Abstract
Treponema denticola has been recognized as an important oral pathogen of the “red complex” bacterial consortium that is associated with the pathogenesis of endodontal and periodontal diseases. However, little is known about the virulence of T. denticola due to its recalcitrant genetic system. The difficulty in genetically manipulating oral spirochetes is partially due to the lack of antibiotic resistance cassettes that are useful for gene complementation following allelic replacement mutagenesis. In this study, a kanamycin resistance cassette was identified and developed for the genetic manipulation of T. denticola ATCC 35405. Compared to the widely used ermF-ermAM cassette, the kanamycin cassette used in the transformation experiments gave rise to additional antibiotic-resistant T. denticola colonies. The kanamycin cassette is effective for allelic replacement mutagenesis as demonstrated by inactivation of two open reading frames of T. denticola, TDE1430 and TDE0911. In addition, the cassette is also functional in trans-chromosomal complementation. This was determined by functional rescue of a periplasmic flagellum (PF)-deficient mutant that had the flgE gene coding for PF hook protein inactivated. The integration of the full-length flgE gene into the genome of the flgE mutant rescued all of the defects associated with the flgE mutant that included the lack of PF filament and spirochetal motility. Taken together, we demonstrate that the kanamycin resistance gene is a suitable cassette for the genetic manipulation of T. denticola that will facilitate the characterization of virulence factors attributed to this important oral pathogen.
INTRODUCTION
Treponema denticola, a Gram-negative, spiral-shaped, obligate anaerobe, is a virulent and invasive spirochete that is associated with advanced periodontal disease (1–3). In addition, T. denticola, along with other “red complex” bacteria, contributes to endodontal infection both in humans and in experimental animals (4–6). Although T. denticola primarily cause infections of the oral cavity, they have also been associated with the development of cardiovascular disease (7–10) and other systemic conditions (11, 12). A number of putative pathogenic attributes that include: the establishment of a stable biofilm, invasive motility, and the production of various proteases and toxins, have been studied in vitro (4, 13–16). Moreover, the use of animal models (17) have revealed additional insight into their pathogenesis (14, 18–22). Efforts to further our understanding of T. denticola biology have been impeded by the fastidious nature and specific genetic restriction modification system of this oral spirochete, particularly in the widely used strain T. denticola ATCC 35405, which has a sequenced and fully annotated genome (23, 24, 25).
Several attempts have been made to develop a reproducible genetic system to study T. denticola (26–29). The ermF-ermAM cassette, originally developed for the mutagenesis of another oral pathogen Porphyromonas gingivalis (27), has frequently been used as a selective marker to inactivate T. denticola genes (30–33). The ermF-ermAM cassette conferred erythromycin resistance in both Escherichia coli and T. denticola (27, 34). Recently, a simplified ermAM cassette has been shown to be effective in the mutagenesis of T. denticola (34). In addition, coumermycin and chloramphenicol cassettes have been used in a different strain, T. denticola ATCC 33520, for the transformation of shuttle plasmids that generally do not replicate in T. denticola 35405. Only erythromycin and gentamicin cassettes have been suitable for the manipulation of the sequenced and annotated strain, T. denticola 35405 (29, 35, 36). The development of a reliable antibiotic cassette is essential for the genetic study of this important oral pathogen since complementation has been problematic.
We screened a variety of antibiotic resistance cassettes and found a kanamycin resistance gene that is a reliable selective marker for T. denticola 35405 and demonstrated the utility of a kanamycin resistance cassette in the mutagenesis of T. denticola genes. Furthermore, we provided evidence this cassette can be used in the complementation of a nonmotile flgE mutant resulting in a fully restored periplasmic flagellum (PF), enabling a motile phenotype (37).
MATERIALS AND METHODS
Culture and growth conditions of T. denticola.
T. denticola ATCC 35405 (American Type Culture Collection, Manassas, VA) cells were inoculated in new spirochete medium (NOS) supplemented with 10% heat-inactivated rabbit serum and incubated at 35°C in an anaerobic chamber (Coy Laboratory Products, Inc., Ann Arbor, MI) with an atmosphere of 80% nitrogen, 10% carbon dioxide, and 10% hydrogen (38).
DNA manipulation and construction of plasmids.
T. denticola genomic DNA was extracted by using an UltraClean microbial DNA isolation kit (catalog no. 12224-50; MO BIO Laboratories, Inc., Carlsbad, CA). All PCR products were amplified by using GoTaq Green master mix (Promega, Madison, WI) except where specifically noted. All primers used were listed in Table 1 and synthesized by Sigma-Aldrich with normal desalting purification. Plasmids and PCR products were purified by using a QIAprep spin miniprep kit and a QIAquick gel extraction kit (Qiagen, Inc., Valencia, CA), respectively. Larger-scale plasmid purification was conducted using the HiPure plasmid midiprep kit (Invitrogen, San Diego, CA). All restriction digestion enzymes were purchased from Promega. DNA sequence validation of various plasmid constructs were performed by the DNA sequencing facility at Genomics Core Laboratories, The University of Alabama at Birmingham, Birmingham, AL.
TABLE 1.
Primers used in this study
aphA, aminoglycoside phosphate hydrolase; cat, chloramphenicol acetyltransferase; gat, gentamicin acetyltransferase.
The sites for restriction digestion enzymes are underlined.
All plasmids and strains constructed in the present study are listed in Table 2. Experimental procedures used to construct each plasmid and bacterial strain are described below. The ErmF promoter was used to drive the expression of different antibiotic cassettes that were cloned into a pGEM-T vector. In brief, the ErmF promoter (P1) was PCR amplified by using the primer set of Promoter1-KpnI-BamHI-F and Promoter1-AgeI-XhoI-XbaI-R and cloned into the pGEM-T vector at the XbaI and SpeI sites of the pGEM-T vector. Various antibiotic resistance genes (ARGs) were subsequently ligated into this plasmid backbone using two restriction enzyme sites, AgeI and XhoI, to generate constructs harboring different antibiotic resistance cassettes, including two kanamycin resistance genes (aphA1 [GenBank accession no. NP_478154] and aphA2 [GenBank accession WP_004614937]; Clontech Laboratories, Inc., Mountain View, CA), a chloramphenicol acetyltransferase gene (cat [GenBank accession no. NP_052903]), and a gentamicin resistance gene (gat [GenBank accession no. P23181] synthesized by GenScript [Piscataway, NJ]; the sequence was optimized according to T. denticola 35405 codon usage), respectively (plasmids I, II, III, and IV; see Table 2 and Fig. 1A).
TABLE 2.
