Abstract
Recently, a positional cloning study proposed that haplotypes at the calpain-10 locus (CAPN10) are associated with increased risk of type 2 diabetes, or non–insulin-dependent diabetes mellitus, in Mexican Americans, Finns, and Germans. To inform the interpretation of the original mapping results and to look for evidence for the action of natural selection on CAPN10, we undertook a population-based genotyping survey of the candidate susceptibility variants. First, we genotyped sites 43, 19, and 63 (the haplotype-defining variants previously proposed) and four closely linked SNPs, in 561 individuals from 11 populations from five continents, and we examined the linkage disequilibrium among them. We then examined the ancestral state of these sites by sequencing orthologous portions of CAPN10 in chimpanzee and orangutan (the identity of sites 43 and 19 was further investigated in a limited sample of other great apes and Old World and New World monkeys). Our survey suggests larger-than-expected differences in the distribution of CAPN10 susceptibility variants between African and non-African populations, with common, derived haplotypes in European and Asian samples (including one of two proposed risk haplotypes) being rare or absent in African samples. These results suggest a history of positive natural selection at the locus, resulting in significant geographic differences in polymorphism frequencies. The relationship of these differences to disease risk is discussed.
Introduction
The calpain-10 locus (CAPN10 [MIM 605286]) encodes an alternatively spliced calpainlike cysteine protease that is ubiquitously expressed (Horikawa et al. 2000; Ma et al. 2001). A combined linkage and case-control study of Mexican Americans (MAs) proposed that variants in CAPN10 contribute to risk of type 2 diabetes, or non–insulin-dependent diabetes mellitus (MIM 125853) (Horikawa et al. 2000); these variants will be referred to as “susceptibility variants.” Several aspects of the results were unanticipated, however. For example, the calpain 10 protein did not appear to play an obvious role in the mediation of the secretion and/or action of insulin. Moreover, the polymorphisms most strongly associated with disease incidence and evidence for linkage were located in intronic regions. Finally, the genotype most strongly associated with the evidence for linkage—that is, SNP-43 G/G (or 1/1)—was not significantly associated with increased risk of diabetes in the MA patient sample. Instead, increased risk was observed for multisite haplotypes centered on the CAPN10 gene, including site 43. Patterns of allelic association in MAs suggested that the risk haplotypes could be defined by genotyping a subset of three variants, at sites 43, 19, and 63. A group of five additional variants, at sites 56, 59, 48, 30, and 65, were found to be in perfect linkage disequilibrium (LD) with site 19 in the screening set of 10 MAs; because these six sites showed a two-haplotype structure, the individual effects that they had on diabetes risk could not be distinguished. Analysis of the three haplotype-defining sites suggested that the 1/1-1/2-2/1 genotype at these sites (composed of haplotypes 1-1-2 and 1-2-1) conferred the greatest risk of disease, a relationship that was observed in two independent groups of MAs (odds ratios [ORs] 2.80 and 3.58, respectively), as well as in Finns (OR 2.55) and Germans (OR 4.97) (Horikawa et al. 2000). On this basis, the gene has been proposed as a candidate susceptibility locus, explaining up to 14% of the risk of type 2 diabetes in MAs and 4% of that in populations of European origin.
Subsequent studies on the role that CAPN10 plays in diabetes susceptibility in other populations have been difficult to interpret. Studies in nondiabetic Pima Indians revealed that homozygotes for the G allele at SNP-43 have reduced muscle CAPN10 mRNA levels and insulin resistance (Baier et al. 2000). However, G/G homozygotes did not have significantly increased risk of type 2 diabetes. Studies in Oji-Cree Indians also showed that G/G homozygotes at SNP-43 did not have an increased risk of type 2 diabetes (Hegele et al. 2001). In African Americans, however, G/G homozygotes at SNP-43 were found to have a significantly increased risk of type 2 diabetes (Garant et al. 2002). Whether any of these studies provide evidence for or against replication of the original findings is unclear, since the proposed susceptibility model suggested that diabetes risk was conferred by a combination of haplotypes defined by three variants (including SNP-43). A smaller number of studies have reported results for at least these three CAPN10 variants. No overtransmission of the high-risk haplotypes to patients was observed in U.K. families, but the rare allele at the tightly linked SNP-44 was overtransmitted (Evans et al. 2001). In a different study of U.K. patients, nondiabetic individuals with the high-risk haplotype combination had significantly elevated fasting and 2-h plasma glucose levels, as well as decreased insulin-secretory response (Lynn et al. 2002). Studies of Samoans revealed no significant association between the high-risk haplotype combination and type 2 diabetes (Tsai et al. 2001). However, the haplotype combinations with the lowest (1-1-2/2-2-1) and the highest (1-1-2/1-2-1) risks in MAs were, respectively, the lowest and among the highest combinations in Samoans. Whereas the magnitude of the difference in risk was ∼7-fold in MAs, it was only ∼3.25-fold in Samoans. These composite results may be explained by a complex (yet not well-understood) model in which diabetes susceptibility is conferred by extended multisite haplotypes whose frequency and composition may vary across human populations.
Despite the ambiguities concerning specific susceptibility variants, the potential role that CAPN10 plays in diabetes etiology makes this locus particularly interesting for study from the point of view of the understanding of human metabolic adaptations. The so-called “thrifty-genotype” hypothesis (Neel 1962), which posits that individuals in some human populations are genetically adapted to the survival of periodic famine, additionally predicts that genes such as CAPN10 may preserve, in their associated polymorphism, evidence of selection for increased and/or more efficient storage of metabolic resources. It is interesting to consider whether such effects are, in fact, evident at this gene. A major advantage of population genetic analysis is that a signature of natural selection may be identified even if the exact variant or variants that contribute to fitness (i.e., metabolic) differences are unknown. In fact, because this signature dissipates with increasing distance from the selected site or sites, it may provide information on the approximate location of the advantageous variants, facilitating an independent evaluation of the significance of previously mapped variants. An important caveat, of course, is that the diabetes phenotype and the hypothetical, advantageous “thrifty phenotype” may not overlap precisely (e.g., age at onset, penetrance, etc. may differ). Furthermore, since diabetes is a complex polygenic trait, it is also likely that the hypothetical advantageous phenotype will be due to variants in multiple genes, all of which could potentially bear the signature of positive natural selection.
