Abstract
Accumulated evidence indicates that PtdIns5P, one of the seven phosphoinositides, found now to be constitutively present in yeast, plants and metazoa, serves as a signaling molecule to modulate pleiotropic cellular functions in both the nucleus and the cytoplasm. The enzymatic routes in biogenesis of basal PtdIns5P have remained incompletely understood. The role for candidate kinase PIKfyve that is principally involved in PtdIns(3,5)P2 production, has been questioned. In this review article we scrutinize the past obstacles that prevented the definitive implication of PIKfyve in PtdIns5P biosynthesis from PtdIns and focus on the recent pharmacological and genetic advancements that now make this conclusion well supported. We further summarize our current knowledge of the diverse stimuli modulating PtdIns5P levels, binding partners and regulated cellular process, with particular reference to the available mechanistic insights for the relevant signaling pathways.
Keywords: PtdIns5P, PIKfyve, MTM/MTMRs, PI4-phosphatases, PIP4-kinases, Insulin-regulated glucose transport, Actin cytoskeleton remodeling, Membrane trafficking, Stress responses, Akt
Introduction
The phosphorylated products of phosphatidylinositol (PtdIns),1 collectively called phosphoinositides (PIs), are eukaryotic cell membrane- anchored phospholipids that serve as versatile signaling molecules regulating diverse and essential cellular processes. PIs are produced by reversible phosphorylation that engages three out of the five hydroxyls in the inositol headgroup, i.e., D3, D4 and/or D5, yielding seven species: PtdIns3P, PtdIns4P, PtdIns5P, PtdIns(3,4)P2, PtdIns(3,5)P2, PtdIns(4,5)P2 and PtdIns(3,4,5)P3 [1]. PIs are heterogeneously localized within cells, where in conjunction with a membrane protein, they serve as co-receptors for specific membrane targeting of cytosolic effector proteins [2–4]. PI metabolism is tightly controlled in time and space by the action of various kinases, phosphatases and phospholipases, each exhibiting specific intracellular localization [5–7].
Levels of the seven PIs vary significantly among each other and among cell types. As a rule, PtdIns(4,5)P2 and PtdIns4P are the high-abundance PIs, whereas the higher D3-phosphorylated species, i.e., PtdIns(3,4)P2, PtdIns(3,5)P2 and PtdIns(3,4,5)P3 are typically at the lowest or even undetectable levels in non-stimulated cells [1]. Although PtdIns5P is certainly not the lowest-abundance PI, its natural occurrence was the last to be identified [8]. This is related to the fact that the PtdIns5P elution characteristics by HPLC (high-pressure liquid chromatography), the principal method for separation and quantitation of individual PIs, are similar to those of PtdIns4P. Moreover, with levels of PtdIns4P being substantially greater than those of PtdIns5P (> one order of magnitude in mammalian cells), even when detected on the chromatograms, PtdIns5P represents a small peak or a shoulder within the descending arm of the PtdIns4P peak [8,9]. Continuing efforts for optimization of the PtdIns5P separation by HPLC [8,10] as well as the development of an alternative indirect enzymatic approach [11] have largely circumvented the detection limitations. These advancements have led to the conclusion that PtdIns5P is constitutively present in several species separated by a great evolutionary distance, including unicellular yeast [12], plants, such as Chlamidomonas and Arabidopsis [13,14], Drosophila [15] and mammals [16]. More importantly, PtdIns5P has been found to be acutely regulated upon physiological and pathophysiological challenges, indicating that in addition to a housekeeping function it serves as a signaling molecule in its own right, modulating cellular responses to various stressors, hormones and growth factors.
Newly developed tools for PIKfyve research, such as knockout (KO) mouse models and chemical inhibitors, have shed light as to how the bulk of PtdIns5P is being synthesized [17–19]. PIKfyve is the primary kinase responsible for PtdIns(3,5)P2 production [20]. Its involvement in direct PtdIns5P biosynthesis from PtdIns has been proposed early on [9,21,22], but such a role remained a matter of debate [16,20]. However, several recent findings suggest direct PIKfyve-catalyzed PtdIns5P synthesis from PtdIns. Thus, upon Pikfyve gene disruption, PtdIns5P is decreased similarly [18], or even preferentially to PtdIns(3,5)P2 [19] despite vastly greater amounts of the former vs. the latter. Concordantly, upon PIKfyve chemical inhibition by the YM203616 compound, intracellular PtdIns5P is more severely reduced than PtdIns(3,5)P2 [17]. This review article critically evaluates the existing literature on PtdIns5P biogenesis and intracellular functions, focusing on new developments. For details on the metabolism and specific functions of the other PIs, the reader is referred to recent reviews [5,16,23– 26].
Enzymatic routes in intracellular PtdIns5P production
The enzymology of PtdIns5P in mammals is rather complex (Fig. 1) and perhaps the reason for the difficulty in reaching consensus for the principal pathway by which basal PtdIns5P is made. Intriguingly, the most obvious route for its intracellular production, i.e., direct phosphorylation of the abundant PtdIns, a pathway documented in biogenesis of the other monophosphorylated PIs, has not been embraced for years, despite the significant amount of accumulated evidence implicating PIKfyve [20]. New data from PIKfyve KO mouse models, however, reinforce this role and together with previous data, indicate that PIKfyve-catalyzed synthesis from PtdIns is the major route for constitutive PtdIns5P [17– 19,22]. Turnover of PtdIns(3,5)P2 and PtdIns(4,5)P2 by myotubularin/myotubularin-related D3-phosphatases (MTM/MTMRs) [27] or PtdIns(4,5)P2 4-phosphatases [28], respectively, may also serve as plausible pathways in response to certain stimuli. Below, I critically analyze the available data for the contribution for each of these enzymatic routes in PtdIns5P biogenesis.
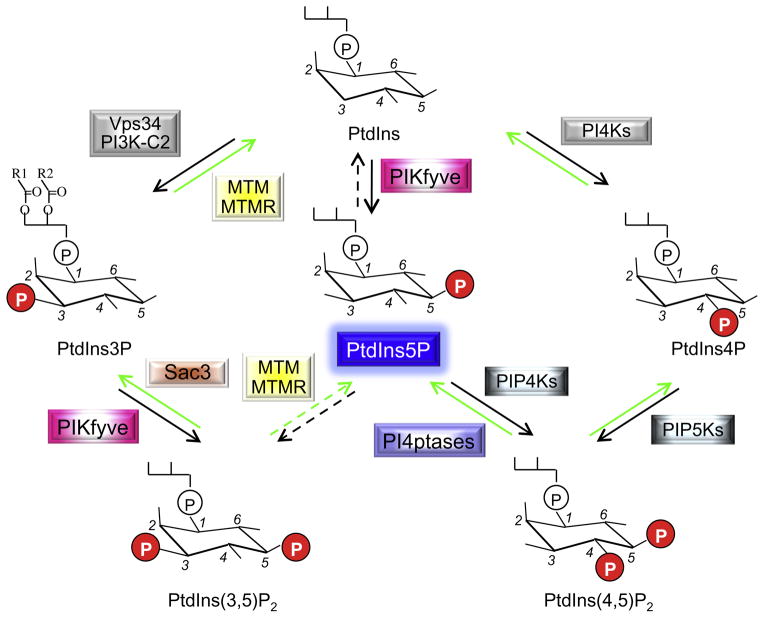
Fig. 1.
Enzymatic reactions by which Ptdns5P is produced and consumed. Indicated are the relevant enzymes and PIs participating in the reactions discussed in this review. The PI molecule includes inositol 1-phosphate (a hydrophilic portion exposed to the cytosol), bound via the phosphate group to 1-R1, 2-R2 diacylglycerol (a hydrophobic membrane-anchored portion). R1/R2, fatty acid; P, phosphate; PtdIns, phosphatidylinositol; PI, phosphoinositide; K, kinase; PIKfyve, phosphoinositide 5-kinase, five-domain containing; MTM/MTMRs, myotubularin/myotubularin-related D3-phosphatases; Sac3, Sac domain-containing D5-phosphatase.
Direct synthesis from PtdIns by PIKfyve
PIKfyve-produces PtdIns5P in vitro from PtdIns apart from MTM/MTMRs
PIKfyve was first implicated in PtdIns5P biosynthesis based on in vitro data from our laboratory, which revealed that native or recombinant PIKfyve preparations purified from various mammalian or insect cells synthesize PtdIns5P from PtdIns and PtdIns(3,5)P2, from PtdIns3P in the presence of [γ-32P]ATP [9,29–31]. Substrate side chains play a key role in the efficiency of the in vitro kinase assay, with naturally occurring acyl chains being greatly preferred over the synthetic di-C16 [9]. Therefore, in studies conducted in our laboratory we typically employ natural PtdIns substrate preparations from soybean or liver (Avanti Polar Lipids), as superior substrates for assaying PIKfyve lipid kinase in vitro [9]. However, because the native PtdIns substrates are contaminated by native PtdIns3P, PtdIns(3,5)P2 synthesis along with PtdIns5P is also observed. Interestingly, under these conditions PtdIns(3,5)P2 production proceeds with a similar or even greater efficiency compared to the usage of synthetic PtdIns3P [9] (see Fig. 2A). Similar in vitro assays have also been conducted by an independent laboratory that used our mouse PIKfyve clone expressed by yeast plasmid [32]. Agreeably, our laboratory and that of Dr. Bob Michelle did not observe PtdIns5P when synthetic PtdIns3P substrate preparations were used in the in vitro assays, despite the great amounts of produced 32P-PtdIns(3,5)P2 [9,29,32] (illustrated in Fig. 2A). Lack of coproduced PtdIns5P along with PtdIns(3,5)P2 has been observed using PtdIns3P substrates with either di-C16 or di-C18 side chains-from multiple vendors (Matreya; Echelon or Michelle’s lab) in in vitro assays conducted by different laboratories [9,29,32]. Thus, the experimental data in these studies had firmly demonstrated that PIKfyve-catalyzed PtdIns5P synthesis in vitro is strictly dependent on the presence of PtdIns substrates.