Plasmids and strains used in this study
| Plasmida or strain | Relevant characteristic(s)b | Source or reference |
|---|---|---|
| Plasmids | ||
| pGEM-T | Cloning vector; Ampr | Promega |
| pGEM-T Easy | Cloning vector; Ampr | Promega |
| pSY118 | Donor vector of ermF-ermAM; Ermr | 27 |
| pGEM-T-P1aphA1* (I) | Kanamycin 1 cassette in pGEM-T; Ampr | This study |
| pGEM-T-P1aphA2† (II) | Kanamycin 2 cassette in pGEM-T; Ampr | This study |
| pGEM-T-P1cat‡ (III) | Chloramphenicol cassette in pGEM-T; Ampr | This study |
| pGEM-T-P1gat§ (IV) | Gentamicin cassette in pGEM-T; Ampr | This study |
| pGEM-T Easy-TDE1430 (V) | Containing TDE1430 and its flanking regions; Ampr | This study |
| pGEM-T Easy-TDE1430-ermF-ermAM (VI) | Replacement of TDE1430 by ermF-ermAM; Ampr | This study |
| pGEM-T Easy-TDE1430-P1aphA1 (VII) | Replacement of TDE1430 by P1aphA1; Ampr | This study |
| pGEM-T Easy-TDE1430-P1aphA2 (VIII) | Replacement of TDE1430 by P1aphA2; Ampr | This study |
| pGEM-T Easy-TDE1430-P1cat (IX) | Replacement of TDE1430 by P1cat; Ampr | This study |
| pGEM-T Easy-TDE1430-P1gat (X) | Replacement of TDE1430 by P1gat; Ampr | This study |
| pGEM-T Easy-TDE1430-aphA2 (XI) | Replacement of TDE1430 by aphA2; Ampr | This study |
| pGEM-T Easy-TDE0911-aphA2 (XII) | Replacement of TDE0911 by aphA2; Ampr | This study |
| pGEM-T Easy-TDE1430-P1aphA2-P2flgE (XIII) | For flgE complementation; Ampr | This study |
| Strains | ||
| E. coli XL1-Blue | For subcloning | Stratagene |
| T. denticola ATCC 35405 | Parent strain | ATCC |
| TDE1430(−) | Replacement of TDE1430 by Kanr | This study |
| flgE(−) | Replacement of flgE by Ermr | This study |
| TDE0911(−) | Replacement of TDE0911 by Kanr | This study |
| flgE(−)/flgE(+) | Inactivation of flgE by Ermr and integration of flgE in the TDE1430 locus using Kanr | This study |
GenBank accession numbers: *, NP_478145; †, WP_004614937; ‡, NP_052903; §, P23181. P1, promoter 1; P2, promoter 2. Plasmids are abbreviated using Roman numerals, indicated in parentheses.
Ampr, ampicillin resistance cassette; Ermr, erythromycin resistance cassette; Kanr, kanamycin resistance cassette.
FIG 1.
Schematic diagram of plasmid constructs for targeted mutagenesis and integration of flgE into T. denticola 35405. (A) Plasmid construction for replacement of TDE1430 with antibiotic cassette driven by a promoter. Plasmids (VI, VII, VIII, IX, and X) were constructed by replacement of TDE1430 with different cassettes from plasmids (pSY118, I, II, III, and IV). (B) Plasmid construction for replacement of TDE1430 in-frame with antibiotic cassette without a promoter. Plasmid XI was constructed by replacement of TDE1430 with aphA2. (C) Plasmid construction for replacement of TDE0911 in-frame with antibiotic cassette without a promoter. Plasmid XII was constructed by replacement of TDE0911 with aphA2. (D) Plasmid construction for complementation of flgE. Plasmid XIII was constructed by replacement of TDE1430 with aphA2 and flgE driven by promoters P1 and P2, respectively.
An allelic replacement mutagenesis scheme was used to inactivate genes of interest. The TDE1430 gene was partially deleted and replaced with an appropriate antibiotic resistance gene. In brief, a 4.0-kb fragment containing the full-length TDE1430 gene (1,008 bp) and 5′ (1,570 bp) and 3′ (1,423 bp) flanking regions of the DNA fragment of TDE1430 was PCR amplified using TDE1430-F1 and TDE1430-R1 primers and ligated into pGEM-T Easy to construct pGEM-T Easy-TDE1430 (plasmid V). This plasmid was used as a template in an inverse PCR with the primer pair TDE1430-BamHI-R and TDE1430-XhoI-F to partially delete TDE1430 from amino acid residues 1 to 270 to generate a 6.2-kb inverse PCR product. This construct was then ligated with different antibiotic resistance cassettes, specifically, ermF-ermAM, P1aphA1, P1aphA2, P1cat, and P1gat using the BamHI and XhoI sites (Fig. 1A) in the place of the partially deleted TDE1430 to generate TDE1430 mutant alleles that were replaced with various antibiotic resistance genes (plasmids VI, VII, VIII, IX, and X; Table 2).
To insert a kanamycin resistance gene without a linked promoter into TDE1430 to construct an in-frame fusion of aphA2 with the remaining TDE1430, an inverse PCR was first performed to partially delete TDE1430. Plasmid V was used as a template, and the primer pair TDE1430-AgeI-R and TDE1430-XhoI-F depicted in Fig. 1A was used in the inverse PCR. The inverse PCR product obtained was then ligated with aphA2 by AgeI and XhoI sites to generate plasmid XI (Fig. 1B).
In-frame deletion of TDE0911 was carried out as follows. In brief, the 5′-flanking region (483 bp) of TDE0911 was amplified and ligated with pGEM-T easy at the XhoI and SpeI restriction sites of pGEM-T Easy. The aphA2 was subsequently cloned downstream of this 483-bp fragment at the AgeI and XhoI restriction sites and then ligated with the 3′-flanking region (1,277 bp) of TDE0911 via XhoI and SpeI sites to generate plasmid XII (Fig. 1C).
Complementation of the flgE mutant was performed by integrating the full-length flgE gene driven by the promoter from the fla operon (P2) (39) into the TDE1430 locus. In brief, the P2 promoter was ligated into 3′ end of the aphA2 using the XhoI and XbaI sites (Fig. 1D), followed by addition of the flgE gene using the XbaI and SalI sites. This integration construct was then released from the pGEM-T vector by restriction digestion using BamHI and SalI, which was then used to replace TDE1430 (1 to 270 amino acid residues) from the plasmid V (Fig. 1A) to generate an integration plasmid (XIII) (Fig. 1D). This step was completed by ligation of the BamHI- and SalI-digested integration construct with the inverse PCR product obtained using plasmid V as a template and the primer pair of TDE1430-BamHI-R and TDE1430-XhoI-F (Fig. 1A). All of these plasmid constructs were confirmed by restriction digestion and DNA sequencing. Constructs used for transformation were then released and linearized by NotI from the corresponding plasmids prior to introduction into T. denticola.
