Abstract
Azotobacter vinelandii is a widely studied model diazotrophic (nitrogen-fixing) bacterium and also an obligate aerobe, differentiating it from many other diazotrophs that require environments low in oxygen for the function of the nitrogenase. As a free-living bacterium, A. vinelandii has evolved enzymes and transporters to minimize the loss of fixed nitrogen to the surrounding environment. In this study, we pursued efforts to target specific enzymes and further developed screens to identify individual colonies of A. vinelandii producing elevated levels of extracellular nitrogen. Targeted deletions were done to convert urea into a terminal product by disrupting the urease genes that influence the ability of A. vinelandii to recycle the urea nitrogen within the cell. Construction of a nitrogen biosensor strain was done to rapidly screen several thousand colonies disrupted by transposon insertional mutagenesis to identify strains with increased extracellular nitrogen production. Several disruptions were identified in the ammonium transporter gene amtB that resulted in the production of sufficient levels of extracellular nitrogen to support the growth of the biosensor strain. Further studies substituting the biosensor strain with the green alga Chlorella sorokiniana confirmed that levels of nitrogen produced were sufficient to support the growth of this organism when the medium was supplemented with sufficient sucrose to support the growth of the A. vinelandii in coculture. The nature and quantities of nitrogen released by urease and amtB disruptions were further compared to strains reported in previous efforts that altered the nifLA regulatory system to produce elevated levels of ammonium. These results reveal alternative approaches that can be used in various combinations to yield new strains that might have further application in biofertilizer schemes.
INTRODUCTION
Nutrient requirements are directly linked to biomass production, and any potential increased improvement in the scale of biomass yield will necessitate a proportional increase in the demand for essential nutrients. For all photosynthetic organisms (photoautotrophs such as land plants, algae, and cyanobacteria) with requisite light energy and water, nitrogen is the most limiting and expensive nutrient input for aquaculture and agricultural production alike (1). A majority of our current nitrogen fertilizer production is tied to the burning of fossil fuels to generate ammonia from molecular nitrogen (N2 gas) through the Haber-Bosch process, which accounts for 3 to 5% of world natural gas consumption, or about 1 to 2% of total worldwide energy expenditures (1–3). In developed countries, industrial nitrogen production is accompanied by a huge economic and energetic cost overall (2), while this key nutrient limits agricultural productivity in developing countries, where energy and infrastructure costs prohibit the utilization of the Haber-Bosch process to produce ammonia from atmospheric nitrogen on a large scale.
The development of biological approaches to improve biofertilizers represents a unique opportunity for future generations to lower the potential economic costs and environmental impacts of current fossil-fuel-dependent industrial methods for the production of ammonia-derived fertilizers (4–6). Various laboratories have been investigating the potential of Azotobacter species to provide nitrogen to specific crops for many years, with various degrees of success (5–11). Although many nitrogen-fixing bacteria produce nitrogen in environments requiring very low oxygen, Azotobacter vinelandii has evolved the capability of fixing nitrogen as a free-living aerobe, despite the fact that nitrogenase is inherently sensitive to oxygen. This makes it an ideal organism for coculture with a broader range of plants, as growth in microaerobic or anaerobic environments is not required. The ultimate long-term goal of biofertilizers would be to develop potential alternatives to the energy intensive Haber-Bosch process.
Biofertilizers provide a range of potential benefits versus current industrial nitrogen production routes. In situ biofertilizer production would circumvent and displace transportation costs and associated environmental impacts related to production and distribution of Haber-Bosch-derived industrial fertilizers. Biological assimilation of nutrients in addition to timed-release of nitrogen compounds would mitigate issues associated with agricultural residue runoff from excessive application of industrial fertilizers, leading to eutrophication of nearby water supplies and streams. Both higher land plants and microalgae are known to produce extracellular carbon as a potential source of fixed carbon to support beneficial heterotrophs that make up part of the rhizosphere (5, 12–15).
Current biofuel feedstock crops such as corn for ethanol require substantial amounts of nitrogen inputs. Potential future production of biomass using next-generation feedstocks such as algae promise significant improvements in overall yield that could be orders of magnitude higher than current conventional land plant crops (16, 17). Since a majority of the current nitrogen requirements for the growth of biofuel crops will likely be derived from energy intensive industrial processes such as Haber-Bosch, the energy use efficiency of current biofuel crops has been questioned (18). A significant amount of the energy acquired from corn ethanol and soybean biodiesel or next-generation biofuel crops such as algae may actually need to be diverted back to these industrial processes to supply the energy required for additional industrial nitrogen fixation. Proposed improvements in final biomass yield will require concomitant increases in macronutrient inputs such as nitrogen. In a world where societies will require more of our agricultural resources to feed expanding populations, and significant portions of our agricultural lands are currently being converted from food to fuel crop production, the impacts and requirements of current methods to provide nitrogen for present and future crops will only increase in importance (18–20).
The long-term aim of biofertilizers is to circumvent the energy cost and the associated greenhouse gas emissions tied to the production and distribution of conventional Haber-Bosch derived nitrogen fertilizers. The potential of biofertilizers can be enhanced by identifying alternative methodologies of nitrogen delivery using diazotrophic bacteria to provide a renewable source of nitrogen to meet the growth requirements of the associated photosynthetic species. Although model symbiotic systems between specific plants and nitrogen fixing bacteria are well established (21, 22), these are limited to a small number of commodity crops. This approach is directed toward expanding similar symbiotic relationships to a broader range of crops or next-generation biomass sources.
In the present study, we use various approaches to increase the production of extracellular nitrogen by A. vinelandii. These approaches include rational manipulations designed to convert urea from a common metabolite into a terminal product. Further application of transposon mutagenesis combined with a nitrogen biosensor reveals a specific gene deletion resulting in increased nitrogen release by A. vinelandii. These alterations are compared to results obtained following the reconstruction of strains reported previously to elevate production of ammonium by A. vinelandii following disruption of the regulatory gene product of nifL. These reconstructed strains were used to make comparisons between the results and reported phenotypes of previous constructs, as well as the strains constructed here, to highlight the potential to produce fixed nitrogen species that might be applied to various biofertilizer schemes.
MATERIALS AND METHODS
Bacterial culture.
A. vinelandii DJ was obtained from Dennis Dean (Virginia Tech) and grown on standard B plates (23) unless otherwise specified. Escherichia coli WM3064 was used for conjugation (24, 25) and grown on lysogeny broth (LB) or on BYE medium (B plates containing 5 g of yeast extract/liter).
Algal culture and cell counts.
Cultures of Chlorella sorokiniana UTEX 1602 were obtained from the UTEX culture collection of algae (Austin, TX) and have been maintained for several years by subculturing on solid media (11). Algae strains were cultured in a freshwater medium as described previously (26). Algal cells in solution were measured using a hemocytometer following the directions of the manufacturer (Hausser Scientific, Horsham, PA).
Genetic constructs of Azotobacter vinelandii.
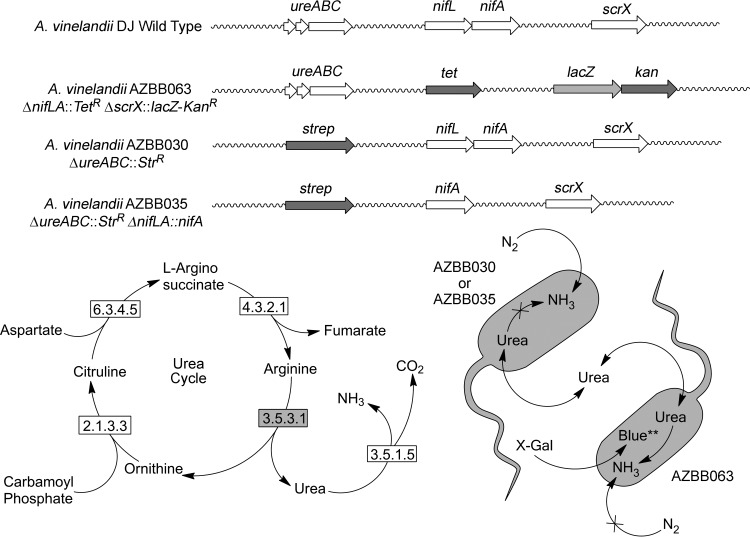
Azotobacter vinelandii AZBB030 and AZBB035 used as initial target strains, and AZBB063 used as a biosensor strain, were constructed as detailed in Table 1 by transforming A. vinelandii DJ with the various plasmids listed in Table 2. The primers used to clone genes or genome segments are listed in Table 3. A graphical representation of the three strains compared to the wild-type A. vinelandii strain is shown at the top of Fig. 2. Following the transposon experiments described below, A. vinelandii AZBB102, AZBB103, AZBB148, and AZBB150 were constructed by using a similar approach to confirm the phenotype found during the transposon experiments or construct strains used as controls. Methods for the manipulation of A. vinelandii have been described previously (23, 27, 28).
TABLE 1.
Mutant strains constructed and/or used in this study
| A. vinelandii strain | Plasmid utilized | Genetic featuresa | Parent strain |
|---|---|---|---|
| DJb | None | Wild type | |
| AZBB010 | pPCRNH3-15 | ΔnifLA::Strr | DJ |
| AZBB020 | pPCRNH3-14 | ΔnifLA::nifA | AZBB010 |
| AZBB023 | pPCRSCRK31 | ΔscrX::lacZ-Kanr | DJ |
| AZBB030 | pPCRURE3 | ΔureABC::Strr | DJ |
| AZBB035 | pPCRURE3 | ΔureABC::Strr and ΔnifLA::nifA | AZBB020 |
| AZBB063 | pPCRNH3-21 | ΔnifLA::Tetr and ΔscrX::lacZ-Kanr | AZBB023 |
| AZBB085 | pEB001 | amtB::Kanr-transposon and ΔureABC::Strr and ΔnifLA::nifA | AZBB035 |
| AZBB086 | pEB001 | amtB::Kanr-transposon and ΔureABC::Strr and ΔnifLA::nifA | AZBB035 |
| AZBB088 | pEB001 | amtB::Kanr-transposon and ΔureABC::Strr | AZBB030 |
| AZBB091 | pEB001 | amtB::Kanr-transposon and ΔureABC::Strr | AZBB030 |
| AZBB093 | pEB001 | amtB::Kanr-transposon and ΔureABC::Strr and ΔnifLA::nifA | AZBB035 |
| AZBB094 | pEB001 | amtB::Kanr-transposon and ΔureABC::Strr and ΔnifLA::nifA | AZBB035 |
| AZBB102 | pPCRAMTBK3 | ΔamtB::Kanr | DJ |
| AZBB103 | pPCRAMTBK3 | ΔamtB::Kanr and ΔureABC::Strr | AZBB030 |
| AZBB148 | pPCRNH3-42 | nifL::Kanr similar to MV376c | AZBB010 |
| AZBB150 | pPCRNH3-43 | nifL::Kanr incorporated slightly upstream of SmaI site | AZBB010 |
| AZBB158 | None | Spontaneous mutation of AZBB148 resulting in Nif+ phenotype | AZBB148 |
| AZBB163 | None | Spontaneous mutation of AZBB150 resulting in Nif+ phenotype | AZBB150 |
TABLE 2.
