Abstract
Objective(s):
Brucellosis is a well-known domestic animal infectious disease, which is caused by Brucella bacterium. GroEL antigen increases Brucella survival and is one of the major antigens that stimulates the immune system. Hence, the objective of the present study was cloning and bioinformatics analysis of GroEL gene.
Materials and Methods:
The full-length open reading frame of this gene was amplified by specific primers and cloned into pTZ57R/T vector. Also, the sequence of this gene in the Brucella melitensis strain Rev 1 was submitted to the NCBI gene bank for the first time. Several prediction software applications were also used to predict B and T-cell epitopes, secondary and tertiary structures, antigenicity ability and enzymatic degradation sites. The used software applications validated experimental results.
Results:
The results of phylogenetic analysis showed that the GroEL sequence had near homology with other species instead of other Brucella spp. The bioinformatics tools used in the current study were validated by the results of four different experimental epitope predictions. Bioinformatics analysis identified eight B and seven T-cell epitopes.
Conclusion:
According to the antigenic ability and proteasomal cleavage sites, four (150-160, 270-285,351-361 and 385-395) common epitopes were predicted for GroEL gene. Bioinformatics analysis showed that these regions had proper epitope characterization and so may be useful for stimulation of cell-mediated and humoral immunity system.
Keywords: Bioinformatics analysis, Brucella melitensis, Heat shock protein 60 kDa, Subunit vaccine
Introduction
Brucellosis is a well-known domestic animal infection which survives within a broad range of eukaryotic cells as a small gram-negative coccobacillus. This disease is characterized by abortion and reduced fertility in animals, and also by chronic infections with symptoms such as undulant fever, arthritis and osteomyelitis in humans (1). GroEL gene encodes an inner membrane protein of Brucella with about 60 kDa molecular weight. This protein belongs to heat shock protein family, and as a chaperonin has an important role in the structure and folding of other proteins. In response to macrophage phagocytosis, Brucella produces the GroEL antigen in order to increase its survival. GroEL is one of the major antigens that stimulates the immune system. Furthermore, this antigen has an important role in disease cycle in humans and animals (2). In a comparative study on two different strains of Brucella, the HSP (GroEL) antigen was introduced as a good candidate for vaccine production and also development of diagnostic kits (3).
Immune system produces antibodies which specifically attach to identified regions of antigens named epitope (4). Epitopes may be classified as B (continuous and discontinuous) and T-cell (MHCI and MHCII) epitopes (5). The continuous or linear epitopes are made up of consecutive amino acids whereas the discontinuous epitopes constitute the spatially folded amino acids which lie far away in the primary sequence (6). T-cell epitopes are antigenic peptide strings recognized by T-cell receptors (7). B and T-cell epitopes could be predicted using computational tools for the vast applications in the area of antibody production, immunodiagnostics, epitope-based vaccine design and selective de-immunization of therapeutic proteins and in autoimmunity (8). Several epitope prediction software applications are currently available. The first generation of these prediction tools was supported by motif-based algorithms (9), antigen’s primary amino acid sequence (10), or other physiochemical protein characteristics. Recently more sophisticated methods using various machine learning based algorithms have been developed based on support vector machines (SVM) (11), hidden markov models (HMM) (12), and artificial neural networks (ANN) (5).
The aim of the present study was cloning, molecular analysis and epitopes prediction of GroEL antigen from B. melitensis strain Rev 1. Finally, B and T-cell epitopes were predicted in order to design an epitope-based vaccine for stimulation of the immune system.
Materials and Methods
Bacterial strains, growth conditions and isolation
In current study, B. melitensis strain Rev 1 was obtained from the Brucella culture collection (Razi Institute, Mashhad, Iran) and cultured as described (13). DNA was extracted using a DNA extraction kit (Bioneer, Korea). The quality and purity of the extracted DNA were analyzed by agarose gel electrophoresis and NanoDrop ND-1000 spectrophotometer (Thermo, USA). Escherichia coli strain DH5α was used as host for cloning, sequencing and maintenance of different DNA fragments. A T/A cloning vector pTZ57R/T (Thermo, USA) was used for cloning and sequencing of the amplified gene.
