SUMMARY
Clostridium difficile infection (CDI) is the leading cause of antimicrobial and health care-associated diarrhea in humans, presenting a significant burden to global health care systems. In the last 2 decades, PCR- and sequence-based techniques, particularly whole-genome sequencing (WGS), have significantly furthered our knowledge of the genetic diversity, evolution, epidemiology, and pathogenicity of this once enigmatic pathogen. C. difficile is taxonomically distinct from many other well-known clostridia, with a diverse population structure comprising hundreds of strain types spread across at least 6 phylogenetic clades. The C. difficile species is defined by a large diverse pangenome with extreme levels of evolutionary plasticity that has been shaped over long time periods by gene flux and recombination, often between divergent lineages. These evolutionary events are in response to environmental and anthropogenic activities and have led to the rapid emergence and worldwide dissemination of virulent clonal lineages. Moreover, genome analysis of large clinically relevant data sets has improved our understanding of CDI outbreaks, transmission, and recurrence. The epidemiology of CDI has changed dramatically over the last 15 years, and CDI may have a foodborne or zoonotic etiology. The WGS era promises to continue to redefine our view of this significant pathogen.
INTRODUCTION
Clostridium difficile Infection
Clostridium difficile is a spore-forming, Gram-positive, anaerobic bacillus found ubiquitously in the environment and the gastrointestinal tracts of humans and animals. C. difficile is a formidable pathogen and currently the leading cause of antimicrobial and health care-associated infectious diarrhea in humans (1). The incidence and severity of C. difficile infection (CDI) present a significant burden to global health care systems due to increasing costs associated with treatment, infection control, disease recurrence, patient length of hospital stay, and mortality, especially among the elderly (2). A recent report from the Centers for Disease Control and Prevention (CDC) ranks C. difficile as the most important antimicrobial-resistant threat to public health in the United States, with 250,000 infections and 14,000 deaths per year and annual excess medical costs (attributable to the cost of extra bed days and associated treatment) totaling $1 billion (3). Another North American study reports that in 2011 alone, the clinical burden of CDI accounted for almost 500,000 infections and 29,000 associated deaths (4).
Originally named Bacillus difficilis due to difficulties in cultivation in vitro, the bacterium was first described in 1935 as a component of the healthy neonatal intestinal microflora (5). Later, investigators verified its toxigenic potential through toxin studies in guinea pigs (6), and in the 1970s, work by John Bartlett et al. identified C. difficile as the cause of antibiotic-associated pseudomembranous colitis (PMC) (7). CDI is a toxin-mediated disease of the colon, with three or more watery, nonbloody stools per 24-h period being the hallmark of symptomatic illness (8). Clinical characteristics of CDI include abdominal pain, cramps, and fever (9), and extraintestinal manifestations are rare (10). CDI is also associated with leukocytosis, hypoalbuminemia, and high serum creatinine levels (8). Disease severity can vary from mild or self-limiting to severe and, in some instances, fatal sequelae, including PMC, toxic megacolon, bowel perforation, and sepsis (7–9). Asymptomatic carriage of C. difficile is also common in health care settings and may play a role in CDI transmission (11).
There are many risk factors for the development of CDI, including comorbidities, surgical and nonsurgical gastrointestinal procedures, duration of hospital stay, admission to an intensive care unit (ICU), immunocompromised status (particularly oncology and hematology patients), and advanced age (>65 years of age) (12, 13). Antimicrobial exposure is the single most important risk factor for the acquisition of CDI due to the disruption and dysbiosis of endogenous colonic microbiota (colonization resistance), allowing C. difficile to colonize and proliferate (12). Almost all antimicrobials have been implicated, especially those with high gut concentrations and activity against bowel flora to which C. difficile is resistant, including clindamycin, penicillin, ampicillin, amoxicillin, cephalosporins, and, for some strains, fluoroquinolones (14, 15).
C. difficile forms spores that are resistant to desiccation, extremes of temperature, and many chemicals and disinfectants (16, 17). Spores are highly transmissible and responsible for contamination of health care environments, often persisting for long periods of time and contributing to the burden of disease (2, 17, 18).
Current treatment options for CDI include antimicrobial therapy (vancomycin, metronidazole, or fidaxomicin) and restoration of colonic microbiota through fecal microbiota transplantation (FMT) (19–21). Phage therapy and treatment with monoclonal antibodies are also active areas of interest (22, 23). In up to 20% of fulminant colitis cases, surgical intervention (subtotal colectomy, resection, and/or ileostomy) is required (24).
CDI is mediated by the production of two large clostridial toxins (LCTs), TcdA and TcdB, which, following expression, inactivate host cell GTP-binding proteins, resulting in actin disassembly, enterocyte apoptosis, and severe inflammation (25–27). In some strains, a third unrelated binary toxin (cytolethal distending toxin [CDT]) is produced. The exact role of CDT in pathogenesis remains unclear; however, it is thought to be involved in epithelial adhesion (25, 27, 28). Additionally, variations in flagella, sporulation factors, and adhesins are thought to play a role in virulence (27, 29, 30).
An optimal diagnostic strategy for laboratory detection of CDI remains controversial (31). Current guidelines recommend PCR-based methods to detect the toxin-encoding genes tcdA and tcdB, either alone or in conjunction with a toxin detection enzyme immunoassay (EIA) (20, 24, 32). Culture of the bacterium from feces does not differentiate toxigenic from nontoxigenic bacteria or asymptomatic carriers from those with CDI; however, it provides an isolate for epidemiological typing.
Methods for Determining Strain Relatedness
Several typing methods have been used to investigate the epidemiology, genetic diversity, and evolution of C. difficile. Some methods are based on macroanalysis of genome architecture (restriction endonuclease analysis [REA] and pulsed-field gel electrophoresis [PFGE]), while some focus on analysis of single regions within the genome (PCR ribotyping and toxinotyping). Sequence-based methods provide differentiation of strains on a single-nucleotide level and can target multiple loci (multilocus sequence typing [MLST] and multilocus variable-number tandem-repeat analysis [MLVA]) or even the entire length of the bacterial genome (whole-genome sequencing [WGS]) (33, 34). Strain nomenclature is often based on one or more of these schemes, as shown by the designation for epidemic strain C. difficile 027/BI/NAP1, where 027 refers to the PCR ribotype (RT), BI refers to the restriction endonuclease group, and NAP1 refers to the North American pulsotype. The applications, limitations, and future perspectives of these techniques have been extensively described elsewhere (33–38), including an excellent 2013 review by Knetsch and colleagues (33).
For C. difficile, PFGE and PCR ribotyping have been most widely adopted methods in North America and the rest of the world, respectively. In PFGE, chromosomal DNA is digested by restriction endonucleases such as SmaI, and DNA fragments are separated in an agarose medium under a pulsed electric current, producing a strain-specific fingerprint or pulsotype (33). PFGE provides a highly discriminatory method for surveillance of CDI outbreaks and tracking of patient transmission events (33, 34). PCR ribotyping exploits the variations in the intergenic spacer region (ISR) located between the 16S and 23S rRNA genes, resulting in a RT-specific set of amplicons after PCR amplification (39). It is worth noting, however, that the use of the word “intergenic” in this context may be misleading, as for some isolates, numerous tRNA genes have been identified in this region (40). The collation of existing and the assignment of new RTs were the responsibilities of the Public Health Laboratory Service Anaerobe Reference Unit in Cardiff, United Kingdom; however, these responsibilities have now moved to the Health Protection Agency-funded C. difficile Ribotype Network (CDRN) based in Leeds, United Kingdom. Currently, there are >600 RTs in the CDRN database (W. Fawley, personal communication). RT nomenclature is under constant review, and recently, there has been a concerted effort to reconcile conventional (agarose-based) and newer (capillary-based) library data.
Toxinotyping is a restriction fragment length polymorphism (RFLP)-based PCR method for differentiating C. difficile strains on the basis of variability in restriction sites in the 19.6-kb pathogenicity locus (PaLoc) (with respect to a reference strain, VPI 10463, toxinotype 0). Currently, 31 variant toxinotypes have been described (types I to XXXI) (41, 42).
MLST is a robust and accurate typing method for identifying clonal relationships among strains of bacteria. For C. difficile, MLST has discriminatory power comparable to that of PCR ribotyping and provides unambiguous data that are easily shared between laboratories (33). Based on an earlier scheme described by Lemee et al. (43), the scheme developed by Griffiths et al. (44), in which sequence types (STs) are assigned based on allelic variants of seven highly conserved housekeeping genes (adk, atpA, dxr, glyA, recA, sodA, and tpi), has been widely adopted for studying CDI epidemiology (44–47). Furthermore, a large, well-curated database (PubMLST [http://pubmlst.org/]) provides simple and rapid ST assignment and allows submission of novel alleles.
MLVA and in silico typing based on WGS (single nucleotide polymorphism [SNP] typing, MLVA, and MLST) currently offer the highest level of bacterial strain discrimination and are powerful tools for studying transmission events in C. difficile (35) and other important global pathogens, including Staphylococcus aureus (48), Mycobacterium tuberculosis (49), and Escherichia coli (50). In the last decade, with the advent of high-throughput or “next-generation” sequencing (NGS) methods such as the Roche 454 and Illumina methods, the number of sequenced C. difficile genomes has risen sharply. To date, several fully “closed” high-quality genomes have been reported (see the section on the C. difficile genome, below); however, of the thousands of individual genomes currently archived in online depositories such as GenBank and the Sequence Read Archive (SRA), the vast majority remain incomplete or “draft” genomes. While adequate for most studies, these draft genomes present some challenges in determining “complete” genomic content, and whole-genome phylogenetic inferences should be made with this limitation in mind (51).
For C. difficile, the NGS era has contributed to significant advances in a number of key areas, many of which are discussed below. WGS has helped to define the architecture, diversity, conservation, and plasticity of the C. difficile genome; describe the mechanisms and forces influencing the evolution of the C. difficile core and pangenome; and provide a robust global phylogeny, particularly of virulent and epidemic lineages. Moreover, WGS has built upon knowledge from MLST studies; improved our understanding of CDI outbreaks, transmission, and recurrence; and further highlighted the potential for zoonotic transmission of C. difficile.
C. DIFFICILE PHYLOGENOMICS AND STRAIN DIVERSITY
The C. difficile Genome
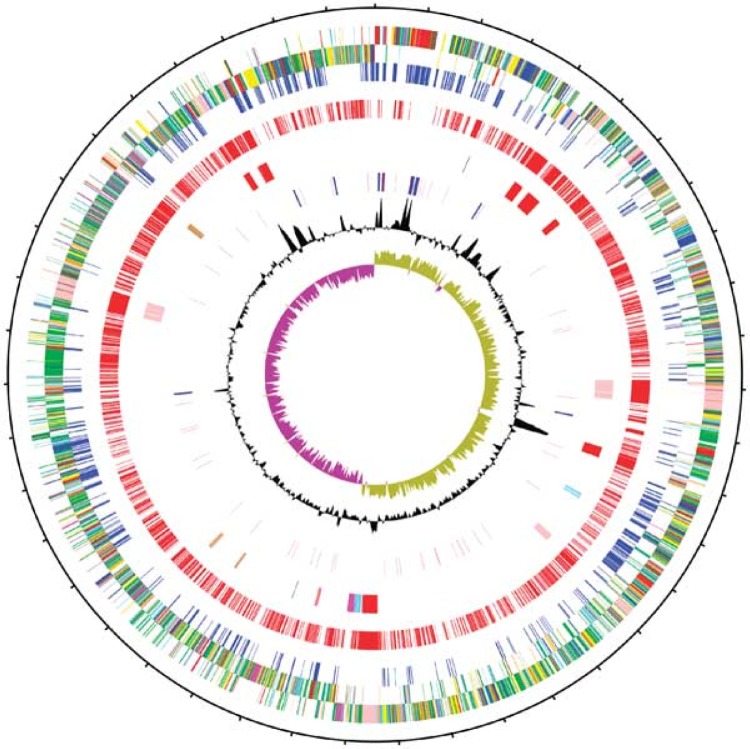
In 2006, Sebaihia and colleagues described the first fully sequenced and annotated closed genome of C. difficile (strain 630; RT012) (52). This virulent, highly transmissible, and multidrug-resistant strain of C. difficile was originally isolated in 1982 from a patient with PMC in Zurich, Switzerland. Sequencing and annotation of strain 630 revealed a large circular chromosome of 4,290,252 bp (4.3 Mb), 3,776 putative protein-coding sequences (CDSs), and a GC content of 29.06% (Fig. 1) (52). A circular plasmid (pCD630) of 7,881 bp containing 11 CDSs was also identified (52). Five years later, Monot and colleagues (53) reannotated the genome of strain 630 by using a combined transcriptomic and proteomic approach to update the putative functions of >500 previously putative or unknown genes. Since this time, several other genomes ranging in size from 4.1 to 4.3 Mbp have been fully sequenced and annotated: CD37 (RT009; isolated in the United States in 1980), M68 (RT017; isolated in Ireland in 2006), CF5 (RT017; isolated in Belgium in 1995), M120 (RT078; isolated in the United Kingdom in 2007), G46 (RT027; isolated in the United Kingdom in 2006) (54), R20291 (RT027; isolated in the United Kingdom in 2006), 196 (RT027; isolated in France in 1985), 2007855 (RT027; isolated in the United States in 2007), and BI1 (RT027; isolated in the United States in 1988) (55–57). Reference genomes such as those of strains 630 and M120 play an important role in the NGS data analysis pipeline. They comprise an unambiguous and contiguous sequence of known nucleotides spanning the entire chromosome and plasmids (if present), therefore providing an extremely high-quality reference for mapping of draft genomes.
FIG 1.
Circular illustration of the 4.3-Mb chromosome of C. difficile strain 630. The concentric circles are as follows (from the outside in): circles 1 and 2, 3,776 putative CDSs (transcribed clockwise and counterclockwise); circle 3, CDSs shared with other sequenced Clostridia (blue); circle 4, CDSs unique to C. difficile (red); circle 5, mobile elements (red/pale red, transposons; pink, prophages; brown, partial prophages/transposons; blue, skin element; magenta, genomic island); circle 6, RNA genes (blue, rRNAs; red, tRNAs; purple, stable RNAs); circles 7 and 8, G+C content/GC deviation (plotted using a 10-kb window). (Reproduced from reference 52 by permission from Macmillan Publishers Ltd.)
As was the case with strain 630, WGS of these C. difficile strains revealed much about the architecture of the C. difficile genome. C. difficile has a highly dynamic and mosaic genome comprising a high proportion (∼11% in strain 630) of mobile genetic elements. These include bacteriophages, group I introns, insertion sequences (IS), sigK intervening (skin) elements, clustered regularly interspersed short palindromic repeat (CRISPR)-cas elements, genomic islands, and transposable and conjugative elements, accompanied by an extensive range of accessory genes (52, 53, 55, 58, 59).
Many of the CDSs identified in the genome of C. difficile are associated with adaptation and proliferation in the gastrointestinal tract (germination, adhesion, and growth) and survival in challenging suboptimal environments (endospore formation) (52, 53). These findings support the view that C. difficile lives within a highly dynamic niche and is able to spend a long time coexisting with its host (52). This is in contrast to the genome of Clostridium botulinum, where many of the genes unique to this species encode proteins associated with rapid killing (cytolysins and neurotoxin) and saprophytic feeding (extracellular proteases and chitinases). Moreover, the genome of C. botulinum is much more stable than that of C. difficile, reflecting the short-lived host association compared to that of C. difficile (60).
Large genomes are typically indicative of a bacterium that is able to adapt to and thrive in multiple, often adverse, environments, as seen with Pseudomonas aeruginosa (genome size, 6.3 Mb) (61). This is also true for C. difficile, with a genome up to 42% larger than those of other closely related clostridial species such as C. bifermentans and C. sticklandii and larger than those of most other Firmicutes (62, 63). This large, complex genome reflects the ability of the bacterium to survive, often for long periods of time, within a diverse range of human, animal, and abiotic environments.
The PaLoc
Encompassing a 19.6-kb region of the chromosome, the PaLoc has received significant attention, as it contains the genes encoding the major virulence factors toxin A and toxin B and thus plays an essential role in the pathogenesis of CDI. The PaLoc is present in all toxigenic strains but absent in nontoxigenic strains, where it is replaced by a 115-bp noncoding and highly conserved region known as the integration site (26, 64, 65). In addition to tcdA and tcdB, which encode toxins A and B, respectively, the PaLoc contains three other genes, tcdR, tcdE, and tcdC, as well as CD630_06620, a putative N-acetylmuramoyl-l-alanine amidase identified during the reannotation of the C. difficile 630 genome (53). While tcdR encodes an RNA polymerase sigma factor, a positive regulator of toxin expression (66), tcdE encodes a protein structurally and functionally similar to holin proteins found in bacteriophages (67), and tcdC is considered to be a negative regulator of toxin expression (68–70). For several years, strains with an aberrant tcdC gene (deletions and premature stop codons) have been linked to hypervirulence, although this has been disputed (71, 72). In addition to tcdC mutations, variations in tcdB (specifically the receptor-binding domain [RBD]) have also been associated with a hypervirulence phenotype (45, 73). These hypervirulence-promoting locus variants are common in certain RTs and have a strikingly congruent association with specific clades (45).