Transformation of T. denticola.
Transformation of T. denticola was conducted as previously described (40, 41). Briefly, 15-μg portions of the various DNA constructs were added to 100 μl of competent cells, followed by incubation on ice for 30 min, followed in turn by electroporation at 1.8 kV, 200 Ω, and 25 µF in a 1-mm cuvette (the observed voltages were between 1,787 and 1,786 V, while the time constant remained at 5.1 ms). The transformation mixture resuspended in 1.5 ml of NOS was transferred to an anaerobic chamber to allow electroporated bacteria to grow overnight at 35°C. Ninety percent of the overnight culture was mixed with 4 ml of prewarmed 3% SeaPlaque low-melting-temperature agarose (Lonza, Rockland, ME), and added to 24.5 ml of prewarmed NOS (37°C) that contained different antibiotics (kanamycin [25 μg/ml] or erythromycin [40 μg/ml]) for plating. In addition, the remaining 10% of the transformation mixture was plated on a different plate to obtain countable plates to enumerate bacterial colonies. The plates were transferred to the anaerobic chamber for 3 to 5 days at 35°C after an initial 3 to 5 h of incubation at room temperature.
Identification of positive colonies.
All colonies grown on the antibiotic plates were enumerated to determine the transformation efficiency. Ten colonies were randomly selected and inoculated in 1.0 ml of NOS medium for 48 h prior to PCR identification. Bacterial cultures were used as PCR templates to determine the insertion of the antibiotic cassette or integration of genes, the loss of genes such as TDE1430 and TDE0911, and the orientation of insertions by the listed PCR primers that flanked the targeted genes (Table 1).
SDS-PAGE analysis of bacterial cell lysates.
One milliliter of each bacterial culture that was grown to an optical density at 600 nm at 0.4 was harvested by centrifugation and washed three times with phosphate-buffered saline (PBS), and a fraction (1/20) of the cell pellets was boiled in sample loading buffer and used for SDS–12% PAGE analysis, followed by Coomassie blue staining (42).
Real-time PCR.
RNAs were isolated from 2-day cultures of T. denticola by TRIzol reagent according to the manufacturer's protocol (Life Technologies, Carlsbad, CA). A total of 1 μg of total DNase-treated RNA was used in a final volume of 20 μl for reverse transcription with a Bio-Rad cDNA synthesis kit according to the manufacturer's protocol. Gene expression was evaluated by quantitative real-time PCR with an iCycler (Bio-Rad) using the iScript one-step RT-PCR kit (SYBR green Bio-Rad). The PCR thermal cycling program was used as described by the manufacturer (Life Technologies). Relative quantification was performed according to the ΔΔCT method (43). The expression levels of flgE and flaA were normalized with those for 16S RNA. All primers used in the real-time PCR analysis (RT-flgE-F, RT-flgE-R, RT-flaA-F, RT-flaA-R, RT-TDE16sRNA-F, and RT-TDE16sRNA-R) are listed in Table 1.
Measurement of colony sizes of T. denticola.
Approximately 200 colonies of each strain were plated after serial dilution to 0.8% agarose swarm plates without antibiotics and incubated for 5 days at 35°C. The sizes of the colonies were determined by measuring photographic images with a phase-contrast microscope.
Transmission electron microscopy.
Electron microscopy studies were carried out as described previously (44). In brief, 0.5 ml of a 3-day culture of T. denticola was washed three times with PBS and resuspended in 0.5 ml of PBS containing 1% Triton X-100. The PF of T. denticola were exposed by vortexing. Three additional washings were used to remove the remaining Triton X-100. The final pellet was resuspended in the same volume of PBS, and 10 μl of each sample was applied to Formvar carbon-coated copper grids (300 mesh; Polysciences, Inc., Warrington, PA). After a 1-min absorption at room temperature, excessive liquid was removed by wicking with filter paper. The grids were then stained with 2% uranyl acetate for 45 s before visualization of T. denticola and the exposed PF using a FEI Tecnai Spirit T12 transmission electron microscope operated at 80 keV (High Resolution Imaging Shared Facility, The University of Alabama at Birmingham, Birmingham, AL).
RESULTS AND DISCUSSION
Screening of antibiotic resistance cassettes for T. denticola 35405.
All of the antibiotics available in our lab were used to examine their effectiveness in inhibiting T. denticola (Table 3). Erythromycin was used as a control since it has been widely used for the genetic manipulation of T. denticola. Similar to erythromycin, six antibiotics were able to inhibit growth of T. denticola except zeocin. Zeocin is usually active aerobically (45–47), which may explain why it was not active under the anaerobic condition. To identify effective antibiotic resistance cassettes, genes coding for various antibiotic resistance cassettes, including a chloramphenicol acetyltransferase gene (cat), two kanamycin resistance genes (aphA1 and aphA2), and a gentamicin resistance gene (gat) were first examined in E. coli. These cassettes were cloned into the pGEM-T vector with the promoter from the ermF-ermAM cassette (P1) that is also functional in Porphyromonas gingivalis and Bacteroides spp. (27). The resulting plasmid constructs were transformed into E. coli XL1-Blue and scored for antibiotic resistance. The chloramphenicol cassette worked well in E. coli. The two kanamycin cassettes also conferred resistance against both kanamycin and G418. However, the gentamicin cassette failed to confer resistance because there was no bacterial growth on the gentamicin plate for cultures that were transformed with the gentamicin cassette (Table 3, column 1, E. coli).
TABLE 3.
Effectiveness of antibiotics in inhibiting T. denticola 35405 and utility of the antibiotic cassettes in inactivating the TDE1430 gene
| Antibiotica | IC (μg/ml) and ACb |
|||||
|---|---|---|---|---|---|---|
| IC (T. denticola)c | AC (pGEM-T-P1/ARG)d | IC (E. coli)e | AC (pGEM-T Easy-TDE1430-P1/ARG)f | IC (E. coli)g | IC (T. denticola)h | |
| Ampicillin | 20–120 | ND | ND | ND | ND | |
| Chloramphenicol | 10–70 | III | 35 | IX | No | No |
| Gentamicin | 5–120 | IV | No | X | No | No |
| Erythromycin | 10–∼100 | VI | 300 | 40 | ||
| G418 | 10–100 | I | 20 | VII | No | No |
| Kanamycin | 10–100 | I | 50 | VII | No | No |
| G418 | 10–100 | II | 20 | VIII | No | No |
| Kanamycin | 10–100 | II | 50 | VIII | 50 | 25 |
| Tetracycline | 10–40 | ND | ND | ND | ND | |
| Zeocin | NI (20–100) | ND | ND | ND | ND | |
All antibiotics were tested in NOS medium.