Key plasmids and relevant derivatives of these plasmids used for the construction of A. vinelandii manipulated strains
| Plasmida | Relevant gene(s) cloned or plasmid manipulationb | Parent vector | Source or reference |
|---|---|---|---|
| pEB001 | Plasmid containing Mariner transposon and transposase | 24 | |
| pBB052 | pUC19 with Kanr from pUC4K in place of Ampr | pUC19 | 50 |
| pBB053 | Removed NdeI site from pUC19 by silent mutation | pUC19 | 25 |
| pBB073 | Moved spectinomycin/streptomycin resistance cassette from pHP45Ω into EcoRI site of pBB052 | pBB052 | 51 |
| pBBTET3 | pUC19 with Tetr in place of Ampr | pUC19 | 25 |
| pPCRKAN4 | Cloned Kan cassette from pBBR1MCS-2 into pBBTET3 | pBBTET3 | 25 |
| pLACZF12 | Cloned lacZ gene from E. coli into pBBTET3 and removed various restriction sites by silent mutation with site-specific mutagenesis | pBBTET3 | This study |
| pPCRSCRK2 | Cloned scrX gene and flanking regions from A. vinelandii into pBB053 | pBB053 | This study |
| pPCRSCRK5 | Removed restriction sites from pPCRSCRK2 by blunt fill-in; performed PCR to remove scrX gene from plasmid | pBB053 | This study |
| pPCRSCRK7 | Performed additional blunt fill-in and site-specific mutagenesis to remove additional restriction sites | pBB053 | This study |
| pPCRSCRK28 | Moved Kan cassette from pPCRKAN4 into pPCRSCRK7, then removed restriction sites by site-specific mutagenesis with silent mutations | pBB053 | This study |
| pPCRSCRK31* | Moved lacZ gene from pLACZF12 into pPCRSCRK28 | pBB053 | This study |
| pPCRNH3-10 | Cloned nifA gene from A. vinelandii into pBB114 | pBB114 | This study |
| pPCRNH3-11 | Cloned nifLA genes and flanking regions from A. vinelandii into pBB053 | pBB053 | This study |
| pPCRNH3-12 | Removed restriction site from pPCRNH3-11 by blunt fill-in | pBB053 | This study |
| pPCRNH3-13 | Performed PCR to remove nifLA genes from pPCRNH3-12 and add XbaI and BamHI sites | pBB053 | This study |
| pPCRNH3-14* | Moved nifA gene from pPCRNH3-10 into pPCRNH3-13 | pBB053 | This study |
| pPCRNH3-15* | Moved Str cassette from pBB073 into pPCRNH3-13 | pBB053 | This study |
| pPCRNH3-21* | Moved Tet cassette pBBTET3 into pPCRNH3-13 | pBB053 | This study |
| pPCRNH3-42* | Removed segment of nifL gene from pPCRNH3-11 and inserted Kan cassette from pPCRKAN4 similar to approach taken to construct pBB369 | pBB053 | 8; this study |
| pPCRNH3-43* | Removed larger segment of nifL gene from pPCRNH3-11 than was done in pPCRNH3-42 and inserted Kan cassette from pPCRKAN4 slightly further upstream of nifA as shown in Fig. 7 | pBB053 | This study |
| pPCRURE1 | Cloned ureABC genes and flanking regions from A. vinelandii into pUC19 | pUC19 | This study |
| pPCRURE2 | Performed PCR to remove ureABC genes from pPCRURE1 | pUC19 | This study |
| pPCRURE3* | Moved Str cassette pBB073 into pPCRURE2 | pUC19 | This study |
| pPCRAMTBK1 | Cloned gene amtB and flanking regions from A. vinelandii into pBB053 | pBB053 | This study |
| pPCRAMTBK2 | Performed PCR to remove gene amtB from pPCRAMTBK1 | pBB053 | This study |
| pPCRAMTBK3* | Moved Kan cassette from pPCRKAN4 into pPCRAMTBK2 | pBB053 | This study |
The sequences of all plasmids in this study are available upon request. Plasmids indicated by an asterisk (*) are completed vectors that were used to transform A. vinelandii.
Tetr, tetracycline resistance; Ampr, ampicillin resistance; Strr, streptomycin resistance; Kanr, kanamycin resistance.
TABLE 3.
Primers used in this study
| Primer | Sequence (5′–3′)a | Purpose |
|---|---|---|
| BBP919 | GACTAGAA TTCGTGCAGA AACATCTCTA CCCGGAAG | ureABC gene and flanking region cloning |
| BBP920 | GACTAAAG CTTGGAACAG AAGACGATGA GGATGC | ureABC gene and flanking region cloning |
| BBP938 | GACTAGGA TCCGTGAAGA TCAGCAGCTT GTCTTTCTC | ureABC gene deletion |
| BBP939 | GACTAGGA TCCAAGAAGG ACCTGATCCA CAACG | ureABC gene deletion |
| BBP1147 | GAGCAAGC TTCATGGTCA GGTCGTGGCC TTC | nifLA gene and flanking region cloning |
| BBP1148 | GACAGGAT CCGGTCAGCA CTTCGGACAC CAC | nifLA gene and flanking region cloning |
| BBP1209 | GACATCTA GACTCTCATA TGGTGCCTCG TCTATCCAAG AAAACC | nifLA gene deletion |
| BBP1210 | GACATCTA GAGGATCCGA CCCTCCGGCA ATGGATG | nifLA gene deletion |
| BBP1149 | GACAGGAT CCATATGAAT GCAACCATCC CTCAGCGCT C | nifA gene cloning |
| BBP1150 | GACATCTA GACTAGATCT TGCGCATGTG GATGTTGAG | nifA gene cloning |
| BBP2099 | NNNGATAT CGGGCATTCC GCCCGACCTG GTGCTG | nifL gene segment deletion 1 |
| BBP2100 | NNNGATAT CGACGACGAT GCCCTGGGCC AGCAACTG | nifL gene segment deletion 1 |
| BBP2101 | NNNGATAT CCCGGAGAAG GCGCTGCCCT G | nifL gene segment deletion 2 |
| BBP2102 | NNNGATAT CCATCAGCAC CCGCGTGGAG AAC | nifL gene segment deletion 2 |
| BBP1674 | NNNTCTAG AGGAAGCTAT CCGACGAGGA CAGCCGAG | scrX gene and flanking region cloning |
| BBP1675 | NNNGAATT CCTGAGCGCA GAATTTAGAT ATTGATACTC ATAGTC | scrX gene and flanking region cloning |
| BBP1710 | NNNATGCA TATGGACTTC CTATTGTTGA CATTATTGGT GG | scrX gene deletion |
| BBP1711 | NNNATGCA TGACAGAATT CAGATCTCAC GCCATAAGCT GTTAGCATTT TTCTTG | scrX gene deletion |
| BBP1669 | NNNTCTAG AGGATCCCAT ATGCATACCA TGATTACGGA TTCACTGGCC GTCG | lacZ gene cloning |
| BBP1671 | NNNAAGCT TGGATCCTAT TTTTGACAC CAGACCAACT GGTAATGGTA G | lacZ gene cloning |
| BBP1967 | NNNAAGCT TCGAAGACAT GGCACTCCGA GGCGTTGGCC AGAC | amtB gene and flanking region cloning |
| BBP1968 | NNNTCTAG AGATAGATTC CCTGCCAGGT CCCCAG | amtB gene and flanking region cloning |
| BBP1969 | NNNAGATC TGGTTACAAC CTCTGAGTGT CGGGAG | amtB gene deletion |
| BBP1970 | NNNAGATC TCAGCGTCAT TGATTATTCT CCTGGGGCG | amtB gene deletion |
| BBP982 | GAAGGGCA GCAGCAGGTA GAGG | ureABC gene deletion confirmation |
| BBP983 | CAGCAGTT CGCGAAGACT GTCGAAG | ureABC gene deletion confirmation |
| BBP1836 | CTCAACGT TCGCCAGGTA TATGCCGAAC TC | scrX gene deletion confirmation |
| BBP1837 | CACATAGG ATGAAACGTC ACCGAGCTTG TTCG | scrX gene deletion confirmation |
| BBP950 | GAGCACAC CCATCACGGT CAGAG | nifLA gene deletion confirmation |
| BBP1322 | GATCTCCA TCGACTCGAT CTTGTCCAGG GTGAAC | nifLA gene deletion confirmation |
| BBP2006 | CACGTGCC AGGAATTCCT CCATG | amtB gene deletion confirmation |
| BBP2007 | CTGTGGAC GATGGCCAGG GACATGGATC | amtB gene deletion confirmation |
Specific restriction enzyme sites added to primers are underlined for clarity.
FIG 2.
Descriptions of various strains constructed for this work. Shown above are simple representations of modifications made to various regions of the A. vinelandii genome to construct the nitrogen biosensor A. vinelandii AZBB063, which results in minimal growth in the absence of extraneously provided nitrogen and yields a blue phenotype when grown in the presence of X-Gal. Additional constructs A. vinelandii AZBB030 and AZBB035 contain ΔureABC::Strr, resulting in the accumulation of minor amounts of urea (Fig. 1). A. vinelandii AZBB035 also contains a deregulated nitrogenase by first removing the nifLA genes and then replacing them with nifA behind the nifL promoter, resulting in a rescued nitrogen fixing phenotype. The bottom left shows the urea cycle and urease enzyme (3.5.1.5) as it has been annotated in A. vinelandii. A known gene for arginase (3.5.3.1) has not been identified in A. vinelandii. The bottom right illustration indicates how urea as a terminal product could potentially support the growth of the nitrogen biosensor A. vinelandii AZBB063 if quantities of urea increased to a level sufficient to meet the nitrogen requirements of this strain. Additional metabolites produced by A. vinelandii AZBB030 and AZBB035 following transposon mutagenesis experiments could also substitute for the urea in this representation, as was found in these studies.
Urea quantitation.
