Abstract
We investigated the response of the hydrocarbon-degrading Mycobacterium vanbaalenii PYR-1 to crude oil from the BP Deepwater Horizon (DWH) spill, using substrate depletion, genomic, and proteome analyses. M. vanbaalenii PYR-1 cultures were incubated with BP DWH crude oil, and proteomes and degradation of alkanes and polycyclic aromatic hydrocarbons (PAHs) were analyzed at four time points over 30 days. Gas chromatography-mass spectrometry (GC-MS) analysis showed a chain length-dependent pattern of alkane degradation, with C12 and C13 being degraded at the highest rate, although alkanes up to C28 were degraded. Whereas phenanthrene and pyrene were completely degraded, a significantly smaller amount of fluoranthene was degraded. Proteome analysis identified 3,948 proteins, with 876 and 1,859 proteins up- and downregulated, respectively. We observed dynamic changes in protein expression during BP crude oil incubation, including transcriptional factors and transporters potentially involved in adaptation to crude oil. The proteome also provided a molecular basis for the metabolism of the aliphatic and aromatic hydrocarbon components in the BP DWH crude oil, which included upregulation of AlkB alkane hydroxylase and an expression pattern of PAH-metabolizing enzymes different from those in previous proteome expression studies of strain PYR-1 incubated with pure or mixed PAHs, particularly the ring-hydroxylating oxygenase (RHO) responsible for the initial oxidation of aromatic hydrocarbons. Based on these results, a comprehensive cellular response of M. vanbaalenii PYR-1 to BP crude oil was proposed. This study increases our fundamental understanding of the impact of crude oil on the cellular response of bacteria and provides data needed for development of practical bioremediation applications.
INTRODUCTION
Spills of crude oil, a highly complex mixture mostly composed of diverse hydrocarbon fractions, have acute and long-term adverse ecological impacts on marine and terrestrial systems that the oil contaminates (1–4). Many studies have investigated bacterial responses to these organic contaminants, since bacterial catabolic activities represent one of the primary processes by which crude oil hydrocarbons are eliminated from the environment (5, 6). Extensive studies have been conducted to elucidate bacterial mechanisms of crude oil hydrocarbon degradation, such as metabolic pathways and their biochemical and genetic bases (5–9). However, these studies have mostly focused on the degradation of individual hydrocarbon components of crude oil, such as alkanes or certain types of aromatic hydrocarbons, whereas little is known about the fate of each of the crude oil hydrocarbon components, the mechanism of its degradation, and how its metabolism is associated with other cellular activities, such as central carbon metabolism, in a complex crude oil. Considering the components of a crude oil, such as that spilled in 2010 by the Deepwater Horizon (DWH) blowout in the Gulf of Mexico, it is highly unlikely that total degradation of crude oil hydrocarbons occurs by the same mechanism as pure-hydrocarbon degradation. Moreover, exposure to crude oil and degradation of crude oil compounds triggers not only metabolic reactions, but also overall organism-wide reactions, on which limited information is available. In recent years, activities of microorganisms and their metabolism with respect to crude oil and its components have been investigated at the level of microbial consortia or microbial communities (10, 11). For example, several research teams have used large-scale high-throughput approaches to perform a comprehensive analysis of the responses of microbial communities and their biodegradative potential in the Gulf of Mexico ecosystem with respect to the DWH oil spill (12–17). However, although these functional-genomics studies provide new insights into the activities of the microorganisms, revealing an overall view of the metabolic and ecological reactions of the microbial community to crude oil, few detailed descriptions of the activity of each bacterial species that responds to the input of crude oil are available. To gain a better understanding of the potential role that a bacterium plays, it is also essential to elucidate mechanisms and features controlling the biology of individual microorganisms during crude oil degradation, such as the expression and succession of key catabolic enzymes and overall cellular physiological changes.
Mycobacterium vanbaalenii PYR-1 (18, 19), an isolate from an oil-contaminated estuary of the Gulf of Mexico, Redfish Bay, near Aransas Pass, TX (20), has been extensively studied to elucidate the mechanisms of hydrocarbon degradation. The bacterium has a versatile metabolic ability to degrade a wide range of hydrocarbon components of crude oil, including high-molecular-weight (HMW) polycyclic aromatic hydrocarbons (PAHs) with four or more fused benzene rings, such as pyrene and fluoranthene, and aliphatic hydrocarbons, such as dodecane and hexadecane (21–30). According to previous metabolic, genetic, biochemical, and functional-genomics studies, the biological properties of the bacterium, such as catabolic activity, cellular structures, and ecophysiology, are suitable for the degradation or transformation of various organic pollutants in the environment (31).
It is important to understand how life forms respond to crude oil to assess its impact on environmental health. In this study, we investigated the dynamics of the microbial response by M. vanbaalenii PYR-1 to exposure to BP crude oil. From a metabolic and ecophysiological standpoint, the bacterium is a relevant biological system to comprehensively study the response to crude oil, which would reveal not only the mechanism involved in the degradation of crude oil hydrocarbons, but also other intrinsic biological reactions in the cellular response to crude oil. We used a combination of chemical and molecular approaches in which cells of M. vanbaalenii PYR-1 were incubated with BP crude oil and degradation of selected aliphatic and aromatic hydrocarbons in the oil were examined. We then analyzed profiles of protein expression during culture incubation of the bacterium with the BP crude oil, using a high-throughput whole-cell proteome analysis. We observed a genome-wide complex cellular response of M. vanbaalenii PYR-1 to the BP crude oil, which dynamically changed during the time of exposure. This study has not only increased the overall understanding of the biological impact of crude oil on a microbial system in the environment, but also yielded important insights into microbial behavior with respect to crude oil exposure. This knowledge may be helpful for advancing practical biotechnological applications.
MATERIALS AND METHODS
Bacterial strain, media, and culture conditions.
M. vanbaalenii PYR-1 was maintained at 30°C in Luria-Bertani (LB) medium or Middlebrook 7H10 medium supplemented with oleic acid-albumin-dextrose-catalase (OADC) enrichment (Remel, Lenexa, KS). The same media were solidified with 1.5% agar to make plates. For incubation of bacterial cells with BP crude oil, supplemented minimal medium (SMM) (32), with or without glucose, was used with slight modifications. DWH-sourced light Louisiana BP crude oil (collected from Deepwater Horizon MC252 on 10 June 2010 with sample identifier [ID] SOB-20100610-26) was obtained from the Gulf Coast Seafood Laboratory, Center for Food Safety and Applied Nutrition, FDA, and was added directly to bacterial culture flasks.
Experimental design and procedures. (i) Utilization of crude oil components for bacterial growth.
Colonies of the bacterium grown on Middlebrook 7H10 agar plates were precultured in a flask containing 100 ml of LB medium with shaking at 200 rpm for approximately a week. The primary culture was then washed twice with 100 ml SMM and resuspended in SMM to obtain secondary cultures with a final optical density at 600 nm (OD600) of 0.05, which were further incubated with the addition of 1 and 2% crude oil. Foam plugs were used for flask stoppers after consideration of ventilation during the period of incubation and nonvolatility of HMW PAHs and n-alkanes (C13 and above). For growth comparison, the same bacterial culture was also grown with and without glucose. Cell growth was determined by measuring the increase in OD600 with a Synergy 2 microplate reader (BioTek Instruments, Winooski, VT). Each kinetic value for optical density is the mean of the results of triplicate independent experiments.
(ii) Degradation test of BP crude oil components.
Cells of M. vanbaalenii PYR-1, precultured for a week in a flask containing SMM supplemented with 1% glucose, were resuspended in 650 ml of the same medium at a concentration equivalent to 0.4 optical-density unit at 600 nm. Twenty milliliters of the cell suspension was then transferred to 125-ml flasks. We prepared 32 flasks, each containing 20 ml of cell suspension, for a 30-day time course crude oil biodegradation experiment. To 20 of the culture flasks, 1% BP crude oil was added and incubated at 30°C with shaking at 200 rpm. The other 12 flasks were used as control samples, which were subjected to the same incubation conditions except that the flasks were autoclaved at 125°C for 15 min and then 1% BP crude oil was added. We collected culture flasks at 0, 3, 6, and 30 days. Control samples for chemical analysis at time point 0 were collected right after the BP crude oil was added to the culture, and abiotic control samples were collected at the same time point, except for day 0. The control sample for proteome analysis was taken just before crude oil addition. The culture flasks were stored at −70°C until they were analyzed.
Analytical methods. (i) Chemicals.
Standard chemicals and internal standards are listed in Table S1 in the supplemental material. Solvents were purchased from Sigma-Aldrich (St. Louis, MO) and were of the highest purity available.
(ii) Extraction of samples.
Samples were extracted by combining 15 ml of a sample (mixed until homogeneous) and 15 ml of methylene chloride in a 60-ml glass bottle and rotating it end over end for 30 min. The extract was then sonicated for 15 min and rotated end over end for an additional 60 min. The phases were allowed to separate, and the methylene chloride extract was divided into two fractions; two-thirds was used for the PAH analysis, and one-third was used for the alkane analysis. The methylene chloride was evaporated to dryness at ∼30°C under nitrogen. The residue was then reconstituted in 200 μl of the appropriate internal-standard solution in methylene chloride.
(iii) Analysis of n-alkanes.
Solvent standards of n-pentacosane, n-dodecane, n-tetradecane, n-hexadecane, n-octadecane, n-eicosane, n-henicosane, n-docosane, n-tricosane, n-tetracosane, n-pentacosane, n-hexacosane, n-heptacosane, n-octacosane, and pentacosane-D52 were prepared in methylene chloride at 125 μg/ml. The retention times of these prepared alkane standards were used to analyze alkanes in the BP crude oil. Two hundred microliters of sample was combined with 200 μl of methylene chloride containing the internal standard pentacosane-D52 at 125 μg/ml. The resulting solution was analyzed with an Agilent 6890 gas chromatography-mass spectrometry (GC-MS) instrument equipped with a capillary column (DB-17MS; 60 by 0.25 mm; 0.25-μm film thickness; Agilent Technologies, Santa Clara, CA). One microliter of sample (split 1:5) was injected on the system. The system was programmed as follows: injector, 280°C; detector, 320°C; auxiliary, 250°C; quadrupole, 150°C; oven started at 40°C and increased at 5°C/min to 320°C and held for 24 min. The mass spectrometer was set in the EI+ mode and scanned from 40 to 400 atomic mass units (amu). Extracted ion chromatograms for the alkanes were used for integration. The analyte and internal-standard peaks were integrated, and the response ratio (the analyte peak area divided by the internal-standard peak area) was calculated. The response ratio for each analyte at day 0 was set to 100%, and all subsequent time points were reported as a percentage of that of day 0.
