Abstract
Childhood acute lymphoblastic leukemia can often be retraced to a pre-leukemic clone carrying a prenatal genetic lesion. Postnatally acquired mutations then drive clonal evolution towards overt leukemia. RAG1-RAG2 and AID enzymes, the diversifiers of immunoglobulin genes, are strictly segregated to early and late stages of B-lymphopoiesis, respectively. Here, we identified small pre-BII cells as a natural subset of increased genetic vulnerability owing to concurrent activation of these enzymes. Consistent with epidemiological findings on childhood ALL etiology, susceptibility to genetic lesions during B-lymphopoiesis at the large to small pre-BII transition is exacerbated by abnormal cytokine signaling and repetitive inflammatory stimuli. We demonstrate that AID and RAG1-RAG2 drive leukemic clonal evolution with repeated exposure to inflammatory stimuli, paralleling chronic infections in childhood.
Introduction
Childhood pre-B acute lymphoblastic leukemia (ALL) can frequently be retraced to a pre-leukemic clone carrying a genetic lesion that was acquired in utero (for example ETV6-RUNX1)1,2. During early childhood, pre-leukemic clones can acquire secondary mutations and evolve towards overt leukemia3. Leukemia cells follow Darwinian trajectories of clonal evolution where sequentially acquired genetic lesions increase or decrease adaptive fitness of pre-leukemic clones, resulting in their positive or negative selection4. While this concept is well established, the mechanism(s) driving clonal evolution are not known. Epidemiological studies suggested a model of clonal evolution that is driven by recurrent infections during childhood4-8. It was hypothesized that delayed exposure of infants to infections in highly protective environments of developed societies cause protracted and excessive immune responses with substantial collateral damage6,8. Early exposure to common infections (such as via day care attendance) mitigated this risk5,8. The systematic introduction of vaccination programs during early childhood reduced the incidence of ALL, presumably by reducing chronic infections and the immune responses that they instigate7. For example, the administration of Haemophilus influenzae B vaccine in early childhood significantly reduced the risk of childhood ALL9,10,11 in three different case-control studies conducted in American, Finnish and Canadian cohorts.
Altered cytokine environments in the context of inflammation eliminate multiple normal pre-B cell clones from the repertoire and favor selective outgrowth of pre-B cell clones that already harbor a pre-leukemic genetic lesion, such as, ETV6-RUNX112. ETV6-RUNX1 was identified as a prenatally-acquired genetic rearrangement13. Additional postnatally-acquired lesions over the course of up to 15 years can give rise to leukemia in this sub-group. While ETV6-RUNX1 is frequently found in umbilical cord B cells and neonatal Guthrie blood spots13, less than 1% of newborn children carrying ETV6-RUNX1 eventually develop ALL14. These studies led to the idea that persistent infection and delayed exposure to strong inflammatory stimuli in childhood may increase the risk of acquiring the post-natal genetic lesions. However, the mechanistic basis of how such pre-leukemic clones evolved remained elusive.
Two classes of enzymes are required for somatic recombination and mutation of immunoglobulin (Ig) genes in B cells: The proteins encoded by recombination activating genes (RAG1 and RAG2) introduce DNA double-strand breaks and recombine V, D and J segments15, AID deaminates cytosines in Ig V and switch regions thereby enabling somatic hypermutation (SHM) and class switch recombination (CSR) of Ig genes16. It is particularly interesting to note that genetic diversification of the antibodies by AID and RAG1-RAG2 is activated in response to infectious and inflammatory stimuli, both in early17 and mature18 B cells. A comprehensive breakpoint analysis19 suggested that some chromosomal rearrangements in human B cell malignancies may result from the cooperation between RAG1-RAG2 and AID. During normal B cell development, however, enzymatic activities of these proteins are normally strictly segregated20.
Here, we report that the above enzymes not only genetically diversify the B-lymphocyte repertoire21, but also contribute to the acquisition of genetic lesions and promote clonal evolution towards leukemia. In this paper, we delineate the molecular mechanism by which the normal physiological process of antibody diversification is subverted in pre-leukemic B cell clones (ETV6-RUNX1) leading to overt disease.
Results
Frequently deleted genes in childhood ALL are targets of AID
RAG1 and RAG2 are constitutively expressed in pro- and pre-B cells and target recombination signal sequences (RSSs) in both Ig and non-Ig genes. Motif analyses of deletion breakpoints in the tumor suppressor genes IKZF122, CRLF2, TBL1XR1, CDKN2A, NR3C1, NR3C2 and BTG123, revealed that RSS motifs in these genes were indeed targeted by RAG1-RAG2.
To test the plausibility of AID’s contribution to RAG1-RAG2-mediated deletions and rearrangements, we measured whether lesions in childhood ALL (data from clinical trial P9906) occur preferentially at genes bound by AID in normal B cells. We identified 40 genes that were recurrently mutated, deleted or rearranged in childhood ALL (207 patients in COG P9906) and plotted the frequency of these lesions against mouse AID (mAID) targeting of these genes in mouse B cells. mAID target genes (4,167) were identified in a previous ChIP-seq study based on increased mAID ChIP-seq tags compared to Aicda−/− B cells24. 18,248 genes (non-mAID targets) did not have significantly higher mAID ChIP-seq tag counts compared to Aicda−/− B cells. Interestingly, frequencies of recurrent genetic lesions correlated significantly with mAID ChIP-seq counts (Fig. 1a). In this analysis, 34 of 40 genes with recurrent lesions in childhood ALL are mAID-targets, thereby pointing to AID’s possible contribution to frequent genetic lesions in childhood ALL. Of the remaining six genes that are non-mAID targets, five (CDKN2A, TBL1XR1, NR3C1, NR3C2 and PBX1)23,25 have breakpoints at known RSS motifs with characteristic N-nucleotide addition at the break junctions, suggesting that lesions at these loci were caused by RAG1-RAG2 activity alone. It is important to note that not all AID targets are syntenic between human and mouse. While this represents a limitation to our approach, disparity between AID targets in humans and mice will only lead to an underestimation of the correlation we observed. Moreover, a recent study identifying hAID targets in B cell lymphoma26, although not quantitative, includes multiple genes from our analysis (PAX5, BTG1, BTG2, BACH2, RHOH, EBF1, TCF3). These studies confirm that AID may be responsible for mutating a substantial percentage of genes found frequently altered in pre-B ALLs.
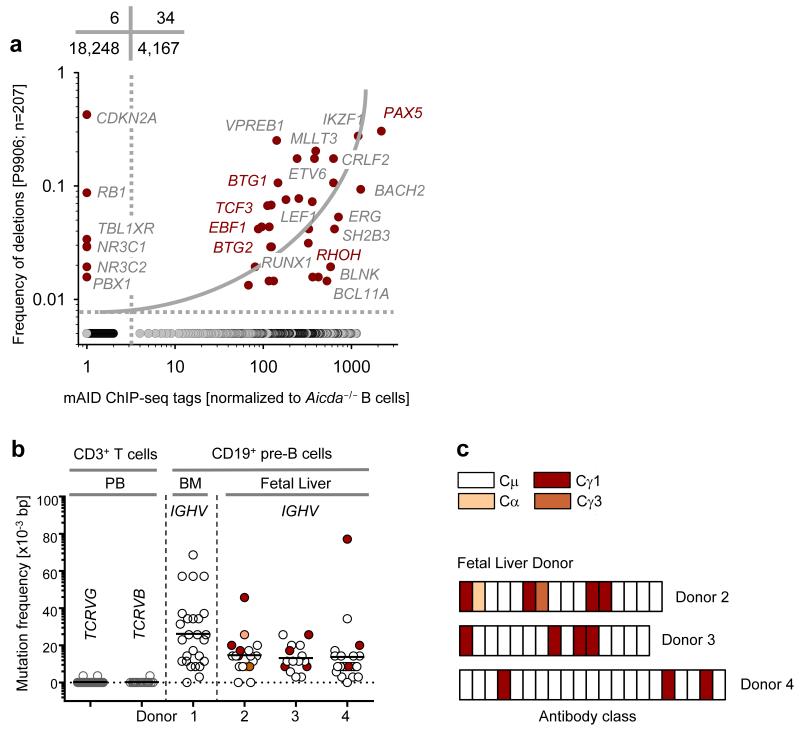
Figure 1. Expression and activity of AID in human B cell precursors and B-lineage ALL.
