Abstract
The first step in the biogenesis of microRNAs is the processing of primary microRNAs (pri-miRNAs) by the microprocessor complex, composed of the RNA binding protein DGCR8 and the ribonuclease type III DROSHA1–4. This initial event requires the recognition of the junction between the stem and the flanking single-stranded RNA of the pri-miRNA hairpin by DGCR8 followed by recruitment of DROSHA, which cleaves the RNA duplex to yield the pre-miRNA product5. While the mechanisms underlying pri-miRNA processing have been elucidated, the mechanism by which DGCR8 recognizes and binds pri-miRNAs as opposed to other secondary structures present in transcripts is not understood. We find that methyltransferase like 3 (METTL3) methylates pri-miRNAs, marking them for recognition and processing by DGCR8. Consistent with this, METTL3 depletion reduced the binding of DGCR8 to pri-miRNAs and resulted in the global reduction of mature miRNAs and concomitant accumulation of unprocessed pri-miRNAs. In vitro processing reactions confirmed the sufficiency of the m6A mark in promoting pri-miRNA processing. Finally, gain-of-function experiments revealed that METTL3 is sufficient to enhance miRNA maturation in a global and non-cell-type specific manner. Our findings reveal that the m6A mark acts as a key post-transcriptional modification that promotes the initiation of miRNA biogenesis.
In our search for post-transcriptional modifications that regulate miRNA processing, we conducted a systematic search for sequence motifs that are over-represented in miRNA-containing regions using the FIRE algorithm6. We observed the over-representation of the GGAC motif in pri-miRNA sequences relative to shuffled sequences (Fig. 1a). This motif is consistent with a previously established recognition sequence RGAC for the RNA methyltransferase enzyme METTL37–9. In contrast to pri-miRNA sequences, this element was not enriched in pre-miRNA sequences, and was actually depleted relative to shuffled sequences (Extended Data Fig. 1a). METTL3 is the catalytic subunit of a multi-component enzyme that methylates RNA, thereby adding the N6-methyladenosine (m6A) mark to eukaryotic RNAs10–13.
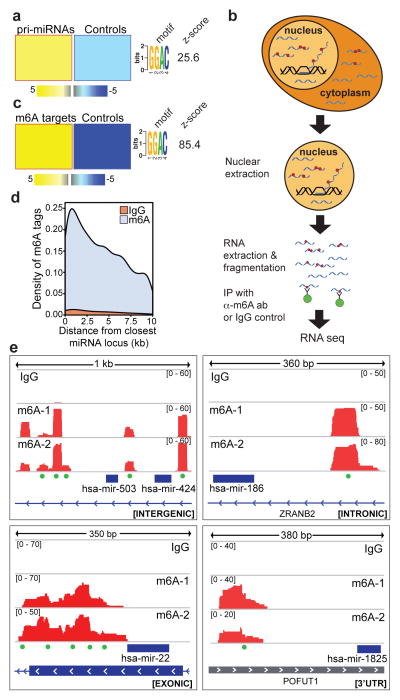
Figure 1. m6A mark is present in pri-miRNA regions.
a, Motif discovery analysis in pri-miRNA sequences using FIRE reveals over-representation of the METTL3 motif; yellow represents over-representation and blue under-representation. The magnitude of the over/under-representation is reflected in the linear scale heat-map shown at the bottom. The z-score is specified on the right. b, Schematic representation of the m6A-seq protocol. c, FIRE motif analysis of m6A peaks compared to controls sequences of the same length. The over-represented motif and their z-score are depicted on the right as in (a). d, Density plot of the abundance of m6A marks and their proximity to given miRNAs within transcripts. Peaks obtained from the IgG immunoprecipitation were used as controls. e, IGV tracks displaying examples of sequencing read clusters from two m6A-seq replicates are shown next to the pre-miRNA genomic loci. The green dots at the bottom of the tracks depict the position of METTL3 consensus motifs.
To determine if the over-representation of the m6A methylation motif in pri-miRNA sequences signifies increased m6A methylated sequences, we conducted m6A-seq8, by immunoprecipitating nuclear RNA from the MDA-MB-231 breast cancer cell line with an anti-m6A antibody followed by RNA seq (Fig. 1b). A search for cis-regulatory elements from m6A-seq revealed a significant enrichment of the METTL3 motif relative to shuffled sequences (Fig. 1c). Furthermore, when we analyzed the density of the peaks in the vicinity of miRNA loci, we found a substantial increase in the density of peaks proximal to pre-miRNA sequences, corresponding to pri-miRNA regions (Fig. 1d). We next inspected individual clusters of reads using the Integrative Genomics Viewer (IGV) software14 and found numerous cases in which there were significant peaks in locations that correspond to pri-miRNAs. These clusters were located in both intergenic and intragenic pri-miRNA sites that contained canonical METTL3 motifs (Fig. 1e). Thus, these results reveal that the m6A modification is enriched within pri-miRNA sequences.
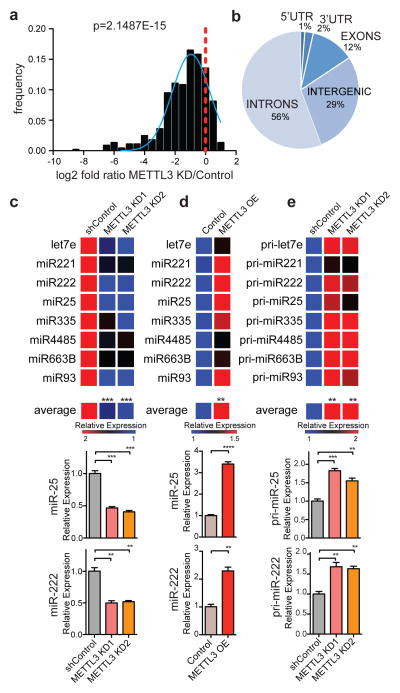
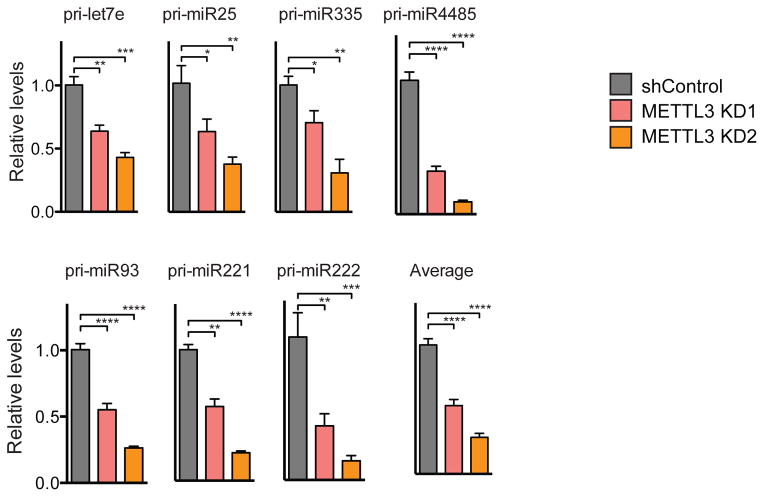
