Graphical abstract
Abbreviations: ZP, zona pellucida; IHP, internal hydrophobic patch; EHP, external hydrophobic patch; TGFR, transforming growth factor-β receptor; Trx, thioredoxin; His6, hexahistidine; EGF, epidermal growth factor; DTT, dithiothreitol; CBB, Coomassie Brilliant Blue; RT, room temperature; DIC, differential interference contrast; MBP, maltose binding protein; THP, Tamm–Horsfall protein
Keywords: Zona pellucida, Egg coat, Fertilization, Extracellular matrix, Interdomain interaction
Highlights
-
•
Chicken ZP1 and ZP3 assemble through strong interactions between their ZP-C domains.
-
•
ZP-C domains of chicken ZP1 and ZP3 are deeply embedded in the egg-coat matrix.
-
•
Chicken ZP1 forms a homocomplex through non-covalent interaction between repeat domains.
-
•
Chicken ZPD is deposited on the interstices of ZP1–ZP3 matrix in the egg coat.
-
•
We propose a model for the architecture of chicken egg-coat matrix from these results.
Abstract
The vertebrate egg coat, including mammalian zona pellucida, is an oocyte-specific extracellular matrix comprising two to six zona pellucida (ZP) glycoproteins. The egg coat plays important roles in fertilization, especially in species-specific interactions with sperm to induce the sperm acrosome reaction and to form the block to polyspermy. It is suggested that the physiological functions of the egg coat are mediated and/or regulated coordinately by peptide and carbohydrate moieties of the ZP glycoproteins that are spatially arranged in the egg coat, whereas a comprehensive understanding of the architecture of vertebrate egg-coat matrix remains elusive. Here, we deduced the orientations and/or distributions of chicken ZP glycoproteins, ZP1, ZP3 and ZPD, in the egg-coat matrix by confocal immunofluorescent microscopy, and in the ZP1–ZP3 complexes generated in vitro by co-immunoprecipitation assays. We further confirmed interdomain interactions of the ZP glycoproteins by far-Western blot analyses of the egg-coat proteins and pull-down assays of ZP1 in the serum, using recombinant domains of ZP glycoproteins as probes. Our results suggest that the ZP1 and ZP3 bind through their ZP-C domains to form the ZP1–ZP3 complexes and fibrils, which are assembled into bundles through interactions between the repeat domains of ZP1 to form the ZP1–ZP3 matrix, and that the ZPD molecules self-associate and bind to the ZP1–ZP3 matrix through its ZP-N and ZP-C domains to form the egg-coat matrix. Based on these results, we propose a tentative model for the architecture of the chicken egg-coat matrix that might be applicable to other vertebrate ones.
1. Introduction
The vertebrate egg coat (zona pellucida in mammals) surrounding the ovulated oocyte is a unique fibrous extracellular matrix [1,2]. The egg coat is involved in the egg–sperm interactions including taxon-specific gamete recognition, induction of sperm acrosome reaction, sperm penetration through the egg coat, and block to polyspermy [1,2]. These gamete interactions are at least partly due to the sperm–egg coat (or sperm–zona) interactions that is mediated by the egg-coat components, i.e. the zona pellucida (ZP) glycoproteins, and the sperm proteins such as zonadhesin [3,4], SED1 [5], ADAM3 [6,7], CRISP1 [8], acrosin/proacrosin [9,10], or the egg cortical-granule protease, ovastacin [11], although physiological contributions of the sperm–zona interactions to normal fertilization remain controversial [12,13]. The ZP glycoproteins are characterized by conserved ∼260-aa-long ZP domain [14] containing two unique immunoglobulin-like folds, named ZP-N and ZP-C domains, respectively [15,16], and classified into totally 6 subfamilies designated ZP1, ZP2/ZPA, ZP3/ZPC, ZP4/ZPB, ZPD and ZPAX, respectively, based on phylogenetic analyses [17,18]. These egg-coat components can also be distinguished by their domain structures in the N-terminal flanking region of the ZP-N domain, and the structural diversities of the ZP-C domain. For example, on the N terminus of the ZP-N domain of both ZP1 and ZP4 homologues, there is an additional ZP-N domain (the N-terminal domain) followed by a trefoil domain, although the N-terminal and trefoil domains of ZP1 but not of ZP4 possess a free Cys residue [19] and a preceding Pro/Thr/Leu-rich linker region, respectively [20–22]. Furthermore, there are three additional ZP-N repeats on the N terminus of the ZP-N domain of ZP2 homologue [19], and a calcium-binding EGF-like domain on that of avian ZPD homologue [23–25]. In addition, the ZP3 homologue possesses the type I ZP-C domain (including the ZP-C subdomain) containing 8 Cys residues [16], and the other homologues possess the type II one containing 6 Cys residues [2,26], except that amphibian ZPD homologue possess an unusual type-II-like ZP-C domain with 4 Cys residues [23–25]. Interestingly, it is suggested that some of genes encoding ZP glycoproteins have independently been lost or become pseudogenes during evolution [18], and consequently, the mature egg coat is constituted of ZP3 and ZP4 in teleost fish [27–29], ZP2, ZP3, ZP4, ZPD and ZPAX in amphibians (Xenopus) [23,30], ZP1, ZP3, ZPD and under detectable levels or faint amounts of ZP2, ZP4 and ZPAX in birds (chicken and quail) [24,25,31]. It is noteworthy that the mammalian egg coat is composed of ZP1, ZP2, ZP3 and ZP4 in some species including hamster, rat and human [21,22,32], ZP2, ZP3 and ZP4 in other ones including pig, cow and dog [33–35], and ZP1, ZP2 and ZP3 only in mouse [18,32,36].
To date, many studies suggest that sugar chains on the ZP glycoproteins play crucial roles in the sperm–zona interactions in cooperation with the polypeptide backbone [16,37–39]. In most cases, it has been shown that ZP glycoproteins individually have abilities to bind to the intact or acrosome-reacted sperm (e.g., ZP2 [40,41] and ZP3 [16,42,43] of mouse and human, and ZP3 and ZPD of chicken [16,24]) and/or to induce sperm acrosome reaction (e.g., ZP3 of mouse [44,45] and ZPD of chicken [24]). While in other cases, for example in cow, ZP3/ZP4 heterodimer but not any individual ZP glycoproteins bind to sperm [34], and in chicken, the dimeric form of ZP1 has much higher activity to induce the acrosome reaction than the monomeric form [24]. These findings imply that the interactions between sperm molecule(s) and ZP glycoproteins are enhanced or regulated by the specific orientations of them including the glycan chains and/or polypeptide backbones in the egg-coat matrix. Therefore, it is necessary to elucidate the molecular arrangements of ZP glycoproteins in the egg coat for understanding how the physiology of the matrix associates with the functional properties of ZP glycoproteins.
Although molecular mechanisms underlying the egg-coat formation or the intermolecular interactions among ZP glycoproteins are still poorly understood, some evidence suggest that the ZP domain is involved in the polymerization of ZP glycoproteins through their internal hydrophobic patch (IHP) in the N terminus of ZP-C domain and the E′–F–G region of ZP-N domain [15,16]. It is also suggested that ZP1 forms disulfide-linked homodimers probably through the above-mentioned free Cys residue of the N-terminal domains [19,44,46], and at least in chicken, the ZP1 homodimer is formed during the interaction with ZP3 [47,48]. Moreover, two models of the egg-coat architecture are proposed based on the results from biochemical, electron/light microscopic and/or gene knockout analyses. That is, the polymers composed of ZP2–ZP3 heterodimer are cross-linked by the ZP1 homodimers in mouse [2,49], and the ZPD-complexes are loosely attached to the insoluble ZP1–ZP3 matrix in chicken [24]. Recently, a series of the first crystal structures have been determined for the ZP-N domain of mouse ZP3 [50], full-length chicken ZP3 containing both ZP-N and the type I ZP-C domains [16], and the type II ZP-C domains of mouse and rat homologues of a non-egg-coat ZP-domain protein, transforming growth factor (TGF)-β receptor type 3 (TGFR-3) [51,52]. These structures provide invaluable information about the configuration of each domain in the ZP-glycoprotein molecules and enable us to examine interdomain interactions being responsible for the egg-coat formation more directly.
In our previous studies, in vitro assays demonstrated that soluble forms of ZP1 in the chicken (i.e. laying hen) serum (serum ZP1) and ZP3 isolated from the egg coat (egg-coat ZP3) interact to form insoluble ZP1–ZP3 complexes or the egg-coat matrix-like filaments [24,47,48]. In these studies, the ZP1–ZP3 complex was not immunoprecipitated by an antibody against the C-terminal region of ZP1 but the one against the other region, suggesting that ZP domain of ZP1 is involved in the ZP1–ZP3 interactions. Furthermore, there were no significant differences in the binding abilities of ZP1 to variously glycosylated isoforms of ZP3 [53], implying that the polypeptide backbone but not the sugar chains of ZP3 is mainly involved in the formation of the ZP1–ZP3 complex. Considering that either ZP domain of chicken ZP1 or ZP3 has only one N-linked glycosylation site [20,54] and that chicken ZP3 has a single O-glycan [16] in the linker region between ZP-N and ZP-C domains, it is expected that the polypeptide backbone of ZP domain in ZP1 and ZP3 plays a major role in the assembly of these molecules into the egg-coat matrix. In the present study, we produced recombinant domains of chicken ZP glycoproteins in Escherichia coli and developed polyclonal antisera against them. Immunofluorescent staining of the egg coat and co-immunoprecipitation assays of the serum ZP1 and the egg-coat ZP3, with the domain-specific antisera, suggest that specific domains of ZP1 and ZP3 might be deeply embedded in the matrix and located at the ZP1–ZP3 interaction interfaces. The distribution of ZPD in the matrix was also determined by the immunofluorescent microscopy. Furthermore, we confirmed the domains of ZP glycoproteins involved in the egg-coat formation by far-Western blotting of the egg-coat proteins and pull-down assays of the serum ZP1 with the recombinant domains of ZP glycoproteins. Finally, we proposed a tentative model for the architecture of the chicken egg-coat matrix that might be true of other vertebrate ones. Investigations of the molecular structure and the underlying formation mechanisms of the egg-coat matrices will provide important insights into the relationship between the physiological functions and supramolecular structures of them, or the cooperative activities of the ZP glycoproteins in the sperm–zona interactions. These insights could be applicable to the reproductive medicine with novel contraceptives based on the inhibition of the sperm–zona interactions as well as with therapies for zona-pellucida related infertilities.