(iv) Analysis of PAH compounds.
Solvent standards of phenanthrene, fluoranthrene, pyrene, and benzo[a]pyrene were prepared in methylene chloride at 150, 100, 20, and 5 μg/ml, respectively. The solvent standards contained the equivalent deuterated internal standards (phenanthrene-D10, fluoranthene-D10, pyrene-D10, and benzo[a]pyrene-D12) at 150, 2, 2, and 10.5 μg/ml, respectively, and were used to confirm retention. Two hundred microliters of sample was combined with 200 μl of methylene chloride containing the appropriate PAH deuterated internal standard (phenanthrene, fluoranthrene, pyrene, or benzo[a]pyrene, at 75, 4, 4, and 1 μg/ml, respectively). The resulting solution was analyzed with an Agilent 6890 GC-MS instrument under the same conditions as for the alkanes. The mass spectrometer was set in the EI+ mode and scanned from 150 to 300 amu. Extracted ion chromatograms for phenanthrene, fluoranthrene, pyrene, and benzo[a]pyrene were used for integration. The analyte and internal-standard peaks were integrated, and the response ratio (the analyte peak area divided by the internal-standard peak area) was calculated. The response ratio for each analyte at day 0 was set to 100%, and all subsequent time points were reported as a percentage of that of day 0.
Proteome analysis. (i) Sample preparation.
The whole-cell proteome of M. vanbaalenii PYR-1 treated with 1% BP crude oil was analyzed as described in our previous studies (33–35). Briefly, proteins of bacterial cells at each time point were extracted using glass bead homogenization. Cell lysates were trichloroacetic acid precipitated and quantified using a Qubit fluorometer (Invitrogen, Carlsbad, CA), and 20 μg of each lysate was loaded onto a 4 to 12% SDS-PAGE gel (Invitrogen). The gel was excised into 40 segments per lane, and gel slices were processed using a robot, ProGest (DigiLab, Marlborough, MA), with the following protocol. Initially, the gels were washed with 25 mM ammonium bicarbonate, followed by acetonitrile. Reduction was performed with 10 mM dithiothreitol at 60°C, followed by alkylation with 50 mM iodoacetamide at room temperature. Digestion was performed with trypsin at 37°C for 4 h, and after quenching with formic acid, the supernatant was analyzed directly without further processing.
(ii) MS analysis.
Gel digests were analyzed by nano-liquid chromatography-tandem MS (LC–MS-MS) with a Waters NanoAcquity high-performance liquid chromatography (HPLC) system interfaced with a ThermoFisher Q Exactive mass spectrometer. Peptides were loaded on a trapping column and eluted over a 75-μm analytical column at 350 nl/min; both columns were packed with Jupiter Proteo resin (Phenomenex, Torrance, CA). The mass spectrometer was operated in a data-dependent mode, with MS and MS-MS performed in the Orbitrap at 70,000-FWHM (full width at half maximum) resolution and 17,500-FWHM resolution, respectively. The 15 most abundant ions were selected for MS-MS.
(iii) Data processing.
Data were searched using a local copy of Mascot with the following parameters: enzyme, trypsin; database, custom M. vanbaalenii (concatenated forward and reverse plus common contaminants); fixed modification, carbamidomethyl (C); variable modifications, oxidation (M), acetyl (protein N-term), deamidation (NQ), and pyro-Glu (N-term Q); mass values, monoisotopic; peptide mass tolerance, 10 ppm; fragment mass tolerance, 0.020 Da; maximum missed cleavages, 2. Mascot DAT files were parsed into the Scaffold software for validation, filtering, and creating a nonredundant list for each sample. The data were filtered at a 1% protein and peptide false-discovery rate (FDR), requiring at least two unique peptides per protein. The proteins that were significantly differently expressed were determined on the basis of the following criteria: (i) the protein was detected as a binary difference, and there were at least 5 spectral counts (SpC) in the unique sample; and (ii) the protein was detected with a 2-fold (up or down) or more change based on dividing the normalized spectral abundance factor (NSAF) values, and there were at least 5 SpC in the higher sample. The quality and quantity of the proteome results were also evaluated with the previous proteomic data for M. vanbaalenii PYR-1. (For detailed information on proteomics and statistical analysis, see the paper by Kweon et al. [35].) In this study, protein abundance was used as a proxy for enzyme activity.
RESULTS AND DISCUSSION
Biodegradation of hydrocarbon components in BP crude oil.
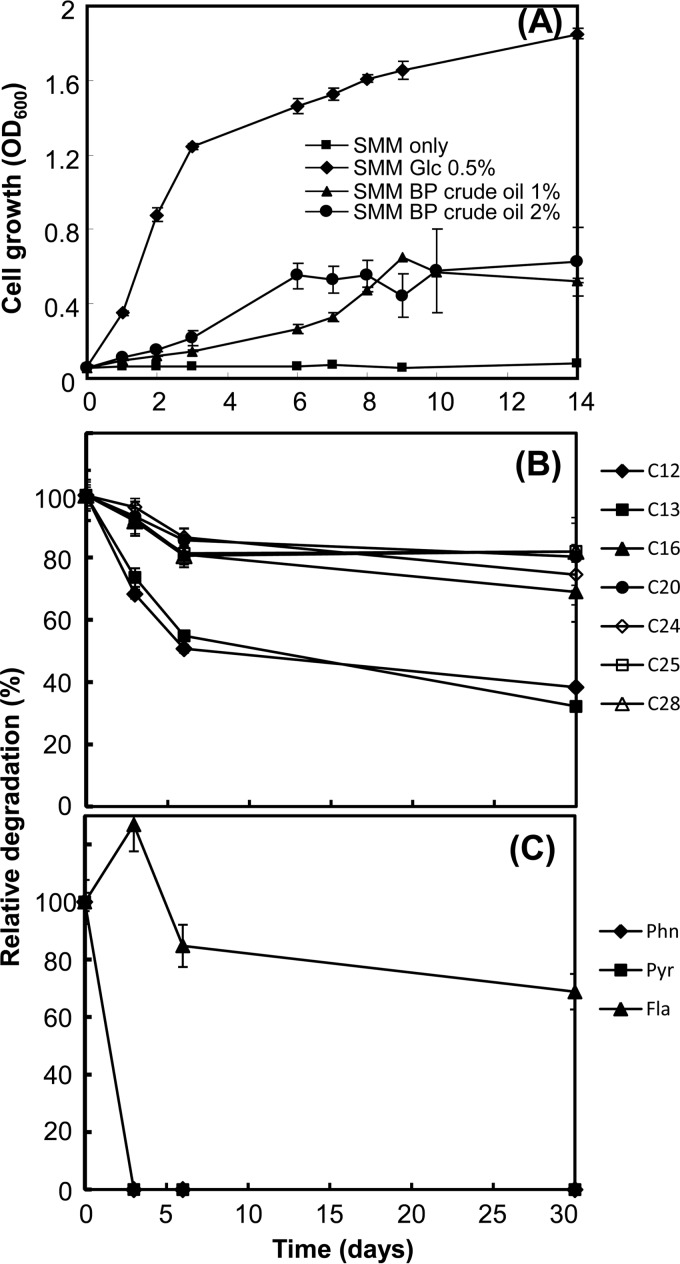
Although M. vanbaalenii PYR-1 has been shown to have versatile ability to degrade a wide range of hydrocarbons, including n-alkanes and PAHs (21–30, 36–38), no investigation has shown whether the bacterium can utilize these hydrocarbons as nutrients for growth when they occur in crude oil. A series of culture experiments were conducted to understand the metabolic behavior of M. vanbaalenii PYR-1 toward BP crude oil. We initially examined whether M. vanbaalenii PYR-1 can grow with BP crude oil as a sole carbon and energy source. As shown in Fig. 1A, during 14 days of incubation, an increase in the turbidity of minimal media with 1 to 2% BP crude oil during the incubation period was observed, which clearly indicates that the bacterium was able to utilize nutrients available in the BP crude oil. We conducted another culture experiment to assess the capacity of M. vanbaalenii PYR-1 to biodegrade BP crude oil components. During a 30-day incubation of the bacterium with BP crude oil, quadruplicate samples from bacterial cultures and abiotic controls were collected for analysis at four time points, days 0, 3, 6, and 30. We then extracted the entire flask contents and identified selected BP crude oil hydrocarbons via GC-MS analysis.
FIG 1.
Growth of M. vanbaalenii PYR-1 in SMM supplemented with 1 or 2% BP crude oil, as well as cultures with no BP crude oil, with or without 0.5% glucose over a 14-day period (A) and biodegradation of n-alkanes (B) and PAHs (C) in liquid cultures of M. vanbaalenii PYR-1 incubated with BP crude oil.
(i) Selective biodegradation of crude oil n-alkanes by M. vanbaalenii PYR-1.
We first obtained a GC-MS chromatographic profile for aliphatic hydrocarbons from the culture extract of M. vanbaalenii PYR-1 grown in a slightly modified supplemented minimal medium (34) containing 1% BP crude oil, which identified 17 peaks for the straight-chain alkanes, ranging from C12 to C28. As shown in Fig. 1B (see Fig. S1A in the supplemental material), the degradation profiles at day 30 showed that decreases in the amount of aliphatic hydrocarbons had occurred for all the alkanes detected during the incubation with BP crude oil. This result is consistent with early studies with this and other strains of environmental mycobacteria on biodegradation of alkanes (36, 39–41). Among these identified alkanes, we selected 7 compounds, dodecane (C12), tridecane (C13), hexadecane (C16), icosane (C20), tetracosane (C24), pentacosane (C25), and octacosane (C28), to monitor differences in degradation by the bacterium with time during incubation with BP crude oil. The amounts of hydrocarbons for controls were calculated from the initial amount of hydrocarbon at time zero, which was further normalized by subtraction of the amount of abiotically removed analytes from that found in the culture at the same time point with dead cells of M. vanbaalenii PYR-1. As shown in Fig. 1B (see Data Set S1 in the supplemental material), M. vanbaalenii PYR-1 eliminated the n-alkanes contained in the BP crude oil at different rates and to different extents. Compared to other aliphatic hydrocarbons, C12 and C13 were degraded most rapidly, with the greatest degradation extents being 62% and 68%, respectively. Although degradation of the other five aliphatic hydrocarbons was observed, the extent of their degradation remained at 20 to 30% after the 30-day incubation, with C28 the lowest (18%). These results indicate that M. vanbaalenii PYR-1 better degraded medium-chain alkanes (C12 and C13) in BP crude oil, showing a relatively narrow substrate specificity of n-alkane degradation pathways. This trend of alkane degradation is also typical of most mycobacteria that metabolize alkanes (42). As discussed below, the chain length dependence that we observed in the degradation of BP crude oil alkanes by M. vanbaalenii PYR-1 is well explained by the protein expression profile and substrate specificity of the n-alkane-degrading enzymes.