To determine if METTL3 plays a role in miRNA processing, we conducted genome-wide miRNA expression profiling of MDA-MB-231 cells expressing a control shRNA as well as cells expressing two independent shRNAs targeting METTL3 (Extended Data Fig. 1b and 1c). METTL3 depletion using independent shRNAs led to a global downregulation of mature miRNAs (p=2.15E-15; Fig. 2a and Extended Data Fig. 1d). Remarkably, most miRNAs (~70%) were downregulated by at least 30%. The miRNAs affected by METTL3 depletion were diverse in terms of their genomic locations, predominantly being intronic (56%), followed by intergenic (29%), exonic (12%) and in untranslated regions (UTRs; 3%), indicating that the effect of METTL3 depletion on miRNA processing was not specific to miRNAs originating from specific locations within or outside of genes (Fig. 2b). We next validated those effects by examining the expression of specific miRNAs by quantitative real time PCR (qRT-PCR; Fig. 2c and Extended Data Fig. 2). Importantly, the effect of METTL3 depletion on miRNA processing was not restricted to a particular cell line and was also observed in multiple cell types (Extended Data Fig. 3). These experiments demonstrate that METTL3 is required for basal expression of the vast majority of miRNAs in these cancerous and non-cancerous cells. To test if METTL3 expression is sufficient to promote miRNA processing, we stably overexpressed METTL3 in MDA-MB-231 cells. METTL3 overexpression was sufficient to significantly increase the expression levels of mature miRNAs (Fig. 2d and Extended Data Fig. 4). We then quantified the expression levels of pri-miRNAs to determine if METTL3 depletion could impact pri-miRNA levels. As expected, we observed a clear and significant up-regulation in the levels of pri-miRNAs upon METTL3 depletion (Fig. 2e and Extended Data Fig. 5a). These effects required enzymatically active METTL3 since they were abrogated upon mutation of the predicted catalytic residues15 (Extended Data Fig. 5c, d). Neither METTL3 depletion nor its overexpression altered the expression, sub-cellular localization, or activity of the Microprocessor complex—consistent with a direct role of METTL3 in pri-miRNA processing (Extended Data Fig. 6). Our findings reveal that pri-miRNAs are marked by the METTL3-dependent m6A modification and that METTL3 expression is required for the appropriate processing of most pri-miRNAs to mature miRNAs.
Figure 2. METTL3 modulates the expression levels of miRNAs.
a, Histogram depicting fold change (log2) in miRNA expression. The ratio of the average value for two independent shRNAs over the average of the two controls is shown. The p-value of the analysis is indicated. b, Pie-chart representation of the genomic locations of miRNAs downregulated upon METTL3 depletion. c, Heat-map representation of qRT-PCR quantification of eight representative mature miRNAs that were affected by METTL3 depletion. Red represents increased expression while blue represents reduced expression. A heat map depicts their aggregate expression change upon METTL3 modulation. At the bottom, bar graphs showing specific examples. d, Heat-map representation of mature miRNA quantification from (c) by qRT-PCR upon METTL3 over-expression (OE). e, Heat-map representing the quantification of pri-miRNA forms of miRNAs from (c–d) by qRT-PCR upon METTL3 depletion. All heat-maps and bar graphs represent a linear scale. Error bars represent s.d. ***, p value <5E-4; **, p value <1E-3.
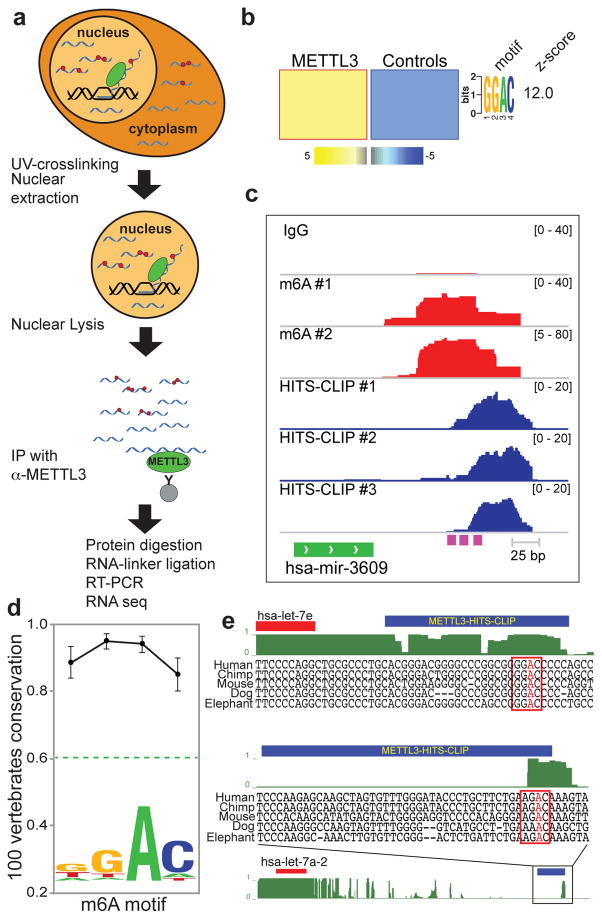
In order to directly implicate METTL3 in the m6A methylation of pri-miRNAs, we conducted high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation16 (HITS-CLIP) experiments using an antibody against endogenous METTL3 (Fig. 3a). Consistent with our findings from m6A-seq, METTL3 HITS-CLIP sequences displayed a significant enrichment of the METTL3 motif (Fig. 3b and Extended Data Fig. 7a). Despite the transient nature of the interaction between METTL3 and its targets, we observed a significant overlap in the miRNAs affected by METTL3 depletion that shared both the METTL3 footprint by HITS-CLIP and the m6A mark by m6A-seq (p-value<2.4E-15; Fig. 3c and Extended Data Fig. 7b). In order to investigate the evolutionary conservation of the METTL3 motif across different species, we used the PhastCons software17 to analyze 30 miRNAs that are conserved among 100 vertebrates (www.targetscan.org) and also contain m6A and/or METTL3 tags. This analysis showed a high degree of conservation for the METTL3 motif among vertebrates (Fig. 3d–e).
Figure 3. METTL3 targets pri-miRNAs for m6A methylation.
a, Schematic representation of the HITS-CLIP protocol used. b, FIRE analysis of the motif of METTL3 HITS-CLIP binding sites. The color scale of the linear scale heat-map is the same as in Fig. 1a. c, An example of sequencing clusters obtained from METTL3 HITS-CLIP (blue) and m6A-seq (red). m6A seq was done in duplicate using IgG as control. METTL3 HITS-CLIP was done in triplicate. The purple boxes at the bottom of the tracks represent conserved METTL3 motifs. d, Average vertebrate conservation of the METTL3 motif of a group of conserved pri-miRNAs using the PhastCons software17. The dotted green line depicts the average conservation of a region of 100nt that surrounds (and includes) the motifs. Error bars represent s.e.m. e, Two examples of pri-miRNA genomic regions containing HITS-CLIP tags are shown. At top, pre-miRNAs are marked in red boxes and METTL3 HITS-CLIP tags in blue boxes. The conserved METTL3 motif is framed in red with the putative methylated adenosine in red.
