Abstract
The sequencing chips and kits of the Ion Torrent Personal Genome Machine (PGM), which employs semiconductor technology to measure pH changes in polymerization events, have recently been upgraded. The quality of PGM sequences has not been reassessed, and results have not been compared in the context of a gene-targeted microbial ecology study. To address this, we compared sequence profiles across available PGM chips and chemistries and with 454 pyrosequencing data by determining error types and rates and diazotrophic community structures. The PGM was then used to assess differences in nifH-harboring bacterial community structure among four corn-based cropping systems. Using our suggested filters from mock community analyses, the overall error rates were 0.62, 0.36, and 0.39% per base for chips 318 and 314 with the 400-bp kit and chip 318 with the Hi-Q chemistry, respectively. Compared with the 400-bp kit, the Hi-Q kit reduced indel rates by 28 to 59% and produced one to seven times more reads acceptable for downstream analyses. The PGM produced higher frameshift rates than pyrosequencing that were corrected by the RDP FrameBot tool. Significant differences among platforms were identified, although the diversity indices and overall site-based conclusions remained similar. For the cropping system analyses, a total of 6,182 unique NifH operational taxonomic units at 5% amino acid dissimilarity were obtained. The current crop type, as well as the crop rotation history, significantly influenced the composition of the soil diazotrophic community detected.
INTRODUCTION
Next-generation sequencing (NGS) technologies have evolved rapidly in the years since pyrosequencing was initially launched in 2005 (1), followed by light-imaging Illumina (2, 3) HiSeq and MiSeq and by the post-light-sequencing Ion Torrent (4, 5) Personal Genome Machine (PGM) and Proton. Sequencing quality (error type and rate) and length have suffered with NGS platforms (3, 6, 7) compared to Sanger sequencing, but the greatly improved sequence yields and reduced cost have resulted in NGS dominating use. The higher error rate has been problematic for microbial ecology, but by parallel sequencing of mock communities of known composition, the type and extent of insertion, deletion, or substitution errors can be characterized. This information can then be used to determine the proper parameters and filters for parsing raw sequence data to minimize the impact of these errors on biological conclusions (8, 9).
To date, sequencing platform comparisons or case studies employing the Ion Torrent PGM were focused mainly on microbial whole-genome sequencing (6, 7, 10, 11) or on bacterial community structure analysis using 16S rRNA gene multiplexing tags (12–14). A few studies used PGM for medical diagnosis with marker genes (15–17). No studies have (i) used ecofunctional genes to evaluate Ion Torrent PGM error profiles with the Sequencing 400 kit (400-bp kit) or the recently released Hi-Q Sequencing kit (Hi-Q kit) or (ii) directly compared the PGM data with those obtained with another sequencing platform.
In this study, we determined the error types and rates and the sequencing output of the Ion Torrent PGM by using the 314 chip with the 400-bp kit and the 318 chip with both the 400-bp and the Hi-Q kits. Mock communities were analyzed to guide the use of appropriate filters for removal of low-quality reads. Sequencing error profiles of the Ion Torrent PGM and Roche 454 platforms were directly compared by using the same samples to determine differences in community structure, diversity indices, and the presence or absence and relative abundances of major operational taxonomic units (OTUs). Lastly, we used the Ion Torrent PGM 318 chip with the Hi-Q kit to sequence nifH amplicons from bulk soil samples as a case study to characterize the nitrogen-fixing bacterial communities in four cropping systems that varied in crop type, rotation, and cover crop.
MATERIALS AND METHODS
Community DNA.
The genomic DNA samples used consisted of a defined community (mock) containing nifH-harboring strains Polaromonas naphthalenivorans CJ2, Desulfitobacterium hafniense DCB-2, and Burkholderia vietnamensis G4; six Oklahoma prairie soil samples from 0 to 15 cm sequenced by 454; and 12 bulk soil samples from four different cropping systems. The Oklahoma grassland samples were from warming treatments (T) and control (C) sites from a tallgrass prairie at the Great Plains Apiaries site (34°58′54″N, 97°31′14″W), where experimental warming has been under way for over a decade (18–20). The DNA of the Oklahoma samples was extracted by using a mechanical-lysis freeze-grinding method (21). The 12 bulk soil samples (G1 to G4, three plots each) were collected at the Great Lakes Bioenergy Research Center intensive site at Arlington, WI (43°18′9.47″N, 89°20′43.32″W), in July 2013. Cropping system G1 was continuous corn, G2 was corn plus a cover crop annually, while G3 and G4 were soybean plus a cover crop and corn plus a cover crop and were in a corn-soybean rotation since 2012. Previously G2 to G4 were in a corn-soybean-canola rotation from 2008 to 2011 (see Table S1 in the supplemental material for the cropping history and Table S2 for soil properties). Six cores (2.5 cm in diameter by 25 cm deep [depth, 0 to 25 cm]) collected from the same plot were sieved (4 mm) and mixed together into one sample. Well-mixed soil (0.3 g) was used for DNA extraction with the Power Soil DNA isolation kit (MO BIO) according to the manufacturer's protocol. The DNA was quantified with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technology) and stored at −20°C for the following application.
nifH library generation and sequencing.
The nifH gene was amplified by using the Poly primers (22) in triplicate PCRs with fusion primers (see Table S3 in the supplemental material). PCR mixtures for both PGM and pyrosequencing contained 1× green buffer (Promega, Madison, WI), 1.8 mM MgCl2 (Promega), 0.5 mM each deoxynucleoside triphosphate (Promega), 500 nM each primer (IDT), 0.1 mg/ml bovine serum albumin (NEB), 2.5 U of Taq polymerase (Promega), and 1 ng/μl template DNA in a 20-μl reaction volume. Amplifications for both PGM and pyrosequencing were performed as follows: 3 min at 95°C; 30 cycles of 45 s at 94°C, 45 s at 61°C (62°C for 454), and 1 min at 72°C; and a final extension for 7 min at 72°C. PCR products were checked on a 1.6% agarose gel by using SYBR Safe gel stain (Invitrogen) and extracted from the gel with the QIAquick gel extraction kit (Qiagen). The eluted DNA was purified again with the Qiagen PCR Purification kit (Qiagen). The mock community was amplified for Ion Torrent sequencing with two barcoded primers to produce M-1 and M-2 samples.
For PGM and pyrosequencing, each purified sample was quantified with the Qubit Fluorometer (Invitrogen). Equivalent DNA of each sample was pooled, and the final concentration was adjusted to >10 ng/μl. For PGM sequencing, the pooled library was sent to the Research Technology Support Facility (RTSF) at Michigan State University (East Lansing) for sequencing on an Ion Torrent PGM (Life Technologies). Briefly, the library was diluted to 26 pM and set up on the OneTouch2 instruments with the Ion PGM Template OT2 400 kit by following the manufacturer's instructions. These templated beads were then purified on the OneTouch ES system. Following enrichment, the beads were loaded onto the PGM chip and sequenced in accordance with the manufacturer's instructions. Sequencing data were processed by the Torrent Suite Software V4.0. To compare PGM chips and sequencing kits, mock (M-1 and M-2) and Oklahoma amplicons were sequenced on a 314 v2 chip with the Ion PGM 400-bp kit first and then sequenced on a 318 v2 chip with both the 400-bp kit and the Ion PGM Hi-Q kit along with the Arlington crop rotation samples. Hi-Q kit sequencing was done in the University of Wisconsin Biotechnology Center by using the same procedures as the RTSF and processed by the synchronized Torrent Suite Software V4.4. The mock community was also sequenced in additional 318 chip runs with the 400-bp kit (M-R2, M-R3, and M-R4) at the RTSF. Pyrosequencing was performed at the Utah State University Center for Integrated Biosystems with the 454 Titanium platform by following the manufacturer's instructions.