To measure extracellular urea concentrations from cultures of A. vinelandii, 1-ml samples were first spun at maximum speed on a microcentrifuge for 1 min (∼21,000 × g), and then the supernatant was removed and stored at −80°C or used immediately. Urea was quantified using the method of Mather and Roland (29) with slight alterations. The reagent was prepared fresh immediately before use by dissolving 10 mg of FeCl3·6H2O in 50 ml of acid solution (8% H2SO4 and 1% H3PO4). Next, 75 mg of diacetyl monoxime was dissolved in 5 ml of H2O and combined with the acid solution. Then, 5 mg of thiosemicarbazide was dissolved in 5 ml of H2O and combined with the acid solution. A standard curve was prepared with a maximum concentration of 1 mM urea. To run the assay, 300 μl of sample or standard was combined with 1 ml of the reagent and mixed thoroughly in a 1.5-ml microcentrifuge tube (polypropylene). Samples were incubated for 20 min at 90°C and then cooled on ice and allowed to sit for 20 min before measuring on a UV-visible spectrophotometer at 520 nm.
Ammonium quantitation.
Ammonium was quantified by several different methods that have been described previously (7, 9, 30, 31) using either fluorescent or colorimetric approaches. For low levels of ammonium obtained from specific culture supernatants, a derivation of the phthalaldehyde method described previously (32, 33) was utilized. In these assays, 500 μl of assay reagent (270 mg of phthalic dicarboxaldehyde dissolved in 5 ml of ethanol and then added to 100 ml of 0.2 M phosphate buffer [pH 7.3] and 50 μl of β-mercaptoethanol) was combined with up to 500 μl of culture supernatant and allowed to react at room temperature for 30 min. Samples were analyzed on a Varian Cary Eclipse fluorescence spectrophotometer using an excitation wavelength of 410 nm and an emission wavelength of 472 nm.
Elemental analysis of cells and supernatants.
Cells were collected by centrifugation at ∼12,000 × g and frozen. Supernatants were separated from the cells for analysis of the remaining solids. Samples were lyophilized and thoroughly mixed using a spatula to ensure a homogeneous mixture. Samples of cells and/or supernatants were analyzed for the percent carbon, hydrogen, and nitrogen at the Stable Isotope Lab in the Geology Department at the University of Minnesota.
Random transposon mutagenesis.
A. vinelandii strains AZBB030 and AZBB035 were transformed by a transposon insertion methodology using Escherichia coli WM3064 and the mariner transposon from plasmid pEB001 (24, 34). Briefly, ∼50 μl of A. vinelandii cells was scraped from a fresh agar plate of cells with a sterile loop and resuspended in 500 μl of sterile phosphate buffer. Separately, ∼50 μl of E. coli WM3064 cells containing the pEB001 plasmid were resuspended in 1 ml of sterile LB broth. Next, 100 μl of the suspended A. vinelandii cells and 20 μl of the suspended E. coli WM3064 cells were combined and mixed with a pipettor and then spotted onto BYE plates supplemented with 100 μM 2,6-diaminopimelic acid (DAP; 50 μl of a 10-mg/ml stock). These cells were incubated overnight at 30°C, transferred with a sterile loop to 100 ml of B medium, and grown overnight at 30°C in a shaker table at 180 rpm. After growth in B medium, 1 ml of culture was removed and pelleted in a microcentrifuge at 12,000 × g, and then all but 100 μl of supernatant was removed. The cells were resuspended in the 100 μl of remaining media, plated onto B plates supplemented with kanamycin (Kan; 3 μg/ml) and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 80 μg/ml), and pretreated with a lawn of A. vinelandii AZBB063. Plates were then incubated at 30°C for several days until colonies formed. Colonies were selected that revealed the blue phenotype, indicating that the integrated transposon had altered the A. vinelandii AZBB030 or AZBB035 target strains so they could support the growth of the nitrogen-dependent (Nif−) phenotype of the A. vinelandii AZBB063 biosensor strain. Colonies that turned blue were transferred to a B plate containing streptomycin (Str; 0.5 μg/ml) to confirm and isolate only the transposon-modified strain and then again checked for the proper blue phenotype by streaking clean isolates onto an X-Gal- and kanamycin-supplemented plate with a lawn of A. vinelandii AZBB063. False-positive colonies were further tested by streaking onto B plates containing streptomycin and X-Gal.
Transposon insertion analysis.
Strains exhibiting the desired phenotype were first purified by streaking individual colonies several times on a B plate supplemented with streptomycin. Cells were then scraped from a clean plate (∼100 μl of cells), and genomic DNA was isolated using a ZR Fungal/Bacterial DNA miniprep kit (Zymo Research, Irvine, CA). Genomic DNA was digested with PstI, purified using a DNA Clean and Concentrator-25 kit (Zymo Research), and ligated using T4 DNA ligase (New England BioLabs, Ipswich, MA). PCR was then run on the ligated DNA using the primer BBP1241 (5′-GACCGCTATCAGGACATAGCGTTG-3′), which aligns near the end of the kan gene used as the selectable marker indicating transposon insertion. This approach relies on the potential of two similar DNA fragments containing the insert and sequence downstream the point of insertion to ligate and orient directionally toward one another. Although this is a rare event, PCR can then be utilized to amplify these segments for sequencing. The PCRs were analyzed by gel electrophoresis to confirm amplification, cleaned using the DNA Clean and Concentrator-25 kit, and sent for Sanger sequencing with the same primer BBP1241. All sequencing products for positive phenotypes identified a segment matching between 200 and 700 bp of genomic sequence from A. vinelandii DJ (35).
Coculture of Azotobacter vinelandii and Chlorella sorokiniana.
A minimal amount of cells from strains of A. vinelandii and C. sorokiniana were spotted onto B plates substituting plant cell culture tested agar (Sigma, P/N A7921) for Bacto agar and grown on a custom light table for several days to make a qualitative assessment of the potential of various strains to support the growth of the algae. Equivalent starting quantities of cells were spotted to the plates. For liquid culture experiments, equivalent quantities of cells were inoculated into 60 ml of B medium in a 125-ml Erlenmeyer flask and grown under a bank of fluorescent lights with 14:10 light-dark cycles, while monitoring numbers of cells/ml daily. The light intensity for both experiments was ∼200 μmol min−1 m2. Liquid cultures were mixed on a shaker table with constant shaking at 160 rpm.
RESULTS
Urea as a terminal nitrogen compound.
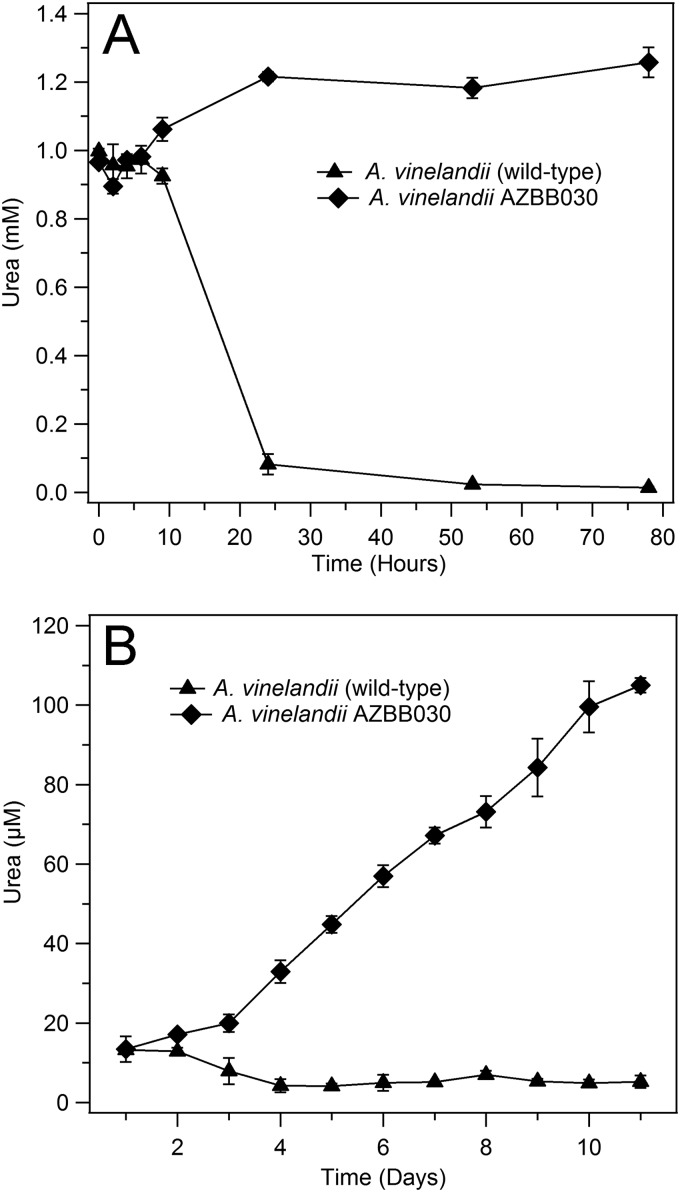
An initial aim pursued in this effort was to modify A. vinelandii metabolism to convert a common nitrogen metabolite into a terminal nitrogen product. Urea was selected as an initial target based on a careful analysis of A. vinelandii metabolic pathways and known enzymes. A. vinelandii contains genes for a known urease enzyme system (ureABC) (35). Substitution of the ureABC operon with a streptomycin antibiotic marker resulted in A. vinelandii AZBB030 that was unable to metabolize extraneously provided urea, even after several days, whereas the wild-type A. vinelandii strain was able to clear as much as 2 to 3 mM urea within 24 h after the start of exponential growth (Fig. 1A), demonstrating that urea had become a terminal product.