Interestingly, there has been very recent direct evidence that toxin synthesis in C. difficile is modulated by an accessory gene regulator (agr) quorum-signaling system distinct from the PaLoc (74). In C. difficile, the agr locus comprises genes encoding a quorum signal generation pathway (agrB and agrD) and genes encoding a quorum response pathway (agrA and agrC) (74). agr-mediated quorum signaling presents an advantageous mechanism for C. difficile toxin production, as there is a fitness advantage through coordinating information about cell density and synchronizing gene expression on a population level rather than on a single-cell level (74). Notably, significant differences in agr locus content between hypervirulent strains (RT027 and RT017 [agrBDAC]) and nonhypervirulent strains (RT012 [agrBD]) have been described, which may suggest a role for quorum sensing in the evolution of virulent lineages (74, 75).
Ultralow Level of Genome Conservation
C. difficile can be defined by its pangenome, the genetic repertoire of the species or “gene pool.” The pangenome is comprised of a core genome (those genes present in all isolates) and an accessory or adaptive genome (genes absent from one or more strains or unique to a particular strain) (76). Scaria et al. (77), using microarray and WGS data derived from a small but diverse collection of clinical and animal strains, estimated that the C. difficile pangenome is comprised of 9,640 CDSs. This figure is comparable to that for Salmonella enterica (n = 9,966) and higher than those for Staphylococcus aureus (n = 4,221) and Streptococcus pneumoniae (n = 3,934) (78). However, this figure is likely an underestimate based on the narrow geographic area (United States and United Kingdom) from which most of the strains originated. Furthermore, as discussed below, C. difficile possesses an “open” genome with extreme levels of plasticity, with access to and frequent exchange with multiple host environments and bacterial gene pools. Consequently, as more C. difficile strains from divergent lineages and diverse animal and environmental sources are sequenced, this estimate will likely increase.
Estimates of the size of the core genome of C. difficile are many orders of magnitude lower (∼600 to 3,000 CDSs) (57, 77, 79). Stabler et al. (73) and Janvilisri et al. (80) showed that much of the core CDSs in C. difficile encode proteins involved in essential cellular processes such as metabolism, biosynthesis, DNA replication, transport, and cell division as well as processes associated with pathogenicity (colonization, adhesion, motility, and antibiotic resistance), indicating their essential role. In addition, many of the core genes show divergent sequences that may indicate host adaption and specificity (80). As with the pangenome, the size of the core genome is influenced by the strains analyzed. Thus, it is probable that as strains belonging to divergent phylogenetic lineages are sequenced, the size of the core genome will likely decrease further.
The contrasting estimates of the sizes of the pan- and core genomes of C. difficile highlight the ultralow levels of genome conservation in this species. To date, three studies have used comparative genomic hybridization (CGH) to measure the size of the genome in C. difficile, with estimates that the amount of shared core genome of C. difficile might be as low as 16%, lower than that of any bacterial species described to date (52, 73, 80). An ultralow level of conservation is rare in bacteria, even in species considered to have high levels of genetic variability, e.g., Campylobacter jejuni (59.2%), Helicobacter pylori (58.5%), Streptococcus pneumoniae (46.5%), and E. coli (∼40.0%) (81–83), and is more typical for phylogenetic distances between genera within a family rather than strains within a species. Such large phylogenetic distances between C. difficile lineages threaten the very definition of C. difficile as a species and support recently suggested taxonomic revisions (84–87).
Taxonomy
The Clostridium group represents an ancient prokaryotic lineage, estimated to have diverged from the bacterial domain 2.34 Ga (billion years) ago, earlier than the Escherichia, Campylobacter, and Helicobacter groups (ca. 1.37 to 1.89 Ga) and around the time when concentrations of molecular oxygen in the atmosphere began to increase (88). Described under the phylum Firmicutes, the class Clostridia incorporates a group of obligately anaerobic, endospore-forming (and thus resistant to desiccation), Gram-positive organisms. Classification of Clostridia was initially made based on these phenotypic characteristics; however, 16S rRNA sequencing showed that the Clostridia were phylogenetically incoherent and required significant taxonomic revision (86). According to the scheme of Collins et al. (86), C. difficile belongs to cluster XI, which represents a taxonomically heterogeneous group more closely related to the non-spore-forming species Peptostreptococcus anaerobius and Eubacterium tenue than the type species of the Clostridia, Clostridium butyricum. Notably, by this scheme, C. difficile does not cluster with many other familiar clostridial species, such as C. botulinum, C. tetani, and C. perfringens, all of which can be pathogenic for humans and animals via toxin-mediated virulence mechanisms.
Bergey's Manual of Systematic Bacteriology (87) now places C. difficile in the Peptostreptococcaceae along with a number of other Clostridium species, including C. bifermentans, C. glycolicum, C. bartlettii, C. sordellii, and C. sticklandii, as well as members of the genera Eubacterium, Peptostreptococcus, Sporacetigenium, and Filifactor. In 2013, further taxonomic revision of the Clostridia was called for, with a proposed name change from Clostridium to Peptoclostridium (85) for C. difficile. Despite NCBI taxonomy adopting this name change, it appears unlikely that the C. difficile community will follow.
Phylogenetics and Molecular Epidemiology
C. difficile has a clonal population structure. In 2004, Lemee et al. (43) conducted the first analysis of C. difficile isolates using MLST. Those authors identified three distinct phylogenetic lineages but noted that geographical affiliation, host species, or a particular phenotype (e.g., strains causing severe disease) was not associated with any particular lineage. In 2006, Stabler and colleagues (73), using DNA microarrays, a Bayesian evolutionary model, and a more diverse C. difficile population, identified four phylogenetic lineages or clades. The majority of STs clustered into a single heterogeneous lineage, but the remaining three represented emergent virulent lineages: RT017 (ST-37), RT027 (ST-1), and a more distantly related group, RT078 (ST-11) (73). Later, the same authors undertook a phylogenetic analysis of C. difficile WGS data and confirmed the lineage topology known at that time (four clades) and, in addition, provided a more in-depth and robust phylogeny (57). Through evaluation of WGS data from six strains representing these four clades, calculations of the evolutionary distance between the clades were made. Based on these and other data, the last common ancestor was estimated to have emerged somewhere between 1.1 and 85 Ma (million years) ago (57). However, it is worth noting that methods for dating bacteria are imperfect and based on models and assumptions about evolutionary rates (in this case divergence of orthologous genes between C. difficile and C. tetani), which may not be entirely accurate.
Using a different MLST scheme, Griffiths and colleagues (44) identified an additional lineage containing toxigenic RT023 (ST22), bringing the total number of clades to five. This population structure has since been confirmed by other studies, including some using WGS (45, 46, 89), and has been summarized in a recent review by Janezic and Rupnik (90). MLST clade 1 represents a highly heterogeneous cluster of toxigenic and nontoxigenic STs (numbering over 100) and RTs, including many of clinical significance, such as RT014 (tcdA positive [A+], tcdB positive [B+], and cdtA and cdtB negative [CDT−]; STs 2, 14, and 49), RT002 (A+ B+ CDT−; ST8), and RT018 (A+ B+ CDT−; ST17), all of which are RTs consistently among the most frequently recovered from patients with CDI (91–94). Clade 2 contains hypervirulent RT027 (A+ B+ CDT+; ST1) and several other RTs of clinical importance, including RT244 (A+ B+ CDT+; ST41) and RT176 (A+ B+ CDT+; ST1) (95, 96). To date, clade 3 has received little interest in comparison, but this clade contains RT023 (A+ B+ CDT+; ST5 and ST22), which has been isolated from humans in Europe (92). Clade 4 contains RT017 (ST37), which has a variant toxin profile (A− B+ CDT−) and is often clindamycin and fluoroquinolone resistant. Despite the absence of toxin A and binary toxin expression, RT017 causes widespread disease; has been associated with outbreaks in Europe (97, 98), North America (99), and Argentina (100); and is responsible for much of the CDI burden in Asia (94). Clade 5, containing RT078 (ST11), has been the focus of much interest because of its significant divergence from the other clades and its association with animals, particularly livestock (101). However, recent MLST and WGS studies have shown that clade 5 is more heterogeneous than first thought, including not only RT078 but also numerous RTs (RT033, RT045, RT066, RT126, RT127, RT237, RT280, RT281, and RT288) from a diverse collection of clinical, animal, and food sources worldwide (46, 102, 103).
Some clade 5 strains show an atypical arrangement of the PaLoc, specifically the genes for LCTs A and B. RT237, which has been recovered from pigs and humans in Australia, is positive for toxin B but negative for toxin A (A− B+) while also possessing binary toxin (CDT+) (104). In RT033 and RT288, CDT is present; however, the entire tcdB gene and the majority of tcdA are absent (41, 105). RT033 was recently isolated from a cluster of six epidemiologically unrelated cases of CDI in France, suggesting that despite the absence of LCTs, there remains a pathogenic potential (105). Additionally, the prevalence and clinical burden of these two LCT-negative, CDT-positive RTs are likely to be underestimated. A recent study by Androga et al. (106) found that current molecular diagnostic assays (which rely on amplification of the toxin A and B genes) fail to detect C. difficile RT033.
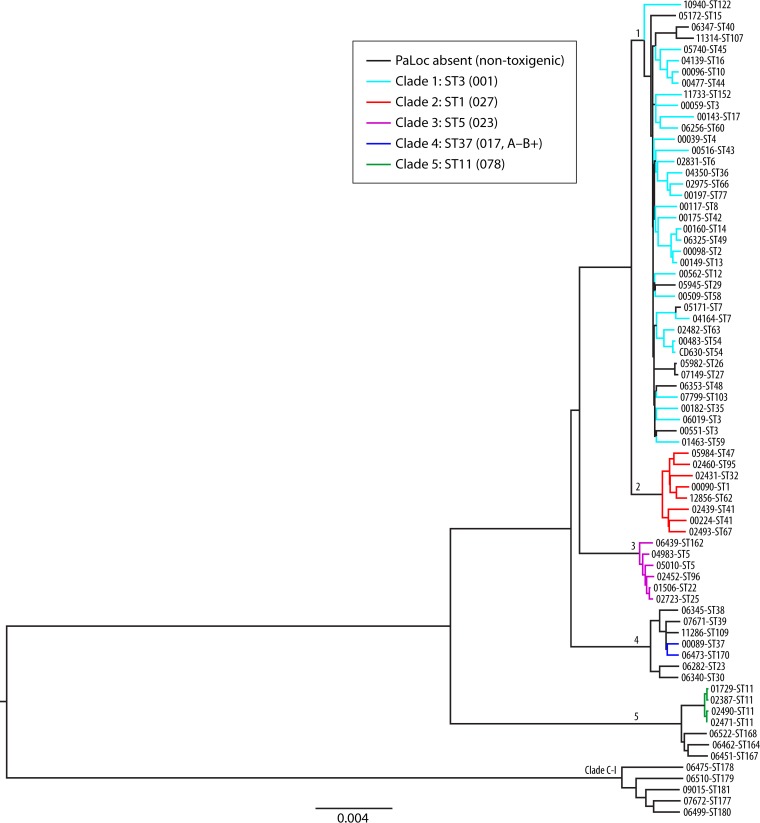
More recently, there have been reports of two novel lineages. One lineage, designated clade 6, contained a single ST (ST122; RT131) and appeared as a sister lineage to clades 1 and 2 (47). Recently, its status has been questioned, as in another study, ST-122 was not an outlier but rather part of heterogeneous clade 1 or possibly a hybrid of clades 1 and 2 (107). The other novel lineage, designated C-I, as it was reminiscent of the cryptic clades of E. coli (107), was highly divergent, entirely nontoxigenic, and potentially a new species or subspecies of C. difficile (107). Figure 2 shows the currently described C. difficile population structure of six clades (107).
FIG 2.
Phylogenetic tree showing representatives of six currently described C. difficile clades and the relationship between toxigenic and nontoxigenic isolates. A maximum likelihood tree was generated from the alignment of 1,426 core genes of 73 C. difficile isolates. Isolates represented extremes of clinical severity, geographic diversity, and toxigenic status. Clades are indicated by their designated number. Nontoxigenic isolates are indicated by black branches. Toxigenic isolates are indicated by branches colored according to clade. The ST and RT (in parentheses) of a well-characterized representative of each clade are indicated. (Reproduced from reference 107 by permission of the Society for Molecular Biology and Evolution.)
MLST demonstrates the high level of genetic diversity within the species and shows that RTs are clade specific (45). Recently, however, the concept of clonal C. difficile lineages has been extended to include more than just RT affiliations. Kurka et al. (108) examined the genomes of strains belonging to 21 different RTs from different MLST lineages, looking at differences in a number of conserved genes, including rpoA and gyrB, encoding RNA polymerase A and gyrase B, respectively. Those authors found that strains with the same sequence deviations in these and many other genes clustered into groups which mirrored the RT diversity inferred by MLST; e.g., all strains of RT126, RT127, and RT033 (clade 5) clustered together. This is interesting, as it shows that RT is indicative of not only differences in the amplified 16S-23S rRNA gene ISR but also specific differences in the nucleotide sequences of a number of conserved genes. Similarly, gene variations in hypervirulent RT027 and RT078 are MLST lineage specific (109).
MECHANISMS SHAPING DIVERSITY AND EVOLUTION IN C. DIFFICILE
The remarkable genetic diversity in the C. difficile genome mirrors the wide variety of phenotypes, ecological adaptations, and physiological versatilities seen in this species. This diversity is a result of the acquisition of foreign DNA coding for novel phenotypes and has been shaped over long and short time periods by mechanisms of lateral gene flux, such as homologous recombination and horizontal gene transfer (HGT) (45, 57, 107, 110).
Transposable Elements
Of the numerous genetic elements found in the genome of C. difficile, many are transposable and can change their position within the genome. Some C. difficile transposons (Tns) are mobilizable, meaning that they rely on complex host-mediated mechanisms for conjugation and full mobility (111, 112). Some elements are self-transmissible and are known as conjugative transposons (CTns) or integrative and conjugative elements (ICEs). CTns are capable of excision, transfer, and integration into the genome of C. difficile and species of other genera through the expression of integrase (int), excisionase (xis), and, in some elements, a site-specific recombinase (tndX) (113). Consequently, there is the potential for various bacteria to acquire new DNA from the highly diverse C. difficile pangenome as well as allowing C. difficile to acquire genes from the intestinal metagenome (114). Many C. difficile mobilizable transposons and CTns have been found throughout different lineages, including those that are separated by large phylogenetic distances, e.g., clades 1 to 4 and clade 5, suggesting insertion of the element prior to clade divergence (45, 57).
The introduction of these elements into the C. difficile genome leads to numerous heritable changes not only in the acquisition of new, possibly advantageous genes but also, as in the case of insertion within an open reading frame (ORF), in gene disruption and phenotypic alteration (112, 114). Dissemination of these elements and their accessory genes by lateral transfer has significantly contributed to the genetic diversity seen in C. difficile and possibly contributed to the success of C. difficile as an opportunistic pathogen (52, 73, 112, 114, 115).
Exposure to antimicrobials has a significant role in the pathogenesis of CDI, and resistance should be considered a virulence factor, as it is in other nosocomial infections such as those caused by extended-spectrum-β-lactamase- and carbapenemase-producing Enterobacteriaceae (12, 116). Unlike these and other related pathogens, resistance to antimicrobials in C. difficile is unusual in that it is mediated predominantly by Tns as opposed to plasmids (52, 112, 114). Through traditional PCR- and, more recently, WGS-based studies, numerous Tns associated with antimicrobial resistance in C. difficile have been described, including Tn916 {tetracycline resistance [tet(M)]} (117), Tn4453a/b (chloramphenicol resistance) (118), Tn5397 [tet(M)] (119), TnB1230 [tet(W)] (120), and Tn5398. Tn5398, first described in 1995, contains two copies of erm(B), which encodes a 23S rRNA methylase conferring the MLS (macrolide-lincosamide-streptogramin B) phenotype, the most common resistance type in C. difficile (112, 121). Tn6215 also carrying erm(B) was reported to be transferred between C. difficile strains by bacteriophage-mediated transduction (115). Tn6218 is a stably integrated transposon with a number of described variants occupying different PaLoc-independent chromosome locations in different RTs (107). In addition to erm(B), some variants of Tn6218 also possess the cfr gene. cfr encodes a methyltransferase that alters binding sites within the bacterial ribosome, resulting in resistance to clindamycin, florfenicol, and chloramphenicol (122). The cfr gene has been found previously in many Gram-positive and -negative bacteria (122), particularly in plasmids of Staphylococcus spp. (123), but this was the first description of it in C. difficile (107). Recently, Marin et al. (124) reported reduced susceptibility to linezolid (MIC, 6 to 16 mg/liter) in 9/891 clinical isolates of C. difficile. These 9 strains belonged to RT001, RT017, and RT078, and they all possessed the cfr gene, which showed 100% sequence identity with a fragment of Tn6218 (124).