Mock community analysis.
To assess the sequencing error type and rate in concert with the read Q (quality) score, the Defined Community Analysis Tool in the FunGene pipeline was adopted to analyze the mock sample reads. This tool compares the sequencing reads with the known corresponding region (reference sequences) from the mock community organisms (9). To this end, filters for passing the raw reads through initial quality filtering were set to a forward primer maximum edit distance of 2, a reverse primer maximum edit distance of 1, a maximum number of N′s of 0, and a minimum sequence length of 300 (excluding primers). To examine the relationship of error rates at each read Q score cutoff, a minimum read Q score of 0 was used to allow reads with any read Q score to pass. Usearch (23) and BLAST (24) were used to detect the small numbers of chimeric and contaminant reads, which were then excluded when the summary file was generated by this tool. The numbers of total substitutions and indels, read Q score distributions, overall error rates (total errors divided by total bases for each read Q score cutoff), and percentages of reads remaining and reads with specific errors that varied by Q score were summarized. The results obtained with this tool further optimized the analysis parameters, such as the Q score quality filter, for the environmental reads.
Data processing.
For both PGM and pyrosequencing, all of the reads obtained from soil DNA were trimmed by the Initial Process tool in the FunGene pipeline (9) (http://fungene.cme.msu.edu/FunGenePipeline/) by using the quality filtering parameters described above, except with a minimum read Q score of 22 to filter out low-quality reads. The resulting filtered reads with primers removed were subjected to Usearch (23) for chimera check in de novo mode. After chimera removal, all samples were translated and frameshift corrected with FrameBot (25) at default settings. The frameshift-corrected protein sequences were aligned with FunGene HMMER3 Aligner and then clustered by RDP mcClust with the complete linkage algorithm. The cluster file was converted to an R-formatted OTU table by the RDP cluster file formatter tool for use by R (version 2.12.0; http://www.r-project.org/) for ordination analysis. Sequences were randomly resampled to 1,814 amino acid sequences per sample for platform comparisons and to 22,616 sequences for the cropping system study.
Platform comparison of Oklahoma samples.
OTUs were generated at amino acid dissimilarities of 0% (OTU0) and 5% (OTU0.05). OTU abundances were Hellinger transformed (square root of relative abundance), and Bray-Curtis and Euclidean dissimilarity matrices (with dummy variable +1) were constructed with PRIMER-v6 (Primer-E Ltd., Plymouth, United Kingdom). Complete linkage clustering was used to construct cluster dendrograms with similarity profile (SIMPROF) testing (26). Permutational multivariate analysis of variance (PERMANOVA) (27) was used to test for significant differences in community structure. Shannon diversity (H′), Margalef's richness (d), Pielou's evenness (J′), and the number of individuals (Sab) were tested for significant differences by one-way analysis of variance (ANOVA) among sequencing platforms with Minitab 16 (Minitab Inc.) with a Tukey correction for multiple comparisons. Representative sequences were generated as the minimum sum of square distances of each OTU0.05. These representative sequences were assigned to a closest match by BLASTp with the FunGene NifH database consisting of 675 hand-curated sequences where the extracted protein corresponded to the amplicon generated by the primers (modified Zehr set) (25).
Nitrogen-fixing communities in soils of different cropping systems.
All of the sequences from the 12 soil samples were clustered at the OTU0.05 level (see Fig. S1 in the supplemental material for the rarefaction curve) and then resampled to 22,616 sequences per sample for calculation of diversity indices (H′ and J′) and downstream analyses. Significant differences in diversity indices were tested by one-way ANOVA with SPSS 18. A heat map of the 30 most abundant OTU0.05s (see Fig. S2) was produced with the labdsv package (version 1.6-1; http://CRAN.R-project.org/package=labdsv). OTU0.05 abundance data were Hellinger transformed and subjected to redundancy analysis (RDA) for ordination under constraint of the four cropping systems with the Vegan package (version 2.0-10; http://CRAN.R-project.org/package=vegan). The significance of the cropping system's influence was assessed by PERMANOVA, and differences between within-sample variances were tested with the permutational dispersion (PERMDISP) test. Read assignment into the taxonomic and NifH clusters was completed with FrameBot when conducting frameshift correction. Soil properties were fitted to the RDA model with the Vegan package.
Nucleotide sequence accession numbers.
Raw sequences from PGM and 454 were deposited in the NCBI Sequence Read Archive under accession number PRJNA266566 and in the European Nucleotide Archive Sequence Read Archive under accession number PRJEB8005 (sample accession numbers ERS629288 to ERS629293), respectively.
RESULTS
PGM sequencing and error type.
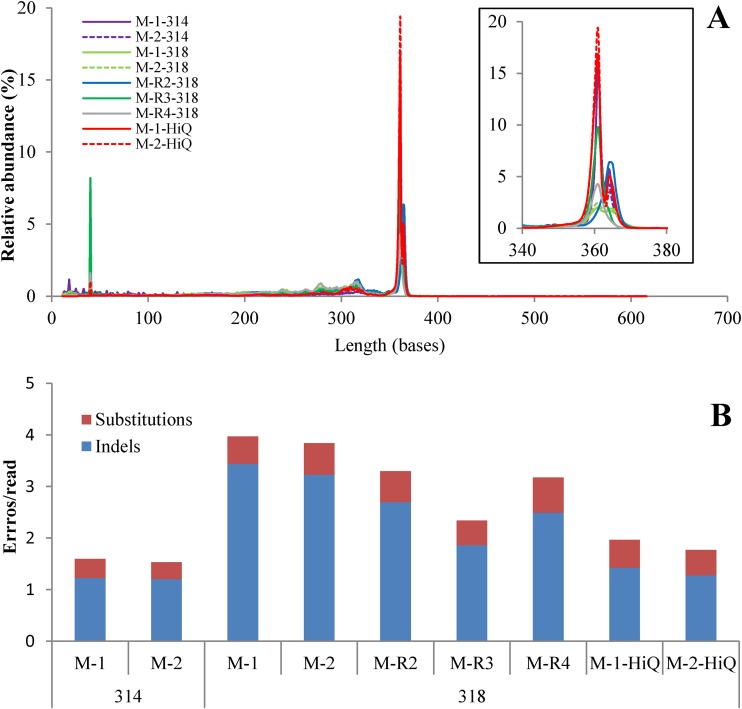
Sequencing resulted in 327,994 raw reads from the PGM 314 chip, while the 318 chip produced an average of 5,588,636 ± 450,701 raw reads per run, an approximately 16-fold increase in yield. Read length distribution was consistent among different runs, as the relative percentage of reads by read length in each run showed a single and highly similar peak at the full nifH amplicon size (360 bp) (Fig. 1A). Full-length reads accounted for 45% of the 314 chip reads, an average of 26% ± 8% of the 318 chip reads with the 400-bp kit, and 60% of the 318 chip reads with the Hi-Q kit (Table 1).
FIG 1.