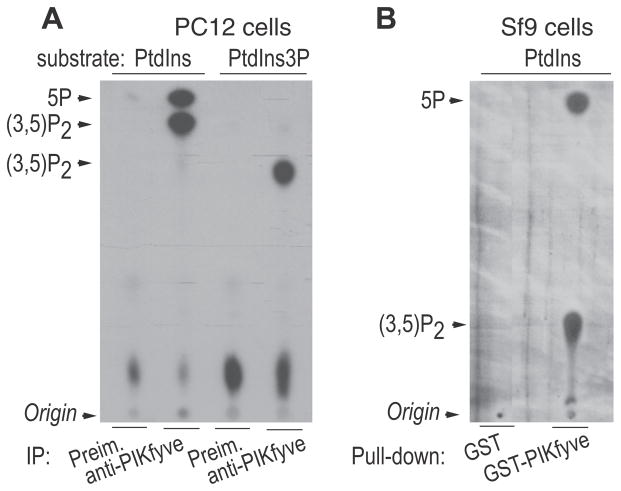
Fig. 2.
In vitro activity of endogenous and recombinant PIKfyve. (A) PIKfyve immune and preimmune sera (Preim.) immunoprecipitates from PC12 cell lysates, adsorbed onto Protein A Sepharose beads were subjected to in vitro lipid kinase assay (15 min) in the presence of 10 μCi of [γ-32P] ATP (50 μM) and 100 mM of indicated substrates, PtdIns (natural form from soybean, Avanti Polar Lipids) and di- C16-PtdIns3P (Echelon Biosciences). Lipids were resolved by TLC using an acidic solvent system [65:35(v/v)1-propanol-2 Maceticacid]. Indicated are PtdIns(3,5)P2 and PtdIns5P products confirmed by HPLC analyses of the radioactive spots. PtdIns(3,5)P2, produced from native PtdIns and synthetic di-C16-PtdIns3P substrates has different mobility due to different side chains in the respective substrates. Only PtdIns(3,5)P2 but not PtdIns5P is generated with the PtdIns3P substrate, ruling out myotubularin-dependent PtdIns5P production through hydrolysis of PtdIns(3,5)P2 during the in vitro reaction, as suggested in Ref. [19]. Modified from Ref. [29]. (B) Lysates from Sf9 cells, infected with baculovirus encoding GST-PIKfyve or control virus were incubated with GSH-agarose beads (1 h). Washed beads were subjected to in vitro kinase assay in the presence of the PtdIns substrate (natural form, Avanti Polar Lipids) under the conditions described in A, plus phosphatase inhibitors Lipids were resolved by TLC using a basic organic solvent system [90:90:20:7 (v/v) chloroform, methanol, water, 30% ammonia]. The radioactive spots were identified by HPLC. PtdIns(3,5)P2 runs faster in A compared to B due to different TLC solvent systems. Similarly to the endogenous, recombinantly produced PIKfyve makes directly PtdIns5P from PtdIns. Modified from Ref. [30].
Given these compelling arguments, a recent experimentally unverified claim, refuting the in vitro PIKfyve 5-kinase activity towards PtdIns substrates [19] is rather peculiar. Because it would serve as a main argument against PIKfyve-catalyzed PtdIns5P production from PtdIns in cellular contexts [19], this claim is important to be carefully examined. In this study [19], previous data by our group for decreases of both PtdIns5P and PtdIns(3,5)P2 in embryonic fibroblasts derived from the heterozygous PIKfyveWT/KO mouse model [18] were confirmed and extended to fibroblasts of PIKfyve gene-trap mice. However, in contrast to our conclusion for impaired direct PtdIns5P synthesis by PIKfyve [18], Weisman’s team hypothesized the role of MTM/MTMRs 3-phosphatases in the observed PtdIns5P decrease along with PtdIns(3,5)P2 [19]. Hence, Zolov et al., interpreted our data from the PIKfyve in vitro assays (Fig. 2) to be a result of MTM/MTMRs, presumably associated/coimmunoprecipitated with endogenous PIKfyve, and catalyzing 32P-PtdIns(3,5)P2 hydrolysis to 32P-PtdIns5P during the in vitro kinase reaction [19]. Unfortunately, however, this interpretation cannot be supported by the published in vitro data summarized above [9,29,30,32]. Furthermore, the interpretation in Zolov et al., [19] also contradicts other reports demonstrating direct in vitro generation of PtdIns5P from PtdIns substrates by a GST-purified PIKfyve recombinant protein, produced by a baculoviral system [30] (Fig. 2B). Under these conditions, such hypothetically associated MTM/MTMRs making 32P-PtdIns5P from 32P-PtdIns(3,5)P2- in vitro would appear to be exceedingly unlikely due to the vastly greater amounts of the recombinant protein relative to the endogenous myotubularin-like insect homologues. Furthermore, the single yeast myotubularin-like phosphatase Ymr1p does not use PtdIns(3,5)P2 substrate [33], an important fact, further challenging the additional data in the yeast system used to support MTM/MTMR’s role [19]. Finally, the view that not PIKfyve but MTM/MTMRs were to be responsible for the in vitro generated PtdIns5P in the PIKfyve kinase assays [19] also faces conspicuous discrepancies with the findings for differential in vitro generation of PtdIns5P vs. PtdIns(3,5)P2 by two PIKfyve point mutants, differing by a single amino acid within the predicted substrate binding pocket. The in vitro kinase potential of these mutants has been compared with identically processed immunoprecipitates from transfected COS cells [34]. The observations that the K2000E mutation renders in vitro synthesis of PtdIns5P less efficient compared to PtdIns(3,5)P2, whereas the K1999E mutation exhibits the opposite effect, is consistent with residue K2000 being selectively required in PtdIns5P vs. PtdIns(3,5)P2 synthesis and vice versa, for the residue K1999 [34]. In this regard, the claims and conclusions in [19] require re-evaluation. Regardless, the above analysis strongly indicates that PIKfyve catalyzes direct conversion of PtdIns to PtdIns5P in vitro preferring native over synthetic substrate preparations and, together with the in vivo data discussed below, imply that PIKfyve produces directly PtdIns5P from PtdIns in a cellular context.
PIKfyve generates directly PtdIns5P in vivo
A great deal of experimental evidence suggests that PIKfyve is the principal enzyme responsible for the constitutive levels of PtdIns5P in intact mammalian cells. The first clue for such a role came in 2002 from findings that several cell types, both mammalian and insect, have higher steady-state levels of PtdIns5P subsequent to infection with PIKfyve adeno- and baculovirus, respectively, as demonstrated by HPLC analyses in radiolabeled cells [22].Corroborating this conclusion, we found by the PtdIns5P mass assay 2-fold higher and 2-fold lower PtdIns5P levels in HEK293 cell lines stably expressing active PIKfyveWT or PIKfyveK1831E, a dominant-negative kinase-deficient form carrying a mutation in the predicted ATP-binding site, respectively [22]. Concordantly, hypo-osmotic shock in 3T3L1 fibroblasts down-regulated both PtdIns5P and PtdIns(3,5)P2 consistent with their concomitant production by the PIKfyve-dependent pathway [22].
More recent data based on PIKfyve pharmacological inhibition or protein knockdown (KD) have generated a new wave of experimental support for the claim that PIKfyve is a major source for biogenesis of PtdIns5P in mammals. For instance, PIKfyve protein depletion or activity inhibition by curcumin or the YM201636 compound [35–37] has resulted in a decline of the PtdIns5P pool in several mammalian cell types as determined by PtdIns5P mass assays [38] and HPLC analyses [17,19]. The notion for PIKfyve-catalyzed PtdIns5P biosynthesis in mammalian cells is further reinforced by recent data from HPLC-based quantitation of steadystate levels of PIs in fibroblasts derived from two independently developed genetically modified mouse models, in which the PIKfyve protein is ablated to a variable degree. Thus, embryonic or skin fibroblasts derived from heterozygous PIKfyveWT/KO [18] or PIKfyve gene-trap (PIKfyveβ-geo/β-geo) mice [19], respectively, exhibit reduction in both PtdIns5P and PtdIns(3,5)P2. Likewise, fibroblasts from the gene-trap mouse model of ArPIKfyve (gene symbol VAC14), the protein that associates with and up-regulates PIKfyve [39], have a decline in both PtdIns5P and PtdIns(3,5)P2 [40].
Whereas this ample amount of experimental evidence is sufficient to convince the biggest skeptics that PIKfyve is the master enzyme responsible for PtdIns5P production in vivo, as suggested near a decade ago [22], these data still do not provide unequivocal support for a direct biosynthesis from PtdIns. Moreover, the documented ability of the MTM/MTMR family members to dephosphorylate PtdIns(3,5)P2 to PtdIns5P along with PtdIns3P to PtdIns in vitro, suggests the possibility for such a reaction to take place in vivo (see Section 2.2 for more detail). However, although MTM/MTMRs might fulfill such a role under certain in vivo conditions, several lines of experimental evidence and quantitative considerations based on simultaneous measurements of PtdIns5P and PtdIns(3,5)P2 indicate that the majority of constitutive PtdIns5P is made in cells directly from PtdIns by PIKfyve, rather than through PtdIns(3,5)P2 synthesis and dephosphorylation by PIKfyve and myotubularins, respectively. Thus, that PtdIns5P is unlikely to be derived from PtdIns(3,5)P2 has been apparent by the observations that steady-state levels of PtdIns5P are many fold higher (from 7 to 50-fold) than those of PtdIns(3,5)P2 [18,40,41]. More recently, it was revealed that low concentrations (160 nM) of the PIKfyve inhibitor YM201636 preferentially prevent formation of PtdIns5P vs. PtdIns(3,5)P2 in several mammalian cells types, such as 3T3L1 adipocytes, HEK293, CHO-T and HEK293, with nearly 2-fold difference, consistent with direct PIKfyve-dependent synthesis of PtdIns5P [17]. Importantly, the decrease of the PIKfyve-inhibitable PtdIns5P under these conditions was found to be quite substantial in all of the tested cell types, by 71–62% [17], indicating a significant portion of the basal PtdIns5P being made by direct biosynthesis from PtdIns. These data are corroborated by findings for a similar preferential decline (by ~40%) of steady-state levels of PtdIns5P vs. PtdIns(3,5)P2 (2.6- and 1.8-fold, respectively, vs. control) in fibroblasts from the gene-trap PIKfyveβ-geo/β-geo mice exhibiting about 10% residual PIKfyve protein [19]. Concordantly, preferential PtdIns5P reduction was also seen in the heterozygous PIKfyve+/β-geo fibroblasts, with steady-state levels of PtdIns5P declining by 30%, whereas those of PtdIns(3,5)P2 remained unaltered [19]. Likewise, data in the ArPIKfyve gene trap mouse model (Vac14β-geo/β-geo) also indicated that, compared to wild-type fibroblasts, PtdIns5P reduction was roughly 50% greater than that of PtdIns(3,5)P2 (PtdIns5P, 0.3% vs. 0.1% in controls;PtdIns(3,5)P2, 0.04% vs. 0.02% in controls [40]. Similarly, in mouse embryonic fibroblasts (MEFs) from heterozygous PIKfyveWT/KO, in which the PIKfyve protein is decreased by ~50%, both PdIns5P and PtdIns(3,5)P2 diminished [18]. In this case, however, the decline of the two lipid products is similar (by 35–39%), consistent with the idea that greater amounts of the remaining PIKfyve might compensate for uneven turnover. Finally, the notion for direct PtdIns5P biogenesis is also supported by the findings that KD of PIKfyve in quiescent or stimulated human BJ fibroblasts reduces steady-state levels of PtdIns5P more effectively than KD of MTMR3 [42]. Notably, in this study, PtdIns(3,5)P2 was not detected due to limitations associated with the used double column in the HPLC separation, which prevents firm conclusions about the probability of PtdIns(3,5)P2 – PtdIns5P conversion. Together, these data and considerations very strongly indicate that the majority of the constitutive PtdIns5P pool is made by direct PIKfyve-catalyzed biosynthesis from PtdIns. Myotubularins may contribute to PtdIns5P production under stimulation condition, as detailed further (Section 2.2).
