012; van Overveld et al., 2003; Larsen et al., 2014). SMCHD1 is a chromatin modifier that binds to the D4Z4 repeat array and is necessary to maintain a repressed D4Z4 chromatin state in somatic cells. Each of 3,3kb large D4Z4 units contains a copy of the double homeobox 4 (DUX4) retrogene (Gabriels et al., 1999). In FSHD1, with the short D4Z4 repeat array, or in FSHD2 with heterozygous SMCHD1 mutations, either cause leads to a partial derepression of the D4Z4 repeat array in somatic cells. This derepression is marked by D4Z4 hypomethylation, a partial loss of repressive chromatin marks and other chromatin changes and the sporadic expression of DUX4 in a small set of muscle nuclei (Balog et al., 2012; Geng et al., 2012; van Overveld et al., 2003; Zeng et al., 2009).
However, either mechanism; D4Z4 short repeat sizes, or SMCHD1 mutations, leading to depression of D4Z4 by themselves are not sufficient to cause FSHD. The D4Z4-embedded DUX4 retrogene requires a polyadenylation signal in the 3′ untranscribed region to produce a stable transcript and hence DUX4 protein in somatic cells (Lemmers et al., 2010a). This DUX4 polyadenylation signal can only be found immediately distal to the last DUX4 copy, but its existence is polymorphic in the population and is only found on 4qA-type chromosomes. 4qB chromosomes, that are equally common, or 10q chromosomes that contain a highly homologous copy of the D4Z4 repeat array, do not have a DUX4 polyadenylation signal (Lemmers et al., 2002; Lemmers et al., 2004; Lemmers et al., 2010a). Thus, digenic inheritance of a short D4Z4 repeat array in cis with a DUX4 polyadenlylation signal in FSHD1, or a heterozygous SMCHD1 mutation and a DUX4 polyadenylation signal in FSHD2, are both mechanisms for the development of FSHD (Lemmers et al., 2010a; Lemmers et al., 2012).
We previously reported that the average repeat array size of the 4qA (i.e. DUX4 producing) chromosome in FSHD2 patients is most often between 11–16 units, which is at the lower end of the repeat size spectrum of 4qA chromosomes in control individuals (Supp. Figure S1) (de Greef et al., 2010; Lemmers et al., 2014). We define FSHD2 patients by a methylation level of <25% measured by Southern blotting at the methylation-sensitive FseI restriction site in D4Z4, a threshold that has been instrumental in identifying SMCHD1 as FSHD2 disease gene (Lemmers et al., 2012; Lemmers et al., 2014; Sacconi et al., 2013). In our recent SMCHD1 mutation screen in affected individuals with FseI methylation levels <25%, we showed that mutations that do not disrupt the open reading frame (ORF) are more deleterious than those that introduce a premature stop codon (Lemmers et al., 2014). This conclusion was based on the difference in D4Z4 hypomethylation for the two types of mutation. We found a significant correlation between the cumulative number of all D4Z4 repeat units in the arrays on chromosomes 4 and 10 and the average methylation of the D4Z4 repeats in controls and FSHD patients. This correlation allowed us to define the Delta1 value, which is the observed CpG methylation level at this site in D4Z4 minus the predicted methylation based on control individuals. The average Delta1 in controls is about zero and ranges between -13% (5% percentile) and 17% (95% percentile), while in carriers of an SMCHD1 mutation this value is about -30% and ranges between -42% (5% percentile) and -22% (95% percentile) (Lemmers et al., 2014). Therefore, Delta1 value represents the methylation defect corrected for the size of the D4Z4 repeat array, emphasizing the strong effect of SMCHD1 mutations on D4Z4 methylation. We also introduced the Delta2 value, which is the observed methylation minus the predicted methylation based on SMCHD1 mutation carriers and reveals the significance of the mutation (Lemmers et al., 2014). SMCHD1 mutations that preserve the ORF most probably result in a dominant negative type mechanism and lead to the lowest Delta1 value, and a negative Delta2 value in somatic cells. ORF disrupting SMCHD1 mutations, seem to be associated with nonsense mediated RNA decay suggesting a haploinsufficiency type mechanism and are associated with lesser degrees of D4Z4 CpG hypomethylation resulting in positive Delta2 values. The observation that both the nature of the SMCHD1 mutation and the size of the D4Z4 repeat array determines the D4Z4 methylation and the disease severity in FSHD2, led to a model in which haploinsufficiency-causing mutations require medium short D4Z4 arrays (most often in the range of 11–16 units) to cause FSHD2, while dominant negative SMCHD1 mutations can also cause FSHD2 with larger 4qA array sizes (Supp. Figure S1) (Lemmers et al., 2014). Thus, while establishing FseI methylation and Delta values are not standard operating procedures in diagnostic settings, these values have contributed to our understanding of FSHD pathogenesis.