With these issues in mind, we undertook a population genetic survey of susceptibility variants that have been proposed by Horikawa et al. (2000). Seven polymorphisms, at sites 44, 43, 56, 59, 19, 30, and 63, were genotyped in 11 human population samples from Africa, Asia, Europe, Oceania, and North and South America, and patterns of LD and haplotype variation among the sites were assessed. The ancestral state of these sites was also inferred by sequencing orthologous portions of CAPN10 in chimpanzee and orangutan. Our results demonstrate striking differences in allele and haplotype frequencies between African and non-African populations, which suggest the impact of natural selection on the CAPN10 gene. The extent to which the inferred selection is consistent with the predictions of the thrifty-genotype hypothesis, however, is less clear.
Subjects, Material, and Methods
Samples
Five hundred eighty-nine unrelated individuals from 11 populations (70 Biaka Pygmies [Allele Frequency Database from Kidd Lab {ALFRED} sample unique identification number {UID} SA000005F], 60 San Francisco Chinese [UID SA000009J], 52 Danes [UID SA000007H], 64 Druze [UID SA000047L], 92 mixed Europeans [UID SA000020C], 38 Japanese [UID SA000010B], 53 Maya [UID SA000013E], 39 Mbuti Pygmies [UID SA000006G], 23 Nasioi [UID SA000012D], 47 Surui [UID SA000014F], and 51 Yakut [UID SA000011C]) were genotyped, and data for 561 of these individuals were analyzed (individuals with three or more sites with missing data were excluded). Levels of missing data ranged from 2% (at site 19) to 15% (at site 44) in the combined sample. Genotypes for successfully assayed samples adhered to Hardy-Weinberg equilibrium (HWE) expectation (after Bonferroni correction), suggesting that dropout was random with respect to genotype (data not shown). Further information regarding these samples can be obtained from the ALFRED Web site.
Genotyping
Seven previously identified sites (Horikawa et al. 2000) were genotyped with one of four methods: fluorescence polarization (sites 56, 59, and 30), PCR-RFLP analysis (site 63), gel-based detection of allelic-length differences (site 19), or direct sequencing (sites 44 and 43). SNPs at sites 56, 59, and 30 (ALFRED UIDs SI000262K, SI000263L, and SI000265N) were genotyped using a modified version of the fluorescence polarization/template-directed dye-terminator incorporation (FP-TDI) assay (SNP Genotyping) (Chen et al. 1999). PCR products encompassing each variant were amplified and then were used for a single-base extension reaction, with the following primers: 30, 5′-TGTATACCCCAAGGATGCAGCAGAGA-3′; 56, 5′-TGTCCTCAGTTTGTGACCTTCCCCT-3′; and 59, 5′-TCATTTTTTTCATACTTAGGATTATTTT-3′. Fluorescence was read by an LJL Analyst fluorescence plate reader. SNP-63 (UID SI000266O) was genotyped using a mismatch-primer/RFLP method. For this assay, a forward primer with an intentional mismatch (5′-AAGGGGGGCCAGGGCCTGACGGGGGTGGCG-3′, with mismatch underlined) was used with an unmodified reverse primer. The modified primer creates a HhaI restriction site in the presence of the C allele, which is then detected by restriction-enzyme digestion. Indel-19 (a simple tandem-repeat insertion/deletion variant with either two or three copies of 32-bp fragment; UID SI000264M), was genotyped by amplifying the repeat region and measuring allele size by gel electrophoresis. A 3.5% agarose gel was used to distinguish 187-bp (three repeats) from 155-bp (two repeats) bands. SNP-44 and SNP-43 (positions 22751 and 22762 [UIDs SI000260I and SI000261J, respectively]) were genotyped by directly sequencing (with the amplification primers and ABI BigDye terminators) a single 680-bp PCR product that encompassed both sites. In most cases, samples were sequenced on one strand only, by use of an ABI 377. Genotypes were determined by visual inspection of sequence chromatograms.
Sequence Analysis of Nonhuman Primates
The amplification primers used to genotype the human samples were also used to amplify and directly sequence homologous regions in one chimpanzee (Pan troglodytes) and one orangutan (Pongo pygmaeus). PCR products were sequenced on both strands, by methods similar to those described above. Further DNA sequence analysis was performed for SNP-43 and Indel-19 with three additional species: bonobo (Pan paniscus), gorilla (Gorilla gorilla), and the Old World monkey, baboon (Papio cynocephalus). The SNP-43 product was also sequenced in the capuchin Cebus apella, a New World monkey.
Haplotype Inference
Haplotype relationships among the genotyped sites were inferred separately for each population sample by the expectation-maximization method through use of the program MLOCUS (Long et al. 1995; Long 1999). A version of the program that accommodates incomplete genotypings (i.e., missing data) was used. Only haplotypes with resultant maximum-likelihood frequencies >0.05 in a given population sample are reported here. Previous analysis has suggested that haplotype frequencies estimated by this algorithm are accurate when they are >5% (Tishkoff et al. 2000). Complete haplotype data have been deposited in the ALFRED database (UID SI000267P).
Statistical Analyses
FST, a measure of allele frequency differences among samples, was estimated per site, for pairs of populations and also the combined African and non-African samples, from the observed allele frequencies by use of Weir's (1996) equation (5.2). Equivalent estimates of FST were generated for the larger data set (Bowcock et al. 1987, 1991a). LD between pairs of sites, measured by the statistic r2, was estimated from the observed genotype frequencies by maximum likelihood (Hill 1974). The significance of r2 was assessed by a likelihood-ratio test under the assumption of HWE. This test was implemented as described by Hudson (2001).