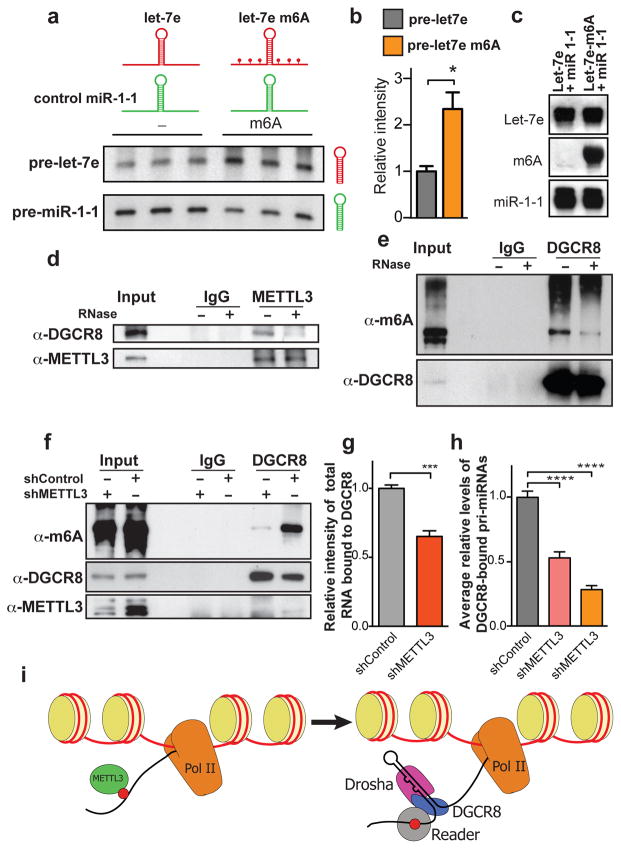
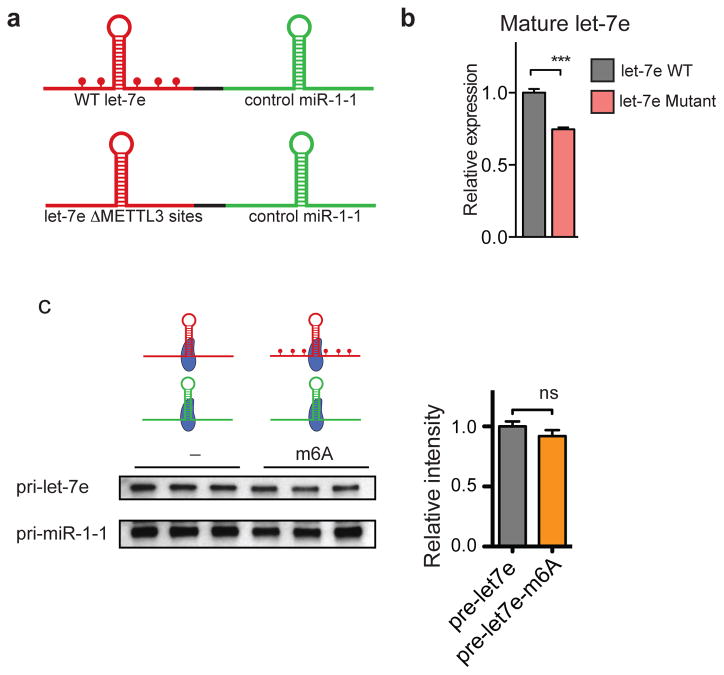
To demonstrate a direct role of the m6A modification in pri-miRNA processing we performed in vitro processing reactions using whole cell extracts from HEK293T cells transfected with DGCR8 and DROSHA18. In this gain-of-function experiment, the extracts were used to process in vitro transcribed pri-miRNAs containing modified N6-methyladenosine or unmodified bases. Consistent with our model, methylated pri-let-7e was more efficiently processed by the microprocessor to produce pre-let7e relative to its un-methylated counterpart as detected by northern blot (Fig. 4a–c). These in vitro experiments suggest that m6A marks in pri-miRNAs are required for efficient processing of pri-miRNAs in vitro. Additionally, we performed a loss-of-function experiment by testing the impact on miRNA processing of mutating select adenosines present in potential METTL3 motifs located in a pri-miRNA. To do this, we modified a previously described reporter construct19. In one reporter, we introduced a wild type version of pri-let-7e. In the other reporter, the adenosines of the METTL3 motifs present in the pri-let-7e region (i.e. 5 out of 18 adenosines present outside the pre-miRNA sequence) were mutated (Extended Data Fig. 8a). Consistent with our findings, mutation of METTL3 motifs in pri-let-7e significantly reduced its processing to the mature form (Extended Data Fig. 8b).
Figure 4. m6A methylation of pri-miRNAs is required for normal processing by DGCR8.
a, In vitro pri-miRNA processing reactions. Pri-let-7e, was transcribed with either modified N6-methyladenosine (depicted in the schematic as red dots) or with unmodified bases. Hybridization intensities were quantified, normalized to the controls and shown in the bar graph (b). c, Input for experiment shown in (a). d, Co-immunoprecipitation of the METTL3-interacting protein DGCR8. Western blot using the indicated antibodies with IgG used as control for the IP. e, Immunoprecipitation of endogenous DGCR8. Western blot using an antibody against m6A methylated RNA. f, Similar as e, but control cells or cells depleted of METTL3 were compared. g, Similar immunoprecipitation as e, however, after immunoprecipitation of DGCR8, a radio-labeled RNA linker was ligated to the RNA bound by DGCR8. Bars represent the average normalized intensity of three biological replicates. ***, p <0.001. h, Similar as g but the pri-miRNA bound by DGCR8 were extracted and quantified by qRT-PCR. The bar graph shows the average level of a panel of miRNAs analyzed and shown in more detail in Extended Data Figure 9. i, Model of METTL3 regulation of miRNA biogenesis. The molecules represented in the schematic are: Histones (yellow), RNA Pol II (orange), METTL3 (green), m6A (red), DGCR8 (blue), DROSHA (pink) and a putative unknown reader of the m6A mark (gray).
Since the m6A mark is added to pri-miRNAs, and we observed an accumulation of unprocessed pri-miRNAs upon METTL3 depletion, we questioned whether METTL3 is required for the engagement of the pri-miRNA by the microprocessor. We first tested whether METTL3 and DGCR8 are part of a complex. Immunoprecipitation of METTL3 from nuclear lysates co-precipitated DGCR8; however this interaction was mediated through RNA, since RNase treatment disrupted this association (Fig. 4d).
