Abstract
Objective(s):
The purpose of this study was to conduct a meta-analysis on human microRNAs (miRNAs) expression data of endometriosis tissue profiles versus those of normal controls and to identify novel putative diagnostic markers.
Materials and Methods:
PubMed, Embase, Web of Science, Ovid Medline were used to search for endometriosis miRNA expression profiling studies of endometriosis. The miRNAs expression data were extracted, and study quality of each article was assessed. The frequently reported miRNAs with consistent regulation were screened out by a meta-profiling algorithm. The putative targets of consistently expressed miRNAs were predicted by using four target prediction tools (TargetScan, PicTar, miRanda, miRDB), and gene ontology pathway enrichment analysis (KEGG and Panther pathways) of the miRNA targets were carried out with GeneCodis web tool.
Results:
A total of 194 related literatures were retrieved in four databases. One hundred and thirty four differentially expressed miRNAs were found in the 12 microRNA expression profiling studies that compared endometriosis tissues with normal tissues, with 28 miRNAs reported in at least two studies, and 9882 candidate genes retrieved for 13 consistently expressed miRNAs. Kyoto encyclopedia of genes and genomes (KEGG) and Panther pathways enrichment analysis showed that endometriosis related differently expressed miRNA targets were mainly enriched in cancer, endocytosis, Wnt signalling pathway, and angiogenesis. It showed that these differently expressed miRNAs and gene are potential biomarkers of endometriosis.
Conclusion:
miRNAs appear to be potent regulators of gene expression in endometriosis and its associated reproductive disorders, raising the prospect of using miRNAs as biomarkers and therapeutic agent in endometriosis.
Keywords: Endometriosis, MicroRNAs, Pathway analysis, Profiling, Target prediction
Introduction
MicroRNAs (miRNAs), a class of short noncoding RNA molecules, are proposed as promising biomarkers for early cancer detection and accurate prognosis, as well as targets for efficient treatment. Some miRNAs are post-transcriptional regulators of gene expression and implicated in central biological processes such as cell proliferation, differentiation and apoptosis (1, 2). Endometriosis is a common disease of reproductive-age women, which has complex pathogenesis. The lesions are extensive, highly invasive and recu-rrent, presenting malignant clinical behavior (3, 4). With the development of antisense technology and gene therapy, miRNA is expected to become a new strategy in endometriosis diagnosis and treatment (5, 6). It has been proved presently that miRNAs reside widely in eukaryotes, and are the largest gene family, participating and regulating various important life processes including cell differentiation, prolife-ration and apoptosis (7). As a method and result of the epigenetic modifications, latest research indicates that miRNA may play important role in the occurrence, development and prognosis of endometriosis, thus providing novel approach for the diagnosis and therapy of endometriosis. MiRNAs are key regulatory elements that control many genes expression and play crucial roles in many biological processes (8). Meta-analysis of mass miRNA expression profiling may uncover potential regulatory mechanisms by which microRNAs result in endometriosis.
Materials and Methods
Search strategies and study selection
PubMed (http://www.ncbi.nlm.nih.gov/pubmed), Embase (http://www.elsevier.com/online-tools/-embase), Web of Science (http://thomsonreuters.com/thomson-reuters-web-of-science/), Ovid Med-line (http://gateway.ovid.com) were used to search for endometriosis miRNA expression profiling studies published from January 2003 to October 2014 (last accessed on 20 October 2014), by means of the key words: endometriosis AND microRNA.
Eligible studies had to meet the following criteria: (a) they were miRNA expression profiling studies in endometriosis patients; (b) they used tissue samples obtained from surgically resected endometriosis and corresponding eutopic or normal tissues for comparison; (c) use of miRNA microarray methods; (d) publishing a cut-off criteria of differentially expressed miRNAs. Therefore, the miRNA profiling studies using the serum samples of endometriosis patients or endometriosis cells in vitro were excluded. Review articles of miRNA expression profiles in endometriosis were also excluded. As repeated efforts can improve reliability and reduce error, valuable candidate miRNAs in this paper are defined as those validated and consistently reported by at least two studies.
