Abstract
Pneumocystis pneumonia remains a common opportunistic infection in the diverse immunosuppressed population. One clear risk factor for susceptibility to Pneumocystis is a declining CD4+ T-cell counts in the setting of HIV/AIDS or primary immunodeficiency. Non-HIV infected individuals taking immunosuppressive drug regimens targeting T-cell activation are also susceptible. Given the crucial role of CD4+ T-cells in host defense against Pneumocystis, we used RNA-sequencing of whole lung early in infection in wild type and CD4-depleted animals as an unbiased approach to examine mechanisms of fungal clearance. In wild type mice, a strong eosinophil signature was observed at day 14 post-Pneumocystis challenge and eosinophils were increased in the bronchoalveolar lavage fluid of wild type mice. Furthermore, eosinophilopoiesis-deficient Gata1tm6Sho/J mice were more susceptible to Pneumocystis infection when compared to BALB/c controls and bone marrow derived eosinophils had in vitro Pneumocystis killing activity. To drive eosinophilia in vivo, Rag1−/− mice were treated with a plasmid expressing IL-5 (pIL5) or an empty plasmid control via hydrodynamic injection. pIL5 treated mice had increased serum IL-5 and eosinophilia in the lung, as well as reduced Pneumocystis burden compared to mice treated with control plasmid. Additionally, pIL5 treatment could induce eosinophilia and reduce Pneumocystis burden in CD4-depleted C57Bl/6 and BALB/c mice, but not eosinophilopoiesis-deficient Gata1tm6Sho/J mice. Taken together, these results demonstrate that an early role of CD4+ T-cells is to recruit eosinophils to the lung and that eosinophils are a novel candidate for future therapeutic development for Pneumocystis pneumonia in the immunosuppressed population.
Introduction
Pneumocystis jirovecii is a host-specific fungal pathogen that causes a diffuse interstitial pneumonia in immunocompromised individuals (1). Pneumocystis remains the most common serious opportunistic infection in the HIV/AIDS population and is a frequent complication in developing countries where combination antiretroviral therapy (cART) and anti-Pneumocystis prophylaxis are difficult to implement (2–7). In developed countries, the incidence of Pneumocystis infection has continued to rise due to the expansion of the immunosuppressed population (8, 9). One study estimates that 75% of cases of Pneumocystis pneumonia are in non-HIV immunosuppressed individuals, such as those receiving immunosuppressive drug therapy for hematologic malignancy and post-transplantation rejection (9). In fact, in the non-HIV infected immunosuppressed population, Pneumocystis tends to have increased morbidity, such as higher rates of mechanical ventilation, and increased mortality compared to the HIV-positive population (10–12).
Given the opportunistic nature of Pneumocystis, much can be gleaned about the host immune response required for clearance of Pneumocystis by examining the populations that are susceptible to infection. The HIV/AIDS population provides the strongest evidence; a clear inverse correlation exists between decreasing CD4+ T-cell counts and increasing susceptibility to Pneumocystis (13–16). The importance of CD4+ T-cells in protecting against Pneumocystis pneumonia has also been demonstrated in patients with genetic immunodeficiencies, such as forms of severe combined immunodeficiency, as well as in animal models of infection (17, 18). Although CD4+ T-cells have been shown to interact with various cell types throughout the course of Pneumocystis infection, such as B cells and macrophages, the ability of other immune cell types to contribute to Pneumocystis clearance in a CD4+ T-cell dependent manner is still an area of active investigation (19–23). Identifying novel cell types that mediate immunity to Pneumocystis could potentially suggest unique pathways for targeted therapeutic development.
To investigate immunologic responses that may mediate clearance of Pneumocystis, we used RNA sequencing of whole lung at day 14 of Pneumocystis murina infection in CD4-depleted (which develop chronic progressive infection) and wild type C57Bl/6 mice (which clear by 4 weeks). This analysis revealed a prominent eosinophil signature in wild type mice compared to CD4-depleted mice. We also observed a substantial increase in recruited eosinophils in the bronchoalveolar lavage of infected CD4 replete mice compared to CD4 depleted mice. Using hydrodynamic injection of a plasmid encoding IL-5, CD4-depleted and Rag1−/− knockout mice receiving pIL5 demonstrated significant eosinophilia in the lung and decreased Pneumocystis burden 14 days post-challenge. Finally, GATA1tm6Sho knockout mice deficient in eosinophilopoiesis had no difference in burden when treated with pIL5. Taken together, this study demonstrates that one role of CD4+ T-cells during Pneumocystis infection is to recruit eosinophils to the lung, which then contribute to clearance of Pneumocystis.
Methods
Mice
C57Bl/6 mice, Rag1−/− knockout mice on a C57Bl/6 background, BALB/c mice, and Gata1tm6Sho/J mice were all ordered from The Jackson Laboratory (24). Mice were all 6–8 week old females and were bred in the Rangos Research Building Animal Facility. All use of laboratory animals was approved and performed in accordance with the University of Pittsburgh Institutional Care and Use Committee.
Pneumocystis infection time course and primary infections
Twenty-five C57Bl/6 female mice were CD4-depleted using weekly intraperitoneal administration of 0.3 mg of GK1.5 monoclonal antibody per mouse and were subsequently challenged with 2.0 × 106/mL Pneumocystis murina cysts using oropharyngeal inoculation as previously described (22, 25, 26). Twenty-five age-matched C57Bl/6 female mice were inoculated at the same time, but were not CD4-depleted. Five mice from each group were then sacrificed at day 0, 3, 7, 10, and 14. Four BALB/c and Gata1tm6Sho/J mice were also inoculated with Pneumocystis oropharyngeally and sacrificed at day 14. Six uninfected BALB/c mice were used as naïve controls.
RNA isolation and qRT-PCR
Lung RNA was purified using Trizol® Reagent (Life Technologies). Briefly, lungs were homogenized and following the addition of chloroform, RNA in the aqueous phase was collected and precipitated in isopropanol. Following centrifugation, the RNA was washed with 75% ethanol, centrifuged again, and then resuspended in nuclease-free water. Following incubation at 55°C, RNA was quantified using a Nanodrop and 1 μg of RNA was converted to cDNA using iScript™ cDNA synthesis kit per manufacturer’s instructions (Bio-Rad). PC burden was then quantified using SsoAdvanced qRT-PCR universal probes supermix (Bio-Rad) using primers and a probe specific for Pneumocystis murina small subunit (SSU) rRNA with a standard curve of known Pneumocystis SSU rRNA concentrations. SSU primer and probe sequences are as follows: Forward: 5′-CATTCCGAGAACGAACGCAATCCT; Reverse: 5′-TCGGACTTGGATCTTTGCTTCCCA; FAM-Probe: 5′-TCATGACCCTTATGGAGTGGGCTACA. Other primers used include: Prg2, Epx, Il5, Clca3, Muc5ac, Muc5b, and Il13 (Applied Biosystems). Prior to sequencing, the RNA was further purified using a Qiagen RNA cleanup kit with DNAse treatment.