(ii) Global biodegradation of crude oil PAHs by M. vanbaalenii PYR-1.
Biodegradation of four PAHs, phenanthrene, pyrene, fluoranthene, and benzo[a]pyrene, was also monitored by measuring the amounts of residual PAHs (Fig. 1C; see Fig. S1B in the supplemental material). The amounts of the four PAHs from dead-cell controls stayed almost the same throughout the incubation period, which indicated no significant abiotic degradation. As shown in Fig. 1C (see Data Set S1 in the supplemental material), the GC-MS analysis showed that the bacterium completely degraded phenanthrene and pyrene. Fluoranthene was also degraded by approximately 31.2% after 30 days of incubation. As a whole, this metabolic capability of M. vanbaalenii PYR-1 is consistent with the results from our prior studies, except for the metabolism of fluoranthene. In our previous biodegradation experiments, M. vanbaalenii PYR-1 always degraded more than 95% of these three PAHs (phenanthrene, pyrene, and fluoranthene) when cultured with single PAH substrates or mixtures of PAHs (33–35, 43–45). Benzo[a]pyrene was not detected in any experimental samples. As discussed below in detail, at the level of PAH metabolism, our proteome analysis revealed that this metabolic disparity from our previous studies in the degradation of the HMW PAH fluoranthene was most likely to have been caused by a differential protein expression pattern, especially of the ring-hydroxylating oxygenase (RHO) systems responsible for the initial hydroxylation of PAH substrates in the ring cleavage process (RCP) functional modules (33, 35).
Proteomic insights into the global cellular response to BP crude oil.
To understand the global cellular response of M. vanbaalenii PYR-1 to BP crude oil, we conducted a whole-cell proteome analysis of the bacterium during culture incubation. We also wanted to examine the mechanistic basis of metabolic activity for the degradation of BP crude oil at the level of genome-wide protein expression. For the proteomic response to be consistent with the degradation of BP crude oil, we used bacterial cultures from the same batches used for the chemical analysis at the same 4 time points, days 0, 3, 6, and 30.
(i) Top-down view of the proteomic data.
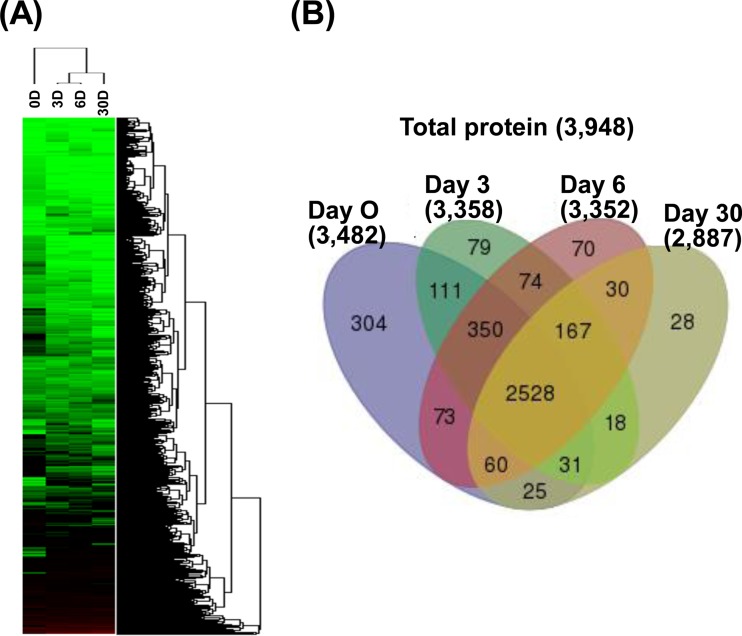
We identified 3,482, 3,358, 3,352, and 2,887 proteins from culture samples from days 0, 3, 6, and 30, respectively, which add up to 3,948 unique proteins in total (Fig. 2; see Data Set S2 in the supplemental material). As shown in Fig. 2A, cluster analysis of the proteomic data set revealed apparent correlations between the profiles of protein expression with respect to crude oil exposure time, indicating the reliability of the proteome results. We identified 2,528 proteins as commonly shared between controls and three other samples, while other proteins were identified either in control (day 0) or crude-oil-exposed (Fig. 2B; see Data Set S2 in the supplemental material) samples. Generally, most of these identified proteins showed similar proportions of clusters of orthologous group (COG) distribution with protein-coding genes (46) based on the genome of M. vanbaalenii PYR-1 (Table 1; see Fig. S2 in the supplemental material).
FIG 2.
Summary of proteins identified in the proteome study. (A) Cluster analysis of proteomic data sets showing the correlation between protein expression profiles and treatment time. D, day(s). (B) Venn diagram showing the numbers of proteins identified in controls (day 0) and in samples at three later time points.
TABLE 1.
Summary of identified proteins classified by COGs
| COG category | Description | No. genome encoded | % of genome | No. of identified proteins | % of identified proteins | No. with >2-fold increase in total | No. with >2-fold increase in common | No. with >2-fold decrease in total | No. with >2-fold decrease in common |
|---|---|---|---|---|---|---|---|---|---|
| A | RNA processing and modification | 1 | 0.02 | 1 | 0.03 | 0 | 0 | 0 | 0 |
| J | Translation, ribosomal structure, and biogenesis | 167 | 2.79 | 153 | 3.88 | 16 | 8 | 27 | 9 |
| K | Transcription | 442 | 7.39 | 319 | 8.08 | 24 | 8 | 29 | 48 |
| L | Replication, recombination, and repair | 234 | 3.91 | 107 | 2.71 | 2 | 1 | 57 | 28 |
| N | Cell motility | 5 | 0.08 | 2 | 0.05 | 0 | 0 | 0 | 0 |
| D | Cell cycle control and cell division | 31 | 0.52 | 25 | 0.63 | 1 | 0 | 5 | 8 |
| M | Cell wall/membrane/envelope biogenesis | 170 | 2.84 | 116 | 2.94 | 14 | 5 | 43 | 13 |
| O | Posttranslational modification, protein turnover, and chaperones | 147 | 2.46 | 105 | 2.66 | 17 | 4 | 31 | 12 |
| T | Signal transduction mechanisms | 215 | 3.60 | 167 | 4.23 | 17 | 6 | 88 | 37 |
| U | Intracellular trafficking, secretion, and vesicular transport | 32 | 0.54 | 19 | 0.48 | 2 | 0 | 5 | 1 |
| V | Defense mechanisms | 47 | 0.79 | 29 | 0.73 | 4 | 1 | 7 | 1 |
| C | Energy production and conversion | 347 | 5.80 | 269 | 6.81 | 38 | 16 | 105 | 51 |
| E | Amino acid transport and metabolism | 352 | 5.89 | 272 | 6.89 | 41 | 11 | 91 | 26 |
| F | Nucleotide transport and metabolism | 93 | 1.56 | 83 | 2.10 | 8 | 1 | 19 | 5 |
| G | Carbohydrate transport and metabolism | 207 | 3.46 | 164 | 4.15 | 21 | 5 | 47 | 18 |
| H | Coenzyme transport and metabolism | 184 | 3.08 | 153 | 3.88 | 24 | 8 | 59 | 13 |
| I | Lipid transport and metabolism | 465 | 7.78 | 374 | 9.47 | 83 | 32 | 124 | 56 |
| P | Inorganic ion transport and metabolism | 260 | 4.35 | 157 | 3.98 | 41 | 21 | 49 | 23 |
| Q | Secondary-metabolite biosynthesis, transport, and catabolism | 436 | 7.29 | 298 | 7.55 | 64 | 20 | 119 | 57 |
| R | General function prediction only | 682 | 11.41 | 513 | 12.99 | 90 | 34 | 215 | 90 |
| S | Function unknown | 326 | 5.45 | 239 | 6.05 | 47 | 9 | 79 | 26 |
| Not in COGs | 1,726 | 28.87 | 831 | 21.05 | 168 | 42 | 237 | 100 | |
| Total | 5,979 | 3,948 | 66.03 | 722 | 232 | 1,546 | 654 |
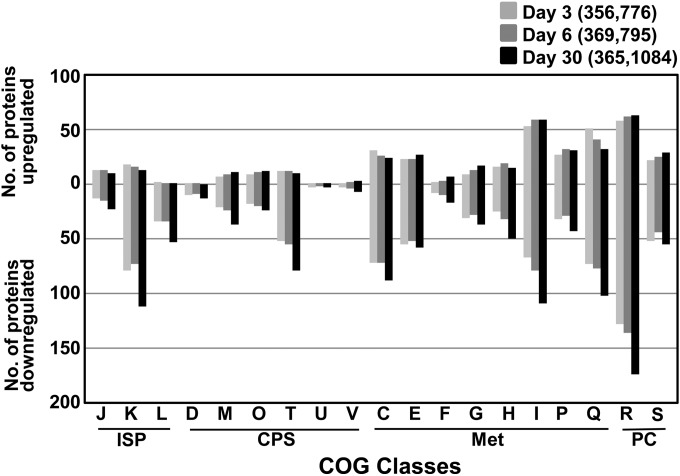
Treatment with BP crude oil had a dynamic effect on the global expression of proteins in M. vanbaalenii PYR-1. We compared the differentially regulated proteins using the criterion of 2-fold change in abundance of expression from day 0. As shown in Fig. S3 and Data Sets S3 and S4 in the supplemental material, 406, 418, and 417 proteins were >2-fold upregulated (651 proteins in total, as shown in Fig. S3A in the supplemental material) and 821, 826, and 1,135 proteins were >2-fold downregulated (1,374 proteins in total, as shown in Fig. S3B in the supplemental material) on days 3, 6, and 30, respectively. Among them, 209 and 549 proteins were commonly up- and downregulated, respectively, at all time points upon BP crude oil treatment. As revealed in the functional analysis of the differentially expressed proteins based on COG categories (Fig. 3; see Fig. S3 in the supplemental material), the overall COG percentage of most of these up- or downregulated proteins in most cases showed a distribution similar to that in the whole proteome, indicating that BP crude oil induced a global cellular response throughout the COG categories. However, some of the COG categories showed more or fewer than the expected proportional number of proteins. For example, the numbers of upregulated proteins belonging to categories Q (secondary-metabolite biosynthesis, transport, and catabolism) and I (lipid transport and metabolism) were higher than proportional numbers of genome-encoded proteins. This indicated that there was a need for upregulation of proteins associated with the COG categories, since many catabolic proteins involved in the degradation of hydrocarbons belong to these categories. As shown in Fig. S4 in the supplemental material, various metabolic pathways were enriched from both up- and downregulated proteins, indicating complex biological processes occurring during BP crude oil incubation. Finally, the proteomic data were further analyzed in terms of the time-dependent effects of BP crude oil on the molecular response of the bacterium using the short time series expression miner (STEM) software (see Fig. S5 and Data Set S2 in the supplemental material) (47).