IC, inhibitory concentration of antibiotic against the indicated strain; AC, antibiotic resistance cassette used for the indicated strain; ARG, antibiotic resistance gene; NI, no inhibition; ND, not determined. Further details regarding each column of data below are provided in the corresponding footnotes. The cassette abbreviations are defined fully in Table 2, column 1.
The inhibitory concentrations of a given antibiotic for T. denticola in NOS medium.
The antibiotic cassette driven by P1 cloned into pGEM-T.
The inhibitory concentrations of antibiotics for E. coli transformed with the antibiotic resistance cassettes from column 3 on an LB agar plate.
The antibiotic cassette driven by P1 cloned into pGEM-T Easy-TDE1430 to replace TDE1430.
The inhibitory concentrations of antibiotics for E. coli transformed with the plasmid from column 5 on an LB agar plate.
Mutagenesis of the TDE1430 gene by transformation of linearized plasmid from column 5 in T. denticola.
To determine whether these cassettes are also functional in T. denticola, the individual cassettes were used to insert into the targeted gene locus TDE1430 and then transformed into E. coli first to determine their antibiotic resistance (Table 3, column 2, in E. coli). The antibiotic resistance testing revealed that only one kanamycin cassette (aphA2) conferred resistance to kanamycin, whereas E. coli transformed with the TDE1430 allele that was disrupted by the other antibiotic cassettes failed to confer resistance to their corresponding antibiotics. It is not clear why most cassettes under the control of P1 were no longer active in E. coli after insertion into the TDE1430 locus. Furthermore, E. coli transformed with the stand alone aphA2 construct conferred resistance to both kanamycin and G418. However, the aphA2 cassette only exhibited resistance to kanamycin but not to G418 when inserted into the TDE1430 region. Constructs in which the TDE1430 allele was disrupted by various antibiotic cassettes were then transformed into T. denticola. Interestingly, only the allele disrupted by aphA2 gave rise to numerous antibiotic-resistant transformants in T. denticola (Table 3). Of note, this was the only construct functional in E. coli. This is also true for the previously characterized erythromycin cassette. These observations suggest the functionality of the cassette integrated into the TDE1430 locus in E. coli can be used to predict the utility in T. denticola. This may simplify our efforts to identify more antibiotic resistance cassettes for this difficult organism.
It is not clear why only one kanamycin resistance gene (aphA2) worked. The aphA1 gene is derived from the pET-SUMO-28 vector that has been used in other bacteria, whereas the aphA2 cassette from either pGBKT7 (Clontech Laboratories) or pCMV-3tag-2 (Agilent Technologies, Santa Clara, CA) has been used in bacteria to confer resistance to kanamycin and mammalian cells to confer resistance to G418. Sequence analysis of these two kanamycin resistance cassettes revealed that they share 35% identity and 52% similarity at the amino acid sequence level; however, only the aphA2 cassette was able to confer resistance to kanamycin in T. denticola, suggesting that a specific sequence is required in T. denticola. The sequences of the gentamicin and chloramphenicol cassettes used in our lab were different from those previously reported that may explain why they were not active in T. denticola despite other reported successes (26, 29, 36, 48).
The kanamycin cassette is effective in the inactivation of TDE1430.
TDE1430 encodes a putative β-1,4-galactosyltransferase, albeit its function in T. denticola ATCC 35405 is unclear. TDE1430 was selected to examine the utility of the kanamycin cassette in the allelic replacement mutagenesis for the following reasons. First, our numerous mutagenesis experiments revealed that inactivation of TDE1430 has the highest efficiency compared to our attempts to inactivate other genes such as flgE, TDE1418 (a probable K5 antigen synthesis protein), TDE1427 (a putative glycosyltransferase gene), TDE1441 (a dTDP-glucose 4,6-dehydratase gene), TDE2349 (a flagellar filament gene), TDE1220 (a Treponema-specific hypothetical gene), and TDE0911. Second, TDE1430 is among a few genes that have been inactivated by the transposon mutagenesis (48). Third, the TDE1430 sequence is resistant to enzymatic digestion by major restriction enzymes in T. denticola. In the genome of T. denticola ATCC 35405, there are three putative type II R-M systems: TDE1268 (TDEI, DpnI), TDE0227/TDE0228 (TDEII, SapI), and TDE0909/TDE0911(TDEIII, Sau96I) that digest exogenous DNA leading to a reduction in transformation efficiency (23, 25). Interestingly, there is no SapI or Sau96I site in the 5′- and 3′-flanking regions of the TDE1430 locus that avoids restriction digestion by these two major enzyme groups. Only three DpnI sites were found located at the terminus of both the 5′-flanking region and the 3′-flanking region (Fig. 1A) that would not destroy the integrity of the TDE1430 deletion construct. This might explain why the transformation efficiency was high when the TDE1430 constructs were used. Thus, TDE1430 was chosen to evaluate the mutagenesis efficiency of the cassette in T. denticola. The mutagenesis efficiency of the kanamycin cassette was compared to the efficiency obtained with the ermF-ermAM cassette. Evaluation of transformation efficiency using the same batch of competent cells revealed the kanamycin cassette interrupted TDE1430 allele yielded much higher efficiency than the ermF-ermAM cassette-interrupted allele (6-fold) (Fig. 2A), demonstrating that the use of the kanamycin cassette is advantageous.
FIG 2.
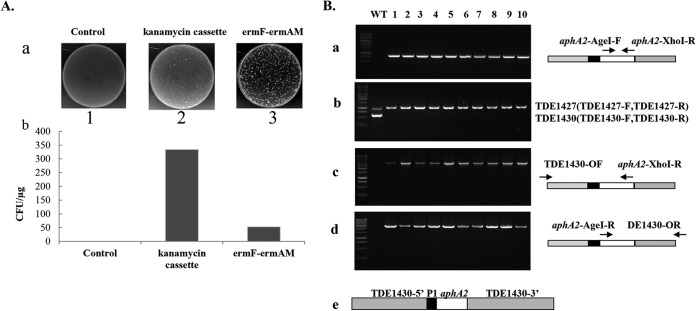
The kanamycin cassette fused with a promoter is more efficient than ermF-ermAM in the activation of TDE1430. (A) Transformation efficiency of the kanamycin cassette. Transformation was conducted as described in Materials and Methods—without DNA (panel a1, Control), with 15 μg of linearized TDE1430P1aphA2 (panel a2), or with TDE1430-ermF-ermAM (panel a3)—and then the samples were plated onto appropriate agarose media to select transformants. (b) Colonies from diluted samples were counted and presented as CFU/μg of input DNA (bars represent the average of three transformation experiments). (B) PCR confirmation of the deletion of TDE1430. Ten colonies were examined by PCR for insertion of the kanamycin cassette (a), loss of TDE1430 (b), and orientation of the kanamycin cassette (c and d). (e) Diagram of the TDE1430 deletion allele represents the structure of plasmid VIII.