(a) Plot showing correlation between common somatic hypermutation targets of mAID and their alteration status in childhood ALL. On plotting the genes which are frequently bound and targeted by mAID24, against genes which are commonly deleted and amplified in childhood ALL (data from clinical trial P9906), a positive correlation was observed using a 2×2 contingency table analysis by Chi-square test with Yates’ correction. (b) SHM frequency of IGH VH region genes in single pre-B cell clones isolated from human bone marrow (n=1) and human fetal liver (n=3). SHM of variable regions of TCRVG and TCRVB measured in these cells were used as negative controls. p-values were calculated using student t-test. (c) A schematic depicting CSR in early human B cells derived from three fetal liver donors.
High AICDA and RAG1 mRNA predict poor ALL patient outcome
If AID allows clonal evolution towards childhood ALL, its expression and activity might correlate with clinical outcomes of patients. To test this possibility, we correlated AICDA mRNA expression in patients at diagnosis with overall and relapse-free survival. ALL patients in two clinical trials (ECOG E2993 and COG P9906) were segregated into two groups based on higher and lower than median AICDA mRNA abundance at the time of diagnosis. The ECOG E2993 trial included 215 patients (106 bone marrow and 109 peripheral blood), and the COG P9906 trial had 207 pediatric patients (131 bone marrow and 76 peripheral blood).
Interestingly, higher than median AICDA expression strongly predicted poor overall patient survival (Supplementary Fig. 1a, left; P=0.026). Likewise, higher than median RAG1 mRNA abundance predicted shorter relapse-free and overall survival of patients with ALL (Supplementary Fig. 1a, center and right). In the COG P9906 trial, 49 patients relapsed after successful initial therapy. Comparing AICDA mRNA expression in matched sample pairs at diagnosis and relapse, for most patients, AICDA mRNA abundance was increased at relapse (Supplementary Fig. 1b). These findings suggest that AICDA expression at diagnosis can predict ALL patient-outcome.
RAG and AID proteins are active in human B cell precursors
B-ALL arises from sites of early B-lymphopoiesis- the fetal liver and the bone marrow during pre- and postnatal development, respectively. RAG enzymes are active in B cell precursors in both sites. Studies in identical twins with concordant ETV6-RUNX1 pre-B ALL indicate clonal origin in cells undergoing RAG-dependent IGH variable region rearrangement and continued recombinant activity during subsequent clonal evolution13,27. To measure AID-activity in human fetal liver and bone marrow B cell precursors, we sorted CD19+ pre-B cells that lack κ and λ immunoglobulin light chains, from fetal liver tissues of three donors and bone marrow from one healthy adult donor. We then cloned and sequenced the IGH variable regions for SHM and constant regions for CSR (Supplementary Tables 1,2). Interestingly, most bone marrow pre-B cell clones expressed IGH with mutated VH regions (26×10−3 bp, Fig. 1b, Supplementary Table 1). Likewise, fetal liver pre-B cells of three donors carried mutated VH regions (14×10−3 bp, Fig. 1b, Supplementary Table 2). To control for reverse transcriptase and Pfu DNA polymerase errors, peripheral blood-derived CD3+ T cells lacking AICDA expression were sorted and, rearranged TCRB and TCRG variable regions were amplified and analyzed in parallel with IGH VH regions of bone marrow and fetal liver pre-B cells. Overall, IGH VH regions from sorted bone marrow and fetal liver pre-B cells carried a significant frequency of somatic mutations (P=3.3×10−18), relative to TCRB and TCRG variable regions from peripheral blood T-lymphocytes (Fig. 1b). We also found a significant fraction of the sorted CD19+ pre-B cells sorted from fetal liver tissues expressing class-switched IGH, containing Cγ3, Cγ1 and Cα constant regions (Fig. 1b,c, Supplementary Table 2). It is important to note that our method of IGH cloning and sequencing was not strictly quantitative. Therefore, we are not in a position to predict the exact fraction of fetal liver pre-B cells that express class-switched IGH constant regions. These findings suggested that AID might be prematurely activated in human bone marrow and fetal liver pre-B cells. While we found genetic traces of AID activity in RAG1-RAG2-expressing pre-B cells, these findings did not establish that AID and RAG1-RAG2 are concurrently active in the same cells.
Small pre-BII cells have increased genetic vulnerability
To elucidate whether and how premature activity of mAID is regulated during early B-lymphopoiesis, we sorted B cell precursors from normal mouse bone marrow as described previously20 by flow cytometry. Pro-B cells (c-kit+ B220+), large pre-BII (CD25+ B220+ FSChi SSClo) and small pre-BII (CD25+ B220+ FSClo SSClo) were isolated. As large and small pre-BII cells carry similar surface markers we used their size differences (FSC) to separate them. We then measured Aicda mRNA abundance in each fraction of early B cell development (Fig. 2a). In comparison to activated mature splenic B cells, Aicda mRNA in bone marrow B cell precursors was generally low or absent. However, Aicda mRNA was increased ~21-fold at the small pre-BII as compared to the large pre-BII stage of early B-lymphopoiesis (Fig. 2a). This transition is characterized by signals from the pre-B cell receptor (pre-BCR) and its intracellular signaling components that are assembled by the pre-BCR linker BLNK28,29. One consequence of pre-BCR signaling at the large to small pre-BII transition is the downregulation of interleukin-7 receptor (IL-7R) surface expression by BLNK activation (Fig. 2b). Attenuation of IL-7R expression reduces downstream STAT5 signaling in small pre-BII cells30. We therefore tested whether Aicda mRNA abundance was increased in small pre-BII cells by abrogating IL-7R-STAT5 signaling using two methods. First, we reconstituted the pre-BCR linker Blnk in Blnk−/− pre-B cells to induce IL-7R downregulation, and measured Aicda mRNA (Supplementary Fig. 2a) and, mAID protein abundance (Fig. 2c). Second, we removed IL-7 from pre-B cell cultures (Fig. 2d, Supplementary Fig. 2b) to force the transition from large pre-BII to small pre-BII stage. In vitro differentiation by both methods induced mAID protein upregulation, demonstrating that active IL-7R signaling in mouse large pre-BII cells safeguards against pre-mature mAID expression.
Figure 2. Late pre-B cells (small pre-BII) represent a natural subset of increased genetic vulnerability.
(a) Wildtype mouse bone marrow was sorted into different fractions of early B cell development20. Quantitative RT-PCR showing Aicda mRNA abundance in each fraction of early B cell development (n=3; mean ± s.d.), as compared to splenic B cells induced with LPS and IL-4 (positive control) and Aicda−/− pre-B cells (negative control). (b) FACS analysis of surface IL-7Rα in Blnk−/− pre-B cells expressing empty GFP vector, Blnk/GFP or BlnkY96F/GFP. (c) Immunoblot depicting increase in mAID protein level upon differentiation of Blnk−/− pre-B cells from large pre-BII to small pre-BII by Blnk reconstitution. (d) mAID protein levels measured by western blotting before (large pre-BII) and after (small pre-BII) IL-7 withdrawal from mouse pre-B cell cultures. (e) Plots showing AICDA mRNA levels (left) and VH mutation frequency (right) in large pre-BII and small pre-BII human pre-B cells, in children with mutation in one of the chains of the IL-7R. Normal human bone marrows were used as negative controls. All p-values were calculated using student t-test. One representative of three experiments is shown for FACS plots and immunoblots.
IL-7R safeguards human pre-B cells from AID activation
We next investigated whether IL-7R signaling protects against pre mature AID expression in human pre-B cells. While IL-7R signaling is essential for B cell development in mice, humans with inherited IL7R deficiency have almost normal B cell counts31, which allows experimental analysis of human B-lymphopoiesis in the absence of IL-7R signaling. We isolated large and small pre-BII cells from bone marrow samples of three patients with biallelic germline mutations in either gene that encode the IL-7R, namely IL7R and IL2RG. One patient harbored an IL7RG28X mutation, the two other patients carried IL2RGQ144X and IL2RGR289X mutations, respectively (Fig. 2e). As controls, we studied sorted large and small pre-BII cells from bone marrow of three healthy donors. As in mouse B-lymphopoiesis, we observed that human pre-B cells upregulate AICDA mRNA by 10- to 25-fold at the large pre-BII to small pre-BII transition. In the three patients lacking functional IL-7R signaling, pre-B cells expressed more AICDA mRNA as compared to the healthy controls (Fig. 2e). No significant difference was observed between large and small pre-BII cell subsets obtained from IL-7R-deficient patients. Amplification and sequencing of IGH VH regions revealed a low frequency of SHM in normal small pre-BII cells. Frequencies of somatic IGH VH region mutations were ~5-fold higher, in both large and small pre-BII cells isolated from patients lacking functional IL-7R (Fig. 2e). Thus, IL-7R safeguards human pre-B cells from premature AID activation.
