Abstract
Objective(s):
FOXP3 gene is an X-linked gene that encodes FOXP3 protein, an essential transcription factor in CD4+CD25+FOXP3+ regulatory T (Treg) cells. We aimed, in the present study, to investigate the association of two FOXP3 polymorphisms, -2383 C/T (rs3761549) and IVS9+459 T/C (rs2280883), with lung cancer.
Materials and Methods:
In a case-control study we analyzed genotypes and alleles frequencies at -2383 C/T and IVS9+459 T/C positions in 156 patients with lung cancer and 156 age and sex matched healthy controls in Southern Iranian population, using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) methods. The data were verified by direct automated DNA sequencing.
Results:
The frequency of -2383 T allele was significantly higher in the patients than in the control group (11.8% versus 5.9%, P-value=0.04, OR=2.13, 95%CI=1.04-4.54). T allele frequency at IVS9+459 T/C position was higher, compared to the controls, in the patients who presented the disease over 55 years old (69.9% versus 59.1%, P-value=0.04, OR=1.61, 95%CI=1.01-2.55) and also in SCLC patients (77.8% versus 59.1%, P-value=0.03, OR=2.42, 95%CI=1.05-5.59). No significant differences were found in the genotypes and haplotypes distributions between the cases and controls. A high degree of linkage disequilibrium was observed between two polymorphisms.
Conclusion:
As the first study dealing with -2383 C/T and IVS9+459 T/C in lung cancer, our data conclusively suggest the association of -2383 T allele with susceptibility to lung cancer in Iranian population. The association of IVS9+459 T allele with susceptibility to lung cancer in old patients suggests the age-dependent effects of FOXP3 gene on cancer occurrence.
Keywords: FOXP3 gene, Gene polymorphism, Lung cancer, PCR-RFLP
Introduction
Lung cancer is one of the most fatal malignant disorders all over the world and most patients are diagnosed at late clinical stages (1). Forkhead box P3 (FOXP3) gene (Gene ID: 50943, Xp11.23) is an X-linked gene that encodes FOXP3 protein, an essential transcription factor in CD4+CD25+FOXP3 regulatory T (Treg) cells (2, 3). Treg cells play an important role in the down-regulation of chronic inflammation, hindering of autoimmune reactions, and maintenance of peripheral immunological tolerance (4, 5). Suppressing immune responses by FOXP3+Treg cells may contribute to tumorigenesis. Upregulation of CD4+CD25+Treg cells expressing FOXP3 in tumor tissues has been reported repeatedly (6-9).
Moreover, the role of FOXP3 as a tumor suppressor gene has been documented (10, 11) and the mutations of this gene has already been reported in cancer patients (12-14).
Regarding the above mentioned dual role of FOXP3, investigation of probable association of FOXP3 gene polymorphisms in cancers may shed light on the molecular pathogenesis of cancer and open new windows to screening of susceptible individuals. To date, few studies, have investigated FOXP3 gene polymorphisms in cancer patients (15-19). In the present study we have investigated the association between two single nucleotide polymorphisms (SNPs) of FOXP3 gene, a promoter SNP; -2383 C/T (rs3761549) and an intronic SNP; IVS9+459 T/C (rs2280883) with susceptibility to lung cancer in a population from the South of Iran.
Material and Methods
In a case-control study, 156 patients with lung cancer and 156 age and sex matched healthy controls without any history of cancer and autoimmune diseases in their first-degree relatives were enrolled. The study was approved by the Ethic Committee of Shiraz University of Medical Sciences. Informed consent was obtained from all participants before sample collection. 126 male and 30 female individuals were included in both groups. The mean age of male patients, male controls, female patients, and female controls were respectively 63.70±11.250, 62.75±10.246, 63.77±11.796, and 64.10±11.681. After taking informed consent from the participants, venous blood sample was collected and genomic DNA was extracted from peripheral blood leukocytes using salting out method (20). Afterwards, the individuals’ genotypes at both positions were determined using PCR-RFLP methods, followed by the band detection on GelRed stained (Biotium, USA) 3% agarose gel (Invitrogen, UK) after electrophoresis.
