SUMMARY
Small RNA-guided protein complexes play an essential role in CRISPR-mediated immunity in prokaryotes. While these complexes initiate interference by flagging cognate invader DNA for destruction, recent evidence has implicated their involvement in new CRISPR memory formation, called priming, against mutated invader sequences. The mechanism by which the target recognition complex mediates these disparate responses—interference and priming—remains poorly understood. Using single-molecule FRET, we visualize how bona fide and mutated targets are differentially probed by E. coli Cascade. We observe that the recognition of bona fide targets is an ordered process that is tightly controlled for high fidelity. Mutated targets are recognized with low fidelity, which is featured by short-lived and PAM- and seed-independent binding by any segment of the crRNA. These dual roles of Cascade in immunity with distinct fidelities underpin CRISPR-Cas robustness, allowing for efficient degradation of bona fide targets and priming of mutated DNA targets.
INTRODUCTION
Clustered regularly interspaced short palindromic repeats (CRISPR) loci are widely spread throughout prokaryotic genomes and provide an inheritable RNA-guided adaptive immune system against bacteriophages and mobile genetic elements (Barrangou, 2013; Charpentier and Marraffini, 2014; Fineran and Charpentier, 2012; Reeks et al., 2013; Samson et al., 2013; Sorek et al., 2013; Westra et al., 2014). In response to invading phages or mobile genetic elements, CRISPR-associated (Cas) proteins integrate small fragments of foreign DNA into the CRISPR array, which are subsequently processed into mature CRISPR RNAs (crRNAs). crRNAs form a complex with one Cas protein (Cas9 from Type II) or multiple Cas proteins (Types I and III), which utilizes the crRNA as a guide to trigger degradation of cognate invading nucleic acids. While it is DNA that is targeted in Types I and II (van der Oost et al., 2014), recent studies suggest that both DNA and RNA are targeted in Type III (Goldberg et al., 2014; Hale et al., 2014; Rouillon et al., 2013; Staals et al., 2013; Tamulaitis et al., 2014). Among the target recognition complexes, Cas9 has been widely applied as a versatile tool for genome engineering in a broad spectrum of organisms (Hsu et al., 2014; Terns and Terns, 2014).
In the CRISPR-Cas/I-E system of Escherichia coli, mature crRNAs are incorporated into Cascade (CRISPR-associated complex for antiviral defense), an eleven-subunit complex comprised of five different Cas proteins (Cse11, Cse22, Cas76, Cas51 and Cas61) (Jore et al., 2011) (Figure 1A). In the CRISPR interference pathway, Cascade generates an R-loop between the crRNA and its double-stranded DNA (dsDNA) target (protospacer), which subsequently leads to target degradation by the nuclease-helicase Cas3 (Mulepati and Bailey, 2013; Sinkunas et al., 2013; Westra et al., 2012b). The first 8 nt (with exception of the 6th nt) of the protospacer, or “seed” region, must be a perfect match for efficient R-loop formation (Kunne et al., 2014). Additionally, R-loop formation requires an immediately neighboring tri-nucleotide protospacer adjacent motif (PAM). This conserved PAM sequence at the seed end of the protospacer is recognized by the Cse1 subunit and is essential for the discrimination between targets and non-targets (Sashital et al., 2012; Westra et al., 2013).
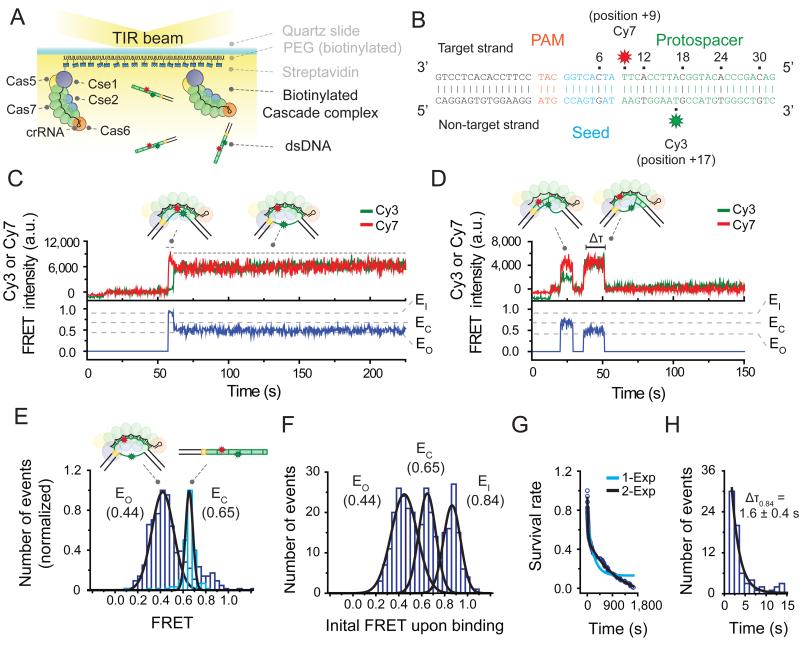
Figure 1. Two binding modes of Cascade revealed by a single-molecule FRET assay.
(A) Schematic of a single-molecule FRET experiment used to monitor binding of Cascade to target DNA substrates. (B) The bona fide target construct consists of a 15 bp flank (black), a PAM (orange), and a protospacer (green), with its seed highlighted in blue. Cy7 (red star) was attached to position +9 of the target strand and Cy3 (green star) to position +17 of the non-target strand. (C) A representative time trace of donor (Cy3, green) and acceptor (Cy7, red) fluorescence and corresponding FRET (blue) exhibiting the long-lived binding of the bona fide target. High FRET (~0.84, named EI for FRET efficiency of an intermediate state) exhibited upon binding is followed by low FRET (~0.44, named EO for FRET efficiency of an open state). DNA was added at time 10 sec. (D) A representative time trace exhibiting the short-lived binding of the bona fide target exhibits two FRET states (EO ~0.44 and EC ~0.65). EC is for FRET efficiency of a closed state). The duration of each state is measured as the dwell time (Δτ). DNA was added at time 10 sec. (E) The FRET distribution of the bona fide target DNA alone (light blue) or after equilibration with immobilized Cascade (purple) with peaks at EC (0.65) and EO (0.44), respectively (derived from Gaussian fit, black line). Data obtained from 5 fields of view each. (F) A histogram of the initial FRET upon binding (average of first 1.5 sec of each event) of the bona fide target exhibits three peaks at FRET = EO (0.44), EC (0.65), EI (0.84) (derived from Gaussian fit, black line). (G) The survival rate of events that start at EI (0.84) was fitted using a single (light blue color) and a double (black color) exponential curve. The double exponential fit resulted in two characteristic times (25.9 and 1040 sec). (H) The dwell time distribution of EI (0.84) state of bona fide target binding with mean ΔτE0.84 (derived from single exponential fit, black line). Error represent standard deviation (3 individual data sets). See also Figure S1 and Table S1.