Results
Geographic Structure of CAPN10 Susceptibility Variants
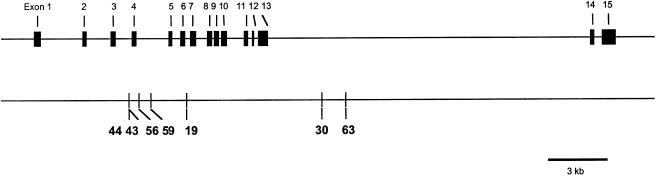
Seven previously identified polymorphic positions, spanning 11.5 kb of CAPN10 (fig. 1), were genotyped: six SNPs (designated sites 44, 43, 56, 59, 30, and 63 by Horikawa et al. [2000]), and one biallelic indel polymorphism (designated site 19 by the same authors). Variation was investigated in 11 population samples: Biaka and Mbuti Pygmies, from Africa; mixed Europeans, Danes, and Druze, from Europe and the Middle East; Chinese, Japanese, and Yakut, from Asia; Nasioi, from Oceania; and Maya and Surui, from North and South America, respectively.
Figure 1.
The human CAPN10 locus on chromosome 2, showing the locations of seven previously identified susceptibility variants surveyed here. The locations of the sites, relative to the numbering in the reference sequence AF158748, are 22751 (site 44), 22762 (site 43), 23325 (site 56), 23928 (site 59), 25830 (site 19), 33021 (site 30), and 34288 (site 63).
All seven sites varied in all 11 population samples; however, the less common allele at sites 56, 59, 19, 30, and 63 was different in the African and non-African populations (table 1; allele frequencies have been deposited in the ALFRED database). The degree of geographic structure, which is reflected in differences in allele frequencies among samples and is estimated by the FST statistic (Weir 1996), was quantified on a site-by-site basis for all pairs of African and non-African populations. FST values, which vary from 0 to 1, increase as allele-frequency differences between population samples become more pronounced. We found average estimates of FST of 0.137, for site 43, and 0.121, for site 44, with the majority of estimates for these two sites being <0.150. In contrast, most FST estimates for the other five variable sites exceeded 0.250. Similarly high FST values were not observed for comparisons including only either African or non-African populations (data not shown).
Table 1.
Allele Frequencies of Genotyped Sites at CAPN10
|
Allele Frequencies among Populations |
||||||||||||
| African |
European |
Asian |
Oceanic |
North and South American |
||||||||
| Site (Allele) | Biaka(n=138) | Mbuti(n=70) | Mixed(n=180) | Danes(n=98) | Druze(n=126) | Chinese(n=110) | Japan(n=72) | Yakut(n=98) | Nasioi(n=44) | Maya(n=94) | Surui(n=92) | Overall(n=1,122) |
| 44 (C) | .02 | .02 | .12 | .18 | .14 | .10 | .10 | .12 | .05 | .06 | .01 | .09 |
| 43 (G) | .91 | .97 | .80 | .63 | .90 | .97 | .87 | .85 | .90 | .69 | .73 | .83 |
| 56 (A) | .75 | .93 | .36 | .34 | .48 | .37 | .23 | .51 | .20 | .39 | .25 | .45 |
| 59 (A) | .68 | .91 | .37 | .36 | .54 | .39 | .34 | .55 | .19 | .38 | .28 | .46 |
| 19 (2) | .12 | .01 | .63 | .69 | .53 | .65 | .70 | .45 | .82 | .62 | .73 | .53 |
| 30 (T) | .73 | .94 | .34 | .33 | .49 | .37 | .36 | .55 | .21 | .38 | .28 | .46 |
| 63 (C) | .31 | .12 | .91 | .92 | .90 | .82 | .79 | .61 | .96 | .71 | .72 | .71 |
Under evolutionary neutrality, variation in allele frequencies across subpopulations is determined by genetic drift alone. Because drift is determined entirely by the demographic properties of the populations, all loci in the genome have the same expected degree of differentiation, and this expectation may be used to detect the action of positive natural selection (Cavalli-Sforza 1966; Lewontin and Krakauer 1973). In other words, if allele-frequency data are available for a set of putatively neutral loci, then an empirical distribution of FST values can be constructed to identify loci with unusual patterns of differentiation due to positive natural selection (Bowcock et al. 1991b; Karl and Avise 1992; Berry and Kreitman 1993; Cavalli-Sforza et al. 1994; Taylor et al. 1995). Such a data set of putatively neutral loci already exists for a representative subset of our population samples: 86 biallelic variants, located throughout the genome, were previously assayed in the exact same samples of Biaka, Mbuti, and Nasioi, and in a large subset of the same sample of Chinese; a similar sample of mixed Europeans was also assayed (Bowcock et al. 1987, 1991a).
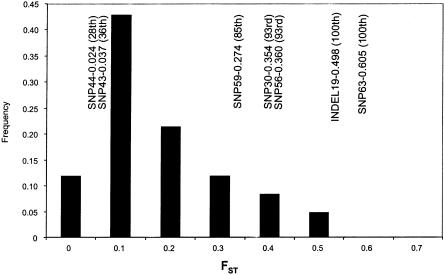
To determine if the degree of interpopulation differentiation at CAPN10 differs from the genomewide pattern, we calculated the FST values for the CAPN10 variants and the 86 biallelic variants with equivalent sets of pooled African versus pooled non-African samples (the six nonoverlapping population samples were disregarded in this comparison). As shown in figure 2, the CAPN10 site-specific estimates were <0.1 for sites 43 and 44, and ranged from 0.274 (site 59) to 0.605 (site 63) for the other five sites. This analysis suggests that the CAPN10 FST values are exceptionally high, with estimates for two (i.e., sites 19 and 63) of the seven genotyped sites that fall above the 100th percentile of the genomewide distribution. Thus, the susceptibility variants at CAPN10 show an unusual pattern of geographic structure. This pattern is not explained by either sampling differences or human demography but may reflect the effects of natural selection on the gene.
Figure 2.
FST values between African and non-African samples at CAPN10, compared to those observed at 86 RFLPs. The distribution of values, estimated for a collection of previously assayed biallelic RFLPs (Bowcock et al. 1987, 1991a), is shown. Above the histogram, the approximate FST values observed for the seven susceptibility variants in this study are indicated. The percentile rank of each site, relative to the observed distribution, is given in parentheses.