Safeguard mechanism of IL-7R against AID activation
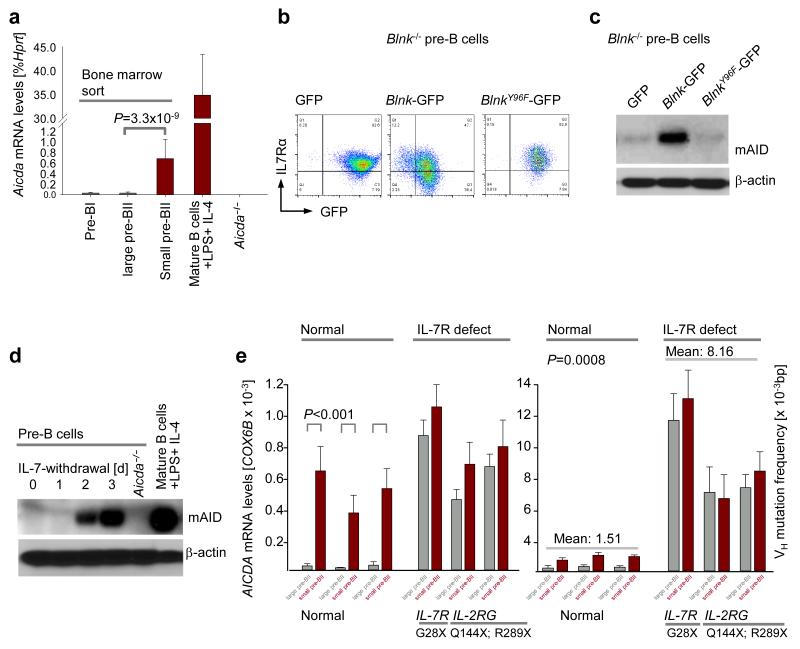
IL-7R-STAT5 signaling in early pre-B cells32 prevents upregulation of Rag1-Rag2 mRNA and recombination of Igk V and J gene segments32,33. Downregulation of IL-7R expression attenuates activities of STAT5 and Akt, and induces activation of Rag1-Rag2 mRNA (Supplementary Fig. 2c). STAT5 recruits the polycomb repressor EZH2 to Rag1-Rag2, while Akt phosphorylates and inactivates FOXO1, a potent transcriptional activator of Rag1 and Rag234. Here we show that inducible, Cre-mediated deletion of Stat5a and Stat5b in large pre-BII cells not only results in activation of RAG1-RAG2 but also upregulation of AID (Fig. 3a). The mechanistic role of Akt-Foxo1 signaling in regulation of Aicda mRNA abundance was tested using genetic loss of function of PTEN, a negative regulator of Akt, and gain-of-function studies for Foxo1. Inducible deletion of Pten results in hyperactivation of Akt and, hence, phosphorylation and inactivation of Foxo transcription factors35. Pre-BCR signaling via BLNK not only decreased IL-7R surface expression but also caused activation of Foxo factors (Fig. 3b). CRE-mediated, inducible ablation of Pten caused near-complete loss of Rag1, Rag2 and Aicda mRNA expression (Fig. 3c-e). In agreement with these findings, a constitutively active form of Foxo1 that is protected against Akt-mediated phosphorylation strongly activates Rag1, Rag2 and Aicda transcription. Interestingly, these effects were further enhanced by removal of IL-7 from cell culture medium (Fig. 3f-h, Supplementary Fig. 2d). In summary, IL-7R signaling protects pre-B cells from premature activation of Aicda through STAT5- and Akt-dependent pathways.
Figure 3. Aicda and Rag1-Rag2 are regulated by the same transcriptional control elements in pre-B cells.
(a) AID protein levels in Stat5fl/fl IL-7-dependent cells transduced with empty ERT2 (no Stat5 deletion) and Cre-ERT2 (with Stat5 deletion) vectors were compared by immunoblotting. (b) Immunoblot for active nuclear FOXO4 after Blnk−/− IL-7-dependent pre-B cells are differentiated from large pre-BII to small pre-BII stage by Blnk reconstitution. (c) Aicda mRNA levels were compared between the empty vector (EV) transduced and Pten deleted Ptenfl/fl pre-B cells 48 hours after tamoxifen induction, by qRT-PCR (n=3, mean ± s.d.). (d-e) Rag1 and Rag2 mRNA levels were measured by qRT-PCR following inducible deletion of Pten in Ptenfl/fl pre-B cells (n=3, mean ± s.d.). (f-h) Mouse IL-7-dependent pre-B cells were retrovirally transduced with either an empty vector or a constitutively active form of Foxo1 (Foxo1CA). The 2 groups of cells were then subject to two conditions each, either they were retained in the presence of IL-7 (large pre-BII) or IL-7 withdrawal was carried out for 24 hours to differentiate them to small pre-BII. (f) Aicda mRNA level was then measured in each case by qRT-PCR (n=3, mean ± s.d.). (g-h) Rag1 and Rag 2 mRNA levels measured by qRT-PCR after retroviral expression of a constitutively active form of Foxo1 (Foxo1CA) or empty vector (EV) in mouse pre-B cells, in the presence or absence of IL-7 (n=3, mean ± s.d.). All p-values were calculated using student t-test.
Concurrent RAG and AID activity in pre-B cell clones
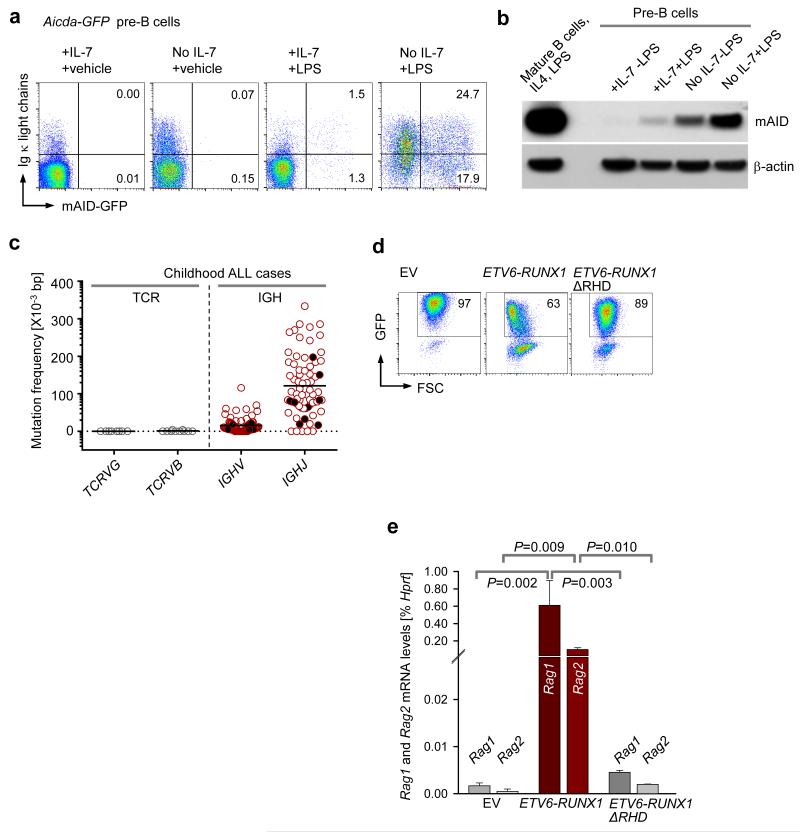
AID is upregulated in mature B cells upon antigen encounter in germinal center, where it introduces SHM in Igh VH regions and promotes CSR of constant regions16,36. Antigen encounter in germinal centers can be mimicked by treatment of mature splenic B cells with lipopolysaccharide (LPS) and IL-4. We tested whether small pre-BII cells respond to inflammatory agents like LPS in a manner similar to mature B cells, by upregulating mAID. To measure mAID-activation in pre-B cells at the single cell level, we studied pre-B cells from an Aicda-GFP reporter mouse strain where GFP gene was fused in frame to Aicda exon 537. Aicda-GFP pre-B cells were propagated in IL-7 and then treated with or without LPS, and in the presence or absence of IL-7 (Fig. 4a, Supplementary Fig. 3a,b). Treatment with LPS induced minimal increases of mAID protein expression. However, when LPS-treatment was combined with IL-7-withdrawal, upregulation of mAID was increased by 20-fold compared to LPS-treatment in the presence of IL-7 (Fig. 4a,b). To measure mAID and RAG1-RAG2 activities in parallel at the single-cell level, we monitored mAID-GFP and de novo surface expression of Igκ light chains after RAG-mediated Vκ-Jκ gene rearrangement (Fig.4a, Supplementary Fig. 3b). Treatment of Aicda-GFP pre-B cells with LPS in the presence of IL-7 induced activation of the Aicda-GFP reporter in 2.8% of cells as compared to 0.01% in LPS-untreated condition. However, combination of LPS-treatment with removal of IL-7 from the cell culture medium increased the fraction of mAID-expressing cells to 42% from 2.8% in the presence of IL-7. Interestingly, majority of Aicda-GFP+ cells also expressed de novo κ light chains (Fig. 4a, Supplementary Fig. 3b), providing direct evidence that mAID expression and RAG-mediated recombination of Vκ- and Jκ-gene segments can occur in the same cells. These findings were confirmed in a second Aicda reporter mouse model, in which mAID drives expression of Cre for excision of a loxP-flanked Stop cassette of an eYPF marker that is located in the Rosa26 locus37 (Supplementary Fig. 3c,d).