2. Results
2.1. Distinctive staining patterns of egg-coat matrix being observed with antisera against individual domains of ZP glycoproteins
To analyze detailed distributions and molecular orientations of ZP1, ZP3 and ZPD in the egg coat, immunofluorescent staining of them with a series of antisera against the distinct domains of ZP glycoproteins (see Section 4) and confocal laser-scanning microscopy were performed (Fig. 2A and B). The filamentous structures with a diameter of approximately 0.8–1.4 μm were observed in the differential-interference-contrast (DIC) images of egg coat (panels 3 and 4 of A). Immunofluorescent staining confirmed that the near surface of the filaments was significantly labeled with the antiserum against repeat domain of ZP1 (anti-ZP1_repeat; panel 2 of A). Interestingly, the anti-ZP1_ZP-N and anti-ZP3_ZP-N antisera showed punctuate staining patterns along the surface of the egg-coat filaments (panels 3 and 6 of B, respectively), and no staining were detected with the anti-ZP1_ZP-C and anti-ZP3_ZP-C antisera (panels 4 and 7 of B, respectively), whereas the near surface of the filaments were labeled with the anti-ZP1_N-terminal, anti-ZP1_trefoil, and anti-ZP3_N-terminal antisera as well as with the anti-ZP1_repeat antiserum (panels 1, 2 and 5 of B, respectively). In addition, the interstices of the egg-coat filaments were labeled with the anti-ZPD_EGF, ZP-N and ZP-C antisera to a similar extent (panels 8–10 of B, respectively). The anti-tag antiserum (see Section 4) was used as a negative control (panel 1 of A).
Fig. 2.
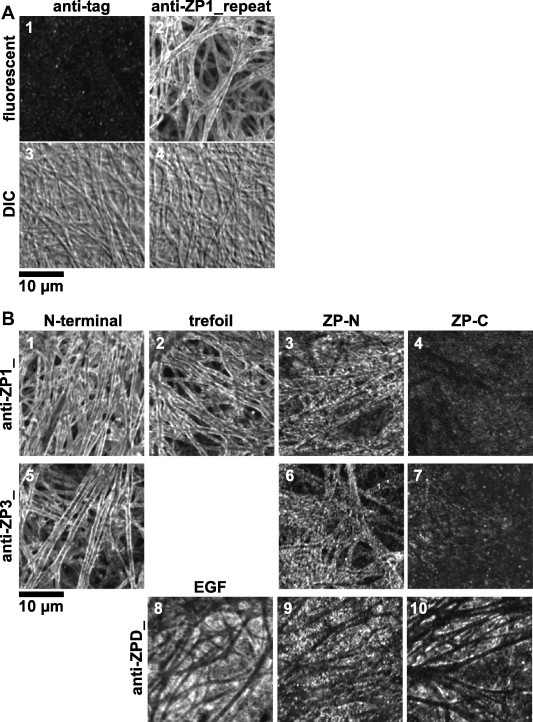
Immunofluorescent microscopy of chicken egg-coat matrix. Small pieces of the egg coat were stained with the mouse anti-tag (panels 1 and 3 of A) and anti-ZP1_repeat antisera (panels 2 and 4 of A), anti-ZP1_N-terminal, trefoil, ZP-N and ZP-C antisera (panels 1–4 of B, respectively), anti-ZP3_N-terminal, ZP-N and ZP-C antisera (panels 5–7 of B, respectively) and anti-ZPD_EGF, ZP-N and ZP-C antisera (panels 8–10 of B, respectively) being produced as described in Section 4 followed by Alexa Fluor® 488 goat anti-mouse IgG, and then observed under a confocal laser-scanning microscope equipped with a 488-nm excitation laser. Light images of differential-interference-contrast (DIC) microscopy are shown (panels 3 and 4 of A). Bar = 10 μm.
2.2. Specific interactions of recombinant domains of ZP1 with egg-coat proteins detected by far-Western blot analyses
To identify domains of ZP1 involved in the ZP1–ZP1 and ZP1–ZP3 interactions in the egg-coat matrix, far-Western blot analyses using a series of recombinant domains of ZP1 (see Section 4) as probes were performed (Fig. 3A). Chicken egg-coat proteins were separated by SDS–PAGE under non-reducing conditions and electroblotted onto nitrocellulose membranes (lanes 1–6) or visualized by CBB staining (lane 7). Then the membranes were incubated with the control protein (lane 1; see Section 4) and the recombinant domains of ZP1 (rZP1_N-terminal, repeat, trefoil, ZP-N and ZP-C) containing the Trx tag (lanes 2–6, respectively; see Section 4), respectively, and subsequently with rabbit anti-Trx antibody. The bands corresponding to monomeric (mo-) and dimeric (di-) forms of ZP1 and the previously-reported anti-ZP1 positive proteins [47,53] (asterisks) on the membrane incubated with the rZP1_repeat (lane 3), and the bands corresponding to ZP3 on the membrane incubated with the rZP1_ZP-C (lane 6), were detected at significantly higher levels than the other bands. Furthermore, the bands corresponding to ZP3 and ZPD on the membrane incubated with the rZP1_repeat (lane 3), and the one corresponding to the anti-ZP1 positive proteins [47,53] (asterisks) on the membrane incubated with the rZP1_ZP-C (lane 6) were also detected, although at weaker levels. On the other hand, all the bands on the membranes incubated with the rZP1_N-terminal, trefoil and ZP-N (lanes 2, 4 and 5, respectively) were detected at levels similar to control (lane 1).
Fig. 3.
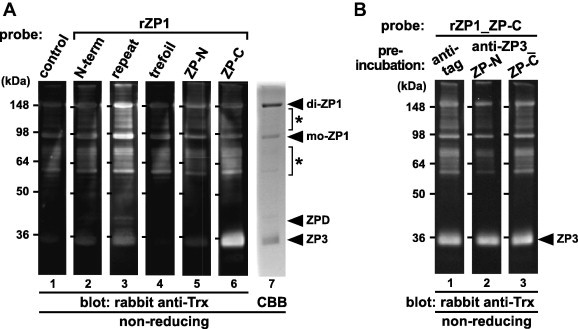
Far-Western blot analyses of the egg-coat proteins with the recombinant domains of ZP1. The egg-coat proteins were separated by SDS–PAGE under non-reducing conditions followed by electroblotting onto nitrocellulose membranes. (A) The blocked membranes were incubated with the control protein (lane 1), recombinant N-terminal (N-term), repeat, trefoil, ZP-N and ZP-C domains of ZP1 (rZP1s) (lanes 2–6, respectively; see Fig. 1) as Trx-tagged probes followed by immunostaining with rabbit anti-Trx antibody. The proteins in the SDS–PAGE gel were also stained with CBB (lane 7). (B) The blocked membranes were preincubated with the mouse anti-tag (lane 1), anti-ZP3_ZP-N and ZP-C antisera (lanes 2 and 3, respectively) being produced as described in Section 4 prior to the incubation with the recombinant ZP-C domain of ZP1 (rZP1_ZP-C), and immunostained with rabbit anti-Trx antibody. Migration positions of dimeric (di-) and monomeric (mo-) ZP1, ZPD and ZP3 (arrowheads), and the previously-reported anti-ZP1 positive proteins (asterisks) are shown on the right. Molecular weights (kDa) are indicated on the left.
In addition, we examined whether the interaction of the rZP1_ZP-C with ZP3 is inhibited by the anti-ZP3_ZP-N and/or ZP-C antisera (Fig. 3B). Far-Western blotting was performed as described above except that the nitrocellulose membranes were preincubated with the mouse anti-tag (lane 1), anti-ZP3_ZP-N (lane 2) and anti-ZP3_ZP-C (lane 3) antisera, respectively, followed by incubation with the rZP1_ZP-C as a probe. Unexpectedly, either bands corresponding to ZP3 on the membranes preincubated with the anti-ZP3_ZP-N (lane 2) or anti-ZP3_ZP-C (lane 3) antiserum was detected similarly to that preincubated with the anti-tag (lane 1).
2.3. Varied efficiency of co-immunoprecipitation of ZP1 in the chicken serum and ZP3 isolated from the egg coat with the antisera against individual domains of ZP glycoproteins
To predict dispositions of the individual domains of ZP1 and ZP3 in the ZP1–ZP3 complexes, co-immunoprecipitation assays of the serum ZP1 and the ZP3 isolated from the egg coat (egg-coat ZP3) with the antisera raised against the domains of ZP1 and ZP3 were performed (Fig. 4A and B). The chicken serum containing ZP1 was mixed with the ZP3 solution prepared as described in Section 4 and incubated to form the ZP1–ZP3 complexes. Antibodies in the antisera against ZP1 (anti-ZP1_N-terminal, repeat, trefoil, ZP-N and ZP-C in A) and ZP3 (anti-ZP3_N-terminal, ZP-N and ZP-C in B) were immobilized on the protein G beads and then incubated with the buffer in the presence (+; odd-numbered lanes, except for lane 11, of A and B) or absence (−; even-numbered lanes of A and B) of the serum-ZP3 mixture. The immunoprecipitates on the beads were subjected to SDS–PAGE under reducing conditions, together with the chicken serum and the ZP3 solution (upper and lower panels in lane 11 of A, respectively), followed by immunoblotting with the anti-ZP1_repeat (anti-ZP1) and anti-ZP3_ZP-C (anti-ZP3) antisera (upper and lower panels, respectively). Immunoglobulin heavy and light chains (single and double asterisks, respectively) were detected by the secondary antibody at similar levels either in the anti-ZP1 or anti-ZP3 immunoprecipitates (A or B, respectively). Antibodies in the anti-ZP1_N-terminal, repeat and ZP-C antisera (lanes 1, 3, and 9 of A, respectively) but not ones in the anti-ZP1_trefoil and ZP-N antisera (lanes 5 and 7 of A, respectively) efficiently co-immunoprecipitated the egg-coat ZP3 (blank arrowheads) with the serum ZP1 (filled arrowheads), while ones in all the anti-ZP1 antisera immunoprecipitated ZP1 at similarly high efficiencies, from the serum–ZP3 mixture. On the other hand, antibodies in the anti-ZP3_N-terminal and ZP-N antisera (lanes 1 and 3 of B, respectively) but not ones in the anti-ZP3_ZP-C antisera (lane 5 of B) efficiently co-immunoprecipitated the egg-coat ZP3 (blank arrowheads) and the serum ZP1 (filled arrowheads) from the serum–ZP3 mixture.
Fig. 4.