The specific primer sequences used for the amplification of a 388 bp fragment of FOXP3 gene including promoter SNP -2383 C/T is followed (21): Forward Primer: 5’-CTG AGA CTT TGG GAC CGT AG-3’, Reverse Primer: 5’-TGC GCC GGG CTT CAT CGA CA-3’ (Takapouzist, Iran). Annealing temperature of PCR thermal program was 60 °C and the PCR thermal program was repeated for 30 cycles. The products underwent digestion, for 16 hr at 65 °C, with BsrI (BseNI) restriction enzyme (NEB, England). This enzyme, independent of -2383 C/T polymorphism, had a cutting site on 388 bp PCR product and cut it into two 127 and 261 bp fragments. In the condition that C nucleotide was present at -2383 position, another cutting site would be generated and 261 bp fragment would be further fragmented into two 184 and 77 bp fragments. Figure 1 illustrates the PCR-RFLP results for -2383 C/T (rs3761549) polymorphism.
Figure 1.
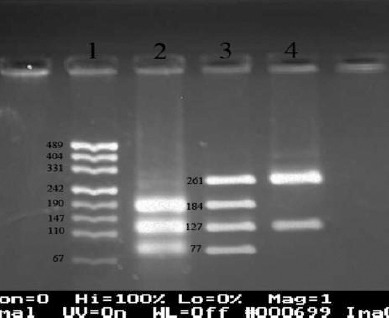
Genotyping of -2383 C/T in FOXP3 gene using polymerase chain reaction-restriction fragment length polymorphism technique with BsrI (BseNI) enzyme, followed by the band detection on gel red-stained 3% agarose gel after electrophoresis. 1: pUC19 marker, 2: CC in females and C in males (184, 127, and77 bp), 3: CT (just in females) (261, 184, 127, and 77 bp), 4: TT in females and T in males (261 and 127 bp)
Following primers were designed and used to amplify a 373 bp fragment around intronic IVS9+459 T/C SNP in FOXP3 gene: forward primer: 5’-ACC ACC ATC CAG GCCAGA GCA-3’, reverse primer: 5’-GTT AGG TGT GGC GCT AGG ATG AAG G-3’ (Takapouzist, Iran). PCR thermal program was repeated 30 cycles with the annealing temperature of 71 °C. The products were then incubated with the restriction enzyme TaaI (HPYCH4III) (Fermentas, Lithuania) for 16 hr at 65 °C. In the condition that C nucleotide was present at the polymorphic site this enzyme would cut 373 bp PCR product into two 120 and 253 fragments. If T nucleotide was present, PCR product would remain intact. Figure 2 illustrates PCR-RFLP results for IVS9+459 T/C (rs2280883) polymorphism. RFLP results were confirmed by direct automated sequencing of PCR products using BigDye terminator chemistry kit (ABI, USA) and 310 genetic analyzer (ABI, USA). Figures 3 and 4 illustrate the electropherograms of two heterozygous individuals for both positions.
Figure 2.
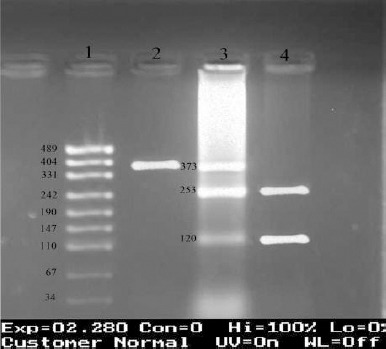
Genotyping of IVS9+459 T/C in FOXP3 gene using polymerase chain reaction-restriction fragment length polymorphism technique with TaaI (HPYCH4III) enzyme, followed by the band detection on gel red-stained 3% agarose gel after electrophoresis. 1: pUC19 marker, 2: TT in females and T in males (373 bp), 3: CT (just in females) (373,253, and 120 bp), 4: CC in females and C in males (253 and 120 bp)
Figure 3.
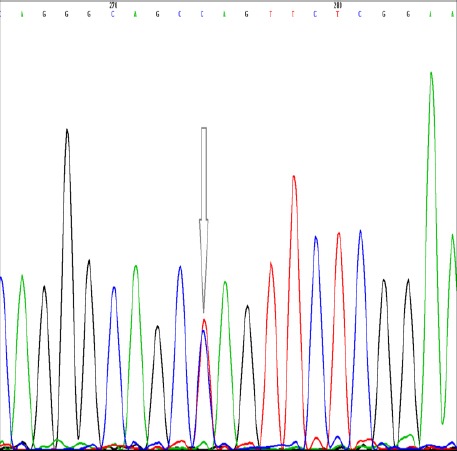
Verifying the heterozygote genotype at position -2383 C/T in FOXP3 gene (rs3761549) by direct automated DNA sequencing using BigDye terminator chemistry kit and 310 genetic analyzer; the heterozygote peak is pointed by arrow
Figure 4.