Data abstraction
Two investigators (SW and YK) independently evaluated and extracted the data with the standard protocol and with all the discrepancies resolved by a third investigator (HX). From the full text and corresponding supplement information, the following eligibility items were collected and recorded for each study: author, journal and year of publication, location of study, selection and characteristics of recruited endometriosis patients, platform of miRNA expression profiling, author defined cut-off criteria of statistically differentially expressed miRNAs and the list of up- and down-regulated miRNA features, and their corresponding fold change. Each included studies comparing miRNA expression between surgically resected endometriosis tissues and eutopic endometrium or normal tissues provided a list of differentially expressed miRNAs.
MiRNA target prediction and pathway enrichment analysis
Consensus targets were defined as genes predicted by at least two algorithms of four target prediction tools, including TargetScan, PicTar, miRanda, and miRDB. Although all three of these articles described the expression profile of miRNAs in endometriosis, they did not systematically predict the biological process and pathway of the identified miRNAs. Therefore, we used GeneCodis web tool (http://genecodis.dacya.ucm.es/)(9, 10) to predict the biological process (GO process) and to perform pathway enrichment analysis (KEGG and Panther pathways) of all miRNAs that were identified as consistently expressed miRNAs in the eligible references.
Results
Selection and overview of the datasets
A total of 194 related literatures were indexed in PubMed, Embase, Web of Science and Ovid database. According to the inclusion criteria and identification of duplicate publication, only 12 publications seemed to meet all of the inclusion criteria and none of the exclusion criteria (Figure 1). The characteristics of these studies are listed in Table 1 in chronological order of the published time (Table 1).
Figure 1.
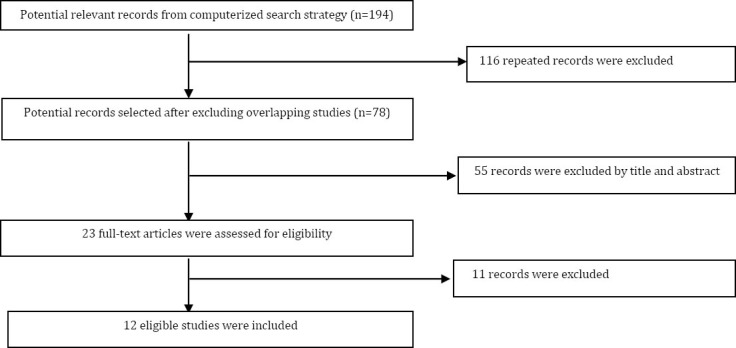
A flow diagram of the literature search and study selection used in this study
Table 1.
Twelve microarray-based human endometriosis miRNA expression profiling studies
| First author (reference) | Year | Region | Assay type | Number of samples* | Cut-off criteria | Up-regulated miRNAs | Down-regulated miRNAs |
|---|---|---|---|---|---|---|---|
| Pan Q (2) | 2007 | USA | MirVana RNA isolation and enrichment kits | 4 pairs+4 N | P<0.05 | 48 | 17 |
| Toloubeydokhti T (11) | 2008 | USA | TaqMan MicroRNA Array | 5 pairs+5 N+4 E | 5 pairs + 5 N+4 E | 1 | 2 |
| Ohlsson Teague EM (12) | 2009 | Australia | qRT-PCR | 7 pairs | P<0.05 FC>1.5 | 14 | 8 |
| Filigheddu N (8) | 2010 | Italy | TaqMan MicroRNA Array y | 13 pairs | P<0.01 FC>2 | 27 | 23 |
| Ramon’ LA (13) | 2011 | Spain | TaqMan assay | 58 pairs+38 N | P<0.05 | 3 | 3 |
| Hawkins SM (14) | 2011 | USA | TaqMan assay | 9 N and 10 E | P<0.01 FC>1.5 | 10 | 12 |
| Petracco R (15) | 2011 | USA | qRT-PCR | 50 N+32 E | P<0.05 | 2 | 0 |
| Dai L (16) | 2012 | China | qRT-PCR | 12 pairs+12 N | P<0.01 | 1 | 0 |
| Liu S (17) | 2012 | China | TaqMan MicroRNA Array | 31 pairs+27 N | P<0.05 | 0 | 1 |
| Lin SC (18) | 2012 | China | qRT-PCR | 10 pairs+37 N+17 E | P<0.05 | 1 | 0 |
| Shen L (19) | 2013 | China | qRT-PCR | 23 pairs+15 N | P<0.05 | 0 | 2 |
| Laudanski P (20) | 2013 | Poland | TaqMan MicroRNA Array | 21E+25N | P<0.05 FC>2 | 2 | 13 |
E= endomeriosis, N=control, pairs= paired eutopic and ectopic endometrial tissues
Determination of the consistently reported miRNAs
A total of 134 differentially expressed miRNAs were obtained in the 12 miRNAs expression profiling studies that compared endometriosis tissues with normal tissues, with 28 miRNAs reported in at least two studies, including 4 consistently reported up-regulated miRNAs(mir-202, mir-365, mir-1 and mir-150), 9 consistently reported down-regulated miRNAs (mir-23b, mir-200a, mir-200b, mir-200c,mir- 15b, mir-106b, mir-196b, mir-141, mir-375) (Table 2). And 15 inconsistently reported direction miRNA (Table 3).
