Abstract
Autophagy delivers cytoplasmic material to lysosomes for degradation. First identified in yeast, the core genes that control this process are conserved in higher organisms. Studies of mammalian cell cultures have expanded our understanding of the core autophagy pathway, but cannot reveal the unique animal-specific mechanisms for the regulation and function of autophagy. Multicellular organisms have different types of cells that possess distinct composition, morphology, and organization of intracellular organelles. In addition, the autophagic machinery integrates signals from other cells and environmental conditions to maintain cell, tissue and organism homeostasis. Here, we highlight how studies of autophagy in flies and worms have identified novel core autophagy genes and mechanisms, and provided insight into the context-specific regulation and function of autophagy.
Keywords: autophagy, aggrephagy, Caenorhabditis elegans, Drosophila melanogaster
Autophagy: moving beyond yeast
Macroautophagy (hereafter referred to as autophagy) is a lysosome-mediated degradation process that involves the engulfment of portions of the cytoplasm in the double-membrane structures, known as autophagosomes. This vesicular delivery of cytoplasmic material to the lysosome was first observed in rat livers following exposure to glucagon [1]. Thirty years later, genetic studies in the yeast Saccharomyces cerevisiae led to the discovery of genes that function during autophagy under nutrient-limiting conditions [2–5]. Autophagosome formation involves a series of sequential membrane remodeling events [6]. Upon induction, a cup-shaped membrane sac, known as the isolation membrane (or phagophore), is formed in the cytoplasm, which further expands to seal and form a double-membrane autophagosome that eventually fuses with lysosomes (vacuoles in yeasts and plants) to form the autolysosome.
Genetic screens in yeast identified a set of autophagy genes, known as ATG genes (see Glossary, Table 1), that are essential for autophagosome formation [7,8]. Remarkably, the core autophagy machinery that is encoded by these genes is conserved between yeast and humans [6]. Many studies of immortalized mammalian cell lines indicate that, as in yeast, nutrient-limiting conditions induce autophagy that is dependent on ATG genes. However, several aspects of the autophagy pathway in higher eukaryotes are distinct from that in yeast. In yeast, all autophagosomes arise from the single perivacuolar preau-tophagosomal structure (PAS). In higher eukaryotes, there is no evidence for a PAS and isolation membranes can be generated simultaneously at multiple sites, implicating more complex sources for autophagosomal membranes. Indeed, the endoplasmic reticulum (ER), mitochondria, plasma membrane, and recycling endosomes have been shown to contribute to autophagosomal membranes [9–13]. In yeast, autophagosomes directly fuse with a single large acidic vacuole for degradation. In higher eukaryotes, nascent autophagosomes undergo a series of maturation processes, in part by fusing with endocytic vesicles, including early and late endosomes and multivesicular bodies (MVBs) [14,15]. The resulting hybrid organelles, called amphisomes, are more acidic and fuse with lysosomes to form degradative autolysosomes. Upon degradation of the engulfed contents, lysosomes are regenerated from autolysosomes, a process known as autophagic lysosome reformation (ALR), to maintain lysosomal homeostasis (Box 1) [16]. Moreover, autophagosome maturation requires an intact endocytic trafficking pathway. The ESCRT pathway, as well as components involved in endocytic vesicle fusion, is essential for proper maturation of the autophagosome [17–20]. Indeed, mutations in the endocytic ESCRT pathway influences autophagy in Drosophila [21], while fusion of autophagosomes with endosomes/lysosomes requires coordinated actions of the SNARE complex, consisting of the autophagosomal Syntaxin 17 (Qa SNARE), SNAP-29 (Qbc SNARE), and the endosomal/lysosomal R-SNARE VAMP8 in mammalian cells and VAMP7 in Drosophila, and the HOPS complex, which interacts with syntaxin 17 [17–20].
Table 1.
Conserved autophagy genes in S. cerevisiae and multicellular organisms
| Function | S. cerevisiae | Human/mammalian | D. melanogaster | C. elegans |
|---|---|---|---|---|
|
Serine/threonine kinase involved in initiation of isolation membrane |
ATG1 |
ULK1, ULK2 |
Atg1 | unc-51 |
| Component of Atg1 complex | ATG13 | Atg13 | Atg13 | atg-13 |
| ATG17 | FIP200 | Atg17 | - | |
| - | ATG101 | Atg101 | epg-9 | |
|
PI3K involved in initiation of isolation membrane |
VPS34 | VPS34 | Vps34 | vps-34 |
| Component of VPS34 complex | VPS15 | VPS15 | Vps15 | vps-15 |
| ATG6 | Beclin1 | Atg6 | bec-1 | |
| ATG14 | ATG14L | Atg14 | epg8 | |
|
Ubiquitin-like protein that conjugates with phosphatidylethanolamine and localizes to isolation membrane |
ATG8 |
GABARAP, LC3, GABARAPL1, GABARAPL2 |
Atg8a, Atg8b |
lgg-1, lgg-2 |
| E1-like enzyme | ATG7 | ATG7 | Atg7 | atg-7 |
| E2-like enzyme | ATG3 | ATG3 | Atg3 | atg-3 |
| Cysteine protease | ATG4 |
ATG4A, ATG4B, ATG4C, ATG4D |
Atg4a, Atg4b |
atg-4.1, atg-4.2 |
|
Ubiquitin-like protein that conjugates with Atg5 |
ATG12 | ATG12 | Atg12 | lgg-3 |
| Conjugates with Atg12 | ATG5 | ATG5 | Atg5 | atg-5 |
| E2-like enzyme | ATG10 | ATG10 | Atg10 | atg-10 |
| Interacts with Atg5-Atg12 conjugate | ATG16 | ATG16L1, ATG16L2 | Atg16 |
atg-16.1, atg-16.2 |
| Effector of PI(3)P | ATG18 |
WIPI1, WIPI2, WIPI3, WIPI4 |
Atg18a, Atg18b |
atg-18, epg-6 |
| ATG18 interacting protein | ATG2 | ATG2 | Atg2 | atg-2 |
|
Transmembrane protein that supplies membrane for autophagosome formation |
ATG9 |
ATG9A, ATG9B |
Atg9 | atg-9 |
Box 1. Autophagy and lysosome homeostasis.
Genes essential for lysosomal function and regeneration are essential for autophagic flux. Loss of function of laat-1, encoding the lysosomal lysine/arginine transporter, causes formation of enlarged, degradation-defective lysosomes in which autophagy substrates accumulate [108]. cup-5, encoding the C. elegans functional ortholog of MLN1/TRPML1, regulates lysosome biogenesis [109]. Loss of function of cup-5 impairs the reformation of lysosomes from endosomelysosome hybrid organelles, which involves sorting and condensation of lysosomal contents, and budding and maturation of nascent lysosomes [110]. In cup-5 mutants, autophagy substrates accumulate in enlarged vacuoles, which show characteristics of endosomes and lysosomes [111]. Loss of autophagy activity partially suppresses the enlarged vacuole accumulation abnormality, and also the embryonic lethality phenotype in cup-5 mutants [111]. Therefore, the basal level of autophagy activity contributes to lysosomal membrane dynamics and cargo processing and thus participates in regulating the size and number of lysosomes.
While studies in cultured mammalian cells have provided an important framework for the evolutionary conservation of autophagy, they fail to consider unique multicellular, animal-specific mechanisms for the regulation and function of autophagy. Cells in multicellular organisms are distinct in composition, morphology, and organization of intracellular organelles and even possess highly specialized organelles; therefore, the autophagic machinery may have evolved to accommodate specific characteristics of each cell type. For example, autophago-somes generated in axons migrate over long distances before reaching lysosomes in the neuron body for fusion [22,23]. Here, we describe how studies in the model organisms Caenorhabditis elegans and Drosophila melanogaster have advanced our understanding of autophagy. Importantly, many of these discoveries are in the context of physiologically relevant scenarios, such as the use of autophagy during development, highlighting how discovery-based investigation of this process using genetics can advance our understanding of autophagy.
Genetic identification of tissue specific variants and metazoan-specific autophagy genes
It has been assumed that the mechanisms controlling autophagy are identical between yeast and humans. Indeed, biochemical purification approaches have been extensively performed to isolate factors that interact with known Atg proteins, including the Atg1/ULK1 complex and the VPS34/Beclin 1 complex, which integrate nutrient status with autophagosome formation [24,25]. However, only a few metazoan-specific autophagy factors that have been isolated by this approach are essential for autophagosome formation under normal physiological conditions [26]. Recent genetic studies in model organisms are expanding the number of autophagy-specific genes that are essential for autophagic flux. For example, the midgut of the fly represents a unique in vivo experimental system to investigate novel autophagy pathways. Furthermore, physiological removal of protein substrates by autophagy, including PGL granules and SQST-1/p62 aggregates, during C. elegans embryogenesis has proved to be an excellent model system to isolate genes essential for autophagy without interference from a change in autophagy regulation [27,28]. The C. elegans embryo is enclosed by a hard eggshell that is impermeable to most solutes, and the development of these embryos relies on degradation of maternally loaded materials, independent of external nutrients. Below, we describe new genes and homologs of genes for autophagy regulation that have been discovered using genetic models.