The mechanism by which Cascade finds its target among the vast amount of DNA in the cell remains elusive. It has been hypothesized that Cascade transiently associates with PAM sequences, interrogating neighboring sequences for a complementary seed, followed by directional R-loop formation (Kunne et al., 2014). A recent single-molecule study has visualized the transient interactions of Cas9 with PAM-rich sequences in real time (Sternberg et al., 2014). Another study with Cascade and Cas9 has shown directional R-loop formation and how PAM and protospacer complementarity influence its stability (Szczelkun et al., 2014). However, it is yet to be shown how the stepwise interaction between PAM, seed and protospacer is coordinated and how off-targeting is avoided during target recognition.
Recent in vivo studies have revealed an additional functionality of CRISPR-Cas immunity. When facing “escape mutants”, previously targeted sequences that bear mutations in their PAM and/or protospacer, Cascade initiates a response called priming wherein the CRISPR-Cas system acquires new spacer sequences from the mutant at an elevated rate to restore immunity (Datsenko et al., 2012; Fineran et al., 2014; Li et al., 2014; Richter et al., 2014). High-throughput plasmid loss assays of a randomized PAM and protospacer library have revealed that priming is a robust process, tolerating up to 13 mutations in the PAM and protospacer sequence (Fineran et al., 2014). Even though Cascade is essential for priming, its role in this process is poorly understood. Intriguingly, biochemical studies have shown that a single point mutation in the PAM or seed sequence leads to a drastic decrease in the binding affinity of Cascade (Semenova et al., 2011). Therefore, it is puzzling how Cascade is able to associate with these mutated substrates despite its low affinity and further, how it distinguishes these mutated substrates from bona fide targets to initiate priming.
Single-molecule fluorescence is a powerful tool for elucidating the intricate mechanistic details of complex protein-nucleic acid interactions (Ha, 2014; Joo et al., 2012; Juette et al., 2014; Robinson and van Oijen, 2013; Schuler and Hofmann, 2013). To dissect Cascade’s two distinct functional roles, we developed a single-molecule FRET assay to monitor the interaction of Cascade with bona fide and mutated substrates. Real-time observation of Cascade-target interactions revealed that an initial recognition complex proceeds to a stable R-loop only if the crRNA makes an extensive match with the target. In addition to this “canonical binding mode”, we identified an alternative binding mode of Cascade that is triggered by partial complementarity to a target. Using an in vivo assay, we validated that this binding mode enables Cascade to probe mutated DNA substrates and consequently initiate priming.
RESULTS
Single-molecule observation of Cascade target binding
For single-molecule measurements, Cascade was labeled with a biotin on the N-terminus of its Cse1 subunit (Figure S1A) and immobilized to the surface of a microscope slide via a biotin-streptavidin linkage (Figure 1A). Dye-labeled dsDNA targets were added to the slide, and individual binding events were imaged in real time with a total-internal-reflection-fluorescence (TIRF) microscope (Figure 1A). DNA constructs consisted of a protospacer, a PAM, and an additional 15 base pair flank (Figure 1B). The target strand (complementary to the crRNA) was labeled with an acceptor dye (Cy7) at protospacer position +9, whereas the non-target strand was labeled with a donor dye (Cy3) at protospacer position +17. These labeling positions yielded a FRET value of ~0.65 (named EC for a FRET state which represents a closed conformation of dsDNA between nt 9 and 17) (Figure 1E) as measured by immobilization of the DNA alone (see Experimental Procedures and Table S1). Control experiments showed that dye labeling of the DNA at protospacer positions +9 and +17 did not appreciably affect the target binding reaction of Cascade (Figure S1F).
We first explored Cascade’s interaction with a bona fide target DNA, a substrate that triggers interference in vivo. This substrate contains a protospacer with perfect complementarity to the crRNA and an interference-permissive PAM (named ‘interfering PAM’) (Figure 1B) (Fineran et al., 2014; Westra et al., 2012b). After equilibration of the DNA with the immobilized Cascade, the measured FRET distribution exhibited one major peak centered at 0.44 (named EO for a FRET state which represents an open conformation of dsDNA between nt 9 and 17), a decrease from the starting value of EC (0.65) (Figure 1E). This decrease in FRET is consistent with the expected open DNA conformation resulting from R-loop formation upon Cascade binding. A similar decrease in FRET was observed upon exchanging the position of the donor and acceptor dyes (Figure S1C) or when Cascade was pre-bound to the DNA prior to immobilization (Figure S1D), indicating that the observed decrease in FRET was not due to a protein- or surface-induced photophysical effect.
Two distinct binding modes of Cascade
Next we characterized the kinetics and structural dynamics of Cascade binding in real time by adding a bona fide target substrate to immobilized Cascade during data acquisition. Interestingly, time trajectories exhibited disparate binding events that varied in their dwell time and FRET value. The dwell time distribution followed a double-exponential decay curve (Figure 1G, a fit in black), suggesting heterogeneity in binding. A histogram of the initial FRET of binding events exhibited three distinct peaks (centered at EO (0.44), EC (0.65), and 0.84) (Figure 1F), which, combined with dwell time analysis, led us to divide the events into two distinct types.
The first type of binding event initiated at a FRET of 0.84, and persisted over the entire duration of our observation time (30 minutes) (Figure 1C) and was therefore considered to be irreversible over the time scale of our experiment (Figure 1G). Interestingly, events of this type did not remain at their initial FRET of 0.84, but exhibited a transition after 1.6 ± 0.4 seconds (Figure 1H) to a final FRET of 0.44 (Figure 1C). This observation is consistent with the single FRET peak centered at 0.44 (EO) observed at equilibrium (Figure 1E). The initial transient state (0.84, named EI for an initial transient state) may represent a target-recognition complex wherein the crRNA interacts with the dsDNA before full displacement of the non-target strand (schematic, Figure 1C). Notably, the FRET of the initial state is higher than that of the DNA alone (EC, 0.65, Figure 1E), likely arising from a subtle conformational change of the dsDNA upon target recognition (e.g. twisting or bending) (Hochstrasser et al., 2014; Westra et al., 2012b).
The observed transition (EI → EO) may represent a previously hypothesized locking process, wherein Cascade slides its Cse2 dimer toward its Cse1 subunit upon target recognition (Szczelkun et al., 2014; Wiedenheft et al., 2011a), ultimately resulting in the displacement of the non-target strand and stable R-loop formation (schematic, Figure 1C). Taken together, considering Cascade’s strong target association and observed conformational change (Semenova et al., 2011; Szczelkun et al., 2014; Wiedenheft et al., 2011a), we interpret the first type of binding event to correspond to Cascade’s canonical mode of target binding that leads to interference in vivo. We therefore refer to this event type as Cascade’s interference mode of binding.
Unlike the interference mode, the second type of binding event was short-lived (25.9 sec, Figure 1G) and exhibited an initial FRET of either EO or EC (Figure 1D). These states were further distinguished from the interference mode as they did not exhibit any kinetic intermediates, nor did they show transitions to other FRET states. As a substrate containing no complementarity (Mut[S1-6]) to the crRNA showed negligible binding (Figure S1E), we speculate that these short binding events (named “non-canonical mode”) arise from sequence-specific interactions wherein the probed region of the target DNA is either opened up in a locally formed R-loop (EO) or remains closed (EC).