Several other putative transposons have also been described. Analysis of the genome of C. difficile strain 630 identified six putative CTns (CTn1, CTn2, CTn4, CTn5, CTn6, and Ctn7) associated with genes encoding efflux pumps and ATP-binding cassette (ABC) transporters, which confer resistance to tetracycline, chloramphenicol, and erythromycin (52, 53). One of these putative CTns, Tn6164, contains aminoglycoside and tetracycline resistance genes. In reference strain M120, Tn6164 is located in a novel 106-kb genetic island that is made up largely of mobile elements from nonclostridial species such as Streptococcus pneumoniae, Enterococcus faecalis, and Thermoanaerobacter sp. (125). The presence of this element was found to vary in porcine and human strains of RT078, and this element was absent from other RTs. Notably, although patient numbers were small, patients infected with strains containing Tn6164 had higher rates of mortality (29% versus 3%) (125). Another CTn of interest is Tn6194, the most common erm(B)-containing element in clinical isolates in European hospitals (126). Tn6194 has been associated with epidemic RT027, particularly in strains belonging to a lineage isolated in the United States and Asia (57). In addition to erm(B), Tn6194 carries genes proposed to function as recombinases and integrases, suggesting the potential to travel laterally between C. difficile strains (intraspecies transfer) (57, 127). This potential has been confirmed by the successful excision and transfer of Tn6194 in strain 630 (114). More recently, Tn6194 has been shown to be capable of inter- and intraspecies transfer. Wasels and colleagues (128) transferred a variant of Tn6194 (identified in the genome of a RT001 strain) into the genomes of strains with two different C. difficile RTs as well as a strain of E. faecalis (128).
630Δerm is a macrolide-susceptible C. difficile mutant derived from strain 630, which has often been used as a tractable strain for genetic analysis of C. difficile. In a recent study by van Eijk et al. (129), WGS revealed that CTn5 is one of several genetic features that might explain the underlying phenotypic differences between 630 and its erythromycin-sensitive derivative. Specifically, it was shown that in 630, CTn5 resides within an adhesin (CD1844); however, in 630Δerm, the same transposon can be found interrupting the methyltransferase gene rumA (129).
Bacteriophages
Another important component of the C. difficile mobilome is bacteriophages (phages). Phages have coevolved with C. difficile over very long periods of time, and phage infection is an inherent part of the natural history and biology of C. difficile (22, 130). Phages are capable of mediating HGT via a process known as transduction, whereby host DNA is packaged into the head particle of a phage and subsequently inserted into the genome of a recipient cell. Consequently, the acquisition of phages by and their loss from the C. difficile genome are significant genetic events that have impacted host evolution (22, 130).
C. difficile carries a diverse collection of phages, including numerous members of the Siphoviridae and Myoviridae families, such as ϕC2, ϕMMP04, ϕCD119, ϕCDHM1, ϕCD38-2, and ϕCD27, ranging in size from 31 to 56 kbp with a GC content not dissimilar to that of the C. difficile genome (28 to 30%) (52, 57, 73, 113, 131–133). Despite the absence of proven virulence factors in C. difficile phage genomes, there is increasing evidence that phages may play a role in C. difficile pathogenesis. All sequenced phages identified in the genome of C. difficile have contained putative integrase genes, suggesting that they have access to the lysogenic life-style (113). Recently, phage ϕC2, which is common to clinical strains of C. difficile, has been shown to mediate the transfer of Tn6215 containing erm(B) between two laboratory strains of C. difficile (115). Viral DNA identical to that of phages (ϕMMP02 and ϕMMP04) found within C. difficile has been recovered from stool samples obtained from patients with CDI, indicating that these phages are induced during infection. Furthermore, the in vitro induction of these phages was increased significantly in the presence of fluoroquinolone antimicrobials, demonstrating how the established CDI risk factor of antimicrobial exposure may influence phage biology and may ultimately promote phage-mediated HGT (113). Notably, Hargreaves et al. (134) describe the presence of agr homologues in the genome of phage ϕCDHM1. In C. difficile, the agr locus is responsible for modulating the expression of 75 genes associated with various cellular functions, such as flagellum assembly and toxin synthesis, particularly during late exponential growth (113). It is hypothesized that the expression of these agr-like genes during phage lysogeny may influence gene expression in the host bacterium through a quorum-signaling mechanism (113, 134). Moreover, phages ϕCD119, ϕCD38-2, and ϕCD27 have been shown to modulate toxin production in C. difficile; however, the genetic basis of these interactions is not yet understood (113, 134).
Few studies to date have investigated the host range of C. difficile phages. Some phages have been shown to infect strains of multiple RTs, and phages have been recovered from human, animal, and environmental populations of C. difficile (131, 132). As a result of the coevolution of phages and their hosts, many species of bacteria, including C. difficile, have developed ways to resist infection. In C. difficile, it was recently shown that strain specificity for phages and the host's ability to resist infection are likely defined by the number, distribution, and diversity of CRISPRs (135). Often associated with cas proteins, the CRISPR-cas system utilizes a series of phage-specific spacers (CRISPR array) to identify and degrade spacer homologues found in phage DNA, a mechanism reminiscent of RNA interference (RNAi) in eukaryotes and hypothesized to be a putative bacterial adaptive immunity system (135, 136). Hargreaves and colleagues (135) recently showed that some phages can evade CRISPRs through polymorphisms in spacer regions. Remarkably, those authors also showed that some C. difficile prophages possess CRISPR arrays of their own, which, if fully functional, present a mechanism that can influence infection by other phages. Given the role of phages in mediating HGT, the interaction between C. difficile CRISPR arrays and phages has undoubtedly impacted the evolution of C. difficile, particularly the extent of HGT.
Homologous Recombination
Homologous recombination is a powerful driver for shaping genetic diversity in a wide variety of bacteria and archaea, ranging from commensal opportunistic pathogens to free-living terrestrial and marine extremophiles (110). The ratio of the nucleotide substitution rate as a result of recombination to that as a result of mutation (r/m) is a measure of the rate of homologous recombination in sequence diversification in bacteria, which varies considerably among species (110). For C. difficile, this rate has been estimated to be 0.2 (110) or slightly higher (0.63 to 1.13) (57). These rates are low compared to those of other gut pathogens such as Vibrio parahaemolyticus (39.8), S. enterica (30.2), and H. pylori (13.6) but comparable to those of other Firmicutes such as E. faecalis (0.6) and S. aureus (0.1) (110). For C. difficile, the r/m is perhaps an underestimation resulting from the geographical segregation of global C. difficile populations, and as many studies have now illustrated, homologous recombination has played a very significant role in shaping the evolution of genes associated with gastrointestinal adaptation and virulence potential in C. difficile.
Many C. difficile genes and associated operons show a mosaic structure that could have arisen only by homologous recombination. In recent years, much attention has been paid to investigating the evolution and phylogeny of these areas of the genome, particularly the PaLoc. Recently, Brouwer et al. (137) demonstrated that the PaLoc is capable of being transferred between C. difficile strains by a conjugation-like mechanism. Those authors were able to demonstrate the transfer of the PaLoc from a toxigenic C. difficile strain (630Δerm) to three nontoxigenic strains (RT009 [CD37], RT138, and RT140). Analysis by a cytotoxic assay revealed that the resulting transconjugants produced toxin B at levels similar to that of the donor strain (137). Analysis of the transconjugants showed that the PaLoc was transferred on variably sized DNA fragments (range, 66 to 272 kbp) and was not contained within an obvious mobile element. Those authors noted that in 630Δerm strains, the regions immediately upstream and downstream of the PaLoc were homologous to regions of chromosomal DNA in nontoxigenic strain CD37, thus facilitating recombination and integration of the PaLoc within this region (137). Such chromosomal transfer of the PaLoc is reminiscent of the high-frequency recombination (Hfr) of bacterial chromosomes mediated by CTns and followed by homologous recombination, as seen in bacteria such as Vibrio cholerae (138) and Bacteroides sp. (139). CTn1, CTn2, and Tn5397 are possible candidates for Hfr-mediated transfer, as they are in close proximity to the PaLoc and are transfer proficient (137). Evidence of PaLoc transfer between strains by this mechanism is thought to be occurring in wild populations and is a key driver of evolution in the C. difficile genome (45, 57). Furthermore, there are elements analogous to the C. difficile PaLoc in other closely related species, notably the tcdA- and tcdB-related toxin genes in C. novyi, C. perfringens, and C. sordellii (140), which suggests that the PaLoc in C. difficile may have arisen by interspecies recombination. When present, the PaLoc is always found at the same chromosomal location (45, 64); however, the presence and composition of the PaLoc vary between isolates and in some instances even among isolates of the same ST (107), again reflecting the genetic diversity within the species.
Recently, the evolutionary history of the PaLoc has been reconstructed (107). Dingle and colleagues compared the core genomes and PaLoc phylogenies of >1,600 toxigenic and nontoxigenic isolates from the United Kingdom and Australia. By using a subset of these isolates with mixed toxigenic statuses representing all previously described clades, the distribution of the PaLoc among the C. difficile population was assessed. Interestingly, the resulting phylogeny (which was based on a maximum likelihood alignment of 1,426 concatenated “core” genes) showed a highly divergent lineage comprised only of nontoxigenic strains (C-I), while the remaining nontoxigenic strains were distributed alongside toxigenic isolates in several clades (107) (Fig. 2).
Remarkably, further analysis of this data set identified 26 independent events of PaLoc acquisition, exchange, and loss, the most recent being ∼30 years ago in clade 1 (107). The overall PaLoc phylogeny showed numerous clade-specific acquisitions, many occurring after clades had diverged. For the most recent instances of PaLoc exchange, several clade-specific homologous recombination events involved very long chromosomal fragments (up to 232 kb). Such large-scale recombination in the C. difficile genome has been reported previously (57). In that instance, several large regions of SNPs were observed throughout the core genome, suggesting recent exchange between RTs, some of which were separated by large phylogenetic distances. Such findings are significant since they indicate that homologous recombination is a key driver of C. difficile (and PaLoc) evolution and, thus, the virulence potential of C. difficile (107).
The 16S-23S ISRs of C. difficile are highly variable, and differences in ISRs define the PCR ribotype (39, 141). Recently, it was proposed that both inter- and intrastrain recombination events have influenced the evolution of, and account for, the heterogeneity and mosaicism seen in the ISR (40). Similar recombination events have been described for rRNA operons in other bacterial species, including E. coli, Haemophilus parainfluenzae, and Vibrio cholerae (40, 142).
Another example of homologous recombination driving genetic diversity can be seen in the C. difficile S layer, a paracrystalline immunodominant cell surface antigen that is the basis of S-layer typing and a component of serological typing in C. difficile (143). The S layer forms an important interface between the bacterium and its host. It is thought that the S layer has a central role in adaptation to life within the gastrointestinal tract and evolves in response host immunological selection (144). The S layer is made up of a number of S-layer proteins (SLPs), principally SlpA. SlpA is encoded by the slpA gene located within a 36.6-kb cell wall protein (cwp) gene cluster. In addition to slpA, the genes secA2 (encoding a secretory protein) and cwp66 (encoding an adhesin) comprise a genetically variable 10-kb cassette (89). Recently, through an analysis of genome sequence data, the extent of cwp cluster diversity has been determined, at the same time providing clues about the evolution of these important loci (89). Analysis of the nucleotide sequences of the 10-kb slpA-cwp66-secA2 cassettes from 58 genetically diverse C. difficile strains revealed 12 distinct stable variants spread across five phylogenetic clades. These findings suggest that frequent and independent horizontal transfer of the cwp cluster has taken place throughout the C. difficile population, a process referred to by the authors of that study as S-layer switching (89). Adding to the diversity in this locus is the finding of a novel cassette in three of the five clades that appeared to encode components of a putative S-layer-glycosylating cluster, the first such finding for a Clostridium species (89).
Forces of Selection
Clonal diversification is influenced by the process of natural selection, whereby synonymous nucleotide changes in CDSs evolve under a neutral model of selection, while mutations that provide a reproductive advantage or “fitness” are fixed in a bacterial lineage by positive or Darwinian selection, and deleterious mutations in the genome are subject to purifying or negative selection (145). To date, a single study by He and colleagues (57) investigated the selective forces acting on the C. difficile genome. To do so, those authors used a robust codon-based substitution model of molecular evolution to analyze CDSs from the nonrepetitive core genomes of 9 C. difficile isolates representing divergent clonal lineages (MLST clades 1 to 5). The calculation of the relative ratio (ω = dN/dS) of nonsynonymous substitutions (dN) and synonymous substitutions (dS) in these core CDSs allowed for inference of signatures of selection. Between highly divergent lineages such as clade 5 (strain M120) and clades 1 to 4 (including strains BI, 630, CD196, CF5, and M68), there was evidence of strong purifying selection (mean ω = 0.08). This paucity of dN in the core genome of divergent strain M120 suggests a long divergence time and further supports its status as an ancient lineage. In contrast, between recently diverged clones (strains representing clades 1 to 4), the value of ω was close to 1, indicating enrichment of dN in the core genome of these lineages. While this may suggest that these core CDSs were under neutral selection, it is probable that purifying selection is somewhat delayed in these recently divergent lineages due to insufficient evolutionary time passing for purging of nonsynonymous substitutions (57). Those authors also noted that selection was not homogenous in the core genome, with 12 CDSs under positive selection. These CDSs encode proteins associated with the bacterial cell surface, membrane, and response regulators and likely reflect the influence of host immune selection on the C. difficile genome (57). It is worth pointing out that this analysis was restricted to the core nonrepetitive genome of a select group of C. difficile clones. It is highly likely that the accessory genome and recombinant/repetitive parts of the core genome that are heavily enriched with SNPs (40, 45, 57, 89, 107, 146, 147) are under various degrees of purifying and Darwinian selection.
THE COMPLEX AND DYNAMIC EPIDEMIOLOGY OF CDI
Evolutionary History of Epidemic Lineage RT027
In the last 2 decades, RT027 (toxinotype III; ST-1 [MLST clade 2]; BI/NAP1; A+ B+ CDT+) has emerged as a major pathogen of humans that has been associated with large, highly publicized outbreaks of CDI, initially in North America and later in Europe (148–150). One notable example was an outbreak of CDI at the Stoke Mandeville hospital in the United Kingdom. In a 3-year period spanning April 2003 to March 2006, 498 patients were diagnosed with CDI while admitted to the hospital, 127 of whom died (151). RT027 possesses a number of attributes that result in a hypervirulent phenotype, including increased production of LCTs (152), the presence of binary toxin (28), higher sporulation rates (153, 154), aberrant forms of tcdC (69), production of toxin B variants with an enhanced spectrum of cytotoxicity (155), and mutations in the DNA gyrase resulting in fluoroquinolone resistance (FQR) (156). Consequently, patients with RT027 infection showed a poor response to treatment and a marked increase in morbidity and mortality (148, 149).
The emergence and dissemination of RT027 brought about a massive change in the global molecular epidemiology of C. difficile. This was likely a result of the culmination of a number of events related to epidemiological, host, and pathogen factors, in particular the selective pressure applied by the extensive use of fluoroquinolone antimicrobials in health care settings as well as human travel (157, 158). WGS has played a significant role in the underlying genetic reasons behind this change. In 2009, Stabler and colleagues (56) undertook a three-way genomic comparison of a nonepidemic “historic” (1985) RT027 C. difficile strain (CD196), a recent epidemic and hypervirulent RT027 strain (R20291), and an RT012 strain (630), the genome of which had been reported previously. Those authors aimed to relate genetic differences in the genomes of these strains to phenotypic differences in antibiotic resistance, toxicity, survival, and motility. The study identified five large genetic regions present within the recent RT027 strain but absent from the preepidemic CD196 counterpart, including transcriptional regulators, a unique phage island, and a two-component regulatory system (56). Those authors looked for the presence of these genetic markers in a larger set of C. difficile RT027 genomes and found that many were acquired very recently, potentially explaining the genetic basis for the emergence of RT027 and its successful dissemination (56). In addition to the acquisition of new genes, numerous point mutations and nucleotide inversions have been identified within or upstream of putative coding regions in epidemic strains of RT027, which likely result in changes in gene functionality and phenotype (59).