Tissue variants in multicellular organisms
Recent studies in multicellular organisms have revealed tissue variants that cannot be observed in yeast studies. The conserved E1 enzyme encoded by Atg7 is not required for autophagy and clearance of mitochondria in the midgut of the fly intestine, even though Atg7 is required for starvation-triggered autophagy in the fat body [29,30]. In addition, neither the E2 Atg3 nor the conjugation/lipidation of the ubiquitin-like protein Atg8a appears to be required for autophagy in midgut cells [29]. Instead, autophagy in midgut cells depends on Uba1 [29]; the E1 used for ubiquitination. Importantly, Uba1 does not appear to function in place of Atg7 based on the failure of Uba1 to charge Atg8a, and genetic impairment of proteasome function fails to affect autophagy in the midgut. By contrast, autophagy and clearance of mitochondria in midgut cells depends on several other core ATGs, including Atg1, Atg2, Atg6, Atg18, and Atg8 [29]. It is particularly interesting that Atg8a is required for this autophagy, where it appears to function in a lipidation-independent manner. These results highlight an important role for ubiquitination in autophagy, which cannot be understood by studying autophagy in yeast.
Mammalian cells also possess variants of the autophagic machinery. In Atg5- and Atg7- deficient mouse cells, autophagosomes/autolysosomes still form and perform autophagy-mediated protein degradation in a manner dependent on Ulk1 and beclin 1 [31]. ULK1/2 is required for the autophagic response to amino acid deprivation and also acute glucose withdrawal, which induces an AMP-activated protein kinase (AMPK)-dependent inactivation of mammalian target of rapamycin (mTOR). However, prolonged glucose starvation in MEFs results in the accumulation of ammonia generated from the catabolism of amino acids that induces autophagy independent of the ULK1/2 complex [32]. It is of great interest to determine whether core autophagy genes are differentially used for autophagosome formation and maturation in different cell types, and developmental contexts during multicellular organism development.
Novel genes discovered in multicellular organisms
Forward genetic screens for genes that are essential for degradation of protein substrates during C. elegans embryogenesis identified highly conserved ATG genes, and also genes that either have no yeast counterparts or are highly divergent from ATG genes, which were named as epg genes (ectopic PGL granules or p62 aggregates). Among the identified epg genes, epg-3, epg-4, epg-5, epg-6, and epg-9 encode components that are absent in S. cerevisiae but exist in higher eukaryotes [28,33,34]. epg-9 encodes the ATG101 homolog [34], which is an integral component of the ULK1/Atg1 complex previously identified using a biochemical approach [26]. Loss of function of epg-3, epg-4, and epg-6 causes a defect at an early step of autophagosome formation, while epg-5 functions at a downstream step in the autophagy pathway; probably in the formation of degradative autolysosomes [28,33]. However, these forward genetic screens may fail to isolate essential autophagy genes that cause lethality when their function is lost. Thus, the list of genes essential for autophagic flux is expected to expand in higher eukaryotes. Further analysis of autophagic machinery in different cell types may allow us to identify tissue-specific factors.
The genes isolated in C. elegans also play a role in the mammalian autophagy pathway. RNAi knockdown of mammalian homologs of epg-3/Vmp1, epg-4/Ei24, epg-5/Epg5, and epg-6/Wipi4 impair autophagic flux [28,33], indicating that they are essential components of the basal autophagy pathway. The endoplasmic reticulum (ER) has been shown to act as a cradle for autophagosome formation [9–13]. EPG-3/VMP1 and EPG-4/EI24 are two ER-localized transmembrane proteins and thus may participate in transforming or reorganizing ER membranes into isolation membranes. Epg5 deficiency blocks the maturation of autophagosomes into degradative autolysosomes, resulting in accumulation of autophagic vacuoles with impaired degradation capacity [35]. Mice deficient in Ei24, Epg5, and Wipi4 (the mammalian Epg6 homolog) have also been generated. Ablation of these autophagy genes in neurons results in the accumulation of ubiqui-tin-positive protein aggregates, p62 aggregates, and elevated levels of LC3, indicating that autophagic flux is impaired [35–37]. Recent human genetic studies have revealed that recessive EPG5 mutations are causally related to the multisystem disorder Vici syndrome [38], while de novo mutations in WIPI4, the homolog of C. elegans epg-6 [33], cause a subtype of neurodegeneration with brain iron accumulation called β-propeller-protein-associated neurodegeneration; also known as static encephalopathy of childhood with neurodegeneration in adulthood [39,40]. Thus, the identification of these genes as regulators of autophagy in worms has helped to define involvement of Epg5, Wipi4, and autophagy in the occurrence of related diseases.
Aggrephagy in C. elegans – hierarchical recruitment of cargo, receptor, and scaffold
While autophagy has been considered a bulk degradation system with little or no selectivity, considerable evidence supports the idea that autophagy can be highly selective, removing damaged organelles, protein aggregates formed by misfolded or disease-related proteins, and invading pathogens [41]. A family of proteins, which simultaneously associate with cargo and Atg8/LC3, has been implicated as the receptor that links cargo with the autophagic machinery [42]. For example, the receptor p62, which contains a self-oligomerization PB1 domain, an LC3-interacting LIR motif and a UBA ubiquitin-binding domain, mediates the aggregation and autophagic degradation of ubiquitinated misfolded proteins [43–45]. The interaction between the cargo, receptor, and LC3/Atg8 is modulated by post-translational modification, which regulates autophagic degradation efficiency. p62 is phosphorylated at serine 403 in the UBA domain, which increases its binding affinity with polyubiquitinated chains [46]. Whether the receptor protein is sufficient to confer selectivity under physiological conditions remains unknown. Here, we summarize current knowledge of autophagic degradation of two distinct protein aggregates during C. elegans embryogenesis, and how it affects our understanding of selective autophagy.
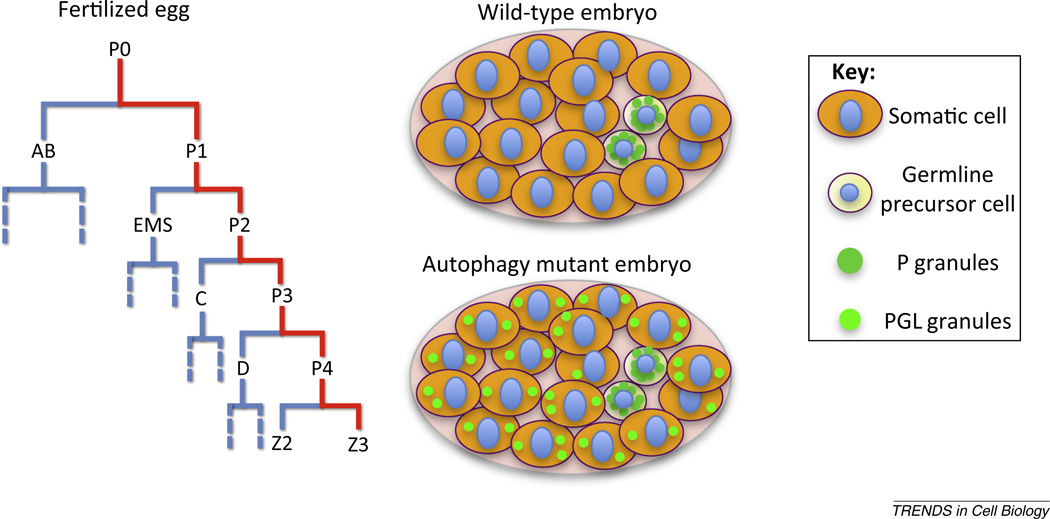
During C. elegans embryogenesis, the maternally derived germline P granule components PGL-1 and PGL-3 are degraded by autophagy in somatic cells [27] (Box 2). In autophagy mutants, PGL-1 and PGL-3 colocalize and accumulate into aggregates in somatic cells, which are named PGL granules. In addition to PGL granules, the C. elegans SQSTM1/p62 homolog SQST-1 is also removed by autophagy during embryogenesis [28]. SQST-1 is expressed in a diffuse pattern at a low level in the cytoplasm in wild-type embryos, but accumulates into a large number of aggregates distinct from PGL granules in autophagy mutants.
Box 2. P granules.
Formation of germ cells in C. elegans involves four sequential asymmetric divisions: a blastomere generates one somatic founder cell and one germline blastomere, P1, P2, P3, and finally P4. P4 then divides equally at the ∼100-cell embryonic stage, giving rise to two germ precursor cells Z2 and Z3, which remain quiescent during embryogenesis and proliferate throughout larval development to generate germ cells [112].