We next sought to determine if DGCR8 interacts with methylated RNA in vivo. To do this, we UV-crosslinked cells, isolated nuclear fractions and immunoprecipitated endogenous DGCR8. After immunoprecipitation of DGCR8 and SDS-PAGE, m6A immunoblotting revealed that DGCR8 indeed interacts with methylated RNA (Fig. 4e). Based on our observations, we predicted that a reduction in METTL3 levels should decrease the levels of methylated RNA bound by DGCR8. Indeed, depleting METTL3 significantly reduced the m6A methylated RNA bound by DGCR8 (Fig. 4f). However, the binding of DGCR8 to methylated RNA is unlikely to be direct since we detected no preference for DGCR8 binding to methylated RNA over unmethylated RNA in vitro (Extended Data Fig. 8c).
Since mRNAs tend to form secondary structures including short hairpins that resemble pri-miRNAs, a potential basis of pri-miRNA methylation might be to confer specificity for, and facilitate the recognition of, pri-miRNA structures by DGCR8. Based on this hypothesis, we would expect that a reduction in the levels of methylated pri-miRNAs would reduce the total amount of RNA recognized and bound by DGCR8. To test this, we immunoprecipitated DGCR8 from control and METTL3 depleted cells and radiolabeled the total RNA bound to DGCR8. Consistent with our model, METTL3 depletion significantly reduced the amount of total RNA (Fig. 4g) as well as pri-miRNAs (Fig. 4h and Extended Data Fig. 9) bound by DGCR8. These findings suggest that the m6A methylation mark allows for the effective recognition of pri-miRNAs by DGCR8 and their subsequent processing to pre-and mature miRNAs.
By using an unbiased approach, we have identified m6A as a novel regulator of miRNA processing. Our findings reveal that METTL3 methylates primary inter- and intragenic miRNAs. The m6A modification facilitates the recognition of pri-miRNA sequences and marks an initiation event in miRNA biogenesis (Fig. 4i). We propose that the m6A mark on transcripts at pri-miRNA sequences initiates a global co-transcriptional program comprising the engagement and processing of primary miRNAs by the microprocessor machinery. The m6A mark thus plays an important role in the nucleus—allowing the microprocessor complex to recognize its specific substrates as opposed to unintended secondary structures. Additionally, altered METTL3 expression in various human malignancies may significantly contribute to the aberrant expression of miRNAs seen in cancer.
METHODS
Tissue culture
MDA-MB-231, HeLa and human umbilical vein endothelial cells (HUVECs) were purchased from ATCC. MDA-MB-231, HEK293T and HeLa cells were propagated in vitro with Dulbecco’s modified Eagles medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% penicillin-streptomycin, 2mM L-Glutamine, 1mM sodium pyruvate, and 2.5 ug/mL fungizone. HUVECs were cultured with Endothelial Cell Growth Media (CC-3162, Lonza). Experiments with HUVEC were conducted with cells from passage 2 to 5.
Stable overexpression and knockdowns
Generation of lentivirus-mediated knockdown and retroviral-overexpressing cells were performed as described previously20. For overexpression studies, human cDNA of METTL3 with C-terminal FLAG tag was cloned into pBabe-Puro retroviral expression vector. For knockdown studies, commercially available shRNA glycerol stocks were purchased (Sigma). Two shRNAs in pLKO lentiviral expression vectors against METTL3 that showed the best knockdown efficiencies (TRCN0000034715, TRCN0000034717) and control shRNA (SHC002) were used for experiments. Due to the loss of knockdown over time, fresh knock down cell-lines were generated after a few passages.
qRT-PCR
Mature miRNAs were quantified by either Taqman microRNA assays (Applied Biosystems) or poly-A tailing of total RNA followed by reverse-transcriptase (RT) reaction with T7 oligo dT (NEB). Quantitative miRNA expression data were acquired and analyzed using a 7900HT Fast Real-Time PCR System (Applied Biosystems). Pri-miRNAs were measured using the TaqMan Pri-miRNA Assay. RNU44, GusB, GAPDH and 18S were used as endogenous controls.
Small RNA isolation
Total RNA from cells was extracted and purified using the mirVana (Applied Biosystems) or Total RNA Purification Kit (Norgen Biotek) according to the manufacturer’s instructions.
m6A Immunoprecipitation and RNA seq
3×107 cells/sample were lysed using LB1 buffer (50 mM Hepes-KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% Triton x-100, and protease inhibitors). The nuclear fraction was then lysed with M-PER buffer (Thermo Scientific) and diluted 10-fold in dilution buffer (50mM Tris-Cl, pH 7.4, 100mM NaCl) before the immunoprecipitation. Rabbit α-m6A antibody (Synaptic Systems) and rabbit IgG control bound to protein A Dynabeads (Invitrogen) were used for the immunoprecipitations. The immunoprecipitated RNA was eluted with N6-methyladenosine (Sigma-Aldrich), ethanol precipitated and resuspended in water. RNA was barcoded using ScriptSeq V2 kit (Epicentre) and sent for sequencing.
m6A analysis
Paired reads were mapped to the human genome (build hg19) using bowtie2 with “sensitive-local” option21. Consistent paired reads were converted into genomic intervals (bed format). ChIPseeqer22 was used to detect statistically significant peaks that were present in the m6A co-precipitation samples and not the IgG control. The following parameters were used: -t 10 –fold_t 2 –minlen 20. FIRE was used for non-discovery analysis, and ChIPseeqer was used for shuffled sequences.
microRNA expression profiling (microarray)
Total RNA extracted from two independent stable knockdowns of METTL3 and two control cells was labeled and hybridized on miRNA microarrays by LC sciences. The arrays have probes for all miRNAs of human (1872 precursors, 2578 mature) available in the miRBase database (Release 20). Of all the probes assayed, those corresponding to 441 miRNAs revealed a signal above background in at least two of the MDA-MB-231 cell lines. Due to the global change in miRNA levels, normalization was done using a set of small ribosomal RNAs present on the platform for use in data normalization. This set of controls contains 6 probes against the 5S ribosomal RNAs and detects 17 RNA species altogether (NR_023363 to NR_023379).
HITS-CLIP
We performed HITS-CLIP as previously described23 with some modifications. After UV crosslinking (400mJ/cm2 of 254nm UV), we isolated nuclear fractions from 3×107 cells/sample using LB1 buffer. The nuclear fraction was then lysed in M-PER buffer and diluted 10 fold in a buffer containing 50 mM Tris-HCl pH 7.4 and 100mM NaCl, before the immunoprecipitation. For the endogenous immunoprecipitation we used 3ug of α-METTL3 rabbit antibody (Bethyl) or rabbit IgG as control bound to protein A dynabeads (Invitrogen). After the immunoprecipitation a 32P-labeled linker was ligated to the RNA on the beads. The samples were resolved using SDS-PAGE, transferred to nitrocellulose membrane and exposed overnight. The films were used to guide the cut of the membrane to extract the radiolabeled RNA. RNA was extracted in 200ul of PK buffer (100mM Tris-Cl pH 7.5, 50mM NaCl and 10mM EDTA) containing proteinase K (4mg/ml), then adding 200ul of PK buffer containing 7M urea and lastly 400 ul of acid phenol:chloroform, each step 20 min at 37°C at 1,000 rpm shaking. The aqueous solution was then precipitated and resuspended in RNase-free water as described in the in vitro processing section. An RNA linker was ligated to the purified RNA at 16°C overnight. The RNA was once again extracted using phenol:chloroform and precipitated as previously described. This new RNA was reverse transcribed and PCR enriched. These PCR amplicons were resolved in a TBE-Urea poly-acrylamide gel and DNA between 90 and 140 bp was extracted. This DNA was further enriched with the addition of Solexa fusion primers and the product was resolved in a 2% metaphor agarose/TBE gel, from which and a DNA between 150–170 bp was extracted. The samples were submitted for high throughput sequencing using an Illumina HiSeq 2000 instrument with 50bp single-end runs.