LD and Haplotype Structure of Susceptibility Variation
Estimates of LD, summarized by the r2 statistic (Hill 1974), were calculated from the observed genotype data for all possible pairs of the seven genotyped sites. The r2 statistic is expected to be 1 when the variation is segregating in a population as two distinct haplotypes. Thus, it is likely to be particularly informative for the susceptibility variants at CAPN10 that showed near-perfect LD (i.e., two-haplotype structure) in the MA screening set. The results for one set of pairwise comparisons (i.e., those with the indel at site 19) are given in table 2. As shown, LD between site 19 and sites 56, 59, and 30 is high (i.e., r2>0.5) in all non-African populations sampled and is particularly high in the Chinese, Native American, and Oceanic samples (perfect LD, i.e., r2=1, was observed among the four sites in the Melanesian Nasioi sample). In the African samples, although r2 values were higher for the 56-59-30 comparisons relative to the 44 and 63 comparisons, none of them exceed 0.5. Thus, LD patterns also differ substantially between the African and non-African samples. Overall, these results suggest extensive LD among many of the variable sites genotyped, including four of five sites with high FST values in the African versus non-African comparison (i.e., 56, 59, 19, and 30). These observations are in broad agreement with the previous observation of near-perfect LD among sites 56, 59, 19, and 30 in a sample of MAs (Horikawa et al. 2000).
Table 2.
Estimates of LD between Each SNP and Indel-19
|
Estimates of LD (r2) among Populations |
|||||||||||
| African |
European |
Asian |
Oceanic |
North and South American |
|||||||
| Site (Distance from Indel-19) | Biaka | Mbuti | Mixed | Danes | Druze | Chinese | Japanese | Yakut | Nasioi | Maya | Surui |
| 44 (3,079 bp) | .003 | .000 | .050* | .283*** | .025 | .083* | .119* | .014 | .012 | .038 | .004 |
| 43 (3,068 bp) | .481*** | .000 | .027 | .226*** | .087** | .018 | .067* | .058 | .062 | .098 | .133** |
| 56 (2,505 bp) | .465*** | .188* | .505*** | .594*** | .891*** | .960*** | .471*** | .846*** | 1.000*** | 1.000*** | 1.000*** |
| 59 (1,902 bp) | .108* | .154 | .835*** | .654*** | .535*** | .920*** | .503*** | .873*** | 1.000*** | .903*** | .776*** |
| 30 (7,191 bp) | .354*** | .238* | .515*** | .951*** | .764*** | .958*** | .707*** | .959*** | 1.000*** | .848*** | .937*** |
| 63 (8,458 bp) | .006 | .112 | .090** | .187** | .123*** | .321*** | .430*** | .383*** | .214* | .649*** | 1.000*** |
P<.05.
P<.01.
P<.001.
The observed patterns of LD can be understood in terms of the underlying haplotype structure of the seven genotyped sites. Haplotype-frequency distributions were inferred for each population sample separately, by use of an implementation of the expectation-maximization (EM) algorithm that takes missing data into account in its haplotype assignments (Long 1999). The haplotypes inferred to occur at a frequency >5% in at least one sample are listed in table 3, and their estimated frequencies are reported only when >5%. By these criteria, 10 haplotypes were inferred in the 11 population samples; the total number of such “common” haplotypes in any single sample varied from two (in the Biaka sample) to seven (in the Druze sample). In most cases, these haplotypes accounted for the vast majority of the chromosomes sampled; slightly more haplotypes at a frequency <5% were inferred to occur in the Biaka and Japanese samples (table 3).
Table 3.
Inferred Haplotypes among Genotyped Sites at CAPN10
|
Frequencies among Populations |
||||||||||||
| African |
European |
Asian |
Oceanic |
North and South American |
||||||||
| Haplotypea | Biaka(n=138) | Mbuti(n=70) | Mixed(n=180) | Danes(n=98) | Druze(n=126) | Chinese(n=110) | Japanese(n=72) | Yakut(n=98) | Nasioi(n=44) | Maya(n=94) | Surui(n=92) | |
| Human: | ||||||||||||
| 1-1-1 | T-G-A-A-2-T-C | .05 | .13 | .06 | .26 | .11 | .10 | .09 | .07 | |||
| C-G-A-A-2-T-C | .07 | .11 | .08 | .08 | .07 | .06 | ||||||
| T-G-G-T-2-G-C | .12 | .06 | ||||||||||
| Sumb | .12 | .11 | .20 | .17 | .34 | .19 | .07 | .16 | .09 | .07 | .00 | |
| 1-1-2 | T-G-A-A-2-T-T | .59 | .82 | .05 | .08 | .09 | .17 | .08 | .29 | .22 | .27 | |
| T-G-G-A-2-T-T | .06 | |||||||||||
| Sumb | .59 | .82 | .05 | .08 | .09 | .17 | .14 | .29 | .00 | .22 | .27 | |
| 1-2-1 | T-G-G-T-3-G-C | .38 | .23 | .31 | .57 | .49 | .25 | .74 | .33 | .41 | ||
| C-G-G-T-3-G-C | .05 | |||||||||||
| T-G-G-A-3-G-C | .05 | |||||||||||
| T-G-A-T-3-T-C | .05 | |||||||||||
| Sumb | .00 | .00 | .43 | .23 | .41 | .57 | .49 | .25 | .74 | .33 | .41 | |
| 2-2-1 | T-A-G-T-3-G-C | .14 | .31 | .06 | .07 | .12 | .05 | .24 | .25 | |||
| Sumb | .00 | .00 | .14 | .31 | .06 | .00 | .07 | .12 | .05 | .24 | .25 | |
| Overall (haplotypes >.05) | .71 | .93 | .82 | .79 | .90 | .93 | .77 | .82 | .88 | .86 | .93 | |
| Risk genotype (1/1-1/2-2/1): | ||||||||||||
| Observed | .03 | .03 | .06 | .06 | .03 | .15 | .03 | .14 | .05 | .15 | .26 | |
| Expectedc | .00 | .00 | .04 | .04 | .07 | .19 | .14 | .15 | .00 | .15 | .22 | |
| Nonhuman: | ||||||||||||
| Chimpanzee | C-G-A-A-3-T-C | |||||||||||
| Orangutan | C-A-A-A-4-T-T | |||||||||||
At sites 44, 43, 56, 59, 19, 30, and 63, respectively. Allele designation at the polymorphic indel (i.e., Indel-19) reflects the number of repeat units.