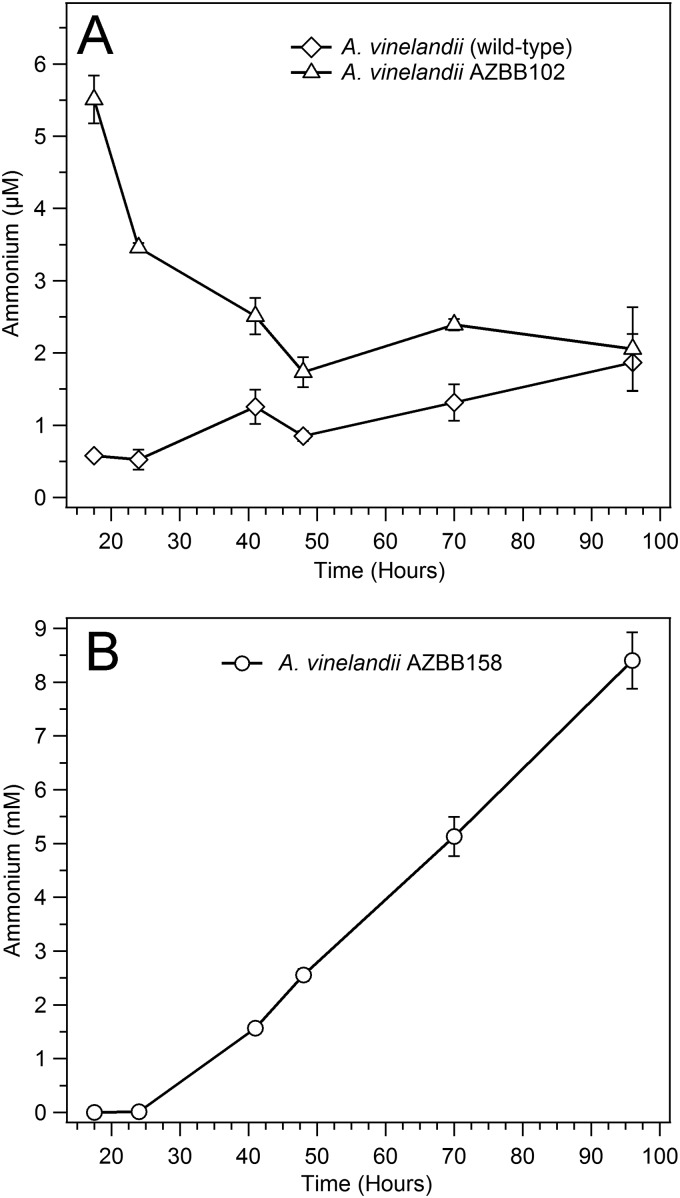
FIG 1.
Urea analysis of key A. vinelandii strains. Shown above are the results of urea analysis of the A. vinelandii wild-type strain and the ΔureABC::Strr strain A. vinelandii AZBB030. (A) Results of an experiment where both strains were provided ∼1.2 mM urea at the time of inoculation, illustrating that A. vinelandii AZBB030 does not metabolize urea to an appreciable amount, while the wild-type strain depletes the urea from the culture during the first 24 h of growth. (B) The urea levels in A. vinelandii AZBB030 increase over time versus the wild-type strain, reaching levels in excess of 100 μM. All results and statistics are calculated based on triplicate samples.
Based on the genome annotation (35), A. vinelandii lacks a known arginase gene but contains all of the remaining genes constituting a functional urea cycle. Further analysis of urea levels in control samples revealed that the ΔureABC::Strr A. vinelandii AZBB030 strain was accumulating urea (Fig. 1B), either through the action of a yet-to-be-characterized arginase enzyme system or through alternative metabolic pathways present in A. vinelandii that yield urea as a by-product. This accumulation of urea in the μM range (∼100 μM after about 10 days of culture under the conditions used here) resulted in only slight improvements supporting the growth of nondiazotrophic strains in coculture but confirmed the potential to increase urea levels using strategies planned for later experiments. The possibility of identifying a potential novel arginase or urea cycle alternatives in A. vinelandii remain points of interest for future studies.
Construction of a nitrogen biosensor strain.
The second goal in this effort was to modify or identify a bacterial strain that could serve as an indicator of extracellular nitrogen production for use in applications to screen transposon mutagenesis experiments. Initially, several strains of bacteria were considered possible candidates for this role, based on physical characteristics such as color that would make them easy to differentiate on plates. However, each of the potential biosensor strains would require incorporation of antibiotic selection markers to be grown together with transposon treated A. vinelandii and did not guarantee that these strains would be able to transport or metabolize all of the potential extracellular forms of nitrogen produced by A. vinelandii. Alternatively, we elected to construct a strain of A. vinelandii that could fulfill this biosensor purpose. First, borrowing from a technique described previously, lacZ from Escherichia coli MG1655 was incorporated into A. vinelandii directly downstream of the scrX promoter (23, 36), resulting in a blue colored phenotype when grown in the presence of X-Gal. Next, we replaced the nitrogenase regulatory genes nifLA with a tetracycline antibiotic marker. These genes regulate expression of nitrogenase when molybdenum is available (8). This modification resulted in a strain that grew extremely slowly in the absence of extraneously provided nitrogen sources such as ammonium or urea (Nif− phenotype) but grew well when a nitrogen source was provided as a component of the medium. Combining these two modifications, this new A. vinelandii AZBB063 met all of the desired requirements of our nitrogen biosensor; requiring extraneous nitrogen for growth, appearing blue when grown on plates containing X-Gal, and having a strong potential to metabolize any nitrogen containing metabolites that might be excreted by a modified or wild-type A. vinelandii strain. A diagram of the alterations made to the A. vinelandii AZBB063 nitrogen biosensor strain versus wild-type A. vinelandii is shown in Fig. 2.
Construction of a ΔnifL strain.
In addition to the ΔureABC::Strr A. vinelandii AZBB030 that accumulates low levels of urea, it was also initially of interest to construct a deregulated nitrogenase strain. Reports more than 2 decades ago by Bali et al. (7) described the construction of an A. vinelandii strain yielding copious quantities of ammonia (as high as 35 mM in later reports [8]) by disrupting the nifL gene but leaving the nifA gene intact behind various promoters. Based on this previous report, we hypothesized that transforming the ΔnifLA::Strr substituted A. vinelandii strain with a plasmid that would incorporate nifA behind the promoter for nifL (ΔnifLA::nifA), should result in a similar phenotype that is deregulated for nitrogenase production. Isolation of A. vinelandii AZBB035 containing nifA in the place of nifLA, along with ΔureABC::Strr, did rescue the nitrogen-fixing phenotype (Nif+) capable of growth on standard B plates but did not result in copious quantities of ammonium being released into the supernatant. Ortiz-Marquez et al. recently constructed a strain using a similar approach to that taken here based on the same concepts from the report by Bali et al. and reported an increase in ammonia production, although not to the same levels as reported by Bali et al. or Brewin et al. (7–9). In each of these cases, the approaches taken to increase ammonia production were slightly different from what was pursued here (see the more detailed analysis of these differences below). When either A. vinelandii AZBB030 or AZBB035 were grown on a plate in the presence of the nitrogen biosensor A. vinelandii AZBB063, extracellular nitrogen produced by either strain (in the form of either urea or ammonium) was not sufficient to support significant growth of the A. vinelandii AZBB063 biosensor strain, in contrast to the results found in experiments described below for other strains.
Transposon mutagenesis experiments to increase nitrogen output.
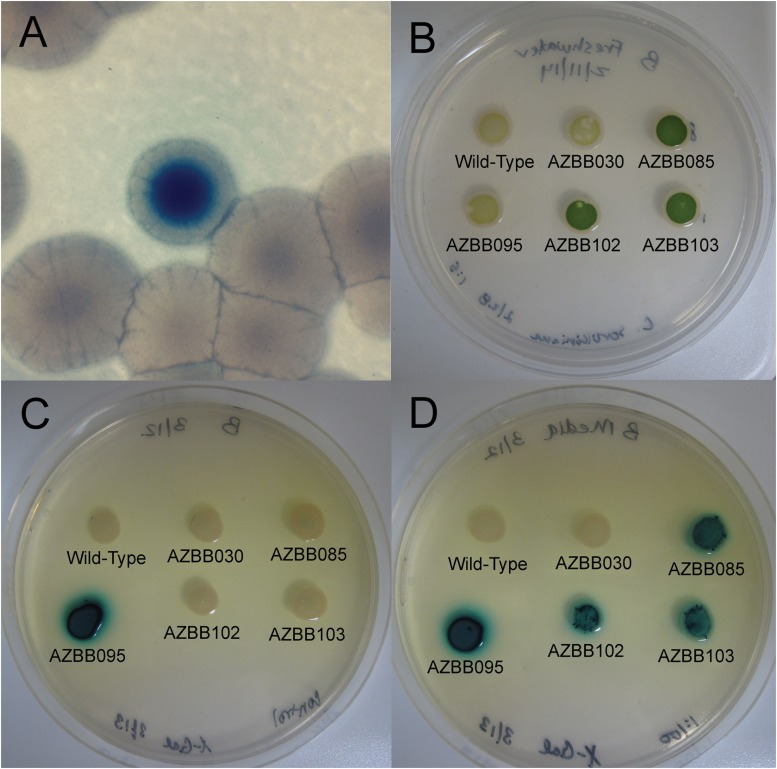
Transposon mutagenesis can be used to deliver random gene disruptions. This approach was utilized here based on the assumption that disrupting specific pathways might result in the accumulation of nitrogen containing metabolites (including urea) that would escape the cell and become available to other organisms in coculture. Alternative disruptions might lead to the loss of metabolites through various transport phenomena. With the target A. vinelandii AZBB030 and AZBB035 and the nitrogen biosensor A. vinelandii AZBB063 strains in hand, the next goal was to use transposon mutagenesis to introduce random gene disruptions and look for a phenotype that results in the production of increased levels of extracellular nitrogen products. Using the techniques described in Materials and Methods to incorporate transposons through random insertions, conjugated cells of A. vinelandii AZBB030 or AZBB035 were spread over a plate containing a lawn of A. vinelandii AZBB063, along with X-Gal and kanamycin, and then grown for several days until colonies developed, indicating insertion of the transposon within the genome. Important to this approach, the initial plate used to select colonies is devoid of added nitrogen compounds, such that any insertions resulting in an undesirable disruption to nitrogen fixation or essential genes would be lost. After several additional days, numerous colonies turned blue, indicating potential extracellular nitrogen production (Fig. 3A). Approximately 15 colonies were isolated (from roughly 3,000 screened) by careful transfer, and the phenotypes were confirmed and tested more thoroughly. After genomic DNA isolation, PCR was performed to amplify the transposon and genomic region where the transposon was incorporated.
FIG 3.
Phenotypes of various A. vinelandii strains constructed or obtained in these studies. (A) Example of the blue phenotype found for a specific colony using the screening technique with the nitrogen biosensor A. vinelandii AZBB063 during the transposon mutagenesis experiment. (B) Various strains of A. vinelandii constructed for the present study and grown as spots on a derivation of B medium with C. sorokiniana to determine whether the strain could provide sufficient nitrogen to support the growth of the green alga. (C) Example of a screen for false-positive strains that had unintentionally obtained the lacZ gene from A. vinelandii AZBB063, resulting in a false-positive phenotype when screened together with A. vinelandii AZBB063. In this experiment, pure strains were grown alone on B medium supplemented with X-Gal. (D) Various strains spotted onto a B plate together with a small amount of A. vinelandii AZBB063 and X-Gal. These results indicate that A. vinelandii AZBB085, AZBB102, and AZBB103 are all able to support the growth of a second strain (either A. vinelandii AZBB063 or C. sorokiniana) that is unable to obtain nitrogen through the action of a functioning internal nitrogenase.