In approximately 15% of the FSHD2 families, identified by phenotype and D4Z4 hypomethylation, we could not identify a mutation in SMCHD1 (Lemmers et al., 2012; Lemmers et al., 2014). In six out of ten of these families, Sanger sequencing of SMCHD1 in the index case did not reveal a causal variant but showed absence of heterozygous SNPs in all of the 48 exons and flanking sequences. These families were subjected to whole exome sequencing and genome wide array analysis.
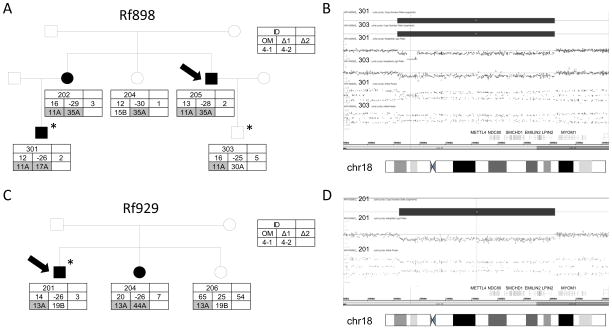
The first family (Rf898) reported in this study came to our attention because of classical features of FSHD, while FSHD1 was excluded by D4Z4 repeat array sizing (Figure 1A and Supp. Table S1). The index case (898.205) was diagnosed at age 49 when he presented with difficulty lifting his arms overhead. In retrospect, he recalls having shoulder difficulties when he was 12 years of age. His physical examination showed a combination of facial weakness, scapular winging and mild foot dorsiflexor weakness as well as mild abdominal muscle weakness. Two additional family members (898.202, 898.301) were moderately affected with similar clinical features. Subject 898.202 was asymptomatic but had definite signs of FSHD with scapular weakness, foot dorsiflexion weakness and mild abdominal muscle weakness when examined at age 65. Her son (898.301) reports symptom onset at age 13 with clear signs of FSHD when examined at age 36 (Supp. Table S1). All carry an 11 units D4Z4 repeat array on a 4qA chromosome containing a DUX4 polyadenylation site. Methylation analysis provided evidence for reduced D4Z4 methylation levels and Delta1 values consistent with FSHD2 in all three affected individuals, but also in two unaffected individuals. Subject 898.303 carries the same 11 unit 4qA allele as his affected father, reports shoulder pain and weakness since the age of 24 but shows no clear signs of weakness on examination at age 36. An EMG study, however, showed myopathic units in his supraspinatus and cervical paraspinal muscles. Individual 898.204 is unaffected at the age of 59, but carries a 35 unit repeat array on a permissive 4qA allele, which appears to be too long to be pathogenic based on previous findings in carriers of a haploinsufficiency-causing SMCHD1 mutation (Lemmers et al., 2014). SMCHD1 mutation analysis by Sanger sequencing failed to identify potentially damaging variants. Whole exome sequencing on 898.301 and 898.303 failed to identify the causative gene, but extended the region of homozygosity into several SMCHD1 neighboring genes, suggestive of a heterozygous deletion of the SMCHD1 locus. Subsequently, we performed a genome wide array analysis in these subjects and identified a 1.2Mb deletion that spans the METTL4, NDC80, SMCHD1, EMILIN2, LPIN2 genes (Figure 1B). This deletion was confirmed in subjects 898.202, 898.204 and 898.205 and this variant was submitted to the Leiden Open Variation Database (http://databases.lovd.nl/shared/variants/SMCHD1). In the second family (Rf929), we identified two FSHD individuals carrying a 13 D4Z4 unit repeat array on 4qA and CpG hypomethylation at D4Z4 suggesting FSHD2 (Figure 1C; Supp. Table S1). The index case’s (929.201) initial complaints were difficulty with his proximal shoulders in his late twenties but was not fully evaluated until age 60 when he presented with facial weakness, shoulder and upper extremity weakness and foot dorsiflexor weakness. One sister (929.204) is also similarly affected whereas his other sister (929.206) is asymptomatic and clinically unaffected. By array analysis in individual 929.201, we identified the same interstitial chromosome 18p deletion including SMCHD1 as was found in family Rf898 (Figure 1D). This 1.2 Mb deletion was confirmed in affected individual 929.204.