HITS-CLIP analysis
The reads were trimmed and clipped using cutadapt (v1.2.1) and were subsequently mapped to the human genome (build hg19) using bowtie221. CIMS package23 was used to detect CLIP clusters with cross-linking induced mutations. FIRE was used for non-discovery analysis, and ChIPseeqer for shuffled sequences.
Density plots analysis
The distance between peaks located in introns to closest miRNAs were measured using closestBed. In R, ggplot was used to generate the density plots.
Co-Immunoprecipitations
Cells were lysed with LB1 and the nuclear fraction was then lysed with M-PER buffer (Thermo Scientific) and diluted 10-fold in dilution buffer. Immunoprecipitations were performed with α-METTL3 rabbit antibody (Bethyl) or α-DGCR8 rabbit antibody (Abcam), previously bound to magnetic Dynabeads Protein A (Life Technologies), in the presence with either RNase A or RNase inhibitor (Promega). The immunoprecipitations were washed twice with high-salt buffer (50 mM Tris-HCl pH 7.4, 300 mM NaCl) followed by two times more wash with low-salt buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl). The following antibodies were used for western blot analysis α-METTL3 (mouse, Novus Biologicals), α-DGCR8 (mouse, Abcam), α-m6A (mouse, Synaptic Systems). For the in vitro binding assay once DGCR8 was immunopurified and still bound to the magnetic beads it was incubated with 1ug of pri-let-7e and 200ng of pri-miR-1-1, in the presence of RNAse inhibitors for 20 min at room temperature, washed twice and eluted with 170 ul of RNA elution buffer (0.3 M sodium acetate, pH 5.5 and 2% SDS). RNA was then extracted with 200ul of acid phenol:chloroform, precipitated and resuspended in RNase-free as described in the in vitro processing section. An aliquot of the elution was then used for northern blot analysis.
For the radiolabeled experiments the cells were UV-crosslinked, the nuclear fraction was lysed with M-PER buffer and diluted 10-fold in dilution buffer. The nuclear extracts were immunoprecipitated with α-DGCR8 antibody or IgG as control. A RNA oligo was labeled with 32P-gamma-ATP (Perkin Elmer) and ligated to the co-immunoprecipitated RNA as in the HITS-CLIP protocol. The samples (triplicates) were loaded in a SDS-PAGE, transferred, and the membrane was exposed overnight or longer if needed. After the image of the radiolabeled RNA was obtained, the same membrane was immunoblotted for DGCR8 for normalization. Quantitation was achieved using ImageJ24.
For the endogenous DGCR8-pri-miRNA binding experiments, similar to the radiolabeled experiments, the cells were UV-crosslinked and the nuclear fraction was lysed with M-PER buffer and diluted in dilution buffer. The nuclear extracts were immunoprecipitated with an anti-DGCR8 antibody or IgG as a control. The RNA was extracted by treatment with 200 ul of proteinase K (4mg/ml) in PK buffer (100mM Tris-Cl pH 7.5, 50mM NaCl and 10mM EDTA) and incubation at 37°C for 20 min with shaking. Then, 200ul 7M urea in PK buffer was added and incubated 20 min more at 37°C with shaking. After that 400ul acid phenol:chloroform (Ambion) was added to the solution, vortexed and centrifuged at maximum speed for 5 min. The aqueous phase was precipitated with 50ul 3M NaOAc pH 5.2, 0.75ul glycoblue and 1mL of 1:1 ethanol:isopropanol at −20°C at least 30 min. The mix was then centrifuged at 4°C at maximum speed for 15 min, and the pellet washed with 80% ethanol and air-dried. The RNA was resuspended in 20ul of RNase-free water and 8 ul were used to produce cDNA by ramdom hexamers (SuperScript III First-Strand Synthesis System, Life Technologies) and qRT-PCR was carried out using specific miRNA primers.
Northern Blots
Non-radioactive northern blots were performed as previously described25. RNA was extracted with mirVana (Applied Biosystems) or Total RNA Purification Kit (Norgen Biotek). DIG-labeled LNA probes against let-7e and miR-1-1 were obtained from EXIQON. Pre-stained marker (DynaMarker) was obtained from BioDynamics Laboratory Inc and Low range ssRNA Ladder from NEB. Mini-protean 10% and 15% pre-cast TBE-Urea gels were purchased from Bio-RAD and TBE buffer from Invitrogen. Samples were prepared using 2x Gel loading buffer (Ambion) and denatured 5min at 95C. Gels were at 200V in 1X TBE buffer and then transfered to a nylon membrane at 80V for 1 hour in 0.5X TBE buffer. Membranes were either chemically crosslinked with EDC (0.753g of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide in 24ml of 125mM solution of 1-methylimidazole pH of 8.0) for mature and pre-miRNAS (2 hours at 65C) or UV crosslinking for pri-miRNAs (240 mJoules). The membrane was then pretreated with hybridization buffer (DIG Easy Hyb Granules, Roche) at 37°C for at least 30 min in hybridization oven. The probe was denatured at 95°C for 1 min and added to the hybridization buffer (37°C overnight). The membrane was washed twice with Low Stringent Buffer at 37°C for 15 min, twice with High Stringent Buffer at 37°C for 5 min and once with Washing Buffer at 37°C for 10 min. Then the membrane was incubated in Blocking Buffer (DIG Wash and block buffer set, Roche) for 3 hrs at room temperature and the anti-Digoxigenin-AP antibody (Roche) was added (1:15,000 dilution) and incubated at room temperature for an extra 30 min. The membrane was washed in DIG Washing buffer four times for 15 min each at room temp. Finally the membrane was incubated in development buffer for 5 min and developed with CDP-Star (Roche) for 5 min and exposed to X-ray films.
