Abstract
Alström Syndrome, a recessive, monogenic ciliopathy caused by mutations in ALMS1, is typically characterized by multi-system involvement including early cone-rod retinal dystrophy and blindness, hearing loss, childhood obesity, type 2 diabetes mellitus, cardiomyopathy, fibrosis and multiple organ failure. The precise function of ALMS1 remains elusive, but roles in endosomal and ciliary transport and cell cycle regulation have been shown. The aim of our study was to further define the spectrum of ALMS1 mutations in patients with clinical features of Alström Syndrome. Mutational analysis in a world-wide cohort of 204 families identified 109 novel mutations, extending the number of known ALMS1 mutations to 239 and highlighting the allelic heterogeneity of this disorder. This study represents the most comprehensive mutation analysis in patients with Alström Syndrome, identifying the largest number of novel mutations in a single study worldwide. Here, we also provide an overview of all ALMS1 mutations identified to date.
Keywords: ALMS1, Alström Syndrome, ciliopathy, SNV
Background
Alström Syndrome (ALMS; MIM# 203800, http://www.ncbi.nlm.nih.gov/omim) is a rare monogenic recessive disorder characterized by a complex array of clinical manifestations, typically beginning in the first year of life. Progressive retinal degeneration is usually noticed in infancy and hearing loss usually presents in the first decade. Obesity generally begins to develop in the first few years of life and the BMI of most young children is >95th centile. Height is normal in early childhood, but growth slows in adolescence and final adult height is usually <5th centile.
Hyperinsulinemia and type 2 diabetes develop in childhood. Dilated cardiomyopathy (DCM) can occur suddenly in infancy (first months of life) due to aberrant differentiation of cardiomyocytes [Shenje et al., 2014; Luow et al., 2014]. Restrictive cardiomyopathy develops slowly in adolescents and adults [Smith et al., 2007]. Neurological disturbances can include absence seizures and febrile epilepsy. In the brain, myelin derangement and cortical reorganization has been documented [Manara et al., 2015]. Children with ALMS have frequent respiratory problems, and pulmonary fibrosis, chronic obstructive respiratory syndrome (COPD), and acute respiratory distress syndrome (ARDS) can develop in later years [Marshall et al., 2005]. There are multiple endocrine disturbances, including hypothyroidism, alterations in the insulin-like growth factor (IGF) system, low testosterone in males and hyperandrogenism in females [Maffei et al. 2002, 2007]. Liver dysfunction begins with hepatosteatosis, and, in a subset of patients, can progress to hepatic fibrosis and cirrhosis. Renal failure develops slowly in late adolescence and adulthood [Izzi et al., 2011]. Fibrotic tissue has been observed in nearly all organs [Marshall et al., 2005]. “The early changes in vision and hearing have tremendous impact on social development (Frölander et al., 2014). Although delay of cognitive development is not a common feature of ALMS, delay in early fine and gross motor developmental milestones is seen in ~27% of affected children. There can be early learning difficulties, delays in language, or in gross or fine motor milestones, which tend, in most case, to resolve as the child ages. However, if one applies the WHO criteria for defining and assessing intellectual disabilities (ID), fewer than 10% of patients meet that criteria. [International Advisory Group for the Revision of ICD-10 Mental and Behavioural Disorders, 2011; personal communication-jdm].
Diagnosis is difficult as affected children often present with only a subset of features early in the course of the disease (Marshall et al., 2005, 2007a). Additionally, there is interfamilial and intrafamilial clinical variability in the phenotypes, including age of onset, and the severity of the disease manifestations, which may be explained by the effects of genetic and/or environmental modifiers.
Located on chromosome 2p13, ALMS1 spans 23 exons and encodes a predicted 461.2-kDa protein of 4,169 amino acids (aa). Exon 1 harbors a polyglutamate tract (aa 13–29) consisting of a (GAG)NGAA(GAG)3 trinucleotide repeat, followed by a stretch of seven alanine residues (aa30–36) [Collin et al., 2002; Hearn et al., 2002]. Exon 8, a 6-kb exon, is also comprised of a large, variable tandem-repeat domain encoding 34 repeats of 45–50 amino acids.
Most pathogenic variants occur downstream from exon 7 and are almost all truncating mutations resulting in the early termination of ALMS1 and a non-functional protein (i.e., nonsense mutations that cause a stop codon or insertions and deletions of one or more nucleotides that cause a frameshift).
ALMS1 localizes to centrosomes and basal bodies of ciliated cells and is expressed in all tissues that are pathologically affected in patients with ALMS. Roles in cell cycle regulation and intraciliary transport [Hearn et al., 2005, Li et al., 2007, Knorz et al., 2010, Collin et al., 2012] have been suggested. Recently, ALMS1 has been shown to contribute to cell migration and extracellular matrix production [Shenje et al., 2014, Louw et al., 2014, Zulato et al., 2011], as well as in the endosomal trafficking of transferrin, GLUT4, and Notch1 [Collin et al., 2012, Favaretto et al., 2014, Leitch et al., 2014]. However, the precise molecular mechanisms underlying the multiple organ pathologies have not been fully elucidated.
The first ALMS1 mutations identified were clustered in exons 8, 10 and 16 [Marshall et al., 2007b, 2011; Joy et al., 2002]. Therefore, early investigations preferentially sequenced these exons. The c.10775delC (p.Thr3592Lysfs*6) mutation in exon 16 was the most frequently identified, with a common founder suggested for persons of British descent [Marshall et al., 2007b]. Subsequently, the wider incorporation of automated sequencing to genotype patients with ALMS has uncovered additional mutations in exon 5 [Paisey et al., 2014; Casey et al., 2014], exon 11 [Taşdemir et al., 2012], exon 12 [Marshall et al.,2007b], exon 18 [Marshall et al., 2007b; Malm et al., 2008], exon 20 [Casey et al., 2014] and intronic regions [Sanyoura,et al., 2014; Ozantürk et al., 2014; Aldahmesh et al., 2009; Bond et al.,2005].
Methods of Subject Ascertainment and Mutation Detection
Study Subjects
Our original cohort consisted of 239 patients from 204 unrelated families with a suspicion of Alström Syndrome who fulfilled the established clinical criterion for a diagnosis of ALMS [Marshall et al., 2007a] but without a molecular diagnosis. Subjects were recruited to The Jackson Laboratory (Bar Harbor, ME, USA), to Padua University (Padua, Italy), or to the University of Strasbourg (Strasbourg, France) through the referral of Alström Syndrome International (ASI) over a period of more than twenty years. The study subjects were ethnically diverse, including multiple kindreds representing North America (59), South America (3), Caribbean (2), Northern Europe (10), Southern Europe (24), Western Europe (23), Eastern Europe (3), Southeastern Europe (25), Oceania (4), East Asia/Middle East (13), Southeast Asia (5), South Asia (6), West Asia (19), and North Africa (6).
Appropriate informed consent was obtained from adult patients and parents/guardians of minors. Clinical histories and medical records were obtained for all affected individuals, but the comprehensiveness of the records was variable from patient to patient. The research was approved by the Institutional Review Board of The Jackson Laboratory, the Padua University Hospital Ethics Committee, and the Ethics Committee of the Strasbourg University Hospital (Comité Consultatif de Protection des Personnes dans la Recherche Biomédicale, CCPPRB).
Mutation Analysis Strategy
Genomic DNA was isolated from blood leukocytes using standard protocols, or DNA samples obtained in a clinical setting were submitted by referring physicians. We adopted a stepwise approach to identify causal variants for each patient. A detailed description of the mutation analysis strategy and methods can be found in Supp. Methods. Mutation nomenclature refers to GenBank reference sequence NM_015120.4; AC074008.5 for ALMS1. Nucleotide and amino acid numbering of mutation sites began at the start codon, ATG (Met) of the open reading frame, originally described by Collin et al.[2002] and Hearn et al.[2002].