Figure 4. Evidence for concurrent activities of RAGs and AID in single pre-B cell clones.
(a) Aicda-GFP pre-B cells37 upregulate expression of AID, RAG1 and RAG2 at small pre-BII in the context of inflammatory signals like LPS (GFP+κLC+ cells) (b) Immunoblot comparing mAID protein levels in large pre-BII and small pre-BII mouse pre-B cells, following LPS exposure. Splenocytes from a wildtype mouse induced with LPS and IL-4 were used as a positive control for mAID expression. (c) SHM frequencies of IGH VH regions in pediatric ALL cases were compared against SHM of TCRG and TCRB in these cases. Leukemia patients displaying ongoing VH replacements are represented by solid black circles. (d-e) Effects of ETV6-RUNX1GFP and its mutant ETV6-RUNX1-ΔRHDGFP on Rag1 and Rag2 expression in mouse pre-B cells were evaluated by overexpression of retroviral vectors encoding these fusion proteins. Empty vector (EV) transduced mouse pre-B cells served as negative control. (d) Representative FACS plots showing the percentages of GFP+ cells in each case on day 2 post transduction. One representative of three replicates is shown. (e) GFP+ cells in (d) were sorted and used to measure Rag1 and Rag2 mRNA levels by qRT-PCR (n=3, mean ± s.d.). All p-values were calculated using student t-test.
Evidence of ongoing RAG and AID activity in human pre-B ALL
Studying IGH VH genes amplified from 72 cases of childhood ALL (13 ETV6-RUNX1, 2 TCF3-PBX1, 3 MLL-rearranged, 1 MYC-IGH, 17 hyperdiploid, 26 with normal or undetermined karyotype and 10 with sporadic chromosomal translocations), we found evidence of ongoing RAG1-RAG2 and AID activity in all subgroups (Supplementary Tables 3-5). Pre-B ALL clones consistently carried somatically mutated VH and JH gene segments, indicative of AID activity (Supplementary Tables 3-5). Interestingly, in 10 of 72 cases, we found subclones in each patient that carried the same defining D-JH junction (Supplementary Tables 3-5). Multiple distinct VH segments diversified these subclones through sequential RAG-mediated VH replacement38. The process of leukemic clonal evolution by cooperative activities of AID (SHM) and RAGs (VH replacement) in a pediatric pre-B ALL patient is shown (Supplementary Fig. 4, Supplementary Table 5). Comparing mutation frequencies in VH and JH segments, we found that JH segments carry a significantly higher load of somatic mutations (102×10−3 bp in JH, 16×10−3 bp in VH; P=0.0007; Supplementary Tables 3-5, Fig. 4c). Multiple rounds of VH replacement, where acquired mutations in VH segments are erased but D-JH junctions remain constant, could explain this unexpected difference. In 7 of 13 ETV6-RUNX1 pre-B ALL cases, we found multiple distinct subclones that had undergone VH replacement, suggesting that RAG enzymes are particularly active in this subset (Supplementary Table 3). This finding is in agreement with a recent study highlighting aberrant activation of RAG-mediated recombination as the primary driver of ETV6-RUNX1 leukemogenesis23. To test if ETV6-RUNX1 activity is linked to deregulated RAG expression, we transduced murine pre-B cells with vectors encoding full-length ETV6-RUNX1, a mutant ETV6-RUNX1 lacking the DNA binding Runt Homology Domain (ΔRHD) and an empty vector (EV) control. Interestingly, full-length ETV6-RUNX1 but not ΔRHD ETV6-RUNX1 induced expression of Rag1-Rag2 in mouse pre-B cells, suggesting that abnormally high recombinase activity in this ALL subset is caused by the ETV6-RUNX1 fusion (Fig. 4d,e). Hence, cooperative AID and RAG activities may explain the mechanism of clonal evolution in the ETV6-RUNX1 leukemias.
Cytosine-deamination at CpGs are enriched in ALL patients
As AID-targets are frequently deleted or mutated in childhood ALL, we analyzed mutational hotspot targeting in 24 patient-derived ALL: 10 ETV6-RUNX1, 4 MLL-AF4, 1 TCF3-PBX1 and 9 cases lacking a recurrent chromosomal translocation (Supplementary Table 6). In agreement with two recent studies23,39, whole exome sequencing revealed a striking enrichment of C>T transitions at CpGs, accounting for 44% of all substitutions. Cytosine-deamination at methyl-CpGs strongly implicates AID-mediated deamination of methyl-CpG19. However, in agreement with previous studies23,38, somatic mutations in human ALL samples did not reveal typical hotspot targeting described for SHM of IGH VH genes (such as RGYW, WRCY)40. AID hotspot targeting typically emerges from a large number of somatic mutations in IGH VH region genes that were non-productively rearranged, hence mutations do not affect the outcome of clonal selection among B cells. The non-Ig gene mutations in ALL cells studied here occur at much lower frequencies and in coding regions, leaving open the possibility that hotspot targeting has been obscured by low mutation frequencies and effects of clonal selection.
Coordinate AID and RAG expression subverts genetic integrity in cord blood B cells
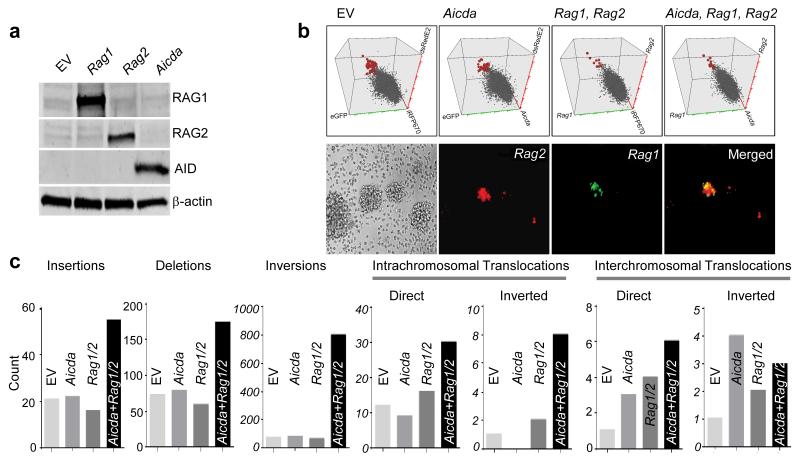
Based on our earlier findings, we propose that pre-mature AID expression in small pre-BII cells represents a vulnerability that exposes early B lymphocyte development to genetic lesions in the context of repeated exposure to inflammatory stimuli (Supplementary Fig. 5). We therefore tested the relevance of RAG and AID cooperation in clonal evolution in normal cord blood-derived human B cells through a genetic gain-of-function experiment. Normal CD19+ B cells were sorted from human cord blood (CB) and immortalized by EBV-containing supernatants from B98-5 cultures. Proliferating CB B cells were then transduced with vectors encoding Aicda (iRFP670), Rag1 (eGFP) and Rag2 (dsRedE2), either alone or in combination or empty vector controls for Aicda, Rag1 and Rag2 vectors (Figs. 5a,b; Supplementary Figs. 6,7). Transduced CB B cells were sorted in each condition and expression of AID, RAG1 and RAG2 and lack thereof was confirmed by immunoblotting (Fig. 5a). Our finding, that AID expression is absent in EBV-infected B cells is consistent with the published literature. EBV suppresses AID expression41, which is the reason why EBV+ B cell lymphomas do not exhibit ongoing SHM and CSR. EBV-mediated suppression of AID is induced by the EBV oncoprotein EBNA242.