PCR amplification
B. melitensis Rev 1 genomic DNA was used as template for amplification of full length open reading frame of GroEL gene using EX Taq DNA polymerase (Takara, Japan). The specific primers were designed according to the B. melitensis 16M as template using Primer Premier 5, according to the available nucleotide sequences on the NCBI GenBank database (Table 1).
Table 1.
The specific primers with restriction sites
| Gene | Primer sequences (5’ → 3’) |
|---|---|
| GroEL | F:5`-ATGGCTGCAAAAGACGTAAAATTCG-3` |
| R:5`-TTATTAGAAGTCCATGCCGCCCATGC-3` |
Polymerase Chain Reaction (PCR) was carried out, using the Personal Cycler™ thermocycler (Biometra, Germany) with the reaction mixture containing 2.5 μl of 10X PCR buffer, 2 μl MgCl2 and 2 μl dNTPs, 0.3 μl of the template DNA (50 to 100 ng/μl), 1.5 μl of primer mixture (5 pmol/μl) and 0.125 U/μl of EX Taq DNA polymerase, and deionized water up to a final volume of 25 μl. The PCR program was performed with an initial denaturation step at 94 °C for 6 min followed by 27 cycles of denaturation at 94 °C for 30 sec, annealing at 58 °C for 30 sec and extension at 72 °C for 45 sec, and a final extension at 72 °C for 10 min.
Cloning and Nucleotide sequences analysis
The PCR products were purified from the agarose gel by Ron’s Agarose Gel Mini prep Kit (BioRon, Germany) according to the manufacturer’s instructions. The purified PCR products were ligated into pTZ57R/T cloning vector by T/A cloning strategy according to the manufacturer’s instructions (Thermo, USA). Escherichia coli DH5α competent cell preparation and transformation steps were followed as described by Sambrook and Russell (14). The recombinant vectors were transformed into competent E. coli DH5α cells. The bacterial clones harboring recombinant plasmid DNA were screened based on their colony PCR and restriction sites enzyme digestion. PCR was used for verification fidelity of E. coli DH5α transformants. The plasmids were purified using the Ron’s Plasmid Mini Kit (BioRon, Germany) and confirmed by restriction sites enzyme digestion. The purified plasmids were subjected to sequencing using Sanger method (Bioneer, South Korea). The obtained nucleotide sequences were analyzed by homology search and aligning them with other GroEL genes using Basic Local Alignment Search Tool (BLAST), CLC Main workbench 5.5 and MEGA 5 software applications, respectively.
Prediction of the secondary and tertiary structures
The secondary structures of candidate genes were analyzed using the improved self-optimized prediction method (SOPMA) software (http://npsapbil.ibcp.fr/cgibin/npsa_automat.pl?page=/NPSA/npsa_sopma.html) (15), with predicting four conformational states (helices, sheets, turns and coils). Also, tertiary structures were predicted using online ligand-binding site prediction server (http://www.sbg.bio.ic.ac.uk) (16).
Servers and software applications for B and T-cell epitopes prediction
B and T-cell epitopes of candidate genes were predicted using different servers and software applications such as: ABCpred, BepiPred, BCPred, SVMTrip and LEPS for B-cell prediction and IEDB, SYFPEITH, PropredI and Propred for T-cell prediction.
Validation bioinformatics analysis approach
In order to confirm our predicted outputs, the results of three different experimental epitope prediction studies were validated by bioinformatics tools, which were used in the present study.
Characterization of epitopes
Final B and T-cell predicted epitopes were evaluated using the VaxiJen 2.0 server (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/-VaxiJen.html) for the alignment-independent prediction of protective antigens. In addition, enzymatic degradation sites, Mass (Da) and pI were determined using the Protein Digest server (http://db.Systemsbiology.net:8080/proteomicsToolkit/proteinDigest.html).
Results
PCR amplification, cloning and nucleotide sequencing analysis
The GroEL gene from B. melitensis Rev 1 was amplified. The accuracy of this fragment was visualized on agarose gel electrophoresis (Figure 1).
Figure 1.