Additionally, using VaRank [Geoffroy et al., 2015] we analyzed next generation sequencing data from 275 patients for rare or frequent Single Nucleotide Variants (SNVs) (See Supp. Methods).
Criteria to Determine the Pathogenicity of New ALMS1 Mutations
Determination of pathogenicity was straight-forward in the case of likely truncating variants – frameshift indels, stop codons gained, and canonical splice site changes. A sequence variation was considered pathogenic when 1) it was not present in control databases (including dbSNP [Sherry et al., 2001]. EVS (Exome variant Server, NHLBI GO Exome Sequencing Project, http://evs.gs.washington.edu/EVS/) and, specifically for the TaGSCAN analysis, in at least 960 randomly selected controls from the Center for Pediatric Genomic Medicine’s TaGSCAN database); 2) it altered a well-conserved amino acid, preferably in a conserved region [Larkin et al., 2007] and 3) it was judged significant in computational prediction tools for novel mutations: PolyPhen-2 analysis [Adzhubei et al., 2010], SIFT [Kumar et al 2009], Mutation Taster [Schwarz et al., 2010] and Condel [Gonzales Perez and Lopes-Bigas, 2011]. A mutation was considered novel if it was not present in the Human Gene Mutation Database (http://www.hgmd.org/) [Stenson et al., 2014], or the dbSNP database, or not previously published.
Database
All novel variants identified in the present study have been submitted to the Euro-WABB online database [Farmer et al., 2013], a locus-specific database (in the Leiden Open Variation Database format; https://lovd.euro-wabb.org/home.php?select_db=ALMS1) listed by the Human Genome Variation Society in the Locus specific Mutation Databases LSDB (www.HGVS.org). We encourage future authors of published reports of ALMS1 mutations to include their findings within this database.
Pathogenic Variants
Novel ALMS1 pathogenic mutations identified in this study are shown in Table 1 and Figure 1. These, along with previously reported ALMS1 mutations, ethnicities, and references are listed together in Supp. Table S1. There are now 239 disease-causing mutations in ALMS1 identified in many diverse ethnic groups. The majority (96%) of the mutations are nonsense and frameshift variations (insertions or deletions) that are predicted to cause premature protein truncation. There is no evidence for triallelic or complex modes of inheritance.
TABLE 1.
109 Novel ALMS1 Mutations
| cDNA Variant | Predicted protein | #Exon | Total affected allelesa | Ethnicity/Regional origin |
|---|---|---|---|---|
| c.454-5T>G | intron 2 | 1 | Oceania | |
| c.592C>T | p.Gln198* | 3 | 1 | N America |
| c.906del | p.Ile302Metfs*30 | 5 | 2 | N Africa |
| c.967G>T | p.Glu323* | 5 | 1 | N America |
| c.1054C>T | p.Arg352* | 5 | 1 | N America |
| c.1613_1614delTC | p.Leu538Glnfs*12 | 8 | 1 | N America |
| c.1628T>A | p.Leu543* | 8 | 1 | N America |
| c.1645delA | p.Thr549Leufs*5 | 8 | 1 | Caribbean |
| c.1674delT | p.559Leufs*37 | 8 | 2 | E Europe |
| c.1897dupT | p.Tyr633Leufs*9 | 8 | 1 | W Europe |
| c.1903C>T | p.Gln635* | 8 | 1 | N America |
| c.2159delG | p.Arg720Lysfs*60 | 8 | 1 | S Europe |
| c.2234C>G | p.Ser745* | 8 | 1 | N America |
| c.2265dupT | p.Glu756* | 8 | 4 | W Asia |
| c.2646dupT | p.Glu883* | 8 | 1 | N America |
| c.2676delT | p.Gly893Aspfs*41 | 8 | 1 | S Europe |
| c.2708C>G | p.Ser903* | 8 | 1 | Oceania |
| c.2778dupT | p.Leu927Serfs*4 | 8 | 1 | E Asia |
| c.2822T>A | p.Leu941* | 8 | 3 | W Europe |
| c.2930_2933dupGAGA | p.Ser979Argfs*13 | 8 | 1 | W Europe |
| c.2967_2970delTGAC | p.Asp990Argfs*38 | 8 | 2 | W Europe |
| c.3019delA | p.Arg1007Glufs*22 | 8 | 1 | N America |
| c.3019dupA | p.Arg1007Lysfs*15 | 8 | 1 | Oceania |
| c.3020delG | p.Arg1007Lysfs*22 | 8 | 1 | S Europe |
| c.3066T>A | p.Tyr1022* | 8 | 2 | N Africa |
| c.3153C>A | p.Tyr1051* | 8 | 1 | S America |
| c.3220_3221delAG | p.Leu1075Glufs*10 | 8 | 1 | E Asia |
| c.3300_3301delAA | p.Lys1103Alafs*16 | 8 | 2 | SE Europe |
| c.3472C>T | p.Gln1158* | 8 | 2 | W Asia |
| c.3518C>G | p.Ser1173* | 8 | 1 | E Asia |
| c.3579C>G | p.Tyr1193* | 8 | 2 | N America |
| c.3583C>T | p.Gln1195* | 8 | 2 | S Asia |
| c.3754C>T | p.Gln1252* | 8 | 1 | Oceania |
| c.3876_3877dupGAAG | p.Lys1293Glufs*13 | 8 | 1 | W Europe |
| c.4039C>T | p.Gln1347* | 8 | 1 | N America |
| c.4145_4146delACTC | p.Tyr1382Phefs*18 | 8 | 1 | E Asia |
| c.4154A>G | p.Ser1383* | 8 | 1 | N Europe |
| c.4180C>T | p.Gln1394* | 8 | 2 | N America |
| c.4205dupT | p.Thr1403Asnfs*2 | 8 | 2 | S Asia |
| c.4213G>T | p.Glu1405* | 8 | 2 | N America |
| c.4274T>A | p.Leu1425* | 8 | 1 | W Europe |
| c.4281_4282dupA | p.Thr1428Asnfs*11 | 8 | 1 | N America |
| c.4363G>T | p.Glu1455* | 8 | 1 | E Asia |
| c.4477G>T | p.Glu1493* | 8 | 1 | S Europe |
| c.4605C>G | Tyr1535* | 8 | 1 | E Asia |
| c.4657delT | p.Ser1553Leufs*15 | 8 | 1 | E Asia |
| c.4718C>G | p.Ser1573* | 8 | 1 | Oceania |
| c.4746C>A | p.Tyr1582* | 8 | 2 | E Asia |
| c.4823delA | p.Lys1608Argfs*9 | 8 | 1 | N America |
| c.4823dupA | p.Thr1609Aspfs*2 | 8 | 1 | W Europe |