How PIKfyve performs dual biosynthesis of PtdIns5P and PtdIns(3,5)P2
As discussed above, PIKfyve is also the principal enzyme for intracellular PtdIns(3,5)P2 production [16]. In the absence of structural information for the PIKfyve catalytic domain, the molecular basis of the two distinct PIKfyve lipid kinase activities and their regulation could only be a matter of speculation. One potential mechanism may operate through substrate recognition by separate amino acid residues of the PIKfyve catalytic domain, selectively coordinating the head-groups in PtdIns or PtdIns3P prior to addition of the D5 phosphate (Fig. 3). Findings that Lys2000 within the predicted PIKfyve substrate binding pocket supports preferentially PtdIns5P over PtdIns(3,5)P2 synthesis, whereas Lys1999’s effect is in an opposite manner, is consistent with such prediction [34]. Accordingly, Lys1999 but not Lys2000 was found to interact, although weakly, with PtdIns3P-containing liposomes in an in vitro binding assay utilizing GST-immobilized PIKfyve peptide fragments [34]. Furthermore, it is also plausible that PtdIns5P and PtdIns(3,5)P2 are produced by distinct subfractions of PIKfyve. It is now clear that the intracellular PtdIns(3,5)P2 synthesis in both yeast and mammalian cells is executed by an evolutionarily-conserved PIKfyve complex that incorporates the ArPIKfyve-Sac3 subcomplex [43,44], the triad typically being referred to as the PAS complex (for the first letters of the three proteins) [45,46]. Thus, it is possible that PtdIns5P is synthesized by a PIKfyve subpopulation, independent of the PAS complex. Such a notion is corroborated by quantitative analyses of the coimmunoprecipitation data from different mammalian cell types, revealing a quite substantial proportion of PIKfyve to be unrelated to the PAS complex [43]. Existence of two distinct PIKfyve pools and/or catalytic residues may explain the data for the differential inhibition of PtdIns5P vs. PtdIns(3,5)P2 production by the YM201636 compound [17], discussed above. Future structural studies combined with molecular modeling will certainly enlighten the basis for the dual enzymatic activities of PIKfyve towards PtdIns3P and PtdIns.
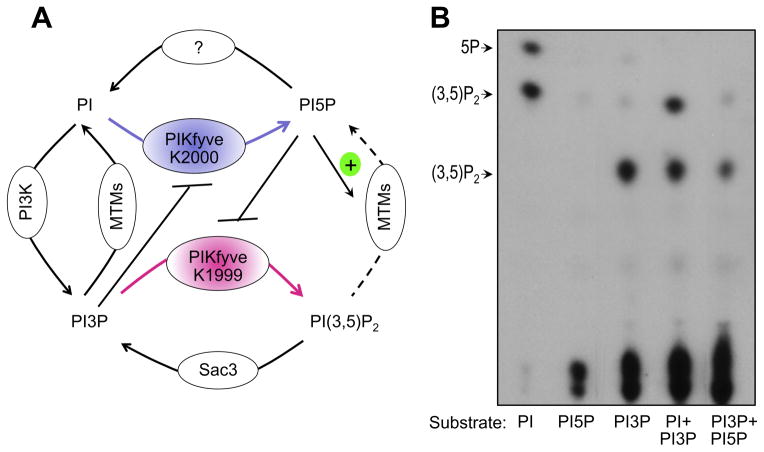
Fig. 3.
Proposed model for PIKfyve dual activity and regulation in vivo. (A) The differential role of K1999 and K2000 residues from the predicted PIKfyve substrate binding pocket in production of PtdIns(3,5P)2 and PtdIns5P, respectively, has been established both in vivo and in vitro [34]. Feed-forward inhibition of PIKfyve-catalyzed PtdIns5P production from PtdIns in the presence of high levels of PtdIns3P was observed previously in vitro [9] and is shown in B. Feedback inhibition of PIKfyve-catalyzed PtdIns(3,5)P2 synthesis from PtdIns3P by high levels of PtdIns5P is suggested based on reduced in vitro synthesis of PtdIns(3,5)P2 in the presence of PtdIns5P. (Sbrissa D & Shisheva A, unpublished observation that is now presented in B). Activation of MTM, MTMR3 and MTM6 by PtdIns5P was observed in vitro [55]. See text for more detail. (B) PIKfyve lipid kinase activity in vitro was determined with the indicated substrates alone or in various combinations, added at a molar ration 1:1, under conditions, specified in Fig. 2A. Presented is the TLC autoradiogram of generated products whose identity was confirmed by HPLC analyses.
Basal PtdIns5P is unlikely to be made by MTM/MTMRs-catalyzed hydrolysis of PtdIns(3,5)P2
Given the considerable amount of evidence indicated above, a legitimate question is why PIKfyve has not been definitively implicated in direct PtdIns5P synthesis [47]. A major obstacle for accepting direct biosynthesis of PtdIns5P by PIKfyve has been an early complementation analysis that found HPLC-undetectable PtdIns5P in a Δfab1 yeast strain (Saccharomyces cerevisiae) ectopically expressing mouse PIKfyve yet the PtdIns(3,5)P2 was elevated [32]. This observation, together with other studies showing no constitutive presence of PtdIns5P in yeast [33,48,49] led to the assumption that like yeast fab1, PIKfyve does not produce PtdIns5P in mammalian cells. However, this assumption is most likely incorrect because of more recent key observations. Thus, the low expression levels of PIKfyve achieved in this initial study were also unable to rescue the hyperosmotic elevation of PtdIns(3,5)P2 in the Δfab1 yeast strain [32]. Subsequent research under more effective PIKfyve expression in the Δfab1 yeast strain has revised this conclusion but information about PtdIns5P under these conditions has not been disclosed [50]. Regardless, a recent study that also revisited this issue, has documented significantly greater PtdIns5P production in wild-type S. cerevisiae upon heterologous expression of human PIKfyve [19]. This study has further documented ~1.7-fold reduction of PtdIns5P in a Δfab1 yeast strain vs. wild type. Together, the available data suggest that PtdIns5P is not only detectable in yeast but also is subject to PIKfyve-dependent regulation.
Whereas these recent data seem to resolve the doubts about mammalian PIKfyve making PtdIns5P in yeast, other results in mammalian cells make the origin of constitutive PtdIns5P still ambiguous. This is related to the potential role of the MTM/MTMRs, a family of D3-phosphatases that in mammals is comprised of 8 enzymatically active forms [51]. Their contribution to PtdIns5P production was first predicted by findings that heterologously expressed mammalian MTMR3 in S. cerevisiae hydrolyzes both PtdIns3P and PtdIns(3,5)P2 thereby increasing PtdIns5P [52]. The following studies revealed that many active MTM/MTMR family members, including MTM1 and MTMR1, 2, 4, 6 or 14, have similar dual specificity, hydrolyzing in vitro both PtdIns3P and PtdIns(3,5)P2 substrates to PtdIns and PtdIns5P, respectively [27,53–58]. These findings have allowed the assumption that MTM/MTMR-catalyzed hydrolysis of PtdIns(3,5)P2 to PtdIns5P may take place in vivo. Unfortunately, this assumption, particularly related to constitutive PtdIns5P production, is highly conjectural.
In fact, several lines of observations from different species imply that in vivo, MTM/MTMRs are unlikely to hydrolyze PtdIns(3,5)P2-to-PtdIns5P at normal conditions. Thus, in mammals, the major enzymatic activity and functions of several MTM/MTMR members, such as MTM1, MTMR2, MTMR3, MTMR6 and MTMR14, are delivered through hydrolysis of PtdIns3P rather than PtdIns(3,5)P2 [59–67]. Reportedly Drosophila mtm acts in a similar manner as does the Caenorhabditis elegance Mtm-1 in supporting phagosome maturation [68,69]. Likewise, in yeast (S.cerevisiae), the single myotubularin phosphatase Ymr1 is also implicated in regulating PtdIns3P, but not PtdIns(3,5)P2 levels [33,59]. For example, yeast strain overexpressing Ymr1 exhibits lower, whereas the Δymr1 strain, higher PtdIns3P, with no changes in PtdIns(3,5)P2 under both conditions [33]. Further limitations related to PtdIns(3,5)P2 turnover to PtdIns5P are also provided by quantitative considerations discussed above. Thus, the fact that basal levels of PtdIns(3,5)P2 are an order of magnitude lower than those of PtdIns5P makes the possibility for the latter being produced from the former highly improbable. Concordantly, knockdown of MTMR3 in quiescent BJ cells does not decrease PtdIns5P, which should be expected if MTMR3 is to hydrolyze PtdIns(3,5)P2 [42]. Furthermore, putative PtdIns(3,5)P2-to-PtdIns5P hydrolysis at basal conditions contradicts data documenting robust PtdIns(3,5)P2 conversion to PtdIns3P in pulse-labeled mouse fibroblasts as assessed by HPLC separation of extracted lipids [70]. Notably, turnover of PtdIns(3,5)P2 to PtdIns3P is well supported by findings in both yeast and mammalian cells, implicating the Sac3 D5-phosphatase that, when complexed with PIKfyve, promotes both synthesis and turnover of PtdIns(3,5)P2 [16,44]. Therefore, fibroblasts from the Sac3 KO mouse model have decreased PtdIns(3,5P)2. Conspicuously, PtdIns(3,5)P2 levels are not restored back to normal upon MTMR2 ablation in the Sac3 KO, as observed in fibroblast from mice with double KO of Sac3 and MTMR2 [12], questioning also MTMR2’s role in hydrolyzing PtdIns(3,5)P2 and contributing to basal PtdIns5P in a measurable outcome.