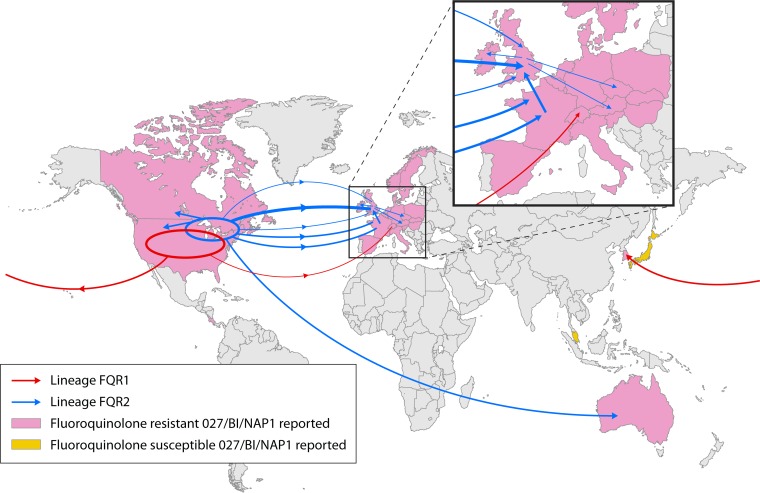
In 2013, He et al. (127) sequenced the genomes of 151 C. difficile RT027 strains collected from 1985 to 2010 with the aim of accurately inferring the population structure. Those authors demonstrated that RT027 acquired fluoroquinolone resistance independently on two separate occasions, resulting in two distinct epidemic lineages (FQR1 and FQR2) with different patterns of global spread (Fig. 3). The FQR1 lineage was thought to have originated in Pittsburgh, PA (earliest isolate dated 2001) and contained epidemic strains associated with severe outbreaks throughout the United States and later sporadic cases in South Korea and Switzerland. The majority of epidemic strains belonged to the FQR2 lineage. FQR2 showed a notable star-like topology in North America, suggestive of a rapid population expansion, likely from a single progenitor clone. Transcontinental dissemination epidemic of FQR2 occurred on no fewer than four occasions in Europe and once in Australia (Fig. 3). The separate acquisitions of FQR and a novel conjugative transposon (CTn5-like, Tn6194) common to both lineages were thought to be the key genetic changes responsible for the rapid emergence and subsequent successful worldwide dissemination of this lineage (127).
FIG 3.
Transcontinental dissemination of epidemic RT027. Shown is the global spread (arrows) of lineages FQR1 and FQR2 inferred from phylogeographic analysis. The width of the arrow is approximately proportional to the number of descendants from each sublineage. The inset shows an enlarged view of transmission in Europe. (Reproduced from reference 127 by permission from Macmillan Publishers Ltd.)
The RT027 lineage is more variable than first thought. Previously presumed to be RT027-REA type BI, three novel and clinically relevant RTs (RT176 [BI-6], RT198 [BI-11], and RT244 [BI-14]), first isolated in the United States between 2001 and 2004, appear to have emerged from the RT027 lineage (96). Of these emergent RTs, RT244 has generated much recent interest, specifically in its pathogenic potential and community acquisition. RT244 infection is associated with a higher mortality rate, and patients with RT244 infection are more likely to develop severe disease and hypoalbuminemia and to have renal impairment (95, 159). WGS of a RT244 strain isolated from a patient who died of severe CDI in Australia revealed a variant toxin B resulting in an enhanced cytopathic effect in vitro (95). In another recent study by Eyre et al. (160), 15 outbreak isolates of RT244 from across Australia were sequenced. All strains were genetically highly related (within 16 single-nucleotide variants [SNVs] of each other), and isolates from a cluster of seven cases from three states differed by just 4 SNVs. However, despite this high degree of genetic similarity, no geographic clustering could be identified, suggesting a single source, possibly in the food chain. Furthermore, these outbreak strains were found to be highly related to a strain isolated from a patient with CDI in the United Kingdom who had recently returned from Australia (160). These findings highlight both the pathological and dissemination potential of RT244 and emphasize the need for ongoing surveillance of strains of this lineage and other newly emergent RTs. The study by Eyre et al. (160) also provided a novel insight into the evolution of this RT. Initially thought to be a relatively recent evolutionary event (96), RT244 and RT027 lineages actually share a relatively ancient common ancestor with current outbreak strains CD196 (RT027) and MDU-064e (RT244), separated by 12,026 SNVs and many hundreds/thousands of years of evolution (160).
Microevolution and Transmission in the Hospital Environment
CDI has traditionally been considered hospital acquired (161, 162), and it was a widely held assumption that much of the C. difficile transmission in hospitals occurs horizontally between symptomatic patients (162, 163). In the past, examination of the molecular epidemiology of CDI outbreaks by MLVA, MLST, and PCR ribotyping has supported this view. However, while MLVA is useful in outbreak investigations, MLST and PCR ribotyping are not sufficiently discriminatory to distinguish between strains or investigate patterns on an ultrafine scale (35, 36, 164, 165). WGS and estimates of the C. difficile molecular clock (within-host mutation rate) have begun to reshape C. difficile surveillance and outbreak investigations (36). Didelot et al. (166) estimated the C. difficile evolutionary rate to be 3.2 × 10−7 mutations per nucleotide per year (95% confidence interval [CI], 1.3 × 10−7 to 5.3 × 10−7 mutations per nucleotide per year), equating to ∼1.4 mutations per genome per year. This estimate of the within-host mutation rate is based on the application of a complex evolutionary model based on coalescent theory to the genomes of serially isolated strains from 91 cases of CDI. Those authors analyzed a total of 486 C. difficile genomes obtained from CDI cases arising in Oxfordshire, United Kingdom, during 2006 to 2010. By combining this estimate with meaningful epidemiological data (hospital admission and patient ward movement), those authors were able to generate a genealogical timeline for pairs of genomes with similar STs and identify plausible transmission events. As expected, these events were all highly associated with pairs of patients sharing the same space and time in hospital. Surprisingly, there was a large proportion of genome pairs, isolated within 30 days of each other and matched by traditional typing, e.g., MLST or PCR ribotyping, that were too distantly related to be direct transmissions. This was an important finding and suggested that transmission between symptomatic patients in hospitals contributed far less to the overall rate of infection than first thought (166).
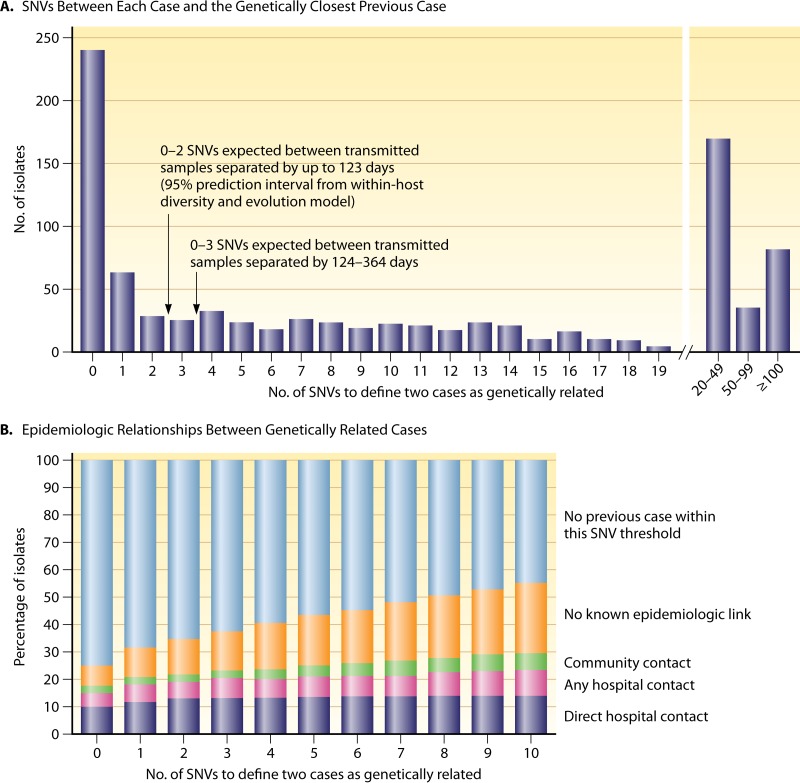
In light of these results, the question remained as to the extent that alternative sources, such as patients with asymptomatic colonization, or community and environmental sources of C. difficile contribute to the overall burden of disease. In a 2013 landmark study by Eyre and colleagues (167), >1,200 isolates of C. difficile obtained from symptomatic patients in Oxford University Hospitals throughout 2007 to 2011 were sequenced, with the aim of identifying the genetic relationship between strains. Those authors began by estimating the evolution rate and mean within-host diversity by evaluating the genomes of isolates from the first and last samples of 145 patients. By applying a coalescent model of evolution and taking into account the interval between the collection times of individual samples, those authors estimated an evolutionary rate of 0.74 SNVs (95% CI, 0.22 to 1.40 SNVs) per genome per year and a mean within-host diversity of 0.30 SNVs (95% CI, 0.13 to 0.42 SNVs). By using these prediction intervals, pairs of isolates separated by <124 days from each other and differing by 0 to 2 SNVs were considered to be a result of direct transmission. A comparison of 957 isolates collected between 2008 and 2011 revealed that just 35% of isolates showed evidence of direct transmission from an earlier case (2007 to 2011), a figure significantly lower than anticipated. Moreover, in one-third of these patients, no plausible epidemiological link could be made (e.g., no contact with another patient in the hospital or in the community) (Fig. 4). Remarkably, 45% of isolates had >10 SNVs, indicating that they were genetically distinct from all other cases (Fig. 4) and likely from a source other than the hospital environment. Importantly, these data demonstrated that genetically diverse sources of C. difficile play a more substantial role in C. difficile transmission than first thought. Although asymptomatic carriage may be an important source of CDI and could account for many unexplained cases (11), these recent findings (11, 166, 167) represent a significant milestone in C. difficile transmission research, as they challenge the prevailing view that horizontal transmission from symptomatic patients is the source of most cases of CDI in health care settings, a concept that is currently the basis for infection control and prevention guidelines (163, 168).
FIG 4.
Diverse sources of C. difficile in the hospital environment. Shown are the genetic variation and epidemiological relationships among 957 isolates obtained from patients with CDI. (A) Numbers of single-nucleotide variants (SNVs) between each sample obtained during the period from 1 April 2008 through 31 March 2011 and the most closely related previous sample obtained after 1 September 2007. (B) Percentages of isolates that were classified as genetically related, according to the different SNV thresholds, along with the epidemiological links between related isolates. (Reproduced from reference 167 with permission from the Massachusetts Medical Society.)
Historically, recurrent or refractory CDI occurs in ∼20 to 25% of CDI patients after treatment of primary infection with metronidazole or vancomycin (169). In the past decade, however, coinciding with increases in the frequency and severity of CDI caused primarily by the emergence of the epidemic RT027 strains, the rates of recurrent CDI have also increased, presenting clinicians with a difficult challenge (169, 170). To better understand the epidemiology of CDI recurrences, Eyre and colleagues applied WGS to 93 paired isolates of C. difficile from patients with recurrent CDI (65 treated with vancomycin and 28 treated with fidaxomicin), with the aim of resolving the nature of the recurrence (171). By using a methodology similar to the one used in their previous study (167), an evolutionary rate of 0.74 SNVs/called genome/year was used to define relapse (≤2 paired SNVs) and reinfection (≥10 SNVs) (171). For 79.6% of participants, there were <2 SNVs between paired isolates, which is indicative of infection with the same strain. Interestingly, for cases of recurrent CDI attributable to RT027, fidaxomicin showed levels of protection comparable to those with vancomycin; however, for non-RT027 strains, fidaxomicin was superior to vancomycin in preventing both reinfection with a new strain and relapse (171).
Animal Reservoirs and Zoonotic Potential
The incidence of community-acquired CDI (CA-CDI) has been increasing globally, in some regions accounting for up to a quarter of all cases (172). Individuals acquiring disease in the community setting do not have the classic risk factors for CDI acquisition and are generally young and healthy, without contact with hospitalized patients and often without prior antimicrobial exposure (173–175). Notably, the genotypes of C. difficile strains acquired in the community differ from those of predominant hospital strains. In particular, RT078 (toxinotype V; ST-11 [MLST clade 5]; NAP7/8; A+ B+ CDT+) has emerged as a significant pathogen associated with the majority of CA-CDI cases in the Northern Hemisphere (1, 37). It is currently among the top three most frequently encountered RTs in European hospitals (92, 93, 176). Furthermore, RT078 shares similar genetic features (binary toxin and tcdC mutations) and disease phenotypes (increased mortality and morbidity) with RT027 (28, 109, 176, 177).
In an attempt to understand the changing epidemiology of CDI in humans, particularly in the community setting, numerous studies have sought to determine if CDI has a foodborne etiology by investigating the prevalence and genotype of C. difficile in animals and food. C. difficile is widely recognized as a commensal and enteric pathogen in a wide range of host species (178–180). To date, C. difficile been recovered from numerous animal sources, including livestock (pigs, piglets, cows, calves, sheep, lambs, goats, and chickens), domestic animals (cats and dogs), equines (horses and foals), wildlife (rabbits, wild birds, shrews, raccoons, feral swine, ostriches, Kodiak bears, zebras, kangaroos, elephants, ibex, tamarin monkeys, and chimpanzees), and marine organisms (bivalve molluscs) (103, 178, 181–196). Many of these studies described differences in prevalence (particularly a decline with age), toxigenic status, antibiotic resistance, clonal lineage, and host susceptibility to disease, as well as differences in veterinary and agricultural practices (178, 180). Furthermore, these studies highlight the ability of C. difficile to adapt to a wide range of host immune systems and gastrointestinal environments, again reflecting the diversity seen in the pangenome.
The predominant strain of C. difficile identified in many of these studies was an RT078 strain. In particular, RT078 is well established in food-producing animals, comprising 75 to 100% of porcine and >90% of bovine isolates (187, 197–199). Detection of C. difficile in livestock has raised concerns that animals are a potential source of CDI in humans and that spores could be transmitted through either direct contact, shedding, or contamination of meat products with fecal material during slaughter (200). RT078 and RT027 strains have been isolated from pork, beef, and chickens in the Northern Hemisphere, with recovery rates varying from 3% in Europe to 42% in North America (201, 202). These data are alarming and provide support for the theory that C. difficile has the potential for zoonotic transmission (178, 201, 203).
Interestingly, RT078 has not been isolated from livestock in Australia (103, 191, 204). However, several other RTs belonging to MLST clade 5 have been found in 7-day-old calves (RT126 [A+ B+ CDT+] and RT127 [A+ B+ CDT+]) and in neonatal pigs (RT033 [A− B− CDT+] and RT237 [A− B+ CDT+]) (103, 204). Albeit in low numbers, these non-RT078 clade 5 RTs have all been isolated from humans with CDI in Australia in recent years and are of emerging One Health importance (205, 206). In the absence of RT078 in Australian livestock, other RTs have become established. In a recent Australian study of C. difficile RTs in neonatal pigs, the predominating RT was found to be RT014 (A+ B+ CDT−) (204). This is noteworthy because RT014 belongs to MLST clade 1, is binary toxin negative, and for many years has been the most common RT isolated from humans with CDI in most geographic regions (91–93, 204).
Characterization of the genetic overlap of C. difficile strains isolated from different reservoirs facilitates a better understanding of possible transmission routes. Several studies have attempted to determine the extent of genetic relatedness between C. difficile isolates of human, animal, and food origins. Initially, isolates of C. difficile sharing the same RT or PFGE pattern (e.g., RT078 or NAP7/8, respectively) were typed by MLVA or MLST, providing greater discriminatory power than non-sequence-based methods. Bakker et al. (207) found that 85% of RT078 isolates of human and porcine origins were genetically related by MLVA. Stabler et al. (46) used MLST to analyze a large collection of isolates (n = 385) from different geographical locations (Europe, North America, and Australia) and sources (humans, food, and animals). Isolates from diverse sources belonged to the same lineage, and many isolates from humans, food, and animals were indistinguishable (46). Most recently, Dutch researchers used whole-genome SNP typing to compare the genomes of 65 C. difficile RT078 isolates of human and porcine origins (208). Analysis of the core genomes of these isolates revealed a total of 401 phylogenetic SNPs, which were used for phylogenetic tree building. The RT078 population-specific mutation rate was estimated to be 2.72 × 10−7 substitutions per site per year (95% CI, 1.43 × 10−7 to 3.99 × 10−7 substitutions per site per year), which is equivalent to 1.1 SNPs per genome per year, a figure comparable with previous estimates (166, 167). Maximum likelihood phylogeny showed isolates of human and porcine origins clustering together. Furthermore, the genomes of pigs and humans harbored identical antimicrobial resistance genes (tetracycline and streptomycin). Notably, these analyses showed a pair of human and pig isolates from the same pig farm in The Netherlands to be indistinguishable (zero SNP differences), suggesting that interspecies transmission had occurred. While this certainly contributes to the theory that CDI is a zoonosis, a common source cannot be ruled out. Moreover, it is possible that zooanthropomorphic (human-to-animal) transmission may have occurred.
CONCLUSIONS AND FUTURE PERSPECTIVES
C. difficile remains a formidable pathogen, and the financial and clinical burden of CDI continues to challenge health care systems the world over. In this review, we have described how sequencing of the genomes of strain 630 and other clinically important reference strains has been a key milestone in unraveling the complexities of this once enigmatic species. We have also described how the application of genomic techniques such as MLST and WGS to robust clinically relevant data sets has significantly advanced our knowledge of both the epidemiology of CDI and the genetics of C. difficile.
The genome of C. difficile is large and genetically diverse, showing remarkable levels of plasticity and ultralow levels of conservation among strains. Complex mechanisms of HGT and recombination between close and distantly clonal lineages, and also between different genera, have had a profound effect on the evolution of many clinically important loci, such as the PaLoc, ISR, S layer, CRISPR-cas, and antimicrobial resistance genes. Many of these mechanisms are mediated by an extensive and diverse collection of transposons and phages that have coevolved with C. difficile over long time periods. These evolutionary events have occurred numerous times and collectively led to the species diversifying into hundreds of strains types spread over at least 6 phylogenetic lineages. WGS and estimates of the in-host mutation rate (molecular clock) have provided novel insights into important aspects of CDI, including ward-based transmission, outbreaks, refractory disease, and the diverse nature of C. difficile in the hospital environment. Moreover, recent studies have used WGS to elucidate the genetic basis of the microevolution and transcontinental dissemination of epidemic RT027 strains and have shown for the first time the possibility of interspecies transmission of C. difficile between pigs and humans.