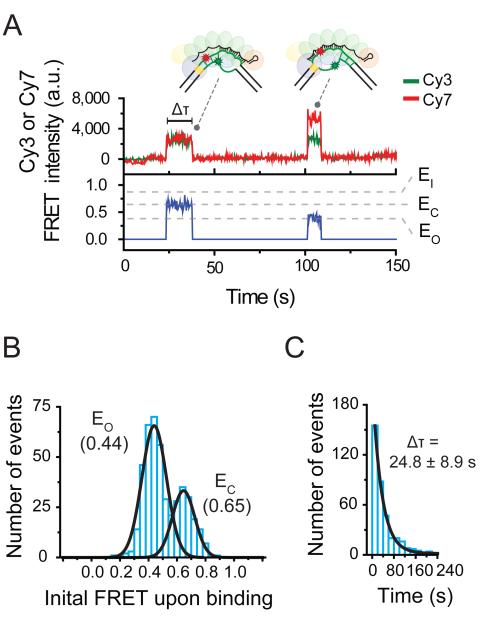
To explore the origin of Cascade’s disparate binding interactions, we first focused on the role of the PAM. We repeated our assay with a DNA substrate containing a point mutation in the PAM (Mut[PAM], Table S2) that represents one of the dominant mutant phenotypes of bacteriophages that escape CRISPR interference (Semenova et al., 2011) and subsequently trigger priming in vivo (Datsenko et al., 2012; Fineran et al., 2014). Notably, while Cascade was still able to interact with Mut[PAM], only binding events characteristic of its non-canonical mode were observed (Figure 2A). A histogram of the initial FRET of each event exhibited only two peaks, centered at EC and EO (Figure 2B), identical to the peak positions observed for the non-canonical binding mode (Figure 1F). In addition, the binding events observed for Mut[PAM] were short-lived, exhibiting a dwell time of 24.8 ± 8.9 seconds (Figure 2C), similar to that of the non-canonical binding mode (Figure 1G). These results indicate that Cascade’s interaction with target substrates through its non-canonical binding mode does not require an interfering PAM.
Figure 2. Short-binding of Cascade to PAM-mutated targets.
(A) A representative time trace exhibiting the short-lived binding of the PAM-mutated target exhibits two FRET states, EO (0.44) and EO (0.65). The duration of each state is measured as the dwell time (Δτ). DNA was added at time 12 sec. (B) A histogram of the initial FRET upon binding (average of first 1.5 sec of each event) of Mut[PAM] exhibits peaks at EO (0.44) and EC (0.65) (derived from Gaussian fit, black line). (C) The dwell time distribution of Mut[PAM] binding events with mean Δτ (derived from single exponential fit, black line). Error represents std (3 individual data sets).
Given the results above, we hypothesize that the observed binding states represent two functional modes of Cascade. The first is the interference mode, in which Cascade binds a bona fide DNA target (i.e. interfering PAM and complementary protospacer) and triggers Cas3-mediated target degradation. The second is the priming mode (non-canonical mode), in which Cascade is able to associate with targets harboring a PAM mutation to initiate primed spacer acquisition.
Structural elements of two distinct binding modes
To investigate the structural elements of Cascade’s two different binding modes, we employed a series of target DNA substrates bearing mutations in their PAM and/or protospacer sequence(s). Recent studies have reported that base pairing between Cascade’s crRNA and the protospacer occurs over five segments of five-nucleotides (segments 1-5) and one segment of two nucleotides (segment 6) (Fineran et al., 2014; Jackson et al., 2014; Mulepati et al., 2014; Zhao et al., 2014). We therefore chose to systematically mutate the protospacer in segments, starting from either the PAM-proximal or PAM-distal end of the protospacer (Figure 3, S2 and Table S2).
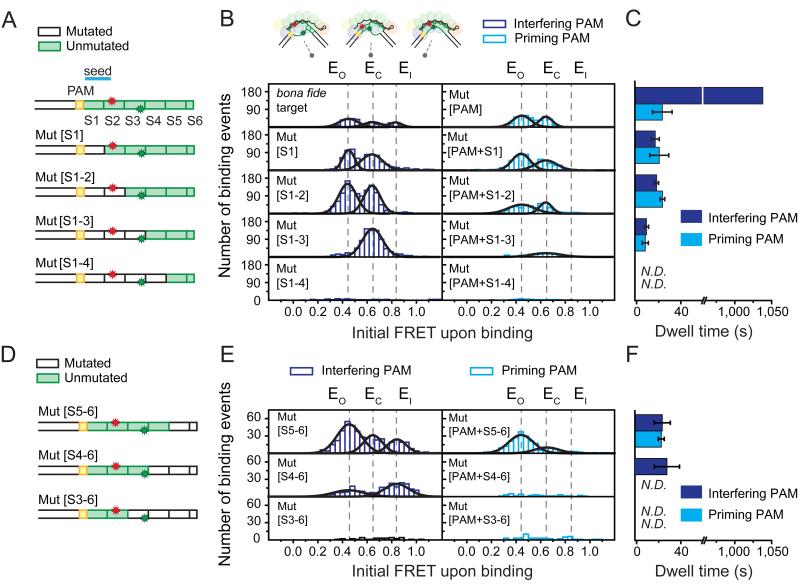
Figure 3. Cascade exhibits non-canonical binding to protospacers with PAM-proximal or PAM-distal segmented mutations.
(A) Schematics of DNA targets in the PAM-proximal mutation series illustrating mutated (white) or unmutated (green) segments (S1-S6) of the protospacer. Mut[S1], Mut[S1-2], Mut[S1-3] and Mut[S1-4] have segments 1, 1-2, 1-3, and 1-4 mutated, respectively. (B) Histograms of the initial FRET upon binding (average of first 1.5 sec of each event) of each PAM-proximal mutant from [A] bearing either an interfering (purple bars, left column) or priming (light blue bars, right column) PAM exhibit peaks (Gaussian fits, black lines) positioned similar to that of the bona fide and Mut[PAM] targets (top row, same as Figures 1F and 2B) at EO (0.44), EC (0.65), or EI (0.84) (dashed black lines). The recorded events are from one field-of-view of the detector. (C) Mean binding dwell time of each PAM-proximal mutant from [A] bearing either an interfering (purple bars) or a priming (light blue bars) PAM (derived from dwell time distributions, see Figure S2A). Error represents std (3 individual data sets). The dwell time of the bona fide target could not be measured accurately due to the photobleaching and thus arbitrarily set 1040 sec to represent the longer characteristic time scale in Figure 1G. (D) Schematics of DNA targets in the PAM-distal mutation series illustrated as in [A]. Mut[S5-6], Mut[S4-6], and Mut[S3-6] have segments 5-6, 4-6, and 3-6 mutated, respectively. (E) Histograms of the initial FRET upon binding of each PAM-distal mutant from [D] displayed in a similar fashion to [B]. (F) Mean binding dwell of each PAM-distal mutant from [D] bearing either an interfering (purple bars) or a priming (light blue bars) PAM (derived from dwell time distributions, see Figure S2A). Error represents std (3 individual data sets). N.D. is “Not Determined”. See also Table S2.