Approximately half of the A. vinelandii colonies isolated contained the blue phenotype when grown in the presence of X-Gal, even in the absence of the nitrogen biosensor A. vinelandii AZBB063 (Fig. 3C). Sequencing revealed that lacZ and the downstream kanamycin cassette from the A. vinelandii AZBB063 (Fig. 2) had been transferred to either A. vinelandii AZBB030 or AZBB035. This false-positive result is likely due to the survival of a small amount of the E. coli WM3064 conjugation strain that introduced lacZ from the A. vinelandii AZBB063 to the nitrogen-fixing target strains following plating on solid medium. The false-positive phenotype was easily differentiated from strains with a true-positive result, which only revealed the blue phenotype when grown together with the A. vinelandii AZBB063 biosensor strain. An alternative construction of the indicator strain separating the kanamycin antibiotic selection marker from the lacZ gene should minimize the potential of these false-positive strains in future experiments. Although this false-positive result is an unfortunate occurrence, the percentage of these strains overall is quite small (<10 total colonies out of several thousand screened) and still allowed for the identification of true-positive colonies, as described below.
The remaining strains yielding the correct blue phenotype when cocultured with the A. vinelandii AZBB063 biosensor strain were also tested by spotting a small amount of each strain together with the green algae Chlorella sorokiniana onto a B plate (devoid of added nitrogen compounds) to determine whether the strain could provide sufficient extracellular fixed nitrogen to support the growth of this algal strain. Figure 3B shows an example where strain A. vinelandii AZBB085 supported the growth of C. sorokiniana while wild-type and the initial target A. vinelandii AZBB030 or AZBB035 did not. A. vinelandii AZBB095, a false-positive strain, was also unable to support the growth of C. sorokiniana (Fig. 3B and C). Testing the potential to support an algal strain is a nice secondary screen, although whether a specific strain of algae can utilize the various extracellular nitrogen compounds produced is likely strain dependent.
Identification of gene deletions resulting in increased extracellular nitrogen.
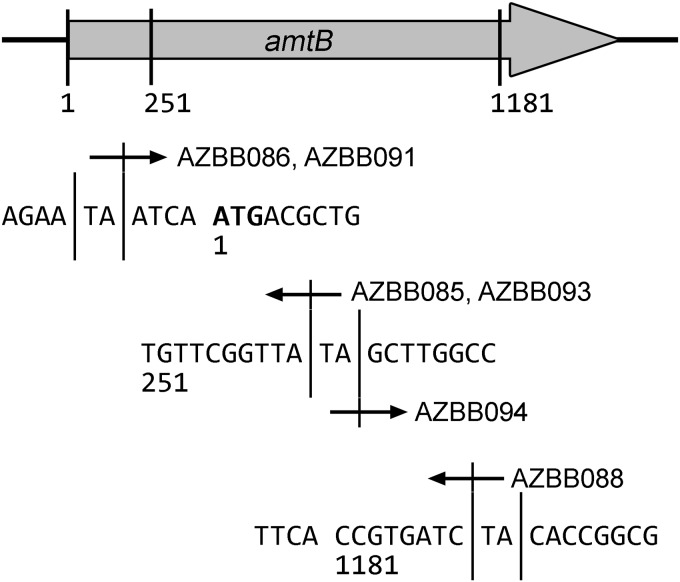
A. vinelandii AZBB085 and five additional strains showed the correct blue phenotype only when spotted on plates with A. vinelandii AZBB063. Each of these strains had the kanamycin cassette transposon inserted into the gene amtB, and this occurred in both A. vinelandii AZBB030 and AZBB035 derived colonies. Although it was possible that some of the colonies could be replicates of the same transposon insertion event, only one of the six cases isolated here contained the same insertion and orientation for the same conjugation experiment (into either A. vinelandii AZBB030 or AZBB035). In all other cases, the insert was either in a different location, or it was inserted in a different direction (Fig. 4). This result indicated a clear potential for developing this phenotype through a targeted substitution of the entire amtB gene. Since it was also feared that the phenotype might potentially be the result of multiple insertions which were not obvious, we developed a strategy to substitute amtB using standard genetic techniques (a double-homologous-recombination approach) without the use of a transposon. The amtB gene has been proposed to play a role in the transport of ammonia or ammonium, although a specific role for amtB in A. vinelandii is still unclear (37–41). Based on this finding and on what is already known about these various genes and the nature of the nitrogen screen, three potential explanations were considered to explain the nitrogen secreting phenotype following amtB disruption. First, deleting amtB might result in increased extracellular ammonium if loss of this gene hindered the ability of the strain to recover ammonia or ammonium that leaks from the cell by natural processes, as has been described in other strains (42). Second, amtB might play a role in the transport of an alternative nitrogen compound, which requires the amtB still present in the A. vinelandii AZBB063 biosensor strain to transport this compound back into the cell. Finally, there was the remote possibility that amtB could also play a role in urea uptake in A. vinelandii, although the likelihood of this was considered low. Any of these explanations would still support a successful application of this approach to yield a strain of A. vinelandii with increased biofertilizer potential. To rule out the last option, amtB gene deletion/replacements were developed for both the ΔureABC::Strr strain and also wild-type A. vinelandii.
FIG 4.
Insertions into amtB resulting in nitrogen production phenotypes. At the top of the figure is a drawing of the amtB gene region of A. vinelandii. The three different regions where insertions were found in the present study during the nitrogen production screen are marked on the diagram, and the sequence of each region is shown below. Specific TA base regions where insertions were made are marked with dividers on either side, and arrows are included depicting the region that was sequenced immediately outside the transposon inverted repeat when sequenced with a primer that complemented a segment within to the kanamycin selection marker from within the transposon. Specific strain numbers of A. vinelandii from Table 1 are labeled beside each arrow.
Construction of an amtB deletion/replacement A. vinelandii strain.
Using the same approach that was taken to generate A. vinelandii strains AZBB030, AZBB035, and AZBB063, we further constructed A. vinelandii strains AZBB102 and AZBB103. A. vinelandii strain AZBB102 contains only ΔamtB::Kanr, whereas AZBB103 contains ΔamtB::Kanr along with ΔureABC::Strr. Both strains resulted in the blue phenotype when grown together with A. vinelandii AZBB063 and also supported the growth of C. sorokiniana when a nitrogen source was not supplemented in the medium (Fig. 3B and D). This indicates that urea accumulation is not required for the observed phenotype, which was further confirmed by determining that urea production and release had not increased versus A. vinelandii AZBB030 and AZBB035 urea levels in these strains (results similar to Fig. 1B).
Analysis of ammonium production by strains AZBB102 and AZBB103.
Since amtB is proposed to be involved in the transport of ammonia or ammonium (38–41), this seemed to be the most probable explanation for supporting the growth of either the algae or the A. vinelandii biosensor strain AZBB063. Cells were grown for as long as a week, while supernatant was removed and ammonium was quantified using several different techniques that have been used previously by ourselves (27, 30) or reported by others (9, 31) to identify ammonium. Previous reports of modifications to the nifLA operon have reported the production of copious amounts of ammonium (7, 8). Our analysis of the supernatant from strain AZBB102 versus A. vinelandii wild type did find elevated levels of ammonium in the media, but levels were in the low micromolar range (Fig. 5A), which is in contrast to the dramatic increases reported for nifLA modifications (7–9).
FIG 5.
Ammonium levels accumulating in A. vinelandii strain supernatants. (A) Results of an analysis of A. vinelandii strain supernatants for the wild type and the ΔamtB::Kanr AZBB102 strain. The levels of ammonium are consistently higher for A. vinelandii AZBB102, although the levels decrease over time and were not found to increase above 10 μM during the entire experiment. The highest levels of ammonium were found early during the growth, while the strain was still in the exponential stage of growth. (B) The levels of ammonium found in the nifLA disruption based on the previously described construction (8) resulted in a phenotype that accumulated significant amounts of ammonium, although these levels showed a lag following inoculation before increasing after about 24 h of growth. The averages and standard deviations shown are the results for triplicate samples. All cultures were grown at 22°C while shaking at 160 rpm.
These results indicate that deletion or disruption of the amtB gene in A. vinelandii results in a slow release of ammonium into the media. Although the levels of ammonium detected in A. vinelandii strain AZBB102 were low, these were consistently and significantly higher than what was found in the wild-type strain control (Fig. 5A). A further elemental analysis of carbon, hydrogen, and nitrogen (CHN analysis) of supernatants that were collected and lyophilized did not find significant amounts of nitrogen accumulating in the supernatants, although an analysis of the dry cell materials did find elevated nitrogen levels (3.63% ± 0.11% nitrogen for strain AZBB102 versus 2.36% ± 0.05% for the wild type, an increase of ca. 50%) under the growth conditions tested here. This indicates that the low level of ammonium found from the analysis described in Fig. 5A is the most likely source of the nitrogen produced by A. vinelandii AZBB102 and not an alternative form of nitrogen.
Potential of A. vinelandii AZBB103 to support the growth of the green alga C. sorokiniana.
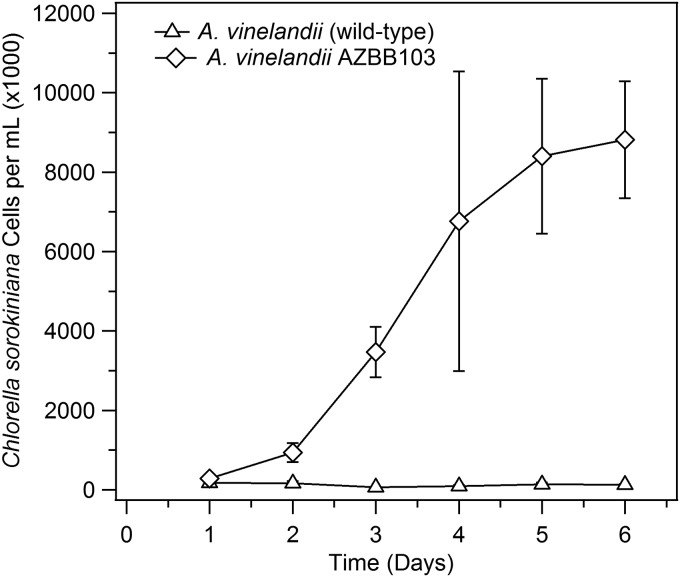
The approach using coculture on solid media (Fig. 3B) is a simple screen that provides a qualitative assessment of the ability of A. vinelandii strains to support algae or other organisms. In the absence of specific methods of quantitation (or if identification of the nitrogen compound responsible for supporting algal growth were unclear), cell growth of a nondiazotroph in coculture can be utilized to estimate the potential of the strain as a biofertilizer. Cocultures of A. vinelandii AZBB103 and C. sorokiniana were grown in a simple B medium devoid of added nitrogen under a bank of fluorescent lights along with a control containing the algae and A. vinelandii wild type to determine levels of algal cells that might be supported by the extracellular nitrogen being released when sufficient sugar is provided in the medium. Figure 6 shows the results of the coculture of the ΔamtB::Kanr and ΔureABC::Strr strain A. vinelandii AZBB103 with C. sorokiniana. As can be seen, the levels of algae cells increased dramatically when cultured with A. vinelandii AZBB103 versus the A. vinelandii wild-type strain (a similar result was found with A. vinelandii AZBB102). The improvement in the yield was >50-fold after only a few days of growth.