Read length distributions and error types produced by different chips and sequencing kits. (A) Raw read length distribution of mock community sequences among runs. The inset is a magnification of the region between 340 and 380 bases. (B) Indels and substitutions of full-length reads in different PGM runs. 314 and 318 are PGM chip types. M-1 and M-2 were amplified from a mock community at the same time with different barcodes and sequenced by both the 314 and 318 chips with the 400-bp kit and by the 318 chip with the Hi-Q kit. M-R2, -R3, and -R4 were individually amplified and sequenced in other 318 chip runs with the 400-bp kit.
TABLE 1.
Summary statistics of the mock community data from different PGM runsa
| PGM chip, no. of raw reads/run, and sample | No. of raw reads/sample | % of full-length reads | % of reads with indels | % of reads with substitutions | Read Q score of 22 |
|
|---|---|---|---|---|---|---|
| Error rate/base (%) | % of reads remaining | |||||
| 314 | ||||||
| 327,994 | ||||||
| M-1 | 28,516 | 46 | 58 | 22 | 0.37 | 84 |
| M-2 | 26,928 | 44 | 57 | 23 | 0.35 | 85 |
| 318 | ||||||
| 5,344,439 | ||||||
| M-1 | 60,727 | 18 | 94 | 33 | 0.70 | 27 |
| M-2 | 63,569 | 18 | 93 | 32 | 0.72 | 29 |
| 4,975,072, M-R2 | 77,587 | 30 | 91 | 39 | 0.64 | 46 |
| 5,616,033, M-R3 | 67,826 | 35 | 77 | 21 | 0.49 | 65 |
| 5,879,455, M-R4 | 72,790 | 19 | 87 | 37 | 0.62 | 47 |
| 6,128,179 | ||||||
| M-1-HiQ | 141,066 | 59 | 67 | 31 | 0.40 | 65 |
| M-2-HiQ | 118,208 | 60 | 63 | 30 | 0.37 | 70 |
Sample names without the letter R were from the first run of each chip with the 400-bp kit or the 318 chip with the Hi-Q kit (HiQ), while the number following the letter R represents the run number with the 318 chip and the 400-bp kit.
Mock community analyses with the Defined Community Analysis Tool showed that the number of errors per full-length read (Fig. 1B) and the percentage of reads with errors (Table 1) varied with the chip type, independent runs, and sequencing chemistry. The numbers of indels and substitutions per full-length read were higher in 318 chip runs with the 400-bp kit (averages of 2.60 ± 0.43 and 0.58 ± 0.1, respectively) and much lower in 314 (1.22 ± 0.35) and 318 (1.35 ± 0.52) chip runs with the Hi-Q kit (Fig. 1B). Thus, the Hi-Q kit reduced the indel rate by 28 to 59% (mean, 48%) compared to that obtained with 318 chip runs with the 400-bp kit. In combination with the higher percentage of reads with indels (2.2 to 3.7 times the number of reads with substitutions) (Table 1), this error rate also indicates that indels are the primary source of PGM sequencing errors. Replicate mock communities (M-1 and M-2) exhibited similar error patterns across chips and sequencing chemistries. With the 400-bp kit, 318 chip runs produced more errors than 314 chip runs, even in the best mock run (M-R3, third 318 chip run), while 318 chip runs with the Hi-Q kit reduced the error rate and approached the performance of 314 chip runs with the 400-bp kit.
Error rate and Q score cutoff.
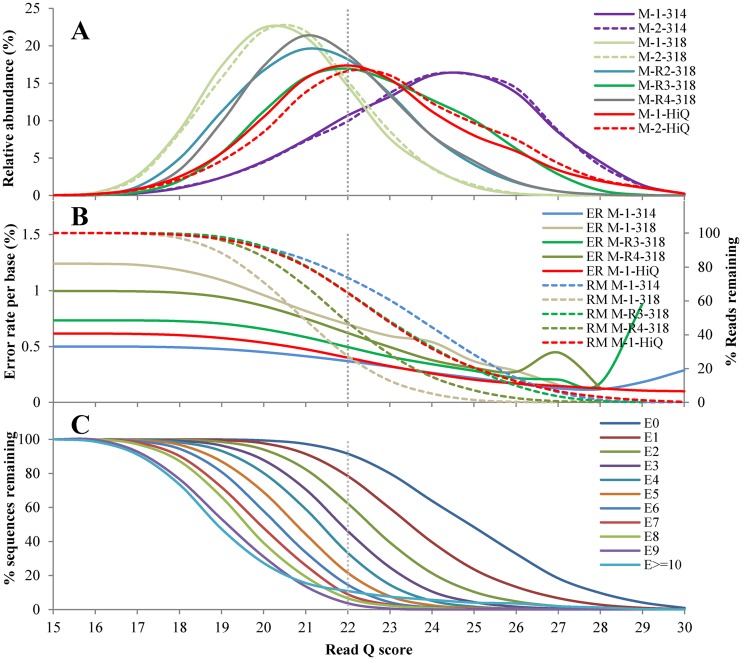
In order to determine the error rate and filtering parameters (e.g., the minimum read Q score) for the environmental samples, mock communities among different runs were further analyzed (Fig. 2). For all mock samples, the read Q score distributions from 314 chip runs peaked at a range of 24 to 25, while they peaked at a range of 20 to 22 in 318 chip runs (Fig. 2A). The Hi-Q sequencing kit displayed the best read Q score distribution in 318 chip runs, with M-R4-318 representing an average sequencing output of 318 chip runs with the 400-bp kit (Fig. 2A). The two replicates (M-1 and M-2) showed a consistent distribution pattern in their individual runs. Both the overall error rate and the percentage of reads remaining decreased with increasing read Q score cutoffs (Fig. 2B). The percentage of reads with the specified number of allowed errors decreased sharply with the read Q score cutoff, taking the Hi-Q kit run as an example (Fig. 2C; see Fig. S3 in the supplemental material for the M-R4-318 run), which showed an overall error rate of 0.40% per base, and 65% of the full-length reads remained when the read Q score was set to 22 (Table 1; Fig. 2B). The percentage of remaining reads was 91% for E0 (reads with 0 error), 78% for E1, 62% for E2, 46% for E3, 33% for E4, 21% for E5, and much less for reads with more errors (Fig. 2C). This indicates that reads containing more errors, especially those with more than three errors (1% of the nifH amplicon length), were mostly excluded from the analysis at a read Q score of 22. Therefore, a read Q score of 22 was chosen for processing of the environmental DNA samples. Overall, the Hi-Q kit allowed the retention of one to seven times more reads than the 318 chip runs with the 400-bp kit (Table 1).
FIG 2.
Read Q score distributions, overall error rates, and percentages of kinds of reads at the read Q scores indicated. (A) Read Q score distributions of whole-length reads of mock (M) communities in different runs. (B) Overall error rates and corresponding percentages of reads remaining by read Q score. ER, error rate; RM, percentage of reads remaining. (C) Percentages of reads with the specified number of errors (E0 to E≥10) by read Q score in M-1-HiQ.
Sequencing platform comparisons of nifH results.