Upon mutation of the first segment of the protospacer (Mut[S1], Figure 3A), which comprises the majority of the seed region, the non-canonical binding mode persisted as binding events exhibited nearly identical FRET values and dwell times to the Mut[PAM] targets (Figure 3B and 3C). The same was observed for a DNA substrate containing both the PAM and seed mutations (Mut[PAM+S1], Figure 3A, 3B, and 3C), indicating that the non-canonical binding mode is largely insensitive to the PAM and seed sequence. This observation is in stark contrast to the canonical binding mode, which requires both an intact seed sequence and an interfering PAM. Remarkably, when the first two PAM-proximal segments, including the entire seed, were mutated (Mut[S1-2]), the non-canonical binding mode was still evident with initial FRET values centered at EO or EC and an average dwell time of 19.6 ± 0.4 seconds (Figure 3B, 3C, and S2A).
Intriguingly, when the first three (Mut[S1-3], Figure 3A) PAM-proximal segments were mutated, the binding events exhibited only one major initial FRET population centered at EC, with an average dwell time of 10.5 ± 1.9 seconds (Figure 3B, 3C and S2A), indicating that these events arise from sequence-specific interactions confined outside of the probed region of the protospacer (segments 4-6, Figure 3A). Removal of complementarity in the first four segments (Mut[S1-4], Figure 3A and 3B) or all segments (Mut[S1-6], Figure S1E) disrupted binding to background levels. Taken together, the series of PAM-proximal mutations indicate that the non-canonical binding mode of Cascade comprises sequence-specific interactions with a minimum requirement of three full segments for target recognition.
The PAM-distal mutation series showed complementary behavior, consistent with the structural features of the non-canonical binding mode observed above (Figure 3D, 3E and 3F). Upon mutation of the last two segments of the protospacer (Mut[S5-6]), the non-canonical binding mode persisted with two peaks centered at EO and EC. When three segments (Mut[S4-6]) were mutated, the non-canonical binding mode exhibited only one peak centered at EC, indicating that these interactions are confined within the probed region (segments 1-3). Further removal of complementarity disrupted binding to background levels, confirming that a minimum of three consecutive segments are required for non-canonical binding.
Besides the non-canonical mode, a fraction of binding events in the PAM-distal mutation series exhibited the signature initial FRET of the interference mode (EI, left column, Figure 3E). Even though this initial FRET was identical to that of the canonical binding mode, binding events were transient and did not exhibit any FRET transitions until dissociation after 24.8 ± 7.3 seconds (Figure 3F and S2C). This state reports on the formation of an interference-like target-recognition complex that cannot be locked and is in line with a previous observation that the PAM-distal region is required for stable R-loop formation in the interference model (Szczelkun et al., 2014).
Finally, to evaluate the role of the PAM in Cascade’s non-canonical binding mode, we repeated both series of protospacer mutations in the presence of the escape-mutant PAM (named ‘priming PAM’, Figure 3 and S2). Overall, mutation of the PAM substantially reduced the number of binding events for each mutant compared to its interfering PAM counterpart (compare columns, Figure 3B and 3E), indicating that the PAM is not strictly required for, but facilitates, non-canonical binding. In addition, EI state observed in Mut[S5-6] and Mut[S4-6] was completely abrogated upon PAM mutation, suggesting that this intermediate requires the coordinated ternary interaction of Cascade with the PAM and the seed.
In summary, our single-molecule results show that the non-canonical binding mode of Cascade is much more robust than its canonical mode, capable of binding a wide variety of mutated targets, yet still exhibiting sequence specificity. Such versatility could facilitate primed spacer acquisition, in which invading DNA variants that harbor mutations in their PAM or protospacer can still be detected by the CRISPR-Cas immune system.
Functional roles of two distinct binding modes
To investigate whether the canonical and non-canonical binding modes of Cascade lead to different functional outcomes, we reconstituted CRISPR interference in vitro. We cloned the segmented mutants that showed binding in our single-molecule experiments into plasmids (Table S3) and tested the plasmids for Cascade-directed degradation by Cas3. Our assay revealed that only the plasmid with a perfectly complementary protospacer accompanied by an interfering PAM led to target degradation, whereas target plasmids containing either an escape PAM mutation and/or segmented mutations proximal or distal to the PAM were unaffected by Cas3 (Figure S3). These results suggest that only Cascade’s canonical binding mode (EI → EO) generates an R-loop structure that supports target degradation by Cas3.
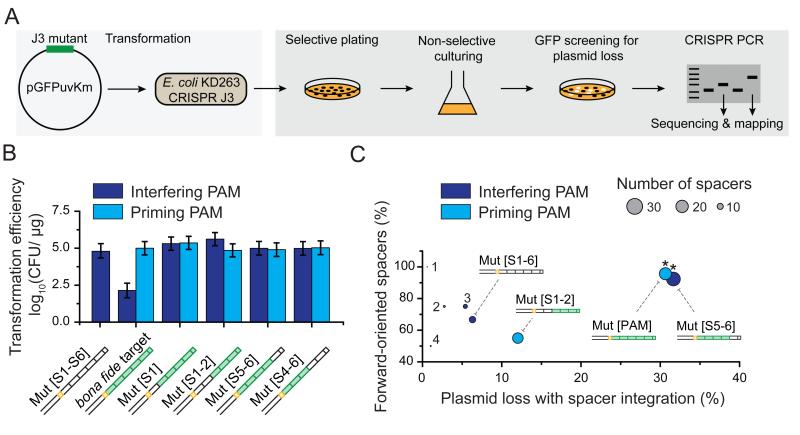
Next, we sought to determine if the non-canonical binding mode of Cascade results in primed spacer acquisition in vivo. To assess primed spacer acquisition, we first transformed the target plasmids with segmented mutations into E. coli containing a targeting CRISPR array plasmid (Figure 4A). Notably, only the target with a perfectly complementary protospacer and interfering PAM led to a reduced transformation efficiency (Figure 4B), confirming that the CRISPR-Cas system exclusively targets the R-loops generated through the canonical binding mode of Cascade. Next, transformants were transferred to non-selective media, which allowed the CRISPR-Cas system to mount a primed response.
Figure 4. Non-canonical binding leads to primed spacer acquisition.
(A) Cartoon representation of the in vivo assay used to determine primed plasmid loss and spacer acquisition. (B) Transformation efficiencies of plasmids harboring different target sequences (see schematics) with an interfering (purple bar) or a priming (light blue bar) PAM. CFU is “Colony-Forming Unit.” Error is std of 3 individual measurements. (C) A two-dimensional bubble plot showing the fraction of forward-oriented spacers acquired versus the percentage of plasmid loss for those targets in [B] that exhibited spacer integration. Circle size represents the total number of spacers that were acquired and circle color represents an interfering (purple) or a priming (light blue) PAM. A star (*) indicates a forward directional bias (relative to random) with a P-value < 1× 10−5 based on binomial statistics. The numbers of 1, 2, 3 and 4 indicate data points from constructs Mut[PAM+S5-6], Mut[PAM+S1], Mut[S4-6] and Mut[PAM+S4-6], respectively. See also Figure S3, Table S1 and Table S3..