| c.4891C>T | p.Gln1631* | 8 | 2 | W Europe N Africa |
| c.4917_4920delTAAA | p.Asn1639Lysfs*4 | 8 | 2 | N America E Asia |
| c.5054_5060delTACCTGA | p.Leu1685* | 8 | 1 | W Europe |
| c.5135T>G | p.Leu1712* | 8 | 2 | S Europe |
| c.5277delC | p.Tyr1760Ilefs*20 | 8 | 1 | N America |
| c.5455C>T | p.Arg1819* | 8 | 2 | SE Europe |
| c.5573_5574del | p. Ser1858Cysfs*5 | 8 | 1 | Oceania |
| c.5575_5576delGT | p.Val1859Hisfs*4 | 8 | 1 | W Asia |
| c.5929C>T | p.Gln1977* | 8 | 1 | N Africa |
| c.6116_6117delTC | p.Pro2040Ilefs*7 | 8 | 3 | SE Europe E Asia |
| c.6169_6170dupAT | p.Leu2058Phefs*17 | 8 | 1 | E Asia |
| c.6288T>A | p.Tyr2096* | 8 | 2 | SE Europe |
| c.6361A>T | p.Lys2121* | 8 | 2 | S Asia |
| c.6421_6427del | p.Ser2141Phefs*20 | 8 | 1 | E Asia |
| c.6486_6489delAACT | p.Thr2163Argfs*4 | 8 | 2 | SE Europe |
| c.6625_6626delA | p.Asn2209Ilefs*2 | 8 | 2 | S America |
| c.6895delG | p.Val2299Trpfs*43 | 8 | 1 | N Europe |
| c.7188_7192delTAGTG | p.Cys2396Trpfs*6 | 8 | 2 | S America |
| c.7376_7379delATAG | p.Asp2459Alafs*18 | 8 | 2 | W Europe |
| c.7378G>T | p.Glu2460* | 8 | 1 | W Europe |
| c.7571_7572delAT | p.His2524Argfs*11 | 9 | 1 | S Europe |
| c.7577_7590delGTGGATACTCCATT | p.Cys2526Phefs*5 | 9 | 2 | W Asia |
| c.7677G>C | p.Lys2559Asn | 9 | 2 | W Europe |
| c.7677+1G>T | p.? | 9 | 1 | SE Europe |
| 5.7kb deletionb | intron 9 | 2 | W Asia | |
| c.7772dupT | p.Thr2592Asnfs*3 | 10 | 1 | N America |
| c.7911dupC | p.Asn2638Glnfs*24 | 10 | 2 | W Asia |
| c.8150_8151insT | p.Ser2719Phefs*7 | 10 | 2 | W Asia |
| c.8352_8355delAGAA | p.Glu2785* | 10 | 1 | N America |
| c.8579C>A | p.Ser2860* | 10 | 2 | N America |
| c.8766C>A | p.Cys2922* | 10 | 2 | Caribbean |
| c.8995A>T | p.Gln2999* | 10 | 1 | N Europe |
| c.9379C>T | p.Gln3127* | 10 | 1 | S Europe |
| c.9517G>T | p.Glu3173* | 10 | 2 | SE Asia |
| c.9801T>A | p.Tyr3267* | 12 | 1 | W Europe |
| deletion of exon 13-16 | 1 | W Asia | ||
| c.10124C>G | p.Ser3375* | 14 | 2 | N Africa |
| c.10265delC | p.Pro3422Glnfs*2 | 15 | 1 | W Europe |
| c.10388-2A>G | intron 15 | 2 | W Asia | |
| c.10483delC | p.Gln3495Lysfs*14 | 16 | 1 | N America |
| c.10535G>A | p.Trp3512* | 16 | 1 | N America |
| c.10561_10564delGACA | p.Asp3521Asnfs*25 | 16 | 2 | S Europe |
| c.10609_10614delinsTCAAA | p.Asp3537Serfs*10 | 16 | 1 | W Europe |
| c.10757dupA | p.Val3587Glyfs*15 | 16 | 1 | W Europe |
| c.10828C>T | p.Gln3610* | 16 | 1 | W Europe |
| c.10830_10831insC | p.Arg3611Glnfs*7 | 16 | 1 | S Europe |
| c.10898dupT | p.Leu3633Phefs*2 | 16 | 2 | N Europe |
| c.10939_10945del | p.Val3647Lysfs*13 | 16 | 1 | N Europe |
| deletion of exon 17 | 17 | 1 | N America | |
| c.11618_11619delCT | p.Ser3873Tyrfs*19 | 17 | 3 | N America S Europe |
| c.11651_11652insGTTA | p.Asn3885Leufs*9 | 17 | 3 | N America |
| c.11654_11657del | p.Asn3885Argfs*11 | 17 | 1 | W Europe |
| c.11703delA | p.Lys3901Asnfs*8 | 18 | 1 | S Europe |
| c.11812dupA | p.Met3938Asnfs*8 | 18 | 1 | SE Europe |
| c.11881dupT | p.Ser3961Phefs*12 | 19 | 1 | W Europe |
| c.12004G>T | p.Glu4002* | 19 | 1 | N America |
| c.12116_12117insT | p.Gln4039Serfs*10 | 19 | 1 | N America |
| c.12365G>A | p.Arg4122Gln | 21 | 1 | N America |
Mutation nomenclature refers to GenBank reference sequence NM_015120.4; AC074008.5 for ALMS1. Nucleotide and amino acid numbering of mutation sites began at the start codon, ATG (Met) of the open reading frame, originally described by Collin et al. [2002] and Hearn et al. [2002]. Ethnicity/Regional origin was designated according to the United Nations Composition of macro geographical (continental) regions, geographical sub-regions, and selected economic and other groupings, http://millenniumindicators.un.org/unsd/methods/m49/m49regin.htm.
Total number of alleles including this cohort and previous reports.
Approximate genomic locations 73715902 in intron 9 to 73721624 in intron 10
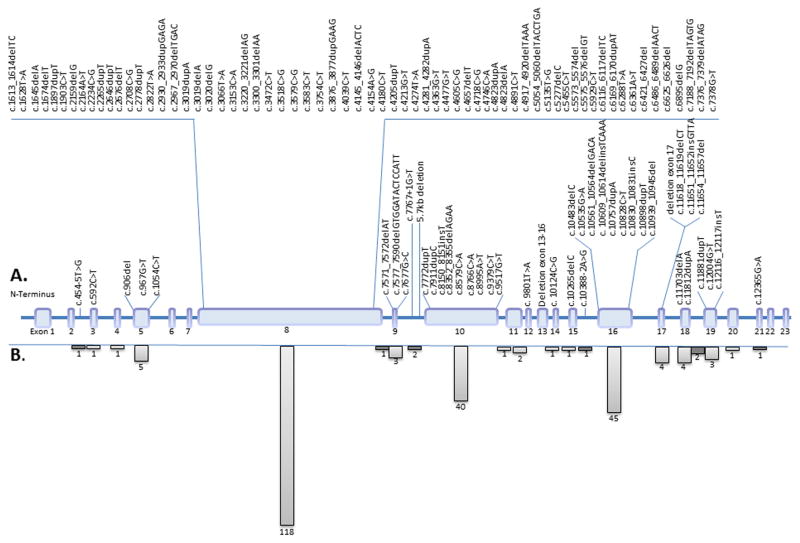
Figure 1.
Novel ALMS1 mutations identified in this report and total previously reported. A. Exonic structure and location of novel ALMS1 mutations. B. Total number of reported substitution, deletion, insertion, and splice-site mutations per exon, including novel mutations and those previously reported in Supp. Table S1. Intronic regions where mutations of unknown significance, but predicted to alter splicing have been identified are shaded in dark gray.
Additionally, deletions removing entire exons and splice-site mutations [Bond et al., 05; Sanyoura et al., 2014], an Alu transposon insertion [Taşkesen et al., 2012] and one balanced translocation [Hearn et al., 2002] have been reported.
Several mutations were found in multiple ethnicities and 3 or more regional origins: c.4156dupA, c.4937C>A, c.5145T>G, c.8164C>T, c.8394_8395insA, c.8782C>T, c.9900dupC, c.10483C>T, c.10790_10791delTG, c.10849G>T, c.10885C>T, c.10975C>T, c.11316_11319delAGAG, c.11385delT, and c.11416C>T (Supp. Table S1). The mutation c.10775delC remains the most common with 24 alleles identified, however, all individuals carrying that particular allele were from the UK or North America.