To further evaluate PGM sequencing, six soil samples that were amplified for nifH and sequenced with the 454 platform were reamplified and sequenced on the PGM with the 314 and 318 chips and the 400-bp kits and with the 318 chip with the Hi-Q kit. After initial quality processing, the number of sequences per sample ranged from 4,831 to 8,175 (454), from 1,997 to 2,967 (PGM 314), from 2,382 to 4,068 (PGM 318), and from 16,270 to 23,868 (Hi-Q). The percentages of chimeras differed significantly (ANOVA, F = 27.74, P < 0.001) among the platforms at 2.15% (454), 8.51% (PGM 314), 5.38% (PGM 318), and 12.31% (Hi-Q). Compared to pyrosequencing, the number of FrameBot-corrected frameshifts was significantly greater for the PGM platform 314 and 318 chips. However, the Hi-Q chemistry resulted in a significant decrease in the number of frameshift errors over the older chemistry with the 318 chip (Table 2). Further analyses were performed at the OTU0 level to discriminate fine differences among platforms and at the OTU0.05 level to illustrate differences that better reflect potential biological conclusions. At both OTU levels, diversity indices were not significantly different among 454 and PGM chip-chemistry combinations, though the Hi-Q kit run generally exhibited the overall lower diversity, number of OTUs, and evenness (Table 3).
TABLE 2.
Frameshift correction by FrameBot for each sequencing platform as a percentage of the total number of dereplicated nifH sequences
| No. of frameshifts | Avg % of total no. of dereplicated nifH sequences ± SDa |
|||
|---|---|---|---|---|
| 454 | PGM 314 | PGM 318 | HiQ | |
| >0 | 27.13 ± 3.60* | 66.29 ± 1.47† | 81.72 ± 0.96‡ | 53.36 ± 1.79§ |
| >1 | 6.23 ± 1.31* | 27.09 ± 1.56† | 44.79 ± 1.47‡ | 15.73 ± 0.53§ |
| >2 | 1.57 ± 0.47* | 8.30 ± 0.65† | 16.71 ± 0.99‡ | 3.15 ± 0.43§ |
| >3 | 0.41 ± 0.21§ | 2.09 ± 0.20† | 4.47 ± 0.13‡ | 0.41 ± 0.22§ |
| >4 | 0.10 ± 0.07§ | 0.53 ± 0.13† | 1.05 ± 0.22‡ | 0.05 ± 0.09§ |
| >5 | 0.02 ± 0.02† | 0.14 ± 0.10‡ | 0.18 ± 0.03‡ | 0.00 ± 0.00† |
Symbols indicate ANOVA groupings with Tukey's correction with grouping according to the number of frameshifts.
TABLE 3.
Diversity indices for OTU0 and OTU0.05 results according to platforma
| Method | Avg diversity index ± SD |
|||||||
|---|---|---|---|---|---|---|---|---|
| OTU0s |
OTU0.05s |
|||||||
| Sab | d | J′ | H′ | Sab | d | J′ | H′ | |
| 454 | 156.5 ± 38.6*† | 20.7 ± 5.1*† | 0.607 ± 0.039 | 3.06 ± 0.33 | 30.8 ± 4.4 | 6.5 ± 0.9 | 0.592 ± 0.054 | 2.03 ± 0.22 |
| PGM 314 | 148.2 ± 15.5*† | 19.6 ± 2.1*† | 0.589 ± 0.051 | 2.94 ± 0.31 | 32.5 ± 3.0 | 6.8 ± 0.1 | 0.532 ± 0.054 | 1.85 ± 0.21 |
| PGM 318 | 178.2 ± 8.4* | 23.6 ± 1.1* | 0.593 ± 0.025 | 3.07 ± 0.14 | 34.7 ± 3.1 | 7.3 ± 0.3 | 0.537 ± 0.024 | 1.90 ± 0.12 |
| HiQ | 135.2 ± 13.9† | 17.9 ± 1.9† | 0.564 ± 0.050 | 2.77 ± 0.30 | 29.0 ± 3.1 | 6.1 ± 0.7 | 0.522 ± 0.054 | 1.75 ± 0.21 |
Sab is the number of OTUs, d is Margalef's richness index, J′ is Pielou's evenness index, and H′ is the Shannon index. Symbols refer to ANOVA groupings with Tukey's adjustment where grouping differences were identified.
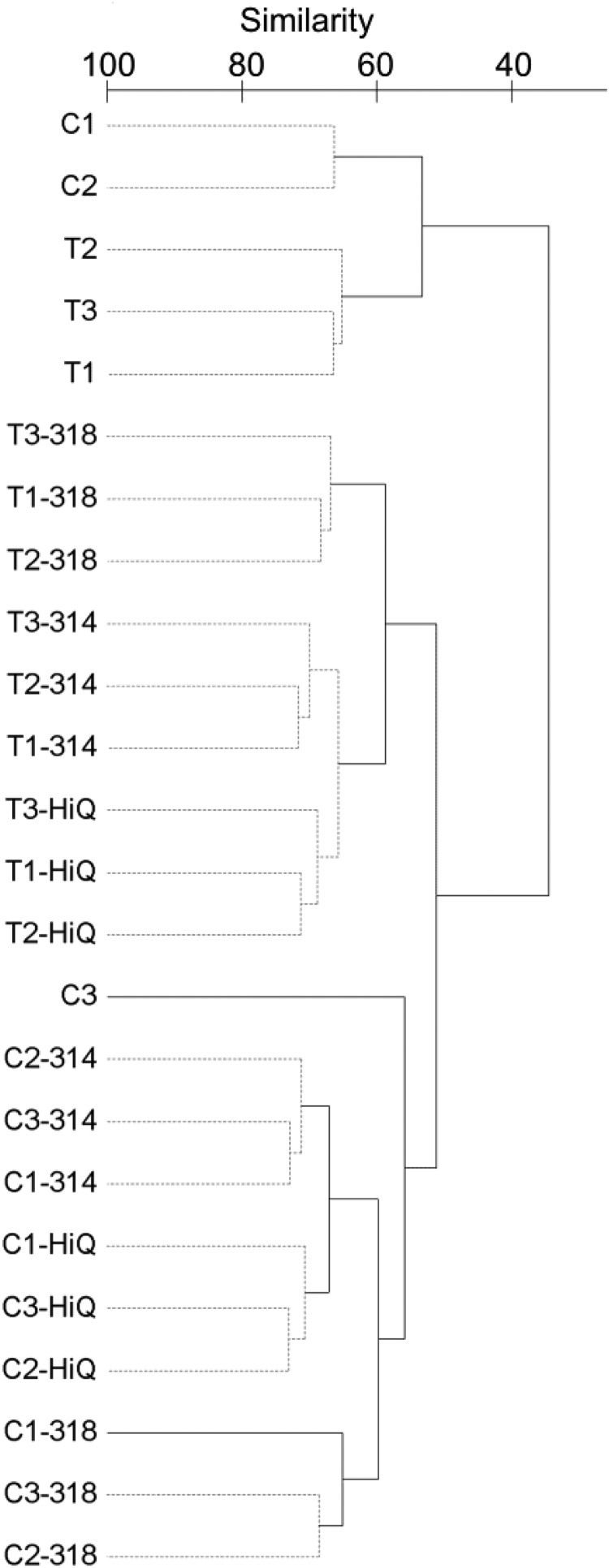
PERMANOVA revealed significant differences in diazotrophic community structure based on OTU0 and OTU0.05 among the 454, PGM 314, PGM 318, and Hi-Q runs. Significant pairwise differences between the Ion Torrent and 454 pyrosequencing platforms were identified on the basis of both Bray-Curtis and Euclidean dissimilarity matrices (Table 4). A cluster dendrogram at OTU0 revealed distinct clustering, first according to site (C versus T) and then by the combination of platform and chemistry (454, PGM 314, PGM 318, and Hi-Q runs) (Fig. 3). The clustering pattern was similar at the OTU0.05 level (see Fig. S4 in the supplemental material). The one exception was sample C3, which clustered separately from the other 454 samples. The larger errors observed in the 454 NifH diversities likely reflect this one outlier. Removal of this outlier from the 454 data set did not affect the significant PERMANOVA comparisons of the platforms.