FIG 6.
Coculture of A. vinelandii AZBB103 and C. sorokiniana. Shown above are the results obtained when A. vinelandii AZBB103 (ΔamtB::Kanr and ΔureABC::Strr) was grown as a coculture with C. sorokiniana cells over a period of 6 days. Cells were counted by using a hemocytometer. A control containing A. vinelandii wild-type grown in coculture with C. sorokiniana was also included for comparison. The large variance seen at day 4 for A. vinelandii AZBB103 is potentially related to cell clumping. After day 4, cell clumps were broken up by repeatedly passage through a pipette tip prior to counting cells. All results and statistics are calculated based on triplicate samples.
Reconstruction of a high ammonium production phenotype.
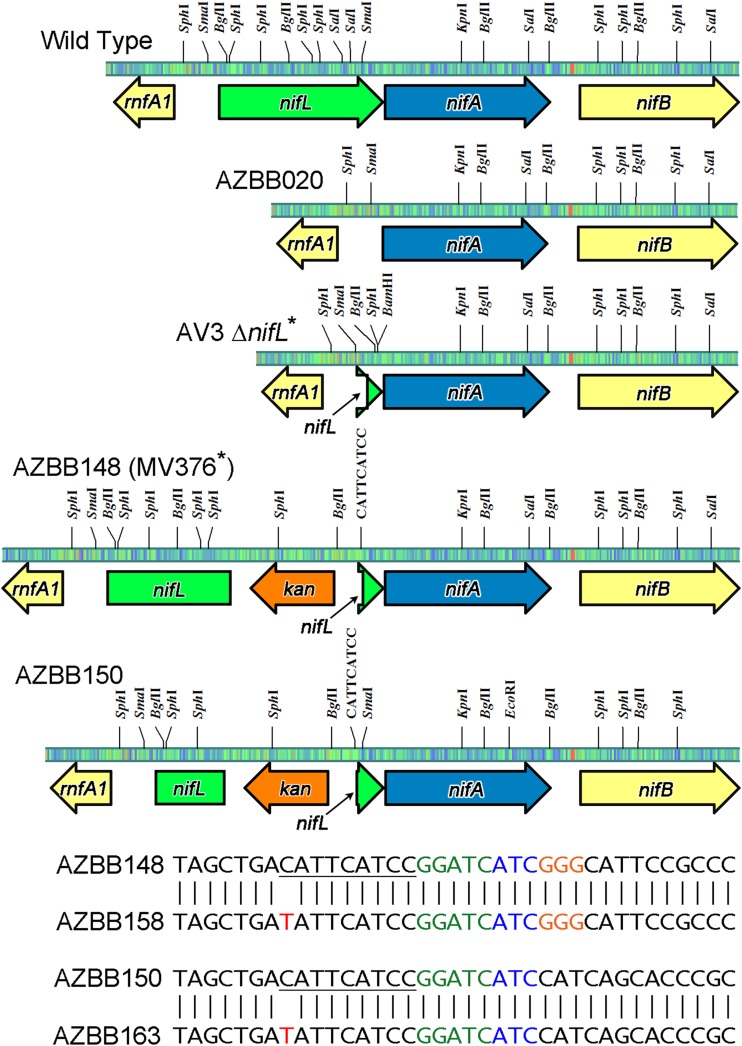
Since the ΔamtB::Kanr ammonium release phenotype differs from what has been reported by others for disruptions to the nifLA operon (7–9), and since our initial efforts to construct a high-ammonium-excreting strain were only partially successful (Nif+ phenotype, but no dramatic release of ammonium to the medium for A. vinelandii AZBB020), we finally pursued efforts to reconstruct an A. vinelandii strain equivalent to what was previously reported by Brewin et al. (8). An attempt to obtain the exact strain reported by Brewin et al. (8) from Martin Drummond was unsuccessful, since the strain was likely lost following his retirement (personal communication from Martin Drummond). Using the approach detailed by Brewin et al., we constructed a plasmid similar to pBB369 (our pPCRNH3-42) and transformed A. vinelandii AZBB010 with this plasmid, isolating kanamycin-resistant colonies (A. vinelandii AZBB148). A. vinelandii AZBB148 was confirmed to have incorporated the kanamycin resistance cassette in the proper location and direction in the genome, but did not yield a Nif+ phenotype. A slightly different strain transformed with plasmid pPCRNH3-43 (A. vinelandii AZBB150) also did not yield a Nif+ phenotype. However, when each of these strains were transferred to B medium devoid of an added nitrogen source and grown as liquid cultures, spontaneous mutants did arise after several days that were Nif+ and accumulated high levels of ammonium in the medium (Fig. 5B). These evolved strains were isolated on solid medium and designated A. vinelandii AZBB158 and AZBB163, respectively. A. vinelandii AZBB158 was grown in B medium similar to what was done for A. vinelandii AZBB102 and A. vinelandii wild-type and was found to yield ammonium at levels approaching 10 mM after 4 days of growth at 22°C, differentiating this with the phenotype found with ΔamtB::Kanr (Fig. 5A). The levels of ammonium measured following depletion of the sucrose from the media reached >20 mM after more than a week of culture at 22°C (∼35 mM at 30°C for A. vinelandii AZBB163). The nifLA regions of both A. vinelandii strains AZBB158 and AZBB163 were amplified, sequenced, and found to contain a point mutation to each residing in the segment of DNA upstream of the kanamycin promoter (Fig. 7). This mutation resided in the same location within the kanamycin cassette in both of the evolved Nif+ strains (AZBB158 and AZBB163), and lies in close proximity to the BamHI restriction site that is used to shuttle the kanamycin cassette between vectors. Both Brewin et al. and Bali et al. used the kanamycin cassette from plasmid pUC4-KIXX (7, 8), which is to the best of our knowledge practically identical to the kanamycin cassette of pPCRKAN4, except for a small segment of DNA just prior to the restriction sites that flank the cassette in each case. As shown in Fig. 7, the kanamycin cassette from pPCRKAN4 contains the sequence (CCCAGTAGCT, where the underlined TAGCT is shown at the beginning of the alignments in Fig. 7) that is derived from Tn5. This CCCAGTAGCT sequence is the same in both the pPCRKAN4 kanamycin cassette and the pUC4-KIXX cassette (pUC4-KIXX cassette sequence kindly provided by Haruyasu Kinashi [43, 44]). Following the CCCAGTAGCT sequence, the pPCRKAN4 contains GACATTCATCC, which is also derived from Tn5, and GGATCATC, which is the product of the filled-in BamHI and EcoRV site ligation product used to incorporate the cassette into the nifL region of both pPCRNH3-42 and pPCRNH3-43. This GACATTCATCC GGATCATC sequence is what differentiates the pPCRKAN4 from the pUC4-KIXX sequence, which contains CGAGAAGCTTCCC downstream of the CCCAGTAGCT sequence and is the remnant of the XhoI, HindIII, and SmaI sites (XhoI also utilizes the CT from the CCCAGTAGCT upstream sequence). Following this sequence, both strain A. vinelandii AZBB148 and MV376 (8) are then followed by GGGCATTCCGCCCG, which is the remainder of the SmaI site and A. vinelandii sequence lying upstream of the nifA gene. Thus, the only difference between our A. vinelandii AZBB148 and A. vinelandii MV376 in this region is the GACATTCATCC GGATCATC sequence that is in place of CGAGAAGCTTCCC, respectively. The modification to A. vinelandii AZBB148 and AZBB150 that resulted in the Nif+ phenotype was GATATTCATCC GGATCATC (where C mutated to the underlined T). We assume that the difference in this short region of sequence is responsible for why we were unable to obtain the high ammonium producing phenotype using our pPCRKAN4 derived kanamycin cassette instead of the cassette obtained from pUC4-KIXX. Further features of this region will be investigated in the future. This highlights the unanticipated importance of the source and size of the kanamycin cassette when attempting to incorporate this cassette. As pointed out previously (8), obtaining the phenotype is directionally dependent on the kanamycin cassette in order to achieve the proper Nif+ and high-ammonium-yielding phenotype and is further evidence that high ammonium production is dependent on a component of the kanamycin cassette that enhances nifA expression.
FIG 7.
Illustration of different approaches to deregulate nitrogen fixation and excrete ammonium through manipulation of the nifLA operon. Shown is an illustration of the nifLA genome region from A. vinelandii (wild type) and various final constructs developed here or reported by others (7–9). The ΔnifLA::nifA construct A. vinelandii AZBB020 resulted in a nitrogen-fixing (Nif+) phenotype but did not increase levels of ammonium in the extracellular space, as initially anticipated. The strain A. vinelandii AV3 ΔnifL constructed by Ortiz-Marquez et al. (9) took a similar approach but left a small segment of nifL intact and resulted in μM levels of ammonium and a Nif+ phenotype (drawn as described previously [9]). Strain A. vinelandii AZBB148 was constructed similar to the approach to construct A. vinelandii strains MV376 and MD367 (7, 8), except that pPCRKAN4 was used as the source of the kanamycin cassette instead of pUC4-KIXX. A. vinelandii AZBB150 was constructed so that this same pPCRKAN4 derived kanamycin cassette would be inserted into nifL slightly further upstream while removing a larger section of the nifL gene. Both A. vinelandii AZBB148 and AZBB150 were found to be Nif− but became Nif+ following a spontaneous mutation to yield A. vinelandii AZBB158 and AZBB163. The location of this mutation (underlined region) is shown in the alignments and is also marked on the illustration for the nifLA region of each of these strains. The sequence shown in green represents the remainder of the polished BamHI site, while the sequence drawn in blue is the remnant of the EcoRV site. The remainder of the SmaI site used here and previously to construct strains MV376 and MD367 (7, 8) is shown in orange for A. vinelandii AZBB148 and AZBB158. The C-to-T mutations for A. vinelandii AZBB158 and AZBB163 are shown in red. The figure was constructed using the program pDRAW32 (AcaClone Software).