Indicates the sum of the frequencies of haplotypes with frequency >.05.
Calculations are based on the sum of the estimated haplotype frequencies as reported in the table.
The 10 haplotypes encompass and subdivide the four main three-site (43-19-63) haplotypes that were identified by Horikawa et al. (2000). In accordance with the genotyping results that were presented in the original association study, the 1-2-1 haplotypes are the most common haplotypes in our non-African samples. However, no 1-2-1 haplotypes were observed at a frequency >5% in either of the African population samples. In addition, no 2-2-1 haplotypes were inferred to occur in the Biaka or Mbuti, despite the fact that this haplotype is found at appreciable frequencies in several non-African samples (e.g., those from Danes, Maya, and Surui). Instead, we found 1-1-2 haplotypes to be much more common in the two African samples than in the non-African populations. This major difference in the underlying haplotype distribution of the two groups of samples is consistent with both the high site-specific FST values that were observed for the African versus non-African comparison and the population-specific patterns of LD.
Clearly, differences in the haplotype structure of CAPN10 variation are pronounced between the African and non-African populations assayed in this study. The consequences of these differences for disease risk are immediately apparent in the population-attributable risk that is implied by the frequencies of the main risk haplotypes (i.e., 1-2-1 and 1-1-2). As shown in table 3, the absence of one of the two candidate risk haplotypes, 1-2-1, in the African samples suggests that very few individuals in these populations possess the 1/1-1/2-2/1 genotype that has previously been proposed to confer the greatest susceptibility to type 2 diabetes in MA and European populations. Therefore, it is possible that CAPN10 is not a major susceptibility locus in African groups or that a different set of CAPN10 variants contributes to risk of type 2 diabetes. The extent to which these inferences are extendable to other populations with a significant African ancestry (e.g., African Americans) awaits further investigation. As observed by Horikawa et al. (2000), the highest frequencies of the main risk genotype are observed among the East Asian and Native American populations. Except for the Japanese sample, the observed frequencies of this genotype were similar to those that were expected on the basis of HWE (table 3). However, the statistical significance of these discordances could not be determined. This is because the expected frequencies of the risk genotype are based on inferred haplotype frequencies and because the currently available methodology for the assessment of such departures is not appropriate for this case.
Ancestral State of Risk Variants
The large differences in haplotype frequencies that we observe between African and non-African populations could be due to the adaptive expansion of certain haplotypes in particular geographic regions. To place our survey in an evolutionary context and to provide some initial insight into the history of genetic change that has occurred at the locus, we genotyped the same seven sites in two closely related species of nonhuman primate: common chimpanzee and orangutan. As shown in table 3, chimpanzee and orangutan were concordant for four (44, 56, 59, and 30) of the seven sites, providing strong evidence for the ancestral state of the polymorphic site within humans at these positions (i.e., C, A, A, and T, respectively). Only 1 of the 10 haplotypes observed in the polymorphism study had the same configuration of alleles at these four sites. This haplotype, the second in the 1-1-1 haplotype group in table 3, also shared identity with chimpanzee at two other sites (43 and 63), differing only in the length of the indel at site 19. These data therefore suggest that the 1-1-1 group of haplotypes is likely to carry the ancestral allele at most of the seven susceptibility variants considered here.
Interestingly, the indel at site 19 is discordant between chimpanzee and orangutan, in which three and four copies of the 32-bp repeat were observed, respectively. Further heterogeneity was uncovered by sequencing site 19 in additional nonhuman primates (see below). Such differences suggest that this repeat is mutationally labile and, thus, that the outgroup sequences may not be a reliable indicator of the ancestral state of the human polymorphism. A similar discordance is observed at sites 43 and 63, but, in these cases, the variants observed in the two outgroups coincide with the two alleles that are present at the site within humans. Both of the latter variants fall at CpG sites, and the observed segregating nucleotides are consistent with the C→T transition that is normally associated with the deamination of a methylated cytosine. At site 43, the high frequency of the G allele in all populations surveyed suggests that the G found in chimpanzee is the true ancestral state at this site. The ancestral state of 63 is more equivocal, because different alleles occur at high frequency in the African and the non-African samples. Despite these discordances, when all genotyped sites are considered, the 1-1-1 and 1-1-2 haplotypes are clearly more similar to the primate outgroup species than are the 1-2-1 and 2-2-1 haplotypes (which are characterized by a large proportion of sites with nonancestral alleles).
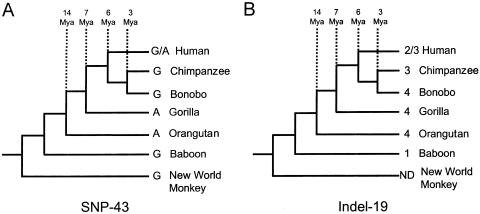
The unexpected discordances observed between chimpanzee and orangutan provoked an extended analysis of sites 43 and 19 in several other primate species, including bonobo, gorilla, baboon, and capuchin. As shown (fig. 3), we observed extensive heterogeneity for both positions among the species examined. At site 43 (fig. 3a), G and A alternated among the great apes, with the G allele (presumed ancestral within humans, as discussed above) predominant in New World and Old World monkeys. This tree requires a minimum of three mutations at site 43. At site 19 (fig. 3b), changes in state appeared to be centered on the human-chimpanzee portion of the phylogeny, although baboon was found to have only a single repeat unit at this site.
Figure 3.