We identified 109 novel mutations in ALMS1 (Table 1; Figure 1) in exons 3, 5, 8, 9, 10, 12, 14, 15, 16, 17, 18, 19, and 21 as well as introns 2, 9, and 15. Additionally, four indel and splice mutations in ALMS1 were detected: c.454-5T>G, c.7677+1G>T, c.10388-2A>G, and c.10609_10614delinsTCAAA.
Several large deletions were also observed. A homozygous 5.7kb deletion with approximate genomic locations 73,715,902 in intron 9 to 73,721,624 in intron 10 was seen in a kindred of Middle Eastern heritage. Another patient of Middle Eastern descent harbored a heterozygous deletion of exons 13-16. Lastly, a patient of German heritage harbored a heterozygous deletion of exon 17. Most of the novel mutations were private mutations identified in only one family (100/109), suggesting that the mutation spectrum is far from being saturated in spite of numerous ALMS1 mutation reports.
Thirty kindreds carried novel homozygous mutations, consistent with the large number of consanguineous unions, while 82 were compound heterozygotes. In four kindreds, only one heterozygous disease-causing allele was identified after complete NGS analysis of ALMS1. No apparent difference in phenotypic expression was observed in patients in whom only one heterozygous mutation has been identified compared to patients with two ALMS1 mutations (data not shown).
Twenty two of the patients who presented with features suggestive of Alström Syndrome received clear alternative diagnoses either subsequently identified by the referring physiciaans or using the TAGScan diagnostic panels (Supp Table S2). Deleterious mutations were identified in Usher Syndrome type 2A (USH2A (4), Bardet Biedl Syndrome (BBS1 (5), BBS2 (1), BBS4 (1), BBS5 (1), BBS9 (2), BBS10 (2), BBS12 (2), Dedicator of Cytokinesis (DOCK7 (2), Myosin heavy chain 7B (MYH7B(1), and Congenital stationary night blindness, CSNBAD3 (1). Although the NGS protocols utilized in this study enrich all exons of ALMS1 and detect nucleotide changes at over 95% of the ALMS1 coding domain, there remain 21 families (10%) in whom we have not yet been able to identify mutations in ALMS1.
In some cases, where no clear pathogenic ALMS1 mutations are found, rare missense variations pose a problem in genetic counseling, as their impact is still unclear. The length of the resulting protein sequence is preserved but the functional consequences of most missense variants that result in a single amino acid residue change in ALMS1 protein is not known. Although missense mutations can render the resulting protein nonfunctional by affecting splice sites or the binding with other proteins, we cautiously questioned some previously reported missense variants given their uncertain pathogenicity. The latest data available for variation frequency such as EVS or ExAC (Exome Aggregation Consortium, http://exac.broadinstitute.org, accessed March 2015) providing an accumulating allele count for several thousand individuals allows for a more critical analysis.
Marshall et al [2007b] described 8 missense variants: p.K1718N, p.S2216R, p.T2458I, p.R2589W, p.V2996G, p.E3500V, p.S3505C and p.I3888T that were predicted to be disease-causing on the basis of SIFT prediction, a BLOSUM45 matrix score, absence of the variant in the general population, and a conserved identity score. All of these variations are either not in the current databases or occur with a very low allele frequency (<0.01%). Previously, others have identified heterozygous missense alterations in ALMS1 (ExAC allele frequencies into parenthesis when available), p.V424I and p.H3882Y (0.15%) [Joy et al., 2002], p.Asn1788Asp (1.443%) and p.Asn2946K (~1%) [Minton et al., 2006], p.S3707C (0.14%) [Sathya-Priya et al., 2015] and p.S2102L (2.56%) [Redin et al., 2012] were identified as potentially disease-causing. However, given that several of these alleles are common in the general population, they should not be included in the cohort of pathogenic ALMS1 mutations without a more thorough analysis.
Single Nucleotide Non-Pathogenic Variants
We also identified 194 SNVs that were unlikely to be pathogenic by assessing their frequency in our cohort of 275 patients and the functional significance of these variants based on the conserved identity of the protein and the severity of the resulting amino acid substitution (Supp. Table S3). It is interesting to note that a frequent SNV in exon 8 (inframe insertion/deletion of a CTC, present in >60% of our patients) can be, under certain circumstances, wrongly interpreted by NGS pipelines (mainly due to bad alignments) and lead to false positive SNVs including frameshifts (i.e. c.1574dup, p.Pro526Serfs*11, representing 25 alleles in our cohort). Therefore we recommend careful examination of any variant identified closely neighboring this SNV (Supp. Figure S1).
Clinical Relevance
Phenotypic variation, such as differences in visual acuity and hearing loss or presence versus absence of cardiomyopathy, and varying hepatic manifestations, is observed in siblings or in unrelated families with the same mutations. This suggests that, besides some possible variability due to the mutation itself, there is interplay between a multitude of potential genetic modifiers, environmental or infectious exposures, and stochastic events leading to the wide variations in age of onset and severity of the ALMS phenotypic spectrum [Collin et al., 2002]. Zumsteg et al. [Zumsteg et al., 2000] speculated that a gene encoding a major modifier of plasma cholesterol levels could be responsible for the phenotypic heterogeneity in lipid metabolism and T2DM in a study of three age-adjusted siblings with ALMS. In three small genotype–phenotype correlation studies, t’Hart et al [t’Hart et al., 2003], Bond et al. [Bond et al., 2005], and Patel et al. [Patel et al., 2006] found no association between the location or type of ALMS1 mutations and type 2 diabetes, BMI, or the occurrence of dilated cardiomyopathy, respectively. Another study suggested a correlation between renal disease and the genomic location of ALMS1 mutation sites. Subjects harboring mutations only in exon 8 appeared to have delayed and milder renal complications, perhaps due to tissue-specific expression of different splice isoforms [Marshall et al., 2007b]. In a recent study of the general population Ichihara et al. found a correlation between the polyglutamine repeat in ALMS1 and early onset myocardial infarct [Ichihara, et al., 2013].
Feasibility of Genotype-Phenotype Correlations
Today, in spite of advances in our knowledge of the spectrum of ALMS1 mutations, we do not yet have evidence for prognostic predictions based on genotype. Since Alström Syndrome is a rare recessively inherited disorder, enumerating individual mutant alleles in patients with various phenotypes has limited value. Indeed, it is the combination of mutated alleles that is important in defining the phenotype. Therefore, to avoid misinterpretation, it would be necessary to include only the homozygous mutations. The majority of mutations identified are not found in homozygosity, thus reducing the number of patients suitable for such analysis.
Additionally, our cohort is very large and ethnically diverse, and patients received inconsistent clinical care. Often a particular phenotype, for instance age of vision loss, diabetes onset, or cardiomyopathy has not been examined in many of these patients.
Alström Syndrome is extremely complex. Most of the features emerge as the children grow. Therefore, when looking for the presence or absence of a particular phenotype, the age of the patient needs to be considered. For example, the average age of onset for diabetes is 16 years [Marshall et al., 2005], so only those patients over the mean age should be included. Likewise, the age of onset of the restrictive cardiomyopathy ranges from 12–40 years. Typically renal disease does not emerge until late adolescence and develops gradually. This limits the number of patients to evaluate with any degree of certainty for each phenotype.
In summary, the possibility of genotype-phenotype correlations should certainly be studied carefully, but further analysis with larger numbers of patients is required. The limitations in current genotype–phenotype studies demonstrate that another major challenge is to determine those factors, other than ALMS1 mutations, contributing to the phenotypic diversity in ALMS.