To confirm the genotype of kanamycin-resistant mutants, we carried out PCR studies. Ten kanamycin-resistant T. denticola colonies were selected randomly to evaluate whether they had mutant genotypes. PCR results indicated that all resistant clones contained the kanamycin cassette (Fig. 2Ba). Furthermore, all resistant clones lost TDE1430 but had TDE1427, a positive control for T. denticola 35405 compared to the wild type (Fig. 2Bb). PCR using the primer pair flanking TDE1430 revealed the insertion of the kanamycin cassette into the TDE1430 locus (Fig. 2Bc and d). These data demonstrate that mutagenesis of TDE1430 using the kanamycin cassette is highly efficient.
The kanamycin cassette without a promoter is also effective in the inactivation of TDE1430.
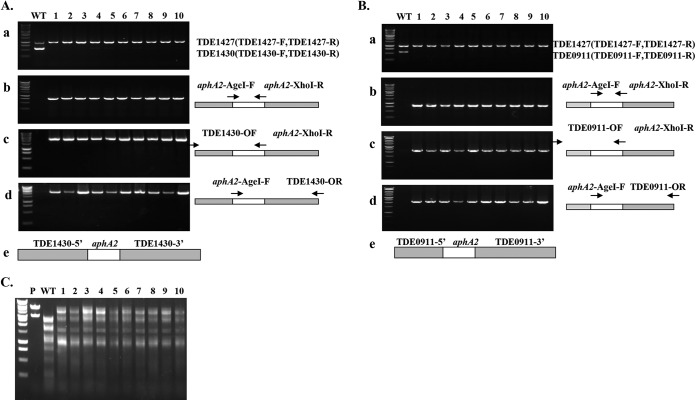
The mutagenesis examined above was carried out using a functional T. denticola promoter to drive the expression of the cassette (Fig. 2Be). In-frame replacement of genes with an antibiotic cassette is another approach to achieve mutagenesis. The advantage of this approach is that the transcription of the antibiotic resistance gene is driven by the native promoter of the mutant gene. To determine whether the cassette is also active without a fused promoter, we deleted TDE1430 from amino acid residues 1 to 270 and fused the kanamycin cassette aphA2 in-frame with the remaining TDE1430 (Fig. 3Ae) and then carried out transformation in T. denticola using the linearized plasmid construct. Ten resistant colonies were then randomly chosen to determine mutagenesis efficiency. PCR analysis revealed that all 10 kanamycin-resistant clones lost the TDE1430 gene (Fig. 3Aa) and had the kanamycin cassette inserted in the intended position at the right orientation (Fig. 3Ab, c, and d), demonstrating the utility of the cassette alone in the mutagenesis of TDE1430.
FIG 3.
Inactivation of TDE1430 and TDE0911 genes by in-frame fusion of the kanamycin cassette. (A) PCR determination of the deletion of TDE1430. Ten clones were identified by colony PCR according to the loss of TDE1430 (a), the insertion of a kanamycin cassette (b), and the direction of cassette insertion compared to the wild type (WT) (c and d). (e) Diagram of the TDE1430 deletion allele represents the structure of plasmid XI. (B) PCR determination of the deletion of TDE0911. Ten clones were identified by colony PCR according to the loss of TDE0911 (a), the insertion of a kanamycin cassette (b), and the direction of cassette insertion compared to the WT (c and d). (e) A diagram of the TDE0911 deletion allele represents the structure of plasmid XII. (C) Restriction digestion patterns influenced by inactivation of TDE0911. The pGEM-T Easy-TDE1430-ermF-ermAM plasmid (VI) was linearized by NotI (lane p) and then subjected to digestion by cell extracts from T. denticola WT and TDE0911 mutant (lanes 1 to 10) and analyzed by agarose gel electrophoresis.
To further examine whether the kanamycin cassette alone is useful for allelic replacement mutagenesis of other genes, we chose TDE0911 as a target. TDE0911 is a type II restriction enzyme that recognizes Sau96I site; thus, inactivation of TDE0911 would change the restriction digestion pattern of DNA and have an apparent phenotype. TDE0911 was readily inactivated using the kanamycin cassette (Fig. 3Be). PCR results showed TDE0911 was no longer amplified by TDE0911-specific primers (Fig. 3Ba); instead, it was replaced by the kanamycin gene (Fig. 3Bb). Furthermore, the cassette was inserted into the intended position with the right orientation (Fig. 3Bc and d). Phenotypically, inactivation of TDE0911 altered the restriction digestion patterns of the plasmid DNA, pGEM T-Easy-TDE1430-ermF-ermAM (Fig. 3C). The plasmid linearized by NotI released two bands: pGEM-T Easy (the lower band) and TDE1430-ermF-ermAM (the higher band) (Fig. 3C, lane P). Treatment of the linearized plasmid with cell lysates of wild-type T. denticola completely eliminated the higher-molecular-weight band and concurrently produced a large number of low-molecular-weight bands. In contrast, treatment with cell lysates from 10 TDE0911 mutants failed to abolish the higher-molecular-weight band, suggesting the inactivation of TDE0911. These results demonstrated the utility of the kanamycin cassette in the inactivation of TDE0911.
Integration of the full-length flgE gene into the T. denticola flgE mutant genome restored expression of flgE.
Because of the existence of the unique restriction and modification system in T. denticola 35405, the strain is extremely difficult to complement by any replicating plasmid. An unmethylated plasmid can be transformed into T. denticola 35405 only when the type II restriction system TDE0911 is inactivated (23). The chromosomal integration has been used successfully to complement a neuraminidase gene mutant (26). Here, we attempted to complement the flgE mutant by integrating the flgE gene into the TDE1430 locus. flgE is the gene for the PF hook protein (FlgE). Deletion of flgE led to complete loss of both PF filament and spirochetal motility (30). Phenotypically, the flgE mutant was nonmotile and no longer swarmed; thus, it was easy to screen if the flgE mutant was complemented and had the ability to translocate and swarm restored when the full-length flgE gene was integrated into the T. denticola chromosome (23, 35).