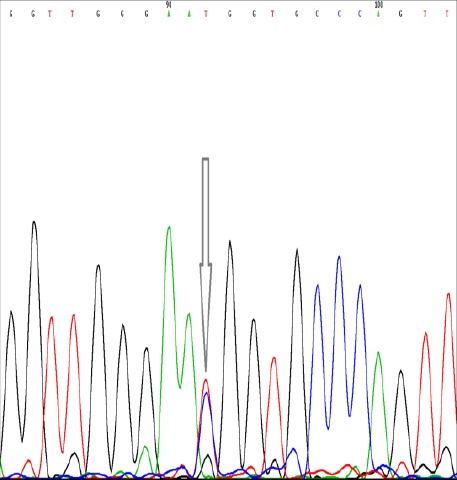
Verifying the heterozygote genotype at position IVS9+459 T/C in FOXP3 gene (rs2280883) by direct automated DNA sequencing using BigDye terminator chemistry kit and 310 genetic analyzer; the heterozygote peak is pointed by arrow
Before statistical analysis, the genotype frequencies were tested for the Hardy-Weinberg equilibrium. SPSS software package (version 18; SPSS Inc, Chicago, IL, USA) was used for analyzing the collected data. Chi-square (X2) statistical test was applied to compare the genotype, allele, and haplotype frequencies between the patients and controls. P-values less than 0.05 were considered statistically significant. All analyses were performed separately for males and females as FOXP3 is an X-linked gene.
The haplotypes was deducted and analyzed using Haploview software package (available at: http://www.broad.mit.edu/mpg/haploview/).
Results
One hundred fifty six patients with lung cancer and 156 age and sex matched controls comprises our study populations. The tumor type in 80.9% of the patients with known tumor type was non-small cell lung cancer (NSCLC) and in 19.1% was small cell lung cancer (SCLC). The tumor type in 4 individuals was not mentioned in their records although they were known cases of lung cancer. Analysis indicated the frequencies of investigated genotypes to be in the Hardy-Weinberg equilibrium. The frequencies of genotypes and alleles corresponding to two FOXP3 SNPs for male and female lung cancer patients and the control groups are illustrated in Table 1. Having two types of lung cancer in our study population (SCLC and NSCLC) and considering the likely age-dependent effect of FOXP3 gene, we also looked at the differences in genotypes and alleles frequencies separately in different tumor types and age groups. Table 1 presents the corresponding data for NSCLC/SCLC patients and for the patients over and under the age of 55 (≥55 y/o and <55 y/o).
Table 1.
The frequencies of genotypes and alleles of FOXP3 polymorphisms in healthy control group and lung cancer patients; the frequencies of genotypes and alleles are also illustrated in patients subdivided by age of cancer onset (<55 y/o and ≥55 y/o) and type of lung cancer (non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC)); description of the data is presented in result section
| FOXP3 locus | Gender | Genotype and allele | Control group n=156 (%) | Lung cancer patients(Total) n=156 (%) | <55 y/o patients n= 36 (%) | ≥55 y/o patients n=120(%) | NSCLC patients n=123(%) | SCLC patients n=29(%) | |
|---|---|---|---|---|---|---|---|---|---|
| -2383 C/T (rs3761549) | Female (n=30) | Genotype | CC | 27 (90.0) | 25 (83.4) | 5 (71.4) | 20 (87.0) | 19 (82.6) | 6 (85.7) |
| CT | 3 (10.0) | 4 (13.3) | 2 (28.6) | 2 (8.7) | 3 (13.0) | 1 (14.3) | |||
| TT | 0.0 | 1 (3.3) | 0.0 | 1 (4.3) | 1 (4.4) | 0.0 | |||
| Allele | C | 57 (95.0) | 54 (90.0) | 12 (85.7) | 42 (91.3) | 41 (89.1) | 13 (92.9) | ||
| T | 3 (5.0) | 6 (10.0) | 2 (14.3) | 4 (8.7) | 5 (10.9) | 1 (7.1) | |||
| Male (n=126) | Genotype | C | 118 (93.7) | 110 (87.3) | 26 (89.7) | 84 (86.6) | 88 (88.0) | 18 (81.8) | |
| T | 8 (6.3) | 16 (12.7) | 3 (10.3) | 13 (13.4) | 12 (12.0) | 4 (18.2) | |||
| Allele | C | 118 (93.7) | 110 (87.3) | 26 (89.7) | 84 (86.6) | 88 (88.0) | 18 (81.8) | ||
| T | 8 (6.3) | 16 (12.7) | 3 (10.3) | 13 (13.4) | 12 (12.0) | 4 (18.2) | |||
| Both genders(total) | Allele | C | 175 (94.1) | 164 (88.2) | 38 (88.4) | 126 (88.1) | 129 (88.4) | 31 (86.1) | |
| T | 11 (5.9) | 22 (11.8) | 5 (11.6) | 17 (11.9) | 17 (11.6) | 5 (13.9) | |||