FIG 3.
Comparative analysis of distributions among differently regulated proteins based on COGs as grouped in Fig. S3 in the supplemental material (see Data Sets S3 and S4 in the supplemental material). ISP, information storage and processing; CPS, cellular processes and signaling; Met, metabolism; PC, poorly characterized. COG category descriptions are provided in Table 1.
(ii) Enzymes responsible for selective degradation of BP crude oil n-alkanes by M. vanbaalenii PYR-1.
Aerobic degradation of alkanes is generally initiated with hydroxylation of a terminal methyl group, producing alcohols, which is followed by further oxidation to aldehydes and then to fatty acids (48). Several types of alkane hydroxylases with different substrate specificities function in the initial hydroxylation of alkane molecules (48). The M. vanbaalenii PYR-1 genome has two genes encoding a membrane-bound AlkB alkane monooxygenase (Mvan_1742 and Mvan_3100), and our proteome analysis confirmed that only one AlkB alkane monooxygenase (Mvan_1742) was upregulated. The gene encoding this monooxygenase enzyme (Mvan_1742) constitutes an alkB gene cluster with other genes, including two rubredoxins (Mvan_1743 and Mvan_1744) and a TetR-type transcriptional regulator, AlkU (Mvan_1745), which is known to activate expression of alkB (49, 50). As shown in Data Set S3 in the supplemental material, all the proteins in this gene cluster were upregulated together, suggesting their functional interaction for alkane hydroxylation. In addition, three putative alcohol dehydrogenases (Mvan_0013, Mvan_0443, and Mvan_5164) and an aldehyde dehydrogenase (Mvan_0352), which are potentially involved in the next two consecutive steps, respectively, in the degradation of alkanes, also showed patterns of upregulation (STEM patterns 17 to 21), although they are positioned distant from each other in the genome of M. vanbaalenii PYR-1. Functional interaction of the series of these enzymes completes the transformation of alkane substrates to the corresponding fatty acids (48). Interestingly, two rubredoxin genes (Mvan_1743 and Mvan_1744) in a row are located immediately downstream of the alkane hydroxylase gene in the alkB cluster, but none of the rubredoxin reductase genes are located close to the alkB cluster in the genome of M. vanbaalenii PYR-1. The rubredoxin gene (Mvan_1743) was upregulated, indicating its functional contribution to electron transfer from rubredoxin reductase(s) to AlkB hydroxylase during terminal hydroxylation of alkanes. We observed expression of the genes encoding ferredoxin (Mvan_0299, Mvan_0485, Mvan_0549, Mvan_0681, Mvan_1767, Mvan_3180, Mvan_4520, Mvan_5031, and Mvan_5309) and ferredoxin reductase (Mvan_0399, Mvan_0400, Mvan_0467, Mvan_1216, Mvan_1290, Mvan_2039, and Mvan_3857), with different regulation patterns, suggesting their potential functional contribution as an electron transfer chain between NAD(P)H and oxygenase components. In this alkB gene cluster, a membrane protein (Mvan_1740) similar to an alkane transporter reported previously (49–51) was also expressed. An additional transporter protein, Mvan_5005, highly similar to the ABC-type lipid transporter and additionally proposed to be involved in alkane transport (50), also had an elevated level of expression. Interestingly, we identified expression of 34 (out of 50 genes) cytochrome P450 oxygenase (CYP) enzymes, 8 of which were upregulated at the protein level, and their involvement in the oxidation of aliphatic hydrocarbons cannot be excluded.
The fatty acids produced from alkane oxidation are further degraded via β-oxidation to acetyl-coenzyme A (CoA), which enters the tricarboxylic acid (TCA) cycle or glyoxylate bypass (40). M. vanbaalenii PYR-1 is highly enriched with genes involved in the multistep β-oxidation, and we found that all the necessary proteins were identified as expressed, with some of them being upregulated (see Data Set S3 in the supplemental material). They included fatty acid-CoA ligase (FadD; Mvan_0131), acyl-CoA dehydrogenase (FadE; Mvan_1974), enoyl-CoA hydratase/isomerase (EchA; Mvan_4579), 3-hydroxyacyl-CoA dehydrogenase (FadB; Mvan_4577), and 3-ketoacyl-CoA thiolase (FadA; Mvan_5067).
(iii) Proteins responsible for PAH metabolism.
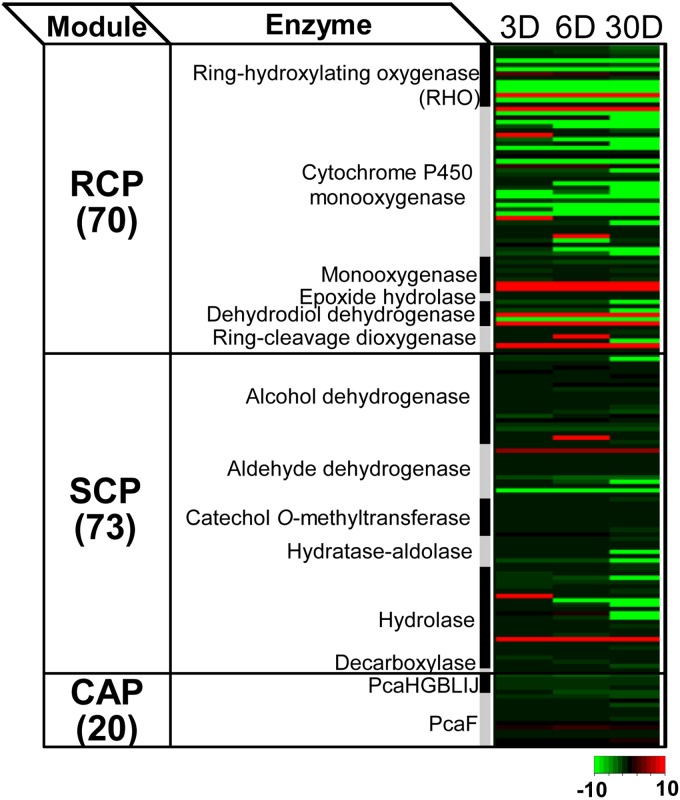
To understand the metabolic response involved in the degradation of BP crude oil PAHs, we investigated proteomic data from the perspective of a PAH metabolic network (PAH-MN). According to the PAH-MN that we previously proposed (33), PAH substrates are degraded by going through a set of interconnected functional processes or modules, termed RCPs, side chain processes (SCPs), and central aromatic processes (CAPs). As shown in Fig. 4, we identified, out of 202 genes initially proposed to be involved in the PAH-MN (33, 52), 163 genes as expressed. Among them, we found that 116 enzymes were up- or downregulated at each time point, which indicates a dynamic metabolic response in the degradation of PAHs (Fig. 4; see Fig. S6 in the supplemental material). Those 163 proteins were functionally diverse, being widely distributed among the three functional modules of the PAH-MN in M. vanbaalenii PYR-1; 70 proteins belonged to the functional module RCP, followed by 73 and 20 enzymes involved in the SCP and CAP, respectively (Fig. 4). As shown in Fig. S6 in the supplemental material, generally, the expression of the PAH-degrading proteins was dependent on functional modules of the PAH-MN. In addition, enzymes responsible for the RCP were more tightly regulated during the incubation period with BP crude oil, which is consistent with our previous results for M. vanbaalenii PYR-1 incubated with single or mixed PAHs (33, 34).
FIG 4.
Heat map presentation of the 163 expressed proteins involved in PAH degradation based on the functional modules of the PAH-MN.
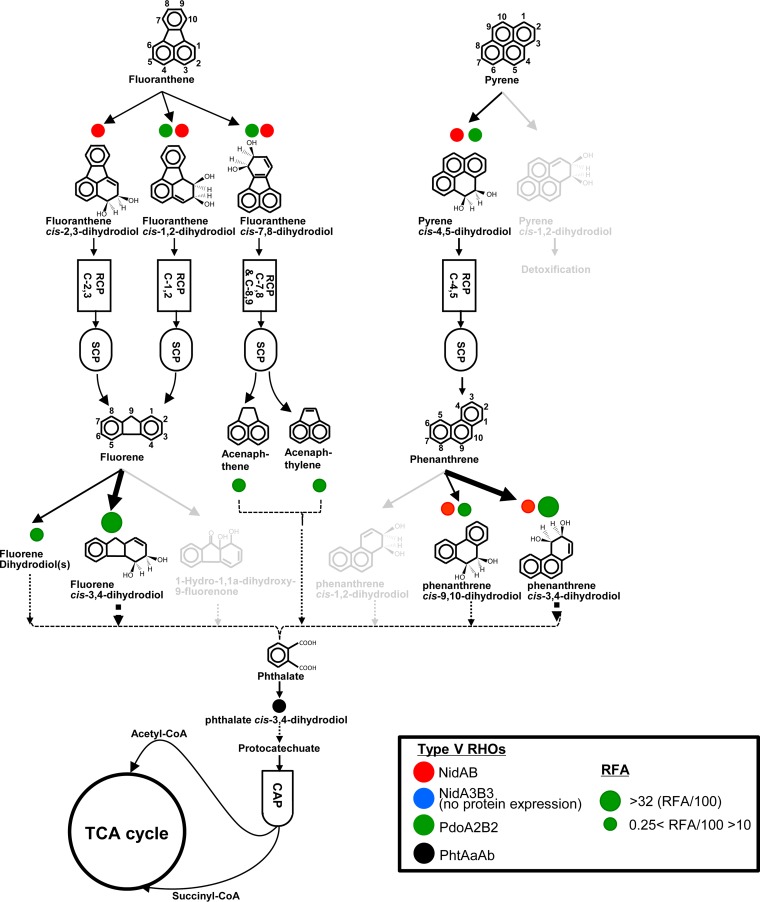
An RHO-centric functional map of the PAH-MN for BP crude oil.