TABLE 4.
PERMANOVA pairwise comparisons of nifH-harboring bacterial community structures according to platforma
| Method | OTU0s |
OTU0.05s |
||||||
|---|---|---|---|---|---|---|---|---|
| Bray-Curtis |
Euclidean |
Bray-Curtis |
Euclidean |
|||||
| t | P | t | P | t | P | t | P | |
| 454-PGM 314 | 2.43 | 0.004 | 2.55 | 0.013 | 3.11 | 0.003 | 2.83 | 0.013 |
| 454-PGM 318 | 2.59 | 0.003 | 2.65 | 0.009 | 3.81 | 0.003 | 3.22 | 0.010 |
| 454-HiQ | 2.43 | 0.003 | 2.50 | 0.012 | 3.05 | 0.007 | 2.83 | 0.014 |
| PGM 314-318 | 1.50 | 0.006 | 1.44 | 0.029 | 2.09 | 0.005 | 1.73 | 0.087 |
| PGM 314-HiQ | 1.26 | 0.072 | 1.12 | 0.111 | 1.22 | 0.174 | 0.98 | 0.494 |
| PGM318-HiQ | 1.79 | 0.004 | 1.61 | 0.003 | 2.49 | 0.008 | 1.98 | 0.011 |
The data shown are for the OTU0 and OTU0.05 results based on Bray-Curtis (+1) and Euclidean (+1) matrices with the univariate t statistics and corresponding P values. Boldface values indicate statistically significant differences.
FIG 3.
Complete linkage clustering of nifH-harboring bacterial community structures at the OTU0 level with SIMPROF analysis at 95% confidence.
The number of singletons identified among the different platforms and chemistries was significantly different at the OTU0 level (ANOVA, F = 4.30, P = 0.02) and ranged from 7.4% (Hi-Q) to 10.4% (PGM 318). At the OTU0.05 level, the trend was reversed, with a minimum of 33.3% singletons for PGM 318 and a maximum of 44.2% singletons with Hi-Q (ANOVA, F = 2.94, P = 0.058). Similarity percentage analysis was used to determine the OTUs generated at OTU0.05 that contributed to the majority of the Bray-Curtis distances among sequencing platforms and chemistries. Five percent (OTU0.05) was chosen in order to reflect the appropriate amino acid dissimilarity that is used to formulate biological conclusions concerning nifH-harboring bacterial communities (25). The number of OTU0.05s that accounted for 50% of the pairwise dissimilarity among platforms was highly constrained to between 8 (454 versus Hi-Q) and 13 (Hi-Q versus PGM 314). This was illustrated by the fact that the top 10 OTU0.05s accounted for 92.2% of all sequences. Of these, significant differences among platforms and chemistries were identified with OTU20 (Geobacter bemidjiensis), OTU12 (Dechloromonas aromatic), and OTU8 (Anaeromyxobacter sp.), which were significantly more abundant (ANOVA, P < 0.05) on the 454 platform, and OTU951 (Opitutaceae TAV2), which was significantly less abundant on the 454 platform. No significant differences among these 10 OTUs were observed among Ion Torrent chips and chemistries.
Metagenomic analyses of crop rotation samples.
A total of 961,674 raw reads were retrieved from the 12 soil samples in a Hi-Q PGM 318 chip run, of which 375,365 sequences (39%) remained with full length at a read Q score cutoff of 22. FrameBot detected and corrected an average of 47% ± 1% of the reads, which had 1.34 ± 0.02 frameshifts per read (Table 5). With 22,616 sequences per sample, 6,182 unique OTU0.05s were generated from the 12 soil samples.
TABLE 5.
Numbers of raw reads, reads that passed the initial quality process, and passing reads without chimeras, FrameBot statistics, and number of OTU0.05s in crop rotation samples on the basis of the 318 chip with the Hi-Q kit
| Sample | No. of raw reads | No. of reads from initial process | No. of reads without chimeras | FrameBot |
No. of OTUsa | ||
|---|---|---|---|---|---|---|---|
| No. of reads passed | % of reads with frameshifts | Avg no. of frameshifts | |||||
| G1-1 | 80,442 | 31,756 | 31,259 | 31,248 | 47 | 1.32 | 1,993 |
| G1-2 | 93,267 | 36,001 | 35,070 | 34,982 | 49 | 1.34 | 2,066 |
| G1-3 | 76,329 | 26,600 | 25,901 | 25,867 | 48 | 1.35 | 2,122 |
| G2-1 | 58,251 | 23,155 | 22,618 | 22,616 | 47 | 1.35 | 2,097 |
| G2-2 | 77,827 | 32,758 | 32,204 | 32,201 | 47 | 1.33 | 2,037 |
| G2-3 | 91,631 | 33,943 | 33,322 | 33,313 | 48 | 1.37 | 1,937 |
| G3-1 | 70,757 | 27,119 | 26,694 | 26,677 | 46 | 1.34 | 1,868 |
| G3-2 | 91,950 | 37,483 | 36,811 | 36,811 | 47 | 1.33 | 1,895 |
| G3-3 | 75,470 | 27,116 | 26,170 | 26,163 | 50 | 1.34 | 2,007 |
| G4-1 | 88,171 | 37,766 | 37,408 | 37,406 | 45 | 1.30 | 1,853 |
| G4-2 | 69,769 | 25,897 | 25,572 | 25,571 | 48 | 1.34 | 1,766 |
| G4-3 | 87,810 | 35,771 | 34,959 | 34,950 | 45 | 1.31 | 1,824 |
At 22,616 reads.
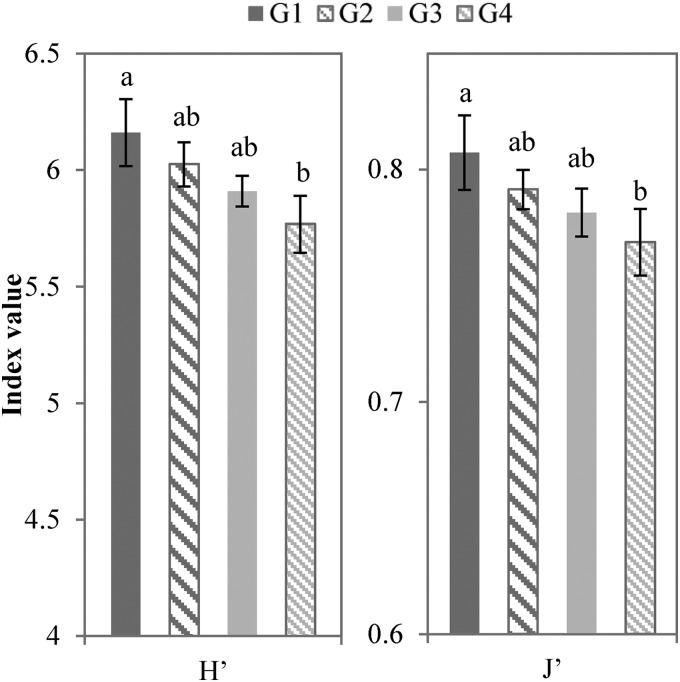
Diversity indices varied substantially among cropping systems, at 5.77 to 6.16 for the Shannon diversity index (H′) and 0.77 to 0.81 for Pielou's evenness index (J′). All of these indices gradually decreased in the order of G1 to G4 (Fig. 4). There were significant differences (ANOVA, P < 0.05) between continuous-corn soils (G1) and the corn soils in rotation and with a cover crop (G4).