Diagnostic Relevance
This study brings the total number of reported ALMS1 disease causing variants to 239. Out of our cohort of 204 families (408 alleles), 357 alleles were identified, achieving a mutation detection rate of 88%, which improves upon previous studies in which mutations were identified in 70% to 75% of Alström families. The large number of private mutations unique to a single kindred underscores the allelic heterogeneity in the disorder.
There is a strong clustering of disease-causing variants in exon 8 (118/239, 49%), exon 10 (40/239, 17%), and exon 16 (45/239, 19%), continuing to support the observation suggesting that these regions represent mutational hotspots for ALMS1. This may be due, in part, to the relative size, although this does not seem to be true for exon 16 [Marshall et al., 2011]. It likely also reflects the screening strategy employed in early studies, in which exons 16, 10, and part of 8 were preferentially selected for complete sequencing. However, an explanation for the disproportionate clustering in these exons is not fully apparent given the extensive use of NGS, which genotypes all coding domain nucleotides.
Certainly, ethnicity contributes significantly to the distribution of mutations, and some generalizations can be made. Among non-consanguineous populations of America and Europe, the prevalence of ALMS is estimated to be approximately 1:1,000,000 [Marshall et al., 2011]. However, evidence exists that the incidence of ALMS increases within populations of high consanguinity or in those that are geographically isolated [Marshall et al., 1997; Aldahmesh et al., 2009; Ozantürk et al., 2014].
Several recurrent mutant alleles have been identified in Northern European populations, including c.10775delC (24/557), c.10483C>T (17/557, and c.11449C>T (15/557), c.11316_11319delAGAG (15/557), and c.11416C>T (10/557). In addition, c.8177_8187, c.8164C>T, and c.10945G>T are common in West Asian/Middle Eastern kindreds. Specifically, c.10945G>T makes up approximately 8.2% of the mutant alleles in this population, c.8164C>T and c.8177_8187 each make up approximately 11%. The common alleles in Europe and the Americas are not generally seen in Chinese and Japanese cohorts. The following variants occurred more than once only in East Asian families: c.6169_6170dupAT, c.10831_10832delAG, and c.11116_11134del19. Therefore, screening for these mutations in patients of West and East Asian descent is a logical first step in mutation detection. Among other European and North/South American patients, however, this strategy fails to identify most of the rare mutant alleles, and whole gene sequencing is required for accurate genotyping.
So far, no disease-causing mutations have been identified in exons 1-2, 6, 7, 13 and 22-23. Variants in these other exons may result in different phenotypes or in no phenotype at all, or alternatively, have deleterious effects on fetal viability.
In four families only one heterozygous disease-causing variant was identified, and in 21 families no disease-causing variant was identified after complete NGS analysis of ALMS1. The “missing” mutated ALMS1 alleles in the 21 families (presumably 42 alleles) and the four families with only one allele identified (four alleles) may harbor other types of molecular lesions including large deletions, chromosomal translocations, promoter/enhancer defects, or splice site alterations, intronic nucleotide variants more than 20 nucleotides from the intron-exon boundary, or variants in a non-exonic regulatory motif, which we have not identified in our screening.
Whole genome sequencing with structural variant detection may identify these missing alleles. Alternatively these patients may have mutations in another disease gene which might be identified through exome or genome sequencing.
Future Prospects
New advances in high throughput genome sequencing will aid in more effective mutational screening for early clinical diagnosis by documenting the high number of private mutations.
Given the emerging importance of disease modifying alleles on the diverse phenotypic expression of other ciliopathies, identifying modifying loci will help elucidate the molecular pathways through which ALMS1 acts. Future studies of discordant sib pairs as well as studies in the mouse models [Collin et al., 2005] are required to understand the disease mechanisms in Alström Syndrome.
Supplementary Material
Acknowledgments
We thank the individuals with Alström Syndrome and their families, who continue to enthusiastically support research efforts. We also thank the colleagues and numerous clinicians and scientists who collaborated with us over many years and provided biological samples and valuable clinical information. Particular thanks go to Robert P. Marshall and Alström Syndrome International. Funding for the Asper Ophthalmics analysis and the TaGSCAN was generously provided by Alström Syndrome International (Mt. Desert, ME), Alström Angels (Lubbock, TX), and Alström Syndrome Canada (Finch, Ontario).
Acknowledgments
Grant Acknowledgments
This work was supported by National Institutes of Health (NIH-HD36878; to J.K.N., J.D.M., and G.B.C.), Programme Hospitalier de Recherche Clinique National Alström 2012 (PHRC N5514 to HD and JM), Italian Ministry of Education, University and Research (MIUR) (PRIN prot. 2005060925_002; and EURO-WABB: an EU rare diseases registry for Wolfram syndrome, Alström syndrome and Bardet-Biedl syndrome project has received funding from the European Union, in the framework of the Health Programme (Grant Agreement Reference: 2010 12 05). to P.M., R.V., G.M., F.F.); Children’s Mercy Hospital, Marion Merrell Dow Foundation, William T. Kemper Foundation, Pat & Gil Clements Foundation, Claire Giannini Foundation, Black & Veatch, and U19HD077693 (to S.F.K). The Jackson Laboratory institutional multimedia, allele typing, and sequencing shared services were supported by US. Public Health Service (PHS), National Institutes of Health (CA34196),
Footnotes
None of the authors have a conflict of interest.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldahmesh MA, Abu-Safieh L, Khan AO, Al-Hassnan ZN, Shaheen R, Rajab M, Monies D, Meyer BF, Alkuraya FS. Allelic heterogeneity in inbred populations: the Saudi experience with Alström syndrome as an illustrative example. Am J Med Genet A. 2009;149(4):662–665. doi: 10.1002/ajmg.a.32753. [DOI] [PubMed] [Google Scholar]
- Aliferis K, Hellé S, Gyapay G, Duchatelet S, Stoetzel C, Mandel J-L, Dollfus H. Differentiating Alstrom from Bardet-Biedl syndrome (BBS) using systematic ciliopathy genes sequencing. Ophthalmic Genetics. 2011:1–5. doi: 10.3109/13816810.2011.620055. [DOI] [PubMed] [Google Scholar]