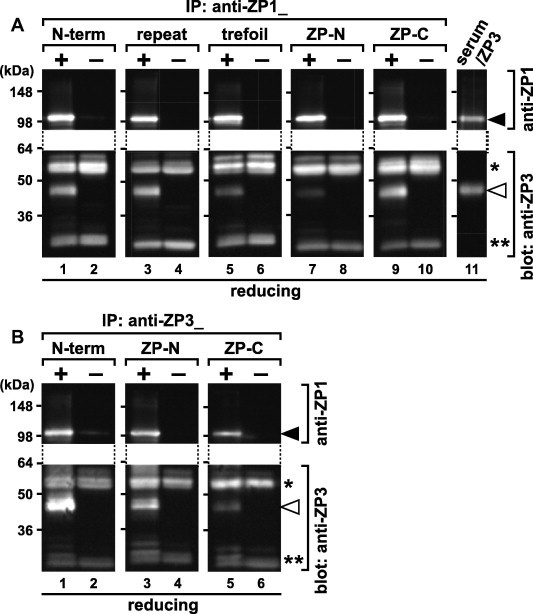
Co-immunoprecipitation assays of ZP1 in the chicken serum and ZP3 isolated from the egg coat. The chicken serum containing ZP1 and the ZP3 solution prepared as described in Section 4 were mixed and incubated to form the ZP1–ZP3 complexes. Protein G magnetic beads coated with the antibodies in the anti-ZP1_N-terminal, repeat, trefoil, ZP-N and ZP-C antisera (A) and the anti-ZP3_N-terminal, ZP-N and ZP-C antisera (B) were incubated in the presence (+; odd-numbered lanes, except for lane 11) or absence (−; even-numbered lanes) of the serum–ZP3 mixture and pulled down by magnetic field. The immunoprecipitates on the beads were subjected to SDS–PAGE under reducing conditions, together with the serum and the ZP3 solution (upper and lower panels in lane 11 of A, respectively) followed by immunoblotting with the anti-ZP1_repeat (anti-ZP1) and anti-ZP3_ZP-C (anti-ZP3) antisera (upper and lower panels, respectively). Migration positions of ZP1 (filled arrowheads), ZP3 (blank arrowheads), and immunoglobulin heavy and light chains (single and double asterisks, respectively) are shown on the right. Molecular weights (kDa) are indicated on the left.
2.4. Interactions of recombinant ZP-N and ZP-C domains of ZP3 with ZP1 in the chicken serum being compared by pull-down assays
To identify the domains of chicken ZP3 involved in the formation of the ZP1–ZP3 complexes, pull-down assays of ZP1 in the chicken serum were performed using the rZP3_ZP-N and ZP-C as probes (Fig. 5A and B, respectively). The beads coated with the rZP3s (lanes 2, 4 and 6) and with the control protein (lanes 3 and 5) were incubated in the presence (lanes 2–5) or absence (serum–; lanes 6) of the chicken serum containing ZP1. Proteins being bound to the beads were released using 4.0% Triton X-100 (TX-100; lanes 2 and 3) and subsequently 2.0% SDS (SDS; lanes 4–6), and subjected to SDS–PAGE under non-reducing conditions, together with the chicken serum (lanes 1), followed by immunoblotting with the anti-ZP1_repeat antiserum (anti-ZP1). ZP1 (arrowheads) was detected in the 2.0% SDS eluates from the beads coated with the rZP3_ZP-C (lane 4 of B) at significantly higher levels than that with the rZP3_ZP-N (lane 4 of A). The anti-ZP1 positive proteins in the serum (single asterisks), and the ones in the recombinant protein solutions (double asterisks) were also detected in the 2.0% SDS eluates. Almost no proteins were detected by the anti-ZP1 in all the 4.0% Triton X-100 eluates (lanes 2 and 3). Since the antisera were raised against the recombinant ZP glycoproteins containing peptides derived from the tag sequence as described in Section 4, the anti-ZP1 antiserum also detected the control protein (triple asterisks).
Fig. 5.
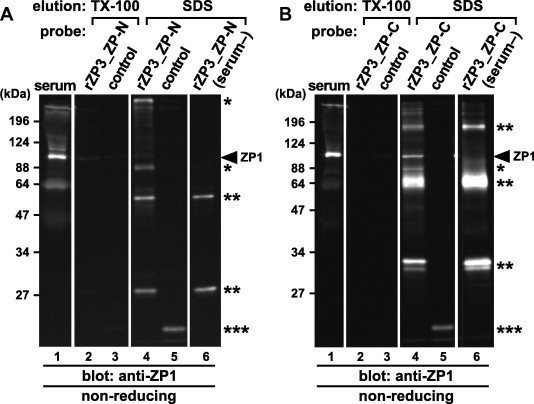
Pull-down assays from the chicken serum containing ZP1 with the recombinant ZP domains of ZP3. Nickel-magnetic beads coated with the control protein (lanes 3 and 5), the recombinant ZP-N (lanes 2, 4 and 6 of A) and ZP-C (lanes 2, 4 and 6 of B) domains of ZP3 (rZP3_ZP-N and ZP-C; A and B, respectively; see Fig. 1) were incubated in the presence (lanes 2–5) or absence (serum–; lanes 6) of the chicken serum containing ZP1 and pulled down by magnetic field followed by subsequent incubation in 4.0% Triton X-100. The proteins released from the beads by the Triton X-100 treatment (TX-100; lanes 2 and 3), and the ones retained on the beads and released by incubation in the SDS–PAGE sample buffer containing 2.0% SDS (SDS; lanes 4–6) were subjected to SDS–PAGE under non-reducing conditions, together with the chicken serum (serum; lanes 1), followed by immunoblotting with the anti-ZP1_repeat antiserum (anti-ZP1). Migration positions of monomeric ZP1 in the serum (arrowheads), the anti-ZP1 positive proteins in the serum that bound to the recombinant-protein coated beads (single asterisks) and in the recombinant ZP-N and ZP-C solutions (double asterisks), and the control protein (triple asterisks) are shown on the right. Molecular weights (kDa) are indicated on the left.
2.5. Specific interactions of recombinant domains of ZPD with egg-coat proteins detected by far-Western blot analyses
To identify domains of ZPD involved in the binding to ZPD and the other ZP glycoproteins, far-Western blot analyses using a series of recombinant domains of ZPD as probes were performed (Fig. 6A). Chicken egg-coat proteins (odd-numbered lanes) and ZPD in the crude ZPD solution (even-numbered lanes) prepared as described in Section 4 were separated by SDS–PAGE under non-reducing conditions and electroblotted onto nitrocellulose membranes (lanes 1–8) or visualized by CBB staining (lanes 9 and 10). Then the membranes were incubated with the control protein (lanes 1 and 2) and the recombinant domains of ZPD (rZPD_EGF; lanes 3 and 4, ZP-N; lanes 5 and 6, and ZP-C; lanes 7 and 8) containing the Trx tag (see Section 4), respectively, and subsequently with rabbit anti-Trx antibody. The bands corresponding to monomeric (mo-) and dimeric (di-) forms of ZP1 on the membrane incubated with the rZPD_ZP-N and ZP-C (lanes 5 and 7, respectively), the band corresponding to one of the previously-reported anti-ZP1 positive proteins [47,53] (asterisk) on the membrane incubated with the rZPD_ZP-N (lane 5), and the band corresponding to ZPD on the membrane incubated with the rZPD_ZP-C (lane 8), were detected at significantly higher levels than the other bands. Furthermore, the bands corresponding to ZPD and ZP3 were detected on the membrane incubated with the rZPD_ZP-N (lane 6) and the rZPD_ZP-C (lane 7), respectively, although at weaker levels. On the other hand, all the bands on the membranes incubated with the rZPD_EGF (lanes 3 and 4) were detected at levels similar to control (lanes 1 and 2).
Fig. 6.
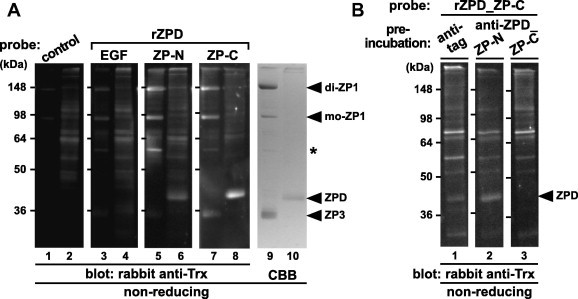
Far-Western blot analyses of the egg-coat proteins with the recombinant domains of ZPD. The proteins in the egg coat (odd-numbered lanes in A) and the crude ZPD solution prepared as described in Section 4 (even-numbered lanes in A and lanes 1–3 in B) were separated by SDS–PAGE under non-reducing conditions followed by electroblotting onto nitrocellulose membranes. (A) The blocked membranes were incubated with the control protein (lanes 1 and 2), recombinant EGF-like (EGF; lanes 3 and 4), ZP-N (lanes 5 and 6) and ZP-C (lanes 7 and 8) domains of ZPD (rZPDs) (see Fig. 1) as Trx-tagged probes followed by immunostaining with rabbit anti-Trx antibody. The proteins in the SDS–PAGE gel were also stained with CBB (lanes 9 and 10). (B) The blocked membranes were preincubated with the mouse anti-tag (lane 1), anti-ZPD_ZP-N and ZP-C (lanes 2 and 3, respectively) antisera being produced as described in Section 4 prior to the incubation with the recombinant ZP-C domain of ZPD (rZPD_ZP-C), and immunostained with rabbit anti-Trx antibody. Migration positions of dimeric (di-) and monomeric (mo-) ZP1, ZPD and ZP3 (arrowheads), and one of the previously-reported anti-ZP1 positive proteins (asterisk) are shown on the right. Molecular weights (kDa) are indicated on the left.
In addition, to predict the binding interface between ZPD molecules involved in the formation of the possible ZPD aggregates, we examined whether the interaction of the rZPD_ZP-C with ZPD is inhibited by the anti-ZPD_ZP-N and/or ZP-C antisera (Fig. 6B). Far-Western blotting were performed as described above except that only the crude ZPD solution were subjected to SDS–PAGE, and that the nitrocellulose membranes were preincubated with the mouse anti-tag (lane 1), anti-ZPD_ZP-N (lane 2) and anti-ZPD_ZP-C (lane 3) antisera, respectively, followed by incubation with the rZPD_ZP-C as a probe. The bands corresponding to ZPD were detected on the membrane preincubated with the anti-tag and anti-ZPD_ZP-N (lanes 1 and 2, respectively), but not on the membrane preincubated with the anti-ZPD_ZP-C (lane 3).