Nonetheless, a recent study indicates that cultured cells of the MTMR2 KO mice display increases in PtdIns(3,5)P2 and decreases in PtdIns5P levels, as measured by HPLC and the mass assay, respectively [12]. Despite the level of variability associated with the usage of different techniques for PtdIns5P/PtdIns(3,5)P2 measurements and in different cell types [12], it should be taken into consideration that the inevitable elevation of PtdIns3P in MTMR2 KO or KD models [63] could increase PIKfyve-catalyzed PtdIns(3,5)P2 production through mass action. Furthermore, elevated PtdIns3P may selectively suppress PtdIns5P production, as inferred by in vitro data for a selective inhibition of PIKfyve-catalyzed synthesis of PtdIns5P from PtdIns, but not of PtdIns(3,5)P2 from PtdIns3P, if the PtdIns and PtdIns3P substrates are present together at a 1:1 M ratio [9]. Support for such a regulatory mechanism (illustrated in Fig. 3A) is aslo provided by observations for a distinct role of residues K1999 and K2000 from the predicted PIKfyve substrate binding pocket in PtdIns5P vs. PtdIns(3,5)P2 synthesis [71]. Whereas this regulatory model requires further investigation, the above analysis of the available data makes the in vivo PtdIns5P production by MTM/MTMRs-catalyzed PtdIns(3,5)P2 hydrolyses, at least under basal conditions, highly improbable. Novel reagents, such as radiolabeled D3-deoxy myo-inositol that is phosphorylation-deficient at 3-OH position, will certainly be of great importance to ultimately define the in vivo contribution of PIKfyve vs. MTM/MTMRs to PtdIns5P production under different conditions.
Hydrolysis of PtdIns(4,5)P2 to PtdIns5P by PI 4-phosphatases
PtdIns5P could also be produced by dephosphorylation of the abundant PtdIns(4,5)P2 (Fig. 1). This pathway has been discovered based on findings that several bacterial pathogens, including Salmonella and Shigella, release SigD and IpgD virulence factors, respectively, that turned out to be homologous to mammalian inositol polyphosphate 4-phosphatases [72–74]. Infection of various mammalian cells with these virulence factors results in profound increases of PtdIns5P due to powerful hydrolysis of PtdIns(4,5)P2 [72,74]. Homology screening of the human genome has led to the identification of two ubiquitously expressed PtdIns(4,5)P2-4-phosphatases, referred to as type I and type II [28]. However, whereas they both produce PtdIns5P from PtdIns(4,5)P2 in vitro, only type I that translocates to the nucleus upon DNA damage is capable of increasing the nuclear pool of Ptdins5P by ~50%, as shown in transfected HeLa cells [28,75]. Given the late endosomal/lysosomal localization of both enzymes, it is likely for this pathway to also be prominent outside the nucleus in certain cell types, in which the endogenous PIKfyve protein levels and/or activity is low. For example, the human osteosarcoma U2OS cell line, in which neither PIKfyve nor myotubularins have been possible to be implicated in constitutive PtdIns5P production [76], is a possible candidate for regulation by PI 4-phosphatases but more work will be required to decipher such a mechanism. At any rate, the endogenous levels of PIKfyve protein in the U2OS cells have not been determined nor were the PtdIns(3,5)P2 basal or stimulated levels detected in the HPLC profiles of the in vivo labeled PIs [76], which prevents firm conclusions.
Enzymes responsible for PtdIns5P removal
Whereas PtdIns5P could be removed by several mechanisms, the obvious enzymatic pathway for its breakdown, i.e., dephosphorylation by a D5-specific phosphatase, remains elusive. In fact, previously implicated phosphatase PTPMT1 in this process is now found to have a different physiological substrate [77]. At any rate, PtdIns5P and PtdIns(3,5)P2 are likely to be hydrolyzed by different phosphatases, and whereas for PtdIns5P, the enzyme(s) is yet to be determined, the Sac3 D5-phosphatase turns over the PtdIns(3,5)P2 pool in mammals [43]. The phosphatases are expected to have different turnover rates towards their respective substrates, which should explains recent observations for a slight difference in the disappearance of PtdIn5P vs. PtdIns(3,5)P2 in fibroblasts, subsequent to chemical inhibition of PIKfyve by YM201636 [19]. A PtdIns5P-removal pathway with a potential physiological significance involves hydrolysis by phospholipase C-δ1 that, under high Ca2+, could acquire preferential specificity for PtdIns5P over PtdIns(4,5)P2 hydrolysis [78]. However, the robustness of this pathway has yet to be explored.
A major role in the PtdIns5P removal pathway is attributed to PIP4Ks that catalyze D4 phosphorylation in PtdIns5P to PtdIns(4,5)P2 [8,79]. Two active isoforms α and β are known, of which the former is ~2000-fold more active that the latter [80]. Whereas the α isoform is cytoplasmic, PIP4Kβ is predominantly nuclear [81,82]. A third, catalytically inactive form, PIP4Kγ, localized on endoplasmic reticulum, is also critical for PtdIns5P levels [10,83] likely acting in intracellular targeting of the active PIP4Kα and/or PIP4Kβ [84]. In addition to alteration of subcellular locales, PIP4Ks may regulate PtdIns5P by transient phosphorylation-dependent inhibition of their enzymatic activities. For example, increased nuclear PtdIns5P under oxidative stress is achieved by p38 MAPK-dependent phosphorylation and inhibition of PIP4Kβ [85]. Similarly, PIP4 Kα might also be inhibited through PKD-catalyzed phosphorylation [86].
PtdIns5P-binding domains and effector proteins
Like the other phosphoinositides, PtdIns5P transduces its physiological effects through associating with specific downstream effector proteins that carry PtdIns5P-recognizing domains [87]. Several effector proteins have been identified, the most well understood functional importance being that of the nuclear ING2 (inhibitor of growth), a member of the protein family of tumor suppressors. ING2 harbors a PHD (plant homeodomain) finger domain, a motif common for many chromatin-regulatory proteins [88] whose interaction with PtdIns5P is proposed to regulate the p53-dependent apoptotic pathway [88,89]. Other PHD domain-containing proteins, such as ING1, WSTF and ACF (ATP-chromatin-remodeling factor), which contain the similar cluster of characteristic lysines, also bind PtdIns5P along with other PIs [89]. Likewise, the PHD-like domain in ATX1, an Arabidopsis homolog of trithorax1 that regulates numerous genes also associates with PtdIns5P [90]. It should be noted, however, that molecular modeling and structural information of the amino acid residues involved in coordinating PtdIns5P reveal a critical role of a polybasic region positioned C-terminally to the PHD domain [91–93].
The mammalian Dok (downstream of tyrosine kinases) adaptor proteins that are tyrosine phosphorylated upon activation of a number of receptor tyrosine kinases, are preferentially expressed in hematopoietic/immune cells [94]. Intriguingly, all seven members of this family contain a PH domain capable of binding PtdIns5P, which is predicted to regulate T cell signaling. The different Dok’s PH domains bind PtdIns5P with various affinity and specificity, with Dok5 exhibiting the highest affinity [95,96].
Proteins, containing FYVE finger domains, known to associate with PtdIns3P, could also bind PtdIns5P to a low degree [97]. As PtdIns5P’s structure is most similar to that of PtdIns3P, this interaction is apparently not surprising. For example, Fgd1, a GDP/GTP exchange factor of the family of cdc42 GTPases that display multiple phosphoinositide binding domains [98],harbors an atypical FYVE finger domain able to strongly associate with PtdIns5P [99].
Septins, a class of evolutionarily conserved GTP-binding filamentous proteins that are responsible for organization of the cellular cytoskeleton have also been reported to bind PIs [100]. Notably, whereas in mammalian cells, the basic residue region within the molecule binds PtdIns(4,5)P2 and PtdIns(3,4,5)P3, yeast septin members show specificity for PtdIns5P [101]. The functional outcome of this association in each case remains to be seen.
There are conflicting data about the PtdIns5P interaction with the PH-GRAM domain that is found in all active and inactive members of MTM/MTMRs [51]. For example, whereas the PH-GRAM domains in MTMR2 and MTMR3 have been reported to bind PtdIns5P [102,103], data from crystallographic analyses of the MTMR2 PH-GRAM domain fail to confirm such an interaction or predict potential PtdIns5P binding [58,104]. Clearly, additional work and approaches [3,105] might be required to elucidate the specificity and functional significance of the PH-GRAM – PtdIns5P interaction.
Intracellular localization of PtdIns5P
Whereas several downstream proteins can associate with PtdIns5P as indicated above, the binding domains derived from these proteins are inadequate as specific cellular probes in analyses of endogenous PtdIns5P distribution. Likewise, antibodies against PtdIns5P are also unavailable. This, together with the several enzymes that might modulate PtdIns5P levels makes the PtdIns5P intracellular localization a challenge. For years, the intracellular PtdIns5P pool has been confined to the nucleus, where it has been demonstrated to be involved in UV stress, apoptosis, transcription activity and the cell cycle [75,85,89,106]. An extranuclear pool of PtdIns5P has been identified only recently. Thus, biochemical fractionation in a mouse insulinoma cell line combined with HPLC separation on a double column revealed substantial PtdIns5P amounts at the plasma membrane as well as in several intracellular membrane compartments such as smooth endoplasmic reticulum and Golgi [10]. However, the ratio between the cytoplasmic and the nuclear pool was not determined in this study. Another study has also addressed the localization by using the PtdIns5P mass assay and postnuclear subcellular fractions derived from baby hamster kidney cells, in which the fractionated compartments have been well characterized [107,108]. Intriguingly, although at a low level, the greatest subfraction of the endogenous post-nuclear PtdIns5P pool was recovered in the fraction of early endosomes [108]. The endosomal fractions also contained the majority of PtdIns5P that has been produced upon transfection of these cells with the virulence factor IpgD but whether this localization is achieved by specific interactions remains to be identified.
Cellular processes and intracellular pathways regulated by PtdIns5P
PtdIns5P regulates various cellular processes in both the nucleus and the cytoplasm. In many instances, PtdIns5P’s role is inferred through the modulation of the enzymes that regulate PtdIns5P levels, such as PIKfyve, PIP4Ks, PI4-phosphatases and the IpgD virulence factor, which inevitably affect levels of the other PIs. With this reservation in mind, below we discuss the cellular processes regulated by PtdIns5P (Fig. 4) and the potential mechanisms. We place emphases on studies where a direct role for PtdIns5P role has been possible to be documented through exogenous delivery into cells.