- Bell CJ, Dinwiddie DL, Miller NA, Hateley SL, Ganusova EE, Mudge J, Langley RJ, Zhang L, Lee CC, Schilkey FD, Sheth V, Woodward JE, et al. Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci Transl Med. 2011;3(65):65ra4. doi: 10.1126/scitranslmed.3001756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J, Flintoff K, Higgins J, Scott S, Bennet C, Parsons J, Mannon J, Jafri H, Rashid Y, Barrow M, Trembath R, Woodruff G, et al. The importance of seeking ALMS1 mutations in infants with dilated cardiomyopathy. J Med Genet. 2005;42:e10. doi: 10.1136/jmg.2004.026617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey J, McGettigan P, Brosnahan D, Curtis E, Treacy E, Ennis S, Lynch SA. Atypical Alstrom syndrome with novel ALMS1 mutations precluded by current diagnostic criteria. Eur J Hum Genet. 2014;57(2):55–59. doi: 10.1016/j.ejmg.2014.01.007. [DOI] [PubMed] [Google Scholar]
- Collin GB, Marshall JD, Ikeda A, So WV, Russell-Eggitt I, Maffei P, Beck S, Boerkoel CF, Sicolo N, Martin M, Nishina PM, Naggert JK. Mutations in ALMS1 cause obesity, type 2 diabetes and neurosensory degeneration in Alström Syndrome. Nat Genet. 2002;31:74–78. doi: 10.1038/ng867. [DOI] [PubMed] [Google Scholar]
- Collin GB, Cyr E, Bronson R, Marshall JD, Gifford EJ, Hicks W, Murray SA, Zheng QY, Smith RS, Nishina PM, Naggert JK. Alms1-disrupted mice recapitulate human Alström syndrome. Hum Mol Genet. 2005;14(16):2323–2333. doi: 10.1093/hmg/ddi235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin GB, Marshall JD, King BL, Milan G, Maffei P, Jagger DJ, Naggert JK. The Alström syndrome protein, ALMS1, interacts with α-actinin and components of the endosome recycling pathway. PLoS One. 2012;7(5):e37925. doi: 10.1371/journal.pone.0037925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat. 2000;15:7–12. doi: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- den Dunnen JT, Antonarakis SE. Nomenclature for the description of human sequence variations. Hum Genet. 2001;109:121–124. doi: 10.1007/s004390100505. [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer A, Aymé S, de Heredia ML, Maffei P, McCafferty S, Mlynarski W, Nunes V, Parkinson K, Paquis-Flucklinger V, Rohayem J, Sinnott R, Tillman V, et al. EURO-WABB: an EU rare diseases registry for Wolfram syndrome, Alstrom syndrome and Bardet-Biedl syndrome. BMC Pediatr. 2013;13(1):130. doi: 10.1186/1471-2431-13-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaretto F, Milan G, Collin GB, Marshall JD, Maffei P, Vettor R, Naggert JK. GLUT4 defects in adipose tissue are the early signs of metabolic alterations in Alms1GT/GT, a mouse model for obesity and insulin resistance. PLosOne. 2014;9(10):e109540. doi: 10.1371/journal.pone.0109540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flintoff KJ, Boute-Benejean O. Novel human pathological mutations. Gene symbol: ALMS1. Disease: Alström syndrome. Hum Genet. 2007;121:645. [PubMed] [Google Scholar]
- Flintoff K, Paisey R. Novel human pathological mutations. Gene symbol: ALMS1. Disease: Alström syndrome. Hum Genet. 2007;121(5):647. [PubMed] [Google Scholar]
- Flintoff KJ, Josifova D. Novel human pathological mutations. Gene symbol: ALMS1. Disease: Alström syndrome. Hum Genet. 2007;121:297. [PubMed] [Google Scholar]
- Flintoff KJ, Pilz D. Novel human pathological mutations. Gene symbol: ALMS1. Disease: Alstrom syndrome. Hum Genet. 2007;121:287. [PubMed] [Google Scholar]
- Frölander HE, Möller C, Marshall JD, Sundqvist A, Rönnåsen B, Falkensson L, Lyxell B. Theory-of-mind in adolescents and young adults with Alström syndrome. Int J Pediatr Otorhinolaryngol. 2014;78(3):530–536. doi: 10.1016/j.ijporl.2013.12.038. [DOI] [PubMed] [Google Scholar]
- Gee HY, Otto EA, Hurd TW, Ashraf S, Chaki M, Cluckey A, Vega-Warner V, Saisawat P, Diaz KA, Fang H, Kohl S, Allen SJ, et al. Whole-exome resequencing distinguishes cystic kidney diseases from phenocopies in renal Ciliopathies. Kidney Int. 2014;85:880–887. doi: 10.1038/ki.2013.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoffroy V, Pizot C, Redin C, Piton A, Vasli N, Stoetzel C, Blavier A, Laporte J, Muller J. VaRank: a simple and powerful tool for ranking genetic variants. PeerJ. 2015;3:e796. doi: 10.7717/peerj.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogi D, Bond J, Long V, Sheridan E, Woods GC. Exudative retinopathy in a girl with Alström syndrome due to a novel mutation. Br J Ophthalmol. 2007;91:983–984. [Google Scholar]
- Gonzalez-Perez A, Lopez-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am J Hum Genet. 2011;88:440. doi: 10.1016/j.ajhg.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearn T, Renforth GL, Spalluto C, Hanley NA, Piper K, Brickwood S, White C, Connolly V, Taylor JFN, Russell-Eggitt I, Bonneau D, Walker M, et al. Mutation of ALMS1, a large gene with a tandem repeat encoding 47 amino acids, causes Alström Syndrome. Nat Genet. 2002;31:79–83. doi: 10.1038/ng874. [DOI] [PubMed] [Google Scholar]
- Hearn T, Spalluto C, Phillips VJ, Renforth GL, Copin N, Hanley NA, Wilson DI. Subcellular localization of ALMS1 supports involvement of centrosome and basal body dysfunction in the pathogenesis of obesity, insulin resistance, and type 2 diabetes. Diabetes. 2005;54:1581–1587. doi: 10.2337/diabetes.54.5.1581. [DOI] [PubMed] [Google Scholar]
- Ichihara S, Yamamoto K, Asano H, Nakatochi M, Sukegawa M, Ichihara G, Izawa H, Hirashiki A, Takatsu F, Umeda H, Iwase M, Inagaki H, et al. Identification of a Glutamic Acid Repeat Polymorphism of ALMS1 as a Novel Genetic Risk Marker for Early-Onset Myocardial Infarction by Genome-Wide Linkage Analysis. Circ Cardiovasc Genet. 2013;6(6):569–578. doi: 10.1161/CIRCGENETICS.111.000027. [DOI] [PubMed] [Google Scholar]
- International Advisory Group for the Revision of ICD-10 Mental and Behavioural Disorders. A conceptual framework for the revision of the ICD–10 classification of mental and behavioural disorders. World Psychiatry. 2011;10:86–92. doi: 10.1002/j.2051-5545.2011.tb00022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzi C, Maffei P, Milan G, Tardanico R, Foini P, Marshall JD, Scolari F. The Case | Familial occurrence of retinitis pigmentosa, deafness, and renal involvement. Kidney Int. 2011;79:691–692. doi: 10.1038/ki.2010.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joy T, Cao H, Black G, Malik R, Charlton-Menys V, Hegele RA, Durrington PN. Alstrom syndrome (OMIM 203800): a case report and literature review. Orphanet J Rare Dis. 2002;2:49. doi: 10.1186/1750-1172-2-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri S, Yoshitake K, Akahori M, Hayashi T, Furuno M, Nishino J, Ikeo K, Tsuneoka H, Iwata T. Whole-exome sequencing identifies a novel ALMS1 mutation (p.Q2051X) in two Japanese brothers with Alström Syndrome. Mol Vis. 2013;19:2393–406. [PMC free article] [PubMed] [Google Scholar]
- Kaya A, Orbak Z, Ҫayır A, Döneray H, Taşdemir Ş, Ozantürk A, Bingöl F. Combined occurrence of Alström Syndrome and bronchiectasis. Pediatrics. 2014;133(3):e780–e783. doi: 10.1542/peds.2013-0284. [DOI] [PubMed] [Google Scholar]