Notwithstanding these advances, numerous areas of study of C. difficile biology are still in their infancy, and there remains much to learn. Further studies are needed to elucidate the complexities of CRISPR-cas elements and the agr locus in C. difficile. With mounting evidence that livestock are a potentially significant reservoir of CDI, further work is needed to investigate the extent and direction of C. difficile transmission between animals and humans as well as genetic exchange between animal and human isolates. In particular, there is a need for a genealogical timeline for the emergence of livestock-associated clones with clinical relevance, such as RT126, RT127, RT237, and RT033, and a greater understanding of their relationship to RT078. There is a need to better understand the overall phylogeny of clade 2, particularly the global epidemiology of RTs other than RT027, and determine their contribution to CA-CDI. By occupying niches within multiple host species, C. difficile is able to access and exchange DNA with an enormously diverse metagenome. Further analysis of the vast array of mobile elements such as phages and CTns present in C. difficile from diverse sources will advance our understanding of C. difficile pathogenesis and diversification of lineages and at the same time expand our knowledge of the C. difficile pangenome. In the coming years, WGS will continue to provide insights into this important pathogen, providing researchers and clinicians with information that can be used to reduce the overall burden of disease caused by C. difficile in humans and animals.
ACKNOWLEDGMENTS
D.R.K., B.E., T.T.P., B.J.C., and T.V.R. declare no conflicts of interest relevant to this article.
We thank Marco R. Oggioni for his critical review of the manuscript.
D.R.K. is a recipient of an Australian postgraduate award from The University of Western Australia.
Biographies

Daniel R. Knight, B.Sc. Hons., completed his undergraduate degree in Biology at University College London, London, United Kingdom. Based initially in London and then in Cambridge, he then spent the next 7 years in contract research, in the laboratory of Dr. Ian Morrissey. His research interests included mechanisms of antimicrobial and biocide resistance, antimicrobial surveillance in bacterial pathogens, the development of novel antimicrobials, bacterial identification, and molecular diagnostics. In 2012, he joined the laboratory of Professor Thomas V. Riley at The University of Western Australia, researching Clostridium difficile infection, and has published several articles on C. difficile molecular epidemiology and diagnostics. Currently in the second year of his Ph.D., his research focus is the genetic diversity and evolution of human and animal isolates of C. difficile. He is a member of the Australian Society for Microbiology (ASM) and the Australian Society for Antimicrobials (ASA) and coordinates the national C. difficile antimicrobial resistance surveillance program.

Briony Elliott, Ph.D., received a bachelor of science with first-class Honors in Microbiology and Environmental Microbiology at The University of Western Australia (UWA). Her Ph.D., also obtained at UWA, focused on the molecular epidemiology of C. difficile infection in Western Australia. She is currently a Research Associate at UWA coordinating C. difficile reference laboratory activities and has active research interests and has published several articles on the evolutionary history and virulence of C. difficile.

Barbara J. Chang, Ph.D., completed her B.Sc. (Honors) in Genetics at the University of Adelaide, Australia, and her Ph.D. in Bacterial Genetics at Monash University, Australia, under the supervision of Professor Bruce Holloway. She then gained a postdoctoral fellowship at the University of Calgary, Canada, working on bacterial biofilms in the laboratory of Professor Bill Costerton. She was appointed as a Senior Tutor at The University of Western Australia and has held various positions there, including Head of the Department of Microbiology, Chair of the Discipline of Microbiology and Immunology, and Associate Dean (Teaching) of the Faculty of Science. She is currently a Professor in the School of Pathology and Laboratory Medicine. Her research interests include molecular studies of virulence mechanisms of bacterial pathogens, including Clostridium difficile, the biology and genetics of bacteriophages, and the identification of novel anti-quorum-sensing agents for use in bacterial biocontrol and therapy. She is a Fellow of the Australian Society for Microbiology.

Timothy T. Perkins, Ph.D., completed his B.Sc. in Biophysics and Chemistry at The University of Western Australia (UWA) and his Ph.D. at the Wellcome Trust Sanger Institute registered at Jesus College, Cambridge University, United Kingdom. He was then employed by Novartis Vaccines and Diagnostics, Siena, Italy, as a research associate working on meningococcal genomics before taking up his present position as Research Assistant Professor at the Marshall Centre for Infectious Diseases and Training at UWA. His research interests are genomics, transcriptomics, molecular biology, and, in particular, how such tools can be utilized to determine the molecular bases of pathogenesis and for rational vaccine design.

Thomas V. Riley, Ph.D., worked for 15 years in diagnostic laboratories before completing a Ph.D. part time at The University of Western Australia (UWA) in 1984. Professor Riley did postdoctoral studies in the Anaerobe Reference Unit of the Public Health Laboratory Service in England and then in the Division of Infectious Diseases at Johns Hopkins University in Baltimore, MD. He returned to Australia in 1987 and was appointed Senior Medical Scientist in Charge of the Department of Clinical Microbiology at Sir Charles Gairdner Hospital in Perth in 1988. In 1993 to 1994, he did a masters degree in Applied Epidemiology at the Australian National University, and in 1995, he was appointed Associate Professor, Department of Microbiology, at UWA and Principal Research Scientist at the Western Australian Centre for Pathology & Medical Research, now PathWest Laboratory Medicine. In 2002, he was awarded a Personal Chair at UWA. Professor Riley is a Fellow of the Royal College of Pathologists, the Australian Society for Microbiology, the American Academy of Microbiology, the Society for Healthcare Epidemiology of America, and the Faculty of Science of the Royal College of Pathologists of Australasia and has published over 300 book chapters and refereed journal articles. He has had a long interest in health care-related infections, particularly the diagnosis, pathogenesis, and epidemiology of Clostridium difficile infection (CDI), and started working on CDI in animals over 20 years ago.
REFERENCES
- 1.Rupnik M, Wilcox MH, Gerding DN. 2009. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol 7:526–536. doi: 10.1038/nrmicro2164. [DOI] [PubMed] [Google Scholar]
- 2.Wiegand PN, Nathwani D, Wilcox MH, Stephens J, Shelbaya A, Haider S. 2012. Clinical and economic burden of Clostridium difficile infection in Europe: a systematic review of healthcare-facility-acquired infection. J Hosp Infect 81:1–14. doi: 10.1016/j.jhin.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/drugresistance/threat-report-2013/. [Google Scholar]
- 4.Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, Wilson LE, Winston LG, Cohen JA, Limbago BM, Fridkin SK, Gerding DN, McDonald LC. 2015. Burden of Clostridium difficile infection in the United States. N Engl J Med 372:825–834. doi: 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall IC, O'Toole E. 1935. Intestinal flora in newborn infants with a description of a new pathogenic anaerobe, Bacillus difficilis. Am J Dis Child 49:390–402. doi: 10.1001/archpedi.1935.01970020105010. [DOI] [Google Scholar]
- 6.Snyder ML. 1937. Further studies on Bacillus difficilis (Hall and O'Toole). J Infect Dis 60:223–231. doi: 10.1093/infdis/60.2.223. [DOI] [Google Scholar]
- 7.Bartlett JG, Moon N, Chang TW, Taylor N, Onderdonk AB. 1978. Role of Clostridium difficile in antibiotic-associated pseudomembranous colitis. Gastroenterology 75:778–782. [PubMed] [Google Scholar]
- 8.Heinlen L, Ballard JD. 2010. Clostridium difficile infection. Am J Med Sci 340:247–252. doi: 10.1097/MAJ.0b013e3181e939d8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gebhard RL, Gerding DN, Olson MM, Peterson LR, McClain CJ, Ansel HJ, Shaw MJ, Schwartz ML. 1985. Clinical and endoscopic findings in patients early in the course of Clostridium difficile-associated pseudomembranous colitis. Am J Med 78:45–48. doi: 10.1016/0002-9343(85)90460-7. [DOI] [PubMed] [Google Scholar]
- 10.Gupta A, Patel R, Baddour LM, Pardi DS, Khanna S. 2014. Extraintestinal Clostridium difficile infections: a single-center experience. Mayo Clin Proc 89:1525–1536. doi: 10.1016/j.mayocp.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 11.Eyre DW, Griffiths D, Vaughan A, Golubchik T, Acharya M, O'Connor L, Crook DW, Walker AS, Peto TE. 2013. Asymptomatic Clostridium difficile colonisation and onward transmission. PLoS One 8:e78445. doi: 10.1371/journal.pone.0078445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bignardi GE. 1998. Risk factors for Clostridium difficile infection. J Hosp Infect 40:1–15. doi: 10.1016/S0195-6701(98)90019-6. [DOI] [PubMed] [Google Scholar]
- 13.Chopra T, Alangaden GJ, Chandrasekar P. 2010. Clostridium difficile infection in cancer patients and hematopoietic stem cell transplant recipients. Expert Rev Anti Infect Ther 8:1113–1119. doi: 10.1586/eri.10.95. [DOI] [PubMed] [Google Scholar]
- 14.Appelbaum PC, Hunter PA. 2000. The fluoroquinolone antibacterials: past, present and future perspectives. Int J Antimicrob Agents 16:5–15. doi: 10.1016/S0924-8579(00)00192-8. [DOI] [PubMed] [Google Scholar]
- 15.Bartlett JG. 2006. Narrative review: the new epidemic of Clostridium difficile-associated enteric disease. Ann Intern Med 145:758–764. doi: 10.7326/0003-4819-145-10-200611210-00008. [DOI] [PubMed] [Google Scholar]
- 16.Rutala WA, Gergen MF, Weber DJ. 2012. Efficacy of different cleaning and disinfection methods against Clostridium difficile spores: importance of physical removal versus sporicidal inactivation. Infect Control Hosp Epidemiol 33:1255–1258. doi: 10.1086/668434. [DOI] [PubMed] [Google Scholar]
- 17.Gerding DN, Muto CA, Owens RC Jr. 2008. Measures to control and prevent Clostridium difficile infection. Clin Infect Dis 46(Suppl 1):S43–S49. doi: 10.1086/521861. [DOI] [PubMed] [Google Scholar]
- 18.Kim KH, Fekety R, Batts DH, Brown D, Cudmore M, Silva J Jr, Waters D. 1981. Isolation of Clostridium difficile from the environment and contacts of patients with antibiotic-associated colitis. J Infect Dis 143:42–50. doi: 10.1093/infdis/143.1.42. [DOI] [PubMed] [Google Scholar]
- 19.Bakken JS, Borody T, Brandt LJ, Brill JV, Demarco DC, Franzos MA, Kelly C, Khoruts A, Louie T, Martinelli LP, Moore TA, Russell G, Surawicz C. 2011. Treating Clostridium difficile infection with fecal microbiota transplantation. Clin Gastroenterol Hepatol 9:1044–1049. doi: 10.1016/j.cgh.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Debast SB, Bauer MP, Kuijper EJ. 2014. European Society of Clinical Microbiology and Infectious Diseases: update of the treatment guidance document for Clostridium difficile infection. Clin Microbiol Infect 20(Suppl 2):1–26. doi: 10.1111/1469-0691.12418. [DOI] [PubMed] [Google Scholar]
- 21.Louie TJ, Miller MA, Mullane KM, Weiss K, Lentnek A, Golan Y, Gorbach S, Sears P, Shue YK. 2011. Fidaxomicin versus vancomycin for Clostridium difficile infection. N Engl J Med 364:422–431. doi: 10.1056/NEJMoa0910812. [DOI] [PubMed] [Google Scholar]
- 22.Hargreaves KR, Clokie MR. 2014. Clostridium difficile phages: still difficult? Front Microbiol 5:184. doi: 10.3389/fmicb.2014.00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerding DN. 2012. Clostridium difficile infection prevention: biotherapeutics, immunologics, and vaccines. Discov Med 13:75–83. [PubMed] [Google Scholar]
- 24.Cohen SH, Gerding DN, Johnson S, Kelly CP, Loo VG, McDonald LC, Pepin J, Wilcox MH. 2010. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA). Infect Control Hosp Epidemiol 31:431–455. doi: 10.1086/651706. [DOI] [PubMed] [Google Scholar]
- 25.Shen A. 2012. Clostridium difficile toxins: mediators of inflammation. J Innate Immun 4:149–158. doi: 10.1159/000332946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, Minton NP. 2010. The role of toxin A and toxin B in Clostridium difficile infection. Nature 467:711–713. doi: 10.1038/nature09397. [DOI] [PubMed] [Google Scholar]
- 27.Voth DE, Ballard JD. 2005. Clostridium difficile toxins: mechanism of action and role in disease. Clin Microbiol Rev 18:247–263. doi: 10.1128/CMR.18.2.247-263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barbut F, Decre D, Lalande V, Burghoffer A, Noussair L, Gigandon A, Espinasse F, Raskine L, Robert J, Mangeol A, Branger C, Petit JC. 2005. Clinical features of Clostridium difficile-associated diarrhoea due to binary toxin (actin-specific ADP-ribosyltransferase)-producing strains. J Med Microbiol 54:181–185. doi: 10.1099/jmm.0.45804-0. [DOI] [PubMed] [Google Scholar]
- 29.Stevenson E, Minton NP, Kuehne SA. 2015. The role of flagella in Clostridium difficile pathogenicity. Trends Microbiol 23:275–282. doi: 10.1016/j.tim.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 30.Cheng V, Yam W, Lam O, Tsang J, Tse E, Siu G, Chan J, Tse H, To K, Tai J. 2011. Clostridium difficile isolates with increased sporulation: emergence of PCR ribotype 002 in Hong Kong. Eur J Clin Microbiol Infect Dis 30:1371–1381. doi: 10.1007/s10096-011-1231-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Planche T, Wilcox M. 2011. Reference assays for Clostridium difficile infection: one or two gold standards? J Clin Pathol 64:1–5. doi: 10.1136/jcp.2010.080135. [DOI] [PubMed] [Google Scholar]
- 32.Swindells J, Brenwald N, Reading N, Oppenheim B. 2010. Evaluation of diagnostic tests for Clostridium difficile infection. J Clin Microbiol 48:606–608. doi: 10.1128/JCM.01579-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knetsch CW, Lawley TD, Hensgens MP, Corver J, Wilcox MW, Kuijper EJ. 2013. Current application and future perspectives of molecular typing methods to study Clostridium difficile infections. Euro Surveill 18(4):pii=20381 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20381. [DOI] [PubMed] [Google Scholar]
- 34.Kuijper EJ, van den Berg RJ, Brazier JS. 2009. Comparison of molecular typing methods applied to Clostridium difficile. Methods Mol Biol 551:159–171. doi: 10.1007/978-1-60327-999-4_13. [DOI] [PubMed] [Google Scholar]
- 35.Eyre DW, Fawley WN, Best EL, Griffiths D, Stoesser NE, Crook DW, Peto TE, Walker AS, Wilcox MH. 2013. Comparison of multilocus variable-number tandem-repeat analysis and whole-genome sequencing for investigation of Clostridium difficile transmission. J Clin Microbiol 51:4141–4149. doi: 10.1128/JCM.01095-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eyre DW, Walker AS. 2013. Clostridium difficile surveillance: harnessing new technologies to control transmission. Expert Rev Anti Infect Ther 11:1193–1205. doi: 10.1586/14787210.2013.845987. [DOI] [PubMed] [Google Scholar]
- 37.Cairns MD, Stabler RA, Shetty N, Wren BW. 2012. The continually evolving Clostridium difficile species. Future Microbiol 7:945–957. doi: 10.2217/fmb.12.73. [DOI] [PubMed] [Google Scholar]
- 38.Collins DA, Elliott B, Riley TV. 2015. Molecular methods for detecting and typing of Clostridium difficile. Pathology 47:211–218. doi: 10.1097/PAT.0000000000000238. [DOI] [PubMed] [Google Scholar]