In vitro pri-miRNA processing
Pri-miRNAs were in vitro transcribed using the T7 based MEGAshortscript kit (Life Technologies) according to the manufacture’s indications. In cases were m6A modifications were added in vitro, N6-methyl-ATP (tri-link) was used in a ratio 4:1 to ATP in the in vitro transcription reaction. Pri-miRNA processing reactions were performed as previously described18. In brief, HEK293T cells were co-transfected plasmids carrying DROSHA and DGCR8 (obtained from Addgene). 48 hours later, cells were lysed in lysis buffer (20 mM Tris-HCl pH 8.0, 100 mM KCl, and 0.2 mM EDTA) and sonicated for 1 min with 5 sec pulses at 30% amplitude. The whole cell was obtained after centrifugetion at 12,000 rpm at 4°C for 15 min. In the case of testing the effect of METTL3 depletion on pri-miRNA processing, cells were not transfected and the endogenous activity was measured. The in vitro processing reaction contained 3 ul of 64 mM MgCl2 solution, 1 ug of let-7e pri-miRNA (wildtype or m6A methylated), 50ng of control pri-miR-1-1, 0.75 ul of RNase inhibitor, 15 ul of HEK293T whole extract and RNase-free water to a final volume of 30 ul. The reactions were incubated at 37°C for 90 min. Then 170 ul of RNA elution buffer (0.3 M sodium acetate, pH 5.5 and 2% SDS) was added to the reaction to dissociate the RNA from the proteins and terminate the reaction. RNA was then extracted with 200 ul of acid phenol:chloroform (Ambion), vortexed and centrifuged for 5 min at room temperature. After extraction the RNA was precipitated at −20°C by adding 20 ul of sodium acetate (3M), 1 ul of glycoblue (Life Technologies), and 800 ul of a mix isopropanol:ethanol (1:1). The mix was then centrifuged at 4°C at maximum speed for 15 min, and the pellet washed with 80% ethanol and air-dried. The RNA was resuspended in 30ul of RNase-free water and 5ul were used for Northern Blot analysis.
Reporter assays
Analysis of pri-miRNA processing using ectopic reporter constructs was performed as previously described19. We used the pri-miRNA reporter construct developed by Bartel and collegues19 and replaced the miRNA control pri-miR-1-1 with a mutant version in which the adenosines of the potential METTL3 motifs were mutated. Then we placed the query miRNA, either wildtype pri-let-7e or its altered version in which the A’s of the putative METTL3 motifs were mutated to T’s, upstream of the pri-miR-1-1 control. We then used these two constructs to transfect HEK293T cells using Lipofectamine 2000. 48 hours later the RNA was extracted and qRT-PCR was performed to test the production of mature let-7e and mature miR-1-1.
Extended Data
Extended Data Figure 1.
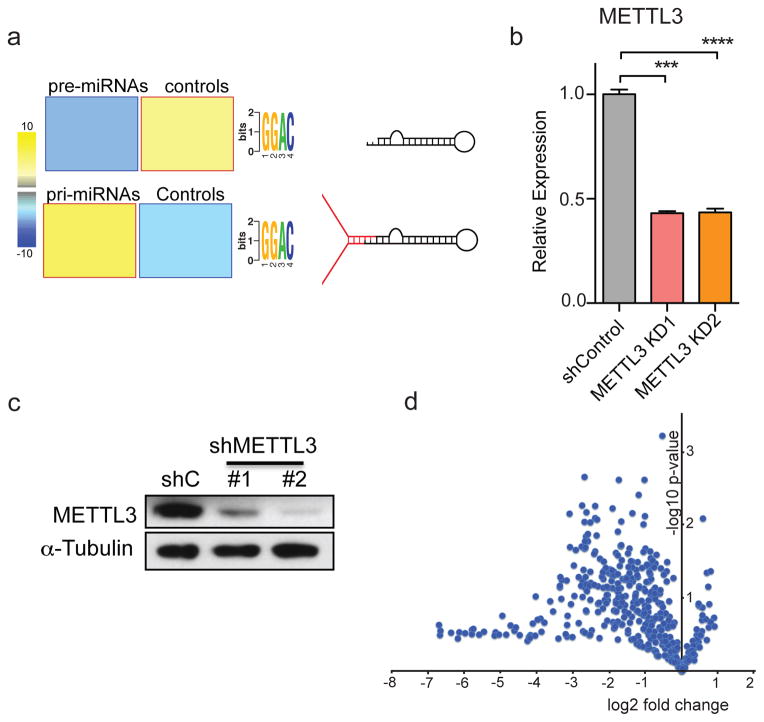
a, Unbiased search for cis-regulatory elements using the FIRE algorithm. FIRE motif discovery analysis of pre-miRNAs and pri-miRNA sequences as well as random sequences of the same length reveals over-representation of the METTL3 motif in pri-miRNAs sequences but not in pre-miRNAs. Yellow represents over-representation, and blue depicts under-representation of the motif. The magnitude of the over/under-representation is represented by the linear scale heat-map on the left. A schematic representation of a pre and a pri-miRNA is shown on the right. b, qRT-PCR and western blot (c) quantifications of METTL3 upon transduction with two independent shRNAs targeting METTL3 (METTL3 KD1 and METTL3 KD2) in MDA-MB-231 cells. Samples were normalized to GAPDH. The data from biological triplicates is shown. Bar graphs represent a linear scale and error bars represent s.d. ***, p-value <5E-4. d, A volcano plot representation of the microarray of miRNAs shown in Figure 2a, where the y-axis represents the −log10 of the p-value, and the x-axis represents the fold change (log2) between the expression levels of the miRNA from the METTL3 depletion (average of two independent shRNAs) versus the average of two control samples.
Extended Data Figure 2.
a, Quantification of representative miRNAs that were affected by METTL3 depletion in MDA-MB-231 cells as measured by qRT-PCR. Expression values were normalized to SNORD44 (RNU44). b, An example of a small RNA that did not display expression level changes upon METTL3 knockdown (SNORD44, small nucleolar RNA) normalized to 18S. All experiments were conducted in biological replicates. Bar graphs represent a linear scale and error bars represent s.e.m.***, p-value <5E-4; **, p-value <1E-3; *, p-value <5E-2.
Extended Data Figure 3.
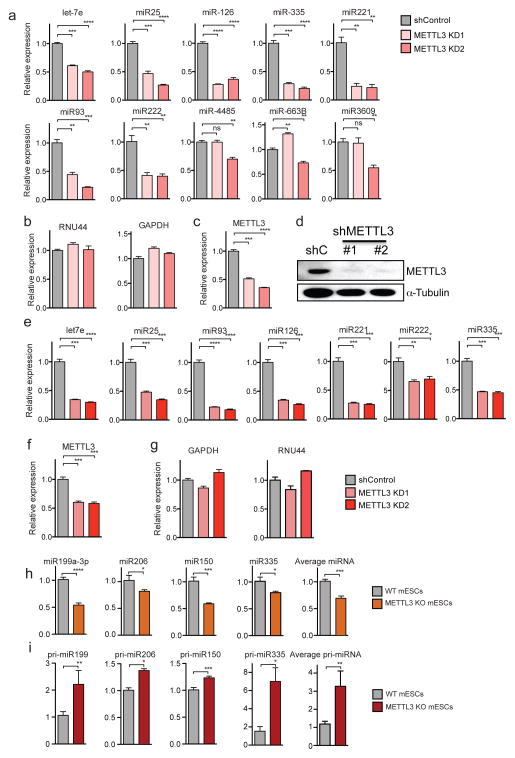
a, qRT-PCR quantification of examples of miRNAs that were modulated upon METTL3 depletion in HeLa cells. Samples were normalized to RNU44. b, Expression levels of genes used for normalization. All experiments were done in biological replicates. c, qRT-PCR and western blot (d) quantifications of METTL3 levels upon transduction with two independent shRNAs targeting METTL3. e, Expression levels of representative miRNAs that were affected by METTL3 depletion in HUVEC cells, as measured by qRT-PCR. Normalization was done by using RNU44 as endogenous control. f, qRT-PCR quantification of METTL3 depletion upon transduction with two independent shRNAs targeting METTL3. g, Quantification of the expression levels of control genes. h and i, Examples of miRNAs affected in mouse embryonic stem cells in which METTL3 has been targeted using CRISPR26, whose expression was measured by qRT-PCR. All experiments were done in biological replicates. Bar graphs represent a linear scale and error bars represent s.d. ***, p value <5E-4; **, p value <1E-3
Extended Data Figure 4.