Table 2.
Consistently reported miRNAs (n = 13) in profiling studies (endometriosis tissues versus eutopic tissues or normal tissues)
| Direction of expression | miRNA name | Number of studies with same direction | Total number of tissue samples | P-value |
|---|---|---|---|---|
| ↑ | mir-202 | 2 | 32 | <0.001 |
| ↑ | mir-365 | 2 | 20 | <0.05 |
| ↑ | mir-1 | 2 | 20 | <0.05 |
| ↑ | mir-150 | 2 | 20 | <0.001 |
| ↓ | mir-23b | 3 | 40 | <0.001 |
| ↓ | mir-200b | 3 | 39 | <0.05 |
| ↓ | mir-200a | 3 | 39 | <0.05 |
| ↓ | mir-200c | 2 | 32 | <0.05 |
| ↓ | mir-141 | 2 | 26 | <0.05 |
| ↓ | mir-375 | 2 | 32 | ≤0.01 |
| ↓ | mir-106b | 2 | 34 | <0.05 |
| ↓ | mir-196b | 2 | 20 | ≤0.05 |
| ↓ | mir-15b | 2 | 64 | <0.05 |
Table 3.
Inconsistently reported miRNAs (n=15) in profiling studies (endometriosis tissues versus normal tissues)
| Direction of expression | miRNA name | Number of studies with same direction | Total number of tissue samples | P-value |
|---|---|---|---|---|
| ↑ | mir-100 | 3 | 39 | ≤0.01 |
| ↓ | mir-100 | 1 | 8 | <0.05 |
| ↑ | mir-17-5p | 1 | 9 | <0.05 |
| ↓ | mir-17-5p | 3 | 79 | <0.05 |
| ↑ | mir-29c | 3 | 39 | <0.01 |
| ↓ | mir-29c | 1 | 8 | <0.05 |
| ↑ | mir-20a | 1 | 49 | <0.05 |
| ↓ | mir-20a | 3 | 78 | ≤0.05 |
| ↑ | mir-126 | 2 | 20 | <0.05 |
| ↓ | mir-126 | 2 | 39 | <0.001 |
| ↑ | mir-145 | 2 | 20 | <0.05 |
| ↓ | mir-145 | 2 | 29 | <0.05 |
| ↑ | mir-125a | 2 | 65 | <0.001 |
| ↓ | mir-125a | 1 | 8 | <0.05 |
| ↑ | mir-99a | 2 | 20 | <0.001 |
| ↓ | mir-99a | 1 | 8 | <0.05 |
| ↑ | mir-143 | 2 | 20 | <0.001 |
| ↓ | mir-143 | 1 | 8 | <0.05 |
| ↑ | mir-23a | 1 | 9 | 0.006 |
| ↓ | mir-23a | 2 | 31 | 0.006 |
| ↑ | mir-222 | 1 | 41 | <0.001 |
| ↓ | mir-222 | 2 | 29 | <0.05 |
| ↑ | mir-199a | 1 | 13 | ≤0.01 |
| ↓ | mir-199a | 2 | 20 | <0.05 |
| ↑ | mir-125b | 1 | 31 | <0.05 |
| ↓ | mir-125b | 1 | 8 | <0.05 |
| ↑ | mir-21 | 1 | 58 | <0.01 |
| ↓ | mir-21 | 1 | 8 | <0.01 |
| ↑ | mir-221 | 1 | 13 | ≤0.01 |
| ↓ | mir-221 | 1 | 8 | <0.05 |
Pathway analysis of miRNA targets
By using four target prediction tools (TargetScan, PicTar, miRanda, and miRDB), 9882 candidate genes were predicted for 13 inconsistently reported miRNAs. And all the predicted candidate genes were analyzed by pathways enriched analysis. KEGG and Panther pathways enrichment analysis showed that 9882 endometriosis-related miRNA targets were mainly cancer-related pathways, endocytosis, Wnt signalling pathway, and angiogenesis (Table 4).