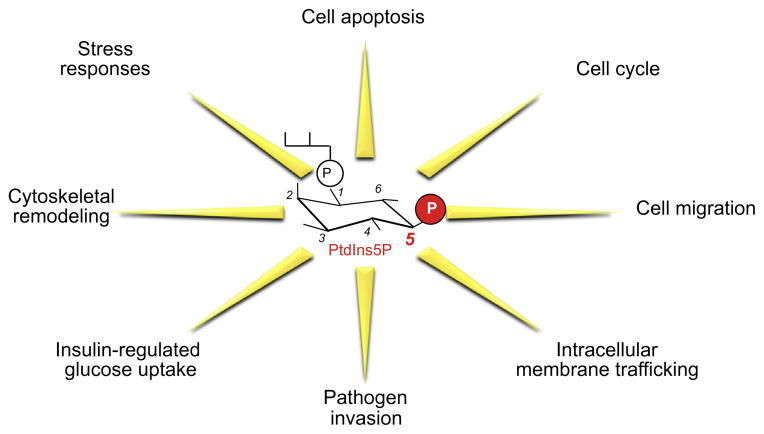
Fig. 4.
Functions of PtdIns5P. Depicted are the main functions of PtdIns5P discussed in this review.
Nuclear functions
The first indication that PtdIns5P plays a role in nuclear function was provided by data for large increases of nuclear PtdIns5P mass during G1 progression of murine erythroleukemia cell proliferation [106]. Several studies in mammalian cells indicate that nuclear PtdIns5P plays a role in apoptosis through increasing p53 activation and stability [75,85,89,92]. For example, the nuclear interaction between PtdIns5P and the tumor supressor ING2 promotes p53 acetylation and cell apoptosis and this is significantly inhibited by overexpression of PIP4Kβ that depletes nuclear PtdIns5P [89,92]. PtdIns5P nuclear levels are reportedly upregulated by a number of stressors, such as UV irradiation, oxidative stress or DNA-damaging agents. This is associated with stress-dependent inhibition of the nuclear PIP4Kβ activity by p38 MAPK phosphorylation [85] and/or nuclear translocation of the PtdIns(4,5)P2 4-phosphatase type 1 [75].
Findings for an interaction of the Cul3-SPOP ubiquitin ligase complex with nuclear PIP4Kβ and stimulation of the complex activity by either of the two PtdIns(4,5)P2-4-phosphatases that increase PtdIns5P has further implicated the latter in nuclear function [109]. The nature of the physiological stimuli that activate this pathway as well as the predicted role of p38 MAPK downstream of PtdIns5P is yet to be determined.
Cytoplasmic functions
Responses to hormone/growth factor cues and environmental stressors
The idea that PtdIns5P has a function outside the nucleus has been supported early on by the observations for acute and transient increases of PtdIns5P in the anucleate platelets upon thrombin stimulation [11]. More detailed information about hormonal regulation and functional significance of cytoplasmic PtdIns5P is provided in the case of insulin challenge in insulin-responsive cells. Thus, PtdIns5P has first been implicated in insulin action in 3T3L1 adipocytes and CHO-T cells expressing the human insulin receptor based on data from our lab for robust and transient increases in PtdIns5P mass levels [110]. Insulin-dependent PtdIns5P increases in CHO-T cells have been confirmed in a recent study [10]. The PtdIns5P rise in CHO-T cells proceeds independently of class IA PI3K activation, evidenced by its resistance to cell pre-treatment with wortmannin, a powerful class IA PI3K inhibitor [110]. Because exogenous di-C16-PtdIns5P, delivered intracellularly by cytoplasmic microinjection has also induced a wortmannin-resistant breakdown of actin stress fibers in CHO-T cells [110], it has been suggested that PtdIns5P is required in the class IA PI3K-independent remodeling of actin filaments, shown to be critical for optimal GLUT4 translocation to the cell surface in response to insulin [111]. Concordantly, exogenously delivered di-C16-PtdIns5P into 3T3L1 adipocytes increased cell surface abundance of ectopically expressed GLUT4 in the absence of insulin [110]. Conversely, sequestering PtdIns5P by the 3×PHD domain derived from various PHD-containing proteins, disabled insulin triggered cell surface gain of ectopically expressed GLUT4 [110]. PtdIns5P increases have also been reproduced in insulin-responsive L6 myotubes but not in L6 myoblasts or other insulin-unresponsive cells [112], suggesting that PtdIns5P might be a key insulin-regulated mediator signaling to GLUT4. Indeed, both GLUT4 translocation and glucose uptake have been found activated upon carrier-mediated delivery of di-C4-PtdIns5P in non-stimulated L6 myoblasts [112]. PtdIns5P-triggered activation of glucose uptake in L6 myoblasts is dependent on the PI3K/Akt pathway, which, together with the wortmannin-resistant PtdIns5P rise suggest that the PI3K/Akt pathway functions downstream of PtdIns5P [110,112].
The role of PIKfyve as the kinase responsible for PtdIns5P upregulation by insulin was first suggested based on observations that PIKfyve produces PtdIns5P both in vivo and in vitro, and that its ectopic expression mimics the effect of exogenously delivered PtdIns5P on F-actin breakdown in CHO-T cells [110]. Findings for insulin-dependent recruitment of a cytosolic subfraction of PIKfyve to intracellular membranes in 3T3L1 adipocytes could mechanistically explain the PtdIns5P increases in this cell type [113]. The predicted role of PIKfyve in acute insulin-dependent PtdIns5P increases has been recently solidified by data for the abolishment of the PtdIns5P increment upon 3T3L1 adipocyte inhibition with the YM201636 compound that preferentially reduces PtdIns5P [17]. Furthermore, the transient nature of the rise suggests that PtdIns5P is removed though PtdIns5P-4-kinases and a putative PtdIns5P phosphatase. Observations for abrogated insulin-regulated PtdIns5P elevation upon ectopic expression of PIP4 Kα in L6 myotubes is in agreement with such a prediction [112]. Because pervanadate, an agent that mimics many insulin responses both in vivo and in vitro via inhibiting tyrosine dephosphorylation [114,115] has also been found to acutely elevate PtdIns5P in HeLa cells [116], a tyrosine phosphorylation step is likely to be required. However, more work is required to shed light onto the molecular events responsible for the acute rise and fall of PtdIns5P by insulin and how these modulations are linked with glucose uptake activation. Whatever the molecular mechanism, it is significant to emphasize recent metabolic data from the newly developed mouse model with Pikfyve specific disruption in skeletal muscle [117], the tissue responsible for the majority of postprandial insulin-stimulated glucose disposal. The observations for whole-body glucose intolerance and insulin resistance due to a primary defect of muscle insulin resistance indicate the physiological role of PtdIns5P and/or PtdIns(3,5)P2 in glucose homeostasis [117].
In addition to insulin receptor, stimulation of T cell receptors by anti-CD3 antibody produces a dramatic rise in the PtdIns5P cytoplasmic pool [95] but the physiological significance of this remains to be determined. Dok protein phosphorylation downstream of PtdIns5P is suggested as a plausible mechanism in T cell activation [95]. PtdIns5P has also been shown to be modestly elevated by stimulation of BJ cells with fibroblast growth factor 1 and related to cell migration through a mechanism that involves PIKfyve and MTMR3 [42].
An extranuclear pool of PtdIns5P has also been implicated in the environmental stress responses in mammalian and plant cells. Thus, acute hypo-osmotic shock in 3T3L1 cells dramatically decreased PtdIns5P [22]. Consistently, hyperosmotic shock in Chlamydomonas increases PtdIns5P [13]. Likewise, in Arabidopsis, the extranuclear PtdIns5P pool is elevated in response to dehydration, leading to gene expression by yet to be established signaling pathway [14]. Recently, oxidative stress has also been shown to acutely increase PtdIns5P in several mammalian cell types, which is linked to Akt activation [76,118]. Together, these data indicate that PtdIns5P is a key mediator in the response mechanisms to multiple hormonal and stress challenges. Details about the upstream regulators and downstream effectors remain to be identified in each case.
Actin remodeling and membrane trafficking
Several studies indicate that PtdIns5P is implicated in actin remodeling. For example, elevation of intracellular PtdIns5P by cytoplasmic microinjection of di-C16-PtdIns5P in CHO-T cells or upon ectopic expression of PIKfyveWT induced a marked F-actin stress fiber breakdown [110]. Conversely, sequestration of endogenous PdIns5P by ectopic expression of a 3×PHD domain abrogated insulin’s ability to trigger actin stress fiber loss. Experimental support for PtdIns5P functioning in F-actin reorganization is also coming from studies in HeLa and NIH-3T3 cells, in which PtdIns5P has been elevated by ectopic expression of IpgD, a bacterial inositol polyphosphate 4-phosphatase [72]. Upon longer expression time, these cells exhibited actin stress fiber disappearance followed by membrane blebbing. Reduced PtdIns5P by knockdown of PIKfyve and MTMR3 protein has also been found to trigger actin fiber remodeling in parallel with reduced cell migration that was restored by delivery of exogenous di-C16 PtdIns5P [42].
Acute actin cytoskeleton rearrangements support many aspects of both inward and outward membrane trafficking. However, only a few studies suggest that PtdIns5P-regulated cytoskeletal remodeling may alter the intracellular trafficking pathways. For example, increased GLUT4 plasma membrane accumulation in both 3T3L1 and L6 myocytes by direct delivery of PtdIns5P, discussed above, is consistent with PtdIns5P’s role in inhibiting GLUT4 endocytosis and/or accelerating exocytosis [110,112]. Corroborating these observations, a comparative genomic approach of PI enzymes and regulators has postulated a role for PtdIns5P in exocytosis [119]. Furthermore, a recent study provides functional information about PtdIns5P’s role in EGFR intracellular trafficking by exploring the ability of IpgD to produce PtdIns5P [108]. The authors have attributed the observed early endosomal retention and prevented lysosomal degradation of EGFR to PtdIns5P-dependent disruption of endosome maturation and/or transport progression from early to late endosomes [120].Whether this conclusion might be correct, it should be emphasized that similar trafficking defects are triggered by intracellular PtdIns(3,5)P2 decline [16,20]. For example, arrested traffic progression of fluid phase cargo from early to late endosomes [121] and inhibited EGFR lysosomal degradation [122] have both been observed by perturbations of PIKfyve activity. Under these conditions, the invariant endosome dilation/cytoplasmic vacuolation is manifested due to a specific decline in PtdIns(3,5)P2 but not in PtdIns5P as shown by direct cytoplasmic delivery of these lipids [34,123]. The cellular mechanisms operating under these conditions are associated in part with enhanced endosome membrane fusion and arrested endosome maturation/vesicle fission [43,124]. Because expression of IpgD might induce cytoplasmic vacuolation, as inferred by the aberrant morphology of illustrated transfected HeLa cells [108], the IpgD-triggered membrane trafficking defects could be related to PIKfyve feedback inhibition by PtdIns5P, markedly elevated on early endosomes under these conditions (>150-fold). Additionally, IpgD-dependent PtdIns5P elevation may activate certain members of MTM/MTMRs [55] to increase PtdIns(3,5)P2 turnover. This prediction is supported by findings for delayed EGFR-lysosomal degradation in COS cells expressing MTM that translocates to late endosomes upon EGF stimulation to hydrolyze EGFR-elevated PtdIns(3,5)P2 and trigger formation of large endosomal vacuoles [54]. The predicted mechanisms (Fig. 3) underlying the trafficking defects under IpgD-elevated PtdIns5P associated with inhibition of PIKfyve-dependent PtdIns(3,5)P2 synthesis and activation of MTM/MTMRs-dependent PtdIns(3,5)P2 hydrolysis warrant further investigation.