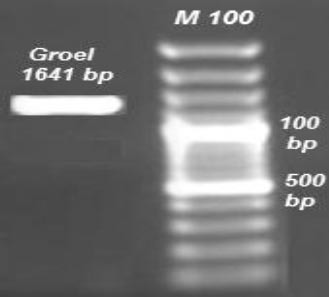
PCR amplification of GroEL (1641 bp) gene
The amplified products were successfully ligated into cloning vector and transformed into competent E. coli DH5α cells. After the selection of positive screen colonies using colony-PCR, the integrity of the recombinant plasmids were confirmed by restriction digestion. The sequencing of the recombinant plasmids was performed. The obtained sequence was analyzed using BLAST and CLC Main workbench 5.5 software program. This sequence was compared and aligned with other chaperone GroEL sequences in different species. Results showed that this sequence had differences with other bacterium and Brucella species (Figure 2). According to the results of the current study a new sequence from strain B. melitensis Rev 1 has been published for the first time on NCBI gene database under accession numbers KJ889017.
Figure 2.
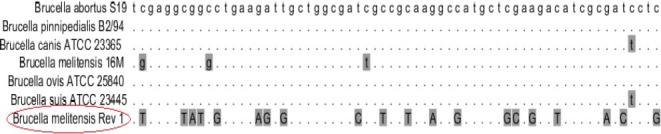
Results of alignment between Brucella melitensis Rev 1 and other Brucella spp. Dark color indicates different nucleotide
Contrary to expectation, the GroEL sequence was found to have near homology with the Yersinia, Salmonella, E. coli and Xenorhabdus instead of other Brucella spp. (Figure 3).
Figure 3.
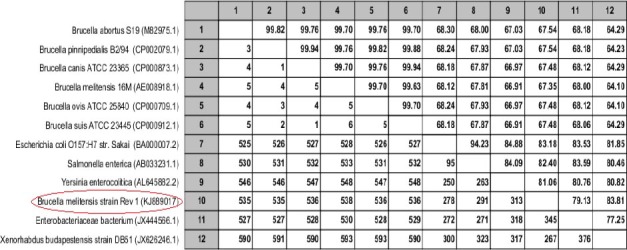
Pairwise comparison between candidate gene and other spp
Upper triangle shows percent identity and down triangle shows differences in each matrix.
In addition, the phylogenic tree was drawn for showing the genetic distance matrix results. This showed that B. melitensis strain Rev 1 had near homology with Xenorhabdus, Yersinia, Salmonella and E. coli, as like as pairwise comparison matrix (Figure 4).
Figure 4.
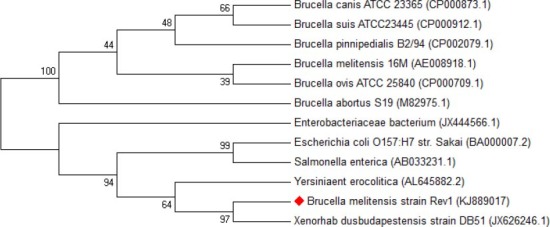
Phylogenic tree between Brucella melitensis Rev 1 and other Brucella spp
Prediction of the secondary structure
In order to assess the antigenic features of the candidate protein, its secondary structure was predicted using SOPMA server software. The results revealed that the proportion of random coils, β turns, α helices and extended strands (β folds) accounted for 22.53, 9.52, 55.31 and 12.64% of the secondary structure, respectively. A greater proportion of α helices corresponded with stability and resistance of protein structure (13).
Validation bioinformatics analysis approach
In order to validate B and T cell prediction software applications used in the present study, three antigens whose epitopes were determined experimentally (http://www.iedb.org), were selected and their epitopes were predicted using the bioinformatics tools used in the present study. The predicted epitopes were compared with the results of experimental research. Results revealed that in sillico predicted epitopes by bioinformatics tools were similar to those found by experimental studies for all of the selected antigens (Table 2).
Table 2.