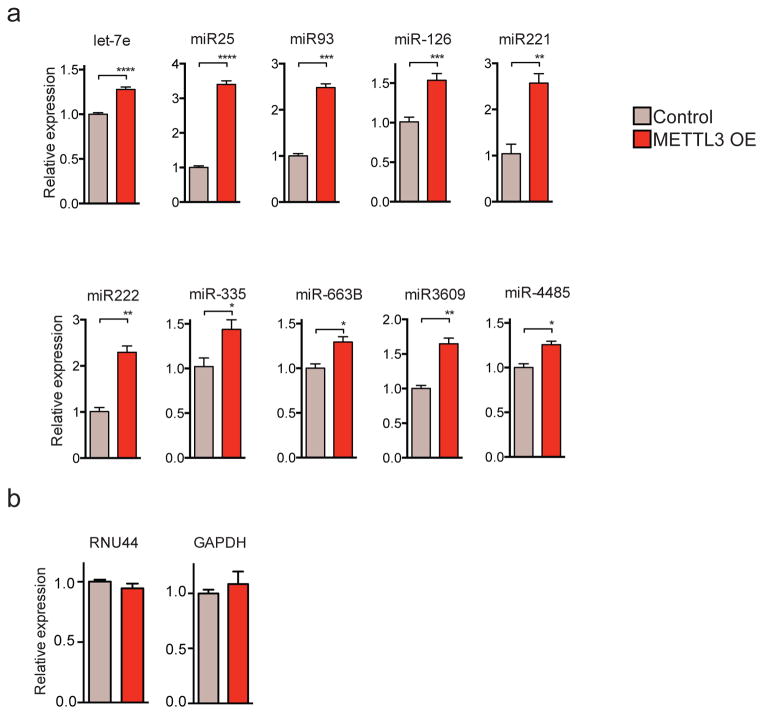
a, qRT-PCR quantification of expression of representative miRNAs modulated upon METTL3 over-expression (METTL3 OE) in MDA-MB-231 cells. Samples were normalized to RNU44. b, qRT-PCR quantification of control RNU44 and GAPDH–genes normalized to 18S. All experiments were done in biological replicates. Bar graphs represent a linear scale and error bars represent s.d. ***, p value <5E-4; **, p value <1E-3.
Extended Data Figure 5.
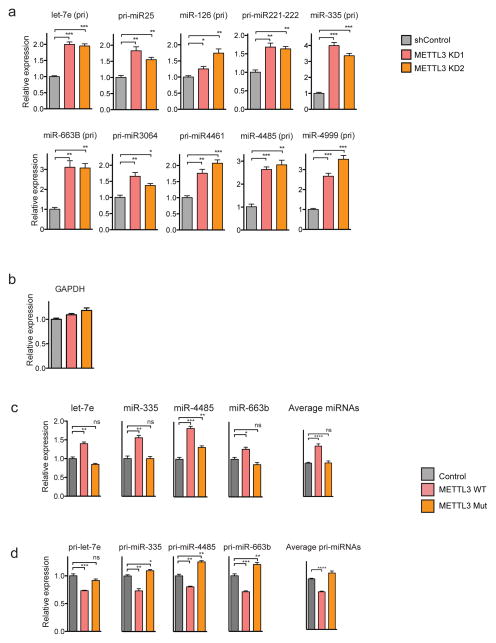
a, qRT-PCR quantification of representative pri-miRNAs that were impacted by METTL3 depletion using two independent hairpins in MDA-MB-231 cells. Expression levels were normalized to GAPDH. b, qRT-PCR quantification of GAPDH, endogenous control. All experiments were done in biological replicates. Quantification of mature (c) and pri-miRNAs (d) upon stable transduction of MDA-MB-231 with either wildtype or a catalytic mutant METTL3. Mature miRNA expression was normalized to RNU44 and pri-miRNAs to GAPDH expression levels. The last bar graph shows the averaged value for all individual miRNAs tested. The experiments were done in biological replicates. Bar graphs represent a linear scale and error bars represent s.d. ****, p value <1E-4, ***, p value <5E-4; **, p value <1E-3; *, p value <5E-2; ns= not significant.
Extended Data Figure 6.
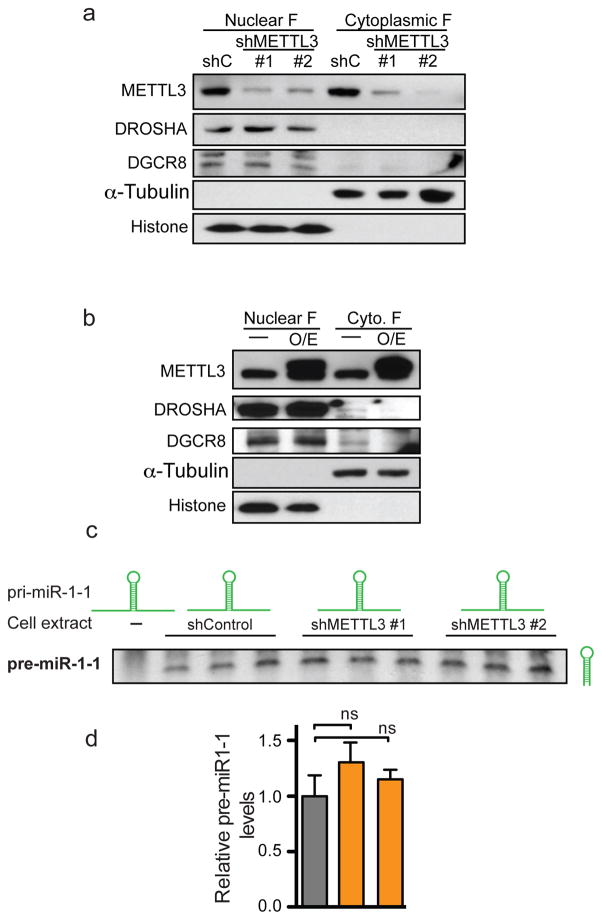
a, Western blot analysis of METTL3, DROSHA and DGCR8 obtained from nuclear and cytoplasmic fractions of cells transduced with two independent shRNAs targeting METTL3 (shMETTL3 #1 and #2) or with an shRNA control (shC). Tubulin and Histone 3 were used as loading controls as well as controls for the efficiency of the fractionation. b, Same as (a), but in this case, lysate from cells overexpressing METTL3 were compared to wildtype control cells. c, In vitro pri-miRNA processing reactions. Whole-cell extracts of control cells or cells depleted of METTL3 with 2 independent shRNAs were used to process in vitro transcribed pri-miR-1-1 to produce pre-miR-1-1 in vitro. Pre-miR-1-1 levels were then analyzed by Northern blot. d, Hybridization intensities of (c) were quantified, normalized by their inputs and shown in a bar graph format. Bar graphs represent a linear scale and error bars represent s.d.