FIG 4.
Shannon diversity index (H′) and Pielou's evenness index (J′) for the four cropping systems. The letters above the columns indicate ANOVA groupings. For visualization, y-axis values do not start at 0. ANOVA was done at a = 0.05, and significant differences (P < 0.05) are indicated by the letters above the bars.
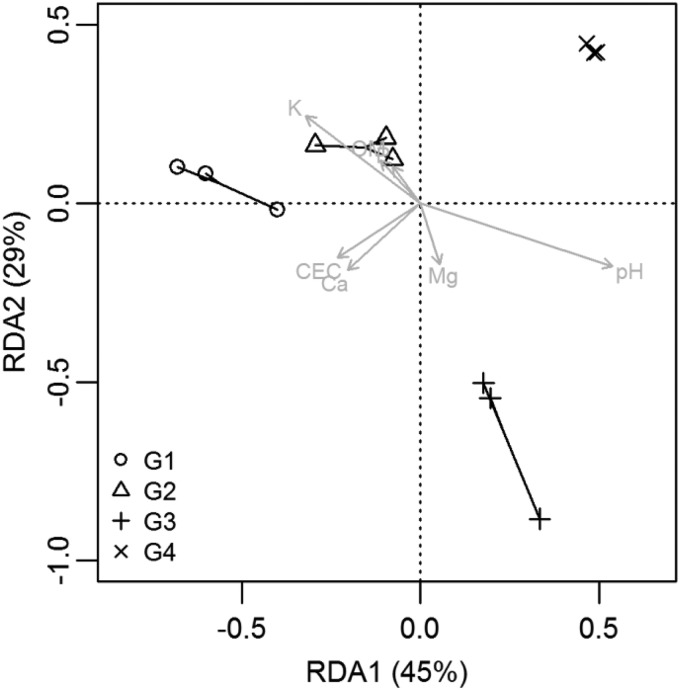
RDA explained 31% of the variation found in the OTU0.05 composition data (Fig. 5), of which 45 and 29% were represented by the first two axes, RDA1 and RDA2, respectively. Overall, the diazotrophic community composition in the four cropping systems was significantly different (PERMANOVA, F = 1.21, P < 0.05; PERMDISP, P = 0.87) (Fig. 5). However, compared with the field that had a more complex cropping history of soybean, a cover crop, and even canola in recent years (G3 and G4), the field with three consecutive years of corn and 1 year of a cover crop (G2) was more similar to the continuous-corn soils (G1). G4 was the most distant from G1, although they were both corn soils at the time samples were taken. This indicates that cropping system changes alter diazotrophic community structure and that crop legacy influences (e.g., rhizosphere, litter chemistry, and quality) can play a larger role than current crop composition. Soil properties were fitted to the model in which pH was significantly linked to the community pattern (P < 0.05).
FIG 5.
RDA of rotation samples constrained by land use. Raw abundance data were Hellinger transformed. The soil environmental variables were fitted to the ordination. Thirty-one percent of the total variance was explained by the factor land use.
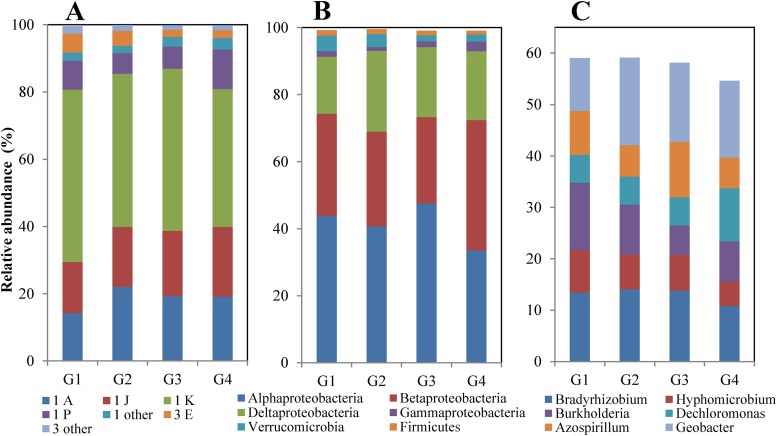
For NifH classification, averages of 94.5, 0.03, and 5.3% of the reads in these four soils belonged to functional NifH clusters I, II, and III, respectively, and a small fraction (0.17%) were NifH-like paralogs. Subclusters 1K, 1A, 1J, 1P, and 3E averaged 46.5, 18.7, 18.3, 8.3, and 3.7% of all reads. G4 samples had relatively higher abundance within 1J and 1P, while G1 had abundant subclusters 1K and 3E and G2 had more 1A (Fig. 6A). Correspondingly, most reads (94.8%) were best matched to sequences found in Proteobacteria composed of Alphaproteobacteria (41.4%), Betaproteobacteria (30.9%), Deltaproteobacteria (20.6%), and a small proportion of Gammaproteobacteria (1.9%) (Fig. 6B). Among these, G4 was characterized by less Alphaproteobacteria but more Betaproteobacteria and Gammaproteobacteria (Fig. 6B). Furthermore, over half of all reads (57.7%) were matched to only six genera (Fig. 6C). The number of read matches to Burkholderia was significantly higher (ANOVA, P < 0.01) in G1 and G2 than in G3 and G4. Outside of the phylum Proteobacteria were Verrucomicrobia (3.1%), Firmicutes (1.4%), and others (0.8% Cyanobacteria, Chlorobi, etc.) in these four cropping systems.
FIG 6.
Compositions of diazotrophic communities in rotation samples. (A) Proportion of nifH subclusters. “Others” means other subclusters with very small proportions. (B) Predominant best matches to taxa at the phylum and class levels. (C) Top six genera. Input data were the average abundances of the cropping systems.
DISCUSSION
In-depth investigations of sequencing profiles and error rates of the Ion Torrent PGM are necessary to identify appropriate quality filters and error correction tools for practical implementation of this system for gene-targeted microbial function studies. The 314 and 318 chips produced comparable read lengths and high sequence yields with more errors than desired, mainly in the form of indels rather than substitutions. This combined error type and frequency differs from errors identified on other sequencing platforms, such as 454 and Illumina MiSeq (3, 28). On the basis of mock community analyses, we found that a read Q score of 22 for both the 314 and 318 chips was best suited for the removal of reads with more than three errors from environmentally sourced nifH amplicons while retaining as many suitable quality reads as possible. The overall error rates over the nifH amplicon lengths were 0.36 and 0.62% error per base for the 314 and 318 chips with the 400-bp kit, respectively. However, the new Hi-Q kit was able to greatly reduce the overall error rate to 0.39% (average of M-1 and M-2) when using the 318 chip, which was contributed by the great reduction in the number of indels. These observed error rates are lower than previous reports for the PGM, which varied from 0.63 to 1.78% (7, 12, 28), and within the recently reported error rate range for paired-end MiSeq amplicon data (e.g., 0.28 to 1.08% [8] and 0.9% [13]), but higher than previously reported for nifH 454 pyrosequencing (0.13%) (9). Despite this error rate, we were able to identify a quality score cutoff to filter high-error reads and yet retain adequate sequencing depth with the Defined Community Analysis tool. The use of FrameBot (25) as a translation and frameshift correction tool overcame the high frequency of frameshift errors, as reflected in the consistent number of OTUs among platforms and the lack of differences in the number of singletons. Some other filters/strategies have been reported to truncate PGM raw sequencing data, including reference-supervised denoising (12) and bidirectional amplicon sequencing and flow order optimization (13).