| IVS9+459 T/C(rs2280883) | Female (n=30) | Genotype | TT | 13 (43.3) | 15 (50.0) | 2 (28.6) | 13 (56.5) | 9 (39.1) | 6 (85.7) |
| TC | 13 (43.3) | 14 (46.7) | 5 (71.4) | 9 (39.1) | 13 (56.5) | 1 (14.3) | |||
| CC | 4 (13.4) | 1 (3.3) | 0.0 | 1 (4.4) | 1 (4.4) | 0.0 | |||
| Allele | T | 39 (65.0) | 44 (73.3) | 9 (64.3) | 35 (76.1) | 31 (67.4) | 13 (92.9) | ||
| C | 21 (35.0) | 16 (26.7) | 5 (35.7) | 11 (23.9) | 15 (32.6) | 1 (7.1) | |||
| Male (n=126) | Genotype | T | 71 (56.3) | 83 (65.9) | 18 (62.1) | 65 (67.0) | 65 (65.0) | 15 (68.2) | |
| C | 55 (43.7) | 43 (34.1) | 11 (37.9) | 32 (33.0) | 35 (35.0) | 7 (31.8) | |||
| Allele | T | 71 (56.3) | 83 (65.9) | 18 (62.1) | 65 (67.0) | 65 (65.0) | 15 (68.2) | ||
| C | 55 (43.7) | 43 (34.1) | 11 (37.9) | 32 (33.0) | 35 (35.0) | 7 (31.8) | |||
| Both genders(total) | Allele | T | 110 (59.1) | 127 (68.3) | 27 (62.8) | 100 (69.9) | 96 (65.8) | 28 (77.8) | |
| C | 76 (40.9) | 59 (31.7) | 16 (37.2) | 43 (30.1) | 50 (34.2) | 8 (22.2) | |||
As illustrated in the Table 1, -2383 C/T (rs3761549) genotype frequencies in male patients were (110 C and 16 T) and in male controls were (118 C and 8 T). The figures for female patients and female controls were (25 CC, 4 CT, and 1 TT) and (27 CC and 3 CT), respectively. Statistical analysis indicated that the frequencies of the genotypes and alleles at this position were not significantly different between the patients and controls neither in the male (P-value=0.09, identical for genotypes and alleles) nor in the female groups (P-value=0.54 for genotypes and 0.30 for alleles), although a trend toward higher frequency of T allele was observed in both male and female patients in comparison to their cognate controls (Table 1). After adding the allele frequencies together in order to do allele analysis in the whole population (both genders); however, T allele frequency observed to be significantly higher in the patients comparing to the control group (11.8% and 5.9% respectively, P-value=0.04, odds ratio (OR)=2.13, 95% CI for OR=1.04-4.54, Table 1). No differences were found between the patients and controls neither in age-related subgroups nor in tumor type-related subgroups.
Regarding the SNP at the locus IVS9+459 T/C (rs2280883), genotype frequencies in male patients and male controls were respectively (83 T and 43 C) versus (71 T and 55 C). These Figures for female patients and female controls were (15TT, 14 TC, and 1 CC) and (13 TT, 13 TC, and 4 CC) respectively. Frequencies of the genotypes and alleles at this position were not significantly different between the patients and the controls neither in the male (P-value=0.12, identical for genotypes and alleles) nor in the female groups (P-value=0.37 for genotypes and 0.32 for alleles). Considering the alleles frequencies altogether (both genders), again no difference was found between the patients and controls (P-value=0.07). Despite of this, the analysis indicated a higher frequency of T allele at position IVS9+459 T/C (rs2280883) in patients who were 55 years old or more (≥55 y/o), but not in those under 55, in comparison to the controls. (69.9% in patients who were 55 years old or more versus 59.1% in control group, P-value=0.04, OR=1.61, 95% CI for OR=1.01-2.55). The T allele at this position was also observed with a higher frequency in SCLC patients, but not in NSCLC, comparing to the control group (77.8% in SCLC patients versus 59.1% in control group, SCLC patients N=29, P-value=0.03, OR=2.42, 95% CI for OR=1.05-5.59).