Figure 1.
A. Pedigree of family Rf898, showing 3 affected individuals (898.202, 898.205 and 898.301) all carrying an 11 unit D4Z4 repeat array on chromosome 4qA (11A) and a partial loss of D4Z4 CpG methylation. The index case (arrow) and the individuals that have been analyzed on the SNP array (asterisk) are indicated. The key indicates the individual ID, the different methylation values (Delta1; Δ1, Delta2; Δ2 and the measured methylation; OM) and the D4Z4 repeat array type and sizes in units on both chromosomes 4 (4-1 and 4-2) B. Array plots of chromosome 18. A 1.2 Mb deletion in 18p11.32p11.31 was detected in patients 898.301 and 898.303 (arr[hg19] 18p11.32p11.31(1,829,674-3,039,186)x1). The deletion is marked by four panels showing from top to bottom: the Copy Number State, the Weighted Log2 Ratio, the Allele Peaks and the genes.
C. Pedigree of family Rf929, showing 2 affected individuals (929.201 and 929.204) both carrying an 13 unit D4Z4 repeat array on chromosome 4qA and a loss of D4Z4 CpG methylation. D. Array analysis of patient 929.201 presenting a 1.2 Mb deletion in 18p11.32p11.31 (arr[hg19] 18p11.32p11.31(1,844,188-3,039,186)x1)).
Both families reside in the same state in the US and although these two families are not known to be related, the observation that they carry the same interstitial 18p deletion suggest otherwise. Detailed SNP analysis on chromosome 18 showed that the SNP difference between nephews 898.301 and 898.303 is 1.8% and the difference between 898.301 (or 898.303) and 929.201 is 4.9%, while the average difference at chromosome 18 between unrelated individuals is usually >15% (CAL Ruivenkamp, personal communication), strongly suggesting that both families are related. Array analysis in the remaining four FSHD2 families that showed absence of heterozygous SNPs in all 48 SMCHD1 exons and flanking sequences did not reveal evidence for a deletion in the SMCHD1 locus.
Although one third of all identified SMCHD1 mutations in FSHD2 most probably lead to haploinsufficiency, SMCHD1 was not known as a dosage sensitive gene. Our current study emphasizes that monosomy of SMCHD1 can cause familial FSHD2, although we cannot rule out that genetic variants in other (currently unknown) epigenetic modifiers of D4Z4 are shared between both families. We identified two families with five affected family members who all carry the same 1.2Mb 18p microdeletion in combination with a D4Z4 repeat array with 11 or 13 units on a 4qA chromosome. This finding also corroborates our previous study in which we show that haploinsufficiency-causing SMCHD1 mutations in general result in positive Delta2 values, suggestive of a less deleterious outcome (Lemmers et al., 2014). The Delta2 value in the 7 carriers of the SMCHD1 deletion varied between 1 and 7%.
None of the other genes within the microdeletion have been reported to be sensitive to hemizygosity. Homozygous mutations in lipin 2 (LPIN2) have been associated with Majeed syndrome, characterized by chronic recurrent multifocal osteomyelitis and congenital dyserythropoietic anaemia (Ferguson et al., 2005). METTL4 (methyltransferase-like 4) encodes for a putative methyltransferase of unknown function while NDC80 encodes a central component of the kinetochore protein complex necessary for proper chromosome segregation and spindle checkpoint signaling (Martin-Lluesma et al., 2002).
Structural and numerical aberrations of the short arm of chromosome 18 are relatively common amongst the chromosomal rearrangement syndromes. The incidence of monosomy 18p is estimated at about 1:50;000 newborns world wide, and although the dysmorphic features in 18p deletion syndrome (18p-) individuals is not very uniform, muscular dystrophy has not been reported (Turleau, 2008). Given the association of SMCHD1 mutations with FSHD and the identification of two families carrying a deletion of SMCHD1, it raises the question whether individuals with 18p- are at increased risk of developing FSHD when also carrying a chromosome 4q that contains a DUX4 polyadenylation signal.