RNA sequencing
Total RNA from mouse whole lung was used to perform RNA sequencing. Each sample was assessed using Qubit 2.0 fluorometer and Agilent Bioanalyzer TapeStation 2200 for RNA quantity and quality. Library preparation was done using Illumina TruSeq Stranded mRNA sample prep kit. The first step in the workflow involves purifying the poly-A containing mRNA molecules using poly-T oligo attached magnetic beads. Following purification, the mRNA is fragmented into small pieces using divalent cations. The cleaved RNA fragments are copied into first strand cDNA using reverse transcriptase and random primers. Strand specificity is achieved by using dUTP in the Second Strand Marking Mix, followed by second strand cDNA synthesis using DNA Polymerase I and RNase H. These cDNA fragments then have the addition of a single ‘A’ base and subsequent ligation of the adapter. The products are then purified and enriched with PCR to create the final cDNA library. The cDNA libraries are validated using KAPA Biosystems primer premix kit with Illumina-compatible DNA primers and Qubit 2.0 fluorometer. Quality is examined using Agilent Bioanalyzer Tapestation 2200. The cDNA libraries will be pooled at a final concentration 1.8pM. Cluster generation and 75 bp paired read single-indexed sequencing was performed on Illumina NextSeq 500’s.
Data Analysis
Raw reads from an Illumina NextSeq 500 in fastq format were trimmed to remove adaptor/primer sequences. Trimmed reads were then aligned using BWA (version 0.5.9, settings aln -o 1 -e 10 -i 5 -k 2 -t 8) against the mouse genomic reference sequence. Additional alignment and post-processing were done with Picard tools (version 1.58) including local realignment and score recalibration (not duplicate marking) to generate a finale genomic aligned set of reads. Reads mapping to the genome were characterized as exon, intron, or intergenic (outside any annotated gene) using the matched annotation for the genomic reference sequence. The remaining unmapped reads from the genomic alignment were then aligned to a splice reference created using all possible combinations of known exons (based on annotation described above) and then categorizing these as known or novel splice events. This aligned data is then used to calculate gene expression by taking the total of exon and known splice reads for each annotated gene to generate a count value per gene. For each gene there is also a normalized expression value generated in two ways: 1) Reads per Mapped Million (RPM), which is calculated by taking the count value and dividing it by the number of million mapped reads, 2) Reads per Mapped Million per Kilobase (RPKM), which is calculated by taking the RPM vale and dividing it by the kilobase length of the longest transcript for each gene. The RPM values are subsequently used for comparing gene expression across samples to remove the bias of different numbers of reads mapped per sample. RPKM values are subsequently used for comparing relative expression of genes to one another to remove the bias of different numbers of mapped reads and different transcript lengths. In addition to gene expression measurements, nucleotide variation was also detected using the GATK (version 1.3–25, -dcov 2000 -stand_call_conf 30.0 -stand_emit_conf 10.0 -A DepthOfCoverage -A BaseCounts -A AlleleBalance), which identified single nucleotide and small insertion/deletion (indel) events using default settings. Mapped exonic reads per WT sample: 44,291,011; 23,432,350; 41,004,408; and 41,308,864. Mapped exonic reads per GK1.5 sample: 23,305,058; 43,821,698; 122,834,770; and 29,805,804. Data were then filtered on a quality score of 20 and probed for eosinophil associated genes: Ear11, Ear5, Ccl8, Ccl24, Ccr3, Prg2, Ccl11, Ccl7, Il13, Ear10, Il5ra, Ear2, Csf2rb, Ccl5, Il5, Ear1, Il4 (did not pass quality filter), and Epx (did not pass quality filter).
Data Availability
The RNA sequencing data contained in this paper is publicly available through the Sequence Read Archive BioProject number: PRJNA276259. Further information can be found at: http://www.ncbi.nlm.nih.gov/bioproject/.
Bronchoalveolar lavage
Wild type C57BL/6, and GK1.5 treated mice infected with Pneumocystis were anesthetized at day 14 post-infection and a 20g Exel Safelet Catheter (Exel International Co.) was inserted in the cricoid cartilage. The needle was then removed and 1 mL aliquots were inserted and removed from the lung using a 1 mL syringe (10 mL total). Bronchoalveolar lavage (BAL) cells were then spun at 300xg for 10 minutes, resuspended in PBS, and counted using Trypan Blue stain. 1×106 cells were then transferred to a round bottom 96-well plate for staining and the remainder of cells was transferred to Trizol® Reagent for RNA isolation (as described above). Naïve (uninfected) mice were also examined as a control.
IL-5 and Eotaxin-1 Luminex on lung homogenate
Lung was collected in PBS containing protease inhibitors (Roche) and homogenized. We used a Bio-Plex Pro™ Assay (23-plex, Bio-Rad) according to the manufacturers recommendations. Briefly, the plate was treated with Bio-Plex assay buffer, followed by vortexing and two washes. Lung homogenates (undilulted), standards, and blanks were then added to the plate and incubated at room temperature for one hour shaking at 850 rpm, covered. Following three washes, detection antibodies were then diluted and added to each well. The plate was incubated as above. Following three washes, diluted SA-PE was added to each well and incubated at room temperature for 20 minutes on shaker. The plate was then washed three times and resuspended in assay buffer and beads were quantified using a Bio-Plex® MAGPIX™ (Bio-Rad).
Flow cytometry
BAL cells or cells from digested lung (1×106 total) were spun at 300xg for 3 minutes, resuspended in PBS, and pelleted once more. Cells were then resuspended in PBS containing 2% heat-inactivated fetal bovine serum and 0.4 μg of anti-CD16/CD32 (eBioscience, clone: 93). Following a 15 minute incubation at 4°C, cells were stained with the following antibodies: SiglecF-PE (BD Pharmigen™, clone: E50-2440), CD11b-APC (BioLegend, clone: M1/70), GR1-PE-Cy7 (BD Pharmigen™, clone:RB6-8C5), CD11c-FITC (eBioscience, clone: N418), and F4/80-APC-e780 (eBioscience, clone: BM8). Following an hour incubation at 4°C, cells were washed with PBS, pelleted, and fixed (BD CytoFix™). Cells were then analyzed using a BD LSRII Flow Cytometer with compensation via OneComp eBeads (eBioscience).
Eosinophil culture and Pneumocystis killing assay
Bone marrow-derived eosinophils were generated using a previously described protocol (27) and per our previous work (28). Briefly, bone marrow was isolated from naïve BALB/c mice and cells plated at 1 × 106 cells/ml in RPMI 1640 containing 20% FBS (Irvine Scientific, Santa Ana, CA), 2 mM Glutamine, 25 mM HEPES, 1X MEM nonessential amino acids, 1 mM sodium pyruvate (all from Life Technologies BRL, Rockville, MD), 50 μM β-mercaptoethanol (Sigma-Aldrich, St. Louis, MO), 100 ng/ml stem cell factor and 100 ng/ml FLT3-L (both from Peprotech). After 4 days, cells were replated in the above media supplemented with 10 ng/ml IL-5. After 10 days, bone marrow cells were fully differentiated into eosinophils. As previously reported (27), samples of 1×105 cells were taken for RNA analysis each time media was changed for real time PCR analysis of Epx for eosinophil development and Mpo (Applied Biosystems) for neutrophil development. In addition, cells were cytospun onto glass slides, Giemsa stained and analyzed for morphology and purity by a murine pathologist in the Comparative Pathology Laboratory at the University of Alabama at Birmingham. On the tenth day, bone marrow-derived eosinophils were enumerated and utilized in experiments. Bone marrow-derived eosinophils (1 × 105) were then co-cultured with 1 × 103 Pneumocystis cysts in 100 μL for 18 h at 37°C and 5% CO2 alone or in the presence of 10 ng/mL of IL-4 and IL-13. Controls included P. murina cultured in the absence of eosinophils as well as in the presence or absence of IL-4 and IL-13. Total RNA was isolated from the contents of each well using TRIZOL LS reagent (Invitrogen, Carlsbad, CA) and Pneumocystis SSU burden was calculated as above. Percent killing was defined as previously described (20).
pIL5 and pCMV hydrodynamic injection
An untagged, murine IL-5 expression vector (pIL5, Origene, MC208784) and an empty vector pCMV6 control (Origene, PS100001) were grown in Mix and go E. coli (Zymo Research) in 200 mL of LB containing kanamycin and were prepared using an EndoFree Plasmid Maxi Kit (Qiagen) per manufacturer’s instructions. Following quantification of vector, 10 μg of vector was added to 2 mL of Ringer’s solution (0.9% NaCl, 0.03% KCL, and 0.016% CaCl2) and injected intravenously via the tail vein within 5 seconds, as previously described (29, 30).