The comparison of the haplotype frequencies emerging from two FOXP3 SNPs between lung cancer patients and healthy control group is shown in Table 2. Statistical analysis indicated no significant difference between the patients and controls. However, Haploview analysis revealed a high degree of linkage disequilibrium between these two FOXP3 gene positions (D’=90).
Table 2.
Frequency of haplotypes emerging from two FOXP3 polymorphisms in patients with lung cancer in comparison to healthy control group
| Gender | FOXP3 haplotype (-2383 C/T, IVS9+459 T/C) | Lung cancer patients n (%) | Control group n (%) | P-value |
|---|---|---|---|---|
| Male (n=126) | CT | 68 (54.0) | 63 (50.0) | 0.17 |
| TT | 15 (11.9) | 8 (6.3) | ||
| CC | 42 (33.3) | 55 (43.7) | ||
| TC | 1 (0.8) | 0.0 | ||
| Female (2n=60) | CT | 38 (63.3) | 36 (60.0) | 0.42 |
| TT | 6 (10.0) | 3 (5.0) | ||
| CC | 16 (26.7) | 21 (35.0) | ||
| TC | 0.0 | 0.0 |
Discussion
As one of the main transcription factor for regulatory T cells, FOXP3 has attracted attention in recent decade. Imbalance of FOXP3 positive regulatory T cells has been widely reported in autoimmune diseases (4). The association of FOXP3 polymorphisms with immune related diseases has already been well documented (22-28). Although there are no functional studies indicating the effects of intronic (IVS9+459 T/C) and promoter (-2383 C/T) polymorphisms in FOXP3 on the gene expression, IVS9+459 T/C has been suggested to affect the splicing of hnRNA and consequently, the FOXP3 gene expression (18). Furthermore, presence of -2383 C/T in the promoter region suggests the likely effect of this SNP on the gene expression. Observing the association between these SNPs with a variety of the diseases (22-28) may verify the functionality associated with these two SNPs. In the present study the genotypes and alleles of -2383 C/T polymorphism observed not to be significantly different between the patients and controls in both male and female groups. The frequency of T allele was, however, observed to be significantly higher in the patients after adding the male and female data together. The OR was 2.13 suggesting the effect of T allele in susceptibility to lung cancer. After dividing the patients based on the age-group and the type of cancer, no significant difference in the genotype/alleles frequencies was found between patients in the subgroups and the controls.
The statistical analysis on IVS9+459 T/C showed no significant difference in the allele and genotype frequencies between the patients and controls in the female group, male group, and the whole people studied (both genders).
After dividing the patients based on their ages (≥55 y/o and <55 y/o) and the type of their cancers (NSCLC, SCLC), a significant difference in allele frequencies was found between the control group and the patients who were ≥55 y/o, as well as patients with SCLC tumor type. The mentioned age group included 120 out of 156 individuals (76.9%) of the whole patients’ population.
There is ample evidence for the difference in the cancer etiopathology between the young and old patients (29, 30). Furthermore the age-dependent changes in immunity have already been well documented (31-33). Overexpression of FOXP3 and the increase in the population of Treg cells in elderly has also been reported in several studies (34-36). Our finding, concordant with these observations, suggests the age-dependent effect of FOXP3 gene in lung cancer, although the underlying reasons need to be clarified. Observing the association of the T allele with SCLC, but not NSCLC might imply the fact that the involved genes and the etiopathology in SCLC and NSCLC might be different (37), although the small number of our patients with SCLC should not be ignored.
The association between -2383 C/T (rs3761549) polymorphism and immune related diseases was already reported in psoriasis in a Chinese population (C allele) (26), hashimoto disease in a Japanese population (C allele) (38), lupus in a Guangxi Zhuang population (T allele) (24), and endometriosis related infertility in a Brazilian population (T allele) (39). Regarding IVS9+459 T/C (rs2280883), there are also reports indicating the associations between this SNP and immune related diseases, including psoriasis in a Chinese population (C allele) (23), idiopathic infertility in a Brazilian population (C allele) (39), primary biliary cirrhosis (PBC) in an American population (T allele) (40), and myasthenia gravis in a Han Chinese population (T allele) (27). In spite of such studies, others could not find significant association between none of these polymorphisms with systemic lupus erythematosus in a Taiwanese population (25) and Graves’ disease and autoimmune Addison’s disease in a British population (21). Furthermore, no relationship was found between IVS9+459 T/C and Crohn’s Disease (CD) in a study on an American population (40).