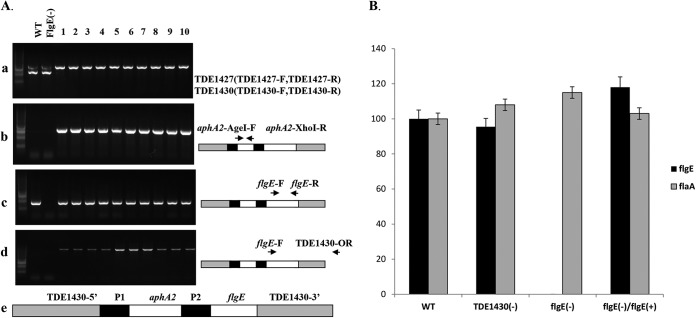
Previously, we generated a flgE mutant by using the ermAF cassette (data not shown), and this mutant had all of the known flgE-deficient phenotypes (31), and was used to carry out chromosomal complementation by the allelic replacement of TDE1430 with a construct that contained the kanamycin cassette and the full-length flgE gene. During our trial, we readily inactivated TDE1430 by allelic-replacement mutagenesis regardless of which functional antibiotic cassettes were used. In addition, the inactivation of TDE1430 did not alter any phenotype of T. denticola that we were interested in studying. Therefore, the TDE1430 locus was chosen as a gene integration site for the chromosomal complementation. PCR analysis of obtained kanamycin-resistant clones revealed the loss of TDE1430 (Fig. 4Aa), the insertion of the kanamycin cassette (Fig. 4Ab), and the flgE gene (Fig. 4Ac) at the appropriate position and direction of the replacement (Fig. 4Ad). Genotypically, the deletion of flgE resulted in the loss of expression of flgE. Furthermore, the complementation restored expression of flgE (Fig. 4B).
FIG 4.
Chromosomal integration of the full-length flgE into T. denticola. The integration construct containing the full-length flgE gene and the kanamycin cassette was integrated into the genome of the flgE mutant. The expressions of flgE and flaA were evaluated. (A) PCR determination of the integration of flgE into the TDE1430 locus. Transformants carrying the integration allele were identified by PCR according to the loss of TDE1430 (a), the insertion of a kanamycin resistance gene (b), the insertion of flgE and the appropriate orientation of the insertion (c) compared to wild type (WT) and the flgE mutant (d). (e) A diagram of the integration allele represents the structure of plasmid XIII. P1, promoter from the ermF cassette; P2, promoter from the fla operon. (B) Restoration of the expression of flgE. The relative expression levels of flgE and flaA were determined by real-time PCR for different T. denticola variants. The data presented here are the means of three independent experiments.
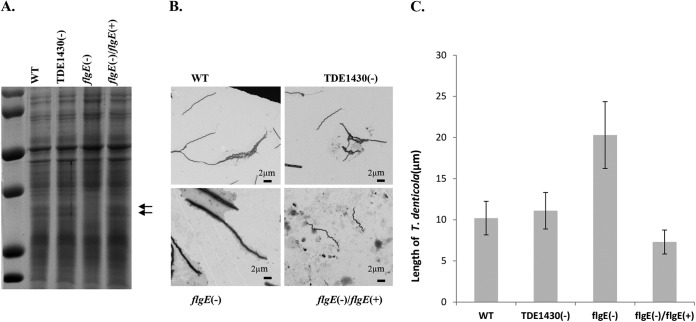
Interestingly, deletion of flgE led to an alteration of the protein profile (loss of 38-kDa FlaA and 35-kDa FlaB PF filament proteins) (44, 49), whereas there was no apparent change in the protein profile when T. denticola TDE1430 was deleted. Complementation of flgE completely restored production of FlaA and FlaB PF filament proteins (Fig. 5A), suggesting the effectiveness of the chromosomal complementation. However, the FlgE, hook protein monomer, could not be detected. This is likely due to the nature of the FlgE protein. FlgE monomers are covalently linked within the hook structure and usually do not readily dissociate for entry into resolving gels (50).
FIG 5.
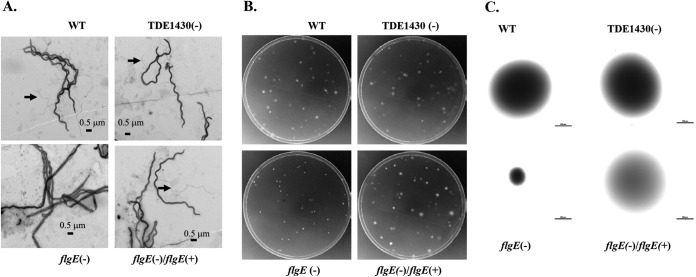
Characterization of T. denticola variants. (A) Protein profile of T. denticola. Spirochetal cell lysates from exponentially grown T. denticola variants were prepared and applied to SDS–12% PAGE, followed by Coomassie blue staining. Arrows represent PF proteins. (B) Electron micrographic examination of T. denticola morphology. Cells were stained with 2% uranyl acetate. (C) Measurement of spirochetal length. Each bar represents the mean length (in μm) of 20 spirochetal cells measured by electronic microscopy.
Complete recovery of PF and swarming ability when flgE was complemented.
Inactivation of TDE1430 did not affect the overall size, PF, or swarming ability of T. denticola. On the other hand, inactivation of flgE altered bacterial shape and size and led to the complete loss of PF. The flgE mutant became elongated and widened and lost the ability to swarm and to maintain a helical shape (31, 32, 35). Complementation of flgE completely restored the wild-type phenotype (Fig. 5B and C and Fig. 6). These results further demonstrated the utility and effectiveness of the kanamycin cassette in the chromosomal complementation.
FIG 6.
Chromosomal integration of flgE restored PF and motility. (A) PF of T. denticola were recovered after the integration of flgE into T. denticola genome. Electron microscopic examination of exposed PF (arrow) from spirochetal cells treated with 1% Triton X-100. Cells were stained with 2% uranyl acetate. (B) T. denticola colonies on agarose plates. Approximately 200 colonies after serial dilution were plated onto 0.8% agarose plates and incubated at 35°C for 5 days. (C) Measurement of representative colonies. Colonies were magnified, and their diameters were measured. Each bar represents 0.5 mm.
In summary, the kanamycin cassette we developed is a useful tool for the genetic manipulation of T. denticola 35405. Higher transformation efficiency mediated by the cassette will help us generate mutants by allelic exchange and facilitate the generation of double mutants when combined with the widely used erythromycin cassette. We also demonstrated the successful integration of a full-length gene allele into the spirochetal genome for chromosomal complementation using this cassette, providing a useful alternative to plasmid complementation that has been proven difficult to establish. Chromosomal integration has been demonstrated in the restoration of a TDE0471 mutant by inserting the full-length TDE0471 into the erythromycin resistance cassette on the chromosome of the mutant using a gentamicin antibiotic cassette (26). Our studies utilized a highly transformable TDE1430 locus to obtain chromosomal complementation. The advantage of our approach is the high transformation efficiency with reproducibility. A disadvantage of this strategy is that the TDE1430 locus was interrupted, which may have unintended side effects when studying bacterial fitness and virulence. This limitation can be overcome by cis-chromosomal complementation. Nevertheless, our studies offer a new and reliable antibiotic cassette to genetically manipulate T. denticola.