Validation of bioinformatics software applications that were used in the present study
| Antigen | Predicted epitopes | Experimental epitopes | Reference |
|---|---|---|---|
| GroEL1 of Yarsinia | 28-42,78-92,178-185,275-290,315-336,430-440,526-545 | 316-326 | 17 |
| Dnak2 of Brucella | 40-67,78-92,210-227,357-370, 523-537, 609-640 | 617-637 | 9 |
| Omp313 of Brucella | 25-28,46-73,122-127,175-182 | 48-74 | 18, 19 |
| SOD 4 of Brucella | 44-50,70-86,134-153,147-165 | 75-86,136-150,149-162 | 20 |
Heat Shock Protein 60,
Heat Shock Protein 70,
Outer Membrane 31,
Sodium Oxide Dismutase
Similar epitopes between predicted epitopes using bioinformatics tools and experimental studies were emboldened and underlined
Prediction of the B and T-cell epitopes
The B- cell and MHCI (A-0101, A0201 and B-2705) and MHCII (DRB1-0101 and DRB1-0401) class of T-cell epitope were predicted using different online software applications. For each software the highest score epitopes were selected as appropriate epitopes. Moreover, eight epitopes for B-cell and seven for T-cell were chosen as final epitopes by identifying those epitopes which had the most conserved sequences in all proposed epitopes (Table 3).
Table 3.
Final B and T-cell epitopes after filtration
| No. | Final B-cell epitope | Final T-cell epitope |
|---|---|---|
| 1 | 30 TLGPKGRNVVL 40 | 160 KLIAEAM 166 |
| 2 | 150 ISANSDETVGKLIAEAM 166 | 150 ISANSDETVGKLIAEAM 166 |
| 3 | 219 ILLVDKKISNI 230 | 205 INKPETGSVELENPYILL 222 |
| 4 | 270 IVKVAAVKAP 279 | 279 PGFGDRR 285 |
| 5 | 351 QQIEDSTSDY 360 | 351 QQIEDSTSDYD 361 |
| 6 | 385 TEVEMKEKR 393 | 385 TEVEMKEKRAR 395 |
| 7 | 453 QIVENAGEEPSVVVNTVK 470 | 418 ALVRVAAALTALT 430 |
| 8 | 526 DDKMDLGA 534 | 470 KGGKGNY 476 |
Characterization of epitopes
The results of protein digest server analysis for determination of mass (Da), pI and enzymatic degradation site are shown in Table 4. Results indicate that most of the predicted epitopes in GroEL were lack of proteasomal cleavage site and no degrade during antigen processing.
Table 4.
Protein digestion analysis of final B-and T-cell epitopes
| B-cell epitopes | Mass (Da) | pI | Undigested enzyme |
|---|---|---|---|
| 30 TLGPKGRNVVL 40 | 1153.39 | 11.0 | Chymotrypsin, Cyanogen Bromide, Iodobenzoate Benzoate, Staph Protease, AspN |
| 150 ISANSDETVGKLIAEAM 166 | 1748.97 | 4.1 | Chymotrypsin, Proline Endopeptidase, Cyanogen Bromide, Iodobenzoate Benzoate, Clostripain |
| 219 ILLVDKKISNI 230 | 1255.56 | 8.6 | Trypsin R, Proline Endopeptidase, Cyanogen Bromide, Clostripain, Iodobenzoate Benzoate, Staph Protease, Chymotrypsin |
| 270 IVKVAAVKAP 279 | 995.27 | 10.0 | Chymotrypsin, Proline Endopeptidase, Cyanogen Bromide, Iodobenzoate Benzoate, Clostripain, Trypsin R, AspN, Staph Protease |
| 351 QQIEDSTSDY 360 | 1185.17 | 3.5 | Trypsin, Chymotrypsin, Proline Endopeptidase, Cyanogen Bromide, Iodobenzoate Benzoate, Clostripain, Trypsin R, Staph Protease, Trypsin K |
| 385 TEVEMKEKR 393 | 1149.33 | 5.9 | Chymotrypsin, Proline Endopeptidase, Iodobenzoate Benzoate, Clostripain, Trypsin K |