- Kingsmore SF, Dinwiddie DL, Miller NA, Soden SE, Saunders CJ for the Children’s Mercy Genomic Medicine Team. Adopting orphans: comprehensive genetic testing of Mendelian diseases of childhood by next-generation sequencing. Expert Rev Mol Diagn. 2011;11(8):855–868. doi: 10.1586/erm.11.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Hanaki K, Kawashima Y, Nagaishi J, Hayashi A, Okada S, Murakami J, Nanba E, Tomonaga R, Kanzaki S. A novel non-sense mutation in Alstrom syndrome: subcellular localization of its truncated protein. Clin Pediatr Endocrinol. 2003;12(2):114. [Google Scholar]
- Knorz VJ, Spalluto C, Lessard M, Purvis TL, Adigun FF, Collin GB, Hanley NA, Wilson DI, Hearn T. Centriolar association of ALMS1 and likely centrosomal functions of the ALMS motif-containing proteins C10orf90 and KIAA1731. Mol Biol Cell. 2010;(21):3617–3629. doi: 10.1091/mbc.E10-03-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocova M, Sukarova-Angelovska E, Kacarska R, Maffei P, Milan G, Marshall JD. The Unique Combination of Dermatological and Ocular Phenotypes in Alström Syndrome: Severe Presentation, Early Onset, and Two Novel ALMS1 Mutations. Br J Dermatol. 2011;164(4):878–880. doi: 10.1111/j.1365-2133.2010.10157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuburović V, Marshall JD, Collin GB, Nykamp K, Kuburović N, Milenković T, Rakić S, Djuric M, Ječmenica J, Milenković S, Naggert JK. Differences in the Clinical Spectrum of Two Adolescent Male Patients with Alström Syndrome. Clin Dysmorphol. 2013;22(1):7–12. doi: 10.1097/MCD.0b013e32835b9017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding nonsynonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. {HYPERLINK: http://www.ncbi.nlm.nih.gov/pubmed/17846036} [DOI] [PubMed] [Google Scholar]
- Leitch CC, Lodh S, Prieto-Echagüe V, Badano JL, Zaghloul NA. Basal body proteins regulate Notch signaling via endosomal trafficking. J Cell Sci. 2014;127(11):2407–2419. doi: 10.1242/jcs.130344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Vega R, Nelms K, Gekakis N, Goodnow C, McNamara P, Wu H, Hong N, Glynne R. A Role for Alstrom Syndrome Protein, Alms1, in Kidney Ciliogenesis and Cellular Quiescence. PLoS Genetics. 2007;3(1):e8. doi: 10.1371/journal.pgen.0030008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Li H, Li H, Xu F, Dong F, Sui R. Novel ALMS1 mutations in Chinese patients with Alström syndrome. Mol Vis. 2013;19:1885–1891. [PMC free article] [PubMed] [Google Scholar]
- Liu L, Dong B, Chen X, Li J, Li Y. Identification of a novel ALMS1 mutation in a Chinese family with Alström syndrome. Eye. 2009;23(5):1210–1212. doi: 10.1038/eye.2008.235. [DOI] [PubMed] [Google Scholar]
- Long PA, Evans JM, Olson TM. Exome sequencing establishes diagnosis of Alström syndrome in an infant presenting with non-syndromic dilated cardiomyopathy. Am J Med Genet A. 2015 Feb 23; doi: 10.1002/ajmg.a.36994. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louw JJ, Corveleyn A, Jia Y, Iqbal S, Boshoff D, Gewilliq M, Peeters H, Moerman P, Devriendt K. Homozygous loss-of-function mutation in ALMS1 causes the lethal disorder mitogenic cardiomyopathy in two siblings. Eur J Med Genet. 2014;57(9):532–535. doi: 10.1016/j.ejmg.2014.06.004. [DOI] [PubMed] [Google Scholar]
- Maffei P, Munno V, Marshall JD, Scandellari C, Sicolo N. The Alström syndrome: is it a rare or unknown disease? Ann Ital Med Int. 2002;17(4):221–228. [PubMed] [Google Scholar]
- Maffei P, Boschetti M, Marshall JD, Paisey RB, Beck S, Resmini E, Collin GB, Naggert JK, Milan G, Vettor R, Minuto F, Sicolo N, Barreca A. Characterization of the IGF system in 15 patients with Alstrom syndrome. Clin Endocrinol. 2007;66(2):269–275. doi: 10.1111/j.1365-2265.2007.02721.x. [DOI] [PubMed] [Google Scholar]
- Malm E, Ponjavic V, Nishina PM, Naggert JK, Hinman EG, Andréasson S, Marshall JD, Möller C. Full–Field Electroretinography and Marked Variability in Clinical Phenotype of Alström Syndrome. Arch Ophthalmol. 2008;126(1):51–57. doi: 10.1001/archophthalmol.2007.28. [DOI] [PubMed] [Google Scholar]
- Manara R, Citton V, Maffei P, Marshall JD, Naggert JK, Milan G, Vettor R, Baglione A, Vitale A, Briani C, Di Salle F, Favaro A. Degeneration and plasticity of the optic pathway in Alström syndrome. Am J Neuroradiol. 2015;36(1):160–165. doi: 10.3174/ajnr.A4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall JD, Ludman MD, Shea SE, Salisbury SR, Willi SM, LaRoche RG, Nishina PM. Genealogy, natural history, and phenotypic features of Alström Syndrome in a large Acadian kindred and three unrelated families. Am J Med Genet. 1997;73:150–161. doi: 10.1002/(sici)1096-8628(19971212)73:2<150::aid-ajmg9>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Marshall JD, Bronson RT, Collin GB, Nordstrom AD, Maffei P, Paisey RB, Carey C, Macdermott S, Russell-Eggitt I, Shea SE, Davis J, Beck S, et al. New Alström Syndrome phenotypes based on the evaluation of 182 cases. Arch Intern Med. 2005;165:675–683. doi: 10.1001/archinte.165.6.675. [DOI] [PubMed] [Google Scholar]
- Marshall JD, Beck S, Maffei P, Naggert JK. Alström Syndrome. Eur J Hum Genet. 2007a;15(12):1193–1202. doi: 10.1038/sj.ejhg.5201933. [DOI] [PubMed] [Google Scholar]
- Marshall JD, Hinman EG, Collin GB, Beck S, Cerqueira R, Maffei P, Milan G, Zhang W, Wilson DI, Hearn T, Tavares P, Vettor R, et al. Spectrum of ALMS1 variants and evaluation of genotype-phenotype correlations in Alström Syndrome. Hum Mutat. 2007b;28(11):1114–1123. doi: 10.1002/humu.20577. [DOI] [PubMed] [Google Scholar]
- Marshall JD, Paisey RB, Carey CM, McDermott S. In: GeneReviews [Internet. Pagon RA, Bird TC, Dolan CR, Stephens K, editors. Seattle (WA): University of Washington, Seattle; 2012. 1993–2003. [updated 2012 May] [Google Scholar]
- Marshall JD, Maffei P, Collin GB, Naggert JK. Alström Syndrome: Genetics and Clinical Overview. Curr Genom. 2011;12(3):225–235. doi: 10.2174/138920211795677912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall JD, Maffei P, Beck S, Barrett TG, Paisey R, Naggert JK. Clinical utility gene card for: Alström Syndrome - update 2013. Eur J Hum Genet. 2013;21(11) doi: 10.1038/ejhg.2013.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani D, Cerutti M, Pezzani L, Maffei P, Milan G, Esposito S. Syndromic obesity: clinical implications of a correct diagnosis. Ital J Pediatr. 2014;40(1):33. doi: 10.1186/1824-7288-40-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton JA, Owen KR, Ricketts CJ, Crabtree N, Shaikh G, Ehtisham S, Porter JR, Carey C, Hodge D, Paisey R, Walker M, Barrett TG. Syndromic obesity and diabetes: changes in body composition with age and mutation analysis of ALMS1 in 12 UK kindreds with Alström Syndrome. J Clin Endocrinol Metab. 2006;91:3110–3116. doi: 10.1210/jc.2005-2633. [DOI] [PubMed] [Google Scholar]