When we compared the sequencing results of 454 with those of PGM for the same DNA samples, the overall composition of the diazotrophic community differed significantly between the sequencing platforms. However, the relationship among sites remained constant, with the C and T samples clustering separately, with the exception of the outlier (C3) in the 454 data. This separate clustering indicates the importance of using not only identical sequencing platforms but also consistent chemistries when comparing community structures. The number of OTUs, richness, evenness, and diversity were the same with the two platforms, but OTU abundances (not the presence or absence of nonsingleton OTUs) caused the differences observed. This indicates that there may be platform-specific influences on the identified abundances of certain lineages, which we confirmed by the BLASTp results obtained with the reference database. These influences may also be artifacts caused by the 1°C difference between the PCR annealing temperatures of the platforms. The PCR used in library preparation for PGM was optimized on the basis of the different Tm values of the PGM fusion primers that indicate another variable to consider when comparing platforms. The DNA was stored at −20°C for 11 months between the 454 and PGM runs; however, it was not thawed during that time, in case storage might contribute to the difference. The significantly higher number of chimeras in the PGM runs, particularly when using the 314 chip, than in the 454 runs impacted the number of sequences available for analyses. Even though the 318 chip with the new Hi-Q chemistry had a markedly higher number of chimeras, the higher sequence output ameliorated the impact on the number of filtered sequences. The high number of sequences with frameshifts (indels) is expected with pyrosequencing and was previously reported for the PGM 314 (10) and PGM 318 (12) chips. However, the new Hi-Q chemistry resulted in a significant decrease in the number of frameshifts from that of the earlier 400-bp kit; though the incidence was still approximately double that of 454. Therefore, indel detection and correction with protein translation are especially necessary for the PGM system to avoid the creation of errant clusters and are particularly important when comparing nucleotide sequences by using low-dissimilarity clusters.
We tested the PGM platform with the Hi-Q kit and our workflow of predefined filters and FrameBot frameshift correction for a typical soil microbial ecology gene-targeted metagenomic application, that of assessing the differences among nifH-harboring soil communities as influenced by four different cropping systems. RDA ordination showed a clear spatial separation of the four soil-crop systems (Fig. 5). This was further supported by the diversity indices H′ and J′, which differed significantly between continuous-corn soils (G1) and soils also currently under corn but with a different crop in preceding years (G4). Land use change alters not only free-living nitrogen-fixing microorganisms (29) but also soil ammonia oxidizer communities in agricultural systems (30), as well as the overall bacterial (31, 32) and other functional gene diversity (33).
The differences among diazotrophic communities documented here were reflected by changes in the relative abundances of taxa or OTUs among cropping systems. For example, OTUs matched to the nearest reference sequences indicated that Proteobacteria was the consistently dominant phylum within all of the cropping systems. The top six proteobacterial closest-match genera accounted for the majority of the available reads, indicating the ubiquity of these diazotrophs. The dominance of Proteobacteria, especially the Alphaproteobacteria, Betaproteobacteria, and Deltaproteobacteria, would be expected in such soils, as found previously (25). Similarly, NifH cluster I, especially subclusters 1K, 1A, 1J, and 1P, was consistently dominant in all samples as well, a pattern identified in other agricultural soils (34).
In summary, our evaluation of PGM for gene-targeted amplicon sequence analyses suggests that the platform provides quality results with the workflow described (frameshift correction with amino acid translation) for data processing. This includes sequencing of a mock community in concert to confirm or tune the quality filter parameters. Although PGM returned relatively high rates of indels that often cause frameshifts, these were detected and corrected in downstream analysis by FrameBot. While clustering and diversity analyses gave similar biological conclusions (e.g., significantly different diazotrophic communities among sites) among the different platforms, chips, and chemistries, significant differences in community composition among platforms were also identified. This suggests that larger-scale conclusions, such as those among sites and treatments, can be resolved independently of the platforms and kits used. However, because of significant differences identified in our finer-resolution analyses (e.g., OTU pairwise comparisons), these comparisons must be constrained to identical PGM chips and sequencing chemistries. The analysis protocol used in this study is broadly transferable to microbial ecology studies of other targeted genes where frameshift correction is possible.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded in part by the DOE Great Lakes Bioenergy Research Center (DOE Office of Science BER DE-FC02-07ER64494), DOE Office of Science grant BER DE-SC0004601, the Chinese Scholarship Council, and the National Natural Science Foundation of China (41376119).
We thank Lawrence Gary Oates for providing the 12 bulk soil samples and their soil metadata.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00111-15.
REFERENCES
- 1.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen Y, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Cheetham RK, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, Rasolonjatovo IMJ, Reed MT, Rigatti R, Rodighiero C, Ross MT, Sabot A, Sankar SV, Scally A, Schroth GP, Smith ME, Smith VP, Spiridou A, Torrance PE, Tzonev SS, Vermaas EH, Walter K, Wu X, Zhang L, Alam MD, Anastasi C, et al. 2008. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu L, Li Y, Li S, Hu N, He Y, Pong R, Lin D, Lu L, Law M. 2012. Comparison of next-generation sequencing systems. J Biomed Biotechnol 2012:251364. doi: 10.1155/2012/251364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rothberg JM, Hinz W, Rearick TM, Schultz J, Mileski W, Davey M, Leamon JH, Johnson K, Milgrew MJ, Edwards M, Hoon J, Simons JF, Marran D, Myers JW, Davidson JF, Branting A, Nobile JR, Puc BP, Light D, Clark TA, Huber M, Branciforte JT, Stoner IB, Cawley SE, Lyons M, Fu Y, Homer N, Sedova M, Miao X, Reed B, Sabina J, Feierstein E, Schorn M, Alanjary M, Dimalanta E, Dressman D, Kasinskas R, Sokolsky T, Fidanza JA, Namsaraev E, McKernan KJ, Williams A, Roth GT, Bustillo J. 2011. An integrated semiconductor device enabling non-optical genome sequencing. Nature 475:348–352. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- 5.Merriman B, Rothberg JM. 2012. Progress in Ion Torrent semiconductor chip based sequencing. Electrophoresis 33:3397–3417. doi: 10.1002/elps.201200424. [DOI] [PubMed] [Google Scholar]