Table 4.
Top ten of the significant GO processes, KEGG pathways and Panther pathways enriched with miRNA targets involved in endometriosis
| GO processes | Process | FDR | Target number |
|---|---|---|---|
| GO:0006355 | Regulation of transcription, DNA-dependent | 5.55032e-182 | 963 |
| GO:0007165 | signal transduction | 2.72922e-125 | 694 |
| GO:0007275 | Multicellular organismal development | 7.66 e -100 | 557 |
| GO:0045944 | Positive regulation transcription of from RNA polymerase II promoter | 4.43532e-86 | 378 |
| GO:0007399 | Nervous system development | 3.20348e-70 | 280 |
| GO:0007155 | Cell adhesion | 8. 37777e-68 | 342 |
| GO:0045893 | Positive regulation of transcription, DNA dependent | 3.78756e -66 | 301 |
| GO:0007268 | Transmission synaptic | 6.22556e-57 | 250 |
| GO:0055085 | Transmembrane transport | 9. 4472e-56 | 353 |
| GO:0000122 | Negative regulation transcription of from RNA polymerase II promoter | 1. 0109e-55 | 263 |
| KEGG pathways | Pathway | FDR | Target number |
|---|---|---|---|
| KEGG:5200 | Pathways in cancer | 3.54381e-53 | 218 |
| KEGG:4144 | Endocytosis | 5.04266e-44 | 144 |
| KEGG:4360 | Axon guidance | 5.91358e-35 | 101 |
| KEGG:4810 | Regulation of actin cytoskeleton | 2.65172e-34 | 140 |
| KEGG:4010 | MAPK signaling pathway | 4.03001e-33 | 162 |
| KEGG:4080 | Neuroactive ligand receptor | 5.56784e-29 | 158 |
| KEGG:4310 | Wnt signaling pathway | 5.87106e-29 | 105 |
| KEGG:4510 | Focal adhesion | 8.49293e-29 | 127 |
| KEGG:4060 | Cytokine signaling pathway | 4.84554 e-27 | 151 |
| KEGG:4062 | Chemokine signaling pathway | 1.23502e-26 | 119 |
| Panther pathways | Pathway | FDR | Target number |
|---|---|---|---|
| P00057 | Wnt signalling pathway | 3.24683e-35 | 173 |
| P00005 | Angiogenesis | 3.83276e-34 | 113 |
| P00034 | Integrin signalling pathway | 4.17619e-29 | 109 |
| P00047 | PDGF signalling pathway | 6.46839e-25 | 90 |
| P00031 | Inflammation mediated by chemokine cytokine signaling pathway | 3.43609e-23 | 119 |
| P00026 | Heterotrimeric G-protein signaling pathway---Gi alpha and Gs, and alpha mediated pathway | 3.32868e-23 | 98 |
| P00012 | Cadherin signalling pathway | 2. 44252e-19 | 88 |
| P00018 | EGF receptor signaling pathway | 2.29847e-18 | 75 |
| P00021 | FGF signaling Pathway | 7.78382e-18 | 72 |
| P00004 | Alzheimer disease --presenilin pathway | 4.27409e-17 | 75 |
Discussion
In this paper, we adopt a meta-analysis to screen endometriosis-related miRNAs from miRNA expression profile data of independent profiling studies, and to obtain conservative target predictions for all miRNAs lists of interest using four up-to-date prediction algorithms (TargetScan, PicTar, PicTar, miRanda and miRDB). And pathway enrichment analysis using different target prediction algorithms were concordant for all of the consistently expressed miRNAs. We found out a meta-signature of four up-regulated and nine down-regulated miRNAs. Although the selected method for miRNA expression meta-analysis relates to analysis of the primary expression parameters, such method was often not possible due to the unavailability of the primary data. A great number of miRNAs known in the present and different technical platforms adopted in certain study make the appropriate synthesis of primary data very complex. Moreover, the relatively small sample sizes of microarray data may have resulted in inconformity of biological consequences. A meta-analysis method could eliminate these disadvantages and directly compare original data extracted from different technical platforms (21).