| 453 QIVENAGEEPSVVVNTVK 470 | 1912.13 | 4.2 | Trypsin, Chymotrypsin, Cyanogen Bromide, Iodobenzoate Benzoate, Clostripain, Trypsin R, Trypsin K, AspN |
| 526 DDKMDLGA 534 | 863.94 | 3.9 | Chymotrypsin, Proline Endopeptidase, Iodose Benzoate, Clostripain, Trypsin R, Staph Protease |
| T-cell epitopes | Mass(Da) | pI | Undigested enzyme |
|---|---|---|---|
| 160 KLIAEAM166 | 774.9 | 6 | Chymotrypsin, Proline Endopeptidase, Cyanogen Bromide, Iodobenzoate Benzoate, Clostripain, Trypsin R, AspN |
| 205 INKPETGSVELENPYILL 222 | 2029.32 | 4.25 | Trypsin, Cyanogen Bromide, Iodobenzoate Benzoate, Clostripain, Trypsin R, Trypsin K, AspN |
| 279 PGFGDRR 285 | 803.88 | 10.0 | Cyanogen Bromide, Iodobenzoate Benzoate, Elastase, Trypsin K, AspN, Staph Protease |
| 385 TEVEMKEKRAR 395 | 1376.59 | 8.26 | Chymotrypsin, Proline Endopeptidase, Iodobenzoate Benzoate, Clostripain,Trypsin. |
| 9.8 | |||
| 418 ALVRVAAALTALT 430 | 1269.55 | 9.7 | Chymotrypsin, Proline Endopeptidase, Cyanogen Bromide, Iodobenzoate Benzoate, Staph Protease, Trypsin K, AspN. |
| 470 KGGKGNY 476 | 722.80 | Chymotrypsin, Proline Endopeptidase, Cyanogen Bromide, Iodobenzoate Benzoate, Staph Protease, Trypsin R, Proline Endopeptidase, AspN, Elastase |
The full length of candidate protein was identified as an antigen by VaxiJen 2.0 server (threshold 0.5) with score 0.52. Also, the antigenicity of the final B and T-cell predicted epitopes was shown in Table 5. Furthermore, the results of VaxiJen 2.0 analysis indicated that six B-cell epitopes and five T-cell epitopes had antigenic ability.
Table 5.
The antigenicity of predicted epitopes
| Final B-cell epitope | score | Final T-cell epitope | score |
|---|---|---|---|
| 30 TLGPKGRNVVL 40 | -0.03* | 160 KLIAEAM 166 | 0. 3* |
| 150 ISANSDETVGKLIAEAM 166 | 0.75 | 205 INKPETGSVELENPYILL 222 | 1.1 |
| 220 ILLVDKKISNI 230 | 0.13* | 279 PGFGDRR 285 | 1.95 |
| 270 IVKVAAVKAP 279 | 0.88 | 351 QQIEDSTSDYD 361 | 0.88 |
| 351 QQIEDSTSDY 360 | 0.85 | 385 TEVEMKEKRAR 395 | 2.03 |
| 385 TEVEMKEKR 393 | 1.95 | 418 ALVRVAAALTALT 430 | 0.3* |
| 453 QIVENAGEEPSVVVNTVK 470 | 0.53 | 470 KGGKGNY 476 | 2.4 |
| 526 DDKMDLGA 534 | 0.7 |
Probable Non-Antigen
According to the final B and T-cell predicted epitopes, common epitopes with the highest vaxiJen score for GroEL antigen were selected (Figure 5 and Table 6). These epitopes may be useful for developing subunit vaccines.
Figure 5.

Schematic predicted epitopes for synthetize (* Start codon; # Stop codon)
Table 6.
Common epitopes with the highest score
| Final common epitopes | Score |
|---|---|
| 150 ISANSDETVGK 160 | 1.95 |
| 270 IVKVAAVKAPGFGDRR 285 | 0.92 |
| 351 QQIEDSTSDYD 361 | 0.88 |
| 385 TEVEMKEKRAR 395 | 2.03 |
After the antigenicity test of the final B and T-cell predicted epitopes, the 3D structure of predicted epitopes with antigenic ability were illustrated using 3D Ligand Site server (Figure 6). 3D structure analysis also showed that all common predicted epitopes were located on the outside of the GroEL antigen molecule.