IL-5 ELISA
Serum IL-5 was quantified using BioLegend ELISA MAX™ Mouse IL-5 ELISA kit per manufacturer’s instructions. Briefly, a 96-well plate was coated with capture antibody and stored overnight at 4°C. Following washes with PBS + 0.05% Tween-20, the plate was blocked with assay diluent for 1 hour at room temperature. Serum samples (diluted 1:20) and IL-5 standard were diluted in assay diluent, added to the plate, and incubate overnight at 4°C. Detection antibody and diluted Avidin-HRP were then added to the plate, with washes in between additions, and the plate was developed with TMB substrate in the dark. Absorbance was then measured at 450 nm.
Lung digestion
The right superior lobe of lung was physically digested using scissors, followed by an hour and a half incubation in collagenase/DNase in a 37°C shaker at 250 rpm. Single cell suspensions were then strained using a 70 μm filter, pelleted, and then resuspended in 10 mL PBS. Following enumeration using Trypan Blue, cells were stained for flow cytometry as described above.
Histology
The left main bronchus was clamped using forceps and 250 μL of 10% formalin was injected into the bronchus. The lung tissue was then submerged in 10% formalin, paraffin-embedded, and processed by the Children’s Hospital of Pittsburgh Histology Core. Sections were then stained from H&E and PAS.
Statistics
All statistics were performed using GraphPad Prism 6. Briefly, an unpaired, two-tailed student’s T test with a p<0.05 considered significant was used for all studies except for the BALB/c pIL-5 treatment. Given the non-Gaussian distribution for the BALB/c pIL-5 treatment, a Mann-Whitney nonparametric rank test was performed with a p<0.05 considered significant. For studies with three groups, a one-way ANOVA with Tukey’s multiple comparisons was used with a p<0.05 considered significant. A Kruskal-Wallis nonparametric test with Dunn’s multiple comparison’s test was used for gene expression analysis in the BALB/c primary challenge experiment with a p<0.05 considered significant. Linear regression was also performed using Prism and Pearson’s correlation coefficient calculations were performed.
Results
RNA sequencing of whole lung identifies an eosinophil signature early in Pneumocystis infection
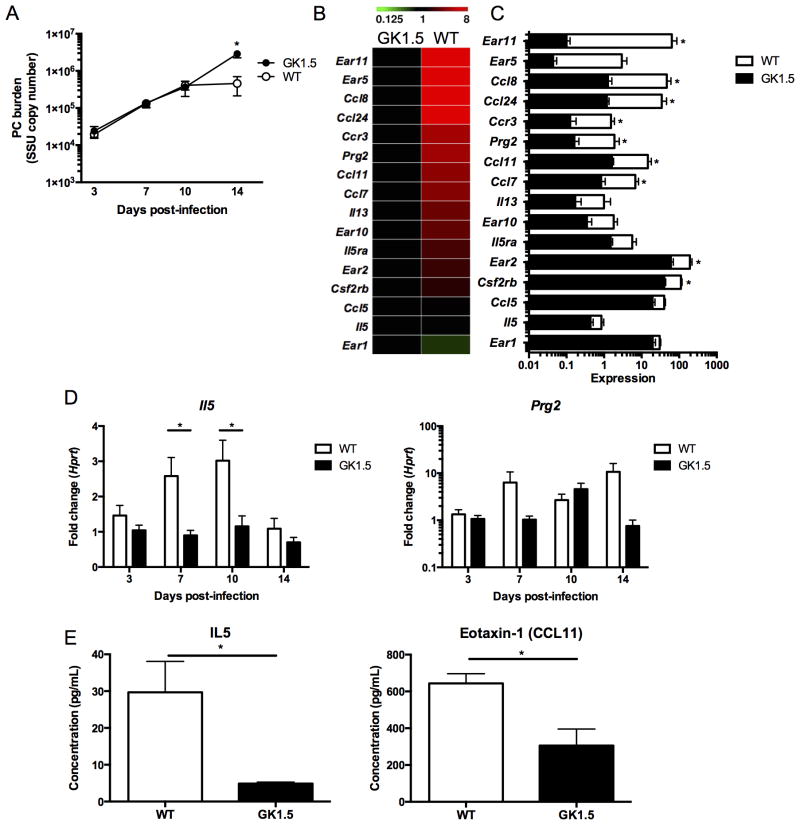
To further understand the role of CD4+ T-cells in Pneumocystis infection, we examined Pneumocystis burden in wild type and GK1.5 treated, CD4-depleted C57/Bl6 mice. In this study, at day 14 using quantitative real time PCR, wild type mice begin to clear infection as CD4-depleted mice had a higher fungal burden at this time point (Figure 1A). As an unbiased approach to investigate potential mechanisms of fungal clearance, we used RNA sequencing of whole lung at this time point to examine the signatures of potential effector cells. Strikingly, several genes associated with eosinophil function and recruitment, such as prg2 (major basic protein), il5ra (IL-5 receptor alpha), ccr3, ccl11 (eotaxin-1), ccl24 (eotaxin-2), were all significantly upregulated at day 14 of infection in wild type animals (Figures 1B and 1C). Another specific eosinophil marker, eosinophil-associated ribonculease 2 (ear2), was also significantly upregulated in wild type animals, while less specific eosinophil-associated ribonucleases (ear5, ear10, ear11) also had higher expression in wild type animals. Importantly, in addition to a robust eosinophil signature at day 14 in wild type animals, IL-5 had significantly higher expression in wild type animals at the transcriptional level at day 7 and day 10 post-infection with Pneumocystis (Figure 1D). Importantly, prg2, an eosinophil associated gene, had a 10-fold increase in expression at day 14 by qRT-PCR, similar to that detected by RNA sequencing (Figure 1D). Furthermore, protein levels of IL-5 and eotaxin-1 (CCL11) were significantly higher in wild type animals at day 14 when compared to CD4-depleted mice (Figure 1E).
Figure 1. RNA sequencing of whole lung shows a prominent CD4-dependent eosinophil signature at day 14 of Pneumocystis infection.
A. Wild type or GK1.5 treated CD4-depleted C57Bl/6 mice were infected with 2.0 × 106 cysts/ml of Pneumocystis and were sacrificed at day 3, 7, 10, or 14 following infection (n=5 at each time point). Pneumocystis burden was calculated by qRT-PCR of the small subunit ribosomal RNA and a significant decrease was seen at day 14 (p<0.01 by student’s t-test). B. RNA sequencing of whole lung RNA at day 14 in GK1.5 treated and wild type mice shows increase in expression in genes associated with eosinophils (n=4 in each group). C. Histogram of expression values from heat map in B with * indicating p<0.05 by student’s t-test. D. Il5 expression over the course of Pneumocystis infection normalized to Hprt and GK1.5 day 3 (fold change) shows increase of il5 at day 7 and 10 in wild type mice (* p<0.05, student’s t-test). Similar to the expression pattern seen by RNA sequencing, a ten-fold increase in Prg2 is seen at day 14 by qRT-PCR (p>0.05). E. IL-5 and Eotaxin-1 (CCL11) protein levels in lung homogenate at day 14 as determined by luminex (* p<0.05).