After two days of cell growth, three mutant constructs (Mut[PAM], Mut[S5-6], Mut[PAM+S1-2]) showed a higher degree of plasmid loss than the negative control construct Mut[S1-6] did (Figure 4C). To identify if these plasmids were lost through primed spacer acquisition, the genomic CRISPR-array was amplified by PCR and amplicons with increased size were sequenced (Figure 4A). In total, 23, 26, and 20 new spacers were obtained that originated from the target plasmids Mut[PAM], Mut[S5-6], Mut[PAM+S1-2], respectively. Sequencing of the genomic CRISPR-array also allowed us to determine whether the acquired spacers showed any strand bias that is typical of the priming process in Type I-E systems (Datsenko et al., 2012; Swarts et al., 2012). Among the three constructs, Mut[PAM] and Mut[S5-6] exhibited bias in spacer acquisition toward the target strand (p-value < 1 × 10−5, Figure 4C), suggesting that these spacers were obtained by primed spacer acquisition. Taken together, the high frequency of plasmid loss and strand bias in the acquired spacers suggests that the non-canonical binding mode acts as a gateway to priming in vivo.
DISCUSSION
Adaptive immune systems are found in both vertebrates and prokaryotes and provide specific defense against invading pathogens. The high specificity of this immunity is important for distinguishing self from non-self (Gandon and Vale, 2014), yet it brings a downside that it can be readily overcome by rapidly evolving pathogens (Koel et al., 2013). However, both vertebrates and prokaryotes have developed sophisticated fail-safe mechanisms to target these pathogens. For example, when vertebrates face invaders bearing mutated antigens, they may still be recognized by a pool of polyclonal antibodies (Purtha et al., 2011). The resulting secondary response proceeds more quickly and efficiently than the primary response, which allows vertebrate hosts to keep pace with their evolving pathogens (Tarlinton and Good-Jacobson, 2013).
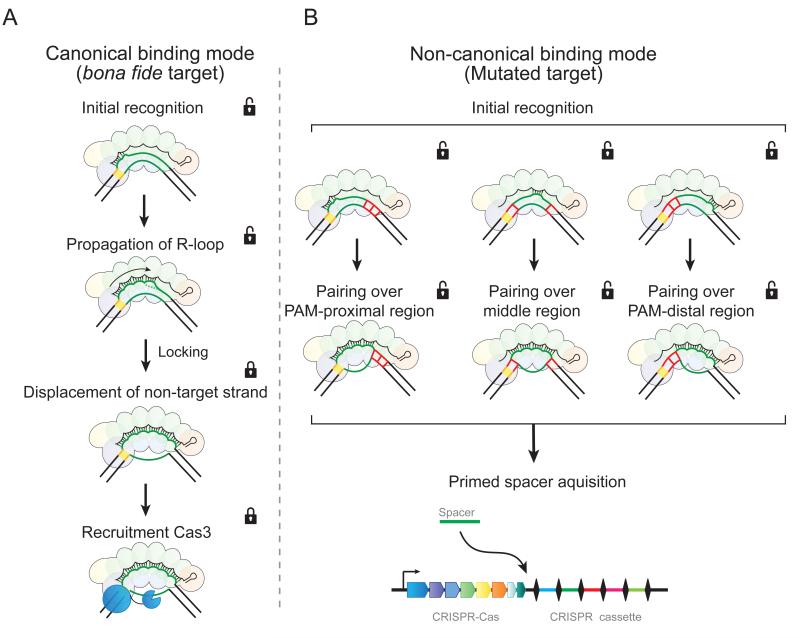
The prokaryotic adaptive immune system faces similar challenges. Rapidly evolving pathogens readily overcome sequence-specific CRISPR-Cas-mediated host defense (Deveau et al., 2008; Semenova et al., 2011), exposing a major limitation to prokaryotic adaptive immunity (Weinberger et al., 2012). However, analogous to vertebrate adaptive immunity, once pre-exposed to an ancestral invader, CRISPR-Cas responds more rapidly and efficiently to future variants then it can to a novel invader (Datsenko et al., 2012; Fineran et al., 2014; Li et al., 2014; Richter et al., 2014; Samson et al., 2013; Tamulaitis et al., 2014; Westra et al., 2012a). Although Cascade was shown to be essential for this “primed” response (Datsenko et al., 2012), the underlying mechanism has remained enigmatic. Here we provide the first insights into this puzzle by showing that Cascade binds mutated targets through a distinct non-canonical mode with low-fidelity compared to the high-fidelity binding mode used for unmutated targets. We show that the canonical, high-fidelity binding mode is a stepwise process that locks, triggering recruitment of nuclease/helicase Cas3 only when all criteria are met, including: an interfering PAM, a matching seed, and pairing of all segments of the crRNA guide. In contrast, the non-canonical, low-fidelity binding mode initiates a downstream pathway that results in rapid spacer acquisition through the priming process (Figure 5).
Figure 5. Two binding modes of Cascade lead to different functional outcomes.
Cascade employs two distinct target-DNA binding modes that trigger (A) interference or (B) priming. (A) In the interference pathway, target recognition initiates from the PAM and PAM-proximal region. R-loop formation then propagates toward the PAM-distal region. When Cascade senses the fully paired structure, it brings this complex into a lower energy state (“locking”) that displaces the non-target strand out of Cascade. This exposed strand is then cleaved by Cas3. (B) In the priming pathway, DNA is probed through brief interactions. PAM recognition facilitates this priming pathway but is not required. The brief interactions may initiate from the PAM-proximal (left), the PAM-distal region (right), or the middle of the protospacer (middle), which becomes stable when paired over 3 or more segments. This non-canonical (“unlocked”) binding mode leads to a unique conformation of the R-loop and signals for primed spacer acquisition.
Protein-mediated high fidelity target recognition
Our single-molecule data demonstrate in real time that high-fidelity target-DNA binding is a multi-step process and occurs in a directional manner from the PAM-proximal to PAM-distal end of the protospacer. Previous studies have shown that the recognition process is initiated when the Cse1 subunit recognizes the PAM (Sashital et al., 2012) and the crRNA hybridizes with the seed sequence. After this initial recognition complex is formed, the R-loop propagates toward the PAM-distal region of the protospacer (Semenova et al., 2011; Szczelkun et al., 2014; Wiedenheft et al., 2011b). When the pairing of the crRNA reaches the PAM distal-end of the protospacer, Cascade senses the fully paired structure and stabilizes this complex into a lower energy state (“locking”) (Szczelkun et al., 2014). This state acts as a flag for the destruction of the target DNA by Cas3 (Mulepati and Bailey, 2013; Sinkunas et al., 2013; Westra et al., 2012b). This stepwise mechanism involves both protein-nucleic acid interactions (Cse1-PAM) and progressive crRNA-DNA base pairing, ensuring efficient and high fidelity recognition, and degradation of targeted DNA.