Figure 6.
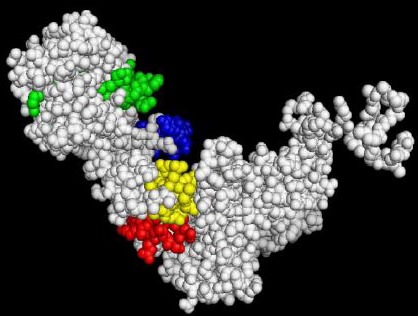
Common predicted epitopes: 150-160, 270-285, 351-361 and 385-395 shown by red, green, blue and yellow colors, respectively
Discussion
Several studies predicted epitopes of desired antigen by computational approaches and used these results in experimental studies with the aim of epitope based vaccine design (21, 17). In this study, GroEl gene of B. melitensis Rev l was candidate for cloning, molecular analysis and epitope prediction. Results showed that all candidate genes successfully cloned, and molecular analysis revealed that GroEL sequence of B. melitensis Rev l is nearly similar to species other than Brucella spp. Because this gene exists in different species and is nearly similar in most of them, although unexpected, had similarity with other Brucella species. Therefore, this gene can be a good candidate for stimulation of immune response against different bacterium species. Comprehensive bioinformatics analyses were conducted on the candidate antigen by online B and T-cell epitope prediction servers. To validate the results of our bioinformatics approaches, we compared computational outputs obtained from different antigens with their experimental results. The experimentally achieved epitopes successfully conformed to the predicted bioinformatics analysis (Table 2).
To prevent degradation of peptide during antigen processing, epitope should lack proteasomal cleavage site (19). In order to, the B and T-cell predicted epitope was analyzed for enzymatic degradation. The proposed epitopes in this study have no proteasomal digestion sites for most cell dominant enzymes (Table 4). The results of epitope prediction showed eight B-cell and seven T-cell epitopes for this antigen. After, examining predicted epitopes according to the proteasomal digestion sites and antigenicity ability, four (150-160, 270-285, 351-361 and 385-395) epitopes became candidates for future research.
Conclusion
The phylogenetic analysis showed that the GroEL sequence had near homology with other species instead of other Brucella spp. According to the antigenic ability and proteasomal cleavage sites, four (150-160, 270-285,351-361 and 385-395) common epitopes were predicted for GroEL gene. Bioinformatics analysis showed that these regions had proper epitope characterization and thus may be useful for stimulation of cell-mediated and humoral immunity systems. Furthermore, in vitro synthesis of determined peptides and experimental validation are essential for using predicted epitope as an effective vaccine against the Brucella pathogen and diagnostic kits; our laboratory has already initiated research in this direction.
Acknowledgment
The present study was funded by INFS (infs.gov.ir) of I.R.I with grant No. 92041813. The authors would like to express their gratitude to INFS for their full support.
References
- 1.Pappas G, Papadimitriou P, Christou L, Akritidis N. Future trends in human brucellosis treatment. Expert Opin Investig Drugs. 2006;15:1141–1149. doi: 10.1517/13543784.15.10.1141. [DOI] [PubMed] [Google Scholar]
- 2.Al Dahouk S, Flèche PL, Noeckler K, Jacques I, Grayon M, Scholz HC, et al. Evaluation of Brucella MLVA typing for human brucellosis. J Microbool Methods. 2007;69:137–145. doi: 10.1016/j.mimet.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 3.Amirmozafari N, Ghazi F, Mostafazadeh A, Mostafaie A, Rajabnia R. Comparison of heat shock response in Brucella obortus and Brucella melitensis. Pak J Biol Sci. 2008;11:188–194. doi: 10.3923/pjbs.2008.188.194. [DOI] [PubMed] [Google Scholar]
- 4.Berzofsky JA. Intrinsic and extrinsic factors in protein antigenic structure. Science. 1985;229:932–940. doi: 10.1126/science.2410982. [DOI] [PubMed] [Google Scholar]