3. Discussion
Structural studies of the egg-coat matrix could provide new knowledge applicable for designing novel therapies for the infertility associated with abnormal morphology of the egg coat and mutations in their unique components, ZP glycoproteins [55–57]. However, despite extensive investigations to elucidate the molecular mechanisms of egg-coat formation and the egg–sperm interactions, a comprehensive understanding of the architecture of vertebrate egg coat remains elusive [58]. In this study, we further mapped the domains of ZP glycoproteins involved in the egg-coat matrix formation using immunohistochemical and biochemical approaches.
To begin to study the interdomain interactions of ZP glycoproteins, we produced the N-terminally Trx-, His6- and S-tagged recombinant domains of chicken ZP1, ZP3 and ZPD (see Fig. 1), which are used to generate mouse polyclonal antisera specific for the individual domains of ZP glycoproteins (see Supplement Fig. S1), and as probes in far-Western blot analyses and pull-down assays. We predicted that the recombinant domains of ZP glycoproteins being expressed in E. coli could be correctly folded for the following reasons. (1) Crystal structure of the ZP-N domain of mouse ZP3 being expressed as a MBP-fusion protein in E. coli Origami B(DE3) (Novagen) [50] shared structural homology with that of full-length chicken ZP3 being expressed in CHO cells [16]. (2) The N-terminal domain of ZP1 is an additional copy of the ZP-N domain [19]. (3) Crystal structures of the type II ZP-C domain of mouse and rat TGFR-3 being expressed in eukaryotic cells [51,52], shared structural homology with that of the natively non-glycosylated type I ZP-C domain of chicken ZP3, except that the ZP-C of ZP3 homologue possesses the type-I specific ZP-C subdomain [16]. (4) Solution structures of the trefoil domain of human pNR-2/pS2 [59] and the EGF-like domain of human fibrillin-1 [60] were determined by NMR of the recombinant proteins being expressed in E. coli. (5) There are neither Cys residue nor N-glycosylation site in the repeat domain of ZP1 and the N-terminal peptide of ZP3 in chicken [20,54]. Therefore, we employed the E. coli strains that facilitate formation of disulfide bonds by carrying the trxB/gor mutations and that enhance the expression of eukaryotic proteins by containing codons rarely used in E. coli to produce active proteins efficiently, and the inclusion bodies of the recombinant proteins for the probes were unfolded and refolded without any reducing agents as described in Section 4. Immunostaining of the chicken egg coat (Fig. 2) and/or co-immunoprecipitation of the serum ZP1 and the egg-coat ZP3 (Fig. 4) revealed that all antibodies raised against the recombinant ZP glycoproteins have high affinities for the natively folded ones. Although some of the recombinant proteins form covalently linked dimer or polymers on the beads in the pull-down assay (double asterisks of Fig. 5), considerable amount of them were remained as monomer (the lowest double asterisks of Fig. 5).
Fig. 1.
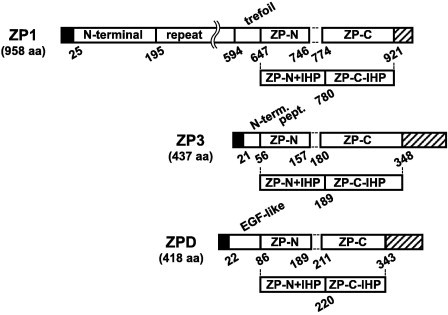
Schematic representation of the domains of chicken ZP glycoproteins being expressed as fusion proteins. Structural and/or functional domains of ZP glycoproteins constituting mature chicken egg coat, i.e., ZP1, ZP3 (GenBank: BAA13760) and ZPD (GenBank: BAD13713), were expressed as N-terminal Trx-, His6-, S-tagged fusion proteins in Escherichia coli as described in Section 4. ZP1 in this study contains an additional 24-aa-long repeat unit in the repeat domain as compared to the currently available sequence (GenBank: CAC16087). Positions of the domains fused to the tags are shown in the schematic diagrams of chicken ZP glycoproteins. Signal peptides and the C-terminal peptides containing furin cleavage sites followed by the external hydrophobic patch (EHP) are indicated as filled and striped rectangles, respectively.
In our previous study, immunohistochemical analyses of chicken egg coat confirmed that the fibrous matrix of chicken egg coat was composed of ZP1 and ZP3, although the distributions of ZP glycoproteins in the matrix remain unclear partly because of the absence of clear images [53]. In the present study, we initially performed immunofluorescence staining of the mature chicken egg coat with the antisera raised against the domains of chicken ZP1, ZP3 and ZPD followed by observation under the confocal laser-scanning microscopy as described in Section 4 (Fig. 2A and B). The optical sections showed that antibodies in the anti-ZP1_N-terminal, trefoil (panels 1 and 2 of Fig. 2B, respectively), repeat (panels 2 and 4 of Fig. 2A) and anti-ZP3_N-terminal (panel 5 of Fig. 2B) antisera bound predominantly to the near surface of the egg-coat filaments. Considering that the core of the filaments showed weak staining with these antisera, and that the ultrastructural studies of mouse and human egg coat showed that their large meshed networks are constructed of three-dimensionally arranged or bundled microfilaments [2,61], these data suggest that the filaments of chicken egg coat might be the tightly packed bundles of the ZP1–ZP3 fibrils, in which the antibodies are prevented from binding to the core. Surprisingly, the antibodies in the anti-ZP1_ZP-N and anti-ZP3_ZP-N antisera (panels 3 and 6 of Fig. 2B, respectively) bound to the surface of the filaments as punctuate patterns, and the ones in the anti-ZP1_ZP-C and anti-ZP3_ZP-C antisera (panels 4 and 7 of Fig. 2B, respectively) did not bind to the filaments. Considering that the reactivity of all antisera generated in this study were confirmed by immunoblotting under non-reducing conditions (Supplement Fig. S1A) and/or co-immunoprecipitation of native ZP glycoproteins (Fig. 4), these results suggest that the ZP1 and ZP3 molecules are arranged to form fibrils in a manner that their ZP-C domains are deeply embedded in the core, the ZP-N domains are located in the medial layer, and that the other regions are exposed on the surface. In our previous study, the antibodies in the antiserum against full-length ZP3 appeared to bind more homogeneously to whole region in the egg-coat filaments [53], probably because the relative abundance of the antibodies against the N-terminal peptide of ZP3 in the antiserum might be insufficient to stain near surface of the filaments intensely. On the other hand, the distribution of ZPD in the egg-coat matrix was visualized in this study. The optical sections showed that antibodies in the anti-ZPD_EGF, ZP-N and ZP-C antisera (panels 8–10 of Fig. 2B, respectively) bound to the thin layer on the interstices of egg-coat filaments with similar affinities. Considering that ZPD is easily released from the egg coat by mechanical treatment as reported previously [24], the results suggest that ZPD forms homopolymers or aggregates to deposit on the interstices of ZP1–ZP3 matrix.
Subsequently, the domains of ZP1 involved in the intermolecular interactions of ZP glycoproteins in the egg-coat matrix were determined by far-Western blot analyses of the egg-coat proteins with the recombinant domains of ZP1 as probes (Fig. 3A and B). The results indicate that the repeat and ZP-C domains of ZP1 bind to ZP1 and ZP3, respectively, and that both of them probably have some affinities to ZPD and/or the previously reported anti-ZP1 positive proteins [47,53] (lanes 3 and 6 of Fig. 3A). Considering that only the repeat domain has a high binding affinity to ZP1 among the five domains of ZP1 (lane 3 of Fig. 3A), the data suggest that ZP1 could form the non-covalently linked homodimer and/or polymer in the matrix through the repeat domain, and the anti-ZP1 positive proteins [47,53] might possess parts of the repeat domain. Unexpectedly, neither the preincubation of ZP3 with the anti-ZP3_ZP-N nor anti-ZP3_ZP-C antiserum inhibited the binding of the rZP1_ZP-C to ZP3 effectively (Fig. 3B), suggesting that the ZP-C domain of ZP1 has greater affinities for the ZP-N and/or ZP-C domains of ZP3 than antibodies in the anti-ZP3_ZP-N and ZP-C antisera.
In our previous study, it is suggested that the ZP1 in the blood of chicken, i.e. laying hen, (serum ZP1) interacts with the ZP3 being expressed in the ovary to form disulfide-linked homodimer and filamentous complexes with ZP3, and that the ZP1–ZP3 complexes probably play an important role as a scaffold of the egg-coat matrix [47,48]. We further determined the domains of ZP glycoproteins involved in the assembly of ZP1–ZP3 complexes, which are probably incorporated into the ZP1–ZP3 matrix during egg-coat formation [47,48]. Co-immunoprecipitation of the serum ZP1 and the egg-coat ZP3 with the antisera against domains of ZP1 and ZP3 (Fig. 4A and B, respectively) suggest that the N-terminal, repeat and ZP-C domains of ZP1, and the N-terminal peptide and the ZP-N domain of ZP3 are exposed on the surface of the ZP1–ZP3 complexes, while the trefoil and ZP-N domains of ZP1, and the ZP-C domain of ZP3 are not. ZP1 was immunoprecipitated by antibodies in all the anti-ZP1 antisera from the serum–ZP3 mixture at similarly high efficiencies (upper panels of Fig. 4A), suggesting that there was an excess of ZP1. On the other hand, pull-down assays from the chicken serum with the recombinant ZP-N and ZP-C domains of ZP3 (Fig. 5A and B, respectively) demonstrate that the serum ZP1 assemble with the egg-coat ZP3 through the interactions with the ZP-C domain of ZP3 to form the 4.0% Triton X-100-resistant rigid complex. Taken together with the above data, these results suggest that ZP1 is bound to ZP3 through their ZP-C domains with unusually high affinities either in the egg coat or the ZP1–ZP3 complexes, and that the ZP-C domain of ZP1 being exposed on the surface of the ZP1–ZP3 complexes might become embedded within the ZP1–ZP3 fibrils during the egg-coat formation. Interestingly, the serum ZP1 did not form intermolecular disulfide bonds by the interaction with the rZP3_ZP-C (lane 4 of Fig. 5B), whereas our previous studies suggest that the serum ZP1 interacts with full-length ZP3 to form disulfide-linked homodimer [47,48], implying that full-length ZP3 possessing both ZP-N and ZP-C domains are required for the ZP1 dimerization. Considering that chicken ZP3 is thought to form a non-covalently linked homodimer through its ZP-N and ZP-C domains belonging to opposite molecules as observed in the crystal structure [16], it is expected that the polymerization of the ZP1–ZP3 complexes through the interaction between ZP3 molecules increases accessibility of ZP1 monomers to form the dimer.