DISCUSSION
The efforts pursued in this work highlight three different approaches to increasing extracellular nitrogen production to enhance the biofertilizer potential of A. vinelandii. Efforts to convert urea into a terminal nitrogen product by deleting a key enzyme involved in nitrogen recycling resulted in μM levels of urea accumulating in the medium and further demonstrated successful limitation of reuptake of the urea nitrogen by A. vinelandii (Fig. 1). The levels of urea nitrogen obtained by deleting the urease are similar to the total nitrogen produced by strain A. vinelandii AV3 (ΔnifL) constructed by Ortiz-Marquez et al. (9) using their alternative approach to produce elevated levels of ammonium.
In addition to new approaches described here, we also reevaluated the potential of nifL disruptions through two additional approaches in this work. Previous reports have described several methods to increase levels of ammonium excreted by A. vinelandii through disruption of the nifL gene from the nifLA operon (7–9). An illustration of various modifications made to the nifLA region here and previously is shown in Fig. 7. Ortiz-Marquez et al. (9) recently published a report applying a derivation of the approach first described by Bali et al. in 1992 (7) that disrupts and deletes a section of the nifL gene while leaving nifA intact. Ortiz-Marquez et al. constructed an almost complete deletion of nifL (9), yielding a strain that also accumulated ammonium in the medium, although Ortiz-Marquez et al. reported ammonium levels of ∼250 μM after 48 h (9), whereas Bali et al. and Brewin et al. had reported levels of ∼10 mM and >25 mM, respectively, over somewhat longer time periods (7, 8). As pointed out by Ortiz-Marquez et al. (9), direct comparisons with the strains obtained by Bali et al. and Brewin et al. are difficult to make since reconstructions of certain strains were unobtainable even by the original authors. Brewin et al. did report confirmation of the original A. vinelandii strain MV376 phenotype described by Bali et al. but reported that other strains were difficult to rescue from their stocks (7, 8, 45). Our initial approach here using a complete deletion of the nifL gene resulted in a Nif+ phenotype but no significant accumulation of ammonium in the growth medium versus the wild type. After applying a directed evolution approach, we were able to isolate a similar phenotype to what was reported using the approach reported by Brewin et al. (8). This required first isolating our initial A. vinelandii strains in the presence of a fixed nitrogen source in the medium and then isolation of a spontaneous Nif+ mutation. These evolved strains resulted in the production of >10 mM ammonium after 4 days of culture (Fig. 5B), similar to the initial reports by Bali et al. (43). A more recent report by Ortiz-Marquez et al. (46) coupled their A. vinelandii ΔnifL strain previously described (9) with point mutations to glutamine synthase to yield ammonium levels up to 7 mM (46), although the long-term stability of their strain may be problematic based upon the potential for recombination events within the cell. The A. vinelandii AZBB158 and AZBB163 are also potentially problematic, since they are prone to contamination by spontaneous cheaters (cells of A. vinelandii that evolve to quit fixing nitrogen and then prey upon the ammonium released by the remaining ammonium excreting cells). However, these results indicate a potential to further improve nitrogen output through further modification of the nifLA operon, even without having to modify glutamine synthase.
Disruption or deletion of the amtB gene identified using the screening technique described here resulted in a low level of ammonium accumulation in the extracellular space (Fig. 5A). This was capable of supporting the growth of nondiazotrophs in coculture when sucrose was provided in the medium to support the growth of the A. vinelandii. Although the overall levels of ammonium quantified in the supernatants were low, they were significantly higher than what was found for the wild-type strain (Fig. 5A). We hypothesize that inclusion of a strain that can utilize the low levels of ammonium may rapidly deplete this nitrogen reservoir, driving additional ammonium across the membrane by diffusion to maintain a constant, but low level of ammonium flux across the membrane.
There is an active debate, as well as contrasting results, concerning the role that amtB plays in the transport of ammonia or ammonium in various bacteria (38–41), including reports of increasing extracellular ammonium when amtB is deleted while also demonstrating that cells can obtain ammonium from the extracellular space even in the absence of amtB (42, 47). The amtB gene has been implicated in the release of ammonium previously in other bacteria. Castorph and Kleiner reported that ammonium transport mutants (Amt−) excreted ammonium in Klebsiella pneumonia (48). Meletzus et al. isolated amtB mutants from A. vinelandii and showed that amtB mutants could grow on limiting ammonium but were unable to transport methylammonium (37). Experiments testing for the loss of ammonium were not described in the Meletzus et al. study, although it was concluded that amtB has no function that is of vital importance in A. vinelandii (37). Our results confirm that deletion of amtB in A. vinelandii did not dramatically affect the growth rate or health of the A. vinelandii but indicate that amtB may be important for minimizing the loss of ammonium to the environment by this species. Another recent report found that ammonium was excreted in an amtB mutant strain of Pseudomonas stutzeri yielding extracellular ammonium levels of ∼6 μM (42), which is very similar to the levels obtained here (Fig. 5A), while a separate report linked upregulation of amtB to the ability of Rhodococcus erythropolis to utilize atmospheric ammonia, with disruption of amtB resulting in a growth defect under nitrogen-limiting conditions (47).
The identification of the amtB disruption demonstrates how the screening approach using a nitrogen biosensor strain can yield new strains producing elevated extracellular nitrogen products regardless of whether the specific nitrogen compound produced was targeted by the approach. This broad screening potential could have further benefits in identifying additional potential “nitrogen shuttle” compounds that might have further utility for providing nitrogen in coculture. A. vinelandii also produces additional nitrogen shuttle compounds such as siderophores that are suitable for supporting the growth of algae in coculture (11), as well as additional extracellular proteins (49). In these studies, the levels of nitrogen released to the culture medium by the amtB disruption were very low (i.e., a low micromolar range) and might be missed by less sensitive assays.
A. vinelandii strains AZBB030, AZBB102, and AZBB103 did not require the introduction of any foreign genes (though antibiotic markers were used under this approach). These disruptions that were shown to increase extracellular nitrogen levels when sufficient sugar is provided to the media by deleting/replacing ureABC and/or amtB could also be constructed as gene deletions using markerless techniques developed in our laboratory (to be published) or through congression approaches (23), resulting in true gene deletions that would not be transgenic but simply gene deficient. Each of these modifications could be coupled to one another or combined with approaches that alter the nifLA operon to enhance total nitrogen output further, similar to the approach taken by Ortiz-Marquez et al. with nifLA alterations coupled to glnA mutations (46). The development of stable genetic constructs that lack foreign genes would yield biofertilizer strains with a greater potential for utilization in algal culture and that could be applied to current conventional agricultural crops as an enhanced biofertilizer as well.
ACKNOWLEDGMENTS
This study is supported by grants (RC-0007-12) from the Initiative for Renewable Energy and the Environment (Institute on the Environment), the MnDRIVE transdisciplinary research initiative through the University of Minnesota based on funding from the state of Minnesota to B.M.B, and the Biotechnology Institute at the University of Minnesota for fellowship funding to L.J.E.
We thank Juan Torrecilla and Peter Nixon for assistance in preliminary urea production assays. We thank Eric Lenneman for assistance in obtaining specific photographs. We thank Haruyasu Kinashi for providing a sequence of the pUC4-KIXX cassette region. We thank the Stable Isotope Laboratory at the University of Minnesota for assistance with CHN analysis.
REFERENCES
- 1.Smith BE. 2002. Structure: nitrogenase reveals its inner secrets. Science 297:1654–1655. [DOI] [PubMed] [Google Scholar]
- 2.International Fertilizer Industry Association. 2008. Food prices and fertilizer markets. International Fertilizer Industry Association, Paris, France. [Google Scholar]
- 3.Smil V. 2001. Enriching the earth: Fritz Haber, Carl Bosch, and the transformation of world food production. MIT Press, Cambridge, MA. [Google Scholar]
- 4.Apte SK, Chaurasia AK. 2011. Improved eco-friendly recombinant Anabaena sp. strain PCC7120 with enhanced nitrogen biofertilizer potential. Appl Environ Microbiol 77:395–399. doi: 10.1128/AEM.01714-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar R, Bhatia R, Kukreja K, Behl RK, Dudeja SS, Narula N. 2007. Establishment of Azotobacter on plant roots: chemotactic response, development and analysis of root exudates of cotton (Gossypium hirsutum L.) and wheat (Triticum aestivum L.). J Basic Microbiol 47:436–439. doi: 10.1002/jobm.200610285. [DOI] [PubMed] [Google Scholar]
- 6.Leaungvutiviroj C, Ruangphisarn P, Hansanimitkul P, Shinkawa H, Sasaki K. 2010. Development of a new biofertilizer with a high capacity for N2 fixation, phosphate and potassium solubilization and auxin production. Biosci Biotechnol Biochem 74:1098–1101. doi: 10.1271/bbb.90898. [DOI] [PubMed] [Google Scholar]
- 7.Bali A, Blanco G, Hill S, Kennedy C. 1992. Excretion of ammonium by a nifL mutant of Azotobacter vinelandii fixing nitrogen. Appl Environ Microbiol 58:1711–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brewin B, Woodley P, Drummond M. 1999. The basis of ammonium release in nifL mutants of Azotobacter vinelandii. J Bacteriol 181:7356–7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ortiz-Marquez JCF, Do Nascimento M, Dublan MDLA, Curatti L. 2012. Association with an ammonium-excreting bacterium allows diazotrophic culture of oil-rich eukaryotic microalgae. Appl Environ Microbiol 78:2345–2352. doi: 10.1128/AEM.06260-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sashidhar B, Podile AR. 2010. Mineral phosphate solubilization by rhizosphere bacteria and scope for manipulation of the direct oxidation pathway involving glucose dehydrogenase. J Appl Microbiol 109:1–12. doi: 10.1111/j.1365-2672.2009.04654.x. [DOI] [PubMed] [Google Scholar]