State changes at SNP-43 and Indel-19 in primates. The nucleotide observed at sites orthologous to SNP-43 in humans is shown in the left-hand tree, whereas the number of 32-bp repeat units present in the region orthologous to Indel-19 is shown in the right-hand tree. Note that trees are not drawn to scale. Mya = million years ago.
The extent to which the observed interspecific differences at these sites reflect mutational processes or, alternatively, rapid adaptive evolution at this locus, is unclear. It is notable that three of the six SNPs that were genotyped in our survey fall in CpG sites (44, 43, and 63). Twenty-three (27%) of the 85 SNPs that were identified by Horikawa et al. (2000) in the immediate vicinity of CAPN10 (sites 16021–50680 of the GenBank reference sequence) fall at CG dinucleotides. A similar survey of polymorphism and divergence at 10 unlinked regions (Frisse et al. 2001; L. Frisse, P. Meyer, and A. Di Rienzo, personal communication) observed approximately the same proportion (24%) of observed variants at CpG sites and found that 18% of CpG-associated SNPs showed a discordance between chimpanzee and orangutan, as we have here observed at sites 43 and 63. Thus, it may be that CpG-associated variants are often discordant between chimpanzee and orangutan.
Discussion
This survey of candidate susceptibility variants at CAPN10 has confirmed several features of the variation reported previously (Horikawa et al. 2000) while simultaneously providing important new insights into the structure of CAPN10 variation within and between human populations. In particular, our results suggest an unusual degree of geographic structure, not identified in the original mapping study and consistent with the effects of natural selection on or near CAPN10. Thus, regardless of whether the role that CAPN10 plays in risk of type 2 diabetes is clarified, the polymorphism data alone strongly suggest that the protein is functionally important and probably harbors adaptive variation.
In line with previous observations (Horikawa et al. 2000), we inferred the presence of a small number of haplotypes defined by the seven genotyped variants, with four three-site haplotypes predominant in the samples genotyped here. We also observed extensive LD among sites 56, 59, 19, and 30 and found appreciable frequencies of the main susceptibility genotype, 1/1-1/2-2/1, in samples of Asian and Native American origin, as has been described elsewhere. Our survey differed from the preliminary study, however, in that we genotyped a larger and more diverse set of aboriginal and nonclinical population samples and in that we examined a larger number of variable sites in those populations. In particular, the inclusion of African population samples allowed us to uncover exceptional, previously unrecognized geographic structure between the African and non-African populations sampled, involving at least two of the three main susceptibility variants identified by Horikawa et al. (2000)—that is, sites 19 and 63. Allele frequencies at 19 and 63 differ considerably among African and non-African samples, with estimates of FST for these variants exceeding all equivalent estimates observed for a collection of 86 biallelic RFLPs sampled throughout the genome (Bowcock et al. 1987, 1991a). This marked population differentiation is accompanied by the presence of extensive LD among sites 56, 59, 19, and 30, in all the non-African populations surveyed. LD among these sites is less pronounced in our African samples.
The discovery of significant LD at sites 19 and 63 implies that their high FST values do not represent independent observations and that their evolutionary history should be investigated in terms of their haplotype structure. Our observations suggest that key differences in the haplotype distribution are present in each set of population samples. The 1-2-1 and 2-2-1 groups of haplotypes, which differ at four of seven sites from the 1-1-1 and 1-1-2 haplotypes, are found at appreciable frequencies only in non-African populations. It is the geographic bifurcation in the distribution of the 1-2-1 and 2-2-1 haplotypes that underlies both the unexpectedly high estimates of FST for those four sites (i.e., 56, 59, 19, and 30) and the high level of LD observed among the same sites in non-African populations. In contrast, site 63 does not show the same pattern of LD as the other four sites, despite the presence of extreme levels of geographic differentiation. Rather, the high FST observed for this site appears to reflect the geographic difference in the distribution of the 1-1-2 class of haplotypes: these haplotypes are very common in the African samples and are present at much lower frequencies outside Africa. Interestingly, despite the previously described association between SNP-43 and the evidence for linkage (Horikawa et al. 2000), neither SNP-43 nor SNP-44, the closely linked variant, show the same patterns of geographic differentiation observed at the other five sites.
Analysis of two nonhuman primate species, chimpanzee and orangutan, suggests that the 1-1-1 group of haplotypes is likely to carry the ancestral allele at most of the sites surveyed here. If so, then the unexpectedly high estimates of FST involving sites 19 and 63 appear to mark the expansion of particular subsets of derived haplotypes in specific geographic regions—that is, the expansion of the 1-1-2 haplotypes within African populations and/or the expansion of the more divergent 1-2-1 and 2-2-1 groups of haplotypes in non-African populations. The observed variation is difficult to explain adequately with any one selective scenario, although patterns are broadly consistent with the population-specific impact of natural selection. One could speculate, for example, that the increase in length of Indel-19 from two to three 32-bp repeats (or another mutation closely linked to it) conferred a fitness advantage such that the 1-2-1 and 2-2-1 haplotypes were selectively favored in populations outside Africa. Another possible scenario would envision a selective advantage associated with the C allele at site 63 in non-African populations. Whatever the exact cause, the effects of selection are strongly implicated: although other loci also exhibit important genetic differences between African and non-African populations (Kidd et al. 1998; Parra et al. 1998), only loci thought to be subject to the effects of natural selection have found estimates of FST that are as large as those observed at CAPN10 (Peterson et al. 1999; Hamblin and Di Rienzo 2000). Fuller examination of the inferred deviation from selective neutrality awaits more-detailed investigation of genetic variation at CAPN10 in these or other closely related populations.