Our study shows how Cascade maintains a strict regime to prevent non-specific cleavage by controlling the pathway toward the proposed locking process (Szczelkun et al., 2014). When Cascade encounters a target with mismatches (e.g. Mut[S5-6], Figure 3F), the initial recognition complex forms, but the R-loop does not propagate throughout the full protospacer (absence of a transition of EI to EO) (Figure S2C). As a result, Cascade will not lock the R-loop, and the initiation complex can disassemble using thermal energy. This process cannot be explained by the thermodynamic properties of base pairing alone since a target with mismatches often form a far larger number of consecutive base pairs than 7 (e.g. Mut[S5-6]), which has been shown to be the minimal number of base pairs required for stable binding (Cisse et al., 2012). Instead, the last step of stepwise recognition (locking) must involve protein-nucleic acid interactions that verify base pairing over the entire protospacer. This model is analogous to the stepwise conformational change observed with Argonaute proteins during its target search process (Schirle et al., 2014; Wang et al., 2009) and contrasts with the low fidelity RecA-mediated target search that does not seem to use protein—nucleic acid interaction in promoting specificity (Chen et al., 2008).
Structural view of the priming mode
The structure of Cascade supports our finding that low-fidelity target-DNA interactions can initiate from any segment of the crRNA (Figure 5B). Cascade is composed of five different Cas protein subunits assembled into a highly interlocked, crRNA-containing protein complex (Jackson et al., 2014; Mulepati et al., 2014; Zhao et al., 2014). The backbone of the complex consists of six Cas7 subunits with a hand-like architecture. Each hand uses its thumb to hold and position the crRNA at 6 nt intervals. As a consequence, every sixth base is flipped out of the plane and is unable to interact with the target DNA. This unusual configuration permits the crRNA to pair with a target in segments of five nucleotides in an underwound, ribbon-like structure (Mulepati et al., 2014). Interestingly, individual segments of the crRNA in the apo-Cascade structure are already pre-ordered in a pseudo A-form helix with their nucleobases facing the solvent (Jackson et al., 2014; Zhao et al., 2014). Structural pre-ordering is a common strategy to facilitate target binding of nucleic acid guided complexes (e.g. Argonaute and RecA) (Chen et al., 2008; Kunne et al., 2014; Wang et al., 2009), and thus the pre-ordered segments of crRNA of Cascade is in line with the idea that low-fidelity interactions can nucleate from any crRNA segment (Figure 5B). Although the low fidelity binding mode leads to relatively short-lived R-loops, it is their distinct conformation that likely signals for a primed spacer acquisition response in the cell.
The DNA recognition mechanism of Cascade contrasts that of Cas9, which has recently been shown to be strictly dependent on the PAM (Sternberg et al., 2014). Furthermore, Cas9 does not base pair its crRNA in segments to the target DNA (Nishimasu et al., 2014) but forms a contiguous double helix, making it more difficult to imagine that PAM-distal regions of Cas9’s crRNA can initiate an interaction with the target DNA. Yet, off-target cleavage analysis of Cas9 during genome editing clearly indicates that Cas9 also tolerates mutations (Kuscu et al., 2014), but whether this leads to a priming response in bacteria with Type II CRISPR-Cas systems remains to be shown.
Mechanisms of the priming mode
Although the interference response of CRISPR immunity is a relatively well-characterized phenomenon, the molecular mechanism of priming remains poorly understood. First, our data shows that Cascade distinguishes mutated targets from bona fide targets using a low-fidelity binding mode that can initiate priming. A recent study showed that priming in E. coli is robust, tolerating up to 13 mutations throughout the 32 nt protospacer and 3 nt PAM (Fineran et al., 2014). Even when mutations were clustered in any of the crRNA defined segments, priming was not abolished. The low-fidelity binding mode of Cascade, in which individual segments may initiate pairing with a target, can explain the reported high tolerance for distributed and clustered mutations in a target during priming. In this mode, Cascade can probe DNA for complementarity to any of its crRNA segments, and extend such an interaction in either direction, thereby achieving sequence-specific detection of targets with limited base complementarity. However, the minimal number of base pairs required for priming (Fineran et al., 2014) insures that detrimental self-priming of the bacterial genome at random sites is unlikely.
Second, we observed that the non-canonical binding mode occurs even for substrates containing an interfering PAM and an intact seed, suggesting that direct interference and priming may occur simultaneously. Indeed, we have previously observed that E. coli is cured from high copy number plasmids by using existing spacers to expand the CRISPR-array with a range of new spacers against the same target (Fineran and Charpentier, 2012; Swarts et al., 2012). For a host this is a highly advantageous strategy, by simultaneously using interference and priming, the CRISPR interference effect is amplified while the chance that invaders evade immunity through point mutations in their protospacers is reduced. Even though it remains to be seen how priming is coordinated in the presence of the remaining Cas protein machinery (Cas1, Cas2 and Cas3), the relatively short time that Cascade spends on a target in the priming mode suggests that other factors might stabilize this relatively weak interaction.
Finally, in CRISPR-Cas/I-E systems priming is a DNA strand dependent process in which approximately 90% of new spacers are integrated from the same strand as the spacer triggering priming (Shmakov et al., 2014). Our results with Mut[PAM] and Mut[S5-6] in Figure 4 are consistent with this strand bias. In contrast, primed spacer acquisition in Type I-B and I-F systems does not exhibit such strand bias (Li et al., 2014; Richter et al., 2014). Interestingly, for protospacers mutated in the PAM and segments 1 and 2 (Mut[PAM+S1-2]), including the seed, we observed a higher degree of spacer acquisition without the typical strand bias, suggesting that these types of targets lead to a priming behavior in which strand specificity is lost.
Conclusion
Faithful copying and decoding of genetic information is central to the most important processes in the cell, including DNA replication (van der Oost et al., 2014), RNA transcription (Xu et al., 2014), and protein translation (Staals et al., 2013). But high fidelity always comes at the cost of reduced processing speed. Here we show how a crRNA guided complex solves this dilemma by employing both high and low fidelity target-DNA recognition modes. While the high fidelity mode ensures destruction of only perfectly matching targets, the low fidelity priming mode enables detection of a whole range of mutated invaders to initiate the priming process. The unique combination of these two properties in a single RNA-guided complex not only makes CRISPR immunity robust, but also reveals versatility of adaptive immunity against rapidly mutating pathogens.
EXPERIMENTAL PROCEDURES
Preparation of Cascade, biotinylated Cascade, and Cas3
Cascade and Cascade lacking subunit Cse1 (CasBCDE) were affinity-purified from E. coli strain BL21(DE3) as described previously (Jore et al., 2011). For biotinylated Cascade, Cascade was site-specifically labeled at the N-terminus of subunit Cse1 with an aldehyde moiety using the formylglycine generating enzyme (FGE) method described previously (Rabuka et al., 2012), reacted with biotin-hydrazide, and purified by size exclusion chromatography. Cas3 was purified by size-exclusion chromatography as described previously (Mulepati and Bailey, 2013) (see protein preparation in Extended Experimental Procedures).