- 11.Villa JA, Ray EE, Barney BM. 2014. Azotobacter vinelandii siderophore can provide nitrogen to support the culture of the green algae Neochloris oleoabundans and Scenedesmus sp. BA032. FEMS Microbiol Lett 351:70–77. doi: 10.1111/1574-6968.12347. [DOI] [PubMed] [Google Scholar]
- 12.Brechignac F, Schiller P. 1992. Pilot CELSS based on a maltose-excreting Chlorella: concept and overview on the technological developments. Adv Space Res 12:33–36. doi: 10.1016/0273-1177(92)90317-Q. [DOI] [PubMed] [Google Scholar]
- 13.Fan B, Carvalhais LC, Becker A, Fedoseyenko D, von Wirén N, Borriss R. 2012. Transcriptomic profiling of Bacillus amyloliquefaciens FZB42 in response to maize root exudates. BMC Microbiol 12:116. doi: 10.1186/1471-2180-12-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lynch JM, Whipps JM. 1990. Substrate flow in the rhizosphere. Plant Soil 129:1–10. doi: 10.1007/BF00011685. [DOI] [Google Scholar]
- 15.Spoehr HA, Milner HW. 1949. The chemical composition of Chlorella; effect of environmental conditions. Plant Physiol 24:120–149. doi: 10.1104/pp.24.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu Q, Sommerfeld M, Jarvis E, Ghirardi M, Posewitz M, Seibert M, Darzins A. 2008. Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J 54:621–639. doi: 10.1111/j.1365-313X.2008.03492.x. [DOI] [PubMed] [Google Scholar]
- 17.Sheehan J, Dunahay T, Benemann J, Roessler P. 1998. A look back at the U.S. Department of Energy's Aquatic Species Program: biodiesel from algae. National Renewable Energy Laboratory, Washington, DC. [Google Scholar]
- 18.Hill J, Nelson E, Tilman D, Polasky S, Tiffany D. 2006. Environmental, economic, and energetic costs and benefits of biodiesel and ethanol biofuels. Proc Natl Acad Sci U S A 103:11206–11210. doi: 10.1073/pnas.0604600103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ortiz-Marquez JC, Do Nascimento M, Zehr JP, Curatti L. 2013. Genetic engineering of multispecies microbial cell factories as an alternative for bioenergy production. Trends Biotechnol 31:521–529. doi: 10.1016/j.tibtech.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 20.Searchinger T, Heimlich R, Houghton RA, Dong F, Elobeid A, Fabiosa J, Tokgoz S, Hayes D, Yu TH. 2008. Use of US croplands for biofuels increases greenhouse gases through emissions from land-use change. Science 319:1238–1240. doi: 10.1126/science.1151861. [DOI] [PubMed] [Google Scholar]
- 21.Ikeda S, Okubo T, Anda M, Nakashita H, Yasuda M, Sato S, Kaneko T, Tabata S, Eda S, Momiyama A, Terasawa K, Mitsui H, Minamisawa K. 2010. Community- and genome-based views of plant-associated bacteria: plant-bacterial interactions in soybean and rice. Plant Cell Physiol 51:1398–1410. doi: 10.1093/pcp/pcq119. [DOI] [PubMed] [Google Scholar]
- 22.Singh RJ, Chung GH, Nelson RL. 2007. Landmark research in legumes. Genome 50:525–537. doi: 10.1139/G07-037. [DOI] [PubMed] [Google Scholar]
- 23.Dos Santos PC. 2011. Molecular biology and genetic engineering in nitrogen fixation. Methods Mol Biol 766:81–92. doi: 10.1007/978-1-61779-194-9_6. [DOI] [PubMed] [Google Scholar]
- 24.Brutinel ED, Gralnick JA. 2012. Anomalies of the anaerobic tricarboxylic acid cycle in Shewanella oneidensis revealed by Tn-seq. Mol Microbiol 86:273–283. doi: 10.1111/j.1365-2958.2012.08196.x. [DOI] [PubMed] [Google Scholar]
- 25.Lenneman EM, Ohlert JM, Palani NP, Barney BM. 2013. Fatty alcohols for wax esters in Marinobacter aquaeolei VT8: two optional routes in the wax biosynthesis pathway. Appl Environ Microbiol 79:7055–7062. doi: 10.1128/AEM.02420-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lenneman EM, Wang P, Barney BM. 2014. Potential application of algicidal bacteria for improved lipid recovery with specific algae. FEMS Microbiol Lett 354:102–110. doi: 10.1111/1574-6968.12436. [DOI] [PubMed] [Google Scholar]
- 27.Barney BM, Yurth MG, Dos Santos PC, Dean DR, Seefeldt LC. 2009. A substrate channel in the nitrogenase MoFe protein. J Biol Inorg Chem 14:1015–1022. doi: 10.1007/s00775-009-0544-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarma R, Barney BM, Hamilton TL, Jones A, Seefeldt LC, Peters JW. 2008. Crystal structure of the L protein of Rhodobacter sphaeroides light-independent protochlorophyllide reductase with MgADP bound: a homologue of the nitrogenase Fe protein. Biochemistry 47:13004–13015. doi: 10.1021/bi801058r. [DOI] [PubMed] [Google Scholar]
- 29.Mather A, Roland D. 1969. The automated thiosemicarbazide-diacetyl monoxime method for plasma urea. Clin Chem 15:393–396. [PubMed] [Google Scholar]
- 30.Barney BM, Igarashi RY, Dos Santos PC, Dean DR, Seefeldt LC. 2004. Substrate interaction at an iron-sulfur face of the FeMo-cofactor during nitrogenase catalysis. J Biol Chem 279:53621–53624. doi: 10.1074/jbc.M410247200. [DOI] [PubMed] [Google Scholar]
- 31.Kanda J. 1995. Determination of ammonium in seawater based on the indophenol reaction with o-phenylphenol (OPP). Water Res 29:2746–2750. doi: 10.1016/0043-1354(95)00149-F. [DOI] [PubMed] [Google Scholar]
- 32.Barney BM, Laryukhin M, Igarashi RY, Lee HI, Dos Santos PC, Yang TC, Hoffman BM, Dean DR, Seefeldt LC. 2005. Trapping a hydrazine reduction intermediate on the nitrogenase active site. Biochemistry 44:8030–8037. doi: 10.1021/bi0504409. [DOI] [PubMed] [Google Scholar]
- 33.Corbin JL. 1984. Liquid chromatographic-fluorescence determination of ammonia from nitrogenase reactions: a 2-min assay. Appl Environ Microbiol 47:1027–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bouhenni R, Gehrke A, Saffarini D. 2005. Identification of genes involved in cytochrome c biogenesis in Shewanella oneidensis, using a modified mariner transposon. Appl Environ Microbiol 71:4935–4937. doi: 10.1128/AEM.71.8.4935-4937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Setubal JC, dos Santos P, Goldman BS, Ertesvåg H, Espin G, Rubio LM, Valla S, Almeida NF, Balasubramanian D, Cromes L, Curatti L, Du Z, Godsy E, Goodner B, Hellner-Burris K, Hernandez JA, Houmiel K, Imperial J, Kennedy C, Larson TJ, Latreille P, Ligon LS, Lu J, Maerk M, Miller NM, Norton S, O'Carroll IP, Paulsen I, Raulfs EC, Roemer R, Rosser J, Segura D, Slater S, Stricklin SL, Studholme DJ, Sun J, Viana CJ, Wallin E, Wang B, Wheeler C, Zhu H, Dean DR, Dixon R, Wood D. 2009. Genome sequence of Azotobacter vinelandii, an obligate aerobe specialized to support diverse anaerobic metabolic processes. J Bacteriol 191:4534–4545. doi: 10.1128/JB.00504-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson DC, Unciuleac MC, Dean DR. 2006. Controlled expression and functional analysis of iron-sulfur cluster biosynthetic components within Azotobacter vinelandii. J Bacteriol 188:7551–7561. doi: 10.1128/JB.00596-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meletzus D, Rudnick P, Doetsch N, Green A, Kennedy C. 1998. Characterization of the glnK-amtB operon of Azotobacter vinelandii. J Bacteriol 180:3260–3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Musa-Aziz R, Chen LM, Pelletier MF, Boron WF. 2009. Relative CO2/NH3 selectivities of AQP1, AQP4, AQP5, AmtB, and RhAG. Proc Natl Acad Sci U S A 106:5406–5411. doi: 10.1073/pnas.0813231106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Musa-Aziz R, Jiang LH, Chen LM, Behar KL, Boron WF. 2009. Concentration-dependent effects on intracellular and surface pH of exposing Xenopus oocytes to solutions containing NH3/NH4+. J Membr Biol 228:15–31. doi: 10.1007/s00232-009-9155-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vo J, Inwood W, Hayes JM, Kustu S. 2013. Mechanism for nitrogen isotope fractionation during ammonium assimilation by Escherichia coli K-12. Proc Natl Acad Sci U S A 110:8696–8701. doi: 10.1073/pnas.1216683110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winkler FK. 2006. Amt/MEP/Rh proteins conduct ammonia. Pflugers Arch 451:701–707. doi: 10.1007/s00424-005-1511-6. [DOI] [PubMed] [Google Scholar]
- 42.Zhang T, Yan Y, He S, Ping SZ, Alam KM, Han Y, Liu X, Lu W, Zhang W, Chen M, Xiang W, Wang X, Lin M. 2012. Involvement of the ammonium transporter AmtB in nitrogenase regulation and ammonium excretion in Pseudomonas stutzeri A1501. Res Microbiol 163:332–339. doi: 10.1016/j.resmic.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 43.Barany F. 1985. Single-stranded hexameric linkers: a system for in-phase insertion mutagenesis and protein engineering. Gene 37:111–123. doi: 10.1016/0378-1119(85)90263-X. [DOI] [PubMed] [Google Scholar]
- 44.Yamamoto S, He Y, Arakawa K, Kinashi H. 2008. γ-butyrolactone-dependent expression of the Streptomyces antibiotic regulatory protein gene srrY plays a central role in the regulatory cascade leading to lankacidin and lankamycin production in Streptomyces rochei. J Bacteriol 190:1308–1316. doi: 10.1128/JB.01383-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blanco G, Drummond M, Woodley P, Kennedy C. 1993. Sequence and molecular analysis of the nifL gene of Azotobacter vinelandii. Mol Microbiol 9:869–879. doi: 10.1111/j.1365-2958.1993.tb01745.x. [DOI] [PubMed] [Google Scholar]
- 46.Ortiz-Marquez JC, Do Nascimento M, Curatti L. 2014. Metabolic engineering of ammonium release for nitrogen-fixing multispecies microbial cell-factories. Metab Eng 23:154–164. doi: 10.1016/j.ymben.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 47.Yoshida N, Inaba S, Takagi H. 2014. Utilization of atmospheric ammonia by an extremely oligotrophic bacterium, Rhodococcus erythropolis N9T-4. J Biosci Bioeng 117:28–32. doi: 10.1016/j.jbiosc.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 48.Castorph H, Kleiner D. 1984. Some properties of a Klebsiella pneumoniae ammonium transport negative mutant (Amt−). Arch Microbiol 139:245–247. [DOI] [PubMed] [Google Scholar]
- 49.Gimmestad M, Steigedal M, Ertesvåg H, Moreno S, Christensen BE, Espín G, Valla S. 2006. Identification and characterization of an Azotobacter vinelandii type I secretion system responsible for export of the AlgE-type mannuronan C-5-epimerases. J Bacteriol 188:5551–5560. doi: 10.1128/JB.00236-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor LA, Rose RE. 1988. A correction in the nucleotide sequence of the Tn903 kanamycin resistance determinant in pUC4K. Nucleic Acids Res 16:358. doi: 10.1093/nar/16.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prentki P, Krisch HM. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]