In our previous study, it is suggested that the third major component of mature chicken egg coat, ZPD, is loosely attached to the ZP1–ZP3 matrix and probably self-associates to form aggregates [24]. In the present study, the domains of ZPD involved in the formation of above-mentioned ZPD homopolymers or aggregates and the binding to the ZP1–ZP3 matrix were also determined by far-Western blot analyses of both the egg-coat proteins and partially purified ZPD with the recombinant domains of ZPD as probes (Fig. 6A and B). The results indicate that both the ZP-N and ZP-C domains of ZPD bind to ZP1 with similarly high affinities (lanes 5 and 7 of Fig. 6A), the ZP-C domain of ZPD binds to ZPD itself with significantly higher affinities than the ZP-N domain (lanes 6 and 8 of Fig. 6A), and that the ZP-C domain of ZPD has some affinities to ZP3 (lane 7 of Fig. 6A). In addition, the binding of the ZP-C domain of ZPD to ZPD itself was significantly inhibited by anti-ZPD_ZP-C antiserum (Fig. 6B). These results suggest that ZPD forms homopolymers perhaps in the head-to-head and tail-to-tail arrangements to be attached to the ZP1–ZP3 matrix. Interestingly, the ZP-N domain of ZPD bound to one of the anti-ZP1 positive proteins [47,53] (asterisks) with significantly higher affinities than the other domains of ZPD (lane 5 of Fig. 6A). Considering that both ZP-N and ZP-C domains of ZPD bind to ZP1 with high affinities, and that the anti-ZP1 positive proteins [47,53] are thought to contain the repeat sequences as discussed above, the ZP-N but not ZP-C domain of ZPD might have a high affinity to the repeat domain of ZP1.
Interestingly, chicken ZPD (GenBank: BAD13713) and human uromodulin (NCBI Reference Sequence: NP_001008390) share the Calcium-binding EGF-like and the ZP domains, although human uromodulin possess two additional EGF-like domains and the uncharacterized D8C domain [62]. Furthermore, a database homology search using NCBI BLAST [63] showed that C58–I362 of the chicken ZPD possesses significant sequence homology with C306–I604 of the human uromodulin (29% identities and 48% positives). These data and the recent reports that the uromodulin or THP plays important roles in the protection from bacterial infection and the regulation of immunosystems in urinary tract [62,64] imply that ZPD may be a close relative of the uromodulin and that ZPD might have functions in the defense of oocyte or early embryo in the egg coat.
In conclusion, the results of this study suggest that ZP1 and ZP3 assemble into probably helical fibrils through strong interactions between their ZP-C domains in a manner that the ZP-C domains are deeply embedded in the core, the ZP-N domains are located in the medial layer, and that the N-terminal, repeat, trefoil domains of ZP1, the N-terminal peptide of ZP3 and probably the glycan chains on the ZP-N/ZP-C linker regions are exposed on the surface. The results also suggest that the ZP1–ZP3 fibrils further assemble into bundles through the intermolecular disulfide bonds between the N-terminal domains of ZP1 and through non-covalent interactions between the repeat domains of ZP1 to form the ZP1–ZP3 matrix. In addition, the results suggest that ZPD forms homopolymers presumably in the head-to-head and tail-to-tail arrangements and attaches to the ZP1–ZP3 fibrils located on the surface of the matrix to form the ZPD aggregates. A tentative model for the architecture of chicken egg coat is shown in Fig. 7, although detailed domain configurations of the ZP glycoproteins, mechanisms of the ZP1 dimerization and involvements of either the above-mentioned non-covalent ZP3 dimer [16] or the previously-reported ZP3 isoforms [53], in the formation of the ZP1–ZP3 fibrils and the ZPD aggregates remain to be elucidated. Our model of the egg-coat architecture is compatible with the mouse model [2,49], and might be applicable to the egg coat of all other vertebrates by replacing the ZP-glycoprotein components. The model presented here might provide new insights into egg-coat formation mechanisms that could have application in research into developing novel approaches to control fertility by modifying the egg-coat architecture for human/veterinary medicine including therapies for infertility associated with the zona-pellucida dysmorphism and/or mutations in ZP glycoproteins [55–57].
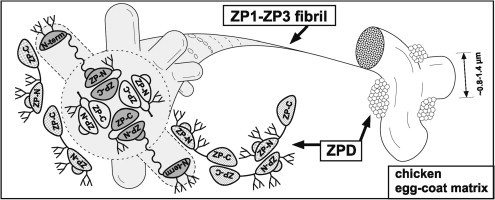
Fig. 7.
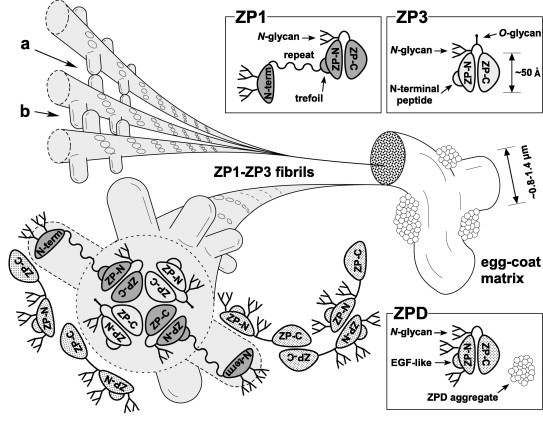
A tentative model for the architecture of the chicken egg-coat matrix. A schematic diagram of the chicken egg-coat matrix architecture was drawn based on the results from the present and previous studies. Chicken ZP1 containing the N-terminal (N-term), repeat, trefoil, ZP-N and ZP-C domains, and N-glycans on both the N-terminal domain and the ZP-N/ZP-C linker region assembles with chicken ZP3 containing the N-terminal peptide, ZP-N and ZP-C domains, and N- and O-glycans on the linker region through the interactions between their ZP-C domains to form the helical ZP1–ZP3 fibrils. The N-terminal, repeat, trefoil domains of ZP1, the N-terminal peptide of ZP3 and above-mentioned glycans are exposed on the surface of the ZP1–ZP3 fibrils to overlie the ZP-N domains. The ZP1–ZP3 fibrils assemble into bundles through both intermolecular disulfide bonds between the N-terminal domains of ZP1 (a), and non-covalent interactions between the repeat domains of ZP1 (b), to form the fibrous ZP1–ZP3 matrix. In addition, chicken ZPD containing the calcium-binding EGF-like (EGF-like), ZP-N and ZP-C domains, and N-glycans on both the ZP-N domain and the ZP-N/ZP-C linker region forms homopolymers in the head-to-head and tail-to-tail arrangements and attaches to the ZP1–ZP3 fibrils located on the surface of the interstices of ZP1–ZP3 matrix to form aggregates. Schematic diagrams of ZP1, ZP3 and ZPD are drawn based on the domain structure of chicken ZP1 and the crystal structure of full-length chicken ZP3 (PDB accession code: 3NK3) [16]. All the putative N-glycosylation sites of the ZP glycoproteins are occupied in this scheme.
4. Materials and methods
All animal procedures were performed according to the guidelines for the care and use of experimental animals of Meijo University. The animal protocols were approved by an institutional animal care and use committee (2013-A-E-1).
4.1. Preparation of ZP3 and ZPD from chicken egg coat, and chicken serum
Chicken egg coat was isolated from the pre-ovulatory mature follicles, and the ZP3 and crude ZPD solutions were prepared as previously described [24], except that the gel-filtration chromatography was performed using ÄKTA prime plus (GE Healthcare, Uppsala, Sweden) equipped with HiPrep™ 16/60 Sephacryl™ S-200 HR column (GE Healthcare), and the pooled fraction containing ZP3 was dialyzed against PBS to be used as the ZP3 solution. The chicken serum containing ZP1 was prepared as previously described [47].
4.2. Protein determination
Protein concentrations were estimated by BCA protein assay kit (Pierce, Rockford, IL) using BSA as the standard, except that the concentration of ZP1 in the chicken serum were determined by densitometric analyses of the immunoblots for the chicken serum with the anti-ZP1_repeat antisera using the below-mentioned control protein as the standard. Amount of the His6-tagged proteins bound to the nickel beads were estimated by densitometric analyses of the CBB-stained gel bands using BSA as the standard.
4.3. Production of recombinant domains of chicken ZP glycoproteins
Complementary DNAs (cDNAs) encoding structural and/or functional domains of ZP glycoproteins (Fig. 1) were amplified by PCR using full-length cDNAs of chicken ZP1 [20], ZP3 (GenBank: D89097) [54] and ZPD (GenBank: AB114441) [24] as templates with primer set listed in the Supplement Table S1. The ZP1 cDNA that we used in this study contains an insertion encoding the additional 24-aa-long repeat unit in the repeat domain as compared to the sequence available in GenBank (GenBank: AJ289697) [20]. The primers in the Supplement Table S1 were designed to amplify cDNAs encoding the N-terminal, repeat, trefoil, ZP-N and ZP-C domains of ZP1 (corresponding to nucleotides +73–582, +583–1707, +1708–1866, +1867–2166 and +2248–2688 of the GenBank: AJ289697, respectively) [20], the N-terminal peptide, ZP-N and ZP-C domains of ZP3 (corresponding to nucleotides +61–165, +166–471 and +538–1041 of the GenBank: D89097, respectively) [54], and the N-terminal region containing Calcium-binding EGF-like (EGF-like), ZP-N and ZP-C domains of ZPD (corresponding to nucleotides +64–255, +256–567 and +631–1026 of the GenBank: AB114441, respectively) [24] with NcoI or BamHI site at the 5′ end, stop codon at the 3′ end, and XhoI or BamHI site immediately after the stop codon. In addition, cDNAs encoding the N-terminal and C-terminal halves of ZP domain (ZP-N + IHP; ZP-N domain followed by the linker region and IHP, and ZP-C − IHP; ZP-C domain lacking the IHP, respectively) in ZP1 (corresponding to nucleotides +1867–2265 and +2266–2688 of GenBank: AJ289697, respectively), ZP3 (corresponding to nucleotides +166–564 and +565–1041 of GenBank: D89097, respectively) and ZPD (corresponding to nucleotides +256–657 and +658–1026 of GenBank: AB114441, respectively) were amplified similarly. The resultant DNA were digested with the restriction enzymes, and inserted between the same restriction sites into pET-32a(+) vector (Novagen®; Merck Millipore, Darmstadt, Germany). The fusion proteins carrying thioredoxin (Trx), hexahistidine (His6) and S tags in this order on the N terminus were expressed in E. coli Rosetta-gami B(DE3) (Novagen) (for trefoil domain of ZP1, ZP-N and ZP-C domains of ZP3, and EGF-like domain of ZPD) or Rosetta 2(DE3) (Novagen) (for other fusion proteins) according to the manufacturer’s instructions. The cell pellet were resuspended in the sonication buffer (PBS containing 0.50 M NaCl and 0.20% Triton X-100) and sonicated as described above followed by centrifugation at 12,000g for 10 min at 4 °C. Soluble fusion proteins in the supernatant (repeat and trefoil domains of ZP1, N-terminal peptide of ZP3, and EGF-like domain of ZPD) were purified by nickel-affinity chromatography, according to the manufacturer’s instructions, dialyzed against PBS and stored at −20 °C before use. The precipitates were washed three times with the inclusion-body (IB) wash buffer (20 mM Tris–HCl, pH7.5, containing 10 mM EDTA and 1.0% Triton X-100) to isolate IBs containing insoluble fusion proteins (N-terminal domain of ZP1, ZP-N and ZP-C domains, the ZP-N + IHP and the ZP-C − IHP of all three ZP glycoproteins). The control protein (vector-encoded protein) was expressed in E. coli BL21(DE3) (Novagen) being transformed with the empty pET-32a(+) vector (Novagen) as a soluble protein, and purified similarly as described above.