Although excessive data are available dealing with FOXP3 gene in autoimmune diseases, only several studies (15-19) deal with the role of FOXP3 polymorphisms in cancer. Chen et al reported a significant higher TT genotype at the IVS9+459 T/C locus in patients with hepatocellular carcinoma comparing to the healthy donors in China (18). This finding is in consistent with our observation of higher significant occurrence of IVS9+459T allele in old lung cancer patients, as well as SCLC patients. In another study by our group(16), no association between -2383 C/T and colorectal cancer was observed, although a trend toward the association of T allele with cancer metastasis was suggested. Chen et al on the other hand reported the association of C allele at -2383 C/T position with hepatocellular carcinoma (18). In the study carried out by Zheng et al another polymorphism in FOXP3 gene, rs2294021, was investigated in Chinese patients with breast cancer (17). The results indicated the association between this SNP and susceptibility to breast cancer. He et al investigated another polymorphism in FOXP3 gene, rs3761548, in NSCLC patients from China, and indicated the relationship between this SNP and the risk of developing NSCLC in the Chinese Han population (19). The discrepancies between the results of different studies may come from the differences in the ethnic origins of the sample populations, differences in disease pathogenesis, and most likely the sample size. Moreover, the dual role of FOXP3 (participating in the development and function of Treg cells from one side (2, 3) and acting as a tumor suppressor gene from other side (10, 11)) should not be ignored.
Statistical analysis of the haplotype frequencies, in the present study, indicated no significant difference between the patients and controls. The wild type CT haplotype (-2383 C, IVS9+459 T) was the most frequent haplotype in both male and female patients. Haploview analysis indicated a high degree of linkage disequilibrium between these two polymorphisms of FOXP3 gene (D’=90). In fact, among 312 subjects, only one mutant TC haplotype (minor allele of both positions) was observed. This finding suggested the existence of an evolutionary pressure which prevented the co-occurrence of minor alleles in an individual, although more investigation is required to verify the functional effect of the observed haplotypes.
Conclusion
In conclusion, the present study revealed the probable role of T allele at -2383 C/T position of FOXP3 gene (rs3761549) in susceptibility to lung cancer. It also indicated the possible involvement of T allele at IVS9+459 T/C locus (rs2280883) in susceptibility to lung cancer in those affected over 55 y/o and who have SCLC tumor type. The limitation of the present study should not, however, be ignored. Increasing the sample size, simultaneous investigation of FOXP3 gene expression, or doing a functional study on the mentioned polymorphisms are required to clearly clarify the role of FOXP3 gene variations in lung cancer.
Acknowledgment
This work was financially supported by a grant from Shiraz Institute for Cancer Research, Shiraz University of Medical Sciences, Shiraz, Iran (ICR-100-500).
References
- 1.Fauci AS, kasper DL. New York: McGraw-Hill; 2008. Principles of Harrison's Internal Medicine. [Google Scholar]
- 2.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. [PubMed] [Google Scholar]
- 3.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 4.Dejaco C, Duftner C, Grubeck-Loebenstein B, Schirmer M. Imbalance of regulatory T cells in human autoimmune diseases. Immunology. 2006;117:289–300. doi: 10.1111/j.1365-2567.2005.02317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Von Boehmer H. Mechanisms of suppression by suppressor T cells. Nat Immunol. 2005;6:338–344. doi: 10.1038/ni1180. [DOI] [PubMed] [Google Scholar]
- 6.Ishibashi Y, Tanaka S, Tajima K, Yoshida T, Kuwano H. Expression of Foxp3 in non-small cell lung cancer patients is significantly higher in tumor tissues than in normal tissues, especially in tumors smaller than 30 mm. Oncol Rep. 2006;15:1315–1319. [PubMed] [Google Scholar]