Although the RCP is a nonproductive process to generate metabolites only for entering the SCP or CAP, this functional module is a crucial stage that determines the substrate range and pathways in the PAH-MN (33–35). In particular, RHOs are the first-step enzyme of RCP modules, and their highly complex epistatic interaction and pleiotropic activity govern the pathway and rate of degradation in the PAH-MN (33–35, 37, 53, 54). Among the 70 expressed RCP enzymes, 9 RHOs (out of 21 genome-predicted RHOs) were expressed upon exposure to BP crude oil, similar to previous proteome studies, in which about 10 RHO systems respond dynamically to pure or mixed PAH substrates (33–35, 44, 45, 55). However, consistent with the different metabolic patterns of the PAH substrates in this study, the profile in the expression of RHO enzymes with respect to BP crude oil was different from those of previous PAH studies. The most notable difference was a lack of upregulation of three type V RHO enzymes, PhtAaAb, NidAB, and NidA3B3, in BP crude oil (56). These three type V oxygenase enzymes use phthalate, pyrene, and fluoranthene, respectively, for their preferred substrates and are mainly involved in the initial hydroxylation of the respective aromatic hydrocarbons (33–35, 44, 45, 55). On the other hand, another type V RHO, Mvan_0546 (PdoA2), and a type X RHO, Mvan_4415, which are mainly involved in the initial oxidation of low-molecular-weight (LMW) PAHs with three or fewer fused benzene rings, such as phenanthrene, fluorene, anthracene, and biphenyl, were consistently upregulated across the incubation period (33–35, 44, 45). To enzymatically understand the different metabolic behavior of M. vanbaalenii PYR-1 toward the PAH substrates in BP crude oil, we reconstructed an RHO-centric functional map of the PAH-MN with pleiotropic and epistatic numerical scores of the expressed RHO enzymes, as proposed previously (35). As shown in Fig. 5, the pleiotropic activity and epistatic interactions of the expressed RHO systems in response to BP crude oil describe the observed metabolic feature: a significant decrease in the degradation of the HMW PAH fluoranthene in BP crude oil compared to pure- or mixed-PAH incubation. Among several possible factors involved in the discrepancy in the metabolism of crude oil PAHs, in particular, lack of upregulation of the two type V Nid systems (NidAB and NidA3B3) appeared to have directly affected the metabolism of BP crude oil HMW PAHs, such as pyrene and fluoranthene. On the basis of the numerically calculated relative functional activity (RFA) of RHO systems (35) for the PAH-MN (see Data Sets S4 and S5 in the supplemental material), among the 9 expressed RHOs, only three RHO systems, PhtAaAb (Mvan_0463/0464), NidAB (Mvan_0487/0488), and PdoA2B2 (Mvan_0546/0547), showed relatively high RFA scores of >5 (RFA/100). This clearly indicates that the three RHO systems were the main oxygenases for the initial ring hydroxylation of PAH substrates in BP crude oil, and their pleiotropic and epistatic hydroxylation activities determined the PAH metabolic quantity and quality in BP crude oil.
FIG 5.
Functional map of the M. vanbaalenii PYR-1 RHO enzymes for the degradation of BP crude oil phenanthrene, pyrene, and fluoranthene. (The map was adapted from our previous reports [34, 35] with modifications.) The arrows indicate degradation pathways, and differences in transformation efficiency are represented by the thickness of the arrows. The light-gray pathways indicate no activation of degradation routes. Circles with color indicate RHO enzymes, with sizes proportional to the degrees of functional contribution.
Fluoranthene degradation in BP crude oil.
The four-ring PAH fluoranthene is a good substrate for the PAH-MN in M. vanbaalenii PYR-1, which is degraded by both mono- and dioxygenation reactions through at least four metabolic routes (33–35). Considering its oxygenation ability toward a series of PAHs (fluoranthene, acenaphthene, acenaphthylene, and fluorene) in the fluoranthene subnetwork of the PAH-MN (33–35), the vertical epistatic and pleiotropic contributions of the NidA3B3 system to fluoranthene metabolism are crucial in terms of the metabolic quantity and quality. This indicates that the pleiotropic and epistatic functional loss of the type V NidA3B3 system could adversely affect the degradation of fluoranthene, as well as other PAHs. As revealed in the RHO-centric functional map for pure fluoranthene (35), the hydroxylation at the C-2,3 positions of fluoranthene, leading to the main C-2,3 dioxygenation pathway, is dependent only on the type V NidA3B3. The lack of epistatic hydroxylation of fluoranthene by other RHO systems indicates no functional redundancy and, finally, no functional robustness of the hydroxylation step for the functional loss of NidA3B3. Considering their low substrate acceptance and low product regiospecificity for the four-ring fluoranthene, functional assistance of other RHO systems for fluoranthene C-2,3 dioxygenation would not be significant. In conclusion, no expression of the type V NidA3B3 in the PAH-MN could have an apparent metabolic impact on fluoranthene degradation in M. vanbaalenii PYR-1, which is consistent with the metabolic pattern observed in this study.
Pleiotropic responsibility of the type V Pdo system.
On the other hand, phenanthrene and pyrene are the PAH substrates dioxygenated with the highest epistatic functional redundancy of RHO systems, including NidAB (Mvan_0487/0488) and PdoA2B2 (Mvan_0546/0547) systems in the PAH-MN (34, 35). Therefore, the observed RHO expression profile—upregulation of the PdoA2B2 system, downregulation of NidAB system, and lack of expression of NidA3B3—seems least likely to be problematic from the standpoint of degradation of the PAH substrates phenanthrene and pyrene in M. vanbaalenii PYR-1, which resulted in complete degradation of phenanthrene and pyrene, in contrast to the significant reduction in the degradation of fluoranthene compared with previous fluoranthene biodegradation studies (33–35, 43–45). In this respect, the epistatic and pleiotropic responsibility of the type V Pdo system seems to be crucial for dioxygenation of BP crude oil PAH substrates, including pyrene and phenanthrene. The type V Pdo system showed the highest RFA value and functional responsibility for at least 10 ring-hydroxylating steps (Fig. 5; see Data Set S5 in the supplemental material).
Other enzymes responsible for degradation of BP crude oil.
In addition to RHO enzymes, other proteins belonging to RCP modules, such as monooxygenase (9 proteins), dihydrodiol dehydrogenase (5 proteins), ring cleavage dioxygenase (6 proteins), and epoxide hydrolase (2 proteins), were also identified as being expressed in response to BP crude oil. Some of the 34 CYPs could also be involved in the initial oxidation of PAHs. In particular, Mvan_5170, encoding CYP150, which was previously implicated in the degradation of PAHs (57), was expressed. We observed the expression of many RCP enzymes, some of which have not been identified in prior proteome studies, which strongly suggests possible functional contributions by these enzymes to the degradation of BP crude oil PAHs.
In the PAH-MN, a side chain is removed from the outputs of the RCP to produce biological precursors, such as pyruvate and substrates for another round of RCPs, in the SCP, and protocatechuate is metabolically connected to the TCA cycle in the CAP (33). As observed in our previous proteomic results, M. vanbaalenii PYR-1 relatively loosely regulates PAH-degrading enzymes responsible for the SCP and CAP modules. Consistent with the previous observation, the expression of SCP enzymes was constitutive over the degradation of BP crude oil aromatic hydrocarbons. Twenty-eight out of the 72 SCP enzymes were stably expressed throughout the experiment, which contrasts with 10 RCP proteins identified as not responsive. Lastly, we identified all the CAP enzymes, PcaHGBLIJF, involved in the β-ketoadipate pathway responsible for the degradation of protocatechuate to succinyl-CoA and acetyl-CoA (Fig. 5; see Data Set S4 in the supplemental material).
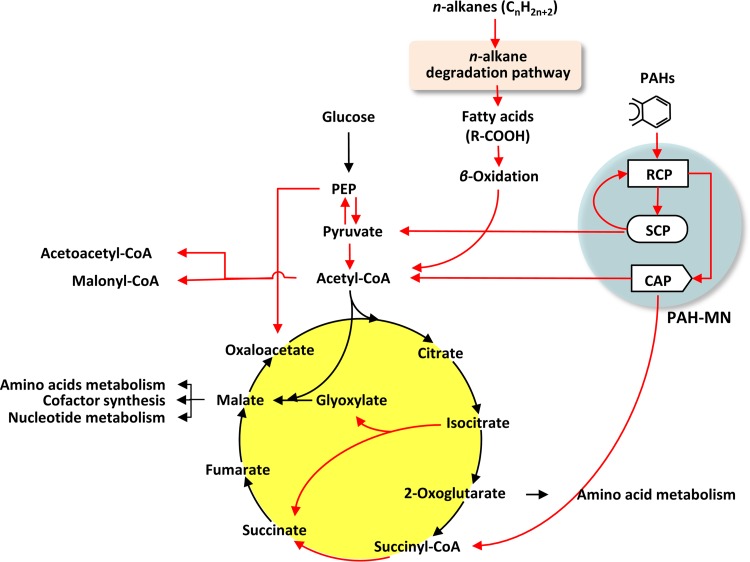
(iv) The global functional metabolic network between the peripheral hydrocarbon pathways and the central carbon pathways.
As discussed above, during growth on BP crude oil, M. vanbaalenii PYR-1 generates several metabolic products connected to the central metabolism, such as pyruvate, acetyl-CoA, and succinyl-CoA, which are the common intermediates of the central carbon metabolism. In BP crude oil metabolism, acetyl-CoA is formed from n-alkane degradation via β-oxidation and from the CAP of the PAH-MN during PAH metabolism, while pyruvate and succinyl-CoA are produced from the SCP and CAP of the PAH-MN, respectively. The proteomic dynamics in M. vanbaalenii PYR-1 showed that these common metabolites bridge the peripheral hydrocarbon breakdown pathways and the central metabolism. To better understand the connectivity of the peripheral hydrocarbon pathways and the corresponding response(s) of the central metabolism, a simplified functional metabolic network was reconstructed on the basis of the metabolic and proteomic information (Fig. 6). This functional metabolic network highlights activated pathways and therefore provides a degree of functional connectivity and activity among the metabolic routes (Fig. 6). In the functional metabolic network, the two peripheral hydrocarbon metabolic pathways, the n-alkane degradation pathway linked to β-oxidation and the PAH-MN, are activated and connected with the central carbon metabolism via pyruvate, acetyl-CoA, and succinyl-CoA. Interestingly, the functional distribution and regulation of the enzymes responsible for peripheral hydrocarbon metabolism appeared to have resulted in the peripheral pathways functioning in concentrating the flux of metabolic intermediates to the central metabolism. This observation further supports our idea about channel management, enhancing input substrate diversity with the controlled production of limited outputs (33). The funnel effects of channel management could decrease the epimetabolome (58) and ensure a more efficient carbon metabolic flow (33). After all, coordinated expression of the enzymes involved in n-alkane degradation and β-oxidation and function-dependent regulation of the PAH-degrading enzymes funnels the metabolic products of the BP crude oil hydrocarbons into the central metabolism. In addition, the central metabolism seems to respond in a way to accommodate channel management of the peripheral hydrocarbon metabolism. Overall, the functional global network indicates that, in M. vanbaalenii PYR-1, n-alkanes and PAH substrates in BP crude oil are ultimately oxidized to CO2 and H2O via the TCA cycle and the respiratory chain and used as a carbon source of biosynthetic precursors.