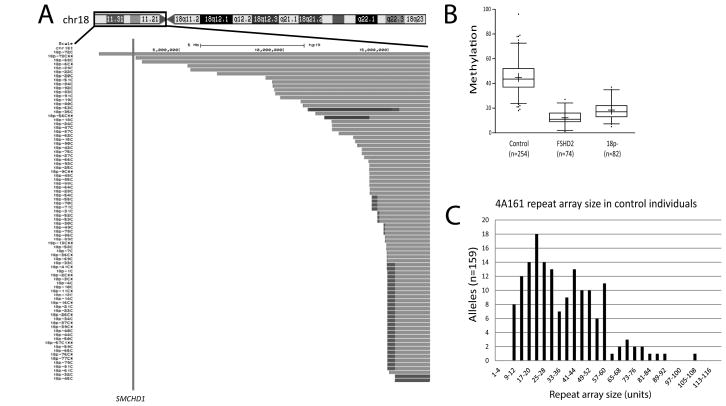
To address this possibility, analysis of CGH array data from 83 individuals with 18p-, showed that SMCHD1 was hemizygous in all but one case (Sebold et al., 2015). Subsequent CpG methylation analysis of the cases with SMCHD1 deletion confirmed a reduced methylation at D4Z4 in 72/82 cases (Figure 2A and B), suggesting that individuals with 18p- have a similar degree of D4Z4 chromatin relaxation as individuals with FSHD2. Previously, we showed that 4qA chromosomes (containing a DUX4 polyadenylation signal) and 4qB chromosomes (without DUX4 polyadenylation signal) are almost equally common in the Caucasian and Asian populations, while 4qA is the most common chromosome 4 variant in the African population (Lemmers et al., 2010b). This suggests that about 25% of 18p- individuals carry two 4qB chromosomes and are therefore not at risk for developing FSHD. In two recent studies, we analyzed the D4Z4 repeat array size and haplotype distribution in control individuals (Lemmers et al., 2014; Schaap et al., 2013). These results showed that 20 out of 159 independent 4A161 chromosomes, the most common risk allele for FSHD in the Caucasian population accounting for 79% of all 4qA alleles (Lemmers et al., 2010b), had repeat arrays of ≤16 units, which gives an estimated incidence of 12.6% (Figure 2C). Because half of the chromosomes 4 are of the 4qB variant, about 12.6% (1 in 8) of individuals in the Caucasian population with 18p- carry a medium-short 4qA array and might therefore be at risk for FSHD. To investigate this possibility, we performed accurate D4Z4 repeat array sizing in nineteen individuals with 18p- that were selected on the basis that they carried a 4A161 allele and had D4Z4 hypomethylation and we found that two individuals carried an array ≤16 units and may have an increased risk of developing FSHD (Supp. Figure S2 and Supp. Table S2). Individual 18p-14 carries a 13 unit repeat array and is 27 years old, while 18p-37 carries a 15 unit repeat and is 24 years. At this time, we do not have information about these individuals clinical status. Contractions on other 4qA alleles, such as 4A159, 4A168 and 4A166H, may also confer a risk to FSHD (Lemmers et al., 2010a). However, these alleles are much less common than the 4A161 allele (Lemmers et al., 2010b), and therefore only increase the FSHD risk in the 18p- patient population by approximately 1.5%.
Figure 2.
A. Chromosome content of individuals with 18p-. The black box around the chromosome ideogram across the top of the figure indicates the region of the chromosome shown below. The study number of each participant is aligned to the left of their data. The intact region of the chromosome is shown as individual grey bars. The breakpoint region is shown in darker grey and regions of duplication are shown as black. The vertical line indicates the location of the SMCHD1 gene. B. The methylation value measured at D4Z4 for control individuals (disomic 18p), individuals that carry an SMCHD1 mutation (FSHD2) and 18p- individuals that have a deletion of the SMCHD1 gene (18p-). C. Distribution of D4Z4 repeat array sizes on 159 independent 4A161 chromosomes in European control individuals (Lemmers et al., 2014; Schaap et al., 2013).