- 7.Zhang HY, Sun H. Up-regulation of Foxp3 inhibits cell proliferation, migration and invasion in epithelial ovarian cancer. Cancer Lett. 2010;287:91–97. doi: 10.1016/j.canlet.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 8.Zeng C, Yao Y, Jie W, Zhang M, Hu X, Zhao Y, et al. Up-regulation of Foxp3 participates in progression of cervical cancer. Cancer Immunol Immunother. 2013;62:481–487. doi: 10.1007/s00262-012-1348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dimitrakopoulos FI, Papadaki H, Antonacopoulou AG, Kottorou A, Gotsis AD, Scopa C, et al. Association of FOXP3 expression with non-small cell lung cancer. Anticancer Res. 2011;31:1677–1683. [PubMed] [Google Scholar]
- 10.Zuo T, Wang L, Morrison C, Chang X, Zhang H, Li W, et al. FOXP3 is an X-linked breast cancer suppressor gene and an important repressor of the HER-2/ErbB2 oncogene. Cell. 2007;129:1275–1286. doi: 10.1016/j.cell.2007.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, Liu R, Ribick M, Zheng P, Liu Y. FOXP3 as an X-linked tumor suppressor. Discov Med. 2010;10:322–328. [PMC free article] [PubMed] [Google Scholar]
- 12.Li W, Wang L, Katoh H, Liu R, Zheng P, Liu Y. Identification of a tumor suppressor relay between the FOXP3 and the Hippo pathways in breast and prostate cancers. Cancer Res. 2011;71:2162–2171. doi: 10.1158/0008-5472.CAN-10-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, Liu R, Li W, Chen C, Katoh H, Chen GY, et al. Somatic single hits inactivate the X-linked tumor suppressor FOXP3 in the prostate. Cancer Cell. 2009;16:336–346. doi: 10.1016/j.ccr.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zuo T, Liu R, Zhang H, Chang X, Liu Y, Wang L, et al. FOXP3 is a novel transcriptional repressor for the breast cancer oncogene SKP2. J Clin Invest. 2007;117:3765–3773. doi: 10.1172/JCI32538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raskin L, Rennert G, Gruber SB. FOXP3 germline polymorphisms are not associated with risk of breast cancer. Cancer Genet Cytogenet. 2009;190:40–42. doi: 10.1016/j.cancergencyto.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Mojtahedi Z, Erfani N, haghshenas MR, hoseini SV, Ghaderi A. Association of Foxp3/Scurfin germline polymorphisms(C-2383T/rs3761549) with colorectal cancer. Ann Colorectal Res. 2013;1:12–16. [Google Scholar]
- 17.Zheng J, Deng J, Jiang L, Yang L, You Y, Hu M, et al. Heterozygous genetic variations of FOXP3 in Xp11.23 elevate breast cancer risk in Chinese population via skewed X-chromosome inactivation. Hum Mutat. 2013;34:619–628. doi: 10.1002/humu.22284. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y, Zhang H, Liao W, Zhou J, He G, Xie X, et al. FOXP3 gene polymorphism is associated with hepatitis B-related hepatocellular carcinoma in China. J Exp Clin Cancer Res. 2013;32:39. doi: 10.1186/1756-9966-32-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He YQ, Bo Q, Yong W, Qiu ZX, Li YL, Li WM. FoxP3 genetic variants and risk of non-small cell lung cancer in the Chinese Han population. Gene. 2013;531:422–425. doi: 10.1016/j.gene.2013.08.066. [DOI] [PubMed] [Google Scholar]
- 20.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Owen CJ, Eden JA, Jennings CE, Wilson V, Cheetham TD, Pearce SH. Genetic association studies of the FOXP3 gene in Graves’ disease and autoimmune Addison's disease in the United Kingdom population. J Mol Endocrinol. 2006;37:97–104. doi: 10.1677/jme.1.02072. [DOI] [PubMed] [Google Scholar]
- 22.Ban Y, Tozaki T, Tobe T, Jacobson EM, Concepcion ES, Tomer Y. The regulatory T cell gene FOXP3 and genetic susceptibility to thyroid autoimmunity: an association analysis in Caucasian and Japanese cohorts. J Autoimmun. 2007;28:201–207. doi: 10.1016/j.jaut.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 23.Gao L, Li K, Li F, Li H, Liu L, Wang L, et al. Polymorphisms in the FOXP3 gene in Han Chinese psoriasis patients. J Dermatol Sci. 2010;57:51–56. doi: 10.1016/j.jdermsci.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 24.Lan Y, Tang XS, Qin J, Wu J, Qin JM. [Association of transcription factor FOXP3 gene polymorphism with genetic susceptibility to systematic lupus erythematosus in Guangxi Zhuang population] Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2010;27:433–436. doi: 10.3760/cma.j.issn.1003-9406.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 25.Lin YC, Lee JH, Wu AS, Tsai CY, Yu HH, Wang LC, et al. Association of single-nucleotide polymorphisms in FOXP3 gene with systemic lupus erythematosus susceptibility: a case-control study. Lupus. 2011;20:137–143. doi: 10.1177/0961203310382428. [DOI] [PubMed] [Google Scholar]