Many of the PtdIns5P-dependent cytoplasmic functions appear to be transduced by Akt that is activated downstream of elevated Ptdins5P [36,112,120,125,126]. Various mechanisms have been implicated to account for this effect, such as activation of class 1A PI3K [120], inhibition of PtdIns(3,4,5)P3 5-dephosphorylation [126] or inhibition of protein phosphatase 2A that dephosphorylates Akt [127]. Further mechanistic details as to how this is achieved as well as information about the specificity of each of the suggested mechanisms would certainly be of great importance.
Concluding remarks
PtdIns5P has typically been referred to as the most mysterious PI and it certainly continues to deserve such a description. Many old challenges remain unchanged, including its low levels, obstacles for co-detection together with the other PIs and existence of multiple enzymatic pathways by which its levels could be regulated. This, coupled with the fact that specific reagents, such as antibodies or bioreporters to reveal more precisely its intracellular localization remain unavailable, continue to place PtdIns5P as one of the most disadvantaged PI for research. Regardless of these multiple and persisting challenges, a great deal of information has recently accumulated to implicate PtdIns5P in regulating a myriad of cellular processes such as stress/hormone-induced signaling, cytoskeletal dynamics, membrane trafficking, cell migration, cell cycle and apoptosis (Fig. 4). For these or other yet to be discovered versatile features, several pathogens have resorted to invade host cells by elevating PtdIns5P. Due to advancement in the development of genetically modified PIKfyve mouse models, it is now clear that PIKfyve is responsible for the majority of PtdIns5P production. Compelling evidence discussed herein imply direct PtdIns5P biosynthesis from PtdIns at basal conditions as a universal road for the majority of constitutive PtdIns5P in quiescent mammalian cells, rather than PIKfyve/MTMRs-dependent PtdIns(3,5)P2 synthesis/hydrolyses, as proposed by some researchers. However, despite these achievements, many pressing questions remain. How does PIKfyve deliver both PtdIns(3,5)P2 and PtdIns5P biosynthesis? Under what conditions do MTM/MTMRs hydrolyze intracellular PtdIns(3,5)P2 and which one of the active members does so? How is PtdIns5P dephosphorylated? The answer to these and other questions will help distinguish between the roles of PtdIns(3,5)P2 and PtdIns5P in physiological and pathological conditions that are manifested in the mouse models, including systemic metabolic derangement upon muscle-specific PIKfyve ablation [117], preimplantation lethality under global PIKfyve KO [18] and neurodegeneration [16]. Future research with more specific tools will be able to enlighten the ample amount of functional and mechanistic challenges.
Acknowledgments
I am grateful to past and current members of my laboratory, and particularly to Drs. Ogi Ikonomov and Diego Sbrissa for their excellent work and stimulating discussions. I thank Drs. Ellen Tisdale and Steve Cala for their insightful comments and the late Violeta Shisheva for her many years of support. The work described from my laboratory was funded by ADA (7-13-BS-161), JDFI and NIH (DK58058).
Footnotes
Abbreviations used: PtdIns, phosphatidylinositol; PIs, phosphoinositides; HPLC, high-pressure liquid chromatography; KO, knockout; KD, knockdown; PHD, plant homeodomain; ACF, ATP-chromatin-remodeling factor; Dok, downstream of tyrosine kinases.
References
- 1.Shisheva A. Front Biosci. 2003;8:s945–s946. doi: 10.2741/1101. [DOI] [PubMed] [Google Scholar]
- 2.Carlton JG, Cullen PJ. Trends Cell Biol. 2005;15:540–547. doi: 10.1016/j.tcb.2005.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lemmon MA. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 4.Di Paolo G, De Camilli P. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 5.Sasaki T, Takasuga S, Sasaki J, Kofuji S, Eguchi S, Yamazaki M, Suzuki A. Prog LipidRes. 2009;48:307–343. doi: 10.1016/j.plipres.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 6.Vicinanza M, D’Angelo G, Di Campli A, De Matteis MA. EMBO J. 2008;27:2457–2470. doi: 10.1038/emboj.2008.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hakim S, Bertucci MC, Conduit SE, Vuong DL, Mitchell CA. Curr Top Microbiol Immunol. 2012;362:247–314. doi: 10.1007/978-94-007-5025-8_12. [DOI] [PubMed] [Google Scholar]
- 8.Rameh LE, Tolias KF, Duckworth BC, Cantley LC. Nature. 1997;390:192–196. doi: 10.1038/36621. [DOI] [PubMed] [Google Scholar]
- 9.Sbrissa D, Ikonomov OC, Shisheva A. J Biol Chem. 1999;274:21589–21597. doi: 10.1074/jbc.274.31.21589. [DOI] [PubMed] [Google Scholar]
- 10.Sarkes D, Rameh LE. Biochem J. 2010;428:375–384. doi: 10.1042/BJ20100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morris JB, Hinchliffe KA, Ciruela A, Letcher AJ, Irvine RF. FEBS Lett. 2000;475:57–60. doi: 10.1016/s0014-5793(00)01625-2. [DOI] [PubMed] [Google Scholar]
- 12.Vaccari I, Dina G, Tronchere H, Kaufman E, Chicanne G, Cerri F, Wrabetz L, Payrastre B, Quattrini A, Weisman LS, Meisler MH, Bolino A. PLoSGenet. 2011;7:e1002319. doi: 10.1371/journal.pgen.1002319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meijer HJ, Berrie CP, Iurisci C, Divecha N, Musgrave A, Munnik T. Biochem J. 2001;360:491–498. doi: 10.1042/0264-6021:3600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ndamukong I, Jones DR, Lapko H, Divecha N, Avramova Z. PLoS One. 2010;5:e13396. doi: 10.1371/journal.pone.0013396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta A, Toscano S, Trivedi D, Jones DR, Mathre S, Clarke JH, Divecha N, Raghu P. Proc Natl Acad Sci USA. 2013;110:5963–5968. doi: 10.1073/pnas.1219333110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shisheva A. Curr Top Microbiol Immunol. 2012;362:127–162. doi: 10.1007/978-94-007-5025-8_7. [DOI] [PubMed] [Google Scholar]
- 17.Sbrissa D, Ikonomov OC, Filios C, Delvecchio K, Shisheva A. Am J Physiol Cell Physiol. 2012;303:C436–C446. doi: 10.1152/ajpcell.00105.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikonomov OC, Sbrissa D, Delvecchio K, Xie Y, Jin JP, Rappolee D, Shisheva A. J Biol Chem. 2011;286:13404–13413. doi: 10.1074/jbc.M111.222364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zolov SN, Bridges D, Zhang Y, Lee WW, Riehle E, Verma R, Lenk GM, Converso-Baran K, Weide T, Albin RL, Saltiel AR, Meisler MH, Russell MW, Weisman LS. Proc Natl Acad Sci USA. 2012;109:17472–17477. doi: 10.1073/pnas.1203106109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shisheva A. Cell Biol Int. 2008;32:591–604. doi: 10.1016/j.cellbi.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shisheva A. Cell Biol Int. 2001;25:1201–1206. doi: 10.1006/cbir.2001.0803. [DOI] [PubMed] [Google Scholar]
- 22.Sbrissa D, Ikonomov OC, Deeb R, Shisheva A. J Biol Chem. 2002;277:47276–47284. doi: 10.1074/jbc.M207576200. [DOI] [PubMed] [Google Scholar]
- 23.Balla T, Szentpetery Z, Kim YJ. Physiology (Bethesda) 2009;24:231–244. doi: 10.1152/physiol.00014.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayinger P. Biochim Biophys Acta. 2012;1821:1104–1113. doi: 10.1016/j.bbalip.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brill JA, Wong R, Wilde A. Curr Biol. 2011;21:R930–R934. doi: 10.1016/j.cub.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 26.Shisheva A. Am J Physiol Endocrinol Metab. 2008;295:E536–E544. doi: 10.1152/ajpendo.90353.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tronchere H, Laporte J, Pendaries C, Chaussade C, Liaubet L, Pirola L, Mandel JL, Payrastre B. J Biol Chem. 2004;279:7304–7312. doi: 10.1074/jbc.M311071200. [DOI] [PubMed] [Google Scholar]
- 28.Ungewickell A, Hugge C, Kisseleva M, Chang SC, Zou J, Feng Y, Galyov EE, Wilson M, Majerus PW. Proc Natl Acad Sci USA. 2005;102:18854–18859. doi: 10.1073/pnas.0509740102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Y, Lee SA, Kutateladze TG, Sbrissa D, Shisheva A, Prestwich GD. J Am Chem Soc. 2006;128:885–897. doi: 10.1021/ja0554716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sbrissa D, Ikonomov OC, Shisheva A. Biochemistry. 2000;39:15980–15989. doi: 10.1021/bi001897f. [DOI] [PubMed] [Google Scholar]