- 5.Buus S, Lauemøller SL, Worning P, Kesmir C, Frimurer T, Corbet S, et al. Sensitive quantitative predictions of peptide-MHC binding by a ‘Query by Committee’ artificial neural network approach. Tissue Antigens. 2003;62:378–384. doi: 10.1034/j.1399-0039.2003.00112.x. [DOI] [PubMed] [Google Scholar]
- 6.Zhang W, Liu J, Zhao M, Li Q. Predicting linear B-cell epitopes by using sequence-derived structural and physicochemical features. Int J Data Min Bioinform. 2012;6:557–569. doi: 10.1504/ijdmb.2012.049298. [DOI] [PubMed] [Google Scholar]
- 7.Ponomarenko JV, van Regenmortel MHV. Structural Bioinformatics. Second Ed 2009. B-cell epitope prediction. [Google Scholar]
- 8.Chen P, Rayner S, Hu KH. Advances of bioinformatics tools applied in virus epitopes prediction. Virol Sin. 2011;26:1–7. doi: 10.1007/s12250-011-3159-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steere AC, Drouin EE, Glickstein LJ. Relationship between immunity to Borrelia burgdorferi outer-surface protein A (OspA) and Lyme arthritis. Clin Infect Dis. 2011;52:259–265. doi: 10.1093/cid/ciq117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hopp TP, Woods KR. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci USA. 1981;78:3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnes P, Elofsson A. Prediction of MHC class I binding peptides, using SVMHC. BMC Bioinformatics. 2002;3:25. doi: 10.1186/1471-2105-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noguchi H, Kato R, Hanai T, Matsubara Y, Honda H, Brusic V, et al. Hidden Markov model-based prediction of antigenic peptides that interact with MHC class II molecules. J Biosci Bioeng. 2002;94:264–270. doi: 10.1263/jbb.94.264. [DOI] [PubMed] [Google Scholar]
- 13.Delpino MV, Estein SM, Fossati CA, Baldi PC, Cassataro J. Vaccination with Brucella recombinant DnaK and SurA proteins induces protection against Brucella abortus infection in BALB/c mice. Vaccine. 2007;25:6721–6729. doi: 10.1016/j.vaccine.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook JF, Russell DW. 3rd ed. NY: Cold Spring Harbor Laboratory, Cold Spring Harbor; 2001. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 15.Geourjon C, Deléage G. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci. 1995;11:681–684. doi: 10.1093/bioinformatics/11.6.681. [DOI] [PubMed] [Google Scholar]
- 16.Wass MN, Sternberg MJ. Prediction of ligand binding sites using homologous structures and conservation at CASP8. Proteins. 2009;77:147–151. doi: 10.1002/prot.22513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simon GG, Hu Y, Khan AM, Zhou J, Salmon J, Chikhlikar PR, et al. Dendritic cell mediated delivery of plasmid DNA encoding LAMP/HIV-1 Gag fusion immunogen enhances T cell epitope responses in HLA DR4 transgenic mice. PLoS One. 2010;5:e8574. doi: 10.1371/journal.pone.0008574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tabatabai LB, Pugh GW., Jr Modulation of immune responses in Balb/c mice vaccinated with Brucella abortus Cu-Zn superoxide dismutase synthetic peptide vaccine. Vaccine. 1995;12:919–924. doi: 10.1016/0264-410x(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 19.Toes RE, Nussbaum AK, Degermann S, Schirle M, Emmerich NP, Kraft M, et al. Discrete cleavage motifsconstitutive and immunoproteasomes revealed by quantitative analysis of cleavage products. J Exp Med. 2001;194:1–12. doi: 10.1084/jem.194.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vizcaíno N, Zygmunt MS, Verger JM, Grayon M, Cloeckaert A. Localization and characterization of a specific linear epitope of the Brucella DnaK protein. FEMS Microbiol Lett. 1997;154:117–122. doi: 10.1111/j.1574-6968.1997.tb12632.x. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Liu X, Zhu Y, Zhou X, Cao C, Hu X, et al. Bioinformatic prediction of epitopes in the Emy162 antigen of Echinococcus multilocularis. Exp Ther Med. 2013;6:335–340. doi: 10.3892/etm.2013.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]