- 26.Song QH, Shen Z, Xing XJ, Yin R, Wu YZ, You Y, et al. An association study of single nucleotide polymorphisms of the FOXP3 intron-1 and the risk of Psoriasis vulgaris. Indian J Biochem Biophys. 2012;49:25–35. [PubMed] [Google Scholar]
- 27.Zhang J, Chen Y, Jia G, Chen X, Lu J, Yang H, et al. FOXP3 -3279 and IVS9+459 polymorphisms are associated with genetic susceptibility to myasthenia gravis. Neurosci Lett. 2013;534:274–278. doi: 10.1016/j.neulet.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 28.Conteduca G, Rossi A, Megiorni F, Parodi A, Ferrera F, Tardito S, et al. Single nucleotide polymorphisms in the promoter regions of Foxp3 and ICOSLG genes are associated with Alopecia areata. Clin Exp Med. 2014;14:91–97. doi: 10.1007/s10238-012-0224-3. [DOI] [PubMed] [Google Scholar]
- 29.Bryant AS, Cerfolio RJ. Differences in outcomes between younger and older patients with non-small cell lung cancer. Ann Thorac Surg. 2008;85:1735–1739. doi: 10.1016/j.athoracsur.2008.01.031. [DOI] [PubMed] [Google Scholar]
- 30.Skarin AT, Herbst RS, Leong TL, Bailey A, Sugarbaker D. Lung cancer in patients under age 40. Lung Cancer. 2001;32:255–264. doi: 10.1016/s0169-5002(00)00233-6. [DOI] [PubMed] [Google Scholar]
- 31.Weiskopf D, Weinberger B, Grubeck-Loebenstein B. The aging of the immune system. Transpl Int. 2009;22:1041–1050. doi: 10.1111/j.1432-2277.2009.00927.x. [DOI] [PubMed] [Google Scholar]
- 32.Zanussi S, Serraino D, Dolcetti R, Berretta M, De Paoli P. Cancer, aging and immune reconstitution. Anticancer Agents Med Chem. 2013;13:1310–1324. doi: 10.2174/18715206113136660348. [DOI] [PubMed] [Google Scholar]
- 33.Shaw AC, Goldstein DR, Montgomery RR. Age-dependent dysregulation of innate immunity. Nat Rev Immunol. 2013;13:875–887. doi: 10.1038/nri3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan XD, Mao YQ, Zhu LJ, Li J, Xie Y, Wang L, et al. Changes of regulatory T cells and FoxP3 gene expression in the aging process and its relationship with lung tumors in humans and mice. Chin Med J (Engl) 2012;125:2004–2011. [PubMed] [Google Scholar]
- 35.Nishioka T, Shimizu J, Iida R, Yamazaki S, Sakaguchi S. CD4+CD25+Foxp3+T cells and CD4+CD25-Foxp3+T cells in aged mice. J Immunol. 2006;176:6586–6593. doi: 10.4049/jimmunol.176.11.6586. [DOI] [PubMed] [Google Scholar]
- 36.Gregg R, Smith CM, Clark FJ, Dunnion D, Khan N, Chakraverty R, et al. The number of human peripheral blood CD4+CD25high regulatory T cells increases with age. Clin Exp Immunol. 2005;140:540–546. doi: 10.1111/j.1365-2249.2005.02798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar V, Abbas AK. 8th ed. Philadelphia: Saunders; 2012. J.C. a. Robbins and Cotran Pathologic Basis of Disease. [Google Scholar]
- 38.Inoue N, Watanabe M, Morita M, Tomizawa R, Akamizu T, Tatsumi K, et al. Association of functional polymorphisms related to the transcriptional level of FOXP3 with prognosis of autoimmune thyroid diseases. Clin Exp Immunol. 2010;162:402–406. doi: 10.1111/j.1365-2249.2010.04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andre GM, Barbosa CP, Teles JS, Vilarino FL, Christofolini DM, Bianco B. Analysis of FOXP3 polymorphisms in infertile women with and without endometriosis. Fertil Steril. 2011;95:2223–2227. doi: 10.1016/j.fertnstert.2011.03.033. [DOI] [PubMed] [Google Scholar]
- 40.Park O, Grishina I, Leung PS, Gershwin ME, Prindiville T. Analysis of the Foxp3/scurfin gene in Crohn's disease. Ann N Y Acad Sci. 2005;1051:218–228. doi: 10.1196/annals.1361.125. [DOI] [PubMed] [Google Scholar]