- 31.Shisheva A, Sbrissa D, Ikonomov O. Mol Cell Biol. 1999;19:623–634. doi: 10.1128/mcb.19.1.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McEwen RK, Dove SK, Cooke FT, Painter GF, Holmes AB, Shisheva A, Ohya Y, Parker PJ, Michell RH. J Biol Chem. 1999;274:33905–33912. doi: 10.1074/jbc.274.48.33905. [DOI] [PubMed] [Google Scholar]
- 33.Parrish WR, Stefan CJ, Emr SD. Mol Biol Cell. 2004;15:3567–3579. doi: 10.1091/mbc.E04-03-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ikonomov OC, Sbrissa D, Mlak K, Kanzaki M, Pessin J, Shisheva A. J Biol Chem. 2002;277:9206–9211. doi: 10.1074/jbc.M108750200. [DOI] [PubMed] [Google Scholar]
- 35.Jefferies HB, Cooke FT, Jat P, Boucheron C, Koizumi T, Hayakawa M, Kaizawa H, Ohishi T, Workman P, Waterfield MD, Parker PJ. EMBORep. 2008;9:164–170. doi: 10.1038/sj.embor.7401155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ikonomov OC, Sbrissa D, Mlak K, Shisheva A. Endocrinology. 2002;143:4742–4754. doi: 10.1210/en.2002-220615. [DOI] [PubMed] [Google Scholar]
- 37.Ikonomov OC, Sbrissa D, Shisheva A. Biochem Biophys Res Commun. 2009;382:566–570. doi: 10.1016/j.bbrc.2009.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coronas S, Lagarrigue F, Ramel D, Chicanne G, Delsol G, Payrastre B, Tronchere H. Biochem Biophys Res Commun. 2008;372:351–355. doi: 10.1016/j.bbrc.2008.05.062. [DOI] [PubMed] [Google Scholar]
- 39.Sbrissa D, Ikonomov OC, Strakova J, Dondapati R, Mlak K, Deeb R, Silver R, Shisheva A. Mol Cell Biol. 2004;24:10437–10447. doi: 10.1128/MCB.24.23.10437-10447.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Zolov SN, Chow CY, Slutsky SG, Richardson SC, Piper RC, Yang B, Nau JJ, Westrick RJ, Morrison SJ, Meisler MH, Weisman LS. Proc Natl Acad Sci USA. 2007;104:17518–17523. doi: 10.1073/pnas.0702275104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leslie NR, Downes CP. Cell Signal. 2002;14:285–295. doi: 10.1016/s0898-6568(01)00234-0. [DOI] [PubMed] [Google Scholar]
- 42.Oppelt A, Lobert VH, Haglund K, Mackey AM, Rameh LE, Liestol K, Schink KO, Pedersen NM, Wenzel EM, Haugsten EM, Brech A, Rusten TE, Stenmark H, Wesche J. EMBORep. 2013;14:57–64. doi: 10.1038/embor.2012.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sbrissa D, Ikonomov OC, Fu Z, Ijuin T, Gruenberg J, Takenawa T, Shisheva A. J Biol Chem. 2007;282:23878–23891. doi: 10.1074/jbc.M611678200. [DOI] [PubMed] [Google Scholar]
- 44.Ho CY, Alghamdi TA, Botelho RJ. Traffic. 2012;13:1–8. doi: 10.1111/j.1600-0854.2011.01246.x. [DOI] [PubMed] [Google Scholar]
- 45.Sbrissa D, Ikonomov OC, Fenner H, Shisheva A. J Mol Biol. 2008;384:766–779. doi: 10.1016/j.jmb.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ikonomov OC, Sbrissa D, Fenner H, Shisheva A. J Biol Chem. 2009;284:35794–35806. doi: 10.1074/jbc.M109.037515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Devereaux K, Di Paolo G. EMBO Rep. 2013;14:214–215. doi: 10.1038/embor.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gary JD, Wurmser AE, Bonangelino CJ, Weisman LS, Emr SD. J Cell Biol. 1998;143:65–79. doi: 10.1083/jcb.143.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cooke FT, Dove SK, McEwen RK, Painter G, Holmes AB, Hall MN, Michell RH, Parker PJ. Curr Biol. 1998;8:1219–1222. doi: 10.1016/s0960-9822(07)00513-1. [DOI] [PubMed] [Google Scholar]
- 50.Michell RH, Heath VL, Lemmon MA, Dove SK. Trends Biochem Sci. 2006;31:52–63. doi: 10.1016/j.tibs.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 51.Hnia K, Vaccari I, Bolino A, Laporte J. Trends Mol Med. 2012;18:317–327. doi: 10.1016/j.molmed.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 52.Walker DM, Urbe S, Dove SK, Tenza D, Raposo G, Clague MJ. Curr Biol. 2001;11:1600–1605. doi: 10.1016/s0960-9822(01)00501-2. [DOI] [PubMed] [Google Scholar]
- 53.Tosch V, Rohde HM, Tronchere H, Zanoteli E, Monroy N, Kretz C, Dondaine N, Payrastre B, Mandel JL, Laporte J. Hum Mol Genet. 2006;15:3098–3106. doi: 10.1093/hmg/ddl250. [DOI] [PubMed] [Google Scholar]
- 54.Tsujita K, Itoh T, Ijuin T, Yamamoto A, Shisheva A, Laporte J, Takenawa T. J Biol Chem. 2004;279:13817–13824. doi: 10.1074/jbc.M312294200. [DOI] [PubMed] [Google Scholar]
- 55.Schaletzky J, Dove SK, Short B, Lorenzo O, Clague MJ, Barr FA. Curr Biol. 2003;13:504–509. doi: 10.1016/s0960-9822(03)00132-5. [DOI] [PubMed] [Google Scholar]
- 56.Berger P, Bonneick S, Willi S, Wymann M, Suter U. Hum Mol Genet. 2002;11:1569–1579. doi: 10.1093/hmg/11.13.1569. [DOI] [PubMed] [Google Scholar]
- 57.Naughtin MJ, Sheffield DA, Rahman P, Hughes WE, Gurung R, Stow JL, Nandurkar HH, Dyson JM, Mitchell CA. J Cell Sci. 2010;123:3071–3083. doi: 10.1242/jcs.060103. [DOI] [PubMed] [Google Scholar]
- 58.Begley MJ, Taylor GS, Kim SA, Veine DM, Dixon JE, Stuckey JA. Mol Cell. 2003;12:1391–1402. doi: 10.1016/s1097-2765(03)00486-6. [DOI] [PubMed] [Google Scholar]
- 59.Taylor GS, Maehama T, Dixon JE. Proc Natl Acad Sci USA. 2000;97:8910–8915. doi: 10.1073/pnas.160255697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim SA, Taylor GS, Torgersen KM, Dixon JE. J Biol Chem. 2002;277:4526–4531. doi: 10.1074/jbc.M111087200. [DOI] [PubMed] [Google Scholar]
- 61.Chaussade C, Pirola L, Bonnafous S, Blondeau F, Brenz-Verca S, Tronchere H, Portis F, Rusconi S, Payrastre B, Laporte J, VanObberghen E. Mol Endocrinol. 2003;17:2448–2460. doi: 10.1210/me.2003-0261. [DOI] [PubMed] [Google Scholar]
- 62.Srivastava S, Li Z, Lin L, Liu G, Ko K, Coetzee WA, Skolnik EY. Mol Cell Biol. 2005;25:3630–3638. doi: 10.1128/MCB.25.9.3630-3638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cao C, Backer JM, Laporte J, Bedrick EJ, Wandinger-Ness A. Mol Biol Cell. 2008;19:3334–3346. doi: 10.1091/mbc.E08-04-0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vergne I, Roberts E, Elmaoued RA, Tosch V, Delgado MA, Proikas-Cezanne T, Laporte J, Deretic V. EMBOJ. 2009;28:2244–2258. doi: 10.1038/emboj.2009.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taguchi-Atarashi N, Hamasaki M, Matsunaga K, Omori H, Ktistakis NT, Yoshimori T, Noda T. Traffic. 2010;11:468–478. doi: 10.1111/j.1600-0854.2010.01034.x. [DOI] [PubMed] [Google Scholar]
- 66.Razidlo GL, Katafiasz D, Taylor GS. J Biol Chem. 2011;286:20005–20019. doi: 10.1074/jbc.M110.197749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Amoasii L, Hnia K, Chicanne G, Brech A, Cowling BS, Muller MM, Schwab Y, Koebel P, Ferry A, Payrastre B, Laporte J. J Cell Sci. 2013;67:1806–1819. doi: 10.1242/jcs.118505. [DOI] [PubMed] [Google Scholar]
- 68.Ribeiro I, Yuan L, Tanentzapf G, Dowling JJ, Kiger A. PLoS Genet. 2011;7:e1001295. doi: 10.1371/journal.pgen.1001295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu N, Shen Q, Mahoney TR, Neukomm LJ, Wang Y, Zhou Z. PLoS Biol. 2012;10:e1001245. doi: 10.1371/journal.pbio.1001245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Whiteford CC, Brearley CA, Ulug ET. Biochem J. 1997;323(Pt. 3):597–601. doi: 10.1042/bj3230597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sbrissa D, Ikonomov OC, Shisheva A. J Biol Chem. 2002;277:6073–6079. doi: 10.1074/jbc.M110194200. [DOI] [PubMed] [Google Scholar]
- 72.Niebuhr K, Giuriato S, Pedron T, Philpott DJ, Gaits F, Sable J, Sheetz MP, Parsot C, Sansonetti PJ, Payrastre B. EMBOJ. 2002;21:5069–5078. doi: 10.1093/emboj/cdf522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Norris FA, Wilson MP, Wallis TS, Galyov EE, Majerus PW. Proc Natl Acad Sci USA. 1998;95:14057–14059. doi: 10.1073/pnas.95.24.14057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mason D, Mallo GV, Terebiznik MR, Payrastre B, Finlay BB, Brumell JH, Rameh L, Grinstein S. J Gen Physiol. 2007;129:267–283. doi: 10.1085/jgp.200609656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zou J, Marjanovic J, Kisseleva MV, Wilson M, Majerus PW. Proc Natl Acad Sci USA. 2007;104:16834–16839. doi: 10.1073/pnas.0708189104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jones DR, Foulger R, Keune WJ, Bultsma Y, Divecha N. FASEB J. 2013;27:1644–1656. doi: 10.1096/fj.12-218842. [DOI] [PubMed] [Google Scholar]
- 77.Zhang J, Guan Z, Murphy AN, Wiley SE, Perkins GA, Worby CA, Engel JL, Heacock P, Nguyen OK, Wang JH, Raetz CR, Dowhan W, Dixon JE. CellMetab. 2011;13:690–700. doi: 10.1016/j.cmet.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu Y, Bruzik KS, Ananthanarayanan B, Cho W. Bioorg Med Chem. 2003;11:2471–2475. doi: 10.1016/s0968-0896(03)00150-0. [DOI] [PubMed] [Google Scholar]