All the data from the 12 published studies consisted of more than 317 endometriosis and eutopic or normal tissue samples were analyzed directly, and a series of early researches (2, 11, 12, 14) were analyzed with less than ten specimens, which may have unreliable results. Ohlsson Teague et al (22) assessed miRNA expression by microarray analysis in seven paired ectopic and eutopic endometrial tissues and identified 14 up-regulated and eight down-regulated miRNAs in endometriosis tissues. Pan et al (2) identified 48 differently expressed miRNAs in a microarray analysis of endometrium of women with and without endometriosis, and mir-23b down-regulation. Compared with nonendometriosis control endometrium, mir-200b expression level decreased obviously in endometriosis (14).
Angiogenesis is the basis of the occurrence and development in endometriosis. Just as tumor, implantation metastasis of endometriosis is based on new neurovascular formation and proliferation, invasion of extracellular matrix, and lesion formation. Recent studies revealed that miRNA may involve in regulation of angiogenesis, and some miRNAs have the distinction of anti-vasoformation, such as mir-15b, mir-16, mir-221, and mir-222. Interestingly, the results suggest that mir-200 family members (mir-200a, mir-200b, mir-200c, mir-141, which were known to be cancer-related miRNAs down-regulated, while expression of some others (mir-21 (2, 13), mir-199a (2, 13, 16)) were inconsistently reported in the selected studies. Some miRNAs which have been specifically investigated in several studies (such as miR-17-5p, mir-23a,mir-23b (19, 23); mir-20a(18); mir-126(17); mir-135a, mir-135b (15)) were found among both up-regulated and down-regulated. All mirRNA-200 family members (miR-200a, miR-200b, miR-200c, miR-141, miR-429) were found down-regulated, and reached the statistical significance in our analysis, of course, mir-429 was only reported in just one study. Currently, our analysis is limited to comparison of endometriosis and eutopic or normal control tissue. The miR-200 family has been reported to be a fundamental regulator of epithelial-mesenchymal transition, thus enhancing their roles in cancer progression. As a founding member in miR-200 family, miR-200b attracts much focus in carcinogenesis in recent years. Down-regulation of miR-200b has been detected in several malignancies (24-26) and in endometriosis (27). The set of miRNAs with significantly decreased expression levels include all members of the miR-200 family known to be involved in the epithelial to mesenchymal transition process (28).
From the clinical viewpoint, it would be meaningful if the discovery of targets correlated with patient diagnosis, therapy and prognosis. These meta-signature miRNAs and gene-to-behavior pathway affected by them are potential candidates as diagnostic and therapeutic agents in endometriosis. Our analysis also concentrated on the challenges connected with the development of miRNA-based tests and emphasizes to the importance of strict inspection of the results before conducting clinical trials. Perez-Iratxeta et al (29) applied a combination of data mining and gene ontology to develop a scoring system for discovering disease-associated genes based on text descriptions of genetically inherited diseases and functional annotations of genes. The scoring showed that the chance of validating potential gene is high for some diseases. Mohammadi et al (30) used microarray data mining and gene ontology to identify disease-causing genes, and predicted marker genes with high accuracy. It indicates that the above method of comparing data is derived from different organisms in studies of disease and human health. Here we showed some familiar and novel pathways for endometriosis, including the pathways in cancer, endocytosis and axon guidance. New visions were also provided into Wnt signaling, angiogenesis, and integrin signaling pathway. These pathways that regulate stem cell transformation indicate the role of miRNAs in endometriosis cell deregulation and development. Therefore, some of these miRNAs may be selected as diagnostic index or acted as therapeutic agent for endometriosis. MiRNAs appear to be potent regulators of gene expression in endometriosis and its associated reproductive disorders, raising the prospect of using miRNAs as biomarkers and therapeutic agent in endometriosis.
It should be emphasized that there were some limitations in our analysis. In this study, the inclusion of researches was based on different assay types. The total number of tissues from available data was relatively small, and tissues included endometriosis tissue, eutopic endometrium, and normal tissue. We extracted data retrieved from different studies, and. published large prospective researches were unattainable for endometriosis. In addition, there were only 12 eligible studies in our meta-analysis, which may also lead to a bias in the results.
Conclusion
By systematic enrichment analysis, we found that these differently expressed miRNAs and gene are potential biomarkers of endometriosis. Our analysis also highlights the challenges connected with the development of miRNA-based tests and emphasizes the need for rigorous evaluation of the results before proceeding to clinical trials.