ACKNOWLEDGMENTS
This study was partially supported by NIH/NIDCR R03 DE016040 (J.R.) and NIH/NIDCR R21 DE022575 (H.W.).
REFERENCES
- 1.Simonson LG, Goodman CH, Bial JJ, Morton HE. 1988. Quantitative relationship of Treponema denticola to severity of periodontal disease. Infect Immun 56:726–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoshida A, Kawada M, Suzuki N, Nakano Y, Oho T, Saito T, Yamashita Y. 2004. TaqMan real-time polymerase chain reaction assay for the correlation of Treponema denticola numbers with the severity of periodontal disease. Oral Microbiol Immunol 19:196–200. doi: 10.1111/j.0902-0055.2004.00142.x. [DOI] [PubMed] [Google Scholar]
- 3.Holt SC, Ebersole JL. 2005. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the ‘red complex’, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol 2000 38:72–122. doi: 10.1111/j.1600-0757.2005.00113.x. [DOI] [PubMed] [Google Scholar]
- 4.Ishihara K. 2010. Virulence factors of Treponema denticola. Periodontol 2000 54:117–135. doi: 10.1111/j.1600-0757.2009.00345.x. [DOI] [PubMed] [Google Scholar]
- 5.Foschi F, Izard J, Sasaki H, Sambri V, Prati C, Müller R, Stashenko P. 2006. Treponema denticola in disseminating endodontic infections. J Dent Res 85:761–765. doi: 10.1177/154405910608500814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siqueira J, Rôças I, Favieri A, Oliveira J, Santos K. 2001. Polymerase chain reaction detection of Treponema denticola in endodontic infections within root canals. Int Endod J 34:280–284. doi: 10.1046/j.1365-2591.2001.00382.x. [DOI] [PubMed] [Google Scholar]
- 7.Chaparro A, Blanlot C, Ramírez V, Sanz A, Quintero A, Inostroza C, Bittner M, Navarro M, Illanes S. 2013. Porphyromonas gingivalis, Treponema denticola and Toll-like receptor 2 are associated with hypertensive disorders in placental tissue: a case-control study. J Periodontal Res 48:802–809. doi: 10.1111/jre.12074. [DOI] [PubMed] [Google Scholar]
- 8.Chukkapalli SS, Rivera MF, Velsko IM, Lee J-Y, Chen H, Zheng D, Bhattacharyya I, Gangula PR, Lucas AR, Kesavalu L. 2014. Invasion of oral and aortic tissues by oral spirochete Treponema denticola in ApoE−/− mice causally links periodontal disease and atherosclerosis. Infect Immun 82:1959–1967. doi: 10.1128/IAI.01511-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haraszthy V, Zambon J, Trevisan M, Zeid M, Genco R. 2000. Identification of periodontal pathogens in atheromatous plaques. J Periodontol 71:1554–1560. doi: 10.1902/jop.2000.71.10.1554. [DOI] [PubMed] [Google Scholar]
- 10.Romano F, Barbui A, Aimetti M. 2007. Periodontal pathogens in periodontal pockets and in carotid atheromatous plaques. Minerva Stomatol 56:169–179. [PubMed] [Google Scholar]
- 11.Ertugrul AS, Arslan U, Dursun R, Hakki SS. 2013. Periodontopathogen profile of healthy and oral lichen planus patients with gingivitis or periodontitis. Int J Oral Sci 5:92–97. doi: 10.1038/ijos.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li C, Liu J, Tan L, Yu N, Lin L, Geng F, Zhang D, Pan Y. 2013. The sociodemographic characteristics, periodontal health status, and subgingival microbiota of patients with chronic periodontitis and type 2 diabetes mellitus: a case-control study in a Chinese population. J Periodontol 84:1058–1066. doi: 10.1902/jop.2012.120282. [DOI] [PubMed] [Google Scholar]
- 13.Bian X-l, Wang H-t, Ning Y, Lee SY, Fenno JC. 2005. Mutagenesis of a novel gene in the prcA-prtP protease locus affects expression of Treponema denticola membrane complexes. Infect Immun 73:1252–1255. doi: 10.1128/IAI.73.2.1252-1255.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dashper S, Seers C, Tan K, Reynolds E. 2011. Virulence factors of the oral spirochete Treponema denticola. J Dental Res 90:691–703. doi: 10.1177/0022034510385242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fenno JC, Wong GW, Hannam PM, McBride BC. 1998. Mutagenesis of outer membrane virulence determinants of the oral spirochete Treponema denticola. FEMS Microbiol Lett 163:209–215. doi: 10.1111/j.1574-6968.1998.tb13047.x. [DOI] [PubMed] [Google Scholar]
- 16.Edwards A, Dymock D, Jenkinson H. 2003. From tooth to hoof: treponemes in tissue-destructive diseases. J Appl Microbiol 94:767–780. doi: 10.1046/j.1365-2672.2003.01901.x. [DOI] [PubMed] [Google Scholar]
- 17.Graves DT, Kang J, Andriankaja O, Wada K, Rossa C Jr. 2012. Animal models to study host-bacteria interactions involved in periodontitis. Front Oral Biol 15:117–132. doi: 10.1159/000329675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frederick J, Sarkar J, McDowell J, Marconi R. 2011. Molecular signaling mechanisms of the periopathogen, Treponema denticola. J Dental Res 90:1155–1163. doi: 10.1177/0022034511402994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kesavalu L, Sathishkumar S, Bakthavatchalu V, Matthews C, Dawson D, Steffen M, Ebersole JL. 2007. Rat model of polymicrobial infection, immunity, and alveolar bone resorption in periodontal disease. Infect Immun 75:1704–1712. doi: 10.1128/IAI.00733-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kesavalu L, Walker S, Holt S, Crawley R, Ebersole J. 1997. Virulence characteristics of oral treponemes in a murine model. Infect Immun 65:5096–5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orth RH, O'Brien-Simpson N, Dashper S, Reynolds E. 2011. Synergistic virulence of Porphyromonas gingivalis and Treponema denticola in a murine periodontitis model. Mol Oral Microbiol 26:229–240. doi: 10.1111/j.2041-1014.2011.00612.x. [DOI] [PubMed] [Google Scholar]
- 22.Visser M, Ellen R. 2011. New insights into the emerging role of oral spirochaetes in periodontal disease. Clin Microbiol Infect 17:502–512. doi: 10.1111/j.1469-0691.2011.03460.x. [DOI] [PubMed] [Google Scholar]