Eosinophils are present in bronchoalveolar lavage fluid early in infection
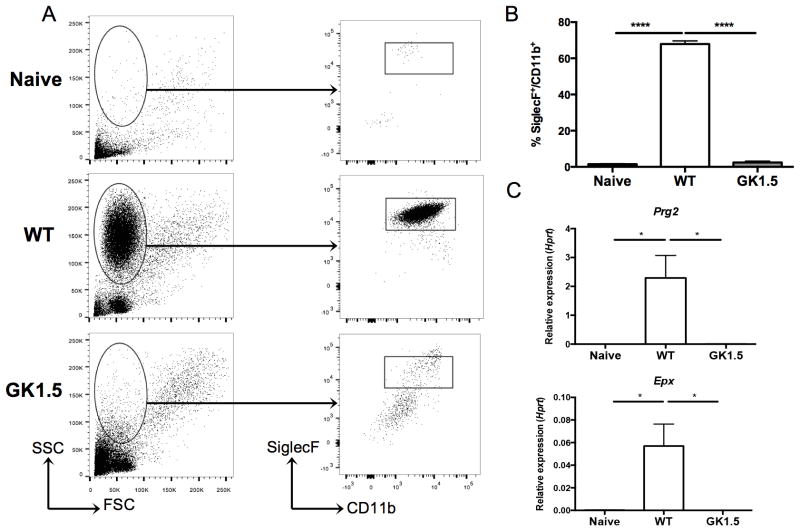
To further clarify the CD4+ T-cell dependent eosinophil response to Pneumocystis infection, we sought to define the cell populations in the bronchoalveolar lavage (BAL) fluid of wild type and CD4-depleted animals at day 14. Cell populations in naïve mice were also analyzed. A population of cells with high side scatter was present in animals with intact CD4+ T-cell responses, but absent in naïve and CD4-depleted animals infected with Pneumocystis (Figure 2A, left panel). After gating on all cells, a population of SiglecF+CD11b+ cells was noted in the wild type animals, but this population was substantially reduced back to naïve levels in animals treated with GK1.5 (Figure 2A, right panel). The SiglecF+CD11b+ population represented over 60% of cells in BAL fluid in wild type mice, while less than 2% of cells were SiglecF+CD11b+ in mice treated with GK1.5 (Figure 2B). Additionally, the RNA from BAL cell pellets was enriched for transcripts associated with eosinophils; Epx and Prg2 expression was nearly 1000-fold higher in BAL cell pellets from wild type mice when compared to naïve and CD4-depleted mice (Figure 2C).
Figure 2. CD4-dependent recruitment of eosinophils to the lung at day 14 of Pneumocystis infection.
A. Bronchoalveolar lavage (BAL) of naïve (uninfected), wild type, and GK1.5 treated CD4-depleted mice 14 days post-inoculation with Pneumocystis shows a large population of cells with high side-scatter in wild type mice (left panel). The cells were gated as shown (left panel), and a SiglecF+CD11b+ population was seen in the wild type, but not the naïve or GK1.5 treated animals (right panel). B. Significant increase in percentage of SiglecF+CD11b+ cells in wild type animals compared to naïve and GK1.5 treated animals (n=4–5, **** p<0.0001 by one-way ANOVA with Tukey’s multiple comparisons). C. qRT-PCR for Epx (top) and Prg2 (bottom) on RNA extracted from BAL cell pellets shows significant increase in expression in wild type animals compared to naïve and GK1.5 treated animals (* p<0.05 by one-way ANOVA with Tukey’s multiple comparisons).
Eosinophils contribute to control of Pneumocystis infection both in vitro and in vivo
To determine the role of eosinophils in Pneumocystis infection, we used a loss-of-function approach and infected eosinophilopoiesis-deficient Gata1tm6Sho/J mice and BALB/c controls. Gata1tm6Sho/J mice had an increased Pneumocystis burden at day 14 post infection compared to control BALB/c mice, while uninfected BALB/c had no detectable Pneumocystis burden (Figure 3A). BALB/c mice had an increase in Epx expression and a modest increase in Prg2 expression compared to Gata1tm6Sho/J mice and BALB/c uninfected controls (Figure 3B). Eosinophils cultured from BALB/c bone marrow also demonstrated anti-Pneumocystis activity in vitro (Figure 3C). Furthermore, bone marrow derived eosinophils displayed increased Pneumocystis killing activity when co-cultured with IL-4 and IL-13 (Figure 3D).
Figure 3. Eosinophils contribute to control of Pneumocystis infection both in vitro and in vivo.
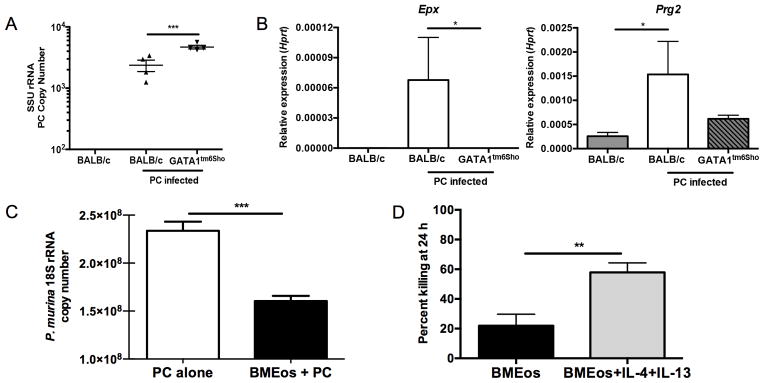
A. BALB/c and Gata1tm6Sho/J knockout mice were infected with Pneumocystis and sacrificed at day 14 post-infection and SSU burden was quantified by qRT-PCR (** p<0.01 by student’s T test). Uninfected BALB/c mice have no detectable Pneumocystis burden. B. qRT-PCR for Epx (left) and Prg2 (right) on RNA from whole lung shows significant increase in BALB/c mice infected with Pneumocystis compared to uninfected BALB/c and infected Gata1tm6Sho/J knockout mice (* p<0.05 by Kruskal-Wallis test with Dunn’s multiple comparisons test). C. Bone marrow derived eosinophils from BALB/c mice demonstrate anti-Pneumocystis activity when co-cultured in vitro for 24 hours at an eosinophil to P. murina cyst ratio of 100:1 (*** p<0.0001, student’s t-test). D. Bone marrow derived eosinophils show enhanced killing activity when co-cultured with Pneumocystis in the presence of 10 ng/ml of IL-4 and IL-13 compared to Pneumocystis alone (** p<0.01 by student’s T test).