Maternally loaded P granules, a specialized type of protein/RNA aggregate, distribute throughout the cytoplasm of newly fertilized embryos. At the subsequent four sequential asymmetric cell divisions, P granules become localized exclusively in the germline blastomeres P1, P2, P3, and P4, and finally in the germ precursor cells Z2 and Z3. P granules are synthesized in germ cells generated from Z2 and Z3 in larval stages with the exception of sperm. Several mechanisms have been shown to contribute to the exclusive partition of P granules into germline blastomeres [113]. P granules directly migrate with the central cytoplasmic flow to the posterior end in P0 and P1 cells or associate with the nucleus in the region destined to be inherited by P3 and P4 when the nuclear membrane breaks down. In addition, P granules remaining in the somatic blastomeres are disassembled or degraded [113] (Figure I).
P granules contain constitutive components that associate with P granules during all development stages and also transient components that interact with P granules in germline blastomeres and disappear in Z2 and Z3. Constitutive components include the RGG box containing RNA binding proteins PGL-1 and PGL-3 and the RNA helicases GLH-1–4. Transient components include the CCCH-type zinc finger proteins PIE-1, MEX-1 and POS-1 [112]. Degradation of these CCCH finger proteins in somatic blastomeres depends on the cullin-dependent proteasome degradation pathway [114]. Removal of PGL-1 and PGL-3 requires autophagy activity [27]. Thus, both UPS and autophagy are involved in degradation of maternally-loaded P granule components in somatic cells during embryogenesis.
Figure I. Lineage diagram showing the generation of germ precursor cells Z2 and Z3 and a cartoon illustrating the distribution of P granules and PGL granules.
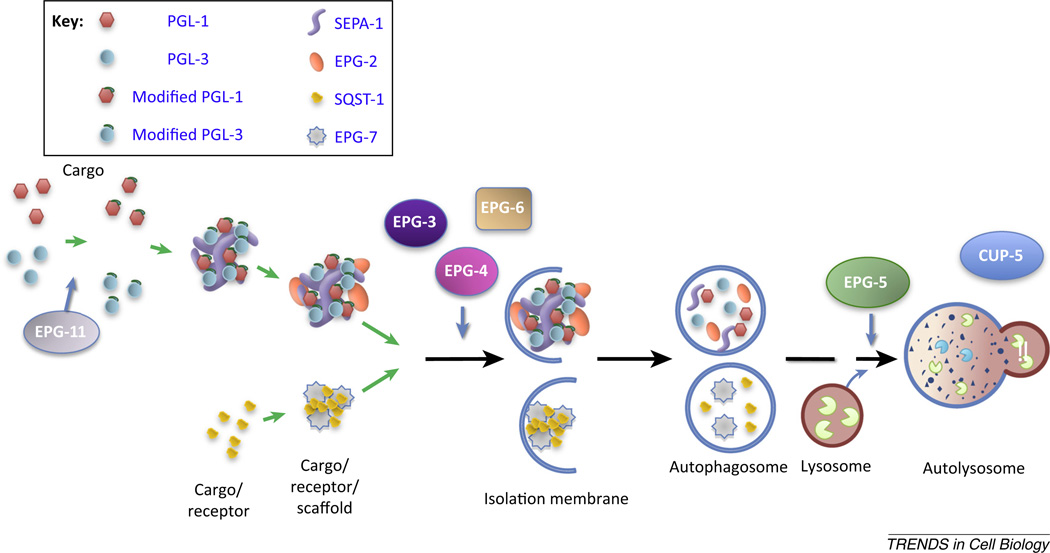
Genetic screens identified the sepa-1 and epg-2 as being required for removal of PGL components, but not for clearance of SQST-1 [27,28] (Figure 1). The self-oligomerizing protein SEPA-1 mediates accumulation of PGL-1 and PGL-3 into aggregates and their degradation[27]. SEPA-1 is also degraded by autophagy. SEPA-1 directly interacts with PGL-3 and LGG-1/Atg8, and thus acts as a receptor for degradation of PGL granules [27]. EPG-2 is required for the removal of PGL-1/PGL-3/SEPA-1 aggregates [28]. EPG-2 forms aggregates that colocalize with PGL granules and is removed by autophagy in a manner that is independent of pgl-1, pgl-3, and sepa-1 [28]. EPG-2 directly interacts with SEPA-1 and also with multiple Atg proteins [47]. Loss of epg-2 function disrupts the colocalization of PGL granules with LGG-1/Atg8 puncta [28]. EPG-2 is proposed to act as a scaffold protein for recruiting ATG proteins to PGL granules.
Figure 1.
Aggrephagy pathway in C. elegans. Selective removal of protein aggregates requires the hierarchical recruitment of receptor and scaffold proteins. Different scaffold proteins mediate the degradation of different cargo/receptor complexes. In degradation of PGL granules, arginine methylated PGL-1 and PGL-3, a process mediated by the C. elegans arginine methyltransferase PRMT1 homolog EPG-11, are recruited into SEPA-1 aggregates, which further associate with the scaffold protein EPG-2. The scaffold protein EPG-7 mediates the degradation of SQST-1. The cargo/receptor/scaffold complex recruits ATG proteins to trigger the formation of surrounding autophagosomal membranes. Genetic screens also identified several metazoan specific autophagy genes that are essential for degradation of both PGL granules and SQST-1 aggregates. The mammalian homologs of these genes are also required for the basal level of autophagy. EPG-3, -4, and -6 are involved in progression of omegasomes to isolation membranes/autophagosomes. EPG-5 is required for the formation of degradative autolysosomes. CUP-5 is essential for lysosomal function. ATG,; EPG,; PGL,; PRMT,; SEPA,; SQST,.
Distinct scaffold proteins appear to be utilized for removal of different autophagy substrates. In degradation of SQST-1 during embryogenesis, the self-oligomerizing protein EPG-7 acts as a scaffold protein, linking SQST-1 aggregates with LGG-1 puncta [48] (Figure 1). EPG-7 itself is degraded by autophagy in an SQST-1-independent manner. EPG-7 directly interacts with SQST-1 and also associates with multiple autophagy proteins, including LGG-1, LGG-3/Atg12, ATG-18, and ATG-9 [48]. In epg-7 mutants, accumulation of SQST-1 aggregates is weaker than in autophagy mutants [48], suggesting that SQST-1 can still be removed with low efficiency; possibly through either its direct interaction with LGG-1 or by non-selective autophagy. At the postlarval stages, SQST-1 aggregates in epg-7 mutants are degraded by starvation-induced autophagy [48]. Therefore, under physiological conditions, in addition to the receptor protein, a family of scaffold proteins confers cargo selectivity and promotes degradation efficiency. The cargo/receptor/scaffold protein complex may act as a PAS-like structure, providing a platform for recruiting core Atg proteins, which then triggers the assembly of surrounding autophagosomal membranes.
The hierarchical requirement of cargo, receptor and scaffold proteins for efficient autophagic degradation could be achieved by either their sequential expression or by post-translational modification of their interactions. In degradation of PGL-1 and PGL-3, SEPA-1 displays a dynamic expression pattern overlapping with EPG-2 [47]. The interaction between the cargo (PGL-1 and PGL-3)/receptor (SEPA-1) complex (PGL granule) with the scaffold protein (EPG-2) is regulated by post-translational arginine methylation of PGL-1 and PGL-3 that is mediated by the C. elegans arginine methyltransferase and PRMT1 homolog, EPG-11 [47]. Loss of epg-11 function causes aggregation of PGL-1 and PGL-3 that is independent of SEPA-1 [47]. Thus, PGL granules in epg-11 mutants show altered composition and/or organization, which in turn impairs the association of SEPA-1 with EPG-2 [47]. Ectopic expression of PGL-3 aggregates impairs the degradation of SEPA-1, but has no effect on the removal of EPG-2 and other autophagy substrates [47]. Thus, regulating the association of the cargo/receptor complex with the scaffold protein prevents congestion of autophagic flux by a specific type of cargo. The hierarchical recruitment of the receptor and scaffold proteins provides multiple regulatory layers for developmental modulation of degradation efficiency.