- 39.Stubbs SL, Brazier JS, O'Neill GL, Duerden BI. 1999. PCR targeted to the 16S-23S rRNA gene intergenic spacer region of Clostridium difficile and construction of a library consisting of 116 different PCR ribotypes. J Clin Microbiol 37:461–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Janezic S, Indra A, Rattei T, Weinmaier T, Rupnik M. 2014. Recombination drives evolution of the Clostridium difficile 16S-23S rRNA intergenic spacer region. PLoS One 9:e106545. doi: 10.1371/journal.pone.0106545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rupnik M. 2008. Heterogeneity of large clostridial toxins: importance of Clostridium difficile toxinotypes. FEMS Microbiol Rev 32:541–555. doi: 10.1111/j.1574-6976.2008.00110.x. [DOI] [PubMed] [Google Scholar]
- 42.Rupnik M, Avesani V, Janc M, von Eichel-Streiber C, Delmee M. 1998. A novel toxinotyping scheme and correlation of toxinotypes with serogroups of Clostridium difficile isolates. J Clin Microbiol 36:2240–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lemee L, Dhalluin A, Pestel-Caron M, Lemeland JF, Pons JL. 2004. Multilocus sequence typing analysis of human and animal Clostridium difficile isolates of various toxigenic types. J Clin Microbiol 42:2609–2617. doi: 10.1128/JCM.42.6.2609-2617.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Griffiths D, Fawley W, Kachrimanidou M, Bowden R, Crook DW, Fung R, Golubchik T, Harding RM, Jeffery KJ, Jolley KA, Kirton R, Peto TE, Rees G, Stoesser N, Vaughan A, Walker AS, Young BC, Wilcox M, Dingle KE. 2010. Multilocus sequence typing of Clostridium difficile. J Clin Microbiol 48:770–778. doi: 10.1128/JCM.01796-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dingle KE, Griffiths D, Didelot X, Evans J, Vaughan A, Kachrimanidou M, Stoesser N, Jolley KA, Golubchik T, Harding RM, Peto TE, Fawley W, Walker AS, Wilcox M, Crook DW. 2011. Clinical Clostridium difficile: clonality and pathogenicity locus diversity. PLoS One 6:e19993. doi: 10.1371/journal.pone.0019993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stabler RA, Dawson LF, Valiente E, Cairns MD, Martin MJ, Donahue EH, Riley TV, Songer JG, Kuijper EJ, Dingle KE, Wren BW. 2012. Macro and micro diversity of Clostridium difficile isolates from diverse sources and geographical locations. PLoS One 7:e31559. doi: 10.1371/journal.pone.0031559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knetsch CW, Terveer EM, Lauber C, Gorbalenya AE, Harmanus C, Kuijper EJ, Corver J, van Leeuwen HC. 2012. Comparative analysis of an expanded Clostridium difficile reference strain collection reveals genetic diversity and evolution through six lineages. Infect Genet Evol 12:1577–1585. doi: 10.1016/j.meegid.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 48.Eyre DW, Golubchik T, Gordon NC, Bowden R, Piazza P, Batty EM, Ip CL, Wilson DJ, Didelot X, O'Connor L, Lay R, Buck D, Kearns AM, Shaw A, Paul J, Wilcox MH, Donnelly PJ, Peto TE, Walker AS, Crook DW. 2012. A pilot study of rapid benchtop sequencing of Staphylococcus aureus and Clostridium difficile for outbreak detection and surveillance. BMJ Open 2:e001124. doi: 10.1136/bmjopen-2012-001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walker TM, Ip CL, Harrell RH, Evans JT, Kapatai G, Dedicoat MJ, Eyre DW, Wilson DJ, Hawkey PM, Crook DW, Parkhill J, Harris D, Walker AS, Bowden R, Monk P, Smith EG, Peto TE. 2013. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis 13:137–146. doi: 10.1016/S1473-3099(12)70277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grad YH, Lipsitch M, Feldgarden M, Arachchi HM, Cerqueira GC, Fitzgerald M, Godfrey P, Haas BJ, Murphy CI, Russ C, Sykes S, Walker BJ, Wortman JR, Young S, Zeng Q, Abouelleil A, Bochicchio J, Chauvin S, Desmet T, Gujja S, McCowan C, Montmayeur A, Steelman S, Frimodt-Moller J, Petersen AM, Struve C, Krogfelt KA, Bingen E, Weill FX, Lander ES, Nusbaum C, Birren BW, Hung DT, Hanage WP. 2012. Genomic epidemiology of the Escherichia coli O104:H4 outbreaks in Europe, 2011. Proc Natl Acad Sci U S A 109:3065–3070. doi: 10.1073/pnas.1121491109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mardis E, McPherson J, Martienssen R, Wilson RK, McCombie WR. 2002. What is finished, and why does it matter. Genome Res 12:669–671. doi: 10.1101/gr.032102. [DOI] [PubMed] [Google Scholar]
- 52.Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeno-Tarrraga AM, Wang HW, Holden MTG, Wright A, Churcher C, Quail MA, Baker S, Bason N, Brooks K, Chillingworth T, Cronin A, Davis P, Dowd L, Fraser A, Feltwell T, Hance Z, Holroyd S, Jagels K, Moule S, Mungall K, Price C, Rabbinowitsch E, Sharp S, Simmonds M, Stevens K, Unwin L, Whithead S, Dupuy B, Dougan G, Barrell B, Parkhill J. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet 38:779–786. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- 53.Monot M, Boursaux-Eude C, Thibonnier M, Vallenet D, Moszer I, Medigue C, Martin-Verstraete I, Dupuy B. 2011. Reannotation of the genome sequence of Clostridium difficile strain 630. J Med Microbiol 60:1193–1199. doi: 10.1099/jmm.0.030452-0. [DOI] [PubMed] [Google Scholar]
- 54.Gaulton T, Misra R, Rose G, Baybayan P, Hall R, Freeman J, Turton J, Picton S, Korlach J, Gharbia S, Shah H. 2015. Complete genome sequence of the hypervirulent bacterium Clostridium difficile strain G46, ribotype 027. Genome Announc 3(2):e00073-15. doi: 10.1128/genomeA.00073-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brouwer MS, Allan E, Mullany P, Roberts AP. 2012. Draft genome sequence of the nontoxigenic Clostridium difficile strain CD37. J Bacteriol 194:2125–2126. doi: 10.1128/JB.00122-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stabler RA, He M, Dawson L, Martin M, Valiente E, Corton C, Lawley TD, Sebaihia M, Quail MA, Rose G, Gerding DN, Gibert M, Popoff MR, Parkhill J, Dougan G, Wren BW. 2009. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol 10:R102. doi: 10.1186/gb-2009-10-9-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He M, Sebaihia M, Lawley TD, Stabler RA, Dawson LF, Martin MJ, Holt KE, Seth-Smith HM, Quail MA, Rance R, Brooks K, Churcher C, Harris D, Bentley SD, Burrows C, Clark L, Corton C, Murray V, Rose G, Thurston S, van Tonder A, Walker D, Wren BW, Dougan G, Parkhill J. 2010. Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc Natl Acad Sci U S A 107:7527–7532. doi: 10.1073/pnas.0914322107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Darling AE, Worden P, Chapman TA, Roy Chowdhury P, Charles IG, Djordjevic SP. 2014. The genome of Clostridium difficile 5.3. Gut Pathog 6:4. doi: 10.1186/1757-4749-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stabler RA, Valiente E, Dawson LF, He M, Parkhill J, Wren BW. 2010. In-depth genetic analysis of Clostridium difficile PCR-ribotype 027 strains reveals high genome fluidity including point mutations and inversions. Gut Microbes 1:269–276. doi: 10.4161/gmic.1.4.11870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sebaihia M, Peck MW, Minton NP, Thomson NR, Holden MT, Mitchell WJ, Carter AT, Bentley SD, Mason DR, Crossman L, Paul CJ, Ivens A, Wells-Bennik MH, Davis IJ, Cerdeno-Tarraga AM, Churcher C, Quail MA, Chillingworth T, Feltwell T, Fraser A, Goodhead I, Hance Z, Jagels K, Larke N, Maddison M, Moule S, Mungall K, Norbertczak H, Rabbinowitsch E, Sanders M, Simmonds M, White B, Whithead S, Parkhill J. 2007. Genome sequence of a proteolytic (group I) Clostridium botulinum strain Hall A and comparative analysis of the clostridial genomes. Genome Res 17:1082–1092. doi: 10.1101/gr.6282807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 62.Wong YM, Juan JC, Gan HM, Austin CM. 2014. Draft genome sequence of Clostridium bifermentans strain WYM, a promising biohydrogen producer isolated from landfill leachate sludge. Genome Announc 2(2):e00077-14. doi: 10.1128/genomeA.00077-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fonknechten N, Chaussonnerie S, Tricot S, Lajus A, Andreesen JR, Perchat N, Pelletier E, Gouyvenoux M, Barbe V, Salanoubat M, Le Paslier D, Weissenbach J, Cohen GN, Kreimeyer A. 2010. Clostridium sticklandii, a specialist in amino acid degradation: revisiting its metabolism through its genome sequence. BMC Genomics 11:555. doi: 10.1186/1471-2164-11-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Braun V, Hundsberger T, Leukel P, Sauerborn M, von Eichel-Streiber C. 1996. Definition of the single integration site of the pathogenicity locus in Clostridium difficile. Gene 181:29–38. doi: 10.1016/S0378-1119(96)00398-8. [DOI] [PubMed] [Google Scholar]
- 65.Fluit AC, Wolfhagen MJHM, Verdonk GPHT, Jansze M, Torensma R, Verhoef J. 1991. Nontoxigenic strains of Clostridium difficile lack the genes for both toxin A and toxin B. J Clin Microbiol 29:2666–2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mani N, Dupuy B. 2001. Regulation of toxin synthesis in Clostridium difficile by an alternative RNA polymerase sigma factor. Proc Natl Acad Sci U S A 98:5844–5849. doi: 10.1073/pnas.101126598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tan KS, Wee BY, Song KP. 2001. Evidence for holin function of tcdE gene in the pathogenicity of Clostridium difficile. J Med Microbiol 50:613–619. [DOI] [PubMed] [Google Scholar]
- 68.Matamouros S, England P, Dupuy B. 2007. Clostridium difficile toxin expression is inhibited by the novel regulator TcdC. Mol Microbiol 64:1274–1288. doi: 10.1111/j.1365-2958.2007.05739.x. [DOI] [PubMed] [Google Scholar]
- 69.Curry SR, Marsh JW, Muto CA, O'Leary MM, Pasculle AW, Harrison LH. 2007. tcdC genotypes associated with severe TcdC truncation in an epidemic clone and other strains of Clostridium difficile. J Clin Microbiol 45:215–221. doi: 10.1128/JCM.01599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carter GP, Douce GR, Govind R, Howarth PM, Mackin KE, Spencer J, Buckley AM, Antunes A, Kotsanas D, Jenkin GA, Dupuy B, Rood JI, Lyras D. 2011. The anti-sigma factor TcdC modulates hypervirulence in an epidemic BI/NAP1/027 clinical isolate of Clostridium difficile. PLoS Pathog 7:e1002317. doi: 10.1371/journal.ppat.1002317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bakker D, Smits WK, Kuijper EJ, Corver J. 2012. TcdC does not significantly repress toxin expression in Clostridium difficile 630 delta Erm. PLoS One 7:e43247. doi: 10.1371/journal.pone.0043247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cartman ST, Kelly ML, Heeg D, Heap JT, Minton NP. 2012. Precise manipulation of the Clostridium difficile chromosome reveals a lack of association between the tcdC genotype and toxin production. Appl Environ Microbiol 78:4683–4690. doi: 10.1128/AEM.00249-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stabler RA, Gerding DN, Songer JG, Drudy D, Brazier JS, Trinh HT, Witney AA, Hinds J, Wren BW. 2006. Comparative phylogenomics of Clostridium difficile reveals clade specificity and microevolution of hypervirulent strains. J Bacteriol 188:7297–7305. doi: 10.1128/JB.00664-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Darkoh C, DuPont HL, Norris SJ, Kaplan HB. 2015. Toxin synthesis by Clostridium difficile is regulated through quorum signaling. mBio 6(2):e02569-14. doi: 10.1128/mBio.02569-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martin MJ, Clare S, Goulding D, Faulds-Pain A, Barquist L, Browne HP, Pettit L, Dougan G, Lawley TD, Wren BW. 2013. The agr locus regulates virulence and colonization genes in Clostridium difficile 027. J Bacteriol 195:3672–3681. doi: 10.1128/JB.00473-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. 2005. The microbial pan-genome. Curr Opin Genet Dev 15:589–594. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 77.Scaria J, Ponnala L, Janvilisri T, Yan W, Mueller LA, Chang YF. 2010. Analysis of ultra low genome conservation in Clostridium difficile. PLoS One 5:e15147. doi: 10.1371/journal.pone.0015147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hall BG, Ehrlich GD, Hu FZ. 2010. Pan-genome analysis provides much higher strain typing resolution than multi-locus sequence typing. Microbiology 156:1060–1068. doi: 10.1099/mic.0.035188-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Forgetta V, Oughton MT, Marquis P, Brukner I, Blanchette R, Haub K, Magrini V, Mardis ER, Gerding DN, Loo VG, Miller MA, Mulvey MR, Rupnik M, Dascal A, Dewar K. 2011. Fourteen-genome comparison identifies DNA markers for severe-disease-associated strains of Clostridium difficile. J Clin Microbiol 49:2230–2238. doi: 10.1128/JCM.00391-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Janvilisri T, Scaria J, Thompson AD, Nicholson A, Limbago BM, Arroyo LG, Songer JG, Grohn YT, Chang YF. 2009. Microarray identification of Clostridium difficile core components and divergent regions associated with host origin. J Bacteriol 191:3881–3891. doi: 10.1128/JB.00222-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu W, Wise M, Tay C, Windsor H, Marshall B, Peacock C, Perkins T. 2014. Comparative analysis of the full genome of the Helicobacter pylori isolate, Sahul64, identifies genes of high divergence. J Bacteriol 196:1073–1083. doi: 10.1128/JB.01021-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Welch RA, Burland V, Plunkett G III, Redford P, Roesch P, Rasko D, Buckles EL, Liou SR, Boutin A, Hackett J, Stroud D, Mayhew GF, Rose DJ, Zhou S, Schwartz DC, Perna NT, Mobley HL, Donnenberg MS, Blattner FR. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci U S A 99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hiller NL, Janto B, Hogg JS, Boissy R, Yu SS, Powell E, Keefe R, Ehrlich NE, Shen K, Hayes J, Barbadora K, Klimke W, Dernovoy D, Tatusova T, Parkhill J, Bentley SD, Post JC, Ehrlich GD, Hu FZ. 2007. Comparative genomic analyses of seventeen Streptococcus pneumoniae strains: insights into the pneumococcal supragenome. J Bacteriol 189:8186–8195. doi: 10.1128/JB.00690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kunin V, Ahren D, Goldovsky L, Janssen P, Ouzounis CA. 2005. Measuring genome conservation across taxa: divided strains and united kingdoms. Nucleic Acids Res 33:616–621. doi: 10.1093/nar/gki181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yutin N, Galperin MY. 2013. A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia. Environ Microbiol 15:2631–2641. doi: 10.1111/1462-2920.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Collins MD, Lawson PA, Willems A, Cordoba JJ, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow JA. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol 44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 87.Ludwig W, Schleifer KH, Whitman WB. 2009. Revised road map to the phylum Firmicutes, p 1–14. In De Vos P, Garrity GM, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer K-H, Whitman WB (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 3 The Firmicutes. Springer, New York, NY. [Google Scholar]
- 88.Sheridan PP, Freeman KH, Brenchley JE. 2003. Estimated minimal divergence times of the major bacterial and archaeal phyla. Geomicrobiol J 20:1–14. doi: 10.1080/01490450303891. [DOI] [Google Scholar]
- 89.Dingle KE, Didelot X, Ansari MA, Eyre DW, Vaughan A, Griffiths D, Ip CL, Batty EM, Golubchik T, Bowden R, Jolley KA, Hood DW, Fawley WN, Walker AS, Peto TE, Wilcox MH, Crook DW. 2013. Recombinational switching of the Clostridium difficile S-layer and a novel glycosylation gene cluster revealed by large-scale whole-genome sequencing. J Infect Dis 207:675–686. doi: 10.1093/infdis/jis734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Janezic S, Rupnik M. 2015. Genomic diversity of Clostridium difficile strains. Res Microbiol 166:353–360. doi: 10.1016/j.resmic.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 91.Foster NF, Collins DA, Ditchburn SL, Duncan CN, van Schalkwyk JW, Golledge CL, Keed ABR, Riley TV. 2014. Epidemiology of Clostridium difficile infection in two tertiary-care hospitals in Perth, Western Australia: a cross-sectional study. New Microbes New Infect 2:64–71. doi: 10.1002/nmi2.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bauer MP, Notermans DW, van Benthem BH, Brazier JS, Wilcox MH, Rupnik M, Monnet DL, van Dissel JT, Kuijper EJ. 2011. Clostridium difficile infection in Europe: a hospital-based survey. Lancet 377:63–73. doi: 10.1016/S0140-6736(10)61266-4. [DOI] [PubMed] [Google Scholar]