FIG 6.
Global functional metabolic network consisting of two major peripheral pathways, the n-alkane pathway via β-oxidation and the PAH-MN, and the TCA cycle for central carbon metabolism. The functional map was reconstructed by integrating the proteomic data with the framework of metabolic pathways (from the KEGG database), in which protein abundance was used as a proxy for enzyme activity. The red arrows represent steps activated in the pathways.
Structural view of the global functional metabolic network.
The integrated view of the top-down and enzyme-centric, bottom-up perspective of the functional metabolic network suggests that global carbon metabolism in M. vanbaalenii PYR-1 is not randomly connected or activated but rather has a scale-free structure and behavior with an apparent connection preference. The scale-free features of the global metabolic network well reflect the functional interactions, showing apparent movement of carbon (carbon flux) between the peripheral and central carbon pathways (Fig. 6). From a structural perspective, acetyl-CoA, pyruvate, phosphoenolpyruvate (PEP), and succinate showed a hub metabolic feature with a relatively high functional connection, indicating active metabolic dynamics, that is, relatively high in-and-out rates. In the functional network, acetyl-CoA showed the highest degree of functionally activated connection, which is three in and two out degrees. Before entering the TCA cycle, the carbon skeletons of n-alkanes and PAHs were degraded to acetyl groups (C2) of the hub metabolite, acetyl-CoA. However, the TCA cycle apparently did not accept most of the acetyl-CoA input. While the two out degrees toward acetoacetyl-CoA and malonyl-CoA were activated by the upregulation of acetyl-CoA acetyltransferases (Mvan_5067, Mvan_5222, and Mvan_5259) and carbamoyl-phosphate synthase (Mvan_4090), respectively, the two out degrees of acetyl-CoA toward the TCA cycle (acetyl-CoA + glyoxylate→malate and acetyl-CoA + oxaloacetate→citrate) were not activated. On the other hand, pyruvate seemed to be mainly converted into both acetyl-CoA and PEP. We observed that the three catalytic enzymes of the pyruvate dehydrogenase complex, pyruvate dehydrogenase (Mvan_1408), dihydrolipoyl transacetylase (Mvan_1409), and dihydrolipoyl dehydrogenase (Mvan_1410), catalyzing the oxidative decarboxylation of pyruvate to acetyl-CoA, were upregulated (STEM pattern 21). Two pyruvate dikinases (EC 2.7.9.2; Mvan_2632 and Mvan_4106), which produce PEP from pyruvate, were also upregulated (STEM pattern 21). Since a PEP carboxylase (Mvan_2707) was also upregulated (STEM pattern 21), the PEP with three active connection degrees could be converted into oxaloacetate, which is the cosubstrate for a pace-making citrate synthase in the first step of the TCA cycle. Citrate synthase catalyzes the condensation of a two-carbon acetate residue from acetyl-CoA and a four-carbon oxaloacetate molecule to form the six-carbon citrate.
Behavioral perspective of the pace-making enzymes in the TCA cycle.
The overall rate of the TCA cycle is controlled by the rate of conversion of pyruvate to acetyl-CoA and by the flux through citrate synthase, isocitrate dehydrogenase, and α-ketoglutarate dehydrogenase. From the perspective of behavior, the pace-making enzymes, citrate synthase (Mvan_5022 and Mvan_5025), isocitrate dehydrogenase (Mvan_3212), and α-ketoglutarate dehydrogenase (Mvan_3964 and Mvan_3965), were constitutively expressed with no upregulation under BP crude oil (see Data Set S2A in the supplemental material). Despite the activation of the production routes of acetyl-CoA, including the oxidative decarboxylation by pyruvate dehydrogenase (Mvan_1408, Mvan_1409, and Mvan_1410), the lack of upregulation of the first step, citrate synthase (Mvan_5022 and Mvan_5025), is intriguing, since the initial citrate condensation would largely determine the overall rate of the TCA cycle in BP crude oil hydrocarbon metabolism. Together with the diversion of acetyl-CoA into acetoacetyl-CoA or malonyl-CoA, induction of the metabolic route producing oxaloacetate from pyruvate via PEP explains that there is no upregulation of the initial pace-making enzyme under BP crude oil metabolism, probably because of a concentration imbalance between the two substrates of citrate synthase, acetyl-CoA and oxaloacetate. This behavioral hypothesis is consistent with the structural perspective of the functional metabolic network, that is, acetyl-CoA shows an activated out-connection degree toward acetoacetyl-CoA and malonyl-CoA, but oxaloacetate has an activated in-connection degree from PEP. Interestingly, the enzyme (Mvan_2707) responsible for the replenishment of oxaloacetate from PEP showed the same expression pattern (STEM profile 21) as the two pyruvate dikinases (Mvan_2632 and Mvan_4106), producing PEP from pyruvate. This coordinated expression pattern between the two steps, pyruvate to PEP and PEP to oxaloacetate, could enhance the replenishment of oxaloacetate.
Glyoxylate bypass and the CO2-releasing decarboxylation route in the TCA cycle.
In contrast to the lack of upregulation in the expression of the pace-making enzymes, among the TCA cycle enzymes, succinyl-CoA synthetase (Mvan_4870) and isocitrate lyase (Mvan_0801) were upregulated. As shown in the functional network, all the upregulated enzymes were concentrated in the steps from the intermediate isocitrate to succinate. In the TCA cycle, isocitrate seemed to be further metabolized by two pathways, the anaplerotic route (glyoxylate bypass) and the CO2-releasing decarboxylation route via α-ketoglutarate and succinyl-CoA. Isocitrate lyase (Mvan_0801) is a hallmark enzyme for the glyoxylate bypass, catalyzing the cleavage of isocitrate into one molecule each of succinate and glyoxylate. However, malate synthase (Mvan_3097), responsible for the condensation of glyoxylate with acetyl-CoA to generate the TCA cycle intermediate malate, showed no change in protein abundance during growth under BP crude oil. Considering that coordinated induction of both isocitrate lyase and malate synthase usually ensures maximal flux through this anaplerotic route, this functional disconnection suggests that the anaplerotic route via glyoxylate and malate is not essential. In agreement with this observation, the gluconeogenesis pathway via oxaloacetate and PEP and several anabolic pathways initiated from malate, such as cofactors and nucleotide metabolism, were not induced (see Data Set S4 in the supplemental material). As explained above, even PEP was used to enhance the concentration of oxaloacetate by upregulation of the PEP carboxylase (EC 4.1.1.31; Mvan_2027). In addition, although the two CO2-releasing decarboxylation enzymes (isocitrate dehydrogenase [Mvan_3212] and α-ketoglutarate dehydrogenase [Mvan_3954 and Mvan_3965]) were not upregulated, succinyl-CoA synthetase (Mvan_4870) in the CO2-releasing decarboxylation route was upregulated. The functional disconnection of the enzymes for the glyoxylate bypass has also been observed in other microorganisms (59–63). As shown in Fig. 6, succinyl-CoA is also produced from the CAP of the PAH-MN, in which the last enzyme, β-ketoadipyl-CoA thiolase, produces succinyl-CoA and acetyl-CoA from β-ketoadipyl-CoA (33, 44, 45).
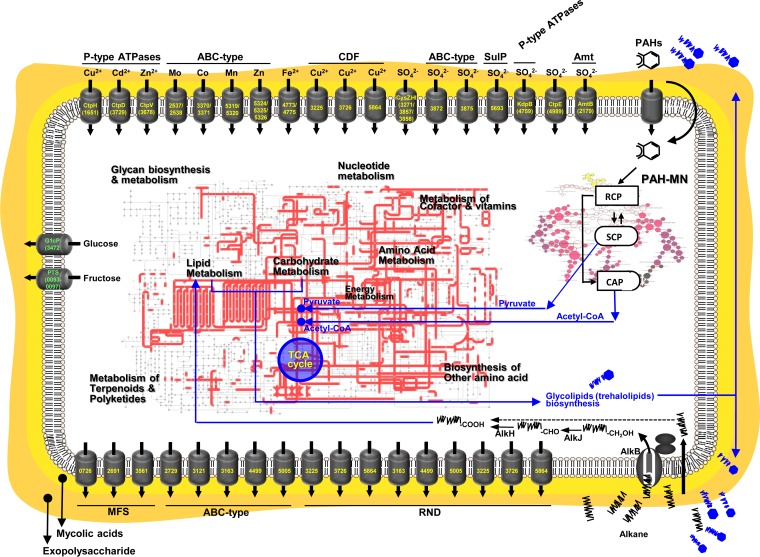
Coordinated cellular responses of M. vanbaalenii PYR-1 to BP crude oil.
Throughout the incubation of M. vanbaalenii PYR-1 with DWH BP crude oil, the bacterium differentially regulated more than 70% (2,735 proteins) of the expressed proteins, whose functions are distributed throughout the COG categories. This indicates that the exposure of M. vanbaalenii PYR-1 to BP crude oil had caused a dynamic, diverse, and complex cellular response. Based on the chemical and proteomic results of this study, we propose an overview of metabolism and other cellular adaptive responses in M. vanbaalenii PYR-1, as shown in Fig. 7.
FIG 7.
Overview of metabolism and transport involved in response to BP crude oil by M. vanbaalenii PYR-1.
Initially, adaptive responses of M. vanbaalenii PYR-1 to BP crude oil are probably triggered by transcriptional regulators. Hundreds of regulatory proteins are involved with expression of metabolic enzymes and other global physiological responses of M. vanbaalenii PYR-1 (see the supplemental material for details). One of the first cellular actions in response to BP crude oil is probably the synthesis of biosurfactant (see the supplemental material for details). When M. vanbaalenii PYR-1 comes into contact with droplets of oil, cells of the bacterium produce biosurfactant. This trehalose-containing glycolipid solubilizes hydrocarbon fractions of oil, which then are delivered into the cytoplasm through membrane transporters, initiating degradation of crude oil hydrocarbons. M. vanbaalenii PYR-1 is able to metabolize both aliphatic and aromatic hydrocarbons at the same time. Degradation of alkanes is probably mainly initiated by the AlkB terminal alkane monooxygenase, which is connected to the β-oxidation of fatty acid degradation. Although M. vanbaalenii PYR-1 metabolizes a broad range of n-alkanes up to chain length C28, it has the highest activity toward medium chain lengths, around C12 to C13. BP crude oil modified the bacterial regulation of enzymes responsible for PAH degradation, which resulted in less degradation of fluoranthene but complete degradation of phenanthrene and pyrene. The aliphatic and aromatic metabolism in M. vanbaalenii PYR-1 involves a sequence of coordinated reactions whereby the channel management enhances input substrate diversity with the controlled production of limited outputs of the peripheral pathways, resulting in concentration of the flux of intermediates to the central metabolism. A suite of transporter proteins involved in nutrient uptake or solutes are upregulated (see the supplemental material for details). Expression of transporters associated with efflux pumps is also upregulated, which is necessary for the efflux of BP crude oil toxic substances, as well as toxic intermediates generated from hydrocarbon metabolism (see the supplemental material for details).