Hydrodynamic injection of IL-5 promotes Pneumocystis clearance in CD4-depleted C57Bl/6 and Rag1−/− mice
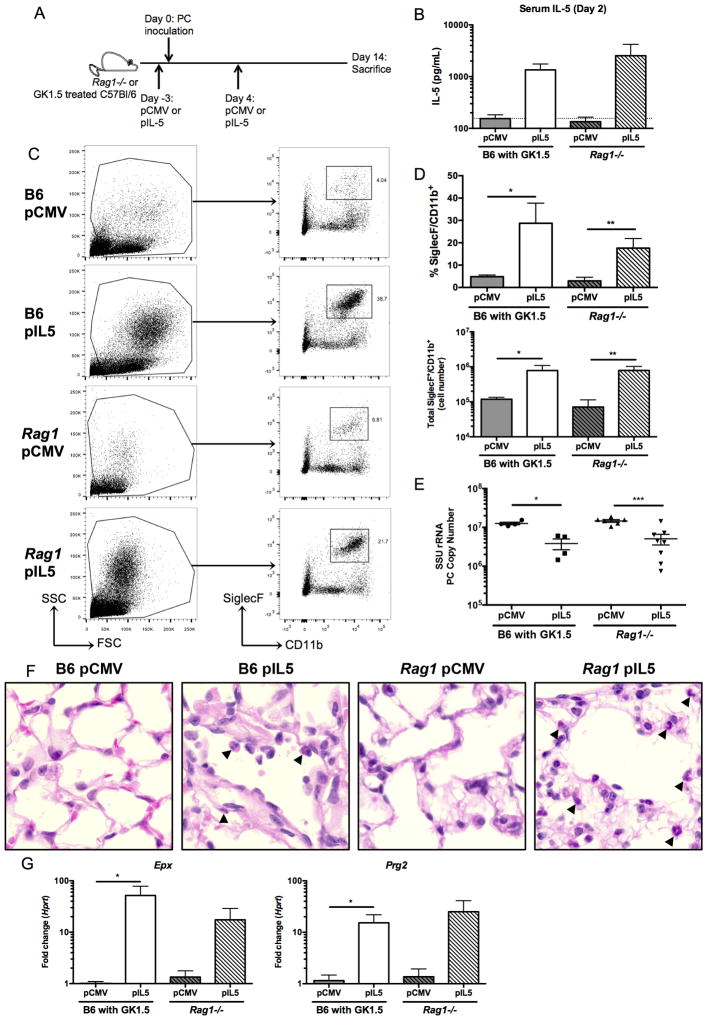
To induce eosinophilia prior to Pneumocystis infection, we employed hydrodynamic injection with either a plasmid expressing IL-5 (pIL5) or an empty plasmid control (pCMV) in C57Bl/6 mice treated with GK1.5 or Rag1−/− mice three days prior to infection (Figure 4A). At day two following infection, mice treated with pIL5 had over a log-fold increase in serum IL-5 (Figure 4B). At day 14 of infection, treatment with pIL5 resulted in an increased abundance of a high side scatter population and a SiglecF+CD11b+ population in both CD4-depleted C57Bl/6 and Rag1−/− mice (Figure 4C). While these populations were present by flow cytometry in the groups treated with pCMV alone (Figure 4C, left panel), C57Bl/6 and Rag1−/− mice treated with pIL5 had significantly more SiglecF+CD11b+ cells as measured by both percentage and total cell number recovered from the lung (Figure 4D).
Figure 4. Treatment of CD4-depleted C57Bl/6 and Rag1−/− mice with pIL5 results in eosinophilia in whole lung and decreased Pneumocystis burden.
A. Schematic showing timing of hydrodynamic injection of either pCMV or pIL5 (day −3 and 4) and Pneumocystis inoculation (day 0) into GK1.5 treated C57Bl/6 (B6) or Rag1−/− mice. B. Serum IL-5 ELISA at day 2 post infection shows log-fold increase in IL-5 in pIL5 treated animals (n=4–7, dotted line represents limit of detection). C. Digested whole lung shows a high side scatter population in the pIL5 groups compared to the pCMV treated groups (left panel). The cells were gated as shown (left panel), and a SiglecF+CD11b+ population was seen in both the B6 and Rag1−/− mice treated with pIL5 at day 14 post infection (right panel). D. pIL5 treatment resulted in a statistically significant increase in percentage (top) and total number (bottom) of SiglecF+CD11b+ cells (* p<0.05, ** p<0.01 by student’s t-test). E. pIL5 treatment results in statistically significant reduction in Pneumocystis burden as measured by qRT-PCR of small subunit ribosomal RNA at day 14 post infection (* p<0.05, *** p<0.001 by student’s t-test). F. H&E staining on paraffin-embedded lung sections demonstrating eosinophils (black arrowheads) in both the B6 and Rag1−/− mice treated with pIL5 at 60X magnification. G. qRT-PCR for Epx (left) and Prg2 (right) on RNA extracted from whole lung shows significant log-fold increase in expression in pIL5 treated animals compared to pCMV treated mice (* p<0.05 by student’s T test).
Strikingly, both the CD4-depleted C57Bl/6 and Rag1−/− mice receiving pIL5 had a statistically significant reduction in Pneumocystis burden by day 14 of infection when compared to mice treated with pCMV (Figure 4E). While the average difference in burden was approximately a half-log in both cohorts, some individual mice had greater than a log-reduction in Pneumocystis burden with pIL5 treatment (Figure 4E). A strong negative correlation existed between total number of eosinophils recruited to the lung and Pneumocystis burden in the C57Bl/6 treated animals (Supplemental Figure 1A, p=0.0003). Recruitment of eosinophils to the lung could also be observed by H&E staining in mice receiving pIL5 (Figure 4F). The pIL5 treated mice also had a ten-fold increase in expression of Epx and Prg2 in whole lung RNA when compared to pCMV treated mice (Figure 4G). Furthermore, although pIL5 treatment increased eosinophilic lung inflammation, this was not associated with Type 2 immune inflammation as measured by goblet cell hyperplasia or mucin expression (as measured by qRT-PCR of Clca3, Muc5ac and PAS staining) or Il13 expression in the pIL5 treated C57Bl/6 mice (Supplemental Figure 2).
IL-5 mediated decrease in Pneumocystis burden is abrogated in eosinophil-deficient Gata1tm6Sho/J mice
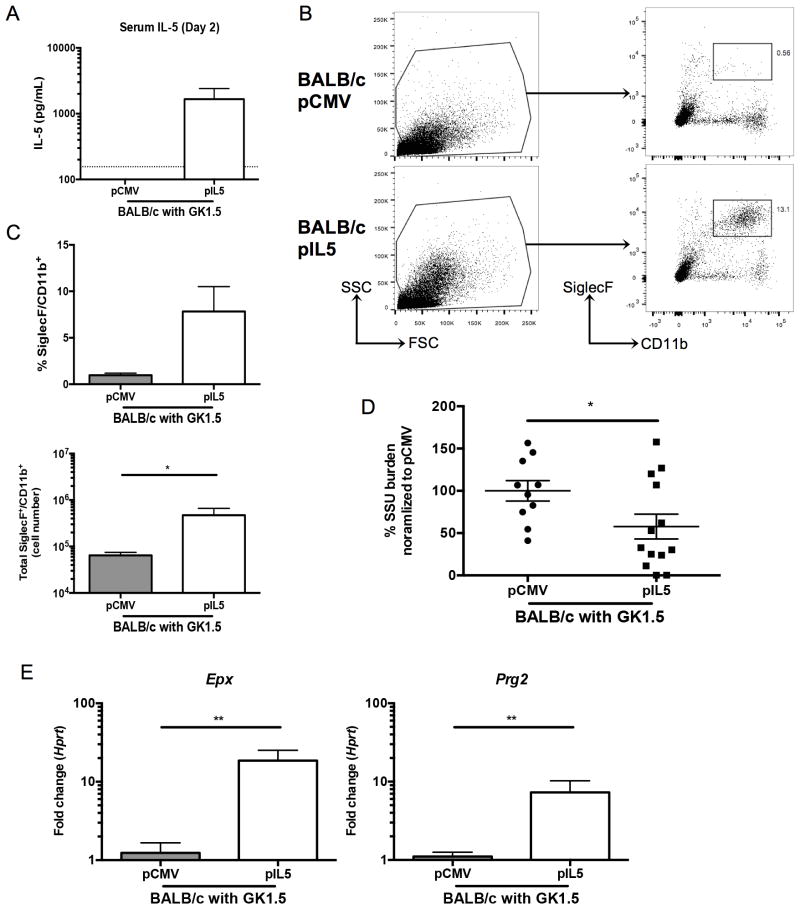
To verify the effect of pIL5 required eosinophilopoiesis, we used a genetic approach. CD4-depleted BALB/c mice were treated with pIL5 as described above and similar levels of IL-5 were induced in the serum (Figure 5A). Similarly, increased eosinophils were noted in the lung digest of pIL5 treated BALB/c mice (Figure 5B). pIL5 treated BALB/c mice had a statistically significant increase in total number and a trend towards higher percentage of SiglecF+/CD11b+ cells compared to pCMV treated BALB/c mice (Figure 5C). Furthermore, upon sacrifice, BALB/c mice treated with pIL5 had nearly a 50% reduction in Pneumocystis burden when compared to pCMV treated animals (Figure 5D, p=0.04). Burden in this case was normalized to the pCMV group as the inoculums over three independent experiments varied; however, a similar reduction in burden was noted with C57Bl/6 and Rag1−/− mice (Figure 4E). Also noteworthy, four pIL5 treated mice had no induction of serum IL-5 and lacked eosinophils in the lung at day 14 by flow cytometry, likely due to technical variation. If only mice that responded to pIL5 treatment were included in the analysis, the mean percentage of Pneumocystis remaining in pIL5 treated mice would be 26% compared to 100% of pCMV treated mice (p<0.0002). The increased Pneumocystis killing in mice that recruited eosinophils to the lungs is also evident in the strong negative correlation that exists between Pneumocystis burden and total SiglecF+CD11b+ recruited cells (Supplemental Figure 1B, p=0.0001). pIL5 treatment was also associated with significant increases in eosinophil associated genes such as Epx and Prg2 (Figure 5E).