Figure 5. Cooperation between RAG1, RAG2 and AID promotes clonal evolution towards pre-B ALL.
(a) Immunoblot depicting protein expression of AID, RAG1 and RAG2 after retroviral overexpression of these vectors in EBV-immortalized human cord blood B cells. (b) Verification of Aicda (iRFP670), Rag1 (eGFP) and Rag2 (dsRedE2) overexpression in EBV transformed human B cells by flow cytometry (top) and verification of Rag1 and Rag2 overexpression in in these cells by fluorescence microscopy (bottom). (c) Whole exome sequencing analyses to compare the total counts of chromosomal abnormalities in empty vector (EV), Aicda, Rag1-Rag2, and Aicda Rag1-Rag2 transduced EBV-transformed human cord blood B cells.
To be able to study genetic lesions at the level of individual clones, single transduced human B cell clones were plated into individual wells on 96-well plates. CB B cell clones were harvested and subjected to whole exome sequencing. CB B cells carrying three empty vector controls (iRFP670, eGFP, dsRedE2), Aicda and two empty vector controls, Rag1, Rag2 and one empty vector control and, Aicda, Rag1 and Rag2, were analyzed. The analysis focused on structural genetic lesions: insertions, deletions, inversions and translocations. Compared to empty vector transduced CB B cells, overexpression of either Aicda alone or Rag1-Rag2 alone only increased the frequency of interchromosomal translocations. However, CB B cells expressing Aicda in combination with Rag1-Rag2 had increased counts of insertions, deletions, inversions and intrachromosomal translocations (Fig. 5c).
RAGs and AID cooperate in ETV6-RUNX1-driven leukemogenesis
We hypothesized that AID and RAGs cooperate and induce secondary genetic lesions to accelerate leukemogenesis of an ETV6-RUNX1 small pre-BII clone in the context of inflammation. To test this, we subjected ETV6-RUNX1 expressing pre-B cells from wildtype mice, Aicda−/−, and Rag1−/− mice to five rounds of IL-7 withdrawal and LPS treatment. During each cycle of IL-7 removal and LPS-treatment a large fraction of stimulated pre-B cells died. Before entering the next round of stimulation, the pre-B cells recovered over the course of ~2 weeks from a small fraction of surviving pre-B cells. The cell death after each cycle of IL-7 withdrawal and LPS treatment occurred independently of the Aicda or Rag1 genotype. This was not surprising because IL-7 withdrawal induces a number of Aicda and Rag-independent signaling pathways that promote apoptosis at the small pre-BII stage B-lymphopoiesis.
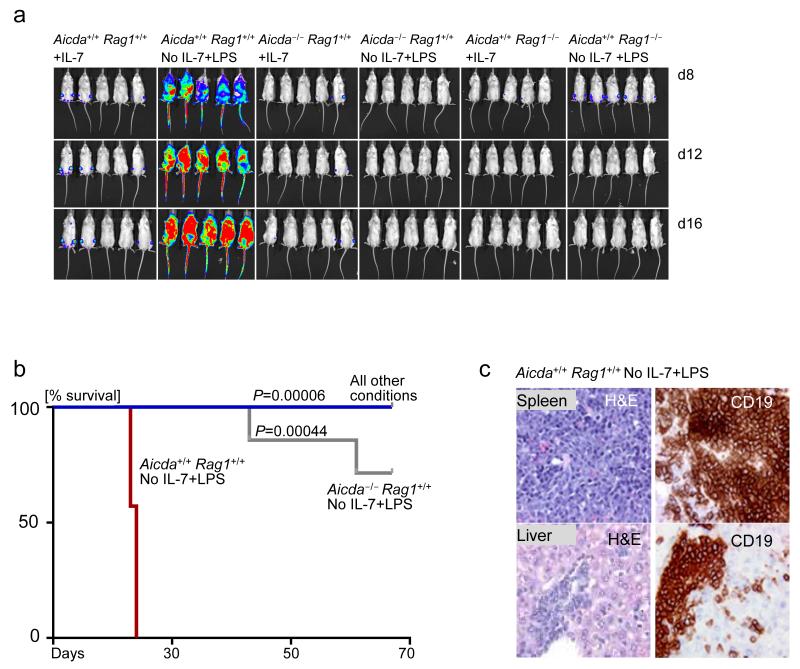
After five cycles, firefly luciferase-labeled pre-B cells were injected intravenously into NOD/SCID mice. All mice receiving wildtype pre-B cells with IL-7 withdrawal and LPS treatment died of leukemia within three weeks. Multiple mice (3-4) in the Aicda+/+ Rag1+/+ -IL-7+LPS group were sacrificed on the same day because they were terminally ill with leukemia. This phenomenon is reflected in the rapid drops of the Kaplan Meier curve for this group. In contrast, the absence of Aicda or Rag1 delayed and abrogated leukemia development respectively, demonstrating that both proteins are required for clonal evolution of ETV6-RUNX1 pre-B cells towards leukemia (Fig. 6a,b, Supplementary Figs. 8,9). In the absence of Aicda, two mice developed leukemia, albeit after a substantially prolonged latency period (Fig. 6a,b). We were able to verify that the sick mice injected with stimulated Aicda+/+ Rag1+/+ pre-B cells indeed died of pre-B ALL (Fig. 6c, Supplementary Figs. 8,9). These findings provide genetic evidence that clonal evolution of pre-leukemic ETV6-RUNX1 pre-B cells in the context of inflammatory/repetitive infectious stimulation requires both AID and RAG activities.
Figure 6. Cooperation between RAGs and AID is required for clonal evolution of pre-leukemic ETV6-RUNX1 B cell precursors.
(a) Luciferase bioimaging of NOD-SCID mice injected with Aicda+/+ Rag1+/+ +IL-7, Aicda+/+ Rag1+/+ No IL-7+LPS, Aicda−/− +IL-7, Aicda−/− No IL-7+LPS, Rag1−/− +IL-7 and Rag1−/− No IL-7+LPS, with all cell types overexpressing ETV6-RUNX1GFP. IL-7 dependent pre-B cells from each group were taken through 5 rounds of IL-7 withdrawal and LPS treatment. 7 mice were used per group. Luciferase bioimages are shown for five mice in each group. (b) Kaplan Meier curves comparing the overall survival percentage of mice in all 6 groups. (c) Verification of pre-B ALL development in the Aicda+/+ Rag1+/+ No IL-7+LPS group by immunohistochemistry. Haematoxylin and Eosin (H&E) staining was carried out to verify lymphocyte infiltration into the liver and spleen of sick mice. Additionally, CD19 immunohistochemistry was carried out to verify that the infiltrating cells represent B-lineage ALL. Immunohistochemistry pictures are shown for one representative mouse out of 7.
Discussion
A recent study demonstrated that aberrant RAG-mediated V(D)J recombination targeting non-Ig genes causes genetic lesions that drive clonal evolution of B-ALL23. Likewise, AID targets non-Ig genes, promotes drug-resistance in B-lineage ALL43,44 and gives rise to AID-dependent chromosomal rearrangements. A comprehensive sequence analysis of 1,700 breakpoints of chromosomal rearrangements in human B cell malignancies suggested that these lesions are caused by cooperation between RAGs and AID19. This was based on the surprising finding that ~70% of chromosomal rearrangements in pro- and pre-B cells involve a break at CpG dinucleotides. Cytosines in CpG dinucleotides are frequently methylated. While AID-mediated cytosine-deamination typically results in conversion to uracil and triggers highly efficient repair by base-excision repair, AID-mediated deamination of methyl-cytosine, e.g. in CpG, results in conversion of cytosine to thymine. The resulting T:G mismatch is more stable than a U:G mismatch, and repair of T:G mismatches is ~2,500 times less efficient45. These T:G mismatches generate a one-base bubble that is recognized by RAGs, resulting in the introduction of a single-strand nick19. AID thus generates multiple one-base bubbles (T:G mismatches) at methylated CpG sequences that form substrates for RAG-mediated cleavage. An important premise for this scenario is that, under some circumstances, B cells express RAGs and AID concomitantly.
Our findings highlight that small pre-BII cells represent a subset of increased genetic vulnerability because they concurrently express AID and RAGs in the presence of strong inflammatory stimuli. The concurrent activation of these enzymes provides an explanation for the thus far elusive origin of childhood leukemia. We formally demonstrate that combined AID and RAG activities are required for clonal evolution from pre-leukemic lesions and that; such cooperative activities potentiate genetic instability in these clones. We elucidate how IL-7R signaling safeguards the early B cell genome from premature AICDA activation. Altered cytokine environments have been previously attributed to the clonal evolution of ETV6-RUNX1-driven pre-B ALLs12. Here, we provide one more example of this phenomenon by which cytokine signaling in pre-B cells can contribute to genomic stability, and hence leukemic transformation.