Acknowledgment
The results described in this paper were part of student thesis. We are grateful to Sibiao SU and Xiaoxiang Wei for investigating their dissertations.
Footnotes
Conflict of interest
The authors declare the manuscript has not been published previously, in any language. This research is not involved any pharmacological agents, devices or medical technology, there is not any interest relating with any funding from or pecuniary interests in companies.
References
- 1.Majid S, Saini S, Dar AA, Hirata H, Shahryari V, Tanaka Y, et al. MicroRNA-205 inhibits Src-mediated oncogenic pathways in renal cancer. Cancer Res. 2011;71:2611–2621. doi: 10.1158/0008-5472.CAN-10-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan Q, Luo X, Toloubeydokhti T, Chegini N. The expression profile of micro-RNA in endometrium and endometriosis and the influence of ovarian steroids on their expression. Mol Hum Rep. 2007;13:797–806. doi: 10.1093/molehr/gam063. [DOI] [PubMed] [Google Scholar]
- 3.Jiang QY, Wu RJ. Growth mechanisms of endometriotic cells in implanted places: a review. Gynecol Endocrinol. 2012;28:562–567. doi: 10.3109/09513590.2011.650662. [DOI] [PubMed] [Google Scholar]
- 4.Vlahos NF, Economopoulos KP, Fotiou S. Endometriosis, in vitro fertilisation and the risk of gynaecological malignancies, including ovarian and breast cancer. Best Pract Res Clin Obstet Gynaecol. 2010;24:39–50. doi: 10.1016/j.bpobgyn.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Suryawanshi S, Vlad AM, Lin HM, Mantia-Smaldone G, Laskey R, Lee M, et al. Plasma microRNAs as novel biomarkers for endometriosis and endometriosis-associated ovarian cancer. Clin Cancer Res. 2013;19:1213–1224. doi: 10.1158/1078-0432.CCR-12-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verit FF, Cetin O. Biomarkers of endometriosis. Fertil Steril. 2013;100:e19. doi: 10.1016/j.fertnstert.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 7.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 8.Chang SH, Hla T. Gene regulation by RNA binding proteins and microRNAs in angiogenesis. Trends Mol Med. 2011;17:650–658. doi: 10.1016/j.molmed.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tabas-Madrid D, Nogales-Cadenas R, Pascual-Montano A. GeneCodis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucl Acids Res. 2012;40:W478–483. doi: 10.1093/nar/gks402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nogales-Cadenas R, Carmona-Saez P, Vazquez M, Vicente C, Yang X, Tirado F, et al. GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res. 2009;37:W317–322. doi: 10.1093/nar/gkp416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toloubeydokhti T, Pan Q, Luo X, Bukulmez O, Chegini N. The expression and ovarian steroid regulation of endometrial micro-RNAs. Reprod Sci. 2008;15:993–1001. doi: 10.1177/1933719108324132. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Ohlsson Teague EM, Van der Hoek KH, Van der Hoek MB, Perry N, Wagaarachchi P, Robertson SA, et al. MicroRNA-regulated pathways associated with endometriosis. Mol Endocrinol. 2009;23:265–275. doi: 10.1210/me.2008-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramon LA, Braza-Boils A, Gilabert-Estelles J, Gilabert J, Espana F, Chirivella M, et al. MicroRNAs expression in endometriosis and their relation to angiogenic factors. Hum Reprod. 2011;26:1082–1090. doi: 10.1093/humrep/der025. [DOI] [PubMed] [Google Scholar]