Figure 5. pIL5 treatment can reduce Pneumocystis burden in CD4-depleted BALB/c mice.
A. Serum IL-5 ELISA at day 2 post infection shows over a log-fold increase in IL-5 in pIL5 treated BALB/c mice (n=10–13, dotted line represents limit of detection). B. Digested whole lung shows a high side scatter population in the pIL5 groups compared to the pCMV treated groups (left panel). The cells were gated as shown (left panel), and a SiglecF+CD11b+ population was seen in the BALB/c mice treated with pIL5 (right panel). C. pIL5 treatment resulted in an increase in percentage (top) and total number (bottom) of SiglecF+CD11b+ cells (* p<0.05, Mann Whitney test). D. pIL5 treatment results in reduction in Pneumocystis burden as measured by qRT-PCR of small subunit ribosomal RNA at day 14 post infection (* p=0.0481 by student’s t-test). E. qRT-PCR for Epx (left) and Prg2 (right) on RNA extracted from whole lung shows significant log-fold increase in expression in pIL5 treated animals compared to pCMV treated mice (** p<0.01 by Mann Whitney test).
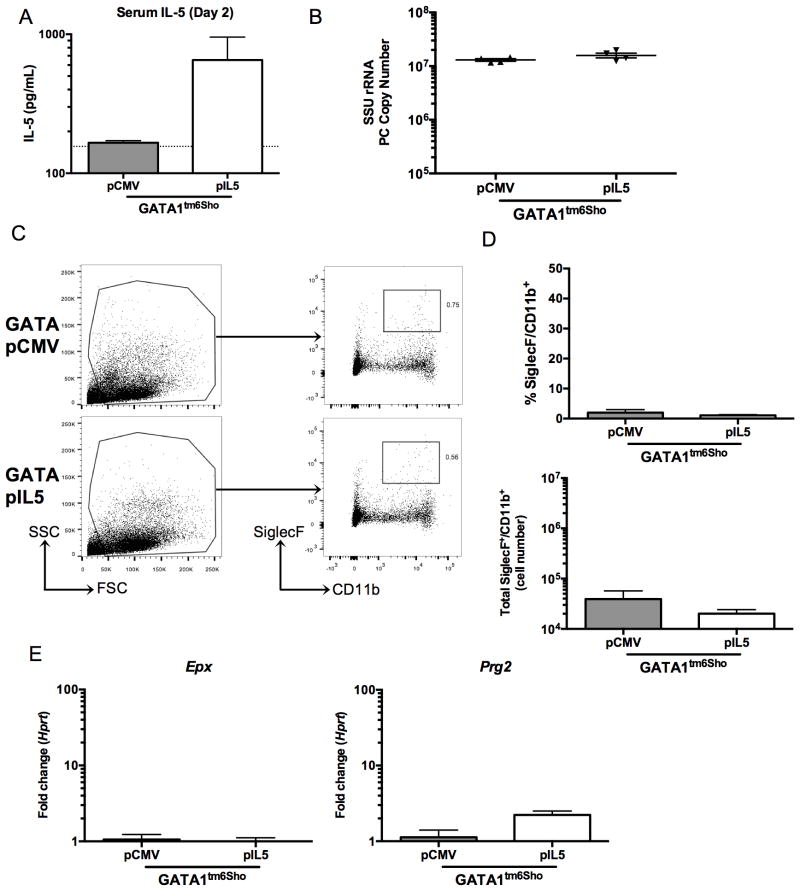
In contrast, Gata1tm6Sho/J mice (on a BALB/c background), mice that are deficient in eosinophilopoiesis, that were CD4-depleted and treated with pIL5 or pCMV as described above failed to show an effect of pIL5 on fungal burden, despite similar levels of IL-5 compared to previous mouse strains (Figure 6A and B). Consistent with no reduction in fungal burden, there was no observable eosinophil recruitment was detected in the lungs of Gata1tm6Sho/J mice, as neither a high side-scatter population nor a SiglecF+CD11b+ was noted by flow cytometry (Figure 6B and C). Furthermore, there was no induction of Epx or Prg2 in the Gata1tm6Sho/J mice treated with pIL5 (Figure 6E).
Figure 6. pIL5 treatment cannot rescue eosinophil-deficient Gata1tm6Sho/J knockout mice.
A. Serum IL-5 ELISA at day 2 post infection shows nearly a log increase in Gata1tm6Sho/J pIL5 treated animals (n=4, dotted line represents limit of detection). B. pIL5 treatment does not reduce Pneumocystis burden at day 14 post infection in Gata1tm6Sho/J mice as measured by qRT-PCR of small subunit ribosomal RNA. C. Digested whole lung shows no difference in high side scatter populations (left panel) or SiglecF+CD11b+ cells (right panel) independent of pIL5 treatment status. D. pIL5 treated Gata1tm6Sho/J mice showed no difference in percentage (top) or total number (bottom) of SiglecF+CD11b+ cells. E. qRT-PCR for Epx (left) and Prg2 (right) on RNA extracted from whole lung shows no increase in eosinophil associated genes in Gata1tm6Sho/J mice.
Discussion
The current study utilized an unbiased RNA sequencing based approach towards evaluating the role of CD4+ T-cells in Pneumocystis infection and suggested that CD4+ T-cells can recruit eosinophils to the lung by day 14 of infection by RNA sequencing. Eosinophils were also shown to have a role in immunity against Pneumocystis in a primary challenge model of eosinophil-deficient mice and in an eosinophil-based Pneumocystis in vitro killing assay. Furthermore, induction of eosinophilia via hydrodynamic injection of pIL5 was capable of reducing Pneumocystis burden in vivo in both CD4-depleted C57Bl/6 and Rag1−/− mice. Finally, the same technique was able to reduce Pneumocystis burden in CD4-depleted BALB/c mice but failed to provide any therapeutic benefit in CD4-depleted Gata1tm6Sho/J mice, further implicating eosinophils as a novel cell population responsible for in vivo antifungal activity.