Extended Data Figure 7.
a, FIRE motif discovery analysis of the METTL3 HITS-CLIP binding sites compared to control sequences; two over-represented versions of the METTL3 motif are shown with a z-score as indicated. The heat-map represents a linear scale. b, Venn diagram representation of the overlap of miRNAs affected by METTL3 depletion and bearing the m6A and/or the METTL3 HITS-CLIP tags within 1kb from any particular miRNA locus. The overlap of miRNAs containing both m6A and METTL3 HITS-CLIP tags is depicted in red, p-value=2.4E-15.
Extended Data Figure 8.
a, Schematic representation of the reporters used to study the role of the METTL3 on pri-miRNA processing. Represented in red is the pri-let-7e sequence and in green, the control pri-miR-1-1. The top reporter contains a wildtype sequence of the pri-let-7e and the potential sites of methylations are depicted as red dots. The reporter on the bottom contains a mutant version of pri-let-7e in which the 5 putative adenines of the METTL3 motif were mutated. b, HEK293T cells were transfected with the reporters depicted in (a), RNA was extracted, and mature miRNA expression quantified by qRT-PCR. The bar graph represents the relative expression levels of mature let-7e normalized to mature miR-1-1. c, In vitro binding assays using immunopurified DGCR8. Samples containing in vitro transcribed pri-let-7e with N6-methyladenosine or unmodified adenosines were incubated with magnetic bead bound-DGCR8, washed, eluted and analyzed by Northern blot. All reactions contained unmodified pri-miR-1-1 as endogenous control. The upper panel shows pri-let-7e and the lower panel pri-miR-1-1. On the right side the bar graph is depicted the average intensity of pri-let-7e normalized by pri-miR-1-1 levels. The experiment was done in biological triplicate. Bar graphs represent a linear scale and error bars represent s.d. ***, p-value <5E-4, ns= not statistically significant.
Extended Data Figure 9.
Immunoprecipitation of endogenous DGCR8 crosslinked to RNA of control cells or cells depleted of METTL3 using two independent shRNAs. After immunoprecipitation, the RNA was extracted and the expression levels of a set of pri-miRNAs were quantified by qRT-PCR. The average quantification is presented in Figure 4h. Bar graphs represent a linear scale and error bars represent s.d. ****, p value <1E-4; ***, p value <1E-3; **, p value <1E-2; *, p value <5E-2.
Acknowledgments
We thank members of the Tavazoie lab as well as S. Kurdistani for comments on previous versions of this manuscript. We thank D. Bartel for suggestions. We thank L. Fish for technical advice. We thank C. Zhao and C. Lai of the Rockefeller Genomics Resource Center for assistance with next-generation RNA-sequencing. We thank H. Molina of the Rockefeller Proteomics Center for his input in proteomics analysis. We thank C. Eicken of LC Sciences for assistance with microarray analysis. We thank H. Chang and P. Batista for generously providing targeted ESCs. C.R.A. was an Anderson Cancer Center Fellow at Rockefeller University. This work was supported by an Era of Hope Department of Defense Award to S.F.T. S.F.T. is a Department of Defense Breast Cancer Collaborative Scholars and Innovators Award recipient.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Competing financial interests
The authors declare no competing financial interests.
RNA-seq data have been deposited in the Gene Expression Omnibus under accession number GSE60213.
Authors Contributions
C.R.A. conceived of the project and designed the experiments and S.F.T. supervised the project. C.R.A. performed the core of the experiments, H.L. generated stable cell lines, performed qRT-PCR reactions and cloning, H.G. performed computational analyses and N.H. provided technical support. C.R.A. and S.F.T. wrote manuscript.
References
- 1.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 2.Gregory RI, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 3.Han J, et al. The Drosha-DGCR8 complex in primary microRNA processing. Genes & development. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Current biology: CB. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Han J, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 6.Elemento O, Slonim N, Tavazoie S. A universal framework for regulatory element discovery across all genomes and data types. Molecular cell. 2007;28:337–350. doi: 10.1016/j.molcel.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei CM, Moss B. Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry. 1977;16:1672–1676. doi: 10.1021/bi00627a023. [DOI] [PubMed] [Google Scholar]
- 8.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 9.Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. Rna. 1997;3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- 11.Sibbritt T, Patel HR, Preiss T. Mapping and significance of the mRNA methylome. Wiley interdisciplinary reviews. RNA. 2013;4:397–422. doi: 10.1002/wrna.1166. [DOI] [PubMed] [Google Scholar]
- 12.Camper SA, Albers RJ, Coward JK, Rottman FM. Effect of undermethylation on mRNA cytoplasmic appearance and half-life. Mol Cell Biol. 1984;4:538–543. doi: 10.1128/mcb.4.3.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carroll SM, Narayan P, Rottman FM. N6-methyladenosine residues in an intron-specific region of prolactin pre-mRNA. Mol Cell Biol. 1990;10:4456–4465. doi: 10.1128/mcb.10.9.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in bioinformatics. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bujnicki JM, Feder M, Radlinska M, Blumenthal RM. Structure prediction and phylogenetic analysis of a functionally diverse family of proteins homologous to the MT-A70 subunit of the human mRNA:m(6)A methyltransferase. Journal of molecular evolution. 2002;55:431–444. doi: 10.1007/s00239-002-2339-8. [DOI] [PubMed] [Google Scholar]
- 16.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome research. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee Y, Kim VN. In Vitro and In Vivo Assays for the Activity of Drosha Complex. 2007;427:87–106. doi: 10.1016/s0076-6879(07)27005-3. [DOI] [PubMed] [Google Scholar]
- 19.Auyeung VC, Ulitsky I, McGeary SE, Bartel DP. Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell. 2013;152:844–858. doi: 10.1016/j.cell.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tavazoie SF, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giannopoulou EG, Elemento O. An integrated ChIP-seq analysis platform with customizable workflows. BMC bioinformatics. 2011;12:277. doi: 10.1186/1471-2105-12-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang C, Darnell RB. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nature biotechnology. 2011;29:607–614. doi: 10.1038/nbt.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nature methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim SW, et al. A sensitive non-radioactive northern blot method to detect small RNAs. Nucleic Acids Res. 2010;38:e98. doi: 10.1093/nar/gkp1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Batista PJ, et al. m(6)A RNA Modification Controls Cell Fate Transition in Mammalian Embryonic Stem Cells. Cell stem cell. 2014;15:707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]