- 93.Freeman J, Vernon J, Morris K, Nicholson S, Todhunter S, Longshaw C, Wilcox MH. 2015. Pan-European longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes. Clin Microbiol Infect 20:248.e9–248.e16. doi: 10.1016/j.cmi.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 94.Collins DA, Hawkey PM, Riley TV. 2013. Epidemiology of Clostridium difficile infection in Asia. Antimicrob Resist Infect Control 2:21. doi: 10.1186/2047-2994-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lim SK, Stuart RL, Mackin KE, Carter GP, Kotsanas D, Francis MJ, Easton M, Dimovski K, Elliott B, Riley TV, Hogg G, Paul E, Korman TM, Seemann T, Stinear TP, Lyras D, Jenkin GA. 2014. Emergence of a ribotype 244 strain of Clostridium difficile associated with severe disease and related to the epidemic ribotype 027 strain. Clin Infect Dis 58:1723–1730. doi: 10.1093/cid/ciu203. [DOI] [PubMed] [Google Scholar]
- 96.Valiente E, Dawson LF, Cairns MD, Stabler RA, Wren BW. 2012. Emergence of new PCR ribotypes from the hypervirulent Clostridium difficile 027 lineage. J Med Microbiol 61:49–56. doi: 10.1099/jmm.0.036194-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kuijper EJ, de Weerdt J, Kato H, Kato N, van Dam AP, van der Vorm ER, Weel J, van Rheenen C, Dankert J. 2001. Nosocomial outbreak of Clostridium difficile-associated diarrhoea due to a clindamycin-resistant enterotoxin A-negative strain. Eur J Clin Microbiol Infect Dis 20:528–534. doi: 10.1007/s100960100550. [DOI] [PubMed] [Google Scholar]
- 98.Drudy D, Harnedy N, Fanning S, Hannan M, Kyne L. 2007. Emergence and control of fluoroquinolone-resistant, toxin A-negative, toxin B-positive Clostridium difficile. Infect Control Hosp Epidemiol 28:932–940. doi: 10.1086/519181. [DOI] [PubMed] [Google Scholar]
- 99.Alfa MJ, Kabani A, Lyerly D, Moncrief S, Neville LM, Al-Barrak A, Harding GK, Dyck B, Olekson K, Embil JM. 2000. Characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile responsible for a nosocomial outbreak of Clostridium difficile-associated diarrhea. J Clin Microbiol 38:2706–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Goorhuis A, Legaria MC, van den Berg RJ, Harmanus C, Klaassen CH, Brazier JS, Lumelsky G, Kuijper EJ. 2009. Application of multiple-locus variable-number tandem-repeat analysis to determine clonal spread of toxin A-negative Clostridium difficile in a general hospital in Buenos Aires, Argentina. Clin Microbiol Infect 15:1080–1086. doi: 10.1111/j.1469-0691.2009.02759.x. [DOI] [PubMed] [Google Scholar]
- 101.Jhung MA, Thompson AD, Killgore GE, Zukowski WE, Songer G, Warny M, Johnson S, Gerding DN, McDonald LC, Limbago BM. 2008. Toxinotype V Clostridium difficile in humans and food animals. Emerg Infect Dis 14:1039–1045. doi: 10.3201/eid1407.071641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Elliott B, Dingle KE, Griffiths D, Eyre DW, Crook D, Riley TV. 2012. Australia—home of Clostridium difficile clade 5?, abstr P53. Abstr 4th Int Clostridium difficile Symp, Bled, Slovenia. [Google Scholar]
- 103.Knight DR, Thean S, Putsathit P, Fenwick S, Riley TV. 2013. Cross-sectional study reveals high prevalence of Clostridium difficile non-PCR ribotype 078 strains in Australian veal calves at slaughter. Appl Environ Microbiol 79:2630–2635. doi: 10.1128/AEM.03951-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Elliott B, Reed R, Chang BJ, Riley TV. 2009. Bacteremia with a large clostridial toxin-negative, binary toxin-positive strain of Clostridium difficile. Anaerobe 15:249–251. doi: 10.1016/j.anaerobe.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 105.Eckert C, Emirian A, Le Monnier A, Cathala L, De Montclos H, Goret J, Berger P, Petit A, De Chevigny A, Jean-Pierre H. 2015. Prevalence and pathogenicity of binary toxin-positive Clostridium difficile strains that do not produce toxins A and B. New Microbes New Infect 3:12–17. doi: 10.1016/j.nmni.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Androga G, McGovern A, Elliott B, Chang BJ, Perkins T, Foster N, Riley TV. 2015. Evaluation of the Cepheid Xpert C. difficile/Epi and Meridian Bioscience illumigene C. difficile assays for detecting Clostridium difficile ribotype 033 strains. J Clin Microbiol 53:973–975. doi: 10.1128/JCM.03297-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dingle KE, Elliott B, Robinson E, Griffiths D, Eyre DW, Stoesser N, Vaughan A, Golubchik T, Fawley WN, Wilcox MH, Peto TE, Walker AS, Riley TV, Crook DW, Didelot X. 2014. Evolutionary history of the Clostridium difficile pathogenicity locus. Genome Biol Evol 6:36–52. doi: 10.1093/gbe/evt204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kurka H, Ehrenreich A, Ludwig W, Monot M, Rupnik M, Barbut F, Indra A, Dupuy B, Liebl W. 2014. Sequence similarity of Clostridium difficile strains by analysis of conserved genes and genome content is reflected by their ribotype affiliation. PLoS One 9:e86535. doi: 10.1371/journal.pone.0086535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Knetsch CW, Hensgens MP, Harmanus C, van der Bijl MW, Savelkoul PH, Kuijper EJ, Corver J, van Leeuwen HC. 2011. Genetic markers for Clostridium difficile lineages linked to hypervirulence. Microbiology 157:3113–3123. doi: 10.1099/mic.0.051953-0. [DOI] [PubMed] [Google Scholar]
- 110.Vos M, Didelot X. 2009. A comparison of homologous recombination rates in bacteria and archaea. ISME J 3:199–208. doi: 10.1038/ismej.2008.93. [DOI] [PubMed] [Google Scholar]
- 111.Adams V, Lyras D, Farrow KA, Rood JI. 2002. The clostridial mobilisable transposons. Cell Mol Life Sci 59:2033–2043. doi: 10.1007/s000180200003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Brouwer MS, Warburton PJ, Roberts AP, Mullany P, Allan E. 2011. Genetic organisation, mobility and predicted functions of genes on integrated, mobile genetic elements in sequenced strains of Clostridium difficile. PLoS One 6:e23014. doi: 10.1371/journal.pone.0023014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Roberts AP, Allan E, Mullany P. 2014. The impact of horizontal gene transfer on the biology of Clostridium difficile. Adv Microb Physiol 65:63–82. doi: 10.1016/bs.ampbs.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 114.Brouwer MS, Roberts AP, Mullany P, Allan E. 2012. In silico analysis of sequenced strains of Clostridium difficile reveals a related set of conjugative transposons carrying a variety of accessory genes. Mob Genet Elements 2:8–12. doi: 10.4161/mge.19297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Goh S, Hussain H, Chang BJ, Emmett W, Riley TV, Mullany P. 2013. Phage ϕC2 mediates transduction of Tn6215, encoding erythromycin resistance, between Clostridium difficile strains. mBio 4(6):e00840-13. doi: 10.1128/mBio.00840-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Davies J, Davies D. 2010. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Roberts AP, Mullany P. 2011. Tn916-like genetic elements: a diverse group of modular mobile elements conferring antibiotic resistance. FEMS Microbiol Rev 35:856–871. doi: 10.1111/j.1574-6976.2011.00283.x. [DOI] [PubMed] [Google Scholar]
- 118.Lyras D, Storie C, Huggins AS, Crellin PK, Bannam TL, Rood JI. 1998. Chloramphenicol resistance in Clostridium difficile is encoded on Tn4453 transposons that are closely related to Tn4451 from Clostridium perfringens. Antimicrob Agents Chemother 42:1563–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mullany P, Pallen M, Wilks M, Stephen JR, Tabaqchali S. 1996. A group II intron in a conjugative transposon from the gram-positive bacterium, Clostridium difficile. Gene 174:145–150. doi: 10.1016/0378-1119(96)00511-2. [DOI] [PubMed] [Google Scholar]
- 120.Spigaglia P, Barbanti F, Mastrantonio P. 2008. Tetracycline resistance gene tet(W) in the pathogenic bacterium Clostridium difficile. Antimicrob Agents Chemother 52:770–773. doi: 10.1128/AAC.00957-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mullany P, Wilks M, Tabaqchali S. 1995. Transfer of macrolide-lincosamide-streptogramin-B (MLS) resistance in Clostridium difficile is linked to a gene homologous with toxin A and is mediated by a conjugative transposon, Tn5398. J Antimicrob Chemother 35:305–315. doi: 10.1093/jac/35.2.305. [DOI] [PubMed] [Google Scholar]
- 122.Kehrenberg C, Schwarz S, Jacobsen L, Hansen LH, Vester B. 2005. A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: methylation of 23S ribosomal RNA at A2503. Mol Microbiol 57:1064–1073. doi: 10.1111/j.1365-2958.2005.04754.x. [DOI] [PubMed] [Google Scholar]
- 123.Kehrenberg C, Schwarz S. 2006. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob Agents Chemother 50:1156–1163. doi: 10.1128/AAC.50.4.1156-1163.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Marin M, Martin A, Alcala L, Cercenado E, Iglesias C, Reigadas E, Bouza E. 2015. Clostridium difficile isolates with high linezolid MICs harbor the multiresistance gene cfr. Antimicrob Agents Chemother 59:586–589. doi: 10.1128/AAC.04082-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Corver J, Bakker D, Brouwer MS, Harmanus C, Hensgens MP, Roberts AP, Lipman LJ, Kuijper EJ, van Leeuwen HC. 2012. Analysis of a Clostridium difficile PCR ribotype 078 100 kilobase island reveals the presence of a novel transposon, Tn6164. BMC Microbiol 12:130. doi: 10.1186/1471-2180-12-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Spigaglia P, Barbanti F, Mastrantonio P. 2011. Multidrug resistance in European Clostridium difficile clinical isolates. J Antimicrob Chemother 66:2227–2234. doi: 10.1093/jac/dkr292. [DOI] [PubMed] [Google Scholar]
- 127.He M, Miyajima F, Roberts P, Ellison L, Pickard DJ, Martin MJ, Connor TR, Harris SR, Fairley D, Bamford KB, D'Arc S, Brazier J, Brown D, Coia JE, Douce G, Gerding D, Kim HJ, Koh TH, Kato H, Senoh M, Louie T, Michell S, Butt E, Peacock SJ, Brown NM, Riley TV, Songer G, Wilcox MH, Pirmohamed M, Kuijper E, Hawkey P, Wren BW, Dougan G, Parkhill J, Lawley TD. 2013. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet 45:109–113. doi: 10.1038/ng.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wasels F, Monot M, Spigaglia P, Barbanti F, Ma L, Bouchier C, Dupuy B, Mastrantonio P. 2014. Inter- and intraspecies transfer of a Clostridium difficile conjugative transposon conferring resistance to MLSB. Microb Drug Resist 20:555–560. doi: 10.1089/mdr.2014.0015. [DOI] [PubMed] [Google Scholar]
- 129.van Eijk E, Anvar S, Browne HP, Leung W, Frank J, Schmitz AM, Roberts AP, Smits W. 2015. Complete genome sequence of the Clostridium difficile laboratory strain 630 Δerm reveals differences from strain 630, including translocation of the mobile element CTn 5. BMC Genomics 16:31. doi: 10.1186/s12864-015-1252-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Siefert JL. 2009. Defining the mobilome. Methods Mol Biol 532:13–27. doi: 10.1007/978-1-60327-853-9_2. [DOI] [PubMed] [Google Scholar]
- 131.Sekulovic O, Garneau JR, Neron A, Fortier LC. 2014. Characterization of temperate phages infecting Clostridium difficile isolates of human and animal origins. Appl Environ Microbiol 80:2555–2563. doi: 10.1128/AEM.00237-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hargreaves KR, Colvin HV, Patel KV, Clokie JJ, Clokie MR. 2013. Genetically diverse Clostridium difficile strains harboring abundant prophages in an estuarine environment. Appl Environ Microbiol 79:6236–6243. doi: 10.1128/AEM.01849-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fortier LC, Moineau S. 2007. Morphological and genetic diversity of temperate phages in Clostridium difficile. Appl Environ Microbiol 73:7358–7366. doi: 10.1128/AEM.00582-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hargreaves KR, Kropinski AM, Clokie MR. 2014. What does the talking? Quorum sensing signalling genes discovered in a bacteriophage genome. PLoS One 9:e85131. doi: 10.1371/journal.pone.0085131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hargreaves KR, Flores CO, Lawley TD, Clokie MR. 2014. Abundant and diverse clustered regularly interspaced short palindromic repeat spacers in Clostridium difficile strains and prophages target multiple phage types within this pathogen. mBio 5(5):e01045-13. doi: 10.1128/mBio.01045-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jansen R, Embden JD, Gaastra W, Schouls LM. 2002. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol 43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 137.Brouwer MS, Roberts AP, Hussain H, Williams RJ, Allan E, Mullany P. 2013. Horizontal gene transfer converts non-toxigenic Clostridium difficile strains into toxin producers. Nat Commun 4:2601. doi: 10.1038/ncomms3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hochhut B, Marrero J, Waldor MK. 2000. Mobilization of plasmids and chromosomal DNA mediated by the SXT element, a constin found in Vibrio cholerae O139. J Bacteriol 182:2043–2047. doi: 10.1128/JB.182.7.2043-2047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Whittle G, Hamburger N, Shoemaker NB, Salyers AA. 2006. A Bacteroides conjugative transposon, CTnERL, can transfer a portion of itself by conjugation without excising from the chromosome. J Bacteriol 188:1169–1174. doi: 10.1128/JB.188.3.1169-1174.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Popoff MR, Bouvet P. 2013. Genetic characteristics of toxigenic Clostridia and toxin gene evolution. Toxicon 75:63–89. doi: 10.1016/j.toxicon.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 141.O'Neill GL, Ogunsola FT, Brazier JS, Duerden BI. 1996. Modification of a PCR ribotyping method for application as a routine typing scheme for Clostridium difficile. Anaerobe 2:205–209. doi: 10.1006/anae.1996.0028. [DOI] [Google Scholar]
- 142.Hashimoto JG, Stevenson BS, Schmidt TM. 2003. Rates and consequences of recombination between rRNA operons. J Bacteriol 185:966–972. doi: 10.1128/JB.185.3.966-972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cerquetti M, Molinari A, Sebastianelli A, Diociaiuti M, Petruzzelli R, Capo C, Mastrantonio P. 2000. Characterization of surface layer proteins from different Clostridium difficile clinical isolates. Microb Pathog 28:363–372. doi: 10.1006/mpat.2000.0356. [DOI] [PubMed] [Google Scholar]
- 144.Spigaglia P, Barketi-Klai A, Collignon A, Mastrantonio P, Barbanti F, Rupnik M, Janezic S, Kansau I. 2013. Surface-layer (S-layer) of human and animal Clostridium difficile strains and their behaviour in adherence to epithelial cells and intestinal colonization. J Med Microbiol 62:1386–1393. doi: 10.1099/jmm.0.056556-0. [DOI] [PubMed] [Google Scholar]
- 145.Kryazhimskiy S, Plotkin JB. 2008. The population genetics of dN/dS. PLoS Genet 4:e1000304. doi: 10.1371/journal.pgen.1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Elliott B, Dingle KE, Didelot X, Crook D, Riley TV. 2014. The complexity and diversity of the pathogenicity locus in Clostridium difficile clade 5. Genome Biol Evol 6:3159–3170. doi: 10.1093/gbe/evu248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Castillo-Ramirez S, Harris SR, Holden MT, He M, Parkhill J, Bentley SD, Feil EJ. 2011. The impact of recombination on dN/dS within recently emerged bacterial clones. PLoS Pathog 7:e1002129. doi: 10.1371/journal.ppat.1002129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Loo VG, Poirier L, Miller MA, Oughton M, Libman MD, Michaud S, Bourgault AM, Nguyen T, Frenette C, Kelly M, Vibien A, Brassard P, Fenn S, Dewar K, Hudson TJ, Horn R, Rene P, Monczak Y, Dascal A. 2005. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N Engl J Med 353:2442–2449. doi: 10.1056/NEJMoa051639. [DOI] [PubMed] [Google Scholar]
- 149.Clements ACA, Magalhaes RJS, Tatem AJ, Paterson DL, Riley TV. 2010. Clostridium difficile PCR ribotype 027: assessing the risks of further worldwide spread. Lancet Infect Dis 10:395–404. doi: 10.1016/S1473-3099(10)70080-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mooney H. 2007. Annual incidence of MRSA falls in England, but C. difficile continues to rise. BMJ 335:958. doi: 10.1136/bmj.39388.597569.DB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Healthcare Commission. 2006. Investigation into outbreaks of Clostridium difficile at Stoke Mandeville Hospital, Buckinghamshire Hospitals NHS Trust. Commission for Healthcare Audit and Inspection, London, United Kingdom: http://www.buckinghamshirehospitals.nhs.uk/healthcarecommision/HCC-Investigation-into-the-Outbreak-of-Clostridium-difficile.pdf. [Google Scholar]