Given the proposal that CAPN10 is a susceptibility locus for type 2 diabetes, a relevant question is whether the selection described above is, in fact, consistent with the predictions of the thrifty-genotype hypothesis. The hypothesis, as first proposed (Neel 1962), suggested that human populations had acquired genetic adaptations allowing them to store and utilize energy more efficiently in times of plentiful food supply, thereby making them more likely to survive periodic famine. The populations that today show the highest incidence of obesity, insulin resistance, and type 2 diabetes (i.e., Amerindians and Pacific Islanders) may have experienced the most pronounced nutritional selection or may have been subject to selective pressures that ended most recently (i.e., when they adopted a Western lifestyle). A simple scenario therefore might envision that these populations harbor the greatest number of susceptibility alleles. If we take, for the sake of argument, the two derived CAPN10 haplotype classes (i.e., 1-2-1 and 2-2-1) as putative “thrifty alleles,” then we do observe higher frequencies of these haplotypes in our Oceanic and Amerindian samples, but differences with respect to the Asian and European samples are not great. Nor is it clear why samples of African origin, which might be expected to have been subject to similar nutritional fluctuations (and are not immune to the effects of type 2 diabetes), would show a significantly different haplotype distribution. Several possible explanations may account for these apparent discrepancies. For example, a large number of loci are likely to affect metabolism, including some that may interact (Cox et al. 1999). Therefore, the postulated selective effects may be distributed across a wide range of genes, and different thrifty alleles at the same locus may predominate in different populations.
Regardless of what evolutionary processes explain the observed differences in haplotype distribution, the differences are pronounced, possibly relevant to disease risk, and certainly relevant to disease mapping. In particular, the absence of the derived 1-2-1 and 2-2-1 classes of haplotypes in the African populations tested means that fewer individuals in these groups are likely to inherit what is proposed to be the main susceptibility genotype (i.e., 1-1-2/1-2-1) at this locus. CAPN10 variation may explain only a small proportion of type 2 diabetes cases observed in such groups, or a different set of susceptibility haplotypes may be acting in populations of African origin. The lack of substantial LD among candidate susceptibility variants in African populations does suggest, however, that populations with similar ancestry could be used to distinguish among the current set of susceptibility variants, whose effects are currently confounded by extensive disequilibrium in MAs. Another important insight to emerge from our survey is the extent to which patterns of LD observed in other populations (reflecting the underlying haplotype distribution) are also present in other Native American and Asian populations. Since the susceptibility genotype is heterozygous, it may have been suspected that the high incidence of diabetes in the recently admixed MA population is due to the combination of two intermediate-frequency haplotypes, each at higher frequencies in Native American and European populations, respectively. That the at-risk haplotypes are found at similar frequencies in both Native American and Asian populations suggests that the recent admixture between European and Native American populations did not enhance the incidence of type 2 diabetes in MAs, at least when variation at this locus alone is considered. This is not inconsistent with the classical finding that prevalence of type 2 diabetes is positively correlated with the Native American admixture proportion in MAs (Chakraborty and Weiss 1986). We also note that admixture, as an artifactual explanation for the observed association between CAPN10 variation and diabetes, is not supported by our results. Since the MA gene pool does not contain substantial African admixture, the exceptional differentiation of allele frequencies between African and non-African populations is unlikely to underlie the observed disease association. Furthermore, associations with insulin resistance have also been reported for a number of populations that did not experience recent admixture (Baier et al. 2000; Lynn et al. 2002).
In conclusion, this survey of CAPN10 susceptibility variants indicates a complex and remarkable geographic structure at the locus. This is consistent with the effects of positive natural selection acting either on the gene itself or on a closely linked site or sites. The thrifty-genotype hypothesis predicts that genes such as CAPN10, which may play a role in the regulation of the secretion and/or action of insulin, may contain within them a signature of the effects of positive natural selection. Although preliminary, our survey suggests that such a signature is indeed present at this locus. As such, these findings may represent a first step toward the testing of the thrifty-genotype hypothesis at the molecular level.
Acknowledgments
We thank L. Frisse and M. Hamblin for discussion and use of unpublished data and K. M. Weiss and T. M. Pollard for their comments on an earlier version of the manuscript. We also thank the Yerkes Primate Center and the Brookfield Zoo for primate DNA samples. This work was supported by National Institutes of Health grants DK20595 and DK56670 (to A.D.), DK55889 (to N.J.C.), DK47486 (to G.I.B.), and GM57672 (to K.K.K.).
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- ALFRED, http://alfred.med.yale.edu/alfred/ (for SNP-44, SNP-43, SNP-56, SNP-59, Indel-19, SNP-30, and SNP-63 [UIDs SI000260I and SI000261J, SI000262K, SI000263L, SI000264M, SI000265N, and SI000266O, respectively] and haplotype data [UID SI000267P])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for non–insulin-dependent diabetes mellitus [MIM 125853] and CAPN10 [MIM 605286])
- SNP Genotyping, http://psy-svr1.bsd.uchicago.edu/geno/snp/SNP.html (for FP-TDI assay)
References
- Baier LJ, Permana PA, Yang X, Pratley RE, Hanson RL, Shen GQ, Mott D, Knowler WC, Cox NJ, Horikawa Y, Oda N, Bell GI, Bogardus C (2000) A calpain-10 gene polymorphism is associated with reduced muscle mRNA levels and insulin resistance. J Clin Invest 106:R69–R73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry A, Kreitman M (1993) Molecular analysis of an allozyme cline: alcohol dehydrogenase in Drosophila melanogaster on the east coast of North America. Genetics 134:869–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowcock AM, Bucci C, Hebert JM, Kidd JR, Kidd KK, Friedlaender JS, Cavalli-Sforza LL (1987) Study of 47 DNA markers in five populations from four continents. Gene Geogr 1:47–64 [PubMed] [Google Scholar]