- 14.Hawkins SM, Creighton CJ, Han DY, Zariff A, Anderson ML, Gunaratne PH, et al. Functional microRNA involved in endometriosis. Mol Endocrinol. 2011;25:821–832. doi: 10.1210/me.2010-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petracco R, Grechukhina O, Popkhadze S, Massasa E, Zhou Y, Taylor HS. MicroRNA 135 regulates HOXA10 expression in endometriosis. J Clin Endocrinol Metab. 2011;96:E1925–1933. doi: 10.1210/jc.2011-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai L, Gu L, Di W. MiR-199a attenuates endometrial stromal cell invasiveness through suppression of the IKKbeta/NF-kappaB pathway and reduced interleukin-8 expression. Mol Hum Reprod. 2012;18:136–145. doi: 10.1093/molehr/gar066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu S, Gao S, Wang XY, Wang DB. Expression of miR-126 and Crk in endometriosis: miR-126 may affect the progression of endometriosis by regulating Crk expression. Arch Gynecol Obstet. 2012;285:1065–1072. doi: 10.1007/s00404-011-2112-6. [DOI] [PubMed] [Google Scholar]
- 18.Lin SC, Wang CC, Wu MH, Yang SH, Li YH, Tsai SJ. Hypoxia-induced microRNA-20a expression increases ERK phosphorylation and angiogenic gene expression in endometriotic stromal cells. J Clin Endocrinol Metab. 2012;97:E1515–1523. doi: 10.1210/jc.2012-1450. [DOI] [PubMed] [Google Scholar]
- 19.Shen L, Yang S, Huang W, Xu W, Wang Q, Song Y, et al. MicroRNA23a and microRNA23b deregulation derepresses SF-1 and upregulates estrogen signaling in ovarian endometriosis. J Clin Endocrinol Metab. 2013;98:1575–1582. doi: 10.1210/jc.2012-3010. [DOI] [PubMed] [Google Scholar]
- 20.Laudanski P, Charkiewicz R, Kuzmicki M, Szamatowicz J, Charkiewicz A, Niklinski J. MicroRNAs expression profiling of eutopic proliferative endometrium in women with ovarian endometriosis. Reprod Biol Endocrinol. 2013;11:78. doi: 10.1186/1477-7827-11-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vosa U, Vooder T, Kolde R, Vilo J, Metspalu A, Annilo T. Meta-analysis of microRNA expression in lung cancer. Int J Cancer J Int du Cancer. 2013;132:2884–2893. doi: 10.1002/ijc.27981. [DOI] [PubMed] [Google Scholar]
- 22.Teague EM, Print CG, Hull ML. The role of microRNAs in endometriosis and associated reproductive conditions. Hum Reprod Update. 2010;16:142–165. doi: 10.1093/humupd/dmp034. [DOI] [PubMed] [Google Scholar]
- 23.Jia SZ, Yang Y, Lang J, Sun P, Leng J. Plasma miR-17-5p, miR-20a and miR-22 are down-regulated in women with endometriosis. Hum Reprod. 2013;28:322–330. doi: 10.1093/humrep/des413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pacurari M, Addison JB, Bondalapati N, Wan YW, Luo D, Qian Y, et al. The microRNA-200 family targets multiple non-small cell lung cancer prognostic markers in H1299 cells and BEAS-2B cells. Int J Oncol. 2013;43:548–560. doi: 10.3892/ijo.2013.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshino H, Enokida H, Itesako T, Tatarano S, Kinoshita T, Fuse M, et al. Epithelial-mesenchymal transition-related microRNA-200s regulate molecular targets and pathways in renal cell carcinoma. J Hum Gen. 2013;58:508–516. doi: 10.1038/jhg.2013.31. [DOI] [PubMed] [Google Scholar]
- 26.He M, Liu Y, Deng X, Qi S, Sun X, Liu G, et al. Down-regulation of miR-200b-3p by low p73 contributes to the androgen-independence of prostate cancer cells. Prostate. 2013;73:1048–1056. doi: 10.1002/pros.22652. [DOI] [PubMed] [Google Scholar]
- 27.Castilla MA, Diaz-Martin J, Sarrio D, Romero-Perez L, Lopez-Garcia MA, Vieites B, et al. MicroRNA-200 family modulation in distinct breast cancer phenotypes. PloS One. 2012;7:e47709. doi: 10.1371/journal.pone.0047709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duns G, van den Berg A, van Dijk MC, van Duivenbode I, Giezen C, Kluiver J, et al. The entire miR-200 seed family is strongly deregulated in clear cell renal cell cancer compared to the proximal tubular epithelial cells of the kidney. Genes Chromosomes Cancer. 2013;52:165–173. doi: 10.1002/gcc.22016. [DOI] [PubMed] [Google Scholar]
- 29.Perez-Iratxeta C, Bork P, Andrade MA. Association of genes to genetically inherited diseases using data mining. Nat Genet. 2002;31:316–319. doi: 10.1038/ng895. [DOI] [PubMed] [Google Scholar]
- 30.Mohammadi A, Saraee MH, Salehi M. Identification of disease-causing genes using microarray data mining and Gene Ontology. BMC Med Genom. 2011;4:12. doi: 10.1186/1755-8794-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]