CD4+ T-cells have been established as crucial mediators to Pneumocystis due to the high incidence of Pneumocystis in HIV/AIDS patients with low CD4+ T-cell counts (1, 5). One clear role of CD4+ T-cells in response to Pneumocystis is to stimulate antibody responses by providing co-stimulatory signals to B cells (19, 23). Antibodies against Pneumocystis are protective; however, at day 14 of infection, prior to antibody production, Pneumocystis burden has already plateaued in wild type animals suggesting an antibody-independent function for CD4 cells (23). As such, CD4+ T-cells appear to recruit eosinophils early in infection while B cells are undergoing maturation into antibody-producing plasma cells and provide preliminary control of Pneumocystis burden. Importantly, IL-5 transcription and eosinophil recruitment appears to be dependent on CD4+ T-cells in this model, as such markers do not appear until after activation of the adaptive immune response at day 7 post-infection.
Eosinophils, classically recognized as mediators of immunity against helminths, have recently been implicated in host defense against a variety of pathogens, including bacterial and viral infections (31). Recently, we have demonstrated that eosinophils contribute to host defense against the fungal pathogen Aspergillus fumigatus through a secretory factor, as eosinophils could mediate fungal killing when separated using transwells (28). The current study extends these findings by demonstrating that eosinophil-deficient mice are more susceptible to Pneumocystis infection and that eosinophils display antifungal activity in vitro. Additionally, treatment of bone marrow derived eosinophils with IL-4 and IL-13 greatly enhanced killing of Pneumocystis, suggesting that Th2 cytokines in the lung may augment eosinophil-dependent antifungal activity.
There are also several lines of evidence that connect Pneumocystis related pathology and eosinophils in both human disease and mouse models. First, eosinophilia in the bronchoalveolar lavage fluid of HIV-positive patients with Pneumocystis pneumonia has been well documented; however, whether a correlation exists between high eosinophil counts and lower Pneumocystis burden in these patients is unknown (32, 33). Secondly, patients with a history of Pneumocystis or bacterial pneumonia had a significantly higher rate of physician-diagnosed asthma, a disease that has a well-established eosinophilic component in regards to pathogenesis (34). Murine models of Pneumocystis have also shown STAT6, a transcription factor required for Th2 responses, is necessary for the development of airway hyperresponsiveness early in the course of infection (23). While eosinophilia was also documented early in this murine model, the study further links a Th2 response, and potentially eosinophilia, with pathology in the context of Pneumocystis infection (23). Additionally studies have shown that CD8+ T-cells may moderate the interactions between CD4+ T-cells and eosinophils, although all three cell types may contribute to Pneumocystis driven pulmonary pathology (35).
However, the current study suggests that eosinophils are more than just the byproduct of a misguided immune response that drives airway hyperresponsiveness and pathology. The current study demonstrates that eosinophils have antifungal effects against Pneumocystis infection in vitro and in vivo and appear to be recruited to the lung by CD4+ T-cells early in infection. Additionally, the role of eosinophils as a potential therapeutic in the setting of HIV/AIDS may warrant further exploration, as IL-5 mediated eosinophilia can provide reduced Pneumocystis burden even in the setting of complete loss of T cells and B cells. Furthermore, such robust eosinophilia actually appeared to mitigate airway pathology (e.g. mucus production) suggesting that Pneumocystis burden may play an equally important role in driving airway hyperresonsivness. These findings provide evidence that the specific pathways responsible for protective and pathologic effects in Pneumocystis pneumonia may be independent and may allow for the targeted use of eosinophil-based treatments for Pneumocystis while avoiding concurrent pathology.
Supplementary Material
Acknowledgments
Grant support: NIAID F30AI114146 (TE), R21HL117090 and RO1HL119770 (C.S.) and NHLBI RO1HL062052 (JKK)
The authors would like to acknowledge Dr. Kong Chen for his assistance in designing the studies in this manuscript and Dr. Mingquan Zheng for his technical assistance. The authors would also like to acknowledge the flow cytometry core at the Children’s Hospital of UPMC in Pittsburgh.
References
- 1.Eddens T, Kolls JK. Pathological and protective immunity to Pneumocystis infection. Semin Immunopathol. 2015;37:153–162. doi: 10.1007/s00281-014-0459-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antiretroviral Therapy Cohort C. Mocroft A, Sterne JA, Egger M, May M, Grabar S, Furrer H, Sabin C, Fatkenheuer G, Justice A, Reiss P, d’Arminio Monforte A, Gill J, Hogg R, Bonnet F, Kitahata M, Staszewski S, Casabona J, Harris R, Saag M. Variable impact on mortality of AIDS-defining events diagnosed during combination antiretroviral therapy: not all AIDS-defining conditions are created equal. Clin Infect Dis. 2009;48:1138–1151. doi: 10.1086/597468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang L, Cattamanchi A, Davis JL, den Boon S, Kovacs J, Meshnick S, Miller RF, Walzer PD, Worodria W, Masur H I. V. a. O. P. S. International, and H. I. V. S. Lung. HIV-associated Pneumocystis pneumonia. Proc Am Thorac Soc. 2011;8:294–300. doi: 10.1513/pats.201009-062WR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malin AS, Gwanzura LK, Klein S, Robertson VJ, Musvaire P, Mason PR. Pneumocystis carinii pneumonia in Zimbabwe. Lancet. 1995;346:1258–1261. doi: 10.1016/s0140-6736(95)91862-0. [DOI] [PubMed] [Google Scholar]
- 5.Morris A, Lundgren JD, Masur H, Walzer PD, Hanson DL, Frederick T, Huang L, Beard CB, Kaplan JE. Current epidemiology of Pneumocystis pneumonia. Emerging infectious diseases. 2004;10:1713–1720. doi: 10.3201/eid1010.030985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tansuphasawadikul S, Pitisuttithum P, Knauer AD, Supanaranond W, Kaewkungwal J, Karmacharya BM, Chovavanich A. Clinical features, etiology and short term outcomes of interstitial pneumonitis in HIV/AIDS patients. The Southeast Asian journal of tropical medicine and public health. 2005;36:1469–1478. [PubMed] [Google Scholar]
- 7.Udwadia ZF, Doshi AV, Bhaduri AS. Pneumocystis carinii pneumonia in HIV infected patients from Mumbai. The Journal of the Association of Physicians of India. 2005;53:437–440. [PubMed] [Google Scholar]