- Bowcock AM, Hebert JM, Mountain JL, Kidd JR, Rogers J, Kidd KK, Cavalli-Sforza LL (1991a) Study of an additional 58 DNA markers in five human populations from four continents. Gene Geogr 5:151–173 [PubMed] [Google Scholar]
- Bowcock AM, Kidd JR, Mountain JL, Hebert JM, Carotenuto L, Kidd KK, Cavalli-Sforza LL (1991b) Drift, admixture, and selection in human evolution: a study with DNA polymorphisms. Proc Natl Acad Sci USA 88:839–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza LL (1966) Population structure and human evolution. Proc R Soc Lond B Biol Sci 164:362–379 [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A (1994) The history and geography of human genes. Princeton University Press, Princeton, NJ [Google Scholar]
- Chakraborty R, Weiss KM (1986) Frequencies of complex diseases in hybrid populations. Am J Phys Anthropol 70:489–503 [DOI] [PubMed] [Google Scholar]
- Chen X, Levine L, Kwok PY (1999) Fluorescence polarization in homogeneous nucleic acid analysis. Genome Res 9:492–498 [PMC free article] [PubMed] [Google Scholar]
- Cox NJ, Frigge M, Nicolae DL, Concannon P, Hanis CL, Bell GI, Kong A (1999) Loci on chromosomes 2 (NIDDM1) and 15 interact to increase susceptibility to diabetes in Mexican Americans. Nat Genet 21:213–215 [DOI] [PubMed] [Google Scholar]
- Evans JC, Frayling TM, Cassell PG, Saker PJ, Hitman GA, Walker M, Levy JC, et al (2001) Studies of association between the gene for calpain-10 and type 2 diabetes mellitus in the United Kingdom. Am J Hum Genet 69:544–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisse L, Hudson RR, Bartoszewicz A, Wall JD, Donfack J, Di Rienzo A (2001) Gene conversion and different population histories may explain the contrast between polymorphism and linkage disequilibrium levels. Am J Hum Genet 69:831–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garant MJ, Kao WHL, Brancati F, Coresh J, Rami TM, Hanis CL, Boerwinkle E, Shuldiner AR (2002) SNP43 of the calcium-activated neutral protease (CAPN10) and the risk of type 2 diabetes in African-Americans: the atherosclerosis risk in communities (ARIC) study. Diabetes 51:231–237 [DOI] [PubMed] [Google Scholar]
- Hamblin MT, Di Rienzo A (2000) Detection of the signature of natural selection in humans: evidence from the Duffy blood group locus. Am J Hum Genet 66:1669–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegele RA, Harris SB, Zinman B, Hanley AJ, Cao H (2001) Absence of association of type 2 diabetes with CAPN10 and PC-1 polymorphisms in Oji-Cree. Diabetes Care 24:1498–1499 [DOI] [PubMed] [Google Scholar]
- Hill WG (1974) Estimation of linkage disequilibrium in randomly mating populations. Heredity 33:229–239 [DOI] [PubMed] [Google Scholar]
- Horikawa Y, Oda N, Cox NJ, Li X, Orho-Melander M, Hara M, Hinokio Y, Lindner TH, Mashima H, Schwarz PE, del Bosque-Plata L, Oda Y, Yoshiuchi I, Colilla S, Polonsky KS, Wei S, Concannon P, Iwasaki N, Schulze J, Baier LJ, Bogardus C, Groop L, Boerwinkle E, Hanis CL, Bell GI (2000) Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet 26:163–175 [DOI] [PubMed] [Google Scholar]
- Hudson RR (2001) Linkage disequilibrium and recombination. In: Balding DJ, Bishop M, and Cannings C (eds) Handbook of statistical genetics. John Wiley & Sons, New York, pp 309–324 [Google Scholar]
- Karl SA, Avise JC (1992) Balancing selection at allozyme loci in oysters: implications from nuclear RFLPs. Science 256:100–102 [DOI] [PubMed] [Google Scholar]
- Kidd KK, Morar B, Castiglione CM, Zhao H, Pakstis AJ, Speed WC, Bonne-Tamir B, Lu RB, Goldman D, Lee C, Nam YS, Grandy DK, Jenkins T, Kidd JR (1998) A global survey of haplotype frequencies and linkage disequilibrium at the DRD2 locus. Hum Genet 103:211–227 [DOI] [PubMed] [Google Scholar]
- Lewontin RC, Krakauer J (1973) Distribution of gene frequency as a test of the theory of the selective neutrality of polymorphisms. Genetics 74:175–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC (1999) Multiple locus haplotype analysis release 2: section on population genetics and linkage. Laboratory of Neurogenetics, National Institute on Alcohol Abuse and Alcoholism, National Institutes of Health, Bethesda, MD [Google Scholar]
- Long JC, Williams RC, Urbanek M (1995) An E-M algorithm and testing strategy for multiple-locus haplotypes. Am J Hum Genet 56:799–810 [PMC free article] [PubMed] [Google Scholar]
- Lynn S, Evans JC, White C, Frayling TM, Hattersley AT, Turnbull DM, Horikawa Y, Cox NJ, Bell GI, Walker M (2002) Variation in the calpain-10 gene affects blood glucose levels in the British population. Diabetes 51:247–250 [DOI] [PubMed] [Google Scholar]
- Ma H, Fukiage C, Kim YH, Duncan MK, Reed NA, Shih M, Azuma M, Shearer TR (2001) Characterization and expression of calpain 10: a novel ubiquitous calpain with nuclear localization. J Biol Chem 276:28525–28531 [DOI] [PubMed] [Google Scholar]
- Neel JV (1962) Diabetes mellitus: a “thrifty” genotype rendered detrimental by “progress”? Am J Hum Genet 14:353–362 [PMC free article] [PubMed] [Google Scholar]
- Parra EJ, Marcini A, Akey J, Martinson J, Batzer MA, Cooper R, Forrester T, Allison DB, Deka R, Ferrell RE, Shriver MD (1998) Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet 63:1839–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RJ, Goldman D, Long JC (1999) Effects of worldwide population subdivision on ALDH2 linkage disequilibrium. Genome Res 9:844–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MF, Shen Y, Kreitman ME (1995) A population genetic test of selection at the molecular level. Science 270:1497–1499 [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Pakstis AJ, Ruano G, Kidd KK (2000) The accuracy of statistical methods for estimation of haplotype frequencies: an example from the CD4 locus. Am J Hum Genet 67:518–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HJ, Sun G, Weeks DE, Kaushal R, Wolujewicz M, McGarvey ST, Tufa J, Viali S, Deka R (2001) Type 2 diabetes and three calpain-10 gene polymorphisms in Samoans: no evidence of association. Am J Hum Genet 69:1236–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir (1996) Genetic data analysis II. Sinauer Associates, Sunderland, MA [Google Scholar]