Novel regulators of autophagy
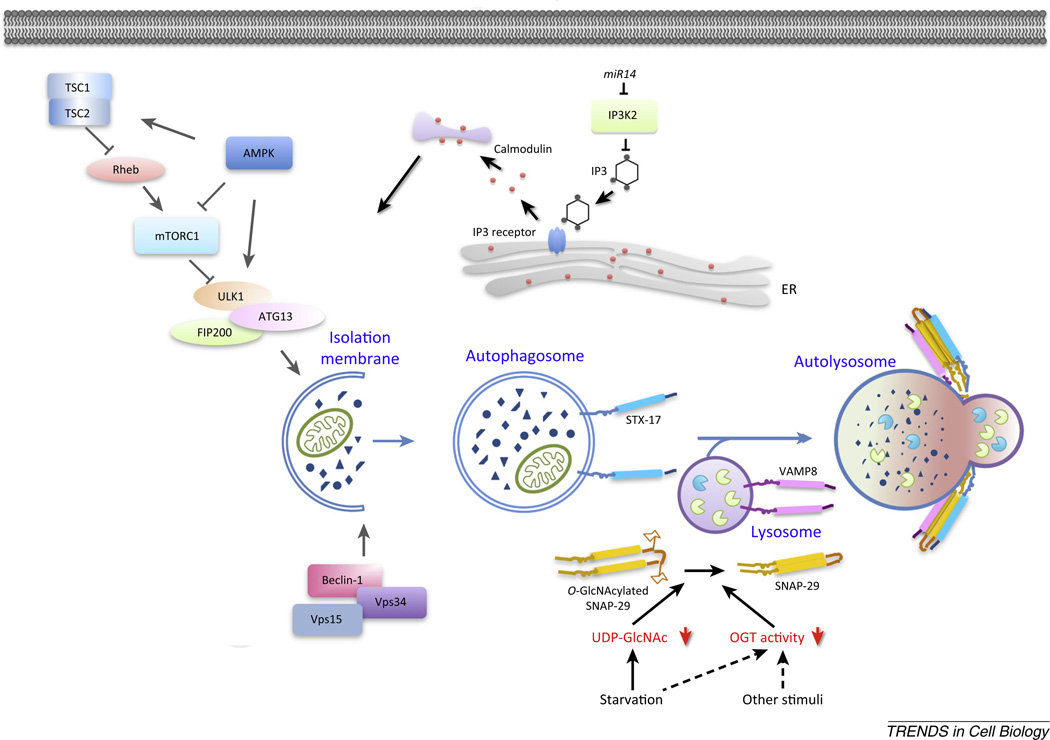
The mammalian target of rapamycin serine/threonine kinase complex 1 (mTORC1) and the VPS34 phosphoinositide 3-kinase (PI3K) complex are the two most extensively studied nodes for integrating the status of nutrients, cellular energy and growth factors with autophagy regulation [24,25] (Figure 2). mTORC1 modulates autophagy by regulating the kinase activity of the ULK1/ Atg1 complex [49,50]. Different systems respond to distinct nutrient stimuli: AMPK responds to the AMP/ATP ratio, while mTOR senses amino acid levels and insulin signaling [25]. The ULK1/Atg1 complex and VPS34 complex control the initiation of autophagosome formation and, consequently, the on-rate of autophagic flux.
Figure 2.
Regulation of autophagy activity. The mTORC1 and the VPS34/PI(3)P regulatory complexes have been shown to be the two most extensively studied nodes for integrating the status of nutrients, cellular energy, and growth factors with the autophagosome initiation. Nutrient status also regulates autophagosome maturation by regulating O-GlcNAylation of SNAP-29. Under starvation conditions, levels of UDP-GlcNAc are decreased, resulting in reduction of O-GlcNAc-modified SNAP-29 levels. Unmodified SNAP-29 forms a more stable SNARE complex with autophagosome-localized Stx17 and lysosomal VAMP8 to mediate the fusion of autophagosomes with endosomes/lysosomes. Starvation and other autophagy stimuli may also directly regulate OGT activity. Autophagy activity is also regulated by calcium signaling. In flies, the miRNA miR14 regulates translation of ip3k2 and IP3 levels in cells to control calcium levels and autophagy. IP3, inositol-1,4,5 trisphosphate; mTORC1, mammalian target of rapamycin serine/threonine kinase complex 1; OGT,; PI(3)P,; SNAP,; SNARE,; Stx,; VAMP,; VPS,.
In higher eukaryotes, cells exhibit tissue-, temporal-and sex-specific fates. The autophagic machinery also needs to integrate signaling from other cells and environmental conditions to maintain cell, tissue, and organism homeostasis. Loss of activity of a gene may elicit various aspects of cellular function that contribute differentially to autophagy regulation. Thus, different cell types may exhibit differences in response to a variety of stresses and signals. For example, components of the AP2 complex, the COG complex, and the ESCRT complex, function in the endocytic pathway and have been previously shown to positively modulate autophagy activity [15]. In C. elegans, RNAi inactivation of these endocytic components results in elevated autophagy activity in the intestine [51], indicating that their partial loss of function may not impair their normal function in the autophagy pathway, but impose a stress on the intestine which in turn activates autophagy.
Autophagy activity can be regulated by transcriptional regulation of essential autophagy genes. The helix–loop–helix protein TFEB is a well-characterized transcription factor that activates genes that are essential for the autophagy–lysosomal pathway [52]. The transcription and nuclear translocation of C. elegans TFEB homolog HLH-30 is induced by starvation, and activates the expression of autophagy genes [53]. Autophagy genes can also be transcriptionally induced by inactivation of different genes associated with developmental processes [54–56], including the transforming growth factor (TGF)-β Sma/Mad pathway and the lin-35 SynMuv B pathway, as well as genes associated with stress-related pathways including the transcription factor PHA-4/FOXA, the transcription factor XBP-1, which mediates the ER stress pathway, and the bZip transcription factor ATFS-1, which regulates the mitochondrial stress pathway [51,57]. Thus, transcriptional regulation of autophagy genes appears to be a widely used mechanism to control autophagy activity by various developmental signals and stresses.
In various neurodegenerative diseases, nondegradative autophagic vacuoles accumulate in neurons [58], indicating that autophagosome maturation may become a rate-limiting step for autophagic flux under certain circumstances. A recent study demonstrated that nutrient status can also impinge on the regulation of autophagosome maturation [59]. Genetic screens in C. elegans identified mutations in ogt-1, encoding O-linked β-N-acetylglucosa-mine (O-GlcNAc) transferase, that promote autophagy activity [59] (Figure 2). OGT knockdown also elevates autophagic flux in mammalian cells in part by facilitating autophagosome maturation. OGT O-GlcNAcylates SNAP-29 at specific amino acids to attenuate its binding affinity to STX17 and VAMP8 [59]. Thus, OGT knockdown enhances the assembly of the STX17/SNAP-29/VAMP8 SNARE complex that mediates the fusion of autophago-somes with endosomes/lysosomes. O-GlcNAc modification-defective SNAP-29 facilitates autophagy activity in mammalian cells and also promotes autophagic degradation of protein aggregates in C. elegans [59]. The level of O-GlcNAcylation has been proposed to serve as a nutrient sensor [60]. Generation of UDP-GlcNAc, the donor for O-GlcNAc addition, is highly responsive to the availability of glucose, fatty acids, uridine, and the amino acid gluta-mine. The levels of UDP-GlcNAc and O-GlcNAcylated SNAP-29 are reduced in C. elegans under starvation conditions. In mammalian cells, levels of UDP-GlcNAc and O-GlcNAcylated SNAP-29 are reduced upon glucose starvation, but not by either serum starvation or rapamycin treatment [59]. Therefore, the discovery of ogt-1 in worms helped to define how SNAP-29 O-GlcNAcylation serves as a cellular mechanism for integrating nutrient availability with autophagosome maturation.
Physiological functions of autophagy
Removal of paternal mitochondria
In animals, offspring inherit mitochondria only from the female parent; a process known as maternal inheritance. Paternal mitochondria and mitochondria DNA (mtDNA) in C. elegans are actively removed by autophagy [61,62]. In wild-type embryos, mitochondria derived from sperm are randomly partitioned into blastomeres at the two- and four-cell stage, but they are quickly eliminated by autophagy and are undetectable at the 64-cell embryonic stage. In autophagy mutants, paternal mitochondria and mtDNA persist in late embryos and larvae. How paternal mitochondria, which are not ubiquitinated, are recognized and degraded by autophagy has yet to be elucidated.
In flies, paternal mtDNA is eliminated by mitochondrial endonuclease G during sperm elongation, and is also discarded from spermatids into waste bags by a cellular remodeling process [63]. Thus, essentially all of the sperm mtDNA is degraded during late spermatogenesis. In the flagellated spermatids, mitochondria undergo structural remodeling; a process involving fusion of all the mitochondria, which further unfold and elongate in close association with the axoneme to form a structure known as the mitochondrial derivative (MD). An unconventional role of autophagy is involved in fragmentation of MDs after fertilization [64]. Multivesicular-body-like vesicles, which are positive for both endocytic markers and Atg8, associate with sperm flagella and contribute to membrane disintegration, MD separation from the axoneme, and breakdown of MDs into smaller fragments. The MD fragments are then enveloped by autophagosomes for degradation in lysosomes. p62 is specifically associated with the ubiquitinated MD, as well as with some of the MD fragments, and plays a role in MD destruction [64].
In the majority of mouse sperm, most of the mtDNA is also eliminated before fertilization. If a sperm containing mtDNA enters the zygote, the mtDNA becomes unevenly distributed in blastomeres during cleavage and eventually persists in only a small fraction of embryonic cells [65]. Sperm mitochondria are labeled by ubiquitin, p62, and LC3 immediately after fertilization, but appear not to be engulfed by autophagosomes [65]. Thus, the role of autophagy in degradation of paternal mitochondria in mice remains elusive.