4.4. Protein refolding
The recombinant proteins were subjected to the unfolding/refolding reactions. The IBs of insoluble fusion proteins were suspended at a concentration of 20 mg wet weight/ml in the IB solubilization buffer (50 mM CAPS, pH 11.0, containing 0.30% N-lauroylsarcosine) without any additive (for the ZP-N + IHP of both ZP1 and ZPD: group A), and the buffer supplemented with 4.5 M urea (for N-terminal domain of ZP1, ZP-N and ZP-C domains of all three ZP glycoproteins, and the ZP-N + IHP of ZP3: group B), 2.0 mM DTT (for the ZP-C − IHPs of both ZP1 and ZPD: group C) or 0.5 mM DTT (for the ZP-C − IHP of ZP3: group D). After incubation at room temperature (RT) for 15 min, the IB solutions containing solubilized fusion proteins were collected by centrifugation of the IB suspension at 10,000g for 10 min at 4 °C and subjected to multi-step dialysis method for refolding. The IB solutions were dialyzed against 250 times the volumes of the sample of the dialysis buffer (20 mM Tris–HCl, pH 8.5) (for the group A) and that supplemented with 2.0 M urea (for the group B), 0.20 mM DTT (for the group C) or 0.050 mM DTT (for the group D) for 16 h at 4 °C and subsequently against same volumes of the buffer (for the group A) and that supplemented with 1.0 M urea (for the group B), 0.10 mM DTT (for the group C) or 0.025 mM DTT (for the group D) for 3 h at 4 °C. The dialysis was continued through two additional changes with the same volumes of the dialysis buffer in the absence of urea and DTT for 3 and 16 h at 4 °C, respectively. The refolded protein solutions were centrifuged at 15,500g for 5 min at 4 °C to remove insoluble materials and stored at −20 °C before use.
4.5. Production of antisera against the domains of chicken ZP glycoproteins
Polyclonal antisera were raised in mice against the above-mentioned recombinant ZP glycoproteins. Mice were immunized and the antisera were collected as previously described [47]. Trx and His6 tags (but not S tag) were removed using Thrombin Cleavage Capture Kit (Novagen) from the recombinant trefoil domain of ZP1, N-terminal peptide of ZP3, and EGF-like and ZP-N domains of ZPD prior to the immunization. The antisera against the N-terminal, repeat and trefoil domains of ZP1 (anti-ZP1_N-terminal, repeat, and trefoil, respectively), the N-terminal peptide of ZP3 (anti-ZP3_N-terminal), the EGF-like domain of ZPD (anti-ZPD_EGF), the N-terminal half of ZP domain in ZP1 and ZP3 (anti-ZP1_ZP-N and anti-ZP3_ZP-N, respectively), the ZP-N of ZPD (anti-ZPD_ZP-N), C-terminal halves of ZP domain in ZP1, ZP3 and ZPD (anti-ZP1_ZP-C, anti-ZP3_ZP-C and anti-ZPD_ZP-C, respectively) and the control protein (anti-tag) were produced. Reactivity of the antisera was confirmed by immunoblotting of ZP glycoproteins being separated by SDS–PAGE under reducing and non-reducing conditions (Supplement Fig. S1).
4.6. Immunofluorescent staining and confocal laser-scanning microscopy of chicken egg coat
Small pieces of isolated egg coat were mounted onto aminosilane-coated glass slides, incubated with PBS containing 0.10% Triton X-100 at RT for 30 min and washed with cold PBS followed by incubation with 2.0% BSA in PBS (2.0% BSA/PBS) at 4 °C overnight for blocking. The BSA-blocked egg coat were incubated with 2.0% BSA/PBS containing the anti-tag (1:1000), the anti-ZP1_N-terminal (1:200), repeat (1:2400), trefoil (1:800), ZP-N (1:20) or ZP-C (1:100), the anti-ZP3_N-terminal (1:10), ZP-N (1:100) or ZP-C (1:200), or the anti-ZPD_EGF (1:100), ZP-N (1:600) or ZP-C (1:400) and washed with PBS followed by final incubation with 2% BSA/PBS containing Alexa Fluor® 488 goat anti-mouse IgG (6.7 μg/ml; Molecular Probes, Eugene, OR) at RT for 30 min in darkness. After washing with PBS and the wash buffer (NET; 150 mM NaCl, 5 mM EDTA, 50 mM Tris–HCl, pH 8.0, 0.05% Triton X-100), the egg coat was observed under the confocal laser-scanning microscopy. Imaging was performed on a Zeiss Axio Obserber.Z microscope equipped with LSM 700 laser scanning confocal optics (Carl Zeiss, Thornwood, NY). A 488-nm diode laser excitation in combination with a 490–635 nm band-pass emission filter was used for imaging the fluorescent probes. Differential-interference-contrast (DIC) microscopy images were taken on the same system.
4.7. Co-immunoprecipitation assay
Protein G magnetic beads (25 μl of 50% slurry; New England Biolabs, Beverly, MA) were prepared according to the manufacturer’s instructions. The beads were incubated for 24 h at 4 °C with gentle agitation in 300 μl of binding buffer (0.1 M sodium phosphate, pH 8.0) with each of the above-mentioned antisera to couple antibodies to the beads, pulled by magnetic field, and washed three times with 500 μl of the binding buffer. On the other hand, the chicken serum containing 5.8 μg of ZP1 was mixed with the ZP3 solution containing 6.4 μg of ZP3 according to the previously defined ZP1/ZP3 ratio [47], incubated at 37 °C for 16 h to form the ZP1–ZP3 complexes, diluted up to 300 μl with the binding buffer, and incubated with the beads, without the antibody coupling for 24 h at 4 °C with gentle agitation. The beads were pulled by magnetic field to remove nonspecifically bound proteins, and the supernatants or 300 μl of the binding buffer were incubated with the above-mentioned antibody-bound beads similarly. The beads were pulled and washed similarly, and half of the immunoprecipitates on the beads were subjected to SDS–PAGE.
4.8. Pull-down assay
PureProteome™ Nickel Magnetic Beads (30 μl of the 75–80 wt% suspension; Merck Millipore, Billerica, MA) were equilibrated in the pull-down binding buffer (50 mM phosphate buffer, pH 8.0, containing 300 mM NaCl) according to the manufacturer’s instructions. The equilibrated beads were resuspended in 1.0 ml of PBS containing 6.0 nM of the His6-tagged proteins or 1.0 ml of 20 mM Tris–HCl, pH 8.5, followed by incubation with gentle agitation for 16 h at 4 °C, pulled by magnetic field and washed 3 times with 150 μl of the pull-down binding buffer. The beads carrying 50 pmol of the fusion proteins or the above-mentioned control protein were incubated in 300 μl of PBS with or without 50% chicken serum, pulled similarly and washed with the pull-down binding buffer as described above. The beads were finally incubated in 10 μl of PBS containing 4.0% Triton X-100 (TX-100) for 60 min at RT and pulled similarly, and 2.0 μl of the TX-100 eluate and one fifth of the beads carrying proteins being retained after the TX-100 treatment were subjected to SDS–PAGE.
4.9. Gel electrophoresis, immunoblotting, and far-Western (ligand) blotting
SDS–PAGE was performed according to the method of Laemmli [65]. The protein samples obtained by the co-immunoprecipitation and pull-down assays, 5.0 μl of the crude ZPD solution, the ZP3 solution containing 1.0 μg of ZP3, 0.5–1.0 μl of the chicken serum and 15 μg wet weight of the egg coat were incubated at 100 °C for 5 min in SDS–PAGE sample buffer (62.5 mM Tris–HCl, pH6.8, containing 2.0% SDS, 0.0020% bromophenol blue, 10% glycerol) in the presence or absence of 42 mM DTT (reducing or non-reducing conditions, respectively) and loaded onto the polyacrylamide gel following removal of the insoluble materials or the beads by centrifugation or magnetic field, respectively. Proteins in the gel were electroblotted onto a nitrocellulose membrane for immunoblotting and far-Western (ligand) blotting. Coomassie Brilliant Blue (CBB) staining, immunoblotting and far-Western blotting were performed as previously described [47], except that, for far-Western blotting, the gelatin-blocked membranes were preincubated in the incubation buffer (NETG; 150 mM NaCl, 5 mM EDTA, 50 mM Tris–HCl, pH8.0, 0.05% Triton X-100, 0.25% gelatin) containing 10.6–82.1 nM of the above-mentioned recombinant proteins as probes at 4 °C for 16 h prior to incubation with primary antibodies. Furthermore, to examine whether the binding affinities of the probes to the protein bands on the membrane were affected by the antisera against the ZP-N and ZP-C domains, the blocked membranes were preincubated with the anti-ZP3_ZP-N (1:50) anti-ZP3_ZP-C (1:1000), anti-ZPD_ZP-N (1:3000) or anti-ZPD_ZP-C (1:2000) at RT for 60 min prior to incubation with the probes. The above-mentioned mouse anti-ZP1_repeat (1:2000), mouse anti-ZP3_ZP-C (1:2000) or rabbit anti-Trx (Anti-Thioredoxin antibody produced in rabbit; Sigma–Aldrich, St. Louis, MO) (1:10,000) were used for primary antibodies, and POD-linked anti-mouse or anti-rabbit IgG (Amersham ECL™ Anti-mouse and Anti-rabbit IgG Peroxidase-linked species specific whole antibody (from sheep or donkey, respectively); GE Healthcare) (1:10,000) were used for secondary antibody, unless otherwise noted.
Author contributions
H.O., T.S., R.S. and H.F. performed the experiments; H.O. analyzed the data; H.O. and T.M. designed the research and experiments; H.O., T.M. and M.U. drafted the paper; H.O. directed the research and wrote the final paper.