Conclusions.
Bacterial metabolism is an organized behavior by which a bacterium obtains the energy and carbon sources to live and reproduce (33, 44, 45). Many research endeavors focus on isolation of oil-consuming microorganisms from oil-contaminated environments and evaluation of their efficiencies in the degradation of crude oil (6–8). Due to its exceptional catabolic abilities, M. vanbaalenii PYR-1 has been extensively studied with respect to metabolism of some of the components of crude oil hydrocarbons, such as PAHs, and discussions have been ongoing about its application in bioremediation (36). As revealed in this study, there was a genome-wide, coordinated cellular response of M. vanbaalenii PYR-1 to BP crude oil, and metabolic evidence confirms the potential of bioremediation for marine and terrestrial systems that the oil has contaminated. However, as revealed in the observed functional differences of PAH-degrading enzymes between pure PAHs and BP crude oil, there is still much to learn about the physiology and biochemistry of oil component biodegradation in vitro, in vivo, in situ, and ex situ. In particular, knowledge gaps between in vitro and in vivo and between in situ and ex situ seriously hinder efforts toward practical bioremediation applications. More systematic research should be conducted to bridge the laboratory and the field in the biodegradation of petroleum components.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dongryeoul Bae and Young-Beom Ahn for critical review of the manuscript.
This work was supported in part by an appointment to the Postgraduate Research Fellowship Program at the National Center for Toxicological Research, administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the U.S. Food and Drug Administration.
The views presented in this article do not necessarily reflect those of the U.S. FDA.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00730-15.
REFERENCES
- 1.Barron MG. 2012. Ecological impacts of the Deepwater Horizon oil spill: implications for immunotoxicity. Toxicol Pathol 40:315–320. doi: 10.1177/0192623311428474. [DOI] [PubMed] [Google Scholar]
- 2.Mendelssohn IA, Andersen GL, Baltz DM, Caffey RH, Carman KR, Fleeger JW, Joye SB, Lin QX, Maltby E, Overton EB, Rozas LP. 2012. Oil impacts on coastal wetlands: implications for the Mississippi River Delta ecosystem after the Deepwater Horizon oil spill. Bioscience 62:562–574. doi: 10.1525/bio.2012.62.6.7. [DOI] [Google Scholar]
- 3.Mishra DR, Cho HJ, Ghosh S, Fox A, Downs C, Merani PBT, Kirui P, Jackson N, Mishra S. 2012. Post-spill state of the marsh: remote estimation of the ecological impact of the Gulf of Mexico oil spill on Louisiana salt marshes. Remote Sens Environ 118:176–185. doi: 10.1016/j.rse.2011.11.007. [DOI] [Google Scholar]
- 4.Silliman BR, van de Koppel J, McCoy MW, Diller J, Kasozi GN, Earl K, Adams PN, Zimmerman AR. 2012. Degradation and resilience in Louisiana salt marshes after the BP-Deepwater Horizon oil spill. Proc Natl Acad Sci U S A 109:11234–11239. doi: 10.1073/pnas.1204922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atlas RM. 1981. Microbial degradation of petroleum hydrocarbons: an environmental perspective. Microbiol Rev 45:180–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leahy JG, Colwell RR. 1990. Microbial degradation of hydrocarbons in the environment. Microbiol Rev 54:305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Hamme JD, Singh A, Ward OP. 2003. Recent advances in petroleum microbiology. Microbiol Mol Biol Rev 67:503–549. doi: 10.1128/MMBR.67.4.503-549.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yakimov MM, Timmis KN, Golyshin PN. 2007. Obligate oil-degrading marine bacteria. Curr Opin Biotechnol 18:257–266. doi: 10.1016/j.copbio.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 9.Cerniglia CE. 1992. Biodegradation of polycyclic aromatic hydrocarbons. Biodegradation 3:351–368. doi: 10.1007/BF00129093. [DOI] [Google Scholar]
- 10.Vilchez-Vargas R, Junca H, Pieper DH. 2010. Metabolic networks, microbial ecology and ‘omics’ technologies: towards understanding in situ biodegradation processes. Environ Microbiol 12:3089–3104. doi: 10.1111/j.1462-2920.2010.02340.x. [DOI] [PubMed] [Google Scholar]
- 11.Stenuit B, Eyers L, Schuler L, Agathos SN, George I. 2008. Emerging high-throughput approaches to analyze bioremediation of sites contaminated with hazardous and/or recalcitrant wastes. Biotechnol Adv 26:561–575. doi: 10.1016/j.biotechadv.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Hazen TC, Dubinsky EA, DeSantis TZ, Andersen GL, Piceno YM, Singh N, Jansson JK, Probst A, Borglin SE, Fortney JL, Stringfellow WT, Bill M, Conrad ME, Tom LM, Chavarria KL, Alusi TR, Lamendella R, Joyner DC, Spier C, Baelum J, Auer M, Zemla ML, Chakraborty R, Sonnenthal EL, D'Haeseleer P, Holman HYN, Osman S, Lu ZM, Van Nostrand JD, Deng Y, Zhou JZ, Mason OU. 2010. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 330:204–208. doi: 10.1126/science.1195979. [DOI] [PubMed] [Google Scholar]
- 13.Mason OU, Hazen TC, Borglin S, Chain PSG, Dubinsky EA, Fortney JL, Han J, Holman HYN, Hultman J, Lamendella R, Mackelprang R, Malfatti S, Tom LM, Tringe SG, Woyke T, Zhou JH, Rubin EM, Jansson JK. 2012. Metagenome, metatranscriptome and single-cell sequencing reveal microbial response to Deepwater Horizon oil spill. ISME J 6:1715–1727. doi: 10.1038/ismej.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.King GM, Kostka JE, Hazen TC, Sobecky PA. 2015. Microbial responses to the Deepwater Horizon oil spill: from coastal wetlands to the deep sea. Annu Rev Mar Sci 7:377–401. doi: 10.1146/annurev-marine-010814-015543. [DOI] [PubMed] [Google Scholar]
- 15.Kostka JE, Prakash O, Overholt WA, Green SJ, Freyer G, Canion A, Delgardio J, Norton N, Hazen TC, Huettel M. 2011. Hydrocarbon-degrading bacteria and the bacterial community response in Gulf of Mexico beach sands impacted by the Deepwater Horizon oil spill. Appl Environ Microbiol 77:7962–7974. doi: 10.1128/AEM.05402-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutierrez T, Singleton DR, Berry D, Yang TT, Aitken MD, Teske A. 2013. Hydrocarbon-degrading bacteria enriched by the Deepwater Horizon oil spill identified by cultivation and DNA-SIP. ISME J 7:2091–2104. doi: 10.1038/ismej.2013.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu ZM, Deng Y, Van Nostrand JD, He ZL, Voordeckers J, Zhou AF, Lee YJ, Mason OU, Dubinsky EA, Chavarria KL, Tom LM, Fortney JL, Lamendella R, Jansson JK, D'Haeseleer P, Hazen TC, Zhou JZ. 2012. Microbial gene functions enriched in the Deepwater Horizon deep-sea oil plume. ISME J 6:451–460. doi: 10.1038/ismej.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khan AA, Kim SJ, Paine DD, Cerniglia CE. 2002. Classification of a polycyclic aromatic hydrocarbon-metabolizing bacterium, Mycobacterium sp. strain PYR-1, as Mycobacterium vanbaalenii sp. nov. Int J Syst Evol Microbiol 52:1997–2002. doi: 10.1099/ijs.0.02163-0. [DOI] [PubMed] [Google Scholar]
- 19.Kim YH, Engesser KH, Cerniglia CE. 2005. Numerical and genetic analysis of polycyclic aromatic hydrocarbon-degrading mycobacteria. Microb Ecol 50:110–119. doi: 10.1007/s00248-004-0126-3. [DOI] [PubMed] [Google Scholar]
- 20.Heitkamp MA, Cerniglia CE. 1988. Mineralization of polycyclic aromatic hydrocarbons by a bacterium isolated from sediment below an oil field. Appl Environ Microbiol 54:1612–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heitkamp MA, Franklin W, Cerniglia CE. 1988. Microbial metabolism of polycyclic aromatic hydrocarbons: isolation and characterization of a pyrene-degrading bacterium. Appl Environ Microbiol 54:2549–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heitkamp MA, Cerniglia CE. 1989. Polycyclic aromatic hydrocarbon degradation by a Mycobacterium sp. in microcosms containing sediment and water from a pristine ecosystem. Appl Environ Microbiol 55:1968–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelley I, Freeman JP, Evans FE, Cerniglia CE. 1991. Identification of a carboxylic acid metabolite from the catabolism of fluoranthene by a Mycobacterium sp. Appl Environ Microbiol 57:636–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moody JD, Freeman JP, Cerniglia CE. 2005. Degradation of benz[a]anthracene by Mycobacterium vanbaalenii strain PYR-1. Biodegradation 16:513–526. doi: 10.1007/s10532-004-7217-1. [DOI] [PubMed] [Google Scholar]
- 25.Moody JD, Fu PP, Freeman JP, Cerniglia CE. 2003. Regio- and stereoselective metabolism of 7,12-dimethylbenz[a]anthracene by Mycobacterium vanbaalenii PYR-1. Appl Environ Microbiol 69:3924–3931. doi: 10.1128/AEM.69.7.3924-3931.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moody JD, Freeman JP, Fu PP, Cerniglia CE. 2004. Degradation of benzo[a]pyrene by Mycobacterium vanbaalenii PYR-1. Appl Environ Microbiol 70:340–345. doi: 10.1128/AEM.70.1.340-345.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moody JD, Doerge DR, Freeman JP, Cerniglia CE. 2002. Degradation of biphenyl by Mycobacterium sp. strain PYR-1. Appl Microbiol Biotechnol 58:364–369. doi: 10.1007/s00253-001-0878-3. [DOI] [PubMed] [Google Scholar]