Autophagic cell death: developmental context matters
Programmed cell death is a genetically regulated process that plays important roles in development and organism health. Schweichel and Merker defined three types of programmed cell death based on morphology: apoptosis, autophagic cell death, and necrosis [66]. Autophagic cell death is characterized by the presence of numerous autophagosomes in dying cells, and little to no role for phagocytes in this process.
The role of autophagy during programmed cell death in Drosophila is context dependent. Autophagic cell death can occur either in parallel, upstream, or downstream, or independent of the cell death executioners known as caspases. In larval salivary glands, both autophagy and caspase activities function in parallel pathways during cell death. Mutations in either single Atg genes, including Atg1, Atg2, Atg3, Atg6, Atg7, Atg8a, Atg12, and Atg18, or impairment of apoptotic caspase proteases results in incomplete degradation of larval salivary glands [67]. By contrast, combined inhibition of both autophagy and caspases increases the suppression of salivary gland degradation [67]. Furthermore, expression of Atg1 is sufficient to induce autophagy in salivary glands that is dependent on downstream core Atg gene function. Importantly, this Atg1-triggered autophagy is sufficient to induce salivary gland cell death in a caspase-independent manner [67].
Recent work indicates that calcium and multiple inositol-1,4,5 trisphosphate (IP3) signaling pathway components, including IP3 kinase 2, IP3 receptor, and calmodulin, regulate autophagy during salivary gland cell death [68] (Figure 2). For calcium signaling to occur, IP3 acts as a secondary messenger that controls calcium release from the ER by binding the IP3 receptor. Furthermore, the miRNA miR14 was found to be necessary and sufficient for autophagy in salivary glands [68], which targets IP3 kinase 2, thus defining a role for calcium signaling in the regulation of autophagy in salivary glands. In addition, the immunoreceptor molecule Draper in Drosophila was found to play a key role in regulating autophagy in dying salivary glands. Its ortholog in C. elegans is the engulfment receptor CED-1, which is best known for being required for recognition of apoptotic cells by phagocytes [69]. Loss of draper prevents autophagy in dying salivary glands, and results in an incomplete larval salivary gland degradation phenotype that is similar to the phenotype observed in Atg gene and miR14 mutants [70]. In sharp contrast to its function in phagocytes, Draper regulates autophagy in a cell-autonomous manner. Draper is known to regulate phagocytosis via engulfment factors and Src kinase [71], and these factors are also required for salivary gland autophagy [70]. The similarities in the mechanisms by which dying cell corpses are cleared by phagocytes and the apparently largely cell autonomous clearance of salivary gland cells by autophagy raises intriguing questions about the evolution of these different modalities of programmed cell death.
The death of the larval midgut cells of the intestine in Drosophila is different from salivary glands. Impairment of caspase function does not influence midgut cell death. By contrast, decreased function of multiple individual Atg genes blocks larval midgut degradation [72]. Additionally, caspase deficiency fails to enhance the Atg mutant phenotype in the midgut [72]. These data indicate that autophagy, and not caspases, is essential for midgut programmed cell death. This novel Atg7- and Atg3-independent autophagy pathway controls programmed cell size reduction of the dying midgut cells [29]. It will be interesting to consider what happens to the recycled material of these dying cells, and if these resources are used to promote the development of forming adult tissues, including the adult intestine.
The Drosophila ovary has also provided valuable insight into our understanding of the relation between autophagy and cell death. During stress-triggered cell death of ovarian support nurse cells, caspases are required for high levels of autophagy that are needed for cell death [73]. At a later stage in ovary development, nurse cells are programmed to die, and this depends on degradation of the inhibitor of apoptosis Bruce by autophagy. The degradation of Bruce enables caspase activation and cell death [74]. Therefore, it appears that autophagy can function either upstream or downstream of caspases in the context of cell death in the fly ovary. These studies highlight the context dependent roles of autophagy during programmed cell death.
Corpse removal in C. elegans
In C. elegans, 131 somatic cells and a large number of germline cells undergo programmed cell death [75]. While autophagy mutants show no defect in germ cell death or the death of six Pn.aap cells (P1, P2, and P9–P12) in the ventral cord of larval animals under physiological conditions [76], the execution of the death program of these cells in mutants with partially compromised caspase activity is suppressed by loss of autophagy activity [76]. Autophagy also contributes to the death of germline cells induced by genotoxic stress, such as γ-ray treatment [76], suggesting a context-dependent role of autophagy in programmed cell death in C. elegans.
Autophagy can also function in the clearance of apoptotic cell corpses. Apoptotic cell corpses are phagocytosed in single-membrane phagosomes that subsequently deliver the corpse to the lysosome for degradation. Phagosomes undergo extensive membrane remodeling and maturation by fusing with endosomes before eventually fusing with lysosomes [77]. During C. elegans embryogenesis, all autophagy genes are involved in cell corpse removal, arguing that the autophagy process itself, rather than additional functions of autophagy genes, is involved [78,79]. Autophagy mutant embryos show reduced PI(3)P levels on cell corpse-containing phagosomes and thus delay phagosome maturation [78]. Autophagy may modulate the formation of different VPS-34 complexes with distinct functions in the endocytic and autophagy pathways. Alternatively, autophagy may maintain phagosomal PtdIns(3)P by fusing PtdIns(3)P-containing autophagic structures with phagosomes.
In contrast to the clearance of cell corpses derived from C. elegans embryogenesis, the two apoptotic cells (QR.aa and QL.aa) generated from the Q neuroblast of L1 larvae are engulfed and degraded by the neighboring hypoder-mal 7 (hyp7) cell in a manner that is independent of the conventional autophagy pathway [80]. ATG-18, EPG-5, and LGG-1, but not ATG-5 and ATG-7, are sequentially recruited to cell corpse-containing phagosomes. In atg-18 and epg-5 mutants, the maturation of cell corpse-containing phagosomes and the formation of phagolysosomes are delayed. unc-51/ATG1 mutants show no defects in clearance of apoptotic Q cell descendants.
Autophagy has distinct functions in cell corpse removal in other systems. LC3 is recruited to apoptotic-cell-containing phagosomes, facilitating their fusion with lysosomes [81]. Recruitment of LC3 depends on the VPS34 complex and the two ubiquitin-like conjugation systems, but in a manner independent of the ULK1 complex [81]. During cavitation in mouse embryoid bodies and chick retinal development, autophagy regulates the clearance of apoptotic cell corpses by generating the cellular ATP that is required to expose the ‘eat-me’ signal; phosphatidylserine on the apoptotic cell surface [82,83]. Thus, depending on the developmental context, autophagy contributes differentially to cell corpse removal.
Despite these findings, the context-dependent roles of autophagy in cell death remain controversial. It is important to empirically determine if autophagy is the mechanism by which cells actually die (cell death by autophagy) or if autophagy is just present during cell death (cell death with autophagy) [84,85].
Cell growth
A recent study in Drosophila indicates that the influence of autophagy on tissue overgrowth depends on the growth inducing stimulus and cell type. Reduced Atg gene function decreased tissue growth in an eye epithelial model of Ras-triggered overgrowth [86]. However, autophagy had the opposite effect on Notch-induced overgrowth, and decreased Atg gene function significantly enhanced the growth of the eye epithelium [86]. In addition, studies of Hippo-triggered overgrowth indicated that autophagy does not influence the growth of the eye epithelium, but does influence Hippo-pathway-triggered overgrowth of glia [86]. These studies indicate that the context of both cell type and growth stimulus may influence how autophagy affects cell growth.
Cell fate specification
In addition to responding to metabolic stress, a recent study showed that, in C. elegans, autophagy modulates miRNA-mediated cell fate specification by degrading AIN-1, a component of the miRNA-induced silencing complex (miRISC) [87]. Autophagy mutants develop normally, but partially suppress the defect in specification of temporal fates of a row of laterally aligned epidermal cells, called seam cells, which are associated with a loss-of-function mutation in components of miRISC. Autophagy mutants also partially rescue the defect in specification of the taste receptor neuron ASEL in miRNA lsy-6 hypomorphic mutants [87]. Thus, besides its widely recognized role in maintaining cell and tissue homeostasis, autophagy participates in cell fate specification during development.
Aging
Autophagy acts as a cell survival mechanism in response to metabolic stresses. During C. elegans development, autophagy regulates survival of L1 larvae under food starvation conditions [88]. When early larvae experience harsh environmental conditions, such as food depletion, high temperature, and/or overcrowding, they enter the dauer diapauses. Autophagy regulates the morphological and metabolic changes during the development of dauer [89]. The extended lifespan caused by germline removal, TOR inhibition, dietary restriction, reduced insulin/ insulin-like growth factor (IGF)-1 signaling, and reduced mitochondrial respiration also depends on autophagy activity [54,89–91].