Acknowledgements
We thank Dr. Ken-ichiro Minato (Meijo University) for helpful instructions for the care and use of laboratory animals. This work is supported in part by Grants-in-Aid for Young Scientists (B) and Scientific Research (C) [Grant Numbers 22780255 and 25450520, respectively] from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Appendix A. Supplementary data
This supplementary data contains supplementary information.
References
- 1.Rankin T., Dean J. The zona pellucida: using molecular genetics to study the mammalian egg coat. Rev. Reprod. 2000;5:114–121. doi: 10.1530/ror.0.0050114. [DOI] [PubMed] [Google Scholar]
- 2.Monné M., Han L., Jovine L. Tracking down the ZP domain: from the mammalian zona pellucida to the molluscan vitelline envelope. Semin. Reprod. Med. 2006;24:204–216. doi: 10.1055/s-2006-948550. [DOI] [PubMed] [Google Scholar]
- 3.Hardy D.M., Garbers D.L. A sperm membrane protein that binds in a species-specific manner to the egg extracellular matrix is homologous to von Willebrand factor. J. Biol. Chem. 1995;270:26025–26028. doi: 10.1074/jbc.270.44.26025. [DOI] [PubMed] [Google Scholar]
- 4.Gao Z., Garbers D.L. Species diversity in the structure of zonadhesin, a sperm-specific membrane protein containing multiple cell adhesion molecule-like domains. J. Biol. Chem. 1998;273:3415–3421. doi: 10.1074/jbc.273.6.3415. [DOI] [PubMed] [Google Scholar]
- 5.Ensslin M.A., Shur B.D. Identification of mouse sperm SED1, a bimotif EGF repeat and discoidin-domain protein involved in sperm–egg binding. Cell. 2003;114:405–417. doi: 10.1016/s0092-8674(03)00643-3. [DOI] [PubMed] [Google Scholar]
- 6.Kim E., Baba D., Kimura M., Yamashita M., Kashiwabara S., Baba T. Identification of a hyaluronidase, Hyal5, involved in penetration of mouse sperm through cumulus mass. Proc. Natl. Acad. Sci. U.S.A. 2005;102:18028–18033. doi: 10.1073/pnas.0506825102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamaguchi R., Yamagata K., Ikawa M., Moss S.B., Okabe M. Aberrant distribution of ADAM3 in sperm from both angiotensin-converting enzyme (Ace)- and calmegin (Clgn)-deficient mice. Biol. Reprod. 2006;75:760–766. doi: 10.1095/biolreprod.106.052977. [DOI] [PubMed] [Google Scholar]
- 8.Maldera J.A., WeigelMunoz M., Chirinos M., Busso D., Raffo F G.E., Battistone M.A., Blaquier J.A., Larrea F., Cuasnicu P.S. Human fertilization: epididymal hCRISP1 mediates sperm–zona pellucida binding through its interaction with ZP3. Mol. Hum. Reprod. 2014;20:341–349. doi: 10.1093/molehr/gat092. [DOI] [PubMed] [Google Scholar]
- 9.Jones R. Evidence for boar sperm proacrosin as a carbohydrate binding protein. Cell Biol. Int. Rep. 1987;11:833. doi: 10.1016/0309-1651(87)90164-0. [DOI] [PubMed] [Google Scholar]
- 10.Baba T., Niida Y., Michikawa Y., Kashiwabara S., Kodaira K., Takenaka M., Kohno N., Gerton G.L., Arai Y. An acrosomal protein, sp32, in mammalian sperm is a binding protein specific for two proacrosins and an acrosin intermediate. J. Biol. Chem. 1994;269:10133–10140. [PubMed] [Google Scholar]
- 11.Burkart A.D., Xiong B., Baibakov B., Jimenez-Movilla M., Dean J. Ovastacin, a cortical granule protease, cleaves ZP2 in the zona pellucida to prevent polyspermy. J. Cell Biol. 2012;197:37–44. doi: 10.1083/jcb.201112094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inoue N., Satouh Y., Ikawa M., Okabe M., Yanagimachi R. Acrosome-reacted mouse spermatozoa recovered from the perivitelline space can fertilize other eggs. Proc. Natl. Acad. Sci. U.S.A. 2011;108:20008–20011. doi: 10.1073/pnas.1116965108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yanagimachi R. Mammalian sperm acrosome reaction: where does it begin before fertilization? Biol. Reprod. 2011;85:4–5. doi: 10.1095/biolreprod.111.092601. [DOI] [PubMed] [Google Scholar]
- 14.Bork P., Sander C. A large domain common to sperm receptors (Zp2 and Zp3) and TGF-beta type III receptor. FEBS Lett. 1992;300:237–240. doi: 10.1016/0014-5793(92)80853-9. [DOI] [PubMed] [Google Scholar]
- 15.Jovine L., Qi H., Williams Z., Litscher E.S., Wassarman P.M. A duplicated motif controls assembly of zona pellucida domain proteins. Proc. Natl. Acad. Sci. U.S.A. 2004;101:5922–5927. doi: 10.1073/pnas.0401600101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han L., Monné M., Okumura H., Schwend T., Cherry A.L., Flot D., Matsuda T., Jovine L. Insights into egg coat assembly and egg-sperm interaction from the X-ray structure of full-length ZP3. Cell. 2010;143:404–415. doi: 10.1016/j.cell.2010.09.041. [DOI] [PubMed] [Google Scholar]
- 17.Spargo S.C., Hope R.M. Evolution and nomenclature of the zona pellucida gene family. Biol. Reprod. 2003;68:358–362. doi: 10.1095/biolreprod.102.008086. [DOI] [PubMed] [Google Scholar]
- 18.Goudet G., Mugnier S., Callebaut I., Monget P. Phylogenetic analysis and identification of pseudogenes reveal a progressive loss of zona pellucida genes during evolution of vertebrates. Biol. Reprod. 2008;78:796–806. doi: 10.1095/biolreprod.107.064568. [DOI] [PubMed] [Google Scholar]
- 19.Callebaut I., Mornon J.P., Monget P. Isolated ZP-N domains constitute the N-terminal extensions of Zona Pellucida proteins. Bioinformatics. 2007;23:1871–1874. doi: 10.1093/bioinformatics/btm265. [DOI] [PubMed] [Google Scholar]
- 20.Bausek N., Waclawek M., Schneider W.J., Wohlrab F. The major chicken egg envelope protein ZP1 is different from ZPB and is synthesized in the liver. J. Biol. Chem. 2000;275:28866–28872. doi: 10.1074/jbc.275.37.28866. [DOI] [PubMed] [Google Scholar]
- 21.Hoodbhoy T., Joshi S., Boja E.S., Williams S.A., Stanley P., Dean J. Human sperm do not bind to rat zonae pellucidae despite the presence of four homologous glycoproteins. J. Biol. Chem. 2005;280:12721–12731. doi: 10.1074/jbc.M413569200. [DOI] [PubMed] [Google Scholar]
- 22.Izquierdo-Rico M.J., Jiménez-Movilla M., Llop E., Pérez-Oliva A.B., Ballesta J., Gutiérrez-Gallego R., Jiménez-Cervantes C., Avilés M. Hamster zona pellucida is formed by four glycoproteins: ZP1, ZP2, ZP3, and ZP4. J. Proteome Res. 2009;8:926–941. doi: 10.1021/pr800568x. [DOI] [PubMed] [Google Scholar]
- 23.Lindsay L.L., Yang J.C., Hedrick J.L. Identification and characterization of a unique Xenopus laevis egg envelope component, ZPD. Dev. Growth Differ. 2002;44:205–212. doi: 10.1046/j.1440-169x.2002.00635.x. [DOI] [PubMed] [Google Scholar]
- 24.Okumura H., Kohno Y., Iwata Y., Mori H., Aoki N., Sato C., Kitajima K., Nadano D., Matsuda T. A newly identified zona pellucida glycoprotein, ZPD, and dimeric ZP1 of chicken egg envelope are involved in sperm activation on sperm–egg interaction. Biochem. J. 2004;384:191–199. doi: 10.1042/BJ20040299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sato T., Kinoshita M., Kansaku N., Tahara K., Tsukada A., Ono H., Yoshimura T., Dohra H., Sasanami T. Molecular characterization of egg envelope glycoprotein ZPD in the ovary of Japanese quail (Coturnix japonica) Reproduction. 2009;137:333–343. doi: 10.1530/REP-08-0057. [DOI] [PubMed] [Google Scholar]
- 26.Jovine L., Darie C.C., Litscher E.S., Wassarman P.M. Zona pellucida domain proteins. Annu. Rev. Biochem. 2005;74:83–114. doi: 10.1146/annurev.biochem.74.082803.133039. [DOI] [PubMed] [Google Scholar]
- 27.Darie C.C., Biniossek M.L., Jovine L., Litscher E.S., Wassarman P.M. Structural characterization of fish egg vitelline envelope proteins by mass spectrometry. Biochemistry. 2004;43:7459–7478. doi: 10.1021/bi0495937. [DOI] [PubMed] [Google Scholar]
- 28.Sano K., Kawaguchi M., Yoshikawa M., Iuchi I., Yasumasu S. Evolution of the teleostean zona pellucida gene inferred from the egg envelope protein genes of the Japanese eel, Anguilla japonica. FEBS J. 2010;277:4674–4684. doi: 10.1111/j.1742-4658.2010.07874.x. [DOI] [PubMed] [Google Scholar]
- 29.Sano K., Kawaguchi M., Watanabe S., Nagakura Y., Hiraki T., Yasumasu S. Inferring the evolution of teleostean ZP genes based on their sites of expression. J. Exp. Zool. B: Mol. Dev. Evol. 2013;320:332–343. doi: 10.1002/jez.b.22507. [DOI] [PubMed] [Google Scholar]
- 30.Lindsay L.L., Wallace M.A., Hedrick J.L. A hatching enzyme substrate in the Xenopus laevis egg envelope is a high molecular weight ZPA homolog. Dev. Growth Differ. 2001;43:305–313. doi: 10.1046/j.1440-169x.2001.00577.x. [DOI] [PubMed] [Google Scholar]
- 31.Mann K. Proteomic analysis of the chicken egg vitelline membrane. Proteomics. 2008;8:2322–2332. doi: 10.1002/pmic.200800032. [DOI] [PubMed] [Google Scholar]
- 32.Lefiévre L., Conner S.J., Salpekar A., Olufowobi O., Ashton P., Pavlovic B., Lenton W., Afnan M., Brewis I.A., Monk M., Hughes D.C., Barratt C.L. Four zona pellucida glycoproteins are expressed in the human. Hum. Reprod. 2004;19:1580–1586. doi: 10.1093/humrep/deh301. [DOI] [PubMed] [Google Scholar]