In conclusion, we report heterozygous deletions of SMCHD1 in two distantly related FSHD2 families and therefore 18p- deletions may constitute a risk factor for FSHD. While other genetic and epigenetic risk factors are likely to contribute to disease penetrance, in order to be at an increased risk of developing FSHD2, two other factors also must be present. First, the presence of a chromosome 4qA containing a DUX4 polyadenylation signal and second, the size of the D4Z4 repeat array with array sizes at the lower end of the normal repeat size spectrum being more sensitive to SMCHD1 hemizygosity than longer arrays.
Supplementary Material
Acknowledgments
The authors thank all individuals with FSHD and 18p- as well as their family members for participating in this study.
The research leading to these results has received funding from the European Community’s Seventh Framework Programme (FP7/2007–2013) under grant agreement n° 2012-305121 “Integrated European –omics research project for diagnosis and therapy in rare neuromuscular and neurodegenerative diseases (NEUROMICS), The Prinses Beatrix Spierfonds (W.OR12-20), Spieren voor Spieren, NIH NIAMS AR066248, NIH NCRR UL1RR024160, the FSHD Global Research Foundation and the Chromosome 18 Registry and Research Society.
References
- Balog J, Thijssen PE, de Greef JC, Shah B, van Engelen BG, Yokomori K, Tapscott SJ, Tawil R, van der Maarel SM. Correlation analysis of clinical parameters with epigenetic modifications in the DUX4 promoter in FSHD. Epigenetics. 2012:7. doi: 10.4161/epi.20001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Greef JC, Lemmers RJ, Camano P, Day JW, Sacconi S, Dunand M, van Engelen BG, Kiuru-Enari S, Padberg GW, Rosa AL, Desnuelle C, Spuler S, Tarnopolsky M, Venance SL, Frants RR, van der Maarel SM, Tawil R. Clinical features of facioscapulohumeral muscular dystrophy 2. Neurology. 2010;75:1548–1554. doi: 10.1212/WNL.0b013e3181f96175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson PJ, Chen S, Tayeh MK, Ochoa L, Leal SM, Pelet A, Munnich A, Lyonnet S, Majeed HA, El-Shanti H. Homozygous mutations in LPIN2 are responsible for the syndrome of chronic recurrent multifocal osteomyelitis and congenital dyserythropoietic anaemia (Majeed syndrome) J Med Genet. 2005;42:551–557. doi: 10.1136/jmg.2005.030759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriels J, Beckers MC, Ding H, De Vriese A, Plaisance S, van der Maarel SM, Padberg GW, Frants RR, Hewitt JE, Collen D, Belayew A. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene. 1999;236:25–32. doi: 10.1016/s0378-1119(99)00267-x. [DOI] [PubMed] [Google Scholar]
- Geng LN, Yao Z, Snider L, Fong AP, Cech JN, Young JM, van der Maarel SM, Ruzzo WL, Gentleman RC, Tawil R, Tapscott SJ. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev Cell. 2012;22:38–51. doi: 10.1016/j.devcel.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M, Rost S, El HN, Ferbert A, Deschauer M, Walter MC, Schoser B, Tacik P, Kress W, Muller CR. Diagnostic approach for FSHD revisited: SMCHD1 mutations cause FSHD2 and act as modifiers of disease severity in FSHD1. Eur J Hum Genet. 2014 doi: 10.1038/ejhg.2014.191. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers RJ, de Kievit P, Sandkuijl L, Padberg GW, van Ommen GJ, Frants RR, van der Maarel SM. Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat Genet. 2002;32:235–236. doi: 10.1038/ng999. [DOI] [PubMed] [Google Scholar]