Similar to worms, autophagy has been shown to play an important role during aging in flies. Loss of the key autophagy genes Atg7 and Atg8a in the entire animal results in decreased lifespan and sensitivity to stress [30,92]. Furthermore, expression of Atg8a in fly brains decreased ubiquitinated protein aggregates and extended lifespan, although it was not clear if autophagy was increased by transgene expression in this study [92]. Interestingly, activation of autophagy in the brain of adult flies by either expression of AMPK or Atg1 was sufficient to induce autophagy in enterocytes of the intestine [93]. Activation of autophagy in enterocytes of the adult intestine facilitates tissue homeostasis and is associated with increased lifespan.
Pleiotropy of autophagy genes
Autophagy also plays a role in nondegradative processes. In yeast, the same set of autophagy genes is involved in the generation of Cvt vesicles that mediate selective transport of two vacuolar enzymes, amino peptidase (Ape)I and a-mannosidase (Ams)I, from the cytoplasm to the vacuole [94,95]. Autophagy is also required for the unconventional secretion of a subset of cytosolic proteins lacking signal peptide sequences, including Acb1 (acyl coenzyme A-binding protein), interleukin (IL)-1β and HMGB1 [96–98]. In this unconventional secretion pathway, it has been suggested that autophagosomes enclose the substrate and then fuse with the plasma membrane for secretion, rather than fusing with lysosome for degradation.
In addition to the nondegradative function of autophagy, a subset of autophagy proteins also function in autophagy-independent processes. UNC-51/Atg1 and its binding partner UNC-14 regulate the axon guidance of many neurons [99,100]. For example, axons are prematurely terminated or are misdirected during circumferential elongation to the dorsal neuron cord in unc-51 and unc-14 mutants in C. elegans. UNC-51 and UNC-14 regulate the subcellular localization of UNC-5; the receptor for the axon-guidance protein Netrin/UNC-6 [101]. Loss of EPG-1 and EPG-9, both of which form a complex with UNC-51 in autophagy, does not result in axon guidance defects [34,102]. Thus, UNC-51, through interaction with distinct binding partners, functions in distinct biological processes.
Atg6 is required for multiple vesicle trafficking pathways and hematopoiesis in Drosophila. Drosophila lacking Atg6 function possess blood cell tumors, and defects in blood cell differentiation [103]. Importantly, autophagy, endocytosis, and protein secretion are all altered in Atg6 mutants. Therefore, it is possible that defective autophagy is responsible for the blood cell defect in flies, but it is also important to consider if altered endocytosis and protein secretion contribute to blood cell tumor development. For example, Atg6 mutant cells appear to take up a marker of fluid phase endocytosis, but retain this marker to a greater extent near the cell cortex than in control cells, raising the possibility that these cells could possess a defect in growth factor receptor downregulation. Similarly, C. elegans bec-1/Atg6 is also involved in endocytic trafficking, such as retrograde transport from endosomes to the Golgi [104].
The autophagy protein Atg9 interacts with dTRAF2/ TRAF6 to regulate oxidative-stress-induced C-Jun N-terminal kinase (JNK) activation and autophagy [105]. Atg9 is known to be required for autophagosome formation in yeast, but the discovery of Atg9 as a mediator of oxidative-stress-triggered autophagosome formation in both Drosophila and mammals advances our understanding of how stress induces catabolism. In addition, Atg9, dTRAF2, and JNK are required for oxidative-stress-induced stem cell renewal in the adult fly intestine, illustrating the important role that this signaling pathway and autophagy play in tissue and organism health.
Components of the two ubiquitin-conjugation systems also act in multiple aspects of infection and host defense that are independent of their role in autophagy [106]. Atg5-Atg12/Atg16L1 complex is required for interferon (IFN)γ to control murine norovirus (MNV) replication by inhibiting formation of the membranous MNV replication complex [107]. The emerging roles of autophagy proteins in a variety of autophagy-independent processes indicate that multiple biological processes previously attributed to dysfunction of autophagy need be re-evaluated.
Concluding remarks
It has been largely assumed that autophagic machinery is evolutionarily conserved in all cells and organisms. Here, we describe that autophagy machinery, in multicellular organisms, not only requires metazoan-specific components but also exhibits tissue specific variants. Furthermore, autophagy has cell-context-specific influences on a variety of biological processes.
Our knowledge of the mechanisms that regulate autophagy in mammalian cells has mainly come from analyses of transformed cell lines. Although studies of transformed cells in culture have expanded our understanding of the core autophagy mechanisms in animals, many of these analyses fail to recognize that cellular metabolism following nutritional resource restriction may be distinct from that during development and adulthood of animals. For example, during glucose deprivation, transformed cell lines undergo a bioenergetic crisis that activates autophagy through AMPK inhibition of mTOR, whereas cells in vivo may use alternative energy sources to maintain cell and tissue homeostasis. Moreover, in multicellular organisms, cell context, intercellular communication, organismal stress, and homeostasis may all affect the regulation and function of this catabolic process. Loss of activity of a gene may affect various aspects of cellular function that contribute differentially to autophagy regulation, and thus the net outcome of autophagy activity may be distinct in different cells, tissues, and developmental and physiological contexts. Thus, caution need be taken to extrapolate the regulation and function of autophagy regulation in transformed cell lines to those in physiological contexts in animals.
We conclude that much remains to be learned about autophagy (Box 3), as different cells can utilize unique regulatory mechanisms to influence autophagy. It is worth noting that distinct autophagy regulation may arise through either unique signaling mechanisms or via the recruitment of distinct cargoes into autophagosomes. Furthermore, multiple types of autophagosomes may exist within a cell, and the net influence of these vesicles and degradative events may influence cell health and fate. Future studies of autophagy in animal model systems will lead to many new discoveries and greatly advance our understanding of autophagy in mammals.
Box 3. Outstanding questions.
Tissue-specific variants of the autophagic machinery. Cells in multicellular organisms show distinct morphology and organization of intracellular organelles. Has the autophagic machinery evolved to accommodate the specific characteristics of each cell type? Are core autophagy genes differentially used for autophagosome formation and maturation in different cell types and developmental contexts? Multiple membrane sources, including the ER, mitochondria, plasma membrane, and endocytic vesicles, have been shown to contribute to autophagosomal membranes. How are these membrane sources coordinately regulated for autophagy induction? Does the source for autophagosomal membranes depend on stress conditions, the degree of autophagy induction and the nature of the substrates removed by autophagy? Can multiple types of autophagosomes with distinct cargoes and possibly membrane sources form within a single cell?
Regulation of autophagy activity in different developmental contexts. Autophagy activity is determined by numerous external and internal factors in the development of multicellular organisms. Do these factors have differential effects in either different cell types or at different developmental stages? How do dynamic changes in lysosomal structure during either development or in response to various stress conditions participate in autophagy regulation? How does the autophagic machinery integrate signaling from other cells and environmental conditions to maintain cell and tissue homeostasis?
Molecular mechanisms for selective autophagy. How is the cargo specifically recognized by the autophagic machinery? Do different cargoes utilize distinct mechanisms to trigger degradation? Do components of ubiquitin-positive aggregates, which vary under different conditions, affect degradation efficiency?
Autophagy and regulation of cell death. Once autophagy is shown to be required for cell death, it is important to determine how this process contributes to either cell killing or clearance. Could killing be regulated by selective clearance of cell survival factors, or by depletion of mitochondria and metabolic substrates to cause metabolic catastrophe? If clearance, it is interesting to consider how recycled resources are used, and could they either influence the immune system or promote the metabolism of other cells and tissues to promote organism health?
Acknowledgments
Research on this subject is supported by the National Basic Research Program of China (2013CB910100, 2011CB910100) to H.Z., the National Natural Science Foundation of China (31421002, 31225018) to H.Z, the National Institutes of Health (GM079431, GM111658, CA159314, AI099708) to E.H.B. E.H.B. is an Ellison Medical Foundation Scholar, and H.Z. is an International Early Career Scientist of the Howard Hughes Medical Institute.
Glossary
- ATG (autophagy-related gene) genes
genes that are involved in selective and non-selective autophagy in yeast. Some ATG genes that are essential for autophagosome formation are evolutionarily conserved. Some ATG genes encode components specific to yeast, such as those for selective autophagy.
- Autophagic cell death
cell death that requires autophagy gene function.
- Autophagic flux
the dynamic nature of autophagy, including autophagosome formation, delivery of the autophagosome and cargo substrates to the lysosome, and degradation of these substrates by the lysosome.
- epg (ectopic PGL granules) genes
genes essential for autophagic degradation of PGL granules and/or the C. elegans SQSTM1 homolog SQST-1 that have no yeast counterparts or that are highly divergent from ATG genes.
- PAS (pre-autophagosomal structure, also known as phagophore assembly site)
a perivacuolar compartment or site where Atg proteins are recruited and where autophagosomes are thought to be generated.
- Scaffold protein
a family of proteins that directly interact with cargo–receptor complexes and multiple Atg proteins to endow cargo selectivity and promote autophagic degradation efficiency. The scaffold protein itself is also removed by autophagy.