- 33.Kudo K., Yonezawa N., Katsumata T., Aoki H., Nakano M. Localization of carbohydrate chains of pig sperm ligand in the glycoprotein ZPB of egg zona pellucida. Eur. J. Biochem./FEBS. 1998;252:492–499. doi: 10.1046/j.1432-1327.1998.2520492.x. [DOI] [PubMed] [Google Scholar]
- 34.Kanai S., Yonezawa N., Ishii Y., Tanokura M., Nakano M. Recombinant bovine zona pellucida glycoproteins ZP3 and ZP4 coexpressed in Sf9 cells form a sperm-binding active hetero-complex. FEBS J. 2007;274:5390–5405. doi: 10.1111/j.1742-4658.2007.06065.x. [DOI] [PubMed] [Google Scholar]
- 35.Blackmore D.G., Baillie L.R., Holt J.E., Dierkx L., Aitken R.J., McLaughlin E.A. Biosynthesis of the canine zona pellucida requires the integrated participation of both oocytes and granulosa cells. Biol. Reprod. 2004;71:661–668. doi: 10.1095/biolreprod.104.028779. [DOI] [PubMed] [Google Scholar]
- 36.Bleil J.D., Wassarman P.M. Mammalian sperm–egg interaction: identification of a glycoprotein in mouse egg zonae pellucidae possessing receptor activity for sperm. Cell. 1980;20:873–882. doi: 10.1016/0092-8674(80)90334-7. [DOI] [PubMed] [Google Scholar]
- 37.Florman H.M., Wassarman P.M. O-linked oligosaccharides of mouse egg ZP3 account for its sperm receptor activity. Cell. 1985;41:313–324. doi: 10.1016/0092-8674(85)90084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Visconti P.E., Florman H.M. Mechanisms of sperm–egg interactions: between sugars and broken bonds. Sci. Signal. 2010;3:pe35. doi: 10.1126/scisignal.3142pe35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clark G.F. The role of carbohydrate recognition during human sperm–egg binding. Hum. Reprod. 2013;28:566–577. doi: 10.1093/humrep/des447. [DOI] [PubMed] [Google Scholar]
- 40.Bleil J.D., Greve J.M., Wassarman P.M. Identification of a secondary sperm receptor in the mouse egg zona pellucida: role in maintenance of binding of acrosome-reacted sperm to eggs. Dev. Biol. 1988;128:376–385. doi: 10.1016/0012-1606(88)90299-0. [DOI] [PubMed] [Google Scholar]
- 41.Avella M.A., Baibakov B., Dean J. A single domain of the ZP2 zona pellucida protein mediates gamete recognition in mice and humans. J. Cell Biol. 2014;205:801–809. doi: 10.1083/jcb.201404025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams Z., Litscher E.S., Jovine L., Wassarman P.M. Polypeptide encoded by mouse ZP3 exon-7 is necessary and sufficient for binding of mouse sperm in vitro. J. Cell. Physiol. 2006;207:30–39. doi: 10.1002/jcp.20532. [DOI] [PubMed] [Google Scholar]
- 43.Bansal P., Chakrabarti K., Gupta S.K. Functional activity of human ZP3 primary sperm receptor resides toward its C-terminus. Biol. Reprod. 2009;81:7–15. doi: 10.1095/biolreprod.108.074716. [DOI] [PubMed] [Google Scholar]
- 44.Wassarman P.M. Zona pellucida glycoproteins. Annu. Rev. Biochem. 1988;57:415–442. doi: 10.1146/annurev.bi.57.070188.002215. [DOI] [PubMed] [Google Scholar]
- 45.Leyton L., Saling P. Evidence that aggregation of mouse sperm receptors by ZP3 triggers the acrosome reaction. J. Cell Biol. 1989;108:2163–2168. doi: 10.1083/jcb.108.6.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takeuchi Y., Cho R., Iwata Y., Nishimura K., Kato T., Aoki N., Kitajima K., Matsuda T. Morphological and biochemical changes of isolated chicken egg-envelope during sperm penetration: degradation of the 97-kilodalton glycoprotein is involved in sperm-driven hole formation on the egg-envelope. Biol. Reprod. 2001;64:822–830. doi: 10.1095/biolreprod64.3.822. [DOI] [PubMed] [Google Scholar]
- 47.Okumura H., Aoki N., Sato C., Nadano D., Matsuda T. Heterocomplex formation and cell-surface accumulation of hen’s serum zona pellucida B1 (ZPB1) with ZPC expressed by a mammalian cell line (COS-7): a possible initiating step of egg-envelope matrix construction. Biol. Reprod. 2007;76:9–18. doi: 10.1095/biolreprod.106.056267. [DOI] [PubMed] [Google Scholar]
- 48.Okumura H., Okajima T., Nadano D., Matsuda T. Association of chicken zona pellucida glycoprotein (ZP) B1 with ZPC induces formation of ZPB1-ZPC fibrous aggregates containing disulfide-bridged ZPB1 dimer. Biochem. Biophys. Res. Commun. 2007;364:682–688. doi: 10.1016/j.bbrc.2007.10.072. [DOI] [PubMed] [Google Scholar]
- 49.Wassarman P.M. Fertilization in mammals. Sci. Am. 1988;259:78–84. doi: 10.1038/scientificamerican1288-78. [DOI] [PubMed] [Google Scholar]
- 50.Monné M., Han L., Schwend T., Burendahl S., Jovine L. Crystal structure of the ZP-N domain of ZP3 reveals the core fold of animal egg coats. Nature. 2008;456:653–657. doi: 10.1038/nature07599. [DOI] [PubMed] [Google Scholar]
- 51.Diestel U., Resch M., Meinhardt K., Weiler S., Hellmann T.V., Mueller T.D., Nickel J., Eichler J., Muller Y.A. Identification of a Novel TGF-beta-Binding Site in the Zona Pellucida C-terminal (ZP-C) Domain of TGF-beta-Receptor-3 (TGFR-3) PLoS ONE. 2013;8:e67214. doi: 10.1371/journal.pone.0067214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin S.J., Hu Y., Zhu J., Woodruff T.K., Jardetzky T.S. Structure of betaglycan zona pellucida (ZP)-C domain provides insights into ZP-mediated protein polymerization and TGF-beta binding. Proc. Natl. Acad. Sci. U.S.A. 2011;108:5232–5236. doi: 10.1073/pnas.1010689108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Okumura H., Fukushima H., Momoda M., Ima Y., Matsuda T., Ujita M. Diverse lectin-binding specificity of four ZP3 glycoprotein isoforms with a discrete isoelectric point in chicken egg coat. Biochem. Biophys. Res. Commun. 2012;424:586–592. doi: 10.1016/j.bbrc.2012.06.157. [DOI] [PubMed] [Google Scholar]
- 54.Takeuchi Y., Nishimura K., Aoki N., Adachi T., Sato C., Kitajima K., Matsuda T. A 42-kDa glycoprotein from chicken egg-envelope, an avian homolog of the ZPC family glycoproteins in mammalian Zona pellucida. Its first identification, cDNA cloning and granulosa cell-specific expression. Eur. J. Biochem./FEBS. 1999;260:736–742. doi: 10.1046/j.1432-1327.1999.00203.x. [DOI] [PubMed] [Google Scholar]
- 55.Pökkylä R.M., Lakkakorpi J.T., Nuojua-Huttunen S.H., Tapanainen J.S. Sequence variations in human ZP genes as potential modifiers of zona pellucida architecture. Fertil. Steril. 2011;95:2669–2672. doi: 10.1016/j.fertnstert.2011.01.168. [DOI] [PubMed] [Google Scholar]
- 56.Margalit M., Paz G., Yavetz H., Yogev L., Amit A., Hevlin-Schwartz T., Gupta S.K., Kleiman S.E. Genetic and physiological study of morphologically abnormal human zona pellucida. Eur. J. Obstet. Gynecol. Reprod. Biol. 2012;165:70–76. doi: 10.1016/j.ejogrb.2012.07.022. [DOI] [PubMed] [Google Scholar]
- 57.Sousa M., da Silva J.T., Silva J., Cunha M., Viana P., Oliveira E., Sa R., Soares C., Oliveira C., Barros A. Embryological, clinical and ultrastructural study of human oocytes presenting indented zona pellucida. Zygote. 2013:1–13. doi: 10.1017/S0967199413000403. [DOI] [PubMed] [Google Scholar]
- 58.Monné M., Jovine L. A structural view of egg coat architecture and function in fertilization. Biol. Reprod. 2011;85:661–669. doi: 10.1095/biolreprod.111.092098. [DOI] [PubMed] [Google Scholar]
- 59.Polshakov V.I., Williams M.A., Gargaro A.R., Frenkiel T.A., Westley B.R., Chadwick M.P., May F.E., Feeney J. High-resolution solution structure of human pNR-2/pS2: a single trefoil motif protein. J. Mol. Biol. 1997;267:418–432. doi: 10.1006/jmbi.1997.0896. [DOI] [PubMed] [Google Scholar]
- 60.Downing A.K., Knott V., Werner J.M., Cardy C.M., Campbell I.D., Handford P.A. Solution structure of a pair of calcium-binding epidermal growth factor-like domains: implications for the Marfan syndrome and other genetic disorders. Cell. 1996;85:597–605. doi: 10.1016/s0092-8674(00)81259-3. [DOI] [PubMed] [Google Scholar]
- 61.Familiari G., Relucenti M., Heyn R., Micara G., Correr S. Three-dimensional structure of the zona pellucida at ovulation. Microsc. Res. Tech. 2006;69:415–426. doi: 10.1002/jemt.20301. [DOI] [PubMed] [Google Scholar]
- 62.Santambrogio S., Cattaneo A., Bernascone I., Schwend T., Jovine L., Bachi A., Rampoldi L. Urinary uromodulin carries an intact ZP domain generated by a conserved C-terminal proteolytic cleavage. Biochem. Biophys. Res. Commun. 2008;370:410–413. doi: 10.1016/j.bbrc.2008.03.099. [DOI] [PubMed] [Google Scholar]
- 63.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Säemann M.D., Weichhart T., Horl W.H., Zlabinger G.J. Tamm–Horsfall protein: a multilayered defence molecule against urinary tract infection. Eur. J. Clin. Invest. 2005;35:227–235. doi: 10.1111/j.1365-2362.2005.01483.x. [DOI] [PubMed] [Google Scholar]
- 65.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This supplementary data contains supplementary information.