- 79.Roberts HF, Clarke JH, Letcher AJ, Irvine RF, Hinchliffe KA. FEBS Lett. 2005;579:2868–2872. doi: 10.1016/j.febslet.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 80.Bultsma Y, Keune WJ, Divecha N. Biochem J. 2010;430:223–235. doi: 10.1042/BJ20100341. [DOI] [PubMed] [Google Scholar]
- 81.Richardson JP, Wang M, Clarke JH, Patel KJ, Irvine RF. Cell Signal. 2007;19:1309–1314. doi: 10.1016/j.cellsig.2007.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ciruela A, Hinchliffe KA, Divecha N, Irvine RF. Biochem J. 2000;346(Pt 3):587–591. [PMC free article] [PubMed] [Google Scholar]
- 83.Itoh T, Ijuin T, Takenawa T. J Biol Chem. 1998;273:20292–20299. doi: 10.1074/jbc.273.32.20292. [DOI] [PubMed] [Google Scholar]
- 84.Clarke JH, Irvine RF. Adv Biol Regul. 2012;52:40–45. doi: 10.1016/j.advenzreg.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 85.Jones DR, Bultsma Y, Keune WJ, Halstead JR, Elouarrat D, Mohammed S, Heck AJ, D’Santos CS, Divecha N. Mol Cell. 2006;23:685–695. doi: 10.1016/j.molcel.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 86.Hinchliffe KA, Irvine RF. Cell Signal. 2006;18:1906–1913. doi: 10.1016/j.cellsig.2006.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kutateladze TG. Curr Top Microbiol Immunol. 2012;362:111–126. doi: 10.1007/978-94-007-5025-8_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Musselman CA, Kutateladze TG. Mol Interv. 2009;9:314–323. doi: 10.1124/mi.9.6.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gozani O, Karuman P, Jones DR, Ivanov D, Cha J, Lugovskoy AA, Baird CL, Zhu H, Field SJ, Lessnick SL, Villasenor J, Mehrotra B, Chen J, Rao VR, Brugge JS, Ferguson CG, Payrastre B, Myszka DG, Cantley LC, Wagner G, Divecha N, Prestwich GD, Yuan J. Cell. 2003;114:99–111. doi: 10.1016/s0092-8674(03)00480-x. [DOI] [PubMed] [Google Scholar]
- 90.Alvarez-Venegas R, Sadder M, Hlavacka A, Baluska F, Xia Y, Lu G, Firsov A, Sarath G, Moriyama H, Dubrovsky JG, Avramova Z. Proc Natl Acad Sci USA. 2006;103:6049–6054. doi: 10.1073/pnas.0600944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pena PV, Musselman CA, Kuo AJ, Gozani O, Kutateladze TG. Magn Reson Chem. 2009;47:352–358. doi: 10.1002/mrc.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang W, Zhang H, Davrazou F, Kutateladze TG, Shi X, Gozani O, Prestwich GD. J Am Chem Soc. 2007;129:6498–6506. doi: 10.1021/ja070195b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kaadige MR, Ayer DE. J Biol Chem. 2006;281:28831–28836. doi: 10.1074/jbc.M605624200. [DOI] [PubMed] [Google Scholar]
- 94.Mashima R, Hishida Y, Tezuka T, Yamanashi Y. Immunol Rev. 2009;232:273–285. doi: 10.1111/j.1600-065X.2009.00844.x. [DOI] [PubMed] [Google Scholar]
- 95.Guittard G, Gerard A, Dupuis-Coronas S, Tronchere H, Mortier E, Favre C, Olive D, Zimmermann P, Payrastre B, Nunes JA. J Immunol. 2009;182:3974–3978. doi: 10.4049/jimmunol.0804172. [DOI] [PubMed] [Google Scholar]
- 96.Guittard G, Mortier E, Tronchere H, Firaguay G, Gerard A, Zimmermann P, Payrastre B, Nunes JA. FEBSLett. 2010;584:2455–2460. doi: 10.1016/j.febslet.2010.04.051. [DOI] [PubMed] [Google Scholar]
- 97.Kutateladze T, Overduin M. Science. 2001;291:1793–1796. doi: 10.1126/science.291.5509.1793. [DOI] [PubMed] [Google Scholar]
- 98.Nakanishi H, Takai Y. J Cell Mol Med. 2008;12:1169–1176. doi: 10.1111/j.1582-4934.2008.00345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sankaran VG, Klein DE, Sachdeva MM, Lemmon MA. Biochemistry. 2001;40:8581–8587. doi: 10.1021/bi010425d. [DOI] [PubMed] [Google Scholar]
- 100.Mostowy S, Cossart P. Nat Rev Mol Cell Biol. 2012;13:183–194. doi: 10.1038/nrm3284. [DOI] [PubMed] [Google Scholar]
- 101.Casamayor A, Snyder M. Mol Cell Biol. 2003;23:2762–2777. doi: 10.1128/MCB.23.8.2762-2777.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lorenzo O, Urbe S, Clague MJ. J Cell Sci. 2005;118:2005–2012. doi: 10.1242/jcs.02325. [DOI] [PubMed] [Google Scholar]
- 103.Berger P, Schaffitzel C, Berger I, Ban N, Suter U. Proc Natl Acad Sci USA. 2003;100:12177–12182. doi: 10.1073/pnas.2132732100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Begley MJ, Taylor GS, Brock MA, Ghosh P, Woods VL, Dixon JE. Proc Natl Acad Sci USA. 2006;103:927–932. doi: 10.1073/pnas.0510006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Narayan K, Lemmon MA. Methods. 2006;39:122–133. doi: 10.1016/j.ymeth.2006.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Clarke JH, Letcher AJ, D’Santos SC, Halstead JR, Irvine RF, Divecha N. Biochem J. 2001;357:905–910. doi: 10.1042/0264-6021:3570905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gorvel JP, Chavrier P, Zerial M, Gruenberg J. Cell. 1991;64:915–925. doi: 10.1016/0092-8674(91)90316-q. [DOI] [PubMed] [Google Scholar]
- 108.Ramel D, Lagarrigue F, Pons V, Mounier J, Dupuis-Coronas S, Chicanne G, Sansonetti PJ, Gaits-Iacovoni F, Tronchere H, Payrastre B. Sci Signal. 2011;4:ra61. doi: 10.1126/scisignal.2001619. [DOI] [PubMed] [Google Scholar]
- 109.Bunce MW, Boronenkov IV, Anderson RA. J Biol Chem. 2008;283:8678–8686. doi: 10.1074/jbc.M710222200. [DOI] [PubMed] [Google Scholar]
- 110.Sbrissa D, Ikonomov OC, Strakova J, Shisheva A. Endocrinology. 2004;145:4853–4865. doi: 10.1210/en.2004-0489. [DOI] [PubMed] [Google Scholar]
- 111.Watson RT, Kanzaki M, Pessin JE. Endocr Rev. 2004;25:177–204. doi: 10.1210/er.2003-0011. [DOI] [PubMed] [Google Scholar]
- 112.Grainger DL, Tavelis C, Ryan AJ, Hinchliffe KA. Pflugers Arch. 2011;462:723–732. doi: 10.1007/s00424-011-1008-4. [DOI] [PubMed] [Google Scholar]
- 113.Shisheva A, Rusin B, Ikonomov OC, DeMarco C, Sbrissa D. J Biol Chem. 2001;276:1869, 11859–11869. doi: 10.1074/jbc.M008437200. [DOI] [PubMed] [Google Scholar]
- 114.Shisheva A, Ikonomov O, Shechter Y. Endocrinology. 1994;134:507–510. doi: 10.1210/endo.134.1.8275968. [DOI] [PubMed] [Google Scholar]
- 115.Shisheva A, Shechter Y. Endocrinology. 1993;133:1562–1568. doi: 10.1210/endo.133.4.8404595. [DOI] [PubMed] [Google Scholar]
- 116.Wilcox A, Hinchliffe KA. FEBS Lett. 2008;582:1391–1394. doi: 10.1016/j.febslet.2008.03.022. [DOI] [PubMed] [Google Scholar]
- 117.Ikonomov OC, Sbrissa D, Delvecchio K, Feng HZ, Cartee GD, Jin JP, Shisheva A. Am J Physiol Endocrinol Metab. 2013;305:E119–E131. doi: 10.1152/ajpendo.00030.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Keune WJ, Jones DR, Bultsma Y, Sommer L, Zhou XZ, Lu KP, Divecha N. Sci Signal. 2012;5:ra86. doi: 10.1126/scisignal.2003223. [DOI] [PubMed] [Google Scholar]
- 119.Lecompte O, Poch O, Laporte J. Trends Biochem Sci. 2008;33:453–460. doi: 10.1016/j.tibs.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 120.Pendaries C, Tronchere H, Arbibe L, Mounier J, Gozani O, Cantley L, Fry MJ, Gaits-Iacovoni F, Sansonetti PJ, Payrastre B. EMBOJ. 2006;25:1024–1034. doi: 10.1038/sj.emboj.7601001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ikonomov OC, Sbrissa D, Foti M, Carpentier JL, Shisheva A. Mol Biol Cell. 2003;14:4581–4591. doi: 10.1091/mbc.E03-04-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.deLartigue J, Polson H, Feldman M, Shokat K, Tooze SA, Urbe S, Clague MJ. Traffic. 2009;10:883–893. doi: 10.1111/j.1600-0854.2009.00915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ikonomov OC, Sbrissa D, Shisheva A. J Biol Chem. 2001;276:26141–26147. doi: 10.1074/jbc.M101722200. [DOI] [PubMed] [Google Scholar]
- 124.Ikonomov OC, Sbrissa D, Shisheva A. Am J Physiol Cell Physiol. 2006;291:C393–C404. doi: 10.1152/ajpcell.00019.2006. [DOI] [PubMed] [Google Scholar]
- 125.Ikonomov OC, Sbrissa D, Dondapati R, Shisheva A. Exp Cell Res. 2007;313:2404–2416. doi: 10.1016/j.yexcr.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Carricaburu V, Lamia KA, Lo E, Favereaux L, Payrastre B, Cantley LC, Rameh LE. Proc Natl Acad Sci USA. 2003;100:9867–9872. doi: 10.1073/pnas.1734038100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ramel D, Lagarrigue F, Dupuis-Coronas S, Chicanne G, Leslie N, Gaits-Iacovoni F, Payrastre B, Tronchere H. Biochem Biophys Res Commun. 2009;387:127–131. doi: 10.1016/j.bbrc.2009.06.139. [DOI] [PubMed] [Google Scholar]