- 8.Maini R, Henderson KL, Sheridan EA, Lamagni T, Nichols G, Delpech V, Phin N. Increasing Pneumocystis pneumonia, England, UK, 2000–2010. Emerging infectious diseases. 2013;19:386–392. doi: 10.3201/eid1903.121151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mikaelsson L, Jacobsson G, Andersson R. Pneumocystis pneumonia--a retrospective study 1991–2001 in Gothenburg, Sweden. J Infect. 2006;53:260–265. doi: 10.1016/j.jinf.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 10.Martin-Garrido I, Carmona EM, Specks U, Limper AH. Pneumocystis pneumonia in patients treated with rituximab. Chest. 2013;144:258–265. doi: 10.1378/chest.12-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monnet X, Vidal-Petiot E, Osman D, Hamzaoui O, Durrbach A, Goujard C, Miceli C, Bouree P, Richard C. Critical care management and outcome of severe Pneumocystis pneumonia in patients with and without HIV infection. Crit Care. 2008;12:R28. doi: 10.1186/cc6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mori S, Sugimoto M. Pneumocystis jirovecii infection: an emerging threat to patients with rheumatoid arthritis. Rheumatology (Oxford) 2012;51:2120–2130. doi: 10.1093/rheumatology/kes244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Centers for Disease C, and Prevention . Pneumocystis pneumonia--Los Angeles. 1981. MMWR. Morbidity and mortality weekly report. 1996;45:729–733. [PubMed] [Google Scholar]
- 14.Walzer PD, Evans HE, Copas AJ, Edwards SG, Grant AD, Miller RF. Early predictors of mortality from Pneumocystis jirovecii pneumonia in HIV-infected patients: 1985–2006. Clin Infect Dis. 2008;46:625–633. doi: 10.1086/526778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colford JM, Jr, Segal M, Tabnak F, Chen M, Sun R, Tager I. Temporal trends and factors associated with survival after Pneumocystis carinii pneumonia in California, 1983–1992. American journal of epidemiology. 1997;146:115–127. doi: 10.1093/oxfordjournals.aje.a009242. [DOI] [PubMed] [Google Scholar]
- 16.Dworkin MS, Hanson DL, Navin TR. Survival of patients with AIDS, after diagnosis of Pneumocystis carinii pneumonia, in the United States. J Infect Dis. 2001;183:1409–1412. doi: 10.1086/319866. [DOI] [PubMed] [Google Scholar]
- 17.Lundgren IS, Englund JA, Burroughs LM, Torgerson TR, Skoda-Smith S. Outcomes and duration of Pneumocystis jiroveci pneumonia therapy in infants with severe combined immunodeficiency. Pediatr Infect Dis J. 2012;31:95–97. doi: 10.1097/INF.0b013e31822db772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casper JT, Ash RA, Kirchner P, Hunter JB, Havens PL, Chusid MJ. Successful treatment with an unrelated-donor bone marrow transplant in an HLA-deficient patient with severe combined immune deficiency (“bare lymphocyte syndrome”) J Pediatr. 1990;116:262–265. doi: 10.1016/s0022-3476(05)82885-9. [DOI] [PubMed] [Google Scholar]
- 19.Lund FE, Hollifield M, Schuer K, Lines JL, Randall TD, Garvy BA. B cells are required for generation of protective effector and memory CD4 cells in response to Pneumocystis lung infection. J Immunol. 2006;176:6147–6154. doi: 10.4049/jimmunol.176.10.6147. [DOI] [PubMed] [Google Scholar]
- 20.Steele C, Marrero L, Swain S, Harmsen AG, Zheng M, Brown GD, Gordon S, Shellito JE, Kolls JK. Alveolar macrophage-mediated killing of Pneumocystis carinii f. sp. muris involves molecular recognition by the Dectin-1 beta-glucan receptor. J Exp Med. 2003;198:1677–1688. doi: 10.1084/jem.20030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shellito J, Suzara VV, Blumenfeld W, Beck JM, Steger HJ, Ermak TH. A new model of Pneumocystis carinii infection in mice selectively depleted of helper T lymphocytes. J Clin Invest. 1990;85:1686–1693. doi: 10.1172/JCI114621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng M, Shellito JE, Marrero L, Zhong Q, Julian S, Ye P, Wallace V, Schwarzenberger P, Kolls JK. CD4+ T cell-independent vaccination against Pneumocystis carinii in mice. J Clin Invest. 2001;108:1469–1474. doi: 10.1172/JCI13826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swain SD, Meissner NN, Siemsen DW, McInnerney K, Harmsen AG. Pneumocystis elicits a STAT6-dependent, strain-specific innate immune response and airway hyperresponsiveness. Am J Respir Cell Mol Biol. 2012;46:290–298. doi: 10.1165/rcmb.2011-0154OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu C, Cantor AB, Yang H, Browne C, Wells RA, Fujiwara Y, Orkin SH. Targeted deletion of a high-affinity GATA-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo. J Exp Med. 2002;195:1387–1395. doi: 10.1084/jem.20020656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng M, Ramsay AJ, Robichaux MB, Norris KA, Kliment C, Crowe C, Rapaka RR, Steele C, McAllister F, Shellito JE, Marrero L, Schwarzenberger P, Zhong Q, Kolls JK. CD4+ T cell-independent DNA vaccination against opportunistic infections. J Clin Invest. 2005;115:3536–3544. doi: 10.1172/JCI26306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beck JM, Warnock ML, Curtis JL, Sniezek MJ, Arraj-Peffer SM, Kaltreider HB, Shellito JE. Inflammatory responses to Pneumocystis carinii in mice selectively depleted of helper T lymphocytes. Am J Respir Cell Mol Biol. 1991;5:186–197. doi: 10.1165/ajrcmb/5.2.186. [DOI] [PubMed] [Google Scholar]
- 27.Dyer KD, Moser JM, Czapiga M, Siegel SJ, Percopo CM, Rosenberg HF. Functionally competent eosinophils differentiated ex vivo in high purity from normal mouse bone marrow. J Immunol. 2008;181:4004–4009. doi: 10.4049/jimmunol.181.6.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lilly LM, Scopel M, Nelson MP, Burg AR, Dunaway CW, Steele C. Eosinophil deficiency compromises lung defense against Aspergillus fumigatus. Infect Immun. 2014;82:1315–1325. doi: 10.1128/IAI.01172-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakagome K, Dohi M, Okunishi K, Tanaka R, Kouro T, Kano MR, Miyazono K, Miyazaki J, Takatsu K, Yamamoto K. IL-5-induced hypereosinophilia suppresses the antigen-induced immune response via a TGF-beta-dependent mechanism. J Immunol. 2007;179:284–294. doi: 10.4049/jimmunol.179.1.284. [DOI] [PubMed] [Google Scholar]
- 30.Ricks D, Chen K, Zheng M, Steele C, Kolls JK. Dectin immunoadhesins and Pneumocystis pneumonia. Infect Immun. 2013;81:3451–3462. doi: 10.1128/IAI.00136-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenberg HF, Dyer KD, Foster PS. Eosinophils: changing perspectives in health and disease. Nat Rev Immunol. 2013;13:9–22. doi: 10.1038/nri3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fleury-Feith J, Van Nhieu JT, Picard C, Escudier E, Bernaudin JF. Bronchoalveolar lavage eosinophilia associated with Pneumocystis carinii pneumonitis in AIDS patients. Comparative study with non-AIDS patients. Chest. 1989;95:1198–1201. doi: 10.1378/chest.95.6.1198. [DOI] [PubMed] [Google Scholar]
- 33.Sadaghdar H, Huang ZB, Eden E. Correlation of bronchoalveolar lavage findings to severity of Pneumocystis carinii pneumonia in AIDS. Evidence for the development of high-permeability pulmonary edema. Chest. 1992;102:63–69. doi: 10.1378/chest.102.1.63. [DOI] [PubMed] [Google Scholar]
- 34.Gingo MR, Wenzel SE, Steele C, Kessinger CJ, Lucht L, Lawther T, Busch M, Hillenbrand ME, Weinman R, Slivka WA, McMahon DK, Zhang Y, Sciurba FC, Morris A. Asthma diagnosis and airway bronchodilator response in HIV-infected patients. J Allergy Clin Immunol. 2012;129:708–714 e708. doi: 10.1016/j.jaci.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swain SD, Meissner NN, Harmsen AG. CD8 T cells modulate CD4 T-cell and eosinophil-mediated pulmonary pathology in pneumocystis pneumonia in B-cell-deficient mice. Am J Pathol. 2006;168:466–475. doi: 10.2353/ajpath.2006.050724. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing data contained in this paper is publicly available through the Sequence Read Archive BioProject number: PRJNA276259. Further information can be found at: http://www.ncbi.nlm.nih.gov/bioproject/.