- SEPA-1 (suppressor of ectopic P granules in autophagy mutants-1)
a C. elegans protein that self-oligomerizes and functions as the receptor for the accumulation of PGL-1 and PGL-3 aggregates and their autophagic degradation.
- SQSTM1/p62 (sequestosome 1)
a self-oligomerizing protein that functions as a receptor linking ubiquitinated proteins to LC3. It is also selectively degraded by autophagy.
References
- 1.Ashford TP, Porter KR. Cytoplasmic components in hepatic cell lysosomes. J. Cell Biol. 1962;12:198–202. doi: 10.1083/jcb.12.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 3.Thumm M, et al. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- 4.Harding TM, et al. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harding TM, et al. Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein. J. Biol. Chem. 1996;271:17621–17624. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- 6.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 7.Nakatogawa H, et al. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 8.Feng Y, et al. The machinery of macroautophagy. Cell Res. 2014;24:24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Axe EL, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hailey DW, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ravikumar B, et al. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat. Cell Biol. 2010;12:747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puri C, et al. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell. 2013;154:1285–1299. doi: 10.1016/j.cell.2013.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Longatti A, et al. TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J. Cell Biol. 2012;197:659–675. doi: 10.1083/jcb.201111079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Longatti A, Tooze SA. Vesicular trafficking and autophagosome formation. Cell Death Differ. 2009;16:956–965. doi: 10.1038/cdd.2009.39. [DOI] [PubMed] [Google Scholar]
- 15.Lamb CA, et al. The autophagosome: origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 2013;14:759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- 16.Yu L, et al. Autophagy termination and lysosome reformation regulated by mTOR. Nature. 2010;465:942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takats S, et al. Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J. Cell Biol. 2013;201:531–539. doi: 10.1083/jcb.201211160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takats S, et al. Interaction of the HOPS complex with Syntaxin 17 mediates autophagosome clearance in Drosophila. Mol. Biol. Cell. 2014;25:1338–1354. doi: 10.1091/mbc.E13-08-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Itakura E, et al. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/ lysosomes. Cell. 2012;151:1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Jiang P, et al. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol. Biol. Cell. 2014;25:1327–1337. doi: 10.1091/mbc.E13-08-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rusten TE, et al. ESCRTs and Fab1 regulate distinct steps of autophagy. Curr. Biol. 2007;17:1817–1825. doi: 10.1016/j.cub.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 22.Maday S, et al. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J. Cell Biol. 2012;196:407–417. doi: 10.1083/jcb.201106120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maday S, Holzbaur EL. Autophagosome biogenesis in primary neurons follows an ordered and spatially regulated pathway. Dev. Cell. 2014;30:71–85. doi: 10.1016/j.devcel.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sengupta S, et al. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol. Cell. 2010;40:310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russell RC, et al. Autophagy regulation by nutrient signaling. Cell Res. 2014;24:42–57. doi: 10.1038/cr.2013.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hosokawa N, et al. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy. 2009;5:973–979. doi: 10.4161/auto.5.7.9296. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, et al. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell. 2009;136:308–321. doi: 10.1016/j.cell.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 28.Tian Y, et al. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell. 2010;141:1042–1055. doi: 10.1016/j.cell.2010.04.034. [DOI] [PubMed] [Google Scholar]
- 29.Chang T-K, et al. Uba1 functions in Atg7- and Atg3-independent autophagy. Nat. Cell Biol. 2013;15:1067–1078. doi: 10.1038/ncb2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Juhasz G, et al. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007;21:3061–3066. doi: 10.1101/gad.1600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishida Y, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 32.Cheong H, et al. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proc. Natl. Acad. Sci. U.S.A. 2011;108:11121–11126. doi: 10.1073/pnas.1107969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu Q, et al. The WD40 repeat PtdIns(3)P-binding protein EPG-6 regulates progression of omegasomes to autophagosomes. Dev. Cell. 2011;21:343–357. doi: 10.1016/j.devcel.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 34.Liang Q, et al. The C. elegans ATG101 homolog EPG-9 directly interacts with EPG-1/Atg13 and is essential for autophagy. Autophagy. 2012;8:1426–1433. doi: 10.4161/auto.21163. [DOI] [PubMed] [Google Scholar]
- 35.Zhao H, et al. Mice deficient in Epg5 exhibit selective neuronal vulnerability to degeneration. J. Cell Biol. 2013;200:731–741. doi: 10.1083/jcb.201211014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao YG, et al. The p53-induced gene Ei24 is an essential component of the basal autophagy pathway. J. Biol. Chem. 2012;287:42053–42063. doi: 10.1074/jbc.M112.415968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao YG, et al. The autophagy gene Wipi4 regulates learning and memory function and axonal homeostasis. Autophagy. 2015 doi: 10.1080/15548627.2015.1047127. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cullup T, et al. Recessive mutations in EPG5 cause Vici syndrome, a multisystem disorder with defective autophagy. Nat. Genet. 2013;45:83–87. doi: 10.1038/ng.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haack TB, et al. Exome sequencing reveals de novo WDR45 mutations causing a phenotypically distinct, X-linked dominant form of NBIA. Am. J. Hum. Genet. 2012;91:1144–1149. doi: 10.1016/j.ajhg.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saitsu H, et al. De novo mutations in the autophagy gene WDR45 cause static encephalopathy of childhood with neurodegeneration in adulthood. Nat. Genet. 2013;45:445–449. doi: 10.1038/ng.2562. [DOI] [PubMed] [Google Scholar]
- 41.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bjørkøy G, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Komatsu M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 45.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 46.Matsumoto G, et al. Serine 403 phosphorylation of p62/ SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol. Cell. 2011;44:279–289. doi: 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- 47.Li S, et al. Arginine methylation modulates autophagic degradation of PGL granules in C. elegans. Mol. Cell. 2013;52:421–433. doi: 10.1016/j.molcel.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 48.Lin L, et al. The scaffold protein EPG-7 links cargo-receptor complexes with the autophagic assembly machinery. J. Cell Biol. 2013;201:113–129. doi: 10.1083/jcb.201209098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hosokawa N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim J, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo B, et al. Genome-wide screen identifies signaling pathways that regulate autophagy during Caenorhabditis elegans development. EMBO Rep. 2014;15:705–713. doi: 10.1002/embr.201338310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Settembre C, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lapierre LR, et al. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat. Commun. 2013;4:2267. doi: 10.1038/ncomms3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hansen M, et al. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Savage-Dunn C. TGF-beta signaling. WormBook. 2005;9:1–12. doi: 10.1895/wormbook.1.22.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu X, et al. Repression of germline RNAi pathways in somatic cells by retinoblastoma pathway chromatin complexes. PLoS Genet. 2012;8:e1002542. doi: 10.1371/journal.pgen.1002542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fay DS, Yochem J. The SynMuv genes of Caenorhabditis elegans in vulval development and beyond. Dev. Biol. 2007;306:1–9. doi: 10.1016/j.ydbio.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boland B, et al. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer’s disease. J. Neurosci. 2008;28:6926–6937. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo B, et al. O-GlcNAc-modification of SNAP-29 regulates autophagosome maturation. Nat. Cell Biol. 2014;16:1215–1226. doi: 10.1038/ncb3066. [DOI] [PubMed] [Google Scholar]
- 60.Slawson C, et al. O-GlcNAc signaling: a metabolic link between diabetes and cancer? Trends Biochem. Sci. 2010;35:547–555. doi: 10.1016/j.tibs.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sato M, Sato K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science. 2011;334:1141–1144. doi: 10.1126/science.1210333. [DOI] [PubMed] [Google Scholar]
- 62.Al Rawi S, et al. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science. 2011;334:1144–1147. doi: 10.1126/science.1211878. [DOI] [PubMed] [Google Scholar]