Our work provides a mechanistic underpinning for the epidemiological findings that altered patterns of infection during early childhood provide a promotional drive to leukemogenesis; especially for subtypes of childhood leukemia initiated in utero via ETV6-RUNX1 fusion or hyperdiploidy6. In future, it may be interesting to investigate if leukemic children with known infectious origins express high levels of AICDA and RAG enzymes. It may also be particularly important to identify the classes of pathogens responsible for overt leukemogenesis in such children. Delineating the molecular bases behind the infectious and inflammatory origins of pre-B ALLs could inform the development of public health policies regarding the benefits of vaccination programs during early childhood for leukemia prevention.
Methods
Patient samples, cord blood and human cell lines
We obtained diagnostic bone marrow samples of individuals with primary pre-B ALL from our collaborators, and human pre-B ALL cell lines from DSMZ, Germany (Supplementary Tables 6,7). These primary pre-B ALL samples, along with some pre-B ALL patient xenografts were used for IGH sequencing studies. Our Primary Patient Xenograft Repository is approved by the University of California, San Francisco (UCSF) and Institutional Review Board of NCI/CTEP and the Eastern Cooperative Oncology Group (ECOG, protocol E2993T5). Human CD19+ cord blood B cells were obtained from AllCells. AllCells procures cord blood subsequent to receiving informed consent from its donors. We have approval to use cord blood cells in our experiments (protocol BU085087).
Mice strains and cell culture
Bone marrow cells were obtained from young female and male mice (less than 6 weeks of age). The following strains of mice were used: Blnk−/−46, Ptenfl/fl47, Aicda-EGFP37, Aicda-Cre X Rosa26-EYFP37, Stat5fl/fl48, Rag1−/−49, Aicda−/−16 and, NOD-SCID (Jackson Laboratories). Bone marrow cells were isolated by flushing the cavities of femur and tibia with PBS. After filtration through a 40-μm filter and depletion of erythrocytes using RBC lysis buffer (BD PharmLyse, BD Biosciences), washed cells were either cryopreserved or utilized for further experiments. Bone marrow cells were cultured with 10 ng/ml IL-7 on either RetroNectin(Takara)-coated non tissue culture dishes or on irradiated OP9 stroma layers. Precursor B cell populations were sorted from mouse bone marrow using the following markers: pro-B cells (c-Kit+ B220+), large pre-BII (CD25+ B220+ FSChi SSClo) and small pre-BII (CD25+ B220+ FSClo SSClo) were separated by flow cytometry using antibodies (Supplementary Table 8) on a FACSAriaII (BD). All pre-B cells were maintained in Iscove’s modified Dulbecco’s medium (IMDM, Invitrogen) with GlutaMAX containing 20% fetal bovine serum, 100 IU/ml penicillin, 100 μg/ml streptomycin, 50 μM 2-mercaptoethanol and 10 ng/ml recombinant mouse IL-7 (Peprotech) at 37 °C in a humidified incubator with 5% CO2. All mouse experiments were approved by the IACUC of the University of California, San Francisco.
In vivo model for inflammatory origins of ETV6-RUNX1-driven leukemia
IL-7-dependent pre-B cells from wild-type, Aicda−/− Rag1+/+, Aicda+/+ Rag1−/− mice were transformed with a retroviral vector encoding ETV6-RUNX1-IRES-GFP (ETV6-RUNX1GFP). Next, these cells were either cultured with 10 ng/ml IL-7 on OP9 plates or subjected to five cycles of IL-7 withdrawal and 2.5 μg/ml LPS treatment. For the latter, each cycle was interrupted by a phase where cells were given 10 ng/ml IL-7 for 1 to 2 days to aid their recovery before the starting the subsequent cycle. We then transduced cells with a retroviral vector encoding the luciferase enzyme and injected cells intravenously into 7 sublethally irradiated (250 cGy) female NOD-SCID mice per group. Recipient mice were monitored for weight loss, hunched posture and inability to move, as indicators of leukemia progression. Recipient mice were imaged for bioluminescence on days 7, 10 and 12 post-injection. Mice were sacrificed after they became terminally ill. For the group that displayed signs of leukemia; immunostaining for CD19 was performed on the bone marrow and spleen of mouse recipients to confirm leukemic disease. We sacrificed mice from all the healthy groups at day 70 post-injection.
Immunoblotting
Cells were lysed in CelLytic buffer (Sigma) supplemented with 1% protease inhibitor cocktail (Pierce). Protein samples were subsequently separated on NuPAGE (Invitrogen) 4–12% Bis-Tris gradient gels and transferred on PVDF membranes (Immobilion, Millipore). For the detection of mouse and human proteins by immunoblot, we used primary antibodies together with the WesternBreeze immunodetection system (Invitrogen). All antibodies used for immunoblotting and their corresponding dilutions are listed in Supplementary Table 8.
Flow cytometry
All antibodies for flow cytometry measurements as well as their respective isotype controls are indicated in Supplementary Table 8. Surface staining antibodies that were PE conjugated were used at a dilution of 1:10 and the ones conjugated with FITC at a dilution of 1:5.
Quantitative RT-PCR
Quantitative real-time PCR carried out with the SYBRGreenER mix from Invitrogen according to standard PCR conditions and an ABI7900HT (Applied Biosystems) real-time PCR system. Primers for quantitative RT-PCR are listed in Supplementary Table 9.
Sequencing for somatic hypermutation of the VH region
Genomic DNA was isolated from the patient samples described in Supplementary Tables 6,7. VH-DJH segments were amplified using the mixture of VH forward primers (VH mix) and JH reverse primers (JH mix) described in Supplementary Table 9. PCR was carried out utilizing the Phusion Hot Start Kit (NEB). PCR fragments were cloned using a Topo TA cloning kit (Invitrogen) and subsequently sequenced unidirectionally in a 96-well format at Eurofins MWG Operon LLC. Comprehensive sequence analysis was used to identify somatic mutations in the VH-DJH junction sequences using IMGT-V Quest (http://www.imgt.org/IMGT_vquest/share/textes/).
Retrovirus production and transduction
Transfections of retroviral constructs and their corresponding empty vector controls (Supplementary Table 10a) were performed using Lipofectamine 2000 (Invitrogen) with Opti-MEM media (Invitrogen). Retroviral supernatants were used to infect murine cells by co-transfecting HEK 293FT cells with the plasmids pHIT60 (gag-pol) and pHIT123 (ecotropic envelope; provided by D.B. Kohn, University of California, Los Angeles, Los Angeles, CA). To infect human cells we replaced pHIT123 with pHIT456 (amphotropic envelope; provided by D.B. Kohn). Cells were cultured in high-glucose DMEM (Invitrogen) with GlutaMAX containing 10% FBS, 100 IU/ml penicillin, 100 μg/ml streptomycin, 25 mM HEPES pH 7.2, 1 mM sodium pyruvate, and 0.1 mM of nonessential amino acids. Serum-free media was replaced after 16 h with growth media containing 10 mM sodium butyrate. After 8 h of incubation, the media was changed back to growth media without sodium butyrate. 24 h later, we harvested the viral supernatants, passed them through a 0.45 μm filter, and centrifuged them twice at 2,000g for 90 min at 32°C on 50 μg/ml RetroNectin-coated non-tissue culture treated 6-well plates. 2–3 × 106 pre–B cells were transduced per well by centrifugation at 600g for 30 min and maintained overnight at 37 °C with 5% CO2 before transferring into culture flasks.
Lentivirus production and transduction
Lentiviral (Supplementary Table 10b) transfections were performed using Lipofectamine 2000 (Invitrogen) with Opti-MEM media (Invitrogen). We produced lentiviral supernatants to infect murine cells by cotransfecting HEK 293FT cells with the plasmids pCD/NL-BH*DDD50 (gag-pol; Addgene 17531) and pMD2.G (VSV-G envelope; Didier Trono, Addgene 12259). Cells were cultivated in high-glucose DMEM (Invitrogen) with GlutaMAX as described above. Culture supernatants were concentrated with Lenti-X™ (Clontech) according to the manufactures recommendation. The concentrate of 3 viruses was incubated for 30 min on 50 μg/ml RetroNectin-coated non-tissue culture treated 6-well plates. 2 × 106 EBV cells were transduced per well by centrifugation at 600g for 30 min and maintained overnight at 37 °C with 5% CO2 before transferring into culture flasks.