- 152.von Eichel-Streiber C, Laufenberg-Feldmann R, Sartingen S, Schulze J, Sauerborn M. 1992. Comparative sequence analysis of the Clostridium difficile toxins A and B. Mol Gen Genet 233:260–268. [DOI] [PubMed] [Google Scholar]
- 153.Moore P, Kyne L, Martin A, Solomon K. 2013. Germination efficiency of clinical Clostridium difficile spores and correlation with ribotype, disease severity and therapy failure. J Med Microbiol 62:1405–1413. doi: 10.1099/jmm.0.056614-0. [DOI] [PubMed] [Google Scholar]
- 154.Burns DA, Heap JT, Minton NP. 2010. The diverse sporulation characteristics of Clostridium difficile clinical isolates are not associated with type. Anaerobe 16:618–622. doi: 10.1016/j.anaerobe.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 155.Lanis JM, Barua S, Ballard JD. 2010. Variations in TcdB activity and the hypervirulence of emerging strains of Clostridium difficile. PLoS Pathog 6:e1001061. doi: 10.1371/journal.ppat.1001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ackermann G, Tang YJ, Kueper R, Heisig P, Rodloff AC, Silva J, Cohen SH. 2001. Resistance to moxifloxacin in toxigenic Clostridium difficile isolates is associated with mutations in gyrA. Antimicrob Agents Chemother 45:2348–2353. doi: 10.1128/AAC.45.8.2348-2353.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Freeman J, Bauer MP, Baines SD, Corver J, Fawley WN, Goorhuis B, Kuijper EJ, Wilcox MH. 2010. The changing epidemiology of Clostridium difficile infections. Clin Microbiol Rev 23:529–549. doi: 10.1128/CMR.00082-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Pépin J, Saheb N, Coulombe M, Alary M, Corriveau M, Authier S, Leblanc M, Rivard G, Bettez M, Primeau V. 2005. Emergence of fluoroquinolones as the predominant risk factor for Clostridium difficile-associated diarrhea: a cohort study during an epidemic in Quebec. Clin Infect Dis 41:1254–1260. doi: 10.1086/496986. [DOI] [PubMed] [Google Scholar]
- 159.De Almeida MN, Heffernan H, Dervan A, Bakker S, Freeman JT, Bhally H, Taylor SL, Riley TV, Roberts SA. 2013. Severe Clostridium difficile infection in New Zealand associated with an emerging strain, PCR-ribotype 244. N Z Med J 126:9–14. [PubMed] [Google Scholar]
- 160.Eyre DW, Tracey L, Elliott B, Slimings C, Huntington PG, Stuart RL, Korman TM, Kotsiou G, McCann R, Griffiths D, Fawley WN, Armstrong P, Dingle KE, Walker AS, Peto TE, Crook DW, Wilcox MH, Riley TV. 2015. Emergence and spread of predominantly community-onset Clostridium difficile PCR ribotype 244 infection in Australia, 2010 to 2012. Euro Surveill 20(10):pii=21059 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=21059. [DOI] [PubMed] [Google Scholar]
- 161.Riley TV. 2004. Nosocomial diarrhoea due to Clostridium difficile. Curr Opin Infect Dis 17:323–327. doi: 10.1097/01.qco.0000136930.83167.9b. [DOI] [PubMed] [Google Scholar]
- 162.McFarland LV, Mulligan ME, Kwok RY, Stamm WE. 1989. Nosocomial acquisition of Clostridium difficile infection. N Engl J Med 320:204–210. doi: 10.1056/NEJM198901263200402. [DOI] [PubMed] [Google Scholar]
- 163.Vonberg RP, Kuijper EJ, Wilcox MH, Barbut F, Tull P, Gastmeier P, van den Broek PJ, Colville A, Coignard B, Daha T, Debast S, Duerden BI, van den Hof S, van der Kooi T, Maarleveld HJ, Nagy E, Notermans DW, O'Driscoll J, Patel B, Stone S, Wiuff C. 2008. Infection control measures to limit the spread of Clostridium difficile. Clin Microbiol Infect 14(Suppl 5):2–20. doi: 10.1111/j.1469-0691.2008.01992.x. [DOI] [PubMed] [Google Scholar]
- 164.Noren T, Akerlund T, Back E, Sjoberg L, Persson I, Alriksson I, Burman LG. 2004. Molecular epidemiology of hospital-associated and community-acquired Clostridium difficile infection in a Swedish county. J Clin Microbiol 42:3635–3643. doi: 10.1128/JCM.42.8.3635-3643.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Walker AS, Eyre DW, Wyllie DH, Dingle KE, Harding RM, O'Connor L, Griffiths D, Vaughan A, Finney J, Wilcox MH, Crook DW, Peto TEA. 2012. Characterisation of Clostridium difficile hospital ward-based transmission using extensive epidemiological data and molecular typing. PLoS Med 9:e1001172. doi: 10.1371/journal.pmed.1001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Didelot X, Eyre DW, Cule M, Ip CL, Ansari MA, Griffiths D, Vaughan A, O'Connor L, Golubchik T, Batty EM, Piazza P, Wilson DJ, Bowden R, Donnelly PJ, Dingle KE, Wilcox M, Walker AS, Crook DW, Peto TE, Harding RM. 2012. Microevolutionary analysis of Clostridium difficile genomes to investigate transmission. Genome Biol 13:R118. doi: 10.1186/gb-2012-13-12-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Eyre DW, Cule ML, Wilson DJ, Griffiths D, Vaughan A, O'Connor L, Ip CL, Golubchik T, Batty EM, Finney JM, Wyllie DH, Didelot X, Piazza P, Bowden R, Dingle KE, Harding RM, Crook DW, Wilcox MH, Peto TE, Walker AS. 2013. Diverse sources of C. difficile infection identified on whole-genome sequencing. N Engl J Med 369:1195–1205. doi: 10.1056/NEJMoa1216064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Surawicz CM, Brandt LJ, Binion DG, Ananthakrishnan AN, Curry SR, Gilligan PH, McFarland LV, Mellow M, Zuckerbraun BS. 2013. Guidelines for diagnosis, treatment, and prevention of Clostridium difficile infections. Am J Gastroenterol 108:478–498. doi: 10.1038/ajg.2013.4. [DOI] [PubMed] [Google Scholar]
- 169.Pepin J, Alary ME, Valiquette L, Raiche E, Ruel J, Fulop K, Godin D, Bourassa C. 2005. Increasing risk of relapse after treatment of Clostridium difficile colitis in Quebec, Canada. Clin Infect Dis 40:1591–1597. doi: 10.1086/430315. [DOI] [PubMed] [Google Scholar]
- 170.Figueroa I, Johnson S, Sambol SP, Goldstein EJ, Citron DM, Gerding DN. 2012. Relapse versus reinfection: recurrent Clostridium difficile infection following treatment with fidaxomicin or vancomycin. Clin Infect Dis 55:S104–S109. doi: 10.1093/cid/cis357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Eyre DW, Babakhani F, Griffiths D, Seddon J, Del Ojo Elias C, Gorbach SL, Peto TE, Crook DW, Walker AS. 2014. Whole-genome sequencing demonstrates that fidaxomicin is superior to vancomycin for preventing reinfection and relapse of infection with Clostridium difficile. J Infect Dis 209:1446–1451. doi: 10.1093/infdis/jit598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Khanna S, Pardi DS. 2012. Community-acquired Clostridium difficile infection: an emerging entity. Clin Infect Dis 55:1741–1742. doi: 10.1093/cid/cis722. [DOI] [PubMed] [Google Scholar]
- 173.Khanna S, Pardi DS, Aronson SL, Kammer PP, Orenstein R, St Sauver JL, Harmsen WS, Zinsmeister AR. 2012. The epidemiology of community-acquired Clostridium difficile infection: a population-based study. Am J Gastroenterol 107:89–95. doi: 10.1038/ajg.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kuntz JL, Chrischilles EA, Pendergast JF, Herwaldt LA, Polgreen PM. 2011. Incidence of and risk factors for community-associated Clostridium difficile infection: a nested case-control study. BMC Infect Dis 11:194. doi: 10.1186/1471-2334-11-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Squire MM, Riley TV. 2012. Clostridium difficile infection: the next big thing! Microbiol Aus 33:163–164. [Google Scholar]
- 176.Goorhuis A, Bakker D, Corver J, Debast SB, Harmanus C, Notermans DW, Bergwerff AA, Dekker FW, Kuijper EJ. 2008. Emergence of Clostridium difficile infection due to a new hypervirulent strain, polymerase chain reaction ribotype 078. Clin Infect Dis 47:1162–1170. doi: 10.1086/592257. [DOI] [PubMed] [Google Scholar]
- 177.Stewart DB, Berg AS, Hegarty JP. 2014. Single nucleotide polymorphisms of the tcdC gene and presence of the binary toxin gene predict recurrent episodes of Clostridium difficile infection. Ann Surg 260:299–304. doi: 10.1097/SLA.0000000000000469. [DOI] [PubMed] [Google Scholar]
- 178.Hensgens MP, Keessen EC, Squire MM, Riley TV, Koene MG, de Boer E, Lipman LJ, Kuijper EJ. 2012. Clostridium difficile infection in the community: a zoonotic disease? Clin Microbiol Infect 18:635–645. doi: 10.1111/j.1469-0691.2012.03853.x. [DOI] [PubMed] [Google Scholar]
- 179.Songer JG, Anderson MA. 2006. Clostridium difficile: an important pathogen of food animals. Anaerobe 12:1–4. doi: 10.1016/j.anaerobe.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 180.Keessen EC, Gaastra W, Lipman LJ. 2011. Clostridium difficile infection in humans and animals, differences and similarities. Vet Microbiol 153:205–217. doi: 10.1016/j.vetmic.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 181.Pasquale V, Romano V, Rupnik M, Capuano F, Bove D, Aliberti F, Krovacek K, Dumontet S. 2012. Occurrence of toxigenic Clostridium difficile in edible bivalve molluscs. Food Microbiol 31:309–312. doi: 10.1016/j.fm.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 182.Zidaric V, Zemljic M, Janezic S, Kocuvan A, Rupnik M. 2008. High diversity of Clostridium difficile genotypes isolated from a single poultry farm producing replacement laying hens. Anaerobe 14:325–327. doi: 10.1016/j.anaerobe.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 183.Weese JS, Wakeford T, Reid-Smith R, Rousseau J, Friendship R. 2010. Longitudinal investigation of Clostridium difficile shedding in piglets. Anaerobe 16:501–504. doi: 10.1016/j.anaerobe.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 184.Weese JS, Finley R, Reid-Smith RR, Janecko N, Rousseau J. 2010. Evaluation of Clostridium difficile in dogs and the household environment. Epidemiol Infect 138:1100–1104. doi: 10.1017/S0950268809991312. [DOI] [PubMed] [Google Scholar]
- 185.Weese JS, Armstrong J. 2003. Outbreak of Clostridium difficile-associated disease in a small animal veterinary teaching hospital. J Vet Intern Med 17:813–816. doi: 10.1111/j.1939-1676.2003.tb02519.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Thean S, Elliott B, Riley TV. 2011. Clostridium difficile in horses in Australia—a preliminary study. J Med Microbiol 60:1188–1192. doi: 10.1099/jmm.0.030908-0. [DOI] [PubMed] [Google Scholar]
- 187.Schneeberg A, Neubauer H, Schmoock G, Grossmann E, Seyboldt C. 2013. Presence of Clostridium difficile PCR ribotype clusters related to 033, 078 and 045 in diarrhoeic calves in Germany. J Med Microbiol 62:1190–1198. doi: 10.1099/jmm.0.056473-0. [DOI] [PubMed] [Google Scholar]
- 188.Rupnik M, Widmer A, Zimmermann O, Eckert C, Barbut F. 2008. Clostridium difficile toxinotype V, ribotype 078, in animals and humans. J Clin Microbiol 46:2146. doi: 10.1128/JCM.00598-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Riley TV, Adams JE, O'Neill GL, Bowman RA. 1991. Gastrointestinal carriage of Clostridium difficile in cats and dogs attending veterinary clinics. Epidemiol Infect 107:659–665. doi: 10.1017/S0950268800049359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Medina-Torres CE, Weese JS, Staempfli HR. 2011. Prevalence of Clostridium difficile in horses. Vet Microbiol 152:212–215. doi: 10.1016/j.vetmic.2011.04.012. [DOI] [PubMed] [Google Scholar]
- 191.Knight DR, Riley TV. 2013. Prevalence of gastrointestinal Clostridium difficile carriage in Australian sheep and lambs. Appl Environ Microbiol 79:5689–5692. doi: 10.1128/AEM.01888-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Indra A, Lassnig H, Baliko N, Much P, Fiedler A, Huhulescu S, Allerberger F. 2009. Clostridium difficile: a new zoonotic agent? Wien Klin Wochenschr 121:91–95. doi: 10.1007/s00508-008-1127-x. [DOI] [PubMed] [Google Scholar]
- 193.Hopman NE, Keessen EC, Harmanus C, Sanders IM, van Leengoed LAMG, Kuijper EJ, Lipman LJ. 2011. Acquisition of Clostridium difficile by piglets. Vet Microbiol 149:186–192. doi: 10.1016/j.vetmic.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 194.Costa MC, Stampfli HR, Arroyo LG, Pearl DL, Weese JS. 2011. Epidemiology of Clostridium difficile on a veal farm: prevalence, molecular characterization and tetracycline resistance. Vet Microbiol 152:379–384. doi: 10.1016/j.vetmic.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 195.Avbersek J, Janezic S, Pate M, Rupnik M, Zidaric V, Logar K, Vengust M, Zemljic M, Pirs T, Ocepek M. 2009. Diversity of Clostridium difficile in pigs and other animals in Slovenia. Anaerobe 15:252–255. doi: 10.1016/j.anaerobe.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 196.Alvarez-Perez S, Blanco JL, Martinez-Nevado E, Pelaez T, Harmanus C, Kuijper E, Garcia ME. 2014. Shedding of Clostridium difficile PCR ribotype 078 by zoo animals, and report of an unstable metronidazole-resistant isolate from a zebra foal (Equus quagga burchellii). Vet Microbiol 169:218–222. doi: 10.1016/j.vetmic.2013.12.018. [DOI] [PubMed] [Google Scholar]
- 197.Debast SB, van Leengoed LA, Goorhuis A, Harmanus C, Kuijper EJ, Bergwerff AA. 2009. Clostridium difficile PCR ribotype 078 toxinotype V found in diarrhoeal pigs identical to isolates from affected humans. Environ Microbiol 11:505–511. doi: 10.1111/j.1462-2920.2008.01790.x. [DOI] [PubMed] [Google Scholar]
- 198.Rodriguez-Palacios A, Stampfli HR, Duffield T, Peregrine AS, Trotz-Williams LA, Arroyo LG, Brazier JS, Weese JS. 2006. Clostridium difficile PCR ribotypes in calves, Canada. Emerg Infect Dis 12:1730–1736. doi: 10.3201/eid1211.051581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Keel K, Brazier JS, Post KW, Weese S, Songer JG. 2007. Prevalence of PCR ribotypes among Clostridium difficile isolates from pigs, calves, and other species. J Clin Microbiol 45:1963–1964. doi: 10.1128/JCM.00224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Rupnik M, Songer JG. 2010. Clostridium difficile: its potential as a source of foodborne disease. Adv Food Nutr Res 60:53–66. doi: 10.1016/S1043-4526(10)60003-4. [DOI] [PubMed] [Google Scholar]
- 201.Weese JS. 2010. Clostridium difficile in food—innocent bystander or serious threat? Clin Microbiol Infect 16:3–10. doi: 10.1111/j.1469-0691.2009.03108.x. [DOI] [PubMed] [Google Scholar]
- 202.Songer JG, Trinh HT, Killgore GE, Thompson AD, McDonald LC, Limbago BM. 2009. Clostridium difficile in retail meat products, USA, 2007. Emerg Infect Dis 15:819–821. doi: 10.3201/eid1505.081071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Rupnik M. 2010. Clostridium difficile: (re)emergence of zoonotic potential. Clin Infect Dis 51:583–584. doi: 10.1086/655693. [DOI] [PubMed] [Google Scholar]
- 204.Knight DR, Squire MM, Riley TV. 2015. Nationwide surveillance study of Clostridium difficile in Australian neonatal pigs shows high prevalence and heterogeneity of PCR ribotypes. Appl Environ Microbiol 81:119–123. doi: 10.1128/AEM.03032-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Squire MM, Riley TV. 2013. Clostridium difficile infection in humans and piglets: a ‘One Health’ opportunity. Curr Top Microbiol Immunol 365:299–314. doi: 10.1007/82_2012_237. [DOI] [PubMed] [Google Scholar]
- 206.Riley TV. 2009. Is Clostridium difficile a threat to Australia's biosecurity? Med J Aust 190:661–662. [DOI] [PubMed] [Google Scholar]
- 207.Bakker D, Corver J, Harmanus C, Goorhuis A, Keessen EC, Fawley WN, Wilcox MH, Kuijper EJ. 2010. Relatedness of human and animal Clostridium difficile PCR ribotype 078 isolates determined on the basis of multilocus variable-number tandem-repeat analysis and tetracycline resistance. J Clin Microbiol 48:3744–3749. doi: 10.1128/JCM.01171-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Knetsch CW, Connor TR, Mutreja A, van Dorp SM, Sanders IM, Browne HP, Harris D, Lipman L, Keessen EC, Corver J, Kuijper EJ, Lawley TD. 2014. Whole genome sequencing reveals potential spread of Clostridium difficile between humans and farm animals in the Netherlands, 2002 to 2011. Euro Surveill 19(45):pii=20954 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20954. [DOI] [PMC free article] [PubMed] [Google Scholar]