- 63.DeLuca SZ, O’Farrell PH. Barriers to male transmission of mitochondrial DNA in sperm development. Dev. Cell. 2012;22:660–668. doi: 10.1016/j.devcel.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Politi Y, et al. Paternal mitochondrial destruction after fertilization is mediated by a common endocytic and autophagic pathway in Drosophila. Dev. Cell. 2014;29:305–320. doi: 10.1016/j.devcel.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 65.Luo SM, et al. Unique insights into maternal mitochondrial inheritance in mice. Proc. Natl. Acad. Sci. U.S.A. 2013;110:13038–13043. doi: 10.1073/pnas.1303231110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schweichel J-U, Merker H-J. The morphology of various types of cell death in prenatal tissues. Teratology. 1973;7:253–266. doi: 10.1002/tera.1420070306. [DOI] [PubMed] [Google Scholar]
- 67.Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–1148. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nelson C, et al. miR-14 regulates autophagy during developmental cell death by targeting ip3-kinase 2. Mol. Cell. 2014;56:376–388. doi: 10.1016/j.molcel.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.MacDonald JM, et al. The Drosophila cell corpse engulfment receptor Draper mediates glial clearance of severed axons. Neuron. 2006;50:869–881. doi: 10.1016/j.neuron.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 70.McPhee CK, et al. Activation of autophagy during cell death requires the engulfment receptor Draper. Nature. 2010;465:1093–1096. doi: 10.1038/nature09127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ziegenfuss JS, et al. Draper-dependent glial phagocytic activity is mediated by Src and Syk family kinase signaling. Nature. 2008;453:935–939. doi: 10.1038/nature06901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Denton D, et al. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr. Biol. 2009;19:1741–1746. doi: 10.1016/j.cub.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hou YC, et al. Effector caspase Dcp-1 and IAP protein Bruce regulate starvation-induced autophagy during Drosophila melanogaster oogenesis. J. Cell Biol. 2008;182:1127–1139. doi: 10.1083/jcb.200712091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nezis IP, et al. Autophagic degradation of dBruce controls DNA fragmentation in nurse cells during late Drosophila melanogaster oogenesis. J. Cell Biol. 2010;190:523–531. doi: 10.1083/jcb.201002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 76.Wang H, et al. Autophagy activity contributes to programmed cell death in Caenorhabditis elegans. Autophagy. 2013;9:1975–1982. doi: 10.4161/auto.26152. [DOI] [PubMed] [Google Scholar]
- 77.Fairn GD, Grinstein S. How nascent phagosomes mature to become phagolysosomes. Trends Immunol. 2012;33:397–405. doi: 10.1016/j.it.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 78.Cheng S, et al. Autophagy genes coordinate with the class II PI/ PtdIns 3-kinase PIKI-1 to regulate apoptotic cell clearance in C. elegans. Autophagy. 2013;9:2022–2032. doi: 10.4161/auto.26323. [DOI] [PubMed] [Google Scholar]
- 79.Huang S, et al. Autophagy genes function in apoptotic cell corpse clearance during C. elegans embryonic development. Autophagy. 2013;9:138–149. doi: 10.4161/auto.22352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li W, et al. Autophagy genes function sequentially to promote apoptotic cell corpse degradation in the engulfing cell. J. Cell Biol. 2012;197:27–35. doi: 10.1083/jcb.201111053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martinez J, et al. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc. Natl. Acad. Sci. U.S.A. 2011;108:17396–17401. doi: 10.1073/pnas.1113421108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Qu X, et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 83.Mellen MA, et al. The autophagic machinery is necessary for removal of cell corpses from the developing retinal neuroepithelium. Cell Death Differ. 2008;15:1279–1290. doi: 10.1038/cdd.2008.40. [DOI] [PubMed] [Google Scholar]
- 84.Baehrecke EH. Autophagy: dual roles in life and death? Nat. Rev. Mol. Cell Biol. 2005;6:505–510. doi: 10.1038/nrm1666. [DOI] [PubMed] [Google Scholar]
- 85.Levine B, Yuan J. Autophagy in cell death: an innocent convict? J. Clin. Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Perez E, et al. Autophagy regulates tissue overgrowth in a context-dependent manner. Oncogene. 2014 doi: 10.1038/onc.2014.285. http://dx.doi.org/10.1038/onc.2014.285. [DOI] [PMC free article] [PubMed]
- 87.Zhang P, Zhang H. Autophagy modulates miRNA-mediated gene silencing and selectively degrades AIN-1/GW182 in C. elegans. EMBO Rep. 2013;14:568–576. doi: 10.1038/embor.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kang C, et al. Dual roles of autophagy in the survival of Caenorhabditis elegans during starvation. Genes Dev. 2007;21:2161–2171. doi: 10.1101/gad.1573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Melendez A, et al. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 90.Tóth ML, et al. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008;4:330–338. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- 91.Lapierre LR, et al. Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr. Biol. 2011;21:1507–1514. doi: 10.1016/j.cub.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Simonsen A, et al. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2007;4:176–184. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- 93.Ulgherait M, et al. AMPK modulates tissue and organismal aging in a non-cell-autonomous manner. Cell Rep. 2014;8:1767–1780. doi: 10.1016/j.celrep.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Scott SV, et al. Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc. Natl. Acad. Sci. U.S.A. 1996;93:12304–12308. doi: 10.1073/pnas.93.22.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hutchins MU, Klionsky DJ. Vacuolar localization of oligomeric alpha-mannosidase requires the cytoplasm to vacuole targeting and autophagy pathway components in Saccharomyces cerevisiae. J. Biol. Chem. 2001;276:20491–20498. doi: 10.1074/jbc.M101150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Duran JM, et al. Unconventional secretion ofAcb1is mediated by autophagosomes. J. Cell Biol. 2010;188:527–536. doi: 10.1083/jcb.200911154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Manjithaya R, et al. Unconventional secretion of Pichia pastoris Acb1 is dependent on GRASP protein, peroxisomal functions, and autophagosome formation. J. Cell Biol. 2010;188:537–546. doi: 10.1083/jcb.200911149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dupont N, et al. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. EMBO J. 2011;30:4701–4711. doi: 10.1038/emboj.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ogura K, et al. Caenorhabditis elegans unc-51 gene required for axonal elongation encodes a novel serine/threonine kinase. Genes Dev. 1994;8:2389–2400. doi: 10.1101/gad.8.20.2389. [DOI] [PubMed] [Google Scholar]
- 100.Ogura K, et al. The UNC-14 protein required for axonal elongation and guidance in Caenorhabditis elegans interacts with the serine/threonine kinase UNC-51. Genes Dev. 1997;11:1801–1811. doi: 10.1101/gad.11.14.1801. [DOI] [PubMed] [Google Scholar]
- 101.Ogura K, Goshima Y. The autophagy-related kinase UNC-51 and its binding partner UNC-14 regulate the subcellular localization of the Netrin receptor UNC-5 in Caenorhabditis elegans. Development. 2006;133:3441–3450. doi: 10.1242/dev.02503. [DOI] [PubMed] [Google Scholar]
- 102.Tian E, et al. epg-1 functions in autophagy-regulated processes and may encode a highly divergent Atg13 homolog in C. elegans. Autophagy. 2009;5:608–615. doi: 10.4161/auto.5.5.8624. [DOI] [PubMed] [Google Scholar]
- 103.Shravage BV, et al. Atg6 is required for multiple vesicle trafficking pathways and hematopoiesis in Drosophila. Development. 2013;140:1321–1329. doi: 10.1242/dev.089490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ruck A, et al. The Atg6/Vps30/Beclin 1 ortholog BEC-1 mediates endocytic retrograde transport in addition to autophagy in C. elegans. Autophagy. 2011;7:386–400. doi: 10.4161/auto.7.4.14391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tang HW, et al. Atg9 interacts with dTRAF2/TRAF6 to regulate oxidative stress-induced JNK activation and autophagy induction. Dev. Cell. 2013;27:489–503. doi: 10.1016/j.devcel.2013.10.017. [DOI] [PubMed] [Google Scholar]
- 106.Subramani S, Malhotra V. Non-autophagic roles of autophagy-related proteins. EMBO Rep. 2013;14:143–151. doi: 10.1038/embor.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hwang S, et al. Nondegradative role of Atg5-Atg12/Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Microbe. 2012;11:397–409. doi: 10.1016/j.chom.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu B, et al. LAAT-1 is the lysosomal lysine/arginine transporter that maintains amino acid homeostasis. Science. 2012;337:351–354. doi: 10.1126/science.1220281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fares H, Greenwald I. Regulation of endocytosis by CUP-5, the Caenorhabditis elegans mucolipin-1 homolog. Nat. Genet. 2001;28:64–68. doi: 10.1038/ng0501-64. [DOI] [PubMed] [Google Scholar]
- 110.Treusch S, et al. Caenorhabditis elegans functional orthologue of human protein h-mucolipin-1 is required for lysosome biogenesis. Proc. Natl. Acad. Sci. U.S.A. 2004;101:4483–4488. doi: 10.1073/pnas.0400709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sun T, et al. CUP-5, the C. elegans ortholog of the mammalian lysosomal channel protein MLN1/TRPML1, is required for proteolytic degradation in autolysosomes. Autophagy. 2011;7:1308–1315. doi: 10.4161/auto.7.11.17759. [DOI] [PubMed] [Google Scholar]
- 112.Strome S. Specification of the germ line. WormBook. 2005;28:1–10. doi: 10.1895/wormbook.1.9.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hird SN, et al. Segregation of germ granules in living Caenorhabditis elegans embryos: cell-type-specific mechanisms for cytoplasmic localisation. Development. 1996;122 doi: 10.1242/dev.122.4.1303. 1303–1313–1312. [DOI] [PubMed] [Google Scholar]
- 114.DeRenzo C, et al. Exclusion of germ plasm proteins from somatic lineages by cullin-dependent degradation. Nature. 2003;424:685–689. doi: 10.1038/nature01887.. [DOI] [PMC free article] [PubMed] [Google Scholar]