Immunohistochemistry
Spleen and liver were obtained from experimental mice were immersed in 20 ml of formalin (VWR) for 24h and transferred to PBS. Paraffin embedding was carried out using standard procedures on a Tissue-TEK VIP processor (Miles Scientific), and 4-μm sections were mounted on Apex superior adhesive slides (Leica Microsystems) and stained on a Ventana BenchMark automated IHC stainer). mCD19 (2E2B6B10) antibody from Abcam was used and the antigen-antibody reaction was detected and visualized using the Ventana iView DAB detection kit, including secondary antibody and other necessary reagents. Immunohistochemistry sections were mounted with mounting medium, antifade reagent (Pro-Long Gold; Invitrogen) was applied, and coverslips were sealed before acquisition of fluorescent images at 25 degrees C on a Zeiss Axiovert 200M inverted confocal microscope with a 40 Plan Neofluor objective using IP Lab 4.0 software (Scanalytics). Photomicrographs were acquired using a Hamamatsu ORCAER HAL100 digital camera (400× original amplification).
Cloning of lentiviral vectors encoding Aicda and Rag1-2
The plasmid pCL6-IRES-dsRedExpress2-wo and pCL6-IRES-iRFP670-wo where generated by replacing the eGFP of pCL6-IRES-eGFP-wo (obtained from H. Hanenberg Heinrich Heine University School of Medicine, Duesseldorf, Germany), with dsRedExpress2 from pDsRed-Express2 (Clontech) or with iRFP670 from pNLS-iRFP670 (V. Verkhusha, Addgene 45466). Rag1 and Rag2 were amplified with Phusion Hot Start II (Thermo Fisher Scientific) from C57BL/6J cDNA and inserted via In-Fusion HD cloning (Clontech) into pCL6-IRES-eGFP-wo and pCL6IEdsRedExpress2-wo vectors respectively. Aicda was amplified with Phusion Hot Start II (Thermo Fisher Scientific) from MIG-AICDA and inserted into pCL6-IRES-iRFP670-wo.
EBV transformation of Cord Blood (CB) CD19+ B Cells
2×106 fresh CD19+ B cells obtained from human cord blood were infected with 1 ml supernatant from B95-8 cells containing EBV (ATCC (VR-1492)) at a 1/4 dilution with RPMI (Invitrogen) containing GlutaMAX containing 20% FBS, 100 IU/ml penicillin, 100 μg/ml streptomycin.
Introduction of Aicda and Rag1-Rag2 in human CD19+ B cells
Lentiviral vectors encoding mouse Aicda (pCL6-Aicda-IRES-iRFP670-wo), Rag1 (pCL6-Rag1-IRES-eGFP-wo), Rag2 (pCL6-Rag2-IRES-dsRedExpress2-wo) and the corresponding empty vector (EV) controls were introduced into EBV-transformed human CD19+ cord blood B cells by the transduction protocol described earlier. Cells were either transduced with EVs, Aicda alone, Rag1-Rag2 combination or both Aicda and Rag1-Rag2. After 4 days, live EBV cord blood B cells, stained with DAPI, that were triple positive for eGFP, iRFP670 and dsRedExpress2 were single-cell sorted into 96-well plates using a 488nm (525/50), 640nm (670/30), 561nm (582/15) and 355nm(450/50) configuration on an BD AriaII Sorter. The DNA of the sorted triple-positive single cells was amplified using REPLI-g Single Cell Kit (Qiagen) according to the manufacturer’s protocol. The resulting DNA was target enriched using SureSelect Human All Exon V5 plus UTR target enrichment (Agilent), paired-end libraries where prepared and Illumina sequenced with 2×100nt paired-end (MOgene). The resulting files were analyzed using GeneSpring (Agilent). The mate status was fixed to exclude reads with an inconsistent mate status. A minimum Indel detection size of 100 bp was applied using the PEMer Indel detection algorithm. The minimum local deviant read coverage used was 11 and the minimum local deviant read fraction was 0.2.
Statistical Analysis
Data are presented as the mean ± s.d. The comparisons for the mean values between two sample groups were made by two-tailed Student’s t-test or Wilcoxon’s rank-sum test using with Graphpad Prism software or R software (R Development Core Team 2009; http://www.r-project.org). For experiments involving transplantation of cells into mice, the minimal number of mice in each group was calculated by using the ‘cpower’ function in R/Hmisc package. The Kaplan-Meier method was used to estimate overall survival and relapse-free survival. Log-rank test was used to compare survival differences between patient groups. The R package ‘survival’ version 2.35-8 was used for the survival analysis. The level of significance was set at P < 0.05.
Supplementary Material
Acknowledgements
We thank J. L. Wiemels, University of California, San Francisco, and C. Gawad, Stanford University, for encouragement and critical discussions. We thank D. B. Kohn, University of California, Los Angeles for giving us the envelope and packaging vectors for lentivirus and retrovirus productions. This work is supported by grants from the NIH/NCI through R01CA137060, R01CA139032, R01CA157644, R01CA169458 and R01CA172558 (to M.M.), the Leukemia and Lymphoma Society (grants 6132-09, 6097-10 and 6221-12 to M.M.), the William Lawrence and Blanche Hughes Foundation, the California Institute for Regenerative Medicine (CIRM; TR2-01816 to MM). M.M. is a scholar of the Leukemia and Lymphoma Society and a Senior Investigator of the Wellcome Trust, UK. MFG and AF are supported by Leukaemia, Lymphoma Research UK and The Wellcome Trust (grant number 105104/Z/14/Z).
Abbreviations
- AICDA
Activation Induced Cytidine Deaminase (gene)
- AID
Activation Induced Cytidine Deaminase (protein)
- RAG
Recombination Activation Gene
- OS
overall survival
- RFS
Relapse Free Survival
- IGH
immunoglobulin heavy chain
- EV
empty vector
- IL
interleukin
- IL-7R
Interleukin 7 receptor
- RSS
recombination signal sequence
- ALL
acute lymphoblastic leukemia
- LPS
lipopolysaccharide
- SHM
somatic hypermutation
- CSR
class switch recombination
- TCR
T cell receptor
- pre-BCR
Pre-B cell receptor
- Foxo
Forkhead Family
- EBV
Epstein-Barr virus
- CB
cord blood
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no conflicting interests.