- 23.Bian J, Li C. 2011. Disruption of a type II endonuclease (TDE0911) enables Treponema denticola ATCC 35405 to accept an unmethylated shuttle vector. Appl Environ Microbiol 77:4573–4578. doi: 10.1128/AEM.00417-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas CM, Nielsen KM. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol 3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 25.Seshadri R, Myers GS, Tettelin H, Eisen JA, Heidelberg JF, Dodson RJ, Davidsen TM, DeBoy RT, Fouts DE, Haft DH. 2004. Comparison of the genome of the oral pathogen Treponema denticola with other spirochete genomes. Proc Natl Acad Sci U S A 101:5646–5651. doi: 10.1073/pnas.0307639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurniyati K, Zhang W, Zhang K, Li C. 2013. A surface-exposed neuraminidase affects complement resistance and virulence of the oral spirochaete Treponema denticola. Mol Microbiol 89:842–856. doi: 10.1111/mmi.12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fletcher HM, Schenkein HA, Morgan RM, Bailey KA, Berry CR, Macrina FL. 1995. Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect Immun 63:1521–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chi B, Chauhan S, Kuramitsu H. 1999. Development of a system for expressing heterologous genes in the oral spirochete Treponema denticola and its use in expression of the Treponema pallidum flaA gene. Infect Immun 67:3653–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bian J, Fenno JC, Li C. 2012. Development of a modified gentamicin resistance cassette for genetic manipulation of the oral spirochete Treponema denticola. Appl Environ Microbiol 78:2059–2062. doi: 10.1128/AEM.07461-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Arakawa S, Deng Q-D, Kuramitsu H. 1999. Characterization of a novel methyl-accepting chemotaxis gene, dmcB, from the oral spirochete Treponema denticola. Infect Immun 67:694–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Ruby J, Charon N, Kuramitsu H. 1996. Gene inactivation in the oral spirochete Treponema denticola: construction of an flgE mutant. J Bacteriol 178:3664–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Limberger RJ, Slivienski LL, Izard J, Samsonoff WA. 1999. Insertional inactivation of Treponema denticola tap1 results in a nonmotile mutant with elongated flagellar hooks. J Bacteriol 181:3743–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lux R, Sim J-H, Tsai JP, Shi W. 2002. Construction and characterization of a cheA mutant of Treponema denticola. J Bacteriol 184:3130–3134. doi: 10.1128/JB.184.11.3130-3134.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goetting-Minesky MP, Fenno JC. 2010. A simplified erythromycin resistance cassette for Treponema denticola mutagenesis. J Microbiol Methods 83:66–68. doi: 10.1016/j.mimet.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chi B, Limberger RJ, Kuramitsu HK. 2002. Complementation of a Treponema denticola flgE mutant with a novel coumermycin A1-resistant T. denticola shuttle vector system. Infect Immun 70:2233–2237. doi: 10.1128/IAI.70.4.2233-2237.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slivienski-Gebhardt LL, Izard J, Samsonoff WA, Limberger RJ. 2004. Development of a novel chloramphenicol resistance expression plasmid used for genetic complementation of a fliG deletion mutant in Treponema denticola. Infect Immun 72:5493–5497. doi: 10.1128/IAI.72.9.5493-5497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Limberger RJ. 2004. The periplasmic flagellum of spirochetes. J Mol Microbiol Biotechnol 7:30–40. doi: 10.1159/000077867. [DOI] [PubMed] [Google Scholar]
- 38.Leschine S, Canale-Parola E. 1980. Rifampin as a selective agent for isolation of oral spirochetes. J Clin Microbiol 12:792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stamm LV, Bergen HL. 1999. Molecular characterization of a flagellar (fla) operon in the oral spirochete Treponema denticola ATCC 35405. FEMS Microbiol Lett 179:31–36. doi: 10.1111/j.1574-6968.1999.tb08703.x. [DOI] [PubMed] [Google Scholar]
- 40.Li H, Kuramitsu H. 1996. Development of a gene transfer system in Treponema denticola by electroporation. Oral Microbiol Immunol 11:161–165. doi: 10.1111/j.1399-302X.1996.tb00352.x. [DOI] [PubMed] [Google Scholar]
- 41.Kuramitsu HK, Chi B, Ikegami A. 2005. Genetic manipulation of Treponema denticola. Curr Protoc Microbiol Chapter 12:Unit 12B.2. doi: 10.1002/9780471729259.mc12b02s00. [DOI] [PubMed] [Google Scholar]
- 42.Ruby J, Lux R, Shi W, Charon N, Dasanayake A. 2008. Effect of glucose on Treponema denticola cell behavior. Oral Microbiol Immunol 23:234–238. doi: 10.1111/j.1399-302X.2007.00417.x. [DOI] [PubMed] [Google Scholar]
- 43.Yuan JS, Reed A, Chen F, Stewart CN. 2006. Statistical analysis of real-time PCR data. BMC Bioinformatics 7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruby JD, Li H, Kuramitsu H, Norris SJ, Goldstein SF, Buttle KF, Charon NW. 1997. Relationship of Treponema denticola periplasmic flagella to irregular cell morphology. J Bacteriol 179:1628–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ehrenfeld GM, Shipley JB, Heimbrook DC, Sugiyama H, Long EC, Van Boom JH, Van der Marel GA, Oppenheimer NJ, Hecht SM. 1987. Copper-dependent cleavage of DNA by bleomycin. Biochemistry 26:931–942. doi: 10.1021/bi00377a038. [DOI] [PubMed] [Google Scholar]
- 46.Hlavová M, Cízková M, Vítová M, Bišová K, Zachleder V. 2011. DNA damage during G2 phase does not affect cell cycle progression of the green alga Scenedesmus quadricauda. PLoS One 6:e19626. doi: 10.1371/journal.pone.0019626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petering DH, Byrnes RW, Antholine WE. 1990. The role of redox-active metals in the mechanism of action of bleomycin. Chem Biol Interact 73:133–182. doi: 10.1016/0009-2797(90)90001-4. [DOI] [PubMed] [Google Scholar]
- 48.Yang Y, Stewart PE, Shi X, Li C. 2008. Development of a transposon mutagenesis system in the oral spirochete Treponema denticola. Appl Environ Microbiol 74:6461–6464. doi: 10.1128/AEM.01424-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Charon NW, Cockburn A, Li C, Liu J, Miller KA, Miller MR, Motaleb M, Wolgemuth CW. 2012. The unique paradigm of spirochete motility and chemotaxis. Annu Rev Microbiol 66:349. doi: 10.1146/annurev-micro-092611-150145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Limberger RJ, Slivienski LL, Samsonoff WA. 1994. Genetic and biochemical analysis of the flagellar hook of Treponema phagedenis. J Bacteriol 176:3631–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]