- Lemmers RJ, Goeman JJ, van der Vliet PJ, van Nieuwenhuizen MP, Balog J, Vos-Versteeg M, Camano P, Ramos Arroyo MA, Jerico I, Rogers MT, Miller DG, Upadhyaya M, et al. Inter-individual differences in CpG methylation at D4Z4 correlate with clinical variability in FSHD1 and FSHD2. Hum Mol Genet. 2014;24:659–669. doi: 10.1093/hmg/ddu486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers RJ, Tawil R, Petek LM, Balog J, Block GJ, Santen GW, Amell AM, van der Vliet PJ, Almomani R, Straasheijm KR, Krom YD, Klooster R, et al. Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat Genet. 2012;44:1370–1374. doi: 10.1038/ng.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers RJ, van der Vliet PJ, Klooster R, Sacconi S, Camano P, Dauwerse JG, Snider L, Straasheijm KR, van Ommen GJ, Padberg GW, Miller DG, Tapscott SJ, et al. A Unifying Genetic Model for Facioscapulohumeral Muscular Dystrophy. Science. 2010a;329:1650–1653. doi: 10.1126/science.1189044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers RJ, Wohlgemuth M, Frants RR, Padberg GW, Morava E, van der Maarel SM. Contractions of D4Z4 on 4qB subtelomeres do not cause facioscapulohumeral muscular dystrophy. Am J Hum Genet. 2004;75:1124–1130. doi: 10.1086/426035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers RJLF, van der Vliet PJ, van der Gaag KJ, Zuninga S, Frants RR, de Knijff P, van der Maarel SM. Worldwide population analysis of the 4q and 10q subtelomeres identifies only four discrete duplication events in human evolution. Am J Hum Genet. 2010b;86:364–377. doi: 10.1016/j.ajhg.2010.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Lluesma S, Stucke VM, Nigg EA. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 2002;297:2267–2270. doi: 10.1126/science.1075596. [DOI] [PubMed] [Google Scholar]
- Padberg GW. Thesis. Leiden University; 1982. Facioscapulohumeral disease. [Google Scholar]
- Sacconi S, Lemmers RJ, Balog J, van der Vliet PJ, Lahaut P, van Nieuwenhuizen MP, Straasheijm KR, Debipersad RD, Vos-Versteeg M, Salviati L, Casarin A, Pegoraro E, et al. The FSHD2 gene SMCHD1 is a modifier of disease severity in families affected by FSHD1. Am J Hum Genet. 2013;93:744–751. doi: 10.1016/j.ajhg.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaap M, Lemmers RJ, Maassen R, van der Vliet PJ, Hoogerheide LF, van Dijk HK, Basturk N, de KP, van der Maarel SM. Genome-wide analysis of macrosatellite repeat copy number variation in worldwide populations: evidence for differences and commonalities in size distributions and size restrictions. BMC Genomics. 2013;14:143. doi: 10.1186/1471-2164-14-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebold C, Soileau B, Heard P, Carter E, O’Donnell L, Hale DE, Cody JD. Whole arm deletions of 18p: Medical and developmental effects. Am J Med Genet A. 2015;167:313–323. doi: 10.1002/ajmg.a.36880. [DOI] [PubMed] [Google Scholar]
- Snider L, Geng LN, Lemmers RJLF, Kyba M, Ware CB, Nelson AM, Tawil R, Filippova GN, van der Maarel SM, Tapscott SJ, Miller DG. Facioscapulohumeral Dystrophy: Incomplete Suppression of a Retrotransposed Gene. PLoS Genet. 2010;6:e1001181. doi: 10.1371/journal.pgen.1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawil R, van der Maarel SM. Facioscapulohumeral muscular dystrophy. Muscle Nerve. 2006;34:1–15. doi: 10.1002/mus.20522. [DOI] [PubMed] [Google Scholar]
- Turleau C. Monosomy 18p. Orphanet J Rare Dis. 2008;3:4. doi: 10.1186/1750-1172-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Overveld PG, Lemmers RJ, Sandkuijl LA, Enthoven L, Winokur ST, Bakels F, Padberg GW, van Ommen GJ, Frants RR, van der Maarel SM. Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat Genet. 2003;35:315–317. doi: 10.1038/ng1262. [DOI] [PubMed] [Google Scholar]
- Wijmenga C, Hewitt JE, Sandkuijl LA, Clark LN, Wright TJ, Dauwerse HG, Gruter AM, Hofker MH, Moerer P, Williamson R, van Ommen GJ, Padberg GW, et al. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat Genet. 1992;2:26–30. doi: 10.1038/ng0992-26. [DOI] [PubMed] [Google Scholar]
- Zeng W, de Greef JC, Chen YY, Chien R, Kong X, Gregson HC, Winokur ST, Pyle A, Robertson KD, Schmiesing JA, Kimonis VE, Balog J, et al. Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD) PLoS Genet. 2009;5:e1000559. doi: 10.1371/journal.pgen.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.