Single-molecule FRET
Biotinylated Cascade complexes were anchored to polyethylene glycol-coated quartz microscope slides by biotin-streptavidin linkage Dye-labeled (Cy3 and Cy7) dsDNA targets (see DNA preparation in Extended Experimental Procedures) were added to the immobilized Cascade complexes and detected by a prism-type TIRF microscope. In a typical field of view, 200-300 molecules were detected. dsDNA targets were excited with a 532 nm laser and fluorescence emissions from Cy3 and Cy7 were separated by dichroic mirrors and imaged onto two halves of a CCD camera after passing through various filters. Imaging buffer consisted of Cascade buffer (50mM HEPES (pH 7.5), 75mM NaCl, 2mM MgCl2), an oxygen scavenging system (1% glucose (v/v), 0.1 mg/mL glucose oxidase (Sigma), 17 μg/μL Catalase (Roche)) to reduce photobleaching, and 1 mM Trolox (Sigma) to reduce photoblinking of the dyes (Rasnik et al., 2006). Imaging was performed at room temperature (23± 2 °C). Fluorescence time traces of individual binding events were identified in recorded movies and subsequently analyzed using custom software developed in IDL and MATLAB, respectively. The FRET value was defined as IA/(ID+IA), where ID and IA represent the fluorescence signals detected in the Cy3 and Cy7 channels, respectively. See single-molecule fluorescence in Extended Experimental Procedures.
Direct interference and priming
Direct interference was assessed by determining the transformation efficiency of target plasmids to E. coli strain KD263 containing pWUR564 (Table S3). Cas gene expression was induced 30 minutes prior to making cells chemically competent. Priming was assessed using plasmid loss assays as previously described (Fineran et al., 2014). Briefly, E. coli transformants containing the target plasmids (pWUR738-747) were grown in LB for 48 hours, plated on LB agar, and imaged under mild UV light. GFP-negative colonies were screened for spacer integration by PCR. Newly acquired spacers were sequenced and were strand specifically mapped onto the target plasmid sequence to verify priming (see direct interference and priming in Extended Experimental Procedures).
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank K. Datsenko, K. Semenova and K. Severinov for providing strain KD263, S. Bailey for providing the Cas3 overexpression construct. S. Sternberg for helpful advice, and C. Buyzerd for optimizing the biotinylation of Cascade. We thank A. Haagsma, M. Docter, P. Tuliniski and I. Geuzebroek for their technical support. This work was supported by the Netherlands Organization for Scientific Research VIDI Grant 864.11.005 and the Beijerinck premium from the Royal Netherlands Academy of Arts and Sciences to S.J.J.B. T.R.B was supported by a Marie Curie International Incoming Fellowship within the seventh European Community Framework Programme. C.J. was funded by the Open Program of the Division for Earth and Life Sciences (822.02.008) of the Netherlands Organization for Scientific Research and by European Research Council under the European Union’s Seventh Framework Programme [FP7/2007-2013] / ERC grant agreement n° [309509]. We thank I. Heo, M. Szczepaniak, and P. Tulinski for critically reading this manuscript.
REFERENCES
- Barrangou R. CRISPR-Cas systems and RNA-guided interference. Wiley interdisciplinary reviews RNA. 2013;4:267–278. doi: 10.1002/wrna.1159. [DOI] [PubMed] [Google Scholar]
- Charpentier E, Marraffini LA. Harnessing CRISPR-Cas9 immunity for genetic engineering. Current opinion in microbiology. 2014;19:114–119. doi: 10.1016/j.mib.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–484. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- Cisse II, Kim H, Ha T. A rule of seven in Watson-Crick base-pairing of mismatched sequences. Nat Struct Mol Biol. 2012;19:623–627. doi: 10.1038/nsmb.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nature communications. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. Journal of bacteriology. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fineran PC, Charpentier E. Memory of viral infections by CRISPR-Cas adaptive immune systems: acquisition of new information. Virology. 2012;434:202–209. doi: 10.1016/j.virol.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Fineran PC, Gerritzen MJ, Suarez-Diez M, Kunne T, Boekhorst J, van Hijum SA, Staals RH, Brouns SJ. Degenerate target sites mediate rapid primed CRISPR adaptation. Proc Natl Acad Sci U S A. 2014;111:E1629–1638. doi: 10.1073/pnas.1400071111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandon S, Vale PF. The evolution of resistance against good and bad infections. Journal of evolutionary biology. 2014;27:303–312. doi: 10.1111/jeb.12291. [DOI] [PubMed] [Google Scholar]
- Goldberg GW, Jiang W, Bikard D, Marraffini LA. Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature. 2014;514:633–637. doi: 10.1038/nature13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T. Single-molecule methods leap ahead. Nat Methods. 2014;11:1015–1018. doi: 10.1038/nmeth.3107. [DOI] [PubMed] [Google Scholar]
- Hale CR, Cocozaki A, Li H, Terns RM, Terns MP. Target RNA capture and cleavage by the Cmr type III-B CRISPR-Cas effector complex. Genes Dev. 2014;28:2432–2443. doi: 10.1101/gad.250712.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser ML, Taylor DW, Bhat P, Guegler CK, Sternberg SH, Nogales E, Doudna JA. CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference. Proc Natl Acad Sci U S A. 2014;111:6618–6623. doi: 10.1073/pnas.1405079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RN, Golden SM, van Erp PB, Carter J, Westra ER, Brouns SJ, van der Oost J, Terwilliger TC, Read RJ, Wiedenheft B. Structural biology. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science. 2014;345:1473–1479. doi: 10.1126/science.1256328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo C, Fareh M, Narry Kim V. Bringing single-molecule spectroscopy to macromolecular protein complexes. Trends Biochem Sci. 2012 doi: 10.1016/j.tibs.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- Juette MF, Terry DS, Wasserman MR, Zhou Z, Altman RB, Zheng Q, Blanchard SC. The bright future of single-molecule fluorescence imaging. Curr Opin Chem Biol. 2014;20:103–111. doi: 10.1016/j.cbpa.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koel BF, Burke DF, Bestebroer TM, van der Vliet S, Zondag GC, Vervaet G, Skepner E, Lewis NS, Spronken MI, Russell CA, et al. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science. 2013;342:976–979. doi: 10.1126/science.1244730. [DOI] [PubMed] [Google Scholar]
- Kunne T, Swarts DC, Brouns SJ. Planting the seed: target recognition of short guide RNAs. Trends in microbiology. 2014;22:74–83. doi: 10.1016/j.tim.2013.12.003. [DOI] [PubMed] [Google Scholar]
- Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol. 2014;32:677–683. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- Li M, Wang R, Zhao D, Xiang H. Adaptation of the Haloarcula hispanica CRISPR-Cas system to a purified virus strictly requires a priming process. Nucleic Acids Res. 2014;42:2483–2492. doi: 10.1093/nar/gkt1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulepati S, Bailey S. In vitro reconstitution of an Escherichia coli RNA-guided immune system reveals unidirectional, ATP-dependent degradation of DNA target. J Biol Chem. 2013;288:22184–22192. doi: 10.1074/jbc.M113.472233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulepati S, Heroux A, Bailey S. Structural biology. Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science. 2014;345:1479–1484. doi: 10.1126/science.1256996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purtha WE, Tedder TF, Johnson S, Bhattacharya D, Diamond MS. Memory B cells, but not long-lived plasma cells, possess antigen specificities for viral escape mutants. The Journal of experimental medicine. 2011;208:2599–2606. doi: 10.1084/jem.20110740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabuka D, Rush JS, deHart GW, Wu P, Bertozzi CR. Site-specific chemical protein conjugation using genetically encoded aldehyde tags. Nat Protoc. 2012;7:1052–1067. doi: 10.1038/nprot.2012.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasnik I, McKinney SA, Ha T. Nonblinking and long-lasting single-molecule fluorescence imaging. Nat Methods. 2006;3:891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- Reeks J, Naismith JH, White MF. CRISPR interference: a structural perspective. The Biochemical journal. 2013;453:155–166. doi: 10.1042/BJ20130316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter C, Dy RL, McKenzie RE, Watson BN, Taylor C, Chang JT, McNeil MB, Staals RH, Fineran PC. Priming in the Type I-F CRISPR-Cas system triggers strand-independent spacer acquisition, bi-directionally from the primed protospacer. Nucleic Acids Res. 2014;42:8516–8526. doi: 10.1093/nar/gku527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A, van Oijen AM. Bacterial replication, transcription and translation: mechanistic insights from single-molecule biochemical studies. Nature reviews Microbiology. 2013;11:303–315. doi: 10.1038/nrmicro2994. [DOI] [PubMed] [Google Scholar]
- Rouillon C, Zhou M, Zhang J, Politis A, Beilsten-Edmands V, Cannone G, Graham S, Robinson CV, Spagnolo L, White MF. Structure of the CRISPR interference complex CSM reveals key similarities with cascade. Mol Cell. 2013;52:124–134. doi: 10.1016/j.molcel.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson JE, Magadan AH, Sabri M, Moineau S. Revenge of the phages: defeating bacterial defences. Nature reviews Microbiology. 2013;11:675–687. doi: 10.1038/nrmicro3096. [DOI] [PubMed] [Google Scholar]
- Sashital DG, Wiedenheft B, Doudna JA. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol Cell. 2012;46:606–615. doi: 10.1016/j.molcel.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirle NT, Sheu-Gruttadauria J, MacRae IJ. Structural basis for microRNA targeting. Science. 2014;346:608–613. doi: 10.1126/science.1258040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler B, Hofmann H. Single-molecule spectroscopy of protein folding dynamics--expanding scope and timescales. Curr Opin Struct Biol. 2013;23:36–47. doi: 10.1016/j.sbi.2012.10.008. [DOI] [PubMed] [Google Scholar]
- Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, van der Oost J, Brouns SJ, Severinov K. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci U S A. 2011;108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S, Savitskaya E, Semenova E, Logacheva MD, Datsenko KA, Severinov K. Pervasive generation of oppositely oriented spacers during CRISPR adaptation. Nucleic Acids Res. 2014;42:5907–5916. doi: 10.1093/nar/gku226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkunas T, Gasiunas G, Waghmare SP, Dickman MJ, Barrangou R, Horvath P, Siksnys V. In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. EMBO J. 2013;32:385–394. doi: 10.1038/emboj.2012.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem. 2013;82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- Staals RH, Agari Y, Maki-Yonekura S, Zhu Y, Taylor DW, van Duijn E, Barendregt A, Vlot M, Koehorst JJ, Sakamoto K, et al. Structure and activity of the RNA-targeting Type III-B CRISPR-Cas complex of Thermus thermophilus. Mol Cell. 2013;52:135–145. doi: 10.1016/j.molcel.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts DC, Mosterd C, van Passel MW, Brouns SJ. CRISPR interference directs strand specific spacer acquisition. PLoS One. 2012;7:e35888. doi: 10.1371/journal.pone.0035888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczelkun MD, Tikhomirova MS, Sinkunas T, Gasiunas G, Karvelis T, Pschera P, Siksnys V, Seidel R. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci U S A. 2014;111:9798–9803. doi: 10.1073/pnas.1402597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamulaitis G, Kazlauskiene M, Manakova E, Venclovas C, Nwokeoji AO, Dickman MJ, Horvath P, Siksnys V. Programmable RNA Shredding by the Type III-A CRISPR Cas System of Streptococcus thermophilus. Mol Cell. 2014;56:506–517. doi: 10.1016/j.molcel.2014.09.027. [DOI] [PubMed] [Google Scholar]
- Tarlinton D, Good-Jacobson K. Diversity among memory B cells: origin, consequences, and utility. Science. 2013;341:1205–1211. doi: 10.1126/science.1241146. [DOI] [PubMed] [Google Scholar]
- Terns RM, Terns MP. CRISPR-based technologies: prokaryotic defense weapons repurposed. Trends in genetics : TIG. 2014;30:111–118. doi: 10.1016/j.tig.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Oost J, Westra ER, Jackson RN, Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nature reviews Microbiology. 2014;12:479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, Patel DJ. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger AD, Wolf YI, Lobkovsky AE, Gilmore MS, Koonin EV. Viral diversity threshold for adaptive immunity in prokaryotes. mBio. 2012;3:e00456–00412. doi: 10.1128/mBio.00456-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra ER, Buckling A, Fineran PC. CRISPR-Cas systems: beyond adaptive immunity. Nature reviews Microbiology. 2014;12:317–326. doi: 10.1038/nrmicro3241. [DOI] [PubMed] [Google Scholar]
- Westra ER, Semenova E, Datsenko KA, Jackson RN, Wiedenheft B, Severinov K, Brouns SJ. Type I-E CRISPR-cas systems discriminate target from non-target DNA through base pairing-independent PAM recognition. PLoS genetics. 2013;9:e1003742. doi: 10.1371/journal.pgen.1003742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra ER, Swarts DC, Staals RHJ, Jore MM, Brouns SJJ, van der Oost J. The CRISPRs, They Are A-Changin’: How Prokaryotes Generate Adaptive Immunity. Annual Review of Genetics. 2012a;46:311–339. doi: 10.1146/annurev-genet-110711-155447. [DOI] [PubMed] [Google Scholar]
- Westra ER, van Erp PB, Kunne T, Wong SP, Staals RH, Seegers CL, Bollen S, Jore MM, Semenova E, Severinov K, et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell. 2012b;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Lander GC, Zhou K, Jore MM, Brouns SJ, van der Oost J, Doudna JA, Nogales E. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011a;477:486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, van Duijn E, Bultema JB, Waghmare SP, Zhou K, Barendregt A, Westphal W, Heck AJ, Boekema EJ, Dickman MJ, et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci U S A. 2011b;108:10092–10097. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Da L, Plouffe SW, Chong J, Kool E, Wang D. Molecular basis of transcriptional fidelity and DNA lesion-induced transcriptional mutagenesis. DNA Repair. 2014;19:71–83. doi: 10.1016/j.dnarep.2014.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Sheng G, Wang J, Wang M, Bunkoczi G, Gong W, Wei Z, Wang Y. Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli. Nature. 2014 doi: 10.1038/nature13733. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.