- 28.Moody JD, Freeman JP, Doerge DR, Cerniglia CE. 2001. Degradation of phenanthrene and anthracene by cell suspensions of Mycobacterium sp. strain PYR-1. Appl Environ Microbiol 67:1476–1483. doi: 10.1128/AEM.67.4.1476-1483.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelley I, Freeman JP, Cerniglia CE. 1990. Identification of metabolites from degradation of naphthalene by a Mycobacterium sp. Biodegradation 1:283–290. doi: 10.1007/BF00119765. [DOI] [PubMed] [Google Scholar]
- 30.Heitkamp MA, Freeman JP, Miller DW, Cerniglia CE. 1991. Biodegradation of 1-nitropyrene. Arch Microbiol 156:223–230. doi: 10.1007/BF00249119. [DOI] [PubMed] [Google Scholar]
- 31.Falkinham JO. 2009. The biology of environmental mycobacteria. Environ Microbiol Rep 1:477–487. doi: 10.1111/j.1758-2229.2009.00054.x. [DOI] [PubMed] [Google Scholar]
- 32.Kieser T, Bibb JM, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, United Kingdom. [Google Scholar]
- 33.Kweon O, Kim SJ, Holland RD, Chen H, Kim DW, Gao Y, Yu LR, Baek S, Baek DH, Ahn H, Cerniglia CE. 2011. Polycyclic aromatic hydrocarbon metabolic network in Mycobacterium vanbaalenii PYR-1. J Bacteriol 193:4326–4337. doi: 10.1128/JB.00215-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim SJ, Song J, Kweon O, Holland RD, Kim DW, Kim JN, Yu LR, Cerniglia CE. 2012. Functional robustness of a polycyclic aromatic hydrocarbon metabolic network examined in a nidA aromatic ring-hydroxylating oxygenase mutant of Mycobacterium vanbaalenii PYR-1. Appl Environ Microbiol 78:3715–3723. doi: 10.1128/AEM.07798-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kweon O, Kim SJ, Kim DW, Kim JM, Kim HL, Ahn Y, Sutherland JB, Cerniglia CE. 2014. Pleiotropic and epistatic behavior of a ring-hydroxylating oxygenase system in the polycyclic aromatic hydrocarbon metabolic network from Mycobacterium vanbaalenii PYR-1. J Bacteriol 196:3503–3515. doi: 10.1128/JB.01945-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bogan BW, Lahner LM, Sullivan WR, Paterek JR. 2003. Degradation of straight-chain aliphatic and high-molecular-weight polycyclic aromatic hydrocarbons by a strain of Mycobacterium austroafricanum. J Appl Microbiol 94:230–239. doi: 10.1046/j.1365-2672.2003.01824.x. [DOI] [PubMed] [Google Scholar]
- 37.Kweon O, Kim SJ, Freeman JP, Song J, Baek S, Cerniglia CE. 2010. Substrate specificity and structural characteristics of the novel Rieske nonheme iron aromatic ring-hydroxylating oxygenases NidAB and NidA3B3 from Mycobacterium vanbaalenii PYR-1. mBio 1:e00135-10. doi: 10.1128/mBio.00135-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim SJ, Kweon O, Cerniglia CE. 2010. Degradation of polycyclic aromatic hydrocarbons by Mycobacterium strains, p 1865–1880. In Timmis KN. (ed), Handbook of hydrocarbon and lipid microbiology, vol 3 Springer, Braunschweig, Germany. [Google Scholar]
- 39.Vila J, Grifoll M. 2009. Actions of Mycobacterium sp. strain AP1 on the saturated- and aromatic-hydrocarbon fractions of fuel oil in a marine medium. Appl Environ Microbiol 75:6232–6239. doi: 10.1128/AEM.02726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Beilen JB, Li Z, Duetz WA, Smits THM, Witholt B. 2003. Diversity of alkane hydroxylase systems in the environment. Oil Gas Sci Technol 58:427–440. doi: 10.2516/ogst:2003026. [DOI] [Google Scholar]
- 41.Perez-de-Mora A, Engel M, Schloter M. 2011. Abundance and diversity of n-alkane-degrading bacteria in a forest soil co-contaminated with hydrocarbons and metals: a molecular study on alkB homologous genes. Microb Ecol 62:959–972. doi: 10.1007/s00248-011-9858-z. [DOI] [PubMed] [Google Scholar]
- 42.van Beilen JB, Funhoff EG. 2007. Alkane hydroxylases involved in microbial alkane degradation. Appl Microbiol Biotechnol 74:13–21. doi: 10.1007/s00253-006-0748-0. [DOI] [PubMed] [Google Scholar]
- 43.Kelley I, Cerniglia CE. 1995. Degradation of a mixture of high-molecular-weight polycyclic aromatic hydrocarbons by a Mycobacterium strain PYR-1. J Soil Contam 4:77–91. doi: 10.1080/15320389509383482. [DOI] [Google Scholar]
- 44.Kim SJ, Kweon O, Jones RC, Freeman JP, Edmondson RD, Cerniglia CE. 2007. Complete and integrated pyrene degradation pathway in Mycobacterium vanbaalenii PYR-1 based on systems biology. J Bacteriol 189:464–472. doi: 10.1128/JB.01310-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kweon O, Kim SJ, Jones RC, Freeman JP, Adjei MD, Edmondson RD, Cerniglia CE. 2007. A polyomic approach to elucidate the fluoranthene degradative pathway in Mycobacterium vanbaalenii PYR-1. J Bacteriol 189:4635–4647. doi: 10.1128/JB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tatusov RL, Galperin MY, Natale DA, Koonin EV. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ernst J, Bar-Joseph Z. 2006. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics 7:191. doi: 10.1186/1471-2105-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rojo F. 2009. Degradation of alkanes by bacteria. Environ Microbiol 11:2477–2490. doi: 10.1111/j.1462-2920.2009.01948.x. [DOI] [PubMed] [Google Scholar]
- 49.Whyte LG, Smits THM, Labbe D, Witholt B, Greer CW, van Beilen JB. 2002. Gene cloning and characterization of multiple alkane hydroxylase systems in Rhodococcus strains Q15 and NRRL B-16531. Appl Environ Microbiol 68:5933–5942. doi: 10.1128/AEM.68.12.5933-5942.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Procopio L, Silva MDPE, van Elsas JD, Seldin L. 2013. Transcriptional profiling of genes involved in n-hexadecane compounds assimilation in the hydrocarbon degrading Dietzia cinnamea P4 strain. Braz J Microbiol 44:633–641. doi: 10.1590/S1517-83822013000200044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abokitse K, Hummel W. 2003. Cloning, sequence analysis, and heterologous expression of the gene encoding a (S)-specific alcohol dehydrogenase from Rhodococcus erythropolis DSM 43297. Appl Microbiol Biotechnol 62:380–386. doi: 10.1007/s00253-003-1310-y. [DOI] [PubMed] [Google Scholar]
- 52.Kim SJ, Kweon O, Jones RC, Edmondson RD, Cerniglia CE. 2008. Genomic analysis of polycyclic aromatic hydrocarbon degradation in Mycobacterium vanbaalenii PYR-1. Biodegradation 19:859–881. doi: 10.1007/s10532-008-9189-z. [DOI] [PubMed] [Google Scholar]
- 53.Khan AA, Wang RF, Cao WW, Doerge DR, Wennerstrom D, Cerniglia CE. 2001. Molecular cloning, nucleotide sequence, and expression of genes encoding a polycyclic aromatic ring dioxygenase from Mycobacterium sp. strain PYR-1. Appl Environ Microbiol 67:3577–3585. doi: 10.1128/AEM.67.8.3577-3585.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim SJ, Kweon O, Freeman JP, Jones RC, Adjei MD, Jhoo JW, Edmondson RD, Cerniglia CE. 2006. Molecular cloning and expression of genes encoding a novel dioxygenase involved in low- and high-molecular-weight polycyclic aromatic hydrocarbon degradation in Mycobacterium vanbaalenii PYR-1. Appl Environ Microbiol 72:1045–1054. doi: 10.1128/AEM.72.2.1045-1054.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim SJ, Jones RC, Cha CJ, Kweon O, Edmondson RD, Cerniglia CE. 2004. Identification of proteins induced by polycyclic aromatic hydrocarbon in Mycobacterium vanbaalenii PYR-1 using two-dimensional polyacrylamide gel electrophoresis and de novo sequencing methods. Proteomics 4:3899–3908. doi: 10.1002/pmic.200400872. [DOI] [PubMed] [Google Scholar]
- 56.Kweon O, Kim SJ, Baek S, Chae JC, Adjei MD, Baek DH, Kim YC, Cerniglia CE. 2008. A new classification system for bacterial Rieske non-heme iron aromatic ring-hydroxylating oxygenases. BMC Biochem 9:11. doi: 10.1186/1471-2091-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brezna B, Kweon O, Stingley RL, Freeman JP, Khan AA, Polek B, Jones RC, Cerniglia CE. 2006. Molecular characterization of cytochrome P450 genes in the polycyclic aromatic hydrocarbon degrading Mycobacterium vanbaalenii PYR-1. Appl Microbiol Biotechnol 71:522–532. doi: 10.1007/s00253-005-0190-8. [DOI] [PubMed] [Google Scholar]
- 58.de Lorenzo V. 2008. Systems biology approaches to bioremediation. Curr Opin Biotechnol 19:579–589. doi: 10.1016/j.copbio.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 59.Chopra T, Hamelin R, Armand F, Chiappe D, Moniatte M, McKinney JD. 2014. Quantitative mass spectrometry reveals plasticity of metabolic networks in Mycobacterium smegmatis. Mol Cell Proteomics 13:3014–3028. doi: 10.1074/mcp.M113.034082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, Dolganov G, Efron B, Butcher PD, Nathan C, Schoolnik GK. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J Exp Med 198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Timm J, Post FA, Bekker LG, Walther GB, Wainwright HC, Manganelli R, Chan WT, Tsenova L, Gold B, Smith I, Kaplan G, McKinney JD. 2003. Differential expression of iron-, carbon-, and oxygen-responsive mycobacterial genes in the lungs of chronically infected mice and tuberculosis patients. Proc Natl Acad Sci U S A 100:14321–14326. doi: 10.1073/pnas.2436197100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith CV, Huang CC, Miczak A, Russell DG, Sacchettini JC, Bentrup KHZ. 2003. Biochemical and structural studies of malate synthase from Mycobacterium tuberculosis. J Biol Chem 278:1735–1743. doi: 10.1074/jbc.M209248200. [DOI] [PubMed] [Google Scholar]
- 63.Micklinghoff JC, Breitinger KJ, Schmidt M, Geffers R, Eikmanns BJ, Bange FC. 2009. Role of the transcriptional regulator RamB (Rv0465c) in the control of the glyoxylate cycle in Mycobacterium tuberculosis. J Bacteriol 191:7260–7269. doi: 10.1128/JB.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.