- Ozantürk A, Marshall JD, Collin GB, Düzenli S, Marshall RP, Candan S, Tos T, Esen I, Taşkesen M, Cayir A, Oztürk S, Ustün I, et al. The phenotypic and molecular genetic spectrum of Alström Syndrome in 45 Turkish kindreds and a literature review of Alström Syndrome in Turkey. J Hum Genet 2015. 2014;60(1):1–9. doi: 10.1038/jhg.2014.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özgül RK, Satman I, Collin GB, Hinman EG, Marshall JD, Kocaman O, Tütüncü Y, Yilmaz T, Naggert JK. Molecular analysis and long-term clinical evaluation of three siblings with Alström syndrome. Clin Genet. 2007;72:351–356. doi: 10.1111/j.1399-0004.2007.00848.x. [DOI] [PubMed] [Google Scholar]
- Paisey RB, Geberhiwot T, Waterson M, Cramb R, Steeds R, Williams K, White A, Hardy C. Modification of severe insulin resistant diabetes in response to lifestyle changes in Alström syndrome. Eur J Med Genet. 2014;57(2–3):71–75. doi: 10.1016/j.ejmg.2013.12.008. [DOI] [PubMed] [Google Scholar]
- Patel S, Minton JAL, Weedon MN, Frayling TM, Ricketts C, Hitman GA, McCarthy MI, Hattersley AT, Walker M, Barrett TG. Common variations in the ALMS1 gene do not contribute to susceptibility to type 2 diabetes in a large white UK population. Diabetologia. 2006;49:1209–1213. doi: 10.1007/s00125-006-0227-2. [DOI] [PubMed] [Google Scholar]
- Pereiro I, Hoskins BE, Marshall JD, Collin GB, Naggert JK, Teresa Piñeiro-Gallego T, Oitmaa E, Katsanis N, Valverde D, Beales PL. Arrayed Primer Extension (APEX) technology simplifies mutation detection in Bardet Biedl and Alström Syndrome. Eur J Hum Genet. 2011;19(4):485–488. doi: 10.1038/ejhg.2010.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñeiro-Gallego T, Cortón M, Ayuso C, Baiget M, Valverde D. Molecular approach in the study of Alström syndrome: Analysis of ten Spanish families. Mol Vis. 2012;18:1794–1802. [PMC free article] [PubMed] [Google Scholar]
- Redin C, Le Gras S, Mhamdi O, Geoffroy V, Stoetzel C, Vincent MC, Chiurazzi P, Lacombe D, Ouertani I, Petit F, Till M, Verloes A, et al. Targeted high-throughput sequencing for diagnosis of genetically heterogeneous diseases: efficient mutation detection in Bardet-Biedl and Alström Syndromes. J Med Genet. 2012;49(8):502–512. doi: 10.1136/jmedgenet-2012-100875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyoura M, Woudstra C, Halaby G, Baz P, Senée V, Guillausseau PJ, Zalloua P, Julier C. A novel ALMS1 splice mutation in a non-obese juvenile-onset insulin-dependent syndromic diabetic patient. Eur J Hum Genet. 2014;22(1):140–143. doi: 10.1038/ejhg.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathya Priya C, Sen P, Umashankar V, Gupta N, Kabra M, Kumaramanickavel G, Stoetzel C, Dollfus H, Sripriya S. Mutation spectrum in BBS genes guided by Homozygosity mapping in an Indian cohort. Clin Genet. 2015;87(2):161–166. doi: 10.1111/cge.12342. [DOI] [PubMed] [Google Scholar]
- Saunders CJ, Miller NA, Soden SE, Dinwiddie DL, Noll A, Alnadi NA, Andraws N, Patterson ML, Krivohlavek LA, Fellis J, Humphray S, Saffrey P, et al. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci Transl Med. 2012;4(154):ra135. doi: 10.1126/scitranslmed.3004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- Sheck L, Al-Taie R, Sharp D, Vincent A. Alström syndrome – an uncommon cause of early childhood retinal dystrophy. BMJ Case Reports. 2011 doi: 10.1136/bcr.06.2011.4388. published online 18 August 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenje LT, Andersen P, Halushka MK, Lui C, Fernandez L, Collin GB, Amat-Alarcon N, Meschino W, Cutz E, Chang K, Yonescu R, Batista DA, et al. Mutations in Alström protein impair terminal differentiation of cardiomyocytes. Nat Commun. 2014;5:3416. doi: 10.1038/ncomms4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha SK, Bhangoo A, Anhalt H, Maclaren N, Marshall JD, Collin GB, Naggert JK, Ten S. Effect of Metformin and Rosiglitazone in a Prepubertal Boy with Alstrom Syndrome. J Pediatr Endocrinol Metab. 2007;20:1045–1052. doi: 10.1515/jpem.2007.20.9.1045. [DOI] [PubMed] [Google Scholar]
- Smith JC, McDonnell B, Retallick C, McEniery C, Carey C, Davies JS, Barrett T, Cockcroft JR, Paisey R. Is arterial stiffening in Alstrom Syndrome linked to the development of cardiomyopathy? Eur J Clin Invest. 2007;37:99–105. doi: 10.1111/j.1365-2362.2007.01759.x. [DOI] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, Shaw K, Phillips A, Cooper DN. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet. 2014;133(1):1–9. doi: 10.1007/s00439-013-1358-4. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taşdemir S, Güzel-Ozantürk A, Marshall JD, Collin GB, Özgül RK, Narin N, Dündar M, Naggert JK. Atypical Presentation and a Novel Mutation in ALMS1: Implications for Clinical and Molecular Diagnostic Strategies for Alström Syndrome. Clin Genet. 2012;83(1):96–98. doi: 10.1111/j.1399-0004.2012.01883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taşkesen M, Collin GB, Evsikov AV, Güzel A, Özgül RK, Marshall JD, Naggert JK. Novel Alu retrotransposon insertion leading to Alström syndrome. Hum Genet. 2012;131(3):407–413. doi: 10.1007/s00439-011-1083-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylan F, Kvarnung M, Lindstrand A, Bui T, Nordgren A, Blennow E, Nordenskjöld M, Nilsson D. Diagnostic exome sequencing can alter a primary clinical diagnosis. ASHG Meeting; Boston MA. October, 2013; 2013. program #2573W. [Google Scholar]
- t’Hart LM, Dekker JM, Heine RJ, Maassen JA. Lack of association between gene variants in the ALMS1 gene and Type 2 diabetes mellitus. Diabetologia. 2003;46:1023–1024. doi: 10.1007/s00125-003-1138-0. [DOI] [PubMed] [Google Scholar]
- Titomanlio L, Buoninconti A, Sperandeo MP, De Brasi D, Pepe A, Andria G, Sebastio G. Alström Syndrome: intrafamilial phenotypic variability in sibs with a novel nonsense mutation of the ALMS1 gene. Clin Genet. 2004;65:156–157. doi: 10.1111/j.0009-9163.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- Wang X, Wang H, Cao M, Li Z, Chen X, Patenia C, Gore A, Abboud EB, Al-Rajhi AA, Lewis RA, Lupski JR, Mardon G, et al. Whole-exome sequencing identifies ALMS1, IQCB1, CNGA3, and MYO7A mutations in patients with Leber congenital amaurosis. Hum Mutat. 2011;32(12):1450–1459. doi: 10.1002/humu.21587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 2010. 2010;26(7):873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zulato E, Favaretto F, Veronese C, Campanaro S, Marshall JD, Romano S, Cabrelle A, Collin GB, Zavan B, Belloni AS, Rampazzo E, Naggert JK, et al. ALMS1-deficient fibroblasts over-express extra-cellular matrix components, display cell cycle delay and are resistant to apoptosis. PLoS One. 2011;6:e19081. doi: 10.1371/journal.pone.0019081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumsteg U, Muller PY, Miserez AR. Alstrom syndrome: confirmation of linkage to chromosome 2p 12–13 and phenotypic heterogeneity in three affected sibs. J Med Genet. 2000;37(7):E8. doi: 10.1136/jmg.37.7.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.