- 6.Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, Pallen MJ. 2012. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol 30:434–439. doi: 10.1038/nbt.2198. [DOI] [PubMed] [Google Scholar]
- 7.Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, Bertoni A, Swerdlow HP, Gu Y. 2012. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 13:341. doi: 10.1186/1471-2164-13-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fish JA, Chai B, Wang Q, Sun Y, Brown CT, Tiedje JM, Cole JR. 2013. FunGene: the functional gene pipeline and repository. Front Microbiol 4:291. doi: 10.3389/fmicb.2013.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bragg LM, Stone G, Butler MK, Hugenholtz P, Tyson GW. 2013. Shining a light on dark sequencing: characterising errors in Ion Torrent PGM data. PLoS Comput Biol 9:e1003031. doi: 10.1371/journal.pcbi.1003031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Veras AAO, de Sá PHCG, Pinheiro KC, Assis das Graças D, Azevedo Baraúna R, Cruz Schneider MP, Azevedo V, Ramos RTJ, Silva A. 2014. Efficiency of Corynebacterium pseudotuberculosis Cp31 genome assembly with the Hi-Q enzyme on an Ion Torrent PGM sequencing platform. J Proteomics Bioinform 7:374–378. doi: 10.4172/jpb.1000342. [DOI] [Google Scholar]
- 12.Salipante SJ, Sengupta DJ, Rosenthal C, Costa G, Spangler J, Sims EH, Jacobs MA, Miller SI, Hoogestraat DR, Cookson BT, McCoy C, Matsen FA, Shendure J, Lee CC, Harkins TT, Hoffman NG. 2013. Rapid 16S rRNA next-generation sequencing of polymicrobial clinical samples for diagnosis of complex bacterial infections. PLoS One 8:e65226. doi: 10.1371/journal.pone.0065226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salipante SJ, Kawashima T, Rosenthal C, Hoogestraat DR, Cummings LA, Sengupta DJ, Harkins TT, Cookson BT, Hoffman NG. 2014. Performance comparison of Illumina and Ion Torrent next-generation sequencing platforms for 16S rRNA-based bacterial community profiling. Appl Environ Microbiol 80:7583–7591. doi: 10.1128/AEM.02206-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pylro VS, Roesch LF, Morais DK, Clark IM, Hirsch PR, Totola MR. 2014. Data analysis for 16S microbial profiling from different benchtop sequencing platforms. J Microbiol Methods 107:30–37. doi: 10.1016/j.mimet.2014.08.018. [DOI] [PubMed] [Google Scholar]
- 15.Cao YY, Qu YJ, Song F, Zhang T, Bai JL, Jin YW, Wang H. 2014. Fast clinical molecular diagnosis of hyperphenylalaninemia using next-generation sequencing-based on a Custom AmpliSeqTM panel and Ion Torrent PGM sequencing. Mol Genet Metab 113:261–266. doi: 10.1016/j.ymgme.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 16.Millat G, Chanavat V, Rousson R. 2014. Evaluation of a new high-throughput next-generation sequencing method based on a custom AmpliSeq library and Ion Torrent PGM sequencing for the rapid detection of genetic variations in long QT syndrome. Mol Diagn Ther 18:533–539. doi: 10.1007/s40291-014-0099-y. [DOI] [PubMed] [Google Scholar]
- 17.Nishio SY, Hayashi Y, Watanabe M, Usami S. 2015. Clinical application of a custom AmpliSeq library and Ion Torrent PGM sequencing to comprehensive mutation screening for deafness genes. Genet Test Mol Biomarkers 19:209–217. doi: 10.1089/gtmb.2014.0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo Y, Wan S, Hui D, Wallace LL. 2001. Acclimatization of soil respiration to warming in a tall grass prairie. Nature 413:622–625. doi: 10.1038/35098065. [DOI] [PubMed] [Google Scholar]
- 19.Zhang W, Parker K, Luo Y, Wan S, Wallace L, Hu S. 2005. Soil microbial responses to experimental warming and clipping in a tallgrass prairie. Glob Chang Biol 11:266–277. doi: 10.1111/j.1365-2486.2005.00902.x. [DOI] [Google Scholar]
- 20.Zhou X, Wan S, Luo Y. 2007. Source components and interannual variability of soil CO2 efflux under experimental warming and clipping in a grassland ecosystem. Glob Chang Biol 13:761–775. doi: 10.1111/j.1365-2486.2007.01333.x. [DOI] [Google Scholar]
- 21.Zhou J, Bruns MA, Tiedje JM. 1996. DNA recovery from soils of diverse composition. Appl Environ Microbiol 62:316–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poly F, Monrozier LJ, Bally R. 2001. Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res Microbiol 152:95–103. doi: 10.1016/S0923-2508(00)01172-4. [DOI] [PubMed] [Google Scholar]
- 23.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 25.Wang Q, Quensen JF, Fish JA, Lee TK, Sun Y, Tiedje JM, Cole JR. 2013. Ecological patterns of nifH genes in four terrestrial climatic zones explored with targeted metagenomics using FrameBot, a new informatics tool. mBio 4:e00592–00513. doi: 10.1128/mBio.00592-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clarke KR, Somerfield PJ, Gorley RN. 2008. Testing of null hypotheses in exploratory community analyses: similarity profiles and biota-environment linkage. J Exp Mar Biol Ecol 366:56–69. doi: 10.1016/j.jembe.2008.07.009. [DOI] [Google Scholar]
- 27.Anderson MJ. 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecol 26:32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x. [DOI] [Google Scholar]
- 28.Jünemann S, Sedlazeck FJ, Prior K, Albersmeier A, John U, Kalinowski J, Mellmann A, Goesmann A, von Haeseler A, Stoye J, Harmsen D. 2013. Updating benchtop sequencing performance comparison. Nat Biotechnol 31:294–296. doi: 10.1038/nbt.2522. [DOI] [PubMed] [Google Scholar]
- 29.Mirza BS, Potisap C, Nusslein K, Bohannan BJM, Rodrigues JLM. 2014. Response of free-living nitrogen-fixing microorganisms to land use change in the Amazon rainforest. Appl Environ Microbiol 80:281–288. doi: 10.1128/AEM.02362-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bissett A, Abell GCJ, Brown M, Thrall PH, Bodrossy L, Smith MC, Baker GH, Richardsson AE. 2014. Land-use and management practices affect soil ammonia oxidiser community structure, activity and connectedness. Soil Biol Biochem 78:138–148. doi: 10.1016/j.soilbio.2014.07.020. [DOI] [Google Scholar]
- 31.Rodrigues JL, Pellizari VH, Mueller R, Baek K, Jesus Eda C, Paula FS, Mirza B, Hamaoui GS Jr, Tsai SM, Feigl B, Tiedje JM, Bohannan BJ, Nusslein K. 2013. Conversion of the Amazon rainforest to agriculture results in biotic homogenization of soil bacterial communities. Proc Natl Acad Sci U S A 110:988–993. doi: 10.1073/pnas.1220608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sul WJ, Asuming-Brempong S, Wang Q, Tourlousse DM, Penton CR, Deng Y, Rodrigues JLM, Adiku SGK, Jones JW, Zhou J, Cole JR, Tiedje JM. 2013. Tropical agricultural land management influences on soil microbial communities through its effect on soil organic carbon. Soil Biol Biochem 65:33–38. doi: 10.1016/j.soilbio.2013.05.007. [DOI] [Google Scholar]
- 33.Paula FS, Rodrigues JLM, Zhou J, Wu L, Mueller RC, Mirza BS, Bohannan BJM, Nusslein K, Deng Y, Tiedje JM, Pellizari VH. 2014. Land use change alters functional gene diversity, composition and abundance in Amazon forest soil microbial communities. Mol Ecol 23:2988–2999. doi: 10.1111/mec.12786. [DOI] [PubMed] [Google Scholar]
- 34.Collavino MM, Tripp HJ, Frank IE, Vidoz ML, Calderoli PA, Donato M, Zehr JP, Aguilar OM. 2014. nifH pyrosequencing reveals the potential for location-specific soil chemistry to influence N2-fixing community dynamics. Environ Microbiol 16:3211–3223. doi: 10.1111/1462-2920.12423. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.