References
- 1.Wiemels JL, et al. Prenatal origin of acute lymphoblastic leukaemia in children. Lancet. 1999;354:1499–1503. doi: 10.1016/s0140-6736(99)09403-9. [DOI] [PubMed] [Google Scholar]
- 2.Greaves MF, Wiemels J. Origins of chromosome translocations in childhood leukaemia. Nat. Rev. Cancer. 2003;3:639–649. doi: 10.1038/nrc1164. [DOI] [PubMed] [Google Scholar]
- 3.Bateman CM, et al. Acquisition of genome-wide copy number alterations in monozygotic twins with acute lymphoblastic leukemia. Blood. 2010;115:3553–3558. doi: 10.1182/blood-2009-10-251413. [DOI] [PubMed] [Google Scholar]
- 4.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilham C, et al. Day care in infancy and risk of childhood acute lymphoblastic leukaemia: findings from UK case-control study. BMJ. 2005;330:1294. doi: 10.1136/bmj.38428.521042.8F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greaves M. Infection, immune responses and the aetiology of childhood leukaemia. Nat. Rev. Cancer. 2006;6:193–203. doi: 10.1038/nrc1816. [DOI] [PubMed] [Google Scholar]
- 7.Greaves M. In: The Hygiene Hypothesis and Darwinian Medicine. Rook GAW, editor. Birkhäuser Basel; 2009. pp. 239–255. at < http://link.springer.com/chapter/10.1007/978-3-7643-8903-1_13>. [Google Scholar]
- 8.Urayama KY, Buffler PA, Gallagher ER, Ayoob JM, Ma X. A meta-analysis of the association between day-care attendance and childhood acute lymphoblastic leukaemia. Int. J. Epidemiol. 2010;39:718–732. doi: 10.1093/ije/dyp378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Auvinen A, Hakulinen T, Groves F. Haemophilus influenzae type B vaccination and risk of childhood leukaemia in a vaccine trial in Finland. Br. J. Cancer. 2000;83:956–958. doi: 10.1054/bjoc.2000.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Groves FD, Sinha D, Kayhty H, Goedert JJ, Levine PH. Haemophilus influenzae type b serology in childhood leukaemia: a case-control study. Br. J. Cancer. 2001;85:337–340. doi: 10.1054/bjoc.2001.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma X, et al. Vaccination history and risk of childhood leukaemia. Int. J. Epidemiol. 2005;34:1100–1109. doi: 10.1093/ije/dyi113. [DOI] [PubMed] [Google Scholar]
- 12.Ford AM, et al. The TEL-AML1 leukemia fusion gene dysregulates the TGF-beta pathway in early B lineage progenitor cells. J. Clin. Invest. 2009;119:826–836. doi: 10.1172/JCI36428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ford AM, et al. Fetal origins of the TEL-AML1 fusion gene in identical twins with leukemia. Proc. Natl. Acad. Sci. U. S. A. 1998;95:4584–4588. doi: 10.1073/pnas.95.8.4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mori H, et al. Chromosome translocations and covert leukemic clones are generated during normal fetal development. Proc. Natl. Acad. Sci. U. S. A. 2002;99:8242–8247. doi: 10.1073/pnas.112218799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 16.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AICDA), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 17.Kumar S, et al. Flexible ordering of antibody class switch and V(D)J joining during B-cell ontogeny. Genes Dev. 2013;27:2439–2444. doi: 10.1101/gad.227165.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu W, et al. Continued RAG expression in late stages of B cell development and no apparent re-induction after immunization. Nature. 1999;400:682–687. doi: 10.1038/23287. [DOI] [PubMed] [Google Scholar]
- 19.Tsai AG, et al. Human chromosomal translocations at CpG sites and a theoretical basis for their lineage and stage specificity. Cell. 2008;135:1130–1142. doi: 10.1016/j.cell.2008.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hardy RR, Hayakawa K. B cell development pathways. Annu. Rev. Immunol. 2001;19:595–621. doi: 10.1146/annurev.immunol.19.1.595. [DOI] [PubMed] [Google Scholar]
- 21.Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 22.Mullighan CG, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–114. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 23.Papaemmanuil E, et al. RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia. Nat. Genet. 2014;46:116–125. doi: 10.1038/ng.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamane A, et al. Deep-sequencing identification of the genomic targets of the cytidine deaminase AICDA and its cofactor RPA in B lymphocytes. Nat. Immunol. 2011;12:62–69. doi: 10.1038/ni.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiemels JL, et al. Site-specific translocation and evidence of postnatal origin of the t(1;19) E2A-PBX1 fusion in childhood acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. U. S. A. 2002;99:15101–15106. doi: 10.1073/pnas.222481199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qian J, et al. B cell super-enhancers and regulatory clusters recruit AID tumorigenic activity. Cell. 2014;159:1524–1537. doi: 10.1016/j.cell.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alpar D, et al. Clonal origins of ETV6-RUNX1(+) acute lymphoblastic leukemia: studies in monozygotic twins. Leukemia. 2014 doi: 10.1038/leu.2014.322. doi:10.1038/leu.2014.322. [DOI] [PubMed] [Google Scholar]
- 28.Fu C, Turck CW, Kurosaki T, Chan AC. BLNK: a central linker protein in B cell activation. Immunity. 1998;9:93–103. doi: 10.1016/s1074-7613(00)80591-9. [DOI] [PubMed] [Google Scholar]
- 29.Geier JK, Schlissel MS. Pre-BCR signals and the control of Ig gene rearrangements. Semin. Immunol. 2006;18:31–39. doi: 10.1016/j.smim.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 30.Johnson K, et al. Regulation of immunoglobulin light-chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling. Immunity. 2008;28:335–345. doi: 10.1016/j.immuni.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 31.Puel A, Ziegler SF, Buckley RH, Leonard WJ. Defective IL7R expression in T(−)B(+)NK(+) severe combined immunodeficiency. Nat. Genet. 1998;20:394–397. doi: 10.1038/3877. [DOI] [PubMed] [Google Scholar]
- 32.Mandal M, et al. Epigenetic repression of the Igk locus by STAT5-mediated recruitment of the histone methyltransferase Ezh2. Nat. Immunol. 2011;12:1212–1220. doi: 10.1038/ni.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duy C, et al. BCL6 is critical for the development of a diverse primary B cell repertoire. J. Exp. Med. 2010;207:1209–1221. doi: 10.1084/jem.20091299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amin RH, Schlissel MS. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat. Immunol. 2008;9:613–622. doi: 10.1038/ni.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herzog S, et al. SLP-65 regulates immunoglobulin light chain gene recombination through the PI(3)K-PKB-Foxo pathway. Nat. Immunol. 2008;9:623–631. doi: 10.1038/ni.1616. [DOI] [PubMed] [Google Scholar]
- 36.Muramatsu M, et al. Specific expression of activation-induced cytidine deaminase (AICDA), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 37.Crouch EE, et al. Regulation of AICDA expression in the immune response. J. Exp. Med. 2007;204:1145–1156. doi: 10.1084/jem.20061952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z, et al. Contribution of Vh gene replacement to the primary B cell repertoire. Immunity. 2003;19:21–31. doi: 10.1016/s1074-7613(03)00170-5. [DOI] [PubMed] [Google Scholar]
- 39.Gawad C, Koh W, Quake SR. Dissecting the clonal origins of childhood acute lymphoblastic leukemia by single-cell genomics. Proc. Natl. Acad. Sci. U. S. A. 2014;111:17947–17952. doi: 10.1073/pnas.1420822111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dörner T, et al. Analysis of the frequency and pattern of somatic mutations within nonproductively rearranged human variable heavy chain genes. J. Immunol. 1997;158:2779–2789. Baltim. Md 1950. [PubMed] [Google Scholar]
- 41.Kurth J, Hansmann M-L, Rajewsky K, Küppers R. Epstein-Barr virus-infected B cells expanding in germinal centers of infectious mononucleosis patients do not participate in the germinal center reaction. Proc. Natl. Acad. Sci. U.S.A. 2003;100:4730–4735. doi: 10.1073/pnas.2627966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tobollik S, et al. Epstein-Barr virus nuclear antigen 2 inhibits AID expression during EBV-driven B-cell growth. Blood. 2006;108:3859–3864. doi: 10.1182/blood-2006-05-021303. [DOI] [PubMed] [Google Scholar]
- 43.Feldhahn N, et al. Activation-induced cytidine deaminase acts as a mutator in BCR-ABL1-transformed acute lymphoblastic leukemia cells. J. Exp. Med. 2007;204:1157–1166. doi: 10.1084/jem.20062662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gruber TA, Chang MS, Sposto R, Müschen M. Activation-induced cytidine deaminase accelerates clonal evolution in BCR-ABL1-driven B-cell lineage acute lymphoblastic leukemia. Cancer Res. 2010;70:7411–7420. doi: 10.1158/0008-5472.CAN-10-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmutte C, Yang AS, Beart RW, Jones PA. Base excision repair of U:G mismatches at a mutational hotspot in the p53 gene is more efficient than base excision repair of T:G mismatches in extracts of human colon tumors. Cancer Res. 1995;55:3742–3746. [PubMed] [Google Scholar]
- 46.Jumaa H, et al. Abnormal development and function of B lymphocytes in mice deficient for the signaling adaptor protein SLP-65. Immunity. 1999;11:547–554. doi: 10.1016/s1074-7613(00)80130-2. [DOI] [PubMed] [Google Scholar]
- 47.Lesche R, et al. Cre/loxP-mediated inactivation of the murine Pten tumor suppressor gene. Genesis. 2002;32:148–149. doi: 10.1002/gene.10036. [DOI] [PubMed] [Google Scholar]
- 48.Liu X, et al. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997;11:179–186. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- 49.Mombaerts P, et al. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 50.Zhang X-Y, La Russa VF, Reiser J. Transduction of bone-marrow-derived mesenchymal stem cells by using lentivirus vectors pseudotyped with modified RD114 envelope glycoproteins. J. Virol. 2004;78:1219–1229. doi: 10.1128/JVI.78.3.1219-1229.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosenfeld C, et al. Phenotypic characterisation of a unique non-T, non-B acute lymphoblastic leukaemia cell line. Nature. 1977;267:841–843. doi: 10.1038/267841a0. [DOI] [PubMed] [Google Scholar]
- 52.Height SE, et al. Analysis of clonal rearrangements of the Ig heavy chain locus in acute leukemia. Blood. 1996;87:5242–5250. [PubMed] [Google Scholar]
- 53.Kumar MS, et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700–2704. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yusuf I, Zhu X, Kharas MG, Chen J, Fruman DA. Optimal B-cell proliferation requires phosphoinositide 3-kinase-dependent inactivation of FOXO transcription factors. Blood. 2004;104:784–787. doi: 10.1182